ABSTRACT
Banana as an important economic crop worldwide, often suffers from serious damage caused by Fusarium oxysporum f. sp. Cubense. Arbuscular mycorrhizal (AM) fungi have been considered as one of the promising plant biocontrol agents in preventing from root pathogens. This study examined the effect of AM fungal inoculation on plant growth and differential expressions of growth- and defense-related genes in banana seedlings. Tissue-cultured seedlings of Brazilian banana (Musa acuminate Cavendish cv. Brail) were inoculated with AM fungus (Rhizophagus irregularis, Ri), and developed good mycorrhizal symbiosis from 4 to 11 weeks after inoculation with an infection rate up to 71.7% of the roots system. Microbial abundance revealed that Ri abundance in banana roots was 1.85×106 copies/ml at 11 weeks after inoculaiton. Inoculation improved plant dry weights by 47.5, 124, and 129% for stem, leaf, and the whole plant, respectively, during phosphate depletion. Among a total of 1411 differentially expressed genes (DEGs) obtained from the transcriptome data analysis, genes related to plant resistance (e.g. POD, PAL, PYR, and HBP-1b) and those related to plant growth (e.g. IAA, GH3, SAUR, and ARR8) were up-regulated in AM plants. This study demonstrates that AM fungus effectively promoted the growth of banana plants and induced defense-related genes which could help suppress wilt disease. The outcomes of this study form a basis for further study on the mechanism of banana disease resistance induced by AM fungi.
KEYWORDS: Banana (Musa), Arbuscular mycorrhiza (AM) fungi, Transcriptome, Differentially expressed genes (DEGs)
Introduction
Bananas are the most high-yield fruit and provide staple food for around 400 million people worldwide.1 Currently, its production is damaged destructively by the soil-born disease induced by Fusarium oxysporum f. sp. Cubense, especially the tropical race 4 (Foc TR4), which resulted in economic, environmental, and safety threat due to excessive fungicide input.2 To respond to the ecological principles of sustainable development, biological control has gained interest in the research of disease resistance mechanism. Studies have shown some microorganisms effectively suppress the pathogen Foc TR4. The endophytic bacterial strain ITBB B5-1 isolated from the rubber tree suppressed the mycelial growth of the pathogenic fungus Foc TR4.4 Application of a bioorganic fertilizer formulated by combining Bacillus amyloliquefaciens NJN-6 with compost significantly decreased Foc TR4 disease incidence by 68.5%6. The glasshouse trial showed that the consortium of Pseudomonas aeruginosa DRB1 and Trichoderma harzianum CBF2 with the bioformulation in pasta granules reduced disease severity (DS) by more than 50 center dot against Foc TR4.5 Our laboratory research found Burkholderia sp. HQB-1 efficiently controlled banana wilt through phenazine-1-carboxylic acid secretion.7 Those discoveries enhance our understanding of biocontrol practices for plant defense.2–4
Arbuscular mycorrhizal (AM) fungi, widely distributed in terrestrial ecosystems, not only improve plant mineral nutrition but also play an important role in controlling soil-borne diseases and regulating soil structur.7, 8 It has been reported that the induction of defense responses was much higher and more rapid in the AM inoculated potatoes than in the un-inoculated treatments in the presence of pathogen infection.9 Besides, a significant reduction of the mosaic disease severity and incidence was observed as a response to the mycorrhizal colonization of the infected tomato.10 AM colonization alleviates F. oxysporum wilt in plants such as asparagus and tomato.11–12 Glomus applied significantly improved banana seedling growth, and offered 100% protection to the ‘Lakatan’ seedlings against Foc TR4, indicating that AM fungi colonization resulted in weak but distinctly enhanced plant resistance to pathogens. However, the underlying mechanism of AMF-induced disease resistance remains elusive.
Previsous studies focus on the analysis of plant physiology and biochemistry following AM fungi inoculation. AM fungi inoculated Medicago sativa plants had higher biomass and nitrogen (N), phosphorus (P) and potassium (K) contents13. Study have found banana roots are also colonized and mount plant vigour which enhances water absorption and mineral nutrient uptake.14 Under natural conditions, AM fungi had a positive effect on banana plant height, chlorophyll content, and leaf N, P, K concentrations.15 However, there are few studies on the molecular mechanism of AM fungi in banana.
In this study, we examined mycorrhizal colonization and abundance in banana roots and analyzed alterations in gene expression following AM fungi inoculation. We hypothesized that AM fungi could induce the defense systems in banana by regulating gene-related expression. To address this, the transcriptome sequencing technique was used to identify differentially expressed genes related to defense and plant growth in mycorrhizal banana plants, and to analyze the disease-related pathways.
Material and methods
Materials
The tissue-cultured seedling with 5 leaf-age in the consistent growth of Brazilian banana (Musa acuminate Cavendish cv. Brail), were provided by Zhangzhou Weitian Biotechnology Co., Ltd. China.
The AM inoculum of Rhizophagus irregularis (Ri) was purchased from Premier Tech Biotechnologies, Riviere-Du-Loup, QC, Canada (https://www.ptagtiv.com/en/products/zone/united-states/). The inoculum contains 4000 spores per ml. It was stored at 4 °C till use.
The substrate was amixture of river sand and soil (v/v, 2:1). The soil was from Huaqiao University campus (Xiamen Campus), and the river sand was purchased from the local market. The sand was washed several times with tap water. The mixed substrate was autoclaved at 121°C for 30 min, then cooled at room temperature prior to potting.
Experimental design
The pot experiment with banana was conducted in aglasshouse of Huaqiao University, China. Acomplete randomized block design was used consisting of two inoculation treatments (with AM fungi inoculaiton, AMF; and without AM fungi inoculaiton, CK), four sampling times (5, 7, 9 and 11 weeks after inoculation) with four replications (pots) per treatment of each harvest.
The inoculum of Rhizophagus irregularis (Ri) was activated in the dark at 25 °C incubators for 30 min. Tissue-cultured banana seedlings were washed with distilled water to remove all medium attached to roots. About 5 ml of Ri solution and 45 ml aseptic water were mixed in a100 ml beaker. The seedling was soaked in the 50 ml suspension for 30 min at room temperature, and the CK was treated by distilled water. The banana seedlings were planted in plastic pots (7cm in width, 7cm in height) filled with 350 g substrates. Each pot contained one seedling. The remaining solution containing AM fungi spores was evenly irrigated into all pots of AM fungi treatment (each pot inoculated with around 1000 spores). The average temperature was 25/15 °C (d/N), with 80% relative humidity in the glasshouse. The seedlings were irrigated with a25 ml phosphorus-free Hoagland solution every two weeks. Eleven weeks later after transplanting, all banana seedlings were harvested for assessments. Seedlings were removed from the soil and divided into above-ground and roots. About 100 fragments of fresh roots were selected for the mycorrhizal infection, and the left was quickly conserved in liquid nitrogen. The dry weights of the above-ground and roots were determined by the conventional method.
Microscopic observation of AM fungi colonization
Banana seedlings were sampled for assessing AM fungal colonization at 5, 7, 9, 11 weeks after inoculation. After washing, root samples were cut into about 1cm in length, and placed in 50 ml centrifuge tubes with FAA (formaldehyde, acetic acid, and 50% ethanol, 13:5:200, v/v/v) for at least 24 h. Then, the roots were cleaned in 20% KOH solution, 90 °C water bath for 1 h. After rinsing with water 3―5 times, the roots were decolorized in alkaline H2O2 (ammonia, 10% H2O2 and water, 1:10:200, v/v/v) for 1.5 h at room temperature. The root sections were rinsed into 5% acetic acid and acidify for 5 min. Afterward, the roots were stained with 5% acetic acid-ink, 66 °C water bath for 30 min. The section covered with alayer of PVLG and used magnified intersections method observation count under amicroscope.
The mycorrhizal colonization rate was calculated as the percentage of infected root sections over the total number of root sections. The infection rate was calculated as follows:
Mycorrhiza colonization rate(%) = number of root segments infected by AM fungi/the total number of root segments × 100%.
Real-time quantitative PCR to detect AM fungi abundance
To isolate high-quality AM fungus DNA, about 1200 spores were selected from the liquid Ri inoculum. The sample was fully ground with mortar and pestle in the presence of liquid nitrogen and the AM fungi DNA was extracted according to fungal genomes DNA instructions. The AM fungal specificity primer references were designed by16, in Table 1.
Table 1.
Two primers used in this experiment
| Sample | Primer name | Primer sequence (5ʹ-3ʹ) | Tm(°C) | Product size(bp) |
|---|---|---|---|---|
| AMF | Intra 1 | GGTGCGATTCTGTGGAGTGTGAGG | 58 | 250 |
| Intra 2 | CAAGCTTTCGGCACCAGAGCAACG |
PCR amplification, the primer specificity was tested in 3% agarose gel. After the target fragment was obtained and purified by gel cutting, and the specificity fragment AMF250 was cloned into the pMD-18-T Vector carrier using Takara pMD-18-T Clone Kit (Takara, Japan]. The Ecoli DH5α was confirmed through Blue-White Screening and PCR to confirm AMF250 expression transformants.
The standard recombinant plasmids carrying AMF250 were extracted, and the plasmid concentration was determined by Implen Ultramicro-spectrophotometer. The SYBR Green for qPCR was used by setting 10Xgradients with the range from 10−1–10−7 copies/mL. The total volume of the system was 25 μl. The AM fungal reaction program was as follows: 30 sdenaturation at 95 °C, followed by 40 cycles at 95 °C 15 sec, 58 °C 90 sec, 72 °C 60 sec, and 72 °C 10 min. The production was dissolved analysis at 95 °C 15 sec, 60 °C 30 sec, 95 °C 15 sec, and each treatment was repeated three times. Based on the standard curve of qPCR, the number of AM fungi was calculated in the tested samples. We tested AM fungi abundance in the banana roots sampled at 5, 7, 9, 11 weeks after inoculation, respectively, and fungal DNA was isolated using the plant DNA extraction kit (Shenggong, Shanghai).
RNA extraction and the library construction for RNA-Seq
The total RNA was extracted from banana roots by RNA Purification Kit (Tiangen Biochemical Technology Co., Ltd.) method. The extent of RNA degradation and pollution was analyzed with agarose gel electrophoresis. The OD 260/280 ratio of the total RNA was checked with Nanodrop to ensure purity. The RNA concentration and integrity were evaluated using Qubit and Agilent 2100, respectively.
The sequencing libraries were constructed after aqualified RNA sample test, and Eukaryotic mRNA was enriched from total RNA using poly-T oligo attached magnetic beads through A-Tcomplementary pairing and mRNA poly-A tail combined way. Soon afterward, the fragmentation buffer was added to interrupt mRNA into short fragments. The first-strand cDNA was synthesized using random hexamers, mRNA as template, and reverse transcriptase, followed by thesecond strand cDNA synthesis using abuffer, dNTPs, DNA polymerase I.The double-stranded cDNA was purified by AMPure XP beads, and then subjected to terminal repair and poly-A tail was extended, and connected with sequencing adapters, choosing suitable fragments. Finally, the PCR amplification was performed, and the final library was obtained.
While the library construction was completed, the RNA concentration of the library was assessed with Qubit 2.0 to preliminary quantify. The insert size was detected using Agilent 2100 and qualified insert size was accurate quantification for effective concentration using the Q-PCR method.
RNA-Seq data analysis
To identify the differential genes induced by AM fungi, the cDNA library was constructed, and the Illumina HiSeqTM 2500 transcriptome analysis was carried out by the platform. The banana genomes were compared with the genomic data of wild banana. Clean Data was mapped to the reference genome using HISAT. The expression level of the gene in each sample was analyzed by HTSeq software using the union model and was estimated as Fragments Per Kilobase of transcript sequence per Millon’s base pairs sequenced (FPKM). Differential expression genes (DEGs) between each chosen sample pairs were detected by DEseq software, and were obtained by screening standard of the gene with |log2 (Fold Change)| > 1 and p-value < 0.05. The identified DEGs were subjected to GO and KEGG enrichment analyses. The KOBAS 2.0 software was used to calculate pathway enrichment of DEGs, we used an FDR ≤ 0.05 to define significantly enriched KEGG pathways and GO terms.
Statistical analysis
Statistical analysis was conducted by the software SPSS 18.0 (IBM, USA). And multiplicity analysis of variance and one-way ANOVA were used to determine the significant differences.
Results
Mycorrhizal colonization
Banana roots of inoculated seedlings were colonized by Ri at 5 to 11 weeks after inoculation (Figure 1). AM fungal sporophyte and mycelium (Figure 1a), the full shape, relatively smooth spores attached the surface of the root, and the hyphal growth along the cell wall (Figure 1b) was observed in root samples of inoculated seedlings under microscope. Moreover, the mycorrhizal infection rates were increased gradually with the culture time, and the maximum infection rate was up to 71.7% after 11 weeks’ pot culture (Figure 2a), indicating Ri and Brazilian banana developed good symbioses. No AM structures were observed in the non-inoculated plants.
Figure 1.
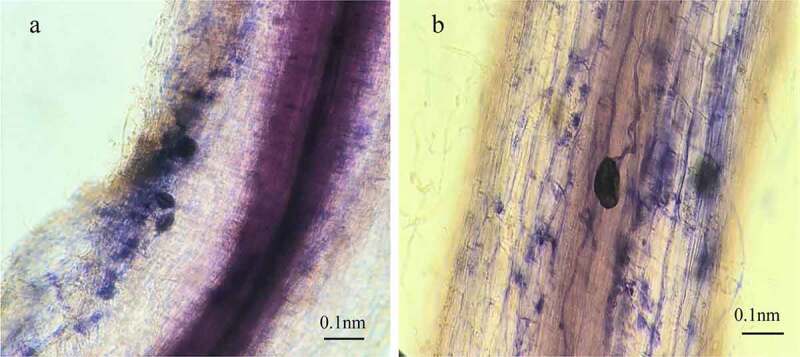
Morphological structure of Brazilian banana roots colonized by AM fungus Rhizophagus irregularis. AM fungal sporophyte and mycelium (a), the full shape, relatively smooth spores attached the surface of the root, and the hyphal growth along the cell wall (b) was observed in root samples of inoculated seedlings under microscope
Figure 2.
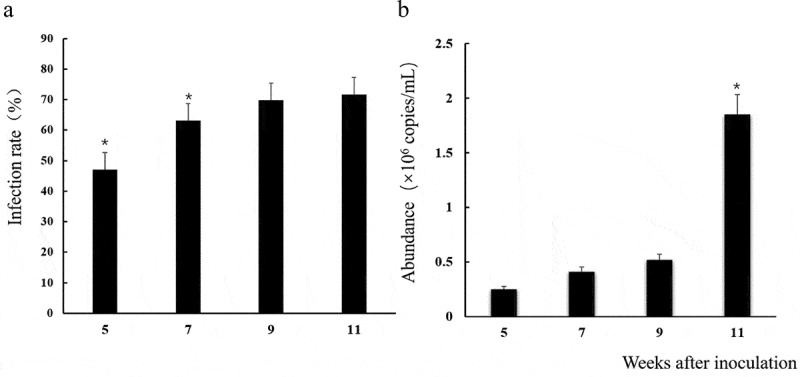
Mycorrhizal infection rates (a) and abundance abundance of Rhizophagus irregularis (b) in Brazilian banana roots at 5, 7, 9 and 11 weeks after inoculation with AM fungus. Error bars represent standard error of the mean. The average expression of four biological replicates is shown in the same time. The t-test was used to calculate significance (*p< .05)
AM fungal abundance in banana roots
The standard recombinant plasmid was diluted 10-fold gradient ranging from 10−1 ~ 10−7 copies/mL (Fig. S1). The amplification standard curve equation of AM fungi was Y= −3.14 X+ 36, R2 = 0.999, E% = 108.4. The amplification curve of the standard fragment of the recombinant plasmid was smooth and atypical S-shaped curve was present, and the interval of each cycle threshold was uniform.
The qPCR results showed that the Ri abundance in banana root was detected effectively at 5 weeks after inoculation, and it increased following the culture time. It was the highest (1.85 × 106 copies/mL) at the 11 weeks, about 7.4 times more than that at 5 weeks (Figure 2b).
Effect of AM inoculation on plant growth
Compared with CK, the growth (the above-ground parts weight, whole plant weight, plant height and maximum root length) of inoculated seedlings was increased significantly (P< .05; Figure 3), and the increments of stem, leaf, and the whole plants were 47.5%, 123.5% and 128.9% respectively (Figure 3a). The average stem diameter of inoculated seedlings was significantly higher than that of CK. Besides, the plant height and the maximum root length were improved by 6.76% and 12.8% in relative to CK (Figure 3b). Dry weight of AM roots was decreased by 55.3% over that of CK (Figure 3a) in that the roots system of CK is more developed than that of AM. The results showed that AM fungi could promote the growth of the above-ground parts in the bananas seedling.
Figure 3.
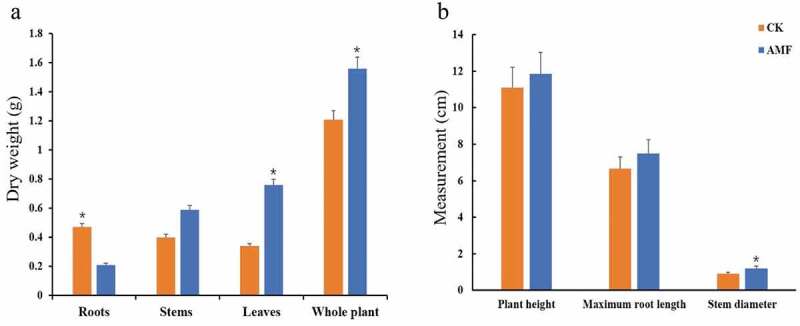
The effect of AM inoculation with Rhizophagus irregularis on biomass accumulations (dry weights) of the root, stem, leave and the whole plant (a) and plant height, maximum root length and stem diameter (b) of Brazilian banana seedlings at 11 weeks after inoculation. Error bars represent standard error of the mean. Per treatment the average expression of four biological replicates is shown. The t-test was used to calculate significance (*p< .05)
Transcriptome analysis
A total of 69,232,109 and 79,591,958 raw reads for the CK and AMF samples were obtained, respectively (Table 2). After removing the low-quality reads, 64,738,040 and 71,307,217 clean reads from the mapped wild banana genome were acquired. The average value of GC content was 50.2%. The percentage of Q20 and Q30 was more than 97 and 92, respectively. The percentage of total mapped reads to total reads was between 76.9 and 85.4, while the multiple mapped reads were ranged from 1.15 to 1.44%.
Table 2.
The summary of sequence analysis of transcriptome
| Sample | Raw reads | Clean reads | Total mapped | Multiple mapped | Q20(%) | Q30(%) | GC (%) |
|---|---|---|---|---|---|---|---|
| CK1 | 72,029,856 | 68,963,152 | 53,007,409 | 792,530 | 97.17 | 92.46 | 50.85 |
| CK2 | 57,703,178 | 55,302,364 | 47,027,339 | 726,840 | 97.44 | 93.12 | 50.43 |
| CK3 | 77,963,292 | 69,948,604 | 59,737,534 | 1006,595 | 97.85 | 94.17 | 50.28 |
| AMF1 | 81,741,046 | 73,378,282 | 59,520,784 | 973,058 | 97.77 | 93.99 | 50.06 |
| AMF2 | 77,138,272 | 68,955,004 | 55,999,100 | 916,177 | 97.88 | 94.29 | 48.71 |
| AMF3 | 79,896,556 | 71,588,366 | 58,093,959 | 942,135 | 97.80 | 94.05 | 49.28 |
A total of 1411 DEGs were obtained, of which 76.25% DEGs were up-regulated and 32.75% were down-regulated (Figure 4, Table S1), emphasizing the complex transcription process of banana roots in response to Ri inoculation.
Figure 4.
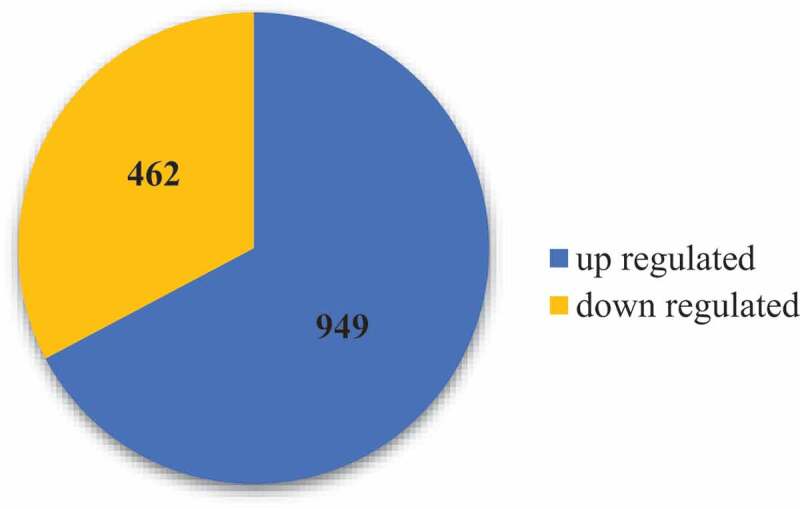
The differentially expressed genes in the root samples between AM colonization and CK
A total of 1101 DEGs (78% of all 1411 DEGs) were annotated to three principal GO categories separately, including biological process (BP), cellular component (CC), and molecular function (MF) (Figure 5). In GO terms that are significantly enriched up-regulated expression genes, among BP, “biological process”, “phosphorus metabolic process” and “cellular protein modification process” were the dominant terms. The top three significantly enriched terms in MF were “small molecule binding”, “iron-binding” and “catalytic activity”. No DEGs were significantly enriched in CC (Fig S2).
Figure 5.
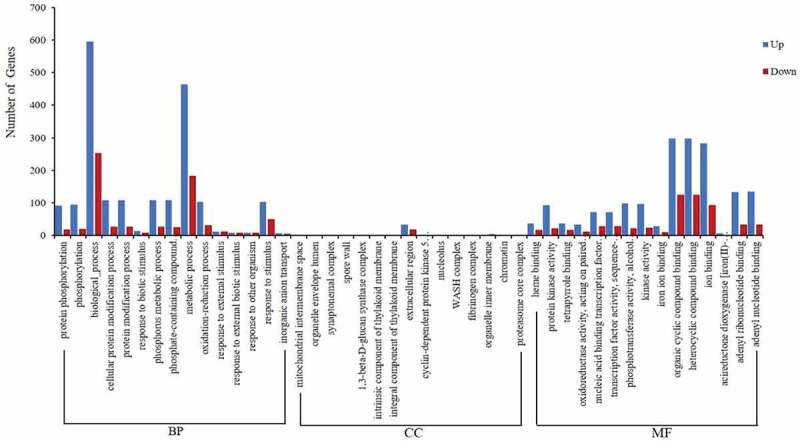
GO classification for the DEGs with inoculation of AM fungi in banana. BP: biological process, CC: cellular component, MF: molecular function
A total of eight significantly enriched KEGG pathways in AM plants (Corrected P< .05) (Figure 6a), of which three pathways, namely Plant hormone signal transduction, Flavonoid biosynthesis, and Phenylpropanoid biosynthesis are normally regarded as disease resistance-related events.
Figure 6.
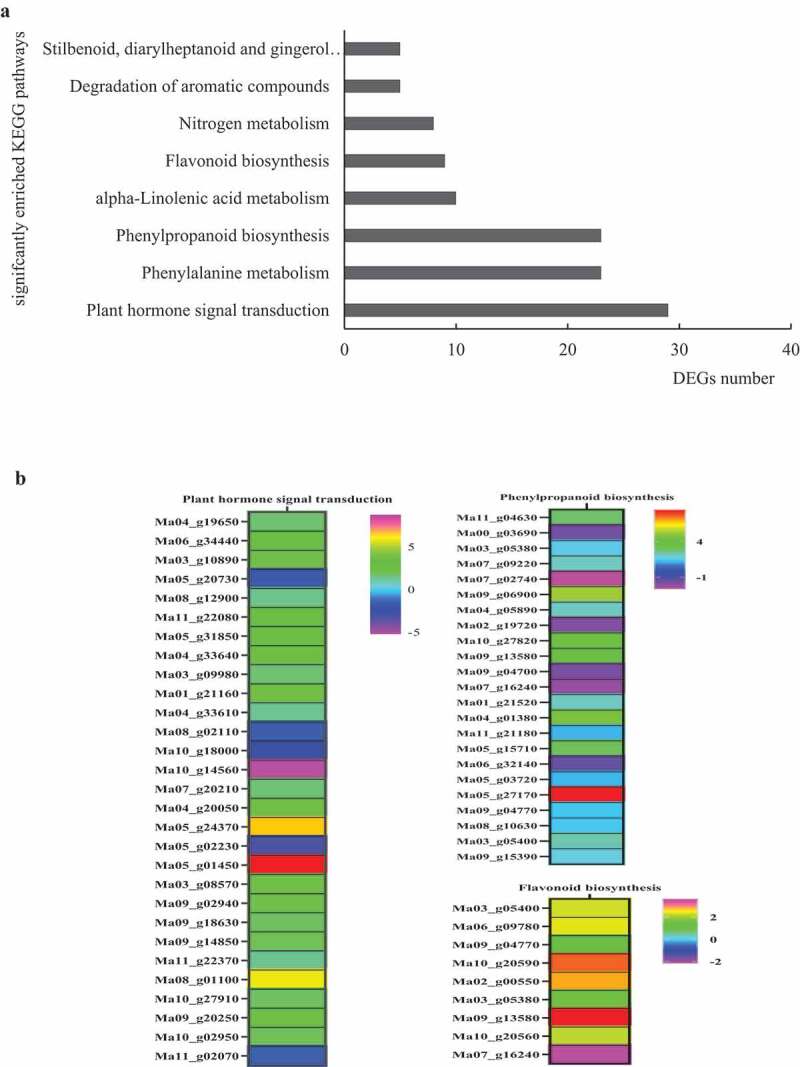
Significantly enriched KEGG pathways of DEGs (a) and three heatmaps of disease resistance-related pathways (b). Clustered heatmaps of log2 fold change values of DEGs
Signal transduction
A total of 29 significant DEGs were involved in plant hormone signal transduction (Figure 6b). The eight genes were enriched in the ABA pathway, including five genes encoding ABA receptor PYR/PYL family (Ma04_g 19650, Ma06_g 34440, Ma11_g 22080, Ma05_g 31850, Ma10_g 27910), and one gene encoding protein phosphatase 2 C (PP2C, Ma11_g22370) were up-regulated rapidly in banana response to the Ri infection. In auxin pathway, four genes respectively encoding auxin-responsive SAUR gene family member (Ma07_g20210), auxin-responsive protein IAA (Ma09_g02940), indole-3-acetic acid-amido synthetase GH3.1 (Ma05_g01450), indole-3-acetic acid-induced protein ARG7 (Ma01_g21160), and two genes encoding auxin-induced protein (Ma05_g24370, Ma09_g14850) were found up-regulated in banana during the Ri infection process (Figure 6b). For the JA pathway, two genes respectively encoding protein TIFY 10B (TIFY10b, Ma03_g09980) and transcription factor (Ma10_g02950) were up-regulated to ahigh level (Figure 6b). Two ET-signaling genes respectively encoding Protein EIN4 (ETR, Ma08_g12900) and Ethylene-responsive transcription factor 1 (ERF1, Ma09_g20250) had high expression levels in banana. In the SA signaling pathway, one gene encoding transcription factor HBP-1b (HBP-1b, Ma03_g10890) was induce higher in the banana stage of Ri infection. Other genes associated with Cytokinine and Gibberellin acid (GA) also responded to Ri. Down-stream in the Cytokinine-signaling pathway was found up-regulated ARR8 gene (Ma03_g08570). For GA signaling, one DELLA protein RGA2 (Ma08_g01100) gene was expressed higher in banana infected by Ri (Figure 6b).
Secondary metabolism
Phenylpropanoid biosynthesis was ametabolic pathway with ahigher number of DEGs including 17 up-regulated genes, and six down-regulated genes. Among the up-regulated genes, four trans-cinnamate 4-monooxygenase genes (Ma05_g15710, Ma03_g05400, Ma09_g04770, Ma09_g13580), three phenylalanine ammonia-lyase genes (PAL, Ma05_g03720, Ma11_g21180, Ma09_g15390), two β-glucosidase genes (Ma04_g01380, Ma09_g06900), one 4–coumarate-CoA ligase 3 gene (4CL, Ma04_g05890), one cytochrome P450 CYP73A (CYP450 73A, Ma03_g05380) gene and one aldehyde dehydrogenase family 2 members (ALDH, Ma05_g27170), were involved in lignin synthesis, indicating plant’s defense ability at cellular level through cell wall reinforce (Figure 6b). Nine genes were found to encode of Peroxidase (POD) family members, of which five genes are up-regulated (Ma08_g10630, Ma07_g09220, Ma01_g21520, Ma11_g04630, Ma10_g27820) in AM plants (Figure 6b). In the flavonoid biosynthesis pathway, eight genes were accumulated abundantly in AM plants, including one of each chalcone synthase (CHS, Mao6_g09780) gene, flavonoid 3ʹ5’-hydroxylase (F3ʹ5 H, Ma02_g00550) gene, curcumin synthase (CURS, Ma10_g20590) gene, and phenylpropanoylacetyl-CoA synthase (Ma10_g20560) gene (Figure 6b).
Discussion
In the present study, AM fungi have ahigher infection rate in banana roots compared to rice, which used the same AM fungi,17 and promoted the above-ground biomass accumulation under phosphate-deficient conditions. These results corroborate previous studies reporting positive effects of plant growth in the presence of AM fungi.15,18 The biocontrol mechanism of AM fungi has been reported in different plant systems (e.g. improved plant nutrition and competition for food). Mycorrhizal colonization increases nutrient absorption and promotes plant growth, both under unstressed conditions or biological and abiotic stress, leading to greater resistance to root-infecting fungi.15,19,20
Identifying key components of transcriptome response to AM fungi using RNA-Seq may stimulate the discovery and annotation of important genes in the plant’s defense response. In this study, we performed acomprehensive transcriptome analysis against AM fungi on the susceptible cultivars Brazilian banana. Compared with the control, 1411 differentially expressed genes (DEGs) were found in the banana roots (Figure 4, Table S1). Eight significantly enriched KEGG pathways (Figure 6), including three disease resistance-related pathways, were identified.
Phenylpropanoid biosynthesis and flavonoid biosynthesis, belonging to secondary metabolism, have been proved to be involved in plant defense response through reinforcement of plant cell walls and phytoalexins synthesis.21 Here, 17 DEGs were identified to involve in the phenylpropanoid pathway. Among them, the PAL gene encodes akey enzyme in this pathway. The isolated PAL from higher plants has been reported to be closely related to the plant resistance.22 Other genes are also related to lignin synthesis. The 4CL is the key rate-limiting enzyme to regulate lignin biosynthesis, which is located in the downstream branch pathway.23 CYP73A24 played acontrolling quantity role in the lignin synthesis pathway.24 POD was mainly distributed in the downstream of the phenylpropanoid biosynthesis pathway, and the corresponding lignin can be synthesized by using the corresponding single-chain alcohols. Thereby the production p-Hydroxyphenyl lignin, Guaiacyl lignin, 5-Hydroxygualacyl lignin, and syring lignin of this pathway increased (Figure 7). Interestingly, the genes including PAL, 4CL, POD, CYP450 showed increased transcript levels and increase accumulation of phenolic compounds in Foc-TR4 treated roots of the resistant and susceptible banana.25,26 ACHS gene and an F3ʹ5’H gene involved in flavonoids biosynthesis accumulated in the roots of Brazilian banana during banana-AM fungi interaction. Taken together, our results suggest that AM fungi can promote the biosynthesis of secondary metabolites to boost plant defense.
Figure 7.
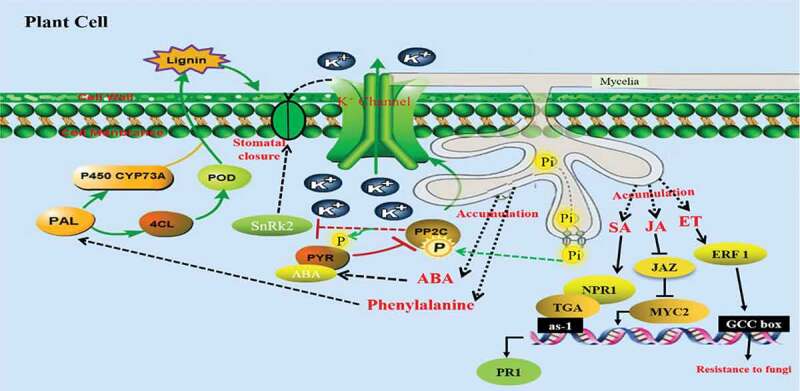
Prediction diagram of defense mechanism in banana against pathogens infection. After banana inoculation with AM, the PAL, 4CL, POD and P450 genes could induce lignin increase in the Phenylpropanoid biosynthesis pathway. AM fungi promote the absorption of phosphorus by plants, PP2C could activate the K+ channel through phosphorylation, and negatively regulate the SnRk2 gene, thereby closing stomata. JAZ proteins are ubiquitinated by interaction with COI1, MYC2 is then released and transcription is activated. NPR1 preferentially presents within the cytoplasm as oligomers while its minor fraction is within the nucleus and interacts with TGA, which binds to PR promoters and enhances PR transcription. ERF transcription factors can bind to the GCC-box cis-acting elements in the promoters of biotic stress-related genes, thereby inducing plants to resist pathogens
Plant hormones such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), and auxin are regarded as playing important roles in mediating plant growth and defense response against biotic stress. The disease resistance and adaptability of plants were enhanced by hormone interactions . In the ABA signaling pathway, we found that five ABA receptor PYR genes were induced higher, and one PP2C gene was up-regulated and two PP2C genes down-regulated. The ABA receptor protein PYR/PYL is normally anegative control type of protein phosphatase 2 C (PP2C).30,31 Some studies reported that ABA and PP2C were involved in the regulation of plant K+ channel, in which PP2C could activate the K+ channel through phosphorylation, resulting in K+ external flow from guard cells and obvious shrinkage of cells, resulting in stomatal closure (Figure 7).32 While PP2C dephosphorylation negatively regulate downstream the SnRk2 gene, also resulting in stomatal closure. Demonstrating that after AM fungus colonization, the host plants activate the host immune system by elicitation of defense-responsive genes. In the auxin signaling pathway, GH3, SAUR and AUX/IAA gene families belong to plant auxin primary reaction genes. Some GH3s can combine with various acyl substrates, such as salicylic acid, jasmonic acid.33,34 Therefore, GH3 protein can maintain the dynamic balance of auxin contents in plants to acertain extent.35,27–29
JA signaling plays regulatory roles in plant development and responses to fungal infection. We observed that TIFY10b was up-regulated, which is JASMONATE-ZIM DOMAIN (JAZ) proteins, as negative regulators of JA signaling. JAZ proteins are repressors of diverse transcription factor (TF) families, including basic helix–loop–helix (bHLH), MYB, ethylene insensitive (EIN), WRKY and so on.36–38 The COI1–JAZ complex also serves as areceptor for the bacterial phytotoxin coronatine (COR), which is ubiquitinated, MYC2 is then released and transcription is activated (Figure 7).39 Upon pathogen attack, mycorrhizal colonization initiates SA signaling pathways. HBP-1b gene belongs to the HBP subfamily of the bZIP transcription factor family, which is the histone gene binding protein. Several studies have shown that the plant histone genes may take part in various stress responses.40 In the ET signaling pathway, ERF transcription factors can bind to the GCC-box cis-acting elements in the promoters of biotic stress-related genes, and mediate these genes to play arole in the response of plants to biotic stress (Figure 7).41,42 Overall, we speculate that the protective effect of the AM symbiosis in banana plants is similar to that of rice and tomato, both are relies on the systemic activation of defense regulatory genes in the absence of pathogen challenge.9,17 The resistance mechanism of mycorrhizal symbiosis-induced defense genes in the process of banana fusarium wilt resistance will be verified in our next experiments.
Our results suggest that the Brazilian banana produced more lignin through the phenylpropanoid biosynthesis induced by AM inoculation, to strengthen the cell wall and resist the invasion of root pathogens. Meanwhile, the banana resistance might indirectly enhanced by the improved plant growth, and the risk of infection was reduced through the plant hormone signal transduction pathway. Follow-up studies will investigate the role of AM in enhancing banana’s resistance to Fusarium oxysporum f.sp. Cubense under controlled environments and in the field.
Conclusion
The present study describes the production of abiological defense mechanism in banana seedlings induced by AM inoculation. The results exhibited that the resistance of Brazilian banana could be enhanced by inducing resistance-related genes under AM treatment. Most importantly, the above genes were closely related to plant hormone signal transduction, flavonoid biosynthesis, and phenylalanine biosynthesis pathway, which were identified to effectively defend the infection of plant pathogen. It provides insights into the host-AM fungi interactions and uncovering mechanisms of AM fungi to increase banana resistance.
Supplementary Material
Funding Statement
The research was supported by the Special Project on Education and Research of Fujian Province (2017N5009), the Promotion Program for Young and Middle-aged Teacher in Science and Technology Research of Huaqiao University (ZQN-YX507), and the Quanzhou City Science & Technology Program of China (2018N003).
Author contributions
ZMY conceived and designed the experiments. LP and ZMY conducted the experiment. LYQ performed data analysis. LP wrote the manuscript. WMY, LJF and YLC revised the manuscript. All authors reviewed and approved the submitted version.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Dusunceli F. Global programme on banana Fusarium wilt disease: protecting banana production from the disease with focus on tropical race 4 (TR4). In: Side event during the committee on world food security. Sheikh Zayed Centre, FAO HQ; 2017. p. 1–10. Friday 13October2017. [Google Scholar]
- 2.Giovanni B, Manoj K, Maria IP, Cabanás CGL, Mercado-Blanco J. Biological Control Agents AgainstFusarium Wilt of Banana. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.00616 [DOI]
- 3.Castillo AG, Puig CJ, Cumagun R. Non-synergistic effect of Trichoderma harzianum and Glomus spp. in reducing infection of fusarium wilt in banana. Pathogens. 2019;8(2):43. doi: 10.3390/pathogens8020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan DG, Fu LL, Han BY, Sun XP, Zheng P, Zhang JM. Identification of an endophytic antifungal bacterial strain isolated from the rubber tree and its application in the biological control of banana Fusarium wilt. Plos One. 2015;10(7):e0131974. doi: 10.1371/journal.pone.0131974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong CKF, Saidi NB, Vadamalai G, Teh CY, Zulperi D. Effect of bioformulations on the biocontrol efficacy, microbial viability and storage stability of aconsortium of biocontrol agents against Fusarium wilt of banana. JAppl Microbiol. 2019;127(2):544–555. doi: 10.1111/jam.14310. [DOI] [PubMed] [Google Scholar]
- 6.Xue C, Penton CR, Shen ZZ, Zhang RF, Huang QW, Li R, Ruan YZ, Shen QR . Manipulating the banana rhizosphere microbiome for biological control of Panama disease. Sci Rep. 2015;5:14596. doi: 10.1038/srep14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu ZZ, Wang MY, Du JP, Huang T, Liu JF, Dong T, Chen YL. Isolation of Burkholderia sp. HQB-1, apromising biocontrol bacteria to protect banana against Fusarium Wilt through Phenazine-1-Carboxylic Acid secretion. Front Microbiol. 2020;11. doi: 10.3389/fmicb.2020.605152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berruti A, Lumini E, Balestrini R, Bianciotto V. Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes. Front Microbiol. 2016;6:01559. doi: 10.3389/fmicb.2015.01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvioli A, Bonfante P. Systems biology and “omics” tools: acooperation for next-generation mycorrhizal studies. Plant Sci. 2013;203:107–114. doi: 10.1016/j.plantsci.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Song Y, Chen D, Lu K, Sun Z, Zeng R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front Plant Sci. 2015;6:00786. doi: 10.3389/fpls.2015.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aseel DG, Rashad YM, Hammad SM. Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against tomato mosaic virus. Sci Rep. 2019;9(1):9692. doi: 10.1038/s41598-019-46281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernández-Montiel L, Rueda-Puente EO, Cordoba-Matson MV, Holguín-Peña JR, Zulueta-Rodríguez R. Mutualistic interaction of rhizobacteria with arbuscular mycorrhizal fungi and its antagonistic effect on Fusarium oxysporum in Carica papaya seedlings. Crop Prot. 2013;47:61–66. doi: 10.1016/j.cropro.2013.01.008. [DOI] [Google Scholar]
- 13.Nahiyan ASM, Matsubara Y. Tolerance to fusarium root rot and changes in antioxidative ability in mycorrhizal asparagus plants. Hort Sci. 2012;47(3):356–360. doi: 10.21273/HORTSCI.47.3.356. [DOI] [Google Scholar]
- 14.Liu M, Zhao ZJ, Chen L, Wang LQ, Ji LZ, Xiao Y. Influences of arbuscular mycorrhizae, phosphorus fertilizer and biochar on alfalfa growth, nutrient status and cadmium uptake. Ecotoxicol Environ Saf. 2020;196:110537. doi: 10.1016/j.ecoenv.2020.110537. [DOI] [PubMed] [Google Scholar]
- 15.Ortas I, Rafique M, Akpinar C, Kacar YA. Growth media and mycorrhizal species effect on acclimatization and nutrient uptake of banana plantlets. Scientia Hortic. 2017;217(15):55–60. doi: 10.1016/j.scienta.2017.01.025. [DOI] [Google Scholar]
- 16.Sahodaran NK, Arun AK, Ray JG. Native arbuscular mycorrhizal fungal isolates (Funneliformis mosseae and Glomus microcarpum) improve plant height and nutritional status of banana plants. Exp Agric. 2019;55(6):924–933. doi: 10.1017/S0014479719000036. [DOI] [Google Scholar]
- 17.Jansa J, Smith FA, Smith E. Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol. 2008;177(3):779–789. doi: 10.1111/j.1469-8137.2007.02294.x. [DOI] [PubMed] [Google Scholar]
- 18.Campos-Soriano L, García-Martínez J, San Segundo B. The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol Plant Pathol. 2012;13(6):579–592. doi: 10.1111/j.1364-3703.2011.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramírez-Flores MR, Bello-Bello E, Rellán-Álvarez R, Sawers RJH, Olalde-Portugal V. Inoculation with the mycorrhizal fungus Rhizophagus irregularis modulates the relationship between root growth and nutrient content in maize (Zea mays ssp. mays L.). Plant Direct. 2019;3(12):e00192. doi: 10.1002/pld3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Declercka S, Risedeb JM, Rufyikiric G, Delvauxc B. Effects of arbuscular mycorrhizal fungi on severity of root rot of bananas caused by Cylindrocladium spathiphylli. Plant Pathol. 2002;51(1):109–115. doi: 10.1046/j.0032-0862.2001.656.x. [DOI] [Google Scholar]
- 21.Radheshyam Y, Pankaj R, Parikshita R, Wusirika R. Bacteria from native soil in combination with arbuscular mycorrhizal fungi augment wheat yield and biofortification. Plant Physiol Biochem. 2020;150:222–223. doi: 10.1016/j.plaphy.2020.02.039. [DOI] [PubMed] [Google Scholar]
- 22.Zhang JF, Chen L, Fu CL, Wang LX, Liu HN, Cheng YZ. Comparative transcriptome analyses of gene expression change striggered by Rhizoctonia solani ag1 ia infection in resistant and susceptible rice varieties. Front Plant Sci. 2017;8:1422. doi: 10.3389/fpls.2017.01422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao QM, Zhu SF, Kachroo P, Kachroo A. Signal regulators of systemic acquired resistance. Front Plant Sci. 2015;6:228. doi: 10.3389/fpls.2015.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberstein M, Eisenstein M, Abeliovich H. Removing allosteric feedback inhibition of tomato 4-coumarate: coAligase by directed evolution. Plant J. 2012;69(1):57–69. doi: 10.1111/j.1365-313X.2011.04770.x. [DOI] [PubMed] [Google Scholar]
- 25.Millar DJ, Long M, Donovan G, Fraser PD, Boudet AM, Danoun S, Bramley PM, Paul Bolwell G. Introduction of sense constructs of cinnamate 4-hydroxylase (CYP73A24) in transgenic tomato plants shows opposite effects on flux into stem lignin and fruit flavonoids. Phytochemistry. 2007;68(11):1497–1509. doi: 10.1016/j.phytochem.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Sun JM, Zhang JH, HuiFang H, Peng LY, Wei SL, Li CS, Zheng SJ, Lu J. Comparative transcriptome analysis reveals resistance-related genes and pathways in Musa acuminate banana ‘Guijiao9ʹ in response to Fusarium wilt. Plant Physiol Biochem. 2019;141:83–94. doi: 10.1016/j.plaphy.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Cenci A, Rouard M, Zhang D, Wang YY, Tang WH, Zheng S-J. Transcriptomic analysis of resistant and susceptible banana corms in response to infection by Fusarium oxysporum f.sp. cubense tropical race 4. Sci Rep. 2019;9(1):8199. doi: 10.1038/s41598-019-44637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derksen H, Rampitsch C, Daayf F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013;207:79–87. doi: 10.1016/j.plantsci.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Shigenaga AM, Berens ML, Tsuda K, Argueso CT. Towards engineering of hormonal crosstalk in plant immunity. Curr Opin Plant Biol. 2017;38:164–172. doi: 10.1016/j.pbi.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Wu Y, Yi G, Peng X, Huang BZ, Liu E, Zhang JJ. Systemic acquired resistance in Cavendish banana induced by infection with an incompatible strain of Fusarium oxysporum f.sp. cubense. JPlant Physiol. 2013;170(11):1039–1046. doi: 10.1016/j.jplph.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of acore signaling network. Annu Rev Plant Biol. 2010;61(1):651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 32.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signaling. Trends Plant Sci. 2010;15(7):395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Lee SC, Lan W, Buchanan BB, Luan S.. Aprotein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proceedings of the National Academy of Sciences of the United States of America; 2009;106:21419–21424. doi: 10.1073/pnas.0910601106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westfall CS, Herrmann J, Chen Q, Wang SP, Jez JM. Modulating plant hormones by enzyme action The GH3 family of acyl acid amido synthetases. Plant Signal Behav. 2010;5(12):1607–1612. doi: 10.4161/psb.5.12.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Shen H, Wang MY, Li Q, He ZH. Salicyloyl-aspartate synthesized by the acetyl-amido synthetase GH3.5 is apotential activator of plant immunity in Arabidopsis. Acta Biochim Biophys Sin (Shanghai). 2013;45(10):827–836. doi: 10.1093/abbs/gmt078. [DOI] [PubMed] [Google Scholar]
- 36.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, SuzaW. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–672. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y, Liang G, Yang S, Yu D. Arabidopsis WRKY57 functions as anode of convergence for Jasmonic Acid– and Auxin-mediated signaling in Jasmonic Acid–induced leaf senescence. Plant Cell. 2014;26(1):230–245. doi: 10.1105/tpc.113.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song SS, Qi TC, Huang H, Ren QC, Wu DW, Chang CQ, Peng W, Liu YL, Peng JR, Xie DX . The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated Stamen development in Arabidopsis. Plant Cell. 2011;23(3):1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, An F, Feng Y, Li PP, Xue L, Mu A, Jiang ZQ, Kim JY, Kim To T, Li W, Zhang XY, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo HW. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 2011;108:12539–12544. doi: 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Yao J, Ke J, Zhang L, Lam LQ, Xin XF, Zhou XE, Chen J, Brunzelle J, Griffin PR, Zhou MG, Xu HE, Melcher K, He SY. Structural basis of JAZ repression of MYC transcription factors in jasmonate signaling. Nature. 2015;525(7568):269–273. doi: 10.1038/nature14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bray E, Shih TY, Moses M, Cohen A, Imai A. Aplant water-deficit induction of atomato H1 histone requires abscisic acid. Plant Growth Regul. 1999;29(1/2):35–46. doi: 10.1023/A:1006264001528. [DOI] [Google Scholar]
- 42.Zarei A, Krbes AP, Younessi P, Montiel G, Champion A, Memelink J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol. 2011;75(4–5):321–331. doi: 10.1007/s11103-010-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


