ABSTRACT
Beyond their high clinical relevance worldwide, flaviviruses (comprising dengue and Zika viruses) are of particular interest to understand the spatiotemporal control of RNA metabolism. Indeed, their positive single-stranded viral RNA genome (vRNA) undergoes in the cytoplasm replication, translation and encapsidation, three steps of the flavivirus life cycle that are coordinated through a fine-tuned equilibrium. Over the last years, RNA methylation has emerged as a powerful mechanism to regulate messenger RNA metabolism at the posttranscriptional level. Not surprisingly, flaviviruses exploit RNA epigenetic strategies to control crucial steps of their replication cycle as well as to evade sensing by the innate immune system. This review summarizes the current knowledge about vRNA methylation events and their impacts on flavivirus replication and pathogenesis. We also address the important challenges that the field of epitranscriptomics faces in reliably and accurately identifying RNA methylation sites, which should be considered in future studies on viral RNA modifications.
KEYWORDS: Flavivirus, viral RNA, RNA modification, RNA methylation, N6-methyladenosine, 2ʹ-O-methylation, cap, dengue virus, Zika virus, West Nile virus
Introduction
Flaviviruses comprise more than 70 known viruses including several that are clinically relevant worldwide such as dengue virus (DENV), Zika virus (ZIKV), West Nile virus (WNV) and yellow fever virus (YFV). Belonging to the Flavivirus genus within the Flaviviridae virus family, these pathogens are enveloped positive strand RNA viruses. Following receptor-mediated endocytosis and envelop fusion, the non-segmented viral RNA genome (vRNA) is released into the cytosol. It is then translated in a cap-dependent manner into a unique large transmembrane polyprotein at the endoplasmic reticulum (ER), which is further cleaved by cellular and viral proteases. This generates ten mature viral proteins: 1- seven nonstructural (NS) proteins, namely NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 which are responsible for vRNA synthesis and amplification, and 2- Capsid (C), Envelop (E) and pre-membrane (prM) structural proteins which drive with vRNA the assembly of new virions at the ER. Viral particles egress through the secretory pathway and become infectious following cleavage of prM by cellular furin in the Golgi apparatus. Infectious particles are then released in the extracellular space and may infect other target cells [1].
Beyond the clinical relevance of flaviviruses due to the unavailability of vaccines or therapeutics for most of them, these pathogens are fascinating from a fundamental RNA biology perspective. With only one open reading frame in vRNA, the flavivirus replication cycle is relatively simple and occurs exclusively in the cytoplasm without any DNA intermediate. Yet, flavivirus replication is generally very efficient in cell culture, highlighting that all the genetic information required to fulfill a complete life cycle is condensed within one single cistron. More specifically, vRNA undergoes multiple fates in the cytoplasm, which cannot occur simultaneously and involve separate machineries. Indeed, vRNA can be: 1- translated by the ribosomes, 2- replicated by the viral RNA-dependent RNA polymerase (RdRp) NS5, and 3- selectively packaged into newly assembled virus particles (Fig. 1). These processes must be regulated in time and space to achieve an efficient viral life cycle. The equilibrium between those key steps is likely orchestrated by the viral replication factories, which derive from the remodelling of the ER and allow spatial segregation of the vRNA in the cytoplasm. In addition, flaviviruses have developed strategies to dampen sensing of vRNA by the pattern recognition receptors (PRR) of the innate immune system [2]. Finally, through XRN1 exonuclease-dependent 5ʹ-3ʹ degradation of vRNA and partial resistance of its 3ʹ untranslated region (UTR), flaviviruses generate a non-coding RNA named subgenomic flaviviral RNA (sfRNA), which is involved in viral pathogenicity, antagonism of host innate immune and stress responses [3–7]. Hence, vRNA is an interesting tool to gain a better understanding of the fundamental aspects of the spatiotemporal regulation of RNA metabolism.
Figure 1.
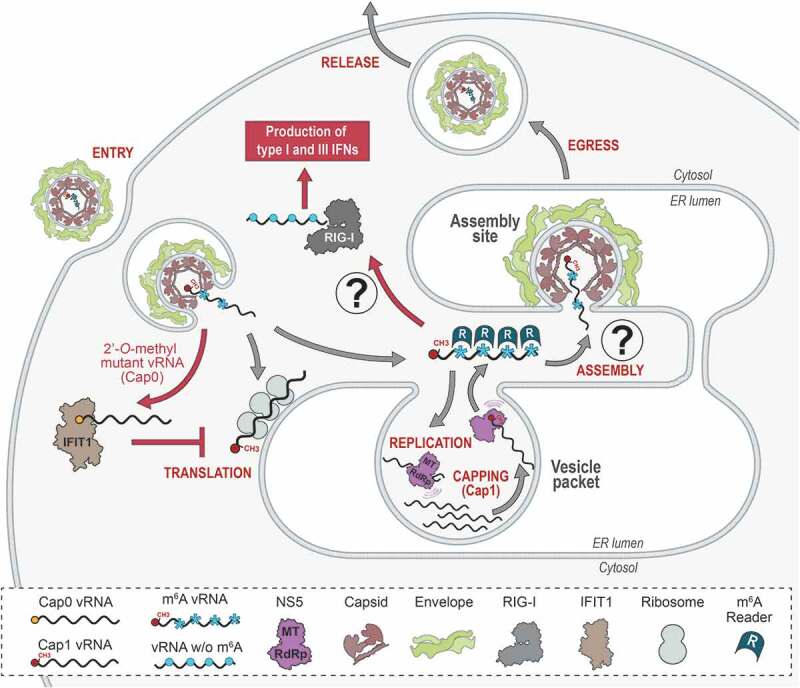
The fates of methylated and non-methylated vRNA species during flavivirus life cycle
Such dynamic control involves RNA modifications. First, the 5ʹ end of the genome is methylated by NS5 during the capping process, which is required for translation initiation. Second, despite well characterized for several decades in the case of transfer RNAs (tRNA) and ribosomal RNAs (rRNA), RNA internal modification has emerged in the recent years as an important process for the posttranscriptional regulation of messenger RNA (mRNA). Notably, mRNA methylation can regulate RNA translation, stability and interactions with proteins sometimes by conferring specific structural conformations [8–10]. Not so surprisingly, vRNA internal methylation has been described for viruses from different families and reported to regulate specific steps of their life cycle as well as to mimic cellular RNA to avoid sensing as non-self RNA by the innate immune system [11–21]. Flaviviruses make no exception and this review explores the current understanding of how flaviviral RNA methylation affects vRNA cytoplasmic fate, life cycle and pathogenesis. We also elaborate on the technical challenges related to confidently identifying and precisely mapping methylation sites in RNAs to consider in future studies.
Methylations of the flavivirus capped vRNA
Capping is likely the first RNA modification event for flaviviral vRNA since it occurs during genome replication. First evidence was obtained in the late 1970s by Wengler et al. who reported the presence of a 2ʹ-O methylated cap1 structure on WNV vRNA molecules purified from hamster and mosquito cells [22]. Similar cap structures were thereafter identified on DENV2 vRNA [23]. One notable feature, however, distinguished flaviviral RNAs from cellular mRNAs, namely the absence of cap2 structures [24].
NS5 MTase, the swiss army knife of flavivirus genome capping
Due to their entirely cytoplasmic life cycle, flaviviruses did not evolve to co-opt the canonical cellular capping machinery of the nucleus but rather incorporated the necessary enzymatic activities into their own replication enzymes. While in eukaryotes cap methylation reactions involve at least five sequential steps and are usually carried out by multiple cellular enzymes [25], vRNA capping is achieved exclusively in the cytoplasm by only two viral proteins, namely NS5 and NS3, reflecting their optimized enzymatic capability. Thus, in addition to its protease activity, NS3 encodes both RNA helicase and RNA 5ʹ triphosphatase (RTPase) activities, first identified in WNV and DENV [26,27]. The former is responsible for unwinding the viral double-stranded RNA involving the neo-synthesized vRNA and the negative strand RNA template, while the latter removes vRNA 5ʹ phosphates. Both activities are required for subsequent cap addition by the RdRp NS5, which remarkably combines three capping enzymatic activities: an atypical RNA guanylyltransferase (GTase) [28–31] and a dual N7- and 2ʹ-O methyltransferase (MTase) [32–34] responsible for the formation of the cap1 structure (Fig. 1, Fig. 2A).
Figure 2.
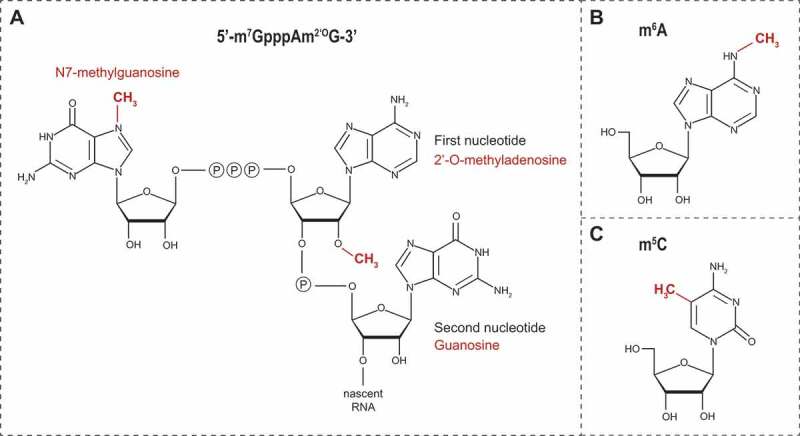
Schematic representation of (A) the flavivirus cap1 structure, (B) N6-methyladenosine (m6A) and (C) 5-methylcytosine (m5C)
Flaviviral NS5 MTases specifically methylate RNAs of viral origin. This selectivity occurs through the recognition of both distinct nucleotides and structural elements in the 5ʹ UTR [32,35]. Thus, the first two transcribed nucleotides after the cap structure are conserved among flaviviruses and consist in an adenine in position +1 and a guanine in position +2 [36,37]. Critical for the N7-methylation and formation of the cap0 structure (m7GpppApGpUp) are the guanine at position +2, the uracil at position +3 as well as the presence of a stem loop structure whose sequence is not relevant for this activity. Subsequently, NS5 modifies vRNA cap0 structure by methylation at the 2ʹ-O position of the ribose of the first adenosine of vRNA to produce the cap1 structure (m7GpppAm2ʹOpGp) (Fig. 2A). This 2ʹ-O-methylation activity requires, in addition to the first adenine and guanine nucleotides, the vRNA to be at least 20 nucleotides long [35]. On the structural level, NS5 substrate preference for flaviviral RNAs is determined by a pocket (constituted by residues I147, G148, E149, and S150), which optimally accommodates the adenine at position +1, and by the presence of a glutamate residue E111, which creates an essential polar interaction with the guanine at position +2 [38]. This nucleotide specificity also influences the subsequent methylations steps [30,32,35,39,40].
Similarly to other viral and cellular RNA MTases [41,42], flaviviral MTases exhibit a conserved, highly positively charged, groove constituted by a K-D-K-E tetrad motif (K61-D146-K182-E218 for WNV and K61-D146-K181-E217 for DENV2) that sequentially orchestrates the cap methylation events [30,32]. Within the tetrad, and likely because of its proximity to the S-adenosylmethionine (SAM) donor site, the aspartate residue D146 is essential to N7-methylation and therefore to virus growth. All other residues of the tetrad are involved in the 2ʹ-O-methylation process and substitutions attenuate or delay viral replication to varying degrees, as measured by the formation of smaller plaques and lower viral titres [30,43].
2ʹ-O-methylation of the flaviviral cap, a mimicry to escape innate immune recognition by IFIT1
Cap1 structures, commonly found in mRNAs, play a crucial role in discerning self from non-self RNAs and preventing the undesired activation the innate immune system. RNAs terminated by a 5ʹ triphosphate usually carry the molecular signature of viral RNAs whose translation is driven by an internal ribosome entry site. These are therefore specifically and efficiently recognized by cytosolic sentinels such as retinoic acid inducible gene I (RIG-I) that initiates the antiviral signalling cascade leading to interferon (IFN) production [44–46]. RIG-I preference for uncapped RNAs is provided by the presence of a histidine residue at position 830 in the triphosphate-RNA-binding pocket, which contributes to the weak binding affinity of methylated RNAs, notably cap1 mRNAs, thereby ensuring tolerance to self-RNAs [47,48].
Flaviviruses have developed a mechanism of 2ʹ-O-methylation of the cap as a strategy to camouflage their genome among cellular mRNAs and escape host antiviral responses. The work of several independent groups confirmed the key function of the 2ʹ-O-methylation on flaviviral cap structures in this regard by characterizing 2ʹ-O-MTase mutant viruses e.g. WNV E218A, WNV K182A, DENV E217A and JEV K61A viruses. In immune-deficient cells such as Vero and BHK-21 cells, the replication competence of the 2ʹ-O mutant viruses is almost similar to that of wild type virus, yet delayed of several hours in some cases [30,49–53]. In contrast, their replication is attenuated in immune-competent cell lines such as A549 cells, primary cells (e.g. mouse embryonic fibroblasts (MEFs), neurons, dendritic cells and macrophages) [50,52–56] and insect cells [30,43,49]. Interestingly, 2ʹ-O-MTase mutant viruses retain their ability to induce an IFN response at levels similar to those of the wild type viruses, however, earlier after infection [52–54]. In vivo, the attenuation of replication [30,50,51,54] correlates with the absence of the virulence and pathogenicity normally observed for wild type viruses, both being restored in the context of IFN-deficient mice lacking the type I IFN receptor (Ifnar1-/-) [49–51,54,55]. Interestingly, 2ʹ-O-MTase mutant viruses showed an increased sensitivity to IFN pretreatment in mice suggesting that non-methylated vRNAs are targeted by IFN-induced effectors [49,50,54].
The IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) [57], also known as p56 and ISG56, is one of the effectors most highly induced in response to IFN and virus infections [57–60]. In their study, Daffis et al. elucidated the role of IFIT proteins in restricting 2ʹ-O-MTase mutant viruses in vivo and showed that ectopic expression of IFIT1 and IFIT2 (ISG54) in MEFs reduced the replication of wild type WNV and abrogated that of WNV E218A mutant virus [54]. While it is clear that IFIT1 senses vRNAs lacking 2ʹ-O-methyls, its restriction mechanism is yet unclear and may involve interactions at two different steps of the translation initiation process. First, IFIT1 and IFIT2 directly interact with the eukaryotic translation initiation factors eIF3E and eIF3C, thereby affecting the stability of the ternary pre-initiation complex and inhibiting cap-dependent translation [61–64]. Second, IFIT1 binds with high affinity to cap0 and 5ʹ triphosphate-terminated RNAs in vitro while this interaction is abrogated by the presence of the cap1 structure. Thus, IFIT1 substrate recognition sorts out uncapped and cap0 RNAs thereby sustaining cap1 RNA translation. In addition, IFIT1 inhibits translation initiation of cap0 RNAs by efficiently outcompeting eIF4E and the cap binding complex eIF4F [65–68] (Fig. 1). Using mobility-shift assays, Kimura et al. confirmed that IFIT1 binds preferentially to uncapped full-length JEV vRNA and selectively inhibits the translation of 2ʹ-O non-methylated (cap0) JEV vRNA [56].
Internal 2ʹ-O-methylation: an additional role for NS5?
Whilst its ability to methylate the cap is more effective, flaviviral NS5 is able to specifically methylate vRNA internal adenosines, in a sequence-unspecific manner [69]. Indeed, in vitro WNV and DENV4 MTases indiscriminately methylate RNAs exclusively composed of adenosines as well as host ribosomal RNAs. With regard to the life cycle, in vitro 2ʹ-O-methylation of internal adenosines does not affect vRNA stability but rather attenuates its translation efficiency and reduces the capacity of the RdRp to elongate vRNA during replication. Liquid chromatography-mass spectrometry (LC-MS) analyses confirmed the existence of internal 2ʹ-O-methyladenosine on genomes purified from DENV1 virions, despite a very low frequency. Most importantly, these internal modifications were not detected in genomes purified from 2ʹ-O-MTase mutant viruses confirming the involvement of NS5 methylation activity [69]. Recently, a large-scale epigenetic analysis by McIntyre et al. reported consistent results for DENV2 and ZIVK genomes [18].
Therapeutic and prophylactic strategies around vRNA cap methylation
NS5 GTase and MTase activities have been the scope of several antiviral therapeutics approaches during the last decades, as well as more recently because of the ZIKV outbreak in the Americas. Compounds developed to block these enzymatic activities include (i) GTP analogues such as BG-323, a thioxothiazolidin-derived inhibitor [70,71], as well as ribavirin 5ʹ triphosphate whose impact on replication [33,72] might rather result from its ability to induce RdRp errors [73–76], (ii) SAM analogues such as sinefungin [31,77–79] and the compounds NSC 12,155 and NSC 12,590 [80], F3043-0013 and F0922-0796 [81], and (iii) non-nucleoside inhibitors [82,83]. However, although the ‘straightforward’ aspect of this therapeutic approach is very appealing, the development of effective and selective methylation inhibitors seems to be very challenging since most of these compounds exhibit half maximal inhibitory concentration (IC50) values in the micromolar range and a relatively low impact on viral replication, notably when replication is already established.
2ʹ-O-MTase mutant viruses have emerged as promising candidates for the development of live-attenuated vaccines with several advantages over conventional live-attenuated viruses, which exhibit reduced replication without fully precluding pathogenic effects (e.g. YFV-17D vaccine [84,85]). This idea emerged from the work by Zhou and colleagues who demonstrated that the WNV E218A 2ʹ-O-MTase mutant virus is attenuated in mice and protects animals from a subsequent challenge with pathogenic wild type WNV [30]. This concept was subsequently applied to the design of JEV, DENV1 and DENV2 live-attenuated vaccines [49,50] and extended more recently to the design of a tetravalent non-chimeric vaccine against all four DENV serotypes [51]. For all approaches, a single dose of 2ʹ-O-MTase mutant viruses induced a protective IFN-based immune response and elicited a CD8+ T cell-specific response against peptides of the non-structural protein that protected both mice and non-human primates from a challenge with the wild type virus [49–51]. With regard to pathogenic effects, JEV 2ʹ-O-MTase mutant virus exhibited a reduced neuroinvasiveness and did not induce any neurological symptoms [49]. Whether these live-attenuated vaccines provide protection in humans without a risk of antibody-dependent enhancement is yet to be addressed.
N6-adenosine methylation of flavivirus vRNA
A methyl group can be added to the nitrogen linked to the position 6 of adenosines (N6) within mRNA (Fig. 2B, m6A), hence conferring several new properties to the modified RNA. This reversible N6-adenosine methylation is the most prevalent internal RNA modification in the cell [86,87]. It primarily occurs in the nucleus within DRAmCH consensus motifs (where D = G, A or U; R = G or A; Am is the methylated adenosine; and H = U, A or C) and is mediated by the MTase-like (METTL) 3 and METTL14, generically called ‘writers’, with the contribution of the cofactors Wilms tumour 1-associated protein (WTAP) and KIAA1429 [88–93]. To a lesser extent, MGAmCK (where M = A or C and K = G or U) and UGAmC sequences are also methylated. The presence m6A in mRNAs confers increased affinity to several cellular RNA-binding proteins referred to as ‘readers’ that regulate the fate of the methylated mRNA [10]. For instance, the most characterized ‘readers’, YTH Domain Family 1 (YTHDF1) and YTHDF2, regulate translation and stability of m6A-containing mRNAs, respectively [94,95]. m6A modification is reversible and can be removed by ‘erasers’, namely demethylases of the AlkB homologues family FTO (ALKBH9) and ALKBH5 [96,97]. Such dynamics suggests the existence of a fine-tuned equilibrium between methylation and demethylation, which may allow a quick and local posttranscriptional response of the cell to environmental and/or metabolic changes. Changes in the m6A-containg mRNA profiles are associated with diseases such as obesity, cancer, neurodevelopment defects and plasticity in brain development [98–104].
Impact of m6A modification on flavivirus life cycle
Taking advantage of m6A-specific antibody-based purification and next-generation RNA sequencing methods (MeRIP-Seq) [86,105], two groups have independently reported the presence of m6A in ZIKV vRNA [14,21]. This observation was expanded to other flaviviruses (DENV, YFV and WNV) as well as to hepatitis C virus (HCV), another Flaviviridae belonging to the Hepacivirus genus [14], suggesting a conserved mechanism across this virus family. The existence of m6A in DENV and ZIKV vRNAs was recently confirmed by oligonucleotide-based purification of vRNA combined with LC-MS-based m6A detection [18].
METTL3/14 knockdown reduced the level of ZIKV vRNA immunopurified with anti-m6A antibodies while the opposite phenotype was observed when the expression of the demethylases was decreased [21]. This demonstrated that m6A modification of ZIKV vRNA is mediated at least partly by cellular proteins in contrast to NS5-mediated 2ʹ-O-methylation. Importantly, the ‘readers’ YTHDF1, YTHDF2 and YTHDF3 are all associated with ZIKV vRNA in a m6A-dependent manner (Fig. 1). Nevertheless, none of these studies mapped the precise methylation sites. Indeed, MeRIP-Seq approaches identify rather large regions of vRNA that are modified (˃ 100 nucleotides) and potential vRNA methylation sites are inferred by the presence of DRACH consensus motifs. Noteworthy, the regions identified in different flaviviruses or in different strains of the same flavivirus do not always fully overlap. For example, regions differ between ZIKV Dakar 41525-DAK strain (African lineage) and Puerto Rican PRVABC59 strain (Asian lineage) [14], the latter being associated with severe symptoms (e.g. neonate microcephaly) observed during the 2015 pandemic. It is thus tempting to hypothesize a link between vRNA N6A methylomic profile and pathogenic features. In contrast, the methylated regions of DENV and YFV vRNAs overlap relatively well suggesting some conservation of methylated regions. Further studies using techniques that allow accurately mapping m6A modifications in vRNA at the single nucleotide resolution will determine in which extent these are conserved across the Flavivirus genus.
In terms of impact on the viral cycle, N6A methylation of ZIKV vRNA limits the production of infectious particles. Indeed, the knockdown of METLL3/14 ‘writers’ as well as YTHDF1-3 ‘readers’ increased extracellular infectious titres [21]. In contrast, decreasing demethylases expression or overexpressing YTHDF proteins altered virus production. Whether the presence of m6A in ZIKV vRNA restricts replication or particle assembly and release is yet unknown. Interestingly, N6A methylation similarly affects HCV production [14]. In contrast to known functions for mRNAs [94,95], m6A modification does not regulate HCV vRNA translation or stability, nor its synthesis but rather acts on virus particle assembly together with YTHDF proteins. Consistently, in HCV-infected cells, YTHDF proteins relocalized around the lipid droplets, where virus assembly takes place [14]. The combined introduction of synonymous mutations into four high confidence DRACH motifs identified in HCV envelope E1 coding sequence increased virus production without altering RNA replication and correlated with a significant enrichment of vRNA in complexes containing HCV structural protein core [14]. Altogether, this suggests that the methylation of this region acts as a negative regulator that is important for genome packaging into assembling virions (Fig. 1). Further studies will have to demonstrate how conserved is this function across the Flaviviridae family and whether it applies to flaviviruses as well. It is though tantalizing to speculate that alterations of vRNA m6A modification profile could influence the equilibrium between the synthesis of vRNA and its packaging into assembling virus particles. Flaviviral vRNA is likely replicated insides vesicular substructure of replication factories, which originate from ER invaginations [106–110]. These so-called vesicle packets (VPs) exhibit an opening that would allow neosynthesized vRNAs to exit and to be targeted either to ribosomes for translation or to assembly complexes for encapsidation. 3D reconstruction of DENV and ZIKV replication factories has revealed that virus particle budding events are juxtaposed to VP pores [106,108]. This suggested that vRNAs exiting VPs might be directly targeted to budding viruses. It will be interesting to assess whether m6A-containing vRNAs are enriched in VPs and whether their demethylation or YTHDF loss-of-binding contribute to the selective packaging of vRNA into assembling particles (Fig. 1). m6A moieties might ‘flag’ vRNAs and contribute to the spatial segregation of the different steps of the life cycle in the cytoplasm.
Several studies have revealed other m6A readers as regulators of flavivirus replication. These include candidates identified in a large-scale analysis of ZIKV and DENV vRNA riboproteomic e.g. IGF2BP1-3, FMR1, hnRNPC, hnRNPG and hnRNPA2/B1 that associate with the vRNA and in some cases modulate viral replication [111–114]. It will be relevant to evaluate whether the regulation of flavivirus replication by these m6A readers occurs in a methylation-dependent manner.
m6A, a possible structural switch regulator?
While these discoveries have broadened our perspective on the regulation of flavivirus life cycle by vRNA m6A modifications, much remains to be explored and understood about the molecular mechanisms involved. Internal N6A methylation alters RNA duplexes with impacts on molecular switches [115–117]. The NH2 group linked to position 6 of adenosine is engaged in pairwise interactions through a hydrogen bond with the neighbouring uridine. Thus, the addition of a methyl to this nitrogen may influence adenosine-uridine base pairing, disturb the hybridization of two strands and hence, local secondary or tertiary structures of the RNA. Alternatively, the secondary structure of the RNA might govern the accessibility of specific regions to ‘writer’, ‘eraser’ and/or ‘reader’ proteins, thus indirectly influencing their posttranscriptional actions. Such alterations are highly relevant for flaviviruses whose vRNA forms long distance intramolecular interactions for its circularization, which is essential for RNA synthesis, and undergoes important structural switches that regulate the equilibrium between the different steps of the life cycle [2,118–120]. While much focus has been paid to vRNA UTRs, there is since recently strong evidence that secondary and tertiary structures (including novel long-range interactions) in the coding region also play important roles in the life cycle of DENV and ZIKV both in cells or in the virions [121–123].
With this in mind, we have compared for the purpose of this review these published structural models with the corresponding epitranscriptomic data described above in order to highlight possible structure/m6A relationships (Fig. 3). The three examples highlighted below reveal interesting features that may merit further investigation. In their MeRIP-Seq analysis mentioned above, Lichinchi et al. identified two contiguous m6A-containing regions in the NS5 coding sequence of ZIKV African strain MR766 vRNA purified from virions (Fig. 3A, nt 8651–8800 in yellow and nt 8904–9073 in light pink) [21]. According to the structural map of the full-length ZIKV MR766 vRNA in cellulo reported by Li et al., these regions are partly located in a highly structured region of the genome with a 90 nucleotide-long imperfect stem (Fig. 3A, upper part of the structure) [123]. Most notably, one strand of this stem (highlighted in grey), which is located right before an unstructured region, makes a long distance interaction with a sequence located 8,000 nucleotides upstream, in the prM coding sequence. Closer analysis of the sequence of the whole region identified seven m6A consensus motifs. This includes two DRACH motifs in the long stem mentioned above (nucleotides circled in red). In addition, one MGACK motif (circled in black) was identified in the unstructured region directly upstream the strand, which hybridizes with the region involved in the NS5/prM RNA interaction. If one considers that the presence of m6A influences local hybridization properties, this suggests that the methylation status of this region influences whether or not this strand makes the stem in NS5 depicted in Fig. 3A or is rather engaged in NS5/prM long-range interaction.
Figure 3.
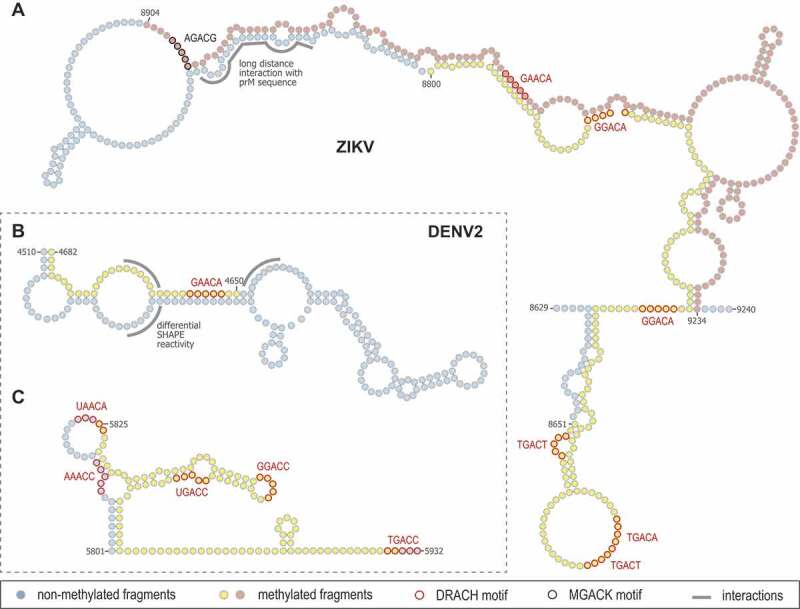
Examples of ZIKV and DENV structural elements with m6A consensus motifs. (A) Two contiguous m6A-containing regions in the NS5 coding sequence of ZIKV MR766 genome (yellow and light pink circles) were identified in the reported secondary structure [14,21,121,123]. The nucleosides in predicted DRACH and MGACK m6A consensus motifs are circled in red and black, respectively. The RNA strand making intramolecular long-range interaction with prM coding region is indicated in grey. (B, C) Two different methylated regions of DENV2 NGC strain (yellow circles) were mapped within two independent reported structures in the NS3 coding sequence of the closely related strain DENV2 S16803 [14,21,121,123]. The nucleosides in predicted DRACH m6A consensus motifs are circled in red. In B, the DRACH motif is located in a 14 nucleotide-long stem flanked by unstructured regions whose SHAPE reactivity (indicated in grey) changed if vRNA was gently extracted from virions
The comparison between the m6A-methylome and the structure of DENV2 vRNA from two independent studies also revealed interesting features [14,121]. Indeed, a methylated region in NS3 coding sequence of the New Guinea C strain partially overlaps with a structured region identified in the genome of the closely related S16803 strain (element 10 in [121]; Fig. 3B, nt 4650–4682 in yellow). This shared region contains a 14 nucleotide-long stem, which includes a consensus DRACH motif, and is flanked by three unstructured sequences whose reactivity changes depending on whether the vRNA structure is analysed in authentic virions (i.e. capsid associated) or following gentle extraction (i.e. protein-free). These differences might reflect a structural switch of this region that would occur during vRNA uncoating following virus entry or during genome encapsidation into assembling particles. In cells, methylation of this stem may affect its stability hence, structurally rearranging the whole region, modifying its interactome and targeting the RNA to specific steps of the life cycle.
Finally, another methylated region is found in a structural element located in NS3 coding sequence (element 14 in [121]; Fig. 3C, nt 5825–5932) and contains 4 DRACH motifs, which all completely or partially overlap with loops of bulges. Another motif is located right downstream this element in an unstructured sequence. Despite still unclear, this suggests that m6A modifications primarily occur in unstructured regions or destabilize neighbouring base pairing. In the case of HCV described above, the four predicted m6A sites in E1 coding sequence which regulates virus production, are located in rather unstructured regions [14,124]. If N6A methylation of vRNA indeed influences its structure, it is conceivable that this will result in direct impacts on structural switches as well as long-range interactions. Whether the three selected examples of a hypothetical methylation/structure relationship for flaviviruses actually exist as m6A switches and are functionally relevant to viral replication needs to be addressed experimentally. It is noteworthy that this comparative exercise did not take into consideration that the reference studies used different cell lines (e.g. Vero vs. Huh7 cells) and vRNAs of different origins (intracellular vRNA vs virion vRNA).
Possible additional functions of flavivirus vRNA N6A methylation
N6A methylation of vRNA may also allow flaviviruses to escape the sensing by the innate immune system. Indeed, for HCV (as well as for human metapneumovirus, a negative strand RNA virus), the presence of internal m6A in vRNA was recently reported to decrease the production of type I IFN in infected cells [17,125]. This correlated with a decreased binding efficiency of the vRNA to RIG-I. While the molecular mechanisms behind this interference remain to be fully elucidated, internal m6A seems to constitute an additional strategy to the cap 2ʹ-O methylation to ‘disguise’ vRNAs as self-RNAs (Fig. 1). Future studies will have to confirm or invalidate this hypothesis.
The fact that flavivirus vRNA does not transit through the nucleus during its life cycle implies that m6A modifications occur in the cytoplasm, in contrast to the co-transcriptional methylation of host mRNAs. While some studies have reported this methylation activity in the cytoplasm, it will be interesting to determine how flaviviruses hijack this cellular activity in the vicinity of replication factories. Interestingly, besides exploiting the N6A-methylation-related machinery to regulate their own genomes, DENV and ZIKV also alter the host N6A methylome [21,126,127]. Notably, infection increased and reduced m6A content in RIO Kinase 3 (RIOK3) and Cold Inducible RNA Binding Protein (CIRBP) mRNAs, respectively, changing the expression profile of the corresponding proteins, which were shown to regulate replication efficiency [126]. These changes in the mRNA methylation profile, likely occurring in the nucleus, were indirect since attributed to the virus-induced ER stress or immune response. Nevertheless, this suggests that flaviviruses hijack the m6A machinery to generate a cellular environment favourable to viral replication.
Other types of RNA methylations potentially involved in flavivirus life cycle
A recent epitranscriptome analysis of affinity-purified vRNAs from several positive strand RNA viruses (including DENV and ZIKV) opened the possibility that flaviviral RNA might actually be decorated by modifications with over 40 types of methylation (including m6A and A2ʹOm) detected by mass spectrometry [18]. While this study made clear that vRNA is modified, the fact that so many types of methylations were detected does not make them directly relevant for replication. Indeed, some were found at very low stoichiometry, and it is conceptually challenging to envision how the viral genome would have evolved to co-opt in the cytoplasm all the cellular machineries required to fulfill this massive epigenetic task, especially considering that vRNA is mostly confined in membranous replication factories. To reconcile this, further accurate mapping and functional studies for each type of nucleoside methylation are needed. Very interestingly, the presence of some methylated nucleotides was specific to either flavivirus. For instance, methylated uridines m3Um and m5Um were detected in DENV vRNA but not in ZIKV vRNA. In contrast, dimethylcytosines m5Cm and m44C were present only within ZIKV vRNA inside virions. This suggests that this modification is not only ZIKV-specific but may also be a marker of encapsidated vRNA since it was absent in genomes purified from infected cells. Whether the methylation landscape of flavivirus influence the different steps of the life cycle or specific aspects of ZIKV pathogenesis need to be fully explored. Furthermore, the authors identified in this study DEAD box protein 6 (DDX6) as a host factor modulating the abundance of m5Cm and m44C as well as ZIKV replication. Future studies will have to evaluate the contribution of m5Cm and m44C to the ZIKV life cycle and whether DDX6 role in viral replication is related to these modifications or rather to its implication in decapping activity, RNA stability in P-bodies and sfRNA metabolism [128,129]. Finally, since m5C (Fig. 2C) was shown to play important roles in viral RNAs of murine leukaemia virus (MLV), human immunodeficiency virus (HIV-1) and Epstein-Barr virus (EBV) [12,15,130,131] and was detected in DENV and ZIKV vRNAs, it will be interesting to further investigate the potential function of this modification in the flavivirus replication cycle.
Technical challenges in flaviviral RNA epigenetics
In relation to the recent attention received by RNA modifications in the RNA field, molecular virology on RNA modifications is still relatively sparse. This is likely due to a number of important technical limitations concerning analytics of RNA modifications and the isolation of pure vRNA in high amounts that we will address in this section. We also highlight additional approaches that have not yet been applied and could be considered for flaviviral RNAs.
Analytical techniques of RNA modifications can be roughly divided into two categories [132,133]. In brief, biophysical methods for the characterization of modified nucleosides such as chromatography, mass spectrometry, or combinations thereof, can identify and quantify RNA modifications in small samples only at the expense of sequence information. A combination of modification and sequence information, commonly obtained from proteomics approaches, is available only for samples of double-digit microgram amounts, i.e. typically for tRNA and rRNA [134,135]. Moreover, as described below, sample purity must be considered in the analysis in order to reliably assign a proper modification to the RNA of interest.
In contrast, there exists an ever-growing array of RNAseq-derived methods that make use of various chemical or biochemical properties of RNA modifications to place them in a sequence context [132]. Results from this so-called ‘modification mapping’, or ‘modification calling’ form the basis for the vast majority of reported viral RNA modification sites. However, these approaches come with their own set of experimental problems. The large diversity of mapping methods makes discussions about advantages and drawbacks impractical at this point. It should be made clear, though, that the different methods result in modification calling data of variegated quality, and that it is typically at the authors’ discretion to set an arbitrary threshold above which a signal is ‘called’ a modification.
Antibody-based detection of viral RNA modifications
Among the various reagents used in modification mapping, antibodies hold a special place, given that the first m6A mapping was performed using antibody affinity enrichment in the 1980s [13,136] before the current surge was triggered by combination with RNAseq [86,87]. However, antibodies recently emerged as problematic [137,138]. Similar to applications in epigenetics, where e.g. single methylations on either nucleobases or amino acids must be discriminated, certain commercial antibodies were found to have been insufficiently validated [139,140]. Most antibody-based mapping reports employ modification calling based on simple enrichment calculations, which identify regions or sequence stretches (generally hundreds nucleotide-long) rather than single nucleotides as mapping data [127]. Hence, the modification calling is necessarily vague. Authors generally look afterwards for consensus methylation motifs to identify putative modified sites and to elaborate validating directed mutagenesis-based experiments. More advanced techniques include a crosslinking step that allows more stringent washing conditions to remove non-specific binders [141].
In the particular case of m6A, it is often assumed (tacitly or explicitly) that signals only originate from activity of the METTL3/14 complex, which is known to depose methylation marks at the DRACH consensus motif, and other potential sites could be ignored. A smart method to validate such presumed m6A sites has been developed already in the 1980s [142]. Within the central consensus, Kane and Beemon mutated the pyrimidine downstream of the methylation target adenosine to a uridine, and found that a DRAUH was no longer methylated. This ‘silent’ mutagenesis approach has been used in several more recent publications on m6A in vRNAs, including Flaviviridae vRNAs [12,14,21] as described above.
Identification of viral RNA modifications from cDNA-affinity purified samples
Sample purity is an often-overlooked problem for such analyses. Indeed, LC-MS-based analyses of mRNA populations purified by oligo-dT affinity frequently pick up marker modifications of contaminating rRNA [143]. Given that viral particles of any kind typically package other host RNAs in a rather unspecific manner [144], an LC-MS analysis of total RNA isolates of viral particles can be expected to contain standard modifications encountered in host tRNA and rRNA fragments [145]. Hence, further highly specific purification approaches e.g. by RNA size fractionation [12] or cDNA affinity are required to reduce non-specific signals [15]. Early work in the 1980s on the detection of modifications in coding sequences relied on in vivo labelling of RNA with 32P and/or L-[methyl-3H]-methionine, respectively. Thus, labelled RNA was affinity-purified e.g. using oligo-dT or cDNA, digested to nucleotides and separated by multidimensional chromatography after which autoradiography of the label allowed quantification [136,146,147]. Many of the RNAs thus investigated were indeed viral coding RNA containing mainly m6A in viruses replicating in the nucleus, e.g. in simian virus 40 [148], adenoviruses [149], herpes virus [150], influenza virus [151] and Rous sarcoma virus [147]. For the latter retrovirus, 13 sites have been individually mapped, constituting a major early contribution to the establishment of the DRACH consensus [146,147]. Other modifications detected in viral RNA using radioactive label included cap structures, ribose methylations, and m5C in low quantities [22,23,152,153].
More examples of cDNA purification-based analysis include the above-mentioned LC-MS analysis of various viral RNAs, including those of ZIKV and DENV [18] and more recently, EBV. Two small RNAs from EBV, namely EBER1 and EBER2, were isolated by cDNA affinity, and m5C was detected by LC-MS in EBER1 at near quantitative stoichiometry [15]. As mentioned above, special attention must be given to the purity of the analysed samples as well as to subsequent validation since heavily modified rRNAs and/or tRNAs, which may be unspecifically co-purified are expected to introduce undesired methylation signals during the detection. In case of EBER1, the presence of m5C was confirmed by bisulfite-seq, which allowed to place the modification at position C145 in this small structured RNA. Of note, the presence of m5C 145 was contingent upon the expression of NSUN2, a tRNA m5C MTase known to act on highly structured tRNA [15].
Other approaches for the identification of viral RNA modifications
The following methods have not yet been applied to study flavivirus epitranscriptomics: (i) Ribomethseq is a mapping method based on protection against alkaline degradation conferred by ribose methylation [154]. Using this approach Ringeard et al. identified 17 distinct 2ʹ-O-methylated residues in the HIV RNA genome and showed that HIV recruits the host methyltransferase FTSJ3, via a complex including the TAR RNA-binding protein to execute internal ribose methylations [155]. Interestingly, absence of ribose methylation resulted in recognition by the PRR melanoma differentiation-associated protein 5 (MDA5) and subsequent induction of IFNs. (ii) Direct RNA sequencing using the nanopore technology was applied to unravel the epitranscriptome of severe acute respiratory syndrome coronaviruses SARS-CoV and most recently SARS-CoV-2 allowing the detection of 41 potential modification sites in the latter [156,157]. (iii) Other methods for modification mapping that yield single nucleotide resolution frequently rely on chemical treatment that achieves a certain discrimination in its reaction with a modified versus unmodified nucleoside [132,133,158,159]. Fortunately for the field of viral RNA modification, these methods perform better on short transcriptomes such as viral RNA genomes than they do on cellular transcriptomes of a complexity in the order of 107. Thus, while many transcriptome-wide studies on modification mapping are subject to controversial findings because the large sequence space gives rise to signal-to-noise problems, short viral RNA genomes can be expected to be suitable and it will be interesting to explore such approach in the case of flaviviral RNA.
Perspectives and conclusions
In addition to being critical for initiating the cap-dependent translation, vRNA methylation has emerged during the last years as a mechanism exploited by flaviviruses to control their life cycle as well as to evade sensing as non-self by the cellular antiviral machinery. While much progress was recently made, several important hypotheses will be scientifically very exciting to address thanks to the advent of new technologies and/or approaches. Regions of the genomes with m6A have been identified but they are generally over 100 nucleotide-long and contain several consensus methylation motifs. Moreover, determination of the m6A methylome mostly relied on the binding of vRNA to anti-m6A antibodies, which may not be completely specific to this modification. Hence, even if functional validation must be made by directed mutagenesis of the consensus motifs, it is not indicative that these sites are actually methylated and it will be critical to accurately identify the modified positions at the single nucleotide resolution. This also applies to 2ʹ-O-methylated internal adenosines as well as to any other modifications for which no role was described at all for flaviviruses although their presence was experimentally evidenced. Deeper characterization of these newly identified modified sites is expected to reveal specific roles in vRNA structure dynamics, notably in riboswitches regulating transitions between vRNA translation, replication and packaging. In the case of m6A in flavivirus vRNA, the step(s) of the life cycle impacted by this modification is unknown. By analogy with HCV, one should anticipate that it is important for the production of virus particles. Future studies will need to address whether unmethylated vRNA is specifically selected for packaging into assembling virions and whether this is related to the architecture of the viral replication compartment or involves specific RNA packaging signals. It will be interesting to also evaluate the regulation by cellular ‘readers’ of methylated vRNA. It is noteworthy that the notion of a ‘reader’ function, seeped into the field from the epigenetic DNA methylation field, implies that the principle function of a modification lies within the selective interaction with a dedicated binding protein. While this has turned out to be a guiding principle in m6A research, the situation is very different for other modifications. Indeed, the examples of tRNA and EBV EBER1/2 modifications [15] pinpoint that given ‘readers’ and ‘writers’ recognize aspects of RNA secondary and/or tertiary structure rather than a sequence motif per se.
Interestingly, methylation of specific vRNA regions may contribute to the evasion of the innate immune response synergistically with the cap1 structure. It will be essential to address whether such molecular mechanisms are conserved across the Flavivirus genus. Alternatively, some structure/activity relationships specific to a given flavivirus may hypothetically contribute to unique aspects of viral tropism and/or pathogenesis (e.g. neurovirulence in ZIKV-infected foetuses). Finally, the methylation status of the noncoding sfRNA, which is abundantly produced in the course of flavivirus infection is completely unknown. Since fragments corresponding to vRNA 3ʹ UTR contained m6A, it is tempting to speculate that the sfRNA is differentially modified as compared to the flaviviral genome. If that is the case, it could be envisioned that specific sfRNA modifications contribute to its biogenesis and/or stability as well as to its functions in virus-induced cell death and antiviral response. Overall, a complete and highly resolved functional landscape of flavivirus RNA methylome, which may be extended to other epigenetic modifications, will hopefully unveil novel molecular mechanisms governing the viral replication cycle and pathogenesis.
Acknowledgments
A.R. and M.H. are funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation: Projects 240245660 – SFB 1129 and 278001972 – TRR 186 to A.R. and SPP1784, HE3397/13-2 and HE3397/14-2 to M.H). The research of L.C.C’s group on flaviviral RNA is supported by grants from Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2016-05584) and Fonds de la Recherche du Québec-Nature et Technologies (FRQNT; 2018-NC-205593). L.C.C receives a research scholarship from Fonds de la Recherche du Québec-Santé (FRQS).
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft[240245660 – SFB 1129; 278001972; SPP1784, HE3397/13-2; HE3397/14-2]; ; Fonds de Recherche du Québec - Nature et Technologies [2018-NC-205593]; Fonds de Recherche du Québec - Santé; Natural Sciences and Engineering Research Council of Canada [RGPIN-2016-05584].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Neufeldt CJ, Cortese M, Acosta EG, et al. Rewiring cellular networks by members of the flaviviridae family. Nat Rev Microbiol. 2018;16(3):125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mazeaud C, Freppel W, Chatel-Chaix L.. The multiples fates of the flavivirus RNA genome during pathogenesis. Front Genet. 2018;9:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bidet K, Dadlani D, Garcia-Blanco MA.. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10(7):e1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chapman EG, Costantino DA, Rabe JL, et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344(6181):307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chapman EG, Moon SL, Wilusz J, et al. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife. 2014;3:e01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pijlman GP, Funk A, Kondratieva N, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4(6):579–591. [DOI] [PubMed] [Google Scholar]
- [7].Manokaran G, Finol E, Wang C, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350(6257):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lewis CJ, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat Rev Mol Cell Biol. 2017;18(3):202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624. [DOI] [PubMed] [Google Scholar]
- [10].Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Courtney DG, Kennedy EM, Dumm RE, et al. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe. 2017;22(3):377–386. e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Courtney DG, Chalem A, Bogerd HP, et al. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio. 2019;10(3):e01209–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gokhale NS, Horner SM. RNA modifications go viral. PLoS Pathog. 2017;13(3):e1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gokhale NS, McIntyre ABR, McFadden MJ, et al. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20(5):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Henry BA, Kanarek JP, Kotter A, et al. 5-methylcytosine modification of an epstein-barr virus noncoding RNA decreases its stability. RNA. 2020;26(8):1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Imam H, Khan M, Gokhale NS, et al. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci U S A. 2018;115(35):8829–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lu M, Zhang Z, Xue M, et al. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol. 2020;5(4):584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McIntyre W, Netzband R, Bonenfant G, et al. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018;46(11):5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xue M, Zhao BS, Zhang Z, et al. Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun. 2019;10(1):4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lichinchi G, Gao S, Saletore Y, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. 2016;1:16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lichinchi G, Zhao BS, Wu Y, et al. Dynamics of human and viral RNA methylation during Zika Virus infection. Cell Host Microbe. 2016;20(5):666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wengler G, Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89(2):423–437. [DOI] [PubMed] [Google Scholar]
- [23].Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96(1):159–165. [DOI] [PubMed] [Google Scholar]
- [24].Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5ʹ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386. [DOI] [PubMed] [Google Scholar]
- [25].Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc Jpn Acad Ser B Phys Biol Sci. 2015;91(8):394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wengler G, Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184(2):707–715. [DOI] [PubMed] [Google Scholar]
- [27].Li H, Clum S, You S, et al. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73(4):3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Egloff MP, Benarroch D, Selisko B, et al. An RNA cap (nucleoside-2ʹ-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21(11):2757–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Issur M, Geiss BJ, Bougie I, et al. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15(12):2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou Y, Ray D, Zhao Y, et al. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bollati M, Milani M, Mastrangelo E, et al. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in flavivirus. J Mol Biol. 2009;385(1):140–152. [DOI] [PubMed] [Google Scholar]
- [32].Ray D, Shah A, Tilgner M, et al. West Nile virus 5ʹ-cap structure is formed by sequential guanine N-7 and ribose 2ʹ-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Benarroch D, Egloff MP, Mulard L, et al. A structural basis for the inhibition of the NS5 dengue virus mRNA 2ʹ-O-methyltransferase domain by ribavirin 5ʹ-triphosphate. J Biol Chem. 2004;279(34):35638–35643. [DOI] [PubMed] [Google Scholar]
- [34].Egloff MP, Decroly E, Malet H, et al. Structural and functional analysis of methylation and 5ʹ-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol. 2007;372(3):723–736. [DOI] [PubMed] [Google Scholar]
- [35].Dong H, Ray D, Ren S, et al. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J Virol. 2007;81(9):4412–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Castle E, Wengler G. Nucleotide sequence of the 5ʹ-terminal untranslated part of the genome of the flavivirus West Nile virus. Arch Virol. 1987;92(3–4):309–313. [DOI] [PubMed] [Google Scholar]
- [37].Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5ʹ-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–299. [DOI] [PubMed] [Google Scholar]
- [38].Zhao Y, Soh TS, Lim SP, et al. Molecular basis for specific viral RNA recognition and 2ʹ-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proc Natl Acad Sci U S A. 2015;112(48):14834–14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dong H, Liu L, Zou G, et al. Structural and functional analyses of a conserved hydrophobic pocket of flavivirus methyltransferase. J Biol Chem. 2010;285(42):32586–32595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chung KY, Dong H, Chao AT, et al. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2ʹ-O methyltransferase activity in dengue virus. Virology. 2010;402(1):52–60. [DOI] [PubMed] [Google Scholar]
- [41].Schnierle BS, Gershon PD, Moss B. Mutational analysis of a multifunctional protein, with mRNA 5ʹ cap-specific (nucleoside-2ʹ-O-)-methyltransferase and 3ʹ-adenylyltransferase stimulatory activities, encoded by vaccinia virus. J Biol Chem. 1994;269(32):20700–20706. [PubMed] [Google Scholar]
- [42].Hager J, Staker BL, Bugl H, et al. Active site in RrmJ, a heat shock-induced methyltransferase. J Biol Chem. 2002;277(44):41978–41986. [DOI] [PubMed] [Google Scholar]
- [43].Dong H, Chang DC, Xie X, et al. Biochemical and genetic characterization of dengue virus methyltransferase. Virology. 2010;405(2):568–578. [DOI] [PubMed] [Google Scholar]
- [44].Hornung V, Ellegast J, Kim S, et al. 5ʹ-triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. [DOI] [PubMed] [Google Scholar]
- [45].Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5ʹ-phosphates. Science. 2006;314(5801):997–1001. [DOI] [PubMed] [Google Scholar]
- [46].Chazal M, Beauclair G, Gracias S, et al. RIG-I recognizes the 5ʹ region of dengue and Zika Virus genomes. Cell Rep. 2018;24(2):320–328. [DOI] [PubMed] [Google Scholar]
- [47].Schuberth-Wagner C, Ludwig J, Bruder AK, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2ʹO-methylated self RNA. Immunity. 2015;43(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Devarkar SC, Wang C, Miller MT, et al. Structural basis for m7G recognition and 2ʹ-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A. 2016;113(3):596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li SH, Dong H, Li XF, et al. Rational design of a flavivirus vaccine by abolishing viral RNA 2ʹ-O methylation. J Virol. 2013;87(10):5812–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zust R, Dong H, Li, XF, et al. Rational design of a live attenuated dengue vaccine: 2ʹ-o-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog. 2013;9(8):e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zust R, Li SH, Xie X et al. Characterization of a candidate tetravalent vaccine based on 2ʹ-O-methyltransferase mutants. PLoS One. 2018;13(1):e0189262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schmid B, Rinas M, Ruggieri A, et al. Live cell analysis and mathematical modeling identify determinants of attenuation of dengue virus 2ʹ-O-methylation mutant. PLoS Pathog. 2015;11(12):e1005345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chang DC, Hoang LT, Mohamed Naim AN, etal. Evasion of early innate immune response by 2ʹ-O-methylation of dengue genomic RNA. Virology. 2016;499:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Daffis S, Szretter KJ, Schriewer J, et al. 2ʹ-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Szretter KJ, Daniels BP, Cho H, et al. 2ʹ-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8(5):e1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kimura T, Katoh H, Kayama H, et al. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5ʹ capped 2ʹ-O unmethylated RNA. J Virol. 2013;87(18):9997–10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mears HV, Sweeney TR. Better together: the role of IFIT protein-protein interactions in the antiviral response. J Gen Virol. 2018;99(11):1463–1477. [DOI] [PubMed] [Google Scholar]
- [58].Der SD, Zhou A, Williams BR, et al. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(26):15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Geiss G, Jin G, Guo J, et al. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276(32):30178–30182. [DOI] [PubMed] [Google Scholar]
- [60].Guo J, Peters KL, Sen GC. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267(2):209–219. [DOI] [PubMed] [Google Scholar]
- [61].Guo J, Hui DJ, Merrick WC, et al. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. Embo J. 2000;19(24):6891–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hui DJ, Bhasker CR, Merrick WC, et al. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J Biol Chem. 2003;278(41):39477–39482. [DOI] [PubMed] [Google Scholar]
- [63].Hui DJ, Terenzi F, Merrick WC, et al. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J Biol Chem. 2005;280(5):3433–3440. [DOI] [PubMed] [Google Scholar]
- [64].Terenzi F, Hui DJ, Merrick WC, et al. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281(45):34064–34071. [DOI] [PubMed] [Google Scholar]
- [65].Habjan M, Hubel P, Lacerda L, et al. Sequestration by IFIT1 impairs translation of 2ʹO-unmethylated capped RNA. PLoS Pathog. 2013;9(10):e1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pichlmair A, Lassnig C, Eberle CA, et al. IFIT1 is an antiviral protein that recognizes 5ʹ-triphosphate RNA. Nat Immunol. 2011;12(7):624–630. [DOI] [PubMed] [Google Scholar]
- [67].Abbas YM, Laudenbach BT, Martinez-Montero S, et al. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2ʹ-O methylations. Proc Natl Acad Sci U S A. 2017;114(11):E2106-E2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kumar P, Sweeney TR, Skabkin MA, et al. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5ʹ-terminal regions of cap0-, cap1- and 5ʹppp- mRNAs. Nucleic Acids Res. 2014;42(5):3228–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dong H, Chang DC, Hua MH, et al. 2ʹ-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012;8(4):e1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bullard KM, Gullberg RC, Soltani E, et al. Murine efficacy and pharmacokinetic evaluation of the flaviviral NS5 capping enzyme 2-thioxothiazolidin-4-one inhibitor BG-323. PLoS One. 2015;10(6):e0130083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Stahla-Beek HJ, April DG, Saeedi BJ, et al. Identification of a novel antiviral inhibitor of the flavivirus guanylyltransferase enzyme. J Virol. 2012;86(16):8730–8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bougie I, Bisaillon M. The broad spectrum antiviral nucleoside ribavirin as a substrate for a viral RNA capping enzyme. J Biol Chem. 2004;279(21):22124–22130. [DOI] [PubMed] [Google Scholar]
- [73].Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98(12):6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Crotty S, Maag D, Arnold JJ, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6(12):1375–1379. [DOI] [PubMed] [Google Scholar]
- [75].Day CW, Smee DF, Julander JG, et al. Error-prone replication of West Nile virus caused by ribavirin. Antiviral Res. 2005;67(1):38–45. [DOI] [PubMed] [Google Scholar]
- [76].Leyssen P, Balzarini J, De Clercq E, et al. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol. 2005;79(3):1943–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Dong H, Ren S, Zhang B, et al. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol. 2008;82(9):4295–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Selisko B, Peyrane FF, Canard B, et al. Biochemical characterization of the (nucleoside-2ʹO)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n). J Gen Virol. 2010;91(Pt 1):112–121. [DOI] [PubMed] [Google Scholar]
- [79].Lim SP, Sonntag LS, Noble C, et al. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011;286(8):6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Brecher M, Chen H, Li Z, et al. Identification and characterization of novel broad-spectrum inhibitors of the flavivirus methyltransferase. ACS Infect Dis. 2015;1(8):340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Stephen P, Baz M, Boivin G, et al. Structural Insight into NS5 of Zika Virus leading to the discovery of MTase inhibitors. J Am Chem Soc. 2016;138(50):16212–16215. [DOI] [PubMed] [Google Scholar]
- [82].Benmansour F, Trist I, Coutard B, et al. Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design. Eur J Med Chem. 2017;125:865–880. [DOI] [PubMed] [Google Scholar]
- [83].Coutard B, Barral K, Lichiere J, et al. Zika virus methyltransferase: structure and functions for drug design perspectives. J Virol. 2017;91(5):e02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Centers for Disease Control and Prevention. Adverse events associated with 17D-derived yellow fever vaccination–United States, 2001-2002. MMWR Morb Mortal Wkly Rep. 2002;51(44):989–993. [PubMed] [Google Scholar]
- [85].Vasconcelos PF, Luna EJ, Galler R, et al. Serious adverse events associated with yellow fever 17DD vaccine in Brazil: a report of two cases. Lancet. 2001;358(9276):91–97. [DOI] [PubMed] [Google Scholar]
- [86].Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. [DOI] [PubMed] [Google Scholar]
- [87].Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3ʹ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the adomet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3(11):1233–1247. [PMC free article] [PubMed] [Google Scholar]
- [89].Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. [DOI] [PubMed] [Google Scholar]
- [90].Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5ʹ sites. Cell Rep. 2014;8(1):284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Yang J, Chen J, Fei X, et al. N6-methyladenine RNA modification and cancer. Oncol Lett. 2020;20(2):1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang L, Song C, Wang N, et al. NADP modulates RNA m6A methylation and adipogenesis via enhancing FTO activity. Nat Chem Biol. 2020;16(12):1394–1402. [DOI] [PubMed] [Google Scholar]
- [101].Shafik AM, Allen EG, Jin P. Dynamic N6-methyladenosine RNA methylation in brain and diseases. Epigenomics. 2020;12(4):371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Du K, Zhang L, Lee T, et al. m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56(3):1596–1606. [DOI] [PubMed] [Google Scholar]
- [103].Li Y, Wang J, Huang C, et al. RNA N6-methyladenosine: a promising molecular target in metabolic diseases. Cell & Bioscience. 2020;10(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Livneh I, Moshitch-Moshkovitz S, Amariglio N, et al. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21(1):36–51. [DOI] [PubMed] [Google Scholar]
- [105].Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, et al. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. [DOI] [PubMed] [Google Scholar]
- [106].Cortese M, Goellner S, Acosta EG, et al. Ultrastructural characterization of Zika Virus replication factories. Cell Rep. 2017;18(9):2113–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Miorin L, Romero-Brey I, Maiuri P, et al. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J Virol. 2013;87(11):6469–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Welsch S, Miller S, Romero-Brey I, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gillespie LK, Hoenen A, Morgan G, et al. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol. 2010;84(20):10438–10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Junjhon J, Pennington JG, Edwards TJ, et al. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol. 2014;88(9):4687–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ooi YS, Majzoub K, Flynn RA, et al. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat Microbiol. 2019;4(12):2369–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Phillips SL, Soderblom EJ, Bradrick SS, et al. Identification of proteins bound to dengue viral RNA in vivo reveals new host proteins important for virus replication. mBio. 2016;7(1):e01865–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Dechtawewat T, Songprakhon P, Limjindaporn T, et al. Role of human heterogeneous nuclear ribonucleoprotein C1/C2 in dengue virus replication. Virol J. 2015;12(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282(42):30497–30508. [DOI] [PubMed] [Google Scholar]
- [115].Liu N, Dai Q, Zheng G, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature. 2015;518(7540):560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Liu N, Zhou KI, Parisien M, et al. N 6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Roost C, Lynch SR, Batista PJ, et al. Structure and thermodynamics of N 6 -methyladenosine in RNA: A spring-loaded base modification. J Am Chem Soc. 2015;137(5):2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Friedrich S, Engelmann S, Schmidt T, et al. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J Virol. 2018;92(6):e01647-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Liu ZY, Li XF, Jiang T, et al. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife. 2016. 5:e17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Villordo SM, Alvarez DE, Gamarnik AV. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA. 2010;16(12):2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Dethoff EA, Boerneke MA, Gokhale NS, et al. Pervasive tertiary structure in the dengue virus RNA genome. Proc Natl Acad Sci U S A. 2018;115(45):11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Huber RG, Lim XN, Ng WC, et al. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Nat Commun. 2019;10(1):1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Li P, Wei Y, Mei M, et al. Integrative analysis of Zika virus genome RNA structure reveals critical determinants of viral infectivity. Cell Host Microbe. 2018;24(6):875–886. e5 [DOI] [PubMed] [Google Scholar]
- [124].Pirakitikulr N, Kohlway A, Lindenbach BD, et al. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol Cell. 2016;62(1):111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Kim GW, Imam H, Khan M, et al. N6-methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem. 2020;295(37):13123-13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Gokhale NS, McIntyre ABR, Mattocks MD, et al. Altered m(6)A modification of specific cellular transcripts affects Flaviviridae infection. Mol Cell. 2020;77(3):542–555. e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].McIntyre ABR, Gokhale NS, Cerchietti L, et al. Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci Rep. 2020;10(1):6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Michalski D, Ontiveros JG, Russo J, et al. Zika virus noncoding sfRNAs sequester multiple host-derived RNA-binding proteins and modulate mRNA decay and splicing during infection. J Biol Chem. 2019;294(44):16282–16296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Ward AM, Bidet K, Yinglin A, et al. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3ʹ UTR structures. RNA Biol. 2011;8(6):1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Eckwahl M, Xu R, Michalkiewicz J, et al. 5-methylcytosine RNA modifications promote retrovirus replication in an ALYREF reader protein-dependent manner. J Virol. 2020;94(13):e00544-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Courtney DG, Tsai K, Bogerd HP, et al. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe. 2019;26(2):217–227. e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18(5):275–291. [DOI] [PubMed] [Google Scholar]
- [133].Motorin Y, Helm M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes (Basel). 2019;10(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Paulines MJ, Wetzel C, Limbach PA. Using spectral matching to interpret LC-MS/MS data during RNA modification mapping. J Mass Spectrom. 2019;54(11):906–914. [DOI] [PubMed] [Google Scholar]
- [135].Yu N, Jora M, Solivio B, et al. tRNA modification profiles and codon-decoding strategies in methanocaldococcus jannaschii. J Bacteriol. 2019;201(9):e00690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Horowitz S, Horowitz A, Nilsen TW, et al. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984;81(18):5667–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Grozhik AV, Olarerin-George AO, Sindelar M, et al. Antibody cross-reactivity accounts for widespread appearance of m(1)A in 5ʹUTRs. Nat Commun. 2019;10(1):5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Helm M, Lyko F, Motorin Y. Limited antibody specificity compromises epitranscriptomic analyses. Nat Commun. 2019;10(1):5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lentini A, Lagerwall C, Vikingsson S, et al. A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat Methods. 2018;15(7):499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Rothbart SB, Dickson BM, Raab JR, et al. An interactive database for the assessment of histone antibody specificity. Mol Cell. 2015;59(3):502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Linder B, Grozhik AV, Olarerin-George AO, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Kane SE, Beemon K. Inhibition of methylation at two internal N6-methyladenosine sites caused by GAC to GAU mutations. J Biol Chem. 1987;262(7):3422–3427. [PubMed] [Google Scholar]
- [143].Legrand C, Tuorto F, Hartmann M, et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27(9):1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Onafuwa-Nuga AA, King SR, Telesnitsky A. Nonrandom packaging of host RNAs in moloney murine leukemia virus. J Virol. 2005;79(21):13528–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Simonova A, Svojanovska B, Trylcova J, et al. LC/MS analysis and deep sequencing reveal the accurate RNA composition in the HIV-1 virion. Sci Rep. 2019;9(1):8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113(1):165–179. [DOI] [PubMed] [Google Scholar]
- [147].Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985;5(9):2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Canaani D, Kahana C, Lavi S, et al. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979;6(8):2879–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Sommer S, Salditt-Georgieff M, Bachenheimer S, et al. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 1976;3(3):749–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Bartkoski MJ Jr., Roizman B. Regulation of herpesvirus macromolecular synthesis. VII. Inhibition of internal methylation of mRNA late in infection. Virology. 1978;85(1):146–156. [DOI] [PubMed] [Google Scholar]
- [151].Narayan P, Ayers DF, Rottman FM, et al. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol Cell Biol. 1987;7(4):1572–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Dubin DT, Stollar V. Methylation of Sindbis virus “26S” messenger RNA. Biochem Biophys Res Commun. 1975;66(4):1373–1379. [DOI] [PubMed] [Google Scholar]
- [153].Dubin DT, Stollar V, Hsuchen CC, et al. Sindbis virus messenger RNA: the 5ʹ-termini and methylated residues of 26 and 42 S RNA. Virology. 1977;77(2):457–470. [DOI] [PubMed] [Google Scholar]
- [154].Motorin Y, Marchand V. Detection and analysis of RNA Ribose 2ʹ-O-methylations: challenges and solutions. Genes (Basel). 2018;9(12):642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Ringeard M, Marchand V, Decroly E, et al. FTSJ3 is an RNA 2ʹ-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019;565(7740):500–504. [DOI] [PubMed] [Google Scholar]
- [156].Kim D, Lee JY, Yang JS, et al. The architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181(4):914–921. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Viehweger A, Krautwurst S, Lamkiewicz K, et al. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29(9):1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Behm-Ansmant I, Helm M, Motorin Y. Use of specific chemical reagents for detection of modified nucleotides in RNA. J Nucleic Acids. 2011;2011:408053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Marchand V, Ayadi L, Ernst FGM, et al. AlkAniline-Seq: profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angew Chem Int Ed Engl. 2018;57(51):16785–16790. [DOI] [PubMed] [Google Scholar]


