ABSTRACT
The intestinal autophagy and barrier function are crucial for maintaining the epithelium homeostasis and tightly regulated through well-controlled mechanisms. RNA-binding proteins (RBPs) and long noncoding RNAs (lncRNAs) modulate gene expression at the posttranscription level and are intimately involved in different physiological processes and diverse human diseases. In this review, we first highlight the roles of several RBPs and lncRNAs in the regulation of intestinal epithelial autophagy and barrier function, particularly focusing on the emerging evidence of RBPs and lncRNAs in the control of mRNA stability and translation. We additionally discuss recent findings that the interactions between RBPs and lncRNAs alter the fate of their target transcripts and thus influence gut epithelium host defense in response to stressful environments. These exciting advances in understanding the posttranscriptional control of the epithelial autophagy and barrier function by RBPs and lncRNAs provide a strong rationale for developing new effective therapeutics based on targeting RBPs and/or lncRNAs to preserve the intestinal epithelial integrity in patients with critical illnesses.
KEYWORDS: RNA-binding proteins, long noncoding RNAs, circRNAs, gut permeability, autophagy, epithelial homeostasis, polyamines, mRNA stability and translation, posttranscriptional regulation
Introduction
The mammalian intestine is colonized with a diverse population of bacteria and exposed to a wide array of luminal noxious substances and pathogens. Although most bacteria perform many beneficial functions, they can threaten host health upon tissue invasion. The intestinal epithelium directly interfaces with these diverse bacteria and noxious substances and acts as the first line of defenses against bacterial penetration and limiting the contact of luminal toxic substances with the subepithelial tissue.1–4 Intestinal epithelial cells (IECs) are connected by apical intercellular junctional complexes, named as tight junctions (TJs) and adherens junctions (AJs), and establish a selectively permeable barrier that prevents even small molecules from leaking between cells.2,3,5 The specialized IECs such as Goblet and Paneth cells secrete mucus and antimicrobial proteins that protect the epithelium from intrusion by luminal noxious substances, allergens, and microbial pathogens.4,6 However, certain luminal pathogens can evade this first line of innate defense and enter epithelial cells. Epithelial cell-intrinsic innate immune responses are therefore necessary to limit the invasion of pathogens and play an important role in maintaining epithelial homeostasis.4,7 Autophagy is an evolutionally conserved process by which cytoplasmic pathogens and unwanted materials are targeted to the lysosome for degradation.7–9 Autophagy activity is regulated via tightly controlled mechanisms, and more than 30 autophagy-related genes (ATGs) have been identified in mammals.8,9 Autophagy is crucial for the recognition and degradation of intracellular pathogens and functions as an innate barrier of infection, whereas its deregulation impairs intestinal epithelial defense and integrity.3,10–12
Posttranscriptional processes, particularly altered mRNA stability and translation by RNA-binding proteins (RBPs) and noncoding RNAs (ncRNAs), are major mechanisms by which IECs control gene expression in response to stressful environments.13–16 After transcription from their genes, mRNAs are subjected to multiple processing and regulatory steps that are tightly controlled by numerous nuclear and cytoplasmic factors. RBPs are a large family of over 2000 proteins that bind to transcripts in all manner of RNA-driven processes and regulate stability and translation of target mRNAs positively or negatively.15–18 The structures and mechanisms that RBPs use to interact with and modulate RNAs are incredibly diverse. RBP associations with different RNAs range from single protein-RNA element interaction to the assembly of multiple RBPs and RNA molecules. Long ncRNAs (lncRNAs) are defined as transcripts spanning >200 nucleotides in length and intimately involved in every level of gene regulation, including chromatin remodeling, transcriptional and posttranscriptional processes, and protein metabolism.19,20 An increasing body of evidence indicates that RBPs and lncRNAs are a novel class of master posttranscriptional regulators of intestinal epithelium homeostasis and that disrupted regulation of RBPs and lncRNAs compromises the intestinal epithelium integrity and contributes to the pathogenesis of various gut mucosal disorders such as inflammatory bowel diseases (IBD), infection, cancers, and sepsis.19,21–23 In this review, we highlight the important roles of several RBPs and lncRNAs in the regulation of intestinal epithelial autophagy and barrier function and further discuss in some detail the mechanisms through which RBPs and lncRNAs and their interactions modulate the stability and translation of target mRNAs.
RBPs in posttranscriptional regulation of autophagy
Control of mRNA stability and translation involves the interaction of specific mRNA sequences (cis element) with specific trans-acting factors including RBPs.17,18 AU-rich elements (AREs) and GU-rich elements (GREs) located at the 3′-untranslated regions (3′-UTRs) of target mRNAs are the best-characterized cis-acting sequences and identified in ~10% of the mRNAs in human and other mammals. RBPs directly interact with AREs and/or GREs or other unknown binding sequences via their special binding domains such as RNA recognition motif (RRM) and dsRNA binding domain and regulate gene expression at the posttranscription level.24,25 Some RBPs have housekeeping functions and interact with different cellular transcripts, but many RBPs interact with specific subsets of mRNAs and regulate gene expression levels in response to pathophysiological stresses. RBPs, including CUG-binding protein 1 (CUGBP1), AU-binding factor 1 (AUF1), tristetraprolin (TTP), BRF1, and KH-domain RNA binding protein (KSRP), enhance the decay of mRNAs and repress translation of target transcripts.16,21 Conversely, the Hu/embryonic lethal and abnormal vision (ELAV) family of RBPs, which consists of three primary neuronal members (HuB, HuC, and HuD) and one ubiquitous member HuR, stabilize mRNAs and stimulate their translation.26 Significant changes in the binding affinity of RBPs for target mRNAs, defects and mutations in their binding regions, and deregulation of RBP expressions and subcellular distribution occur commonly in different human diseases.16,27,28 Using intestinal epithelial tissue-specific knockout mouse models and approaches delivering and/or transfecting RBP transgenes or specific small interfering RNAs (siRNAs), several RBPs are shown to play an essential role in regulating the intestinal epithelial autophagy and barrier function via distinct mechanisms.
HuR regulates autophagy by altering ATG expression
HuR is one of best-studied RBPs and has two N-terminal RRMs through which it binds with high affinity and specificity to AREs located in the 3ʹ-UTRs of labile mRNAs.29–31 In the intestinal epithelium, HuR is distributed predominantly in the nucleus of unstimulated cells but can be rapidly translocated to the cytoplasm where it interacts directly with target mRNAs in response to various stresses, thus altering the gene expression levels. Several studies have shown that HuR regulates autophagy by altering ATG expression.32–34 HuR silencing inhibits autophagosome formation and decreases autophagic flux. Autophagosome is a double-membrane vesicle that contains sequestered cytoplasmic cargo and transports them to the lysosome.35,36 Autophagosome formation depends on products of the Atg genes and is essential for autophagy activation. Mechanistically, HuR directly binds to mRNAs encoding ATG5, ATG12, and ATG16, mostly via their 3ʹ-UTRs, enhances their stability and translation, and increases cellular abundances of ATG proteins. In support of these findings, HuR expression levels positively correlate with the levels of ATG5 and ATG12 in various cancer cells.33
ATG16L1, a product of the Atg16l1 gene, plays an important role in the intestinal epithelium homeostasis partially by interacting with A20 and orchestrating interleukin-22 signaling.37,38 ATG16L1 also inhibits necroptosis in the intestinal epithelium39 and protects against TNF-induced apoptosis during chronic colitis in mice.40 Genome-wide association studies demonstrate the presence of several polymorphisms and mutations in the Atg16l1 gene in patients with IBD and other gut mucosal injury-associated disorders.7,12 We examined changes in the levels of HuR and ATGs in human intestinal mucosa and show that mucosal tissue samples from patients with IBD have reduced abundances of both HuR and ATG16L1. HuR is localized at both the cytoplasm and nucleus in the human intestinal epithelium of control individuals, but these HuR immunoreactive signals in the mucosal tissues from IBD patients decrease remarkably, particularly in the cytoplasm, when compared with those observed in control individuals. Importantly, the decreased levels of HuR in the intestinal mucosa are accompanied by a specific reduction in the levels of ATG16L1 in patients with IBD. In controls, both ATG16L1 and ATG5 are found predominantly in the cytoplasm of the intestinal mucosa, but ATG16L1 levels in the mucosal tissues from IBD patients decrease especially without significant changes in ATG5 content. Notably, the decreased levels of HuR and ATG16L1 are associated with mucosal injury/erosions, inflammation, delayed repair, and gut barrier dysfunction.34,37,38,41
To define the exact function of HuR in the regulation of ATG16L1 expression in vivo, we generated intestinal epithelium tissue-specific HuR knockout (IE-HuR−/-) mice.42 HuR is undetectable in the intestinal mucosa of IE-HuR−/- mice, although it is found at wild-type levels in other tissues and organs such as gastric mucosa, lung, heart, liver, and pancreas. Targeted deletion of HuR in mice does not alter the overall morphology or structure of the small and large intestines. Conditional HuR deletion in mice markedly decreases the levels of ATG16L1 in the small intestinal mucosa, but it fails to alter tissue ATG5 abundance.34 ATG16L2 level in the intestinal mucosa of both IE-HuR−/- and littermate mice are too low to be detected. The basal level of ATG7 in the intestinal mucosa is relatively low but it is also reduced in HuR-ablated mice. Consistently, the levels of autophagy proteins, microtubule-associated protein light chain 3 (LC3)-I and LC3-II, also decrease in the intestinal mucosa of IE-HuR−/- mice relative to control littermates. Targeted deletion of HuR does not alter transcription of the Atg genes, because the levels of all Atg16l1, Atg5, and Atg7 mRNAs in the intestinal mucosa of IE-HuR−/- mice are indistinguishable from those observed in control littermate mice. In addition, IE-HuR−/- mice also exhibit inhibited IEC proliferation and mucosal atrophy of the small intestine and delayed repair of damaged mucosa induced by mesenteric ischemia/reperfusion (I/R) in the small intestine and by dextran sulfate sodium (DSS) in the colon.42,43 Together, these results from experiments conducted in mice and human tissues strongly suggest that HuR plays an important role in the regulation of autophagy in the intestinal epithelium and that deregulation of HuR-mediated ATG16L1 expression disrupts the epithelium-host defense, thus contributing to the pathological process of IBD in human.
HuR is required for Paneth cell function
Paneth cells reside at the bottom of the crypts in the small intestine and are crucial for maintaining homeostasis of the epithelium by engendering host protection from enteric pathogens.6,7 Paneth cells produce abundant antibacterial proteins or peptides, including lysozyme, Reg3 lectins, α-defensin, and phospholipase A2. It has been reported that Paneth cells secrete lysozyme through secretory autophagy to limit bacterial infection of the intestine.44 A recent study reveals that HuR is required for normal Paneth cell function and that HuR-regulated Paneth cells is critical for intestinal epithelial defense.41 Intestinal tissues from IE-HuR−/- mice have reduced numbers of Paneth cells, and Paneth cells exhibit fewer lysozyme granules per cell, compared with tissues from control mice, but there are no effects on differentiation of Goblet cells or enterocytes. This defect in Paneth cell function is associated with repressed autophagic clearance, as shown by the decreased response of LC3 activation to rapamycin in the HuR-deficient epithelium. Importantly, intestinal mucosa from patients with IBD also exhibits reduced levels of HuR and fewer Paneth cells. Lysozyme-positive cells in the ileal mucosa from patients with IBD decrease remarkably compared with those in control patients. In many cases, lysozyme-positive cells are almost completely undetectable in the ileal mucosal samples obtained from IBD patients, along with massive mucosal erosions and inflammation.13,41
HuR deletion causes Paneth cell defects by altering Toll-like receptor 2 (TLR2) activity, since TLR2 serves as an important sensor for autophagy45,46 and IE-HuR−/- mice do not have the apical distribution of TLR2 in the intestinal mucosa as observed in control mice.41 TLR2 is localized primarily at the surface of villi and strongly concentrated along the apical area of the small intestinal mucosa in control mice. However, this apical staining of TLR2 in the small intestinal mucosa of IE-HuR−/- mice disappears completely, associated with an increase in the intensity of TLR2 staining in the basal area of the epithelium. The most striking difference between control littermates and IE-HuR−/- mice is the appearance of many areas of small punctate foci of TLR2 in the latter. These punctate regions are located throughout the cytoplasm in the HuR-deficient intestinal epithelium and are regularly identified in every IE-HuR−/- mouse. Similarly, TLR2 distribution in the colonic mucosa is also compromised by HuR deletion, as indicated by a decrease in the apical TLR2 staining in IE-HuR−/- mice. In an ex vivo model, TLR2 is normally localized at the luminal regions in primarily cultured intestinal organoids isolated from control mice, but this specific distribution of TLR2 is abolished by HuR deletion, as evidence by the fact that TLR2 staining is diffused in HuR-deficient organoids isolated from IE-HuR−/- mice. On the other hand, HuR deletion fails to alter TLR4 subcellular localization in the intestinal mucosa.
To gain a deeper understanding of the abnormalities of TLR signals in IE-HuR−/- mice, TLR expression was examined and shows that HuR deletion in mice does not alter the levels of total TLR proteins in the intestinal mucosa.41 Interestingly, HuR deletion decreases the levels of the endoplasmic reticulum (ER) chaperone canopy3 (CNPY3), a protein necessary for the proper subcellular localization of TLR2 on the IEC plasma membrane to carry out secretory autophagy.47 Reduced levels of CNPY3 by HuR silencing in cultured IECs impair its TLR2 cochaperone function and reduce the apical trafficking for TLR2, leading to an inhibition of TLR2 function. Mechanistically, HuR directly associates with the Cnpy3 mRNA via coding region (CR) but not its 3ʹ-UTR, and this interaction stabilizes Cnpy3 mRNA and enhances its translation. HuR knockout reduces CNPY3 levels and thus inhibits the proper folding, subcellular transportation, and apical distribution of TLR2 in the intestinal epithelium. This regulatory role of HuR in TLR2 subcellular distribution through targeting CNPY3 eventually contributes to the control of IEC autophagy activation and innate immunity. In addition, the HuR-deficient epithelium also exhibits decreased levels of IRGM and beclin-1 but increases NLRX1. Since these proteins also modulate TLR activity and autophagy,45,48 changes in the levels of IRGM, beclin-1, and NLRX1 in IE-HuR−/- mice might be also involved in the mechanisms underlying TLR2 dysfunction and subsequent autophagy inactivation. Recently, HuR is shown to enhance translation of vitamin D receptor that is required for Paneth cell differentiation.49,50
Taken together, the findings obtained from human tissue samples, mice with ablated HuR, intestinal organoids, and cultured IECs suggest a novel model by which HuR plays an essential role in the regulation of intestinal epithelial autophagy under pathophysiological conditions (Figure 1). According to this model, HuR enhances intestinal epithelial autophagy by stimulating the expression of ATGs and promoting Paneth cell function, whereas disrupted HuR activity leads to autophagy inactivation and defective Paneth cells, thus compromising the intestinal epithelial defense and promoting the pathological process of mucosal injury and inflammation.
Figure 1.
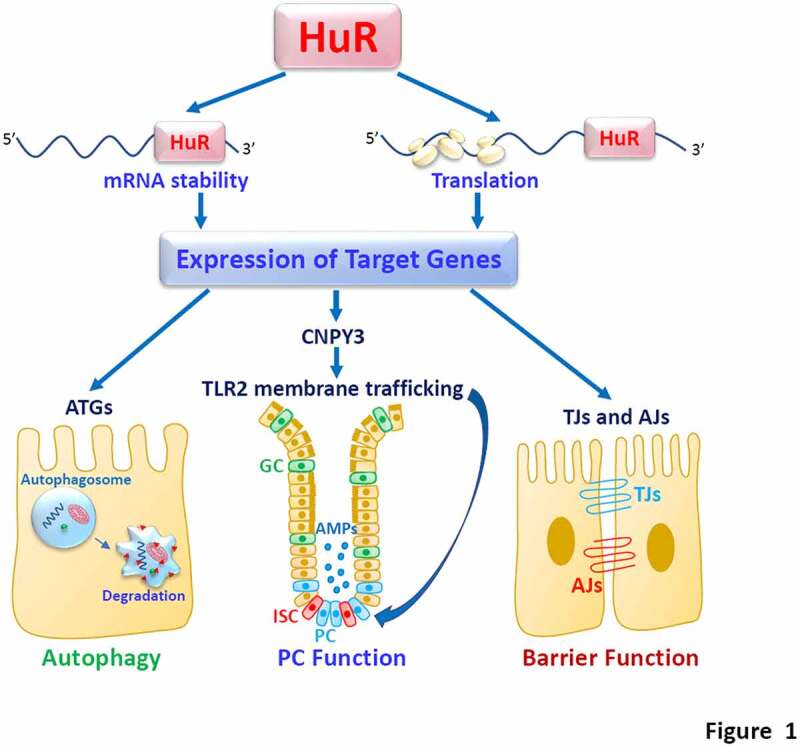
HuR regulates intestinal epithelial defense and barrier by altering stability and translation of its target mRNAs. PC, Paneth cells; GC, Goblet cells; ISC, intestinal stem cells; AMPs, antimicrobial proteins. HuR stimulates epithelial autophagy and barrier function by increasing expression of TJ/AJ and ATGs, but it enhances PC function by manitaining membrane localization of TLR2 via control of CNPY3. HuR activity is tightly regulated by multiple factors, whereas down-regulation of HuR leads to defects in intestinal epithelial autophagy and barrier function
Other RBPs in the regulation of intestinal epithelial autophagy
CUGBP1 binds to a variety of mRNA cis-elements, including GREs and AREs, and enhances mRNA decay and/or represses translation of target transcripts in general.51 CUGBP1 is highly expressed in the intestinal epithelium and its cellular levels and distribution change dramatically in response to stressful environments. Several studies have shown that CUGBP1 is a negative regulator of the intestinal epithelium homeostasis and that CUGBP1 and HuR compete for binding to given mRNAs and regulate target transcripts antagonistically.52,53 Although there is no available evidence showing that CUGBP1 affects the expression of ATGs, CUGBP1 can be involved in the regulation of autophagy indirectly through interaction with HuR.
HuD, ring finger protein 36 (ZFP36)/TTP, and zinc finger protein 423 (ZNF423) also regulate autophagy in other tissues, but their expression levels and functions in the intestinal epithelium remain to be fully investigated. The Atg5 mRNA is a posttranscriptional target of HuD in pancreatic β cells.54 Interaction of HuD with Atg5 mRNA enhances ATG5 translation and thus contributes to the lipidation of LC3 and the formation of LC3-positive autophagosomes. HuD-null mice display lower ATG5 and LC3 levels in pancreatic β cells. ZFP36/TTP promotes Atg16L1 mRNA decay through directly interaction with AREs located at its 3ʹ-UTR in hepatic stellate cells.55 Elevation of the ZFP36/TTP levels inhibits macroautophagy activation by decreasing ATG16L1 and plays a crucial role in regulating ferroptosis. Branched-chain amino transferase 1 (BCAT1) is a key regulatory factor of autophagy in pulmonary artery smooth muscle cells and has recently been identified as a potential therapeutic target for the clinical treatment of lung diseases. The function of BCAT1 is remarkably enhanced by the RBP ZNF423.56 When exposed to hypoxia, ZNF423 is rapidly translocated to the cytoplasm where it binds Bcat1 mRNA via its 3′-UTR and promotes BCAT1 expression, thus enhancing autophagy activation.
RBPs in regulation of the epithelial barrier function
Intercellular junctions are presented at points of cell-cell and cell-matrix contact in all tissues, particularly in epithelia. The intestinal epithelial barrier depends on specialized structures of TJ and AJ complexes that are highly dynamic.2,5 Maintenance of the levels of TJ and AJ proteins is absolutely required for the stability and effectiveness of epithelial barrier structure and function. Several RBPs are intimately implicated in the posttranscriptional control of TJs and AJs in the intestinal epithelial and are crucial for the barrier function.52,57–59
HuR enhances expression of TJs and AJs
HuR binds to several mRNAs encoding TJ proteins including claudin-1, claudin-3, occludin and JAM-1, and AJ protein E-cadherin and enhances the stability and translation of these target transcripts.52,57 Occludin is a transmembrane TJ protein that is necessary for TJ assembly and critical for the maintenance of gut barrier integrity. HuR directly interacts with the 3ʹ-UTR of the occludin mRNA and increases occludin translation with a minor effect on its mRNA stability.52 HuR association with the occludin mRNA is regulated through Chk2-dependent HuR phosphorylation. Decreased HuR phosphorylation by Chk2 silencing or by reduction of Chk2 through polyamine depletion reduces HuR-binding to the occludin mRNA and inhibits occludin translation, whereas Chk2 overexpression induces HuR/occludin mRNA association and promotes occludin expression.
In an in vitro permeability model using differentiated IECs, HuR silencing by transfection with HuR-directed small RNA (siHuR) results in the epithelial barrier dysfunction, as indicated by decreased transepithelial electrical resistance (TEER) and increased paracellular permeability.52,59 Although IE-HuR−/- mice do not exhibit increased gut permeability without any pathological stress, HuR deletion increases the vulnerability of the gut barrier to pathological stress and also inhibits recovery of the barrier functions after treatment with DSS or exposure to cecal ligation and puncture (CLP) and mesenteric I/R.42,43 In mice exposed to CLP, inhibition of HuR binding affinity for mRNAs encoding TJs and AJs by decreasing Chk2-dependent HuR phosphorylation through polyamine depletion reduces the protein levels of TJs and AJs and slows downs the barrier recovery after septic stress.14,60 Consistently, PP2A-associated protein α4 stabilizes HuR through a process involving HuR phosphorylation by IκB kinase α, whereas intestinal epithelial-specific ablation of α4 in mice decreases the levels of HuR, leading to an inhibition of TJ expression and gut barrier dysfunction.61 In addition, HuR also regulates the intestinal barrier function by promoting mucosal renewal, increasing rapid epithelial restitution after acute injury, and protecting IECs against apoptosis. These interesting results have been comprehensively reviewed and nicely summarized in recent publications.5,16
CUGBP1 impairs the intestinal barrier function
CUGBP1 negatively regulates intestinal barrier function by repressing the expression of TJs and AJs.52,59,62 CUGBP1 directly interacts with the mRNAs encoding occludin, claudin-1, and E-cadherin, although it does not bind to the mRNAs encoding claudin-2, claudin-3, claudin-5, ZO-1, and β-catenin. CUGBP1 overexpression specifically inhibits the expression of occludin, claudin-1, and E-cadherin, but fails to alter the levels of other TJs and AJs. This inhibitory effect of CUGBP1 on the expression of given TJs and AJ occurs at the translation level and is mediated through their 3ʹ-UTRs rather than CRs and 5ʹ-UTRs. Ectopically expressed CUGBP1 also damages the epithelial barrier function, as indicated by a decrease in TEER and an increase in paracellular permeability. Interestingly, CUGBP1 and HuR compete for association with the same occludin 3ʹ-UTR and regulate occludin translation competitively.52,59 Increasing the CUGBP1 levels decreases HuR interaction with occludin mRNA and inhibits occludin translation, whereas elevation of HuR levels abolishes CUGBP1 binding to occludin mRNA and enhances occludin translation. Studies using purified GST-HuR or GST-CUGBP1 fusion proteins further show that the occludin 3ʹ-UTR interaction with HuR is progressively increased when increasing concentrations of GST-HuR in the binding reaction mixture, but its binding to CUGBP1 is decreased with increasing GST-HuR levels in the mixture. Moreover, cellular levels of CUGBP1 are tightly controlled by protein kinase C (PKC), HuR, miR-503, and polyamines.60,61,63,64 Activation of PKC increases the stability of CUGBP1 protein through direct phosphorylation.65 Expression of CUGBP1 in IECs is jointly regulated by HuR and miR-503 at the posttranscription level.63 HuR associates with the Cugbp1 mRNA, increases the loading of polyribosomes onto CUGBP1 transcripts, and thereby increases CUGBP1 translation. In contrast, the interaction of miR-503 with the Cugbp1 mRNA inhibits its translation by recruiting the Cugbp1 mRNA to P-bodies where mRNAs are sorted for degradation. Cellular polyamines regulate the CUGBP1 expression by altering the level of cytoplasmic HuR and miR-503 abundances.
Other RBPs in the regulation of intestinal barrier function
AUF1 displays a high affinity for ARE-containing RNAs and poly (U) and is involved in many aspects of cellular functions.66 AUF1 regulates intestinal barrier function via transcription factor JunD that regulates transcription of TJ ZO-1.67 AUF1 binds to JunD mRNA, and this interaction represses JunD expression by destabilizing the JunD transcripts in IECs. Association of AUF1 with JunD mRNA is tightly regulated by polyamines and HuR (stabilizer of JunD mRNA). Polyamines alter JunD mRNA degradation by modulating the competitive binding of HuR and AUF1 to the JunD 3ʹ-UTR.67 Depletion of cellular polyamine increases HuR binding to JunD mRNA but decreases the levels of JunD transcript bound to AUF1, thus stabilizing JunD mRNA. HuR silencing enhances AUF1 interaction with the JunD mRNA, reduces the abundance of HuR/JunD mRNA complexes, and renders the JunD mRNA unstable, thus preventing increases in JunD in polyamine-deficient cells. Decreasing the levels of cellular JunD by AUF1 alters ZO-1 expression and epithelial barrier function.67
RBPs TIA-1 and TIAR are highly expressed in the gut mucosa and inhibit the translation of target mRNAs, especially under conditions of stress-associated cellular damage. TIAR binds the 3ʹ-UTR of the mRNAs encoding translation factors and potently suppresses their translation in response to stressful environments.68 Ectopically expressed TIA-1 and/or TIAR lead to the global inhibition of the cellular translation. Although limited studies are available so far, it has been reported that TIAR directly binds to the ZO-1 mRNA via its 3ʹ-UTR and this interaction represses ZO-1 translation in IECs, resulting in the epithelial barrier dysfunction.69 The inhibitory effect of TIAR on ZO-1 translation is enhanced by increasing JunD through stimulation of TIAR/ZO-1 mRNA association.
LncRNAs in the regulation of autophagy and barrier
LncRNAs are distinct from other well-characterized structural RNAs such as transfer RNAs and ribosomal RNAs and share structural features with mRNAs such as 5ʹ-cap and a 3ʹ-poly(A) tail, but they do not encode recognizable proteins.19,70 Unlike microRNAs (miRNAs), most lncRNAs are poorly conserved among species and dynamically expressed in tissue-, differentiation stage-, and cell type-specific manners. The levels of cellular lncRNAs are altered rapidly in response to pathophysiological stresses and lncRNAs act as molecular scaffolds, decoys or signals and also function through genomic targeting, cis- and trans-regulatory factors, and antisense molecules.71 Generally, nucleus lncRNAs are implicated in gene transcription and chromatin modification, whereas lncRNAs in the cytoplasm regulate the posttranscriptional process through direct interaction with mRNAs, miRNAs, or RBPs. Emerged evidence indicates that lncRNAs regulate the intestinal epithelium homeostasis and are involved in various human diseases.72–74 Here we highlight the importance of several lncRNAs, including H19, uc.173, and SPRY4-IT1, in the control of intestinal epithelial autophagy and barrier function and further discuss their implication in the pathogenesis of various gut mucosal disorders.
H19 disrupt the barrier function and suppresses autophagy
Transcribed from the conserved imprinted H19/igf2 gene cluster, lncRNA H19 plays a role in diverse cell processes and functions.75,76 During embryogenesis, the levels of H19 increase in extraembryonic tissues, the embryo itself, and most fetal tissues but decrease rapidly after birth.77 Increased H19 promotes the expression of imprinted genes and inhibits embryonic placental growth during fetal development.78 In adult tissues, induction in the levels of H19 occurs commonly in a broad spectrum of pathological conditions such as malignancies, inflammation, and after exposure to hypoxia or estrogens.79–82 Target deletion of H19 in mice causes an overgrowth phenotype and increases body weight, whereas transgenic re-expression of the H19 gene prevents the increased growth in mice with ablated H19.77 In the intestinal epithelium, the levels of H19 increase remarkably in patients with IBD and sepsis and murine gut mucosa with inflammation and erosions, which results partially from an increase in the inflammatory cytokine interleukin-22.23
The first evidence demonstrating the importance of H19 in the regulation of intestinal barrier function is from our observations showing that elevation of the H19 levels inhibits expression of ZO-1 and E-cadherin.83 In cultured IECs, ectopically expressed H19 decreases stability and translation of the ZO-1 and E-cadherin mRNAs, leading to a reduction in the levels of ZO-1 and E-cadherin proteins. Further study reveals that the inhibitory effect of H19 on ZO-1 and E-cadherin is mediated by miR-675 that is embedded in H19 exon 1. H19 does not bind to the ZO-1 and E-cadherin mRNAs but it increases miR-675 production. miR-675 directly interacts with the ZO-1 and E-cadherin mRNAs and inhibits their expression posttranscriptionally. Interestingly, HuR binds to H19, inhibits miR-675 processing from H19, and decreases the production of miR-675.83 Ectopic overexpression of HuR rescues the expression of ZO-1 and E-cadherin and prevents the barrier dysfunction in cells overexpressing H19. In contrast, intestinal epithelial tissue-specific deletion of HuR in mice enhances miR-675 production and delays the recovery of the gut barrier function after exposure to mesenteric I/R. H19 also functions as a molecular sponge to decrease the bioavailability of let-7,79 but decreasing the levels of cellular let-7 fails to alter the expression of ZO-1 and E-cadherin in IECs, suggesting that the association of H19 with let-7 plays a little role in H19-induced inhibition of ZO-1 and E-cadherin and subsequent barrier dysfunction. These results indicate that H19 and HuR modulate the expression of ZO-1 and E-cadherin and barrier function antagonistically via control of miR-675 processing from H19.
A recent study further shows that H19 actively participates in the regulation of autophagy and functions of Paneth and Goblet cells in the intestinal epithelium.13 Intestinal mucosal tissue samples from patients with sepsis and septic mice exhibit increased levels of H19, associated with autophagy inactivation and defects in Paneth and Goblet cells. Targeted deletion of the H19 gene in mice increases the function of Paneth and Goblet cells and enhances autophagy in the small intestinal mucosa. The levels of LC3-II, lysozyme, and beclin increase significantly in the H19-deficient epithelium. After exposure to septic stress induced by CLP, H19 deletion protects Paneth and Goblet cells against septic stress, preserves autophagy activation, and promotes gut barrier function. Compared with intestinal organoids isolated from control littermate mice, organoids generated from H19−/- mice exhibit increased numbers of Paneth and Goblet cells and also display increased tolerance to lipopolysaccharide (LPS). Conversely, ectopic overexpression of H19 in cultured IECs prevents rapamycin-induced autophagy and abolishes the rapamycin-induced protection of the epithelial barrier against LPS. Although the exact molecular processes by which H19 regulates autophagy and Paneth and Goblet cells remain largely unknown, available findings indicate that H19 modulates the intestinal epithelium homeostasis through distinct mechanisms (Figure 2). H19 disrupts the barrier function by inhibiting ZO-1 and E-cadherin via miR-675 and impairs the epithelium-host defense by inhibiting autophagy and function of Paneth and Goblet cells. Since H19 also functions as an RNA decoy for miR-34b and let-7 that are also involved in controlling the expression of p53 and other apoptosis-associated proteins,79,84 it is likely that H19 can also modulate the epithelium homeostasis by altering the availability of miR-34b and let-7. On the other hand, HuR blocks the processing of miR-675 from H19 and enhances the epithelial barrier function. Given the fact that intestinal mucosa from patients with critical illnesses exhibits increased H19 but decreased HuR,13,41 these exciting findings shed light on developing the new and effective therapeutics to protect the intestinal epithelium integrity through the intervention of H19 and its regulators such as HuR.
Figure 2.
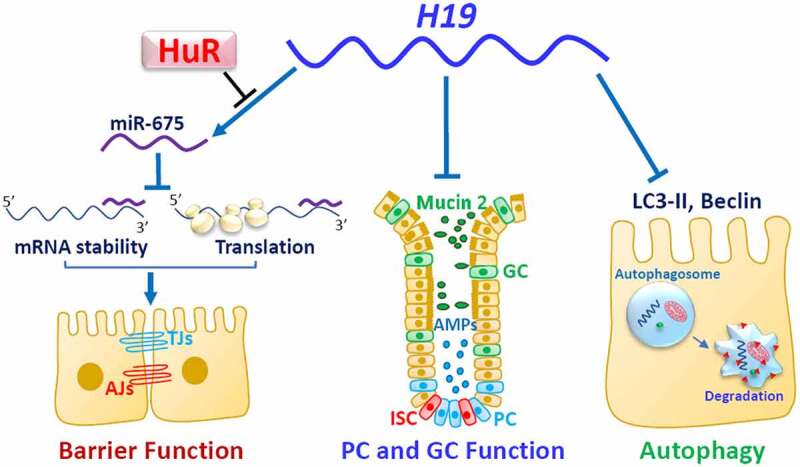
H19 disrupts the intestinal epithelium homeostasis through several distinct mechanisms. H19 impairs the epithelial barrier function by inhibiting expression of ZO-1 and E-cadherin posttranscriptionally through release of miR-675 embedded in H19 exon 1. HuR interacts with H19, prevents miR-675 processing from H19, and enhances the barrier function. Induced H19 also inhibits autophagy and lowers Paneth cell (PC) and Goblet cell (GC) function
Uc.173 enhances the intestinal barrier function
uc.173 is a member of a new class of lncRNAs transcribed from genomic ultraconserved regions (T-UCRs). UCRs are absolutely conserved between orthologous regions of human, rat, and mouse genomes.73 A total of 481 UCRs have been identified and they are widely distributed among mammalian genomes. Genome-wide T-UCR expression profile analysis reveals 21 T-UCRs, including uc.173, differentially expressed in the small intestinal mucosa of fasted mice relative to non-fasted control animals.73 The expression levels of 18 T-UCRs, including uc.173, uc.481, uc.356, uc.138A, uc.46A/45A, uc.141, uc.455(8), uc.475, uc.455(4), and uc.144, decreased in the intestinal mucosa after a 48-h period of food starvation, while the intestinal mucosal levels of 3 T-UCRs, including uc.457(E), uc.477, and uc.457(u), increased in fasted mice. Although there are no comprehensive studies investigating the exact roles of all these T-UCRs in the intestinal epithelial homeostasis yet, uc.173 is shown to be critical for normal gut barrier function.72
The expression patterns of uc.173 in the intestinal epithelium exhibit distinct signature and correlate closely with the status of gut barrier function in response to stressful environments. Intestinal mucosal tissues from patients with IBD display a significant decrease in the levels of uc.173, which is associated with a decrease in the levels of TJ expression and epithelial renewal.72 In cultured IECs, uc.173 silencing specifically inhibits the expression of TJ claudin-1 and weakens epithelial barrier function. Although systemic administration of locked nucleic acid (LNA) to antagonize uc.173 (anti-uc.173) has no acute or sub-chronic toxicities in mice, decreasing the levels of tissue uc.173 by anti-uc.173 increases the vulnerability of the gut barrier to septic stress induced by CLP. Exposure to CLP leads to the gut barrier dysfunction in both anti-uc.173-treated mice and controls, but increased gut permeability following CLP in mice treated with anti-uc.173 is much higher than that observed in control mice. CLP stress also reduces the levels of claudin-1 and claudin-3 in the intestinal mucosa, but inhibition of claudin-1 is amplified by decreasing the levels of uc.173 in anti-uc.173-treated mice. In support of these findings, uc.173 also stimulates the renewal of the small intestinal mucosa, since ectopic uc.173 overexpression increases IEC proliferation and enhances the growth of intestinal organoids.73
Further study shows that uc.173 fails to bind to claudin-1 mRNA but it enhances translation of claudin-1 through interaction with miR-29b.72 miR-29b directly associates with claudin-1 mRNA via its 3ʹ-UTR and inhibits claudin-1 translation. Increasing the levels of uc.173 decreases the binding of miR-29b to claudin-1 mRNA and restores claudin-1 expression in cells overexpressing miR-29b. In addition, miR-29b also potently inhibits renewal of the intestinal mucosa, and elevating miR-29b abundance suppresses mucosal growth and impairs the integrity of the intestinal epithelium.85 On the other hand, uc.173 promotes the growth of the small intestinal mucosa primarily by down-regulating miR-195, a repressor of epithelial homeostasis.73 uc.173 silencing increases miR-195 expression but does not alter the levels of miR-29b and miR-222.73 Elevation of the endogenous miR-195 levels by uc.173 silencing inhibits IEC proliferation, and this effect is almost completely rescued by ectopic transfection of a miR-195 antagomir. Together, the enhancement in intestinal epithelial barrier function by uc.173 results from antagonizing both miR-29b and miR-195 via distinct mechanisms. Since the basal levels of uc.173 in the intestinal mucosa are relatively high and changed remarkably in response to stress, uc.173 plays an essential role in maintaining the intestinal barrier function through interactions with miR-29b and miR-195.
SPRY4-IT1 promotes the epithelial barrier by increasing TJ expression
SPRY4-IT1 (sprouty receptor tyrosine kinase signaling antagonist 4‑intronic transcript 1) is a 706-bp lncRNA that is broadly expressed in various tissues. SPRY4-IT1 is predominantly localized in the cytoplasm and its cellular levels are tightly regulated in response to stress. In the intestinal epithelium, SPRY4-IT1 induces the TJ expression at the posttranscription level, thus enhancing the gut epithelial barrier function.57 The levels of SPRY4-IT1 in the intestinal mucosa from IBD patients decrease significantly, associated with a decrease in the levels of TJs claudin-1, claudin-3, occludin, and JAM-1. In cultured IECs, SPRY4-IT1 silencing specifically inhibits the expression of these TJs but fails to alter the cellular abundance of ZO-1, E-cadherin, α-catenin, and β-catenin. SPRY4-IT1 silencing also disrupts the epithelial barrier function in an in vitro model, which is overcome by overexpression of claudin-1 or occludin. Elevation of the mucosal levels of SPRY4-IT1 by infection with a lentiviral-driven SPRY4-IT expression vector (lenti-SPRY4-IT1) also protects the gut barrier function in mice exposed to CLP. The decreased levels of TJs by CLP are prevented or significantly reduced by increasing SPRY4-IT1 in lenti-SPRY4-IT1-infected mice. Mechanistically, SPRY4-IT1 directly interacts with the mRNAs encoding claudin-1, claudin-3, occludin, and JAM-1, and these interactions stabilize these mRNAs and promote the translation. Moreover, SPRY4-IT1 also physically associates with HuR and promotes the HuR binding to the TJ mRNAs, thus enhancing HuR-mediated stimulation of TJ expression and gut barrier function.
Other lncRNAs in the intestinal epithelial autophagy and barrier function
Gata6 is involved in regulating the intestinal barrier function by altering epithelial renewal via intestinal stem cells (ISCs).86 Target deletion of Gata6 in mice decreases intestinal mucosal growth and disrupts the gut barrier function. Gata6 directly interacts with two subunits of the NURF remodeling complex and recruits the NURF complex onto the Ehf promoter, thus promoting in ISCs the production of EHF, a protein needed for expression of LGR4/5 in ISCs, which in turn stimulates WNT signals. In addition, lncRNAs BANCR, LCPAT1, and DRAIC are also implicated in the regulation of autophagy in other tissues, but their roles in the intestinal epithelium remain unknown. BANCR activates autophagy in papillary thyroid carcinoma cells and tissue,87 and LCPAT1 is found to accelerate the autophagic flux in lung cancer cells.88 LCPAT1 levels increase remarkably in human lung cancer tissues, whereas LCPAT1 knockdown decreases ATG expression and alleviates autophagy activation induced by rapamycin in lung cancer cells. DRAIC is recently identified as a regulator of autophagic flux in MCF-7 breast cancer cells.89 In an autophagy-independent manner, DRAIC silencing inhibits transition from G1 to S-phase during the cell cycle by modulating the activity of ULK1.
Circular RNAs in intestinal autophagy and barrier
Circular RNAs (circRNAs) are a class of widespread and diverse endogenous RNAs that are often expressed in a tissue- and developmental stage-specific manner.90 Opposite to linear RNAs, circRNAs are covalently closed-loop structures without 5ʹ to 3ʹ ends (Figure 3). For many years, circRNAs have only received a little attention, because most classic methods only specifically detect RNA molecules with polyadenylated tails. With the rapid development of new techniques, thousands of circularized transcripts have been identified in various mammalian tissues. Most circRNAs are believed to fulfill noncoding roles,90 although some circRNAs endogenous to Drosophila and human encode proteins.91 circRNAs harbor single or multiple miRNA-binding sites, and some harbor binding sites for multiple miRNAs.92,93 Several circRNAs are shown to interact with and ‘sponge’ miRNAs to decrease the number of freely available miRNAs. For example, circRS-7 contains multiple sites for miR-7, enabling it to sequester miR-7 and thereby decreasing its availability to target mRNAs bearing miR-7 binding sites.94 Similarly, circSRY functions as a sponge for miR-138, while circITCH can interact with miR-7, miR-17, and miR-214.92,95–97 circRNAs also bind to RBPs to jointly regulate gene expression synergistically or antagonistically.98,99 It has been reported that HuR associates with many circRNAs and regulates their biological functions in human cervical carcinoma HeLa cells.100 Differential expression of circRNAs during disease progression suggests the importance of circRNAs in human pathologies.101
Figure 3.
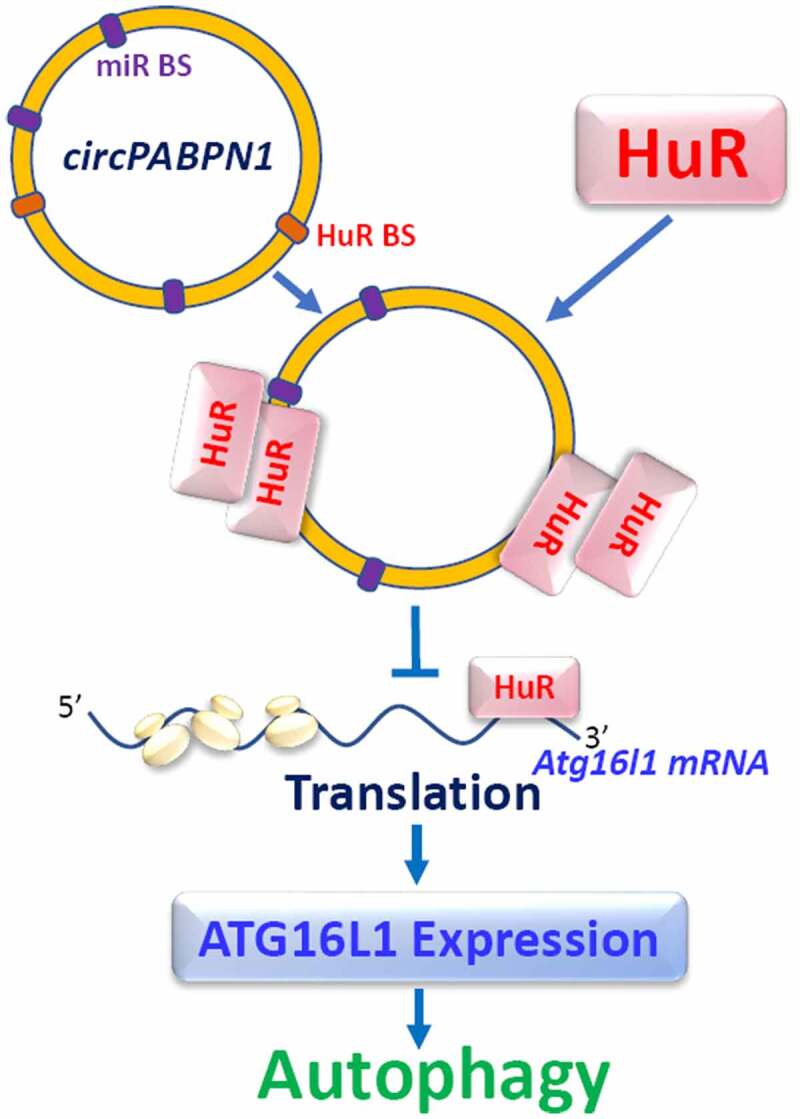
circPABPN1 regulates ATG16L1 expression through interaction with HuR. miR-BS, microRNA binding site; HuR-BS, HuR binding site. circPABPN1 does not directly bind to Atg16l1 mRNA, but it interacts with HuR, forms circPABPN1/HuR complex, and reduces availability of HuR for Atg16l1 mRNA, thus inhibiting ATG16L1 expression
CircPABPN1 is recently shown to regulate intestinal epithelial autophagy by altering ATG16L1 expression via interaction with HuR.34 CircPABPN1 is derived from the PABPN1 gene and was initially shown to regulate PABPN1 expression by altering HuR binding to PABPN1 mRNA.100 Consistent with the findings in HeLa cells, circPABPN1 interacts with HuR in cultured IECs, and elevation of circPABPN1 levels specifically abolishes the binding of HuR to Atg16l1 mRNA and inhibits the expression of ATG16L1 without affecting the expression levels of ATG5 or HuR. Moreover, ectopically expressed HuR partially rescues ATG16L1 expression in cells overexpressing circPABPN1, whereas HuR silencing and circPABPN1 overexpression synergistically inhibit ATG16L1 expression. Since there are no potential binding sites for circPABPN1 in the Atg16l1 mRNA, it is likely that circPABPN1 represses ATG16L1 translation by inhibiting HuR binding to Atg16l1 transcript (Figure 3). Interestingly, human intestinal mucosal tissues from patients with IBD exhibit increased levels of circPABPN1 and decreased HuR abundances, along with decreased ATG16L1 and autophagy inactivation. Given the fact that HuR targets multiple transcripts, circPABPN1 can also regulate different mRNAs through interaction with HuR to alter the intestinal epithelial autophagy and barrier function.
Conclusions and future perspectives
Intestinal epithelial autophagy and intact barrier are essential for maintaining the epithelium homeostasis and health. Disruption of the autophagy and barrier function facilitates the entrance of luminal pathogens into the bloodstream and deteriorates the progression of many diseases such as IBD, sepsis, Alzheimer’s disease, and even aging. Regulation of mRNA stability and translation by RBPs and lncRNAs represents an important layer of complexity governing the gut epithelial defense and barrier function in response to stressful environments. RBPs and lncRNAs have a vast spectrum of biological functions in the intestinal epithelium through interactions with target mRNAs, although the exact roles of most circRNAs in the gut mucosa have not been fully investigated yet. The results summarized here provide evidence that several RBPs and lncRNAs expressed highly in the intestinal epithelium participate in a wide variety of cellular processes and play an important role in intestinal autophagy and barrier function under various pathophysiological conditions. Among RBPs, HuR enhances the epithelium defense by increasing ATG expression and promoting Paneth cell function and it sustains integrity and effectiveness of the barrier function by increasing the expression of TJs and AJs. HuR stabilizes and/or promotes translation of mRNAs encoding ATGs, TJs, AJs, and other barrier-promoting/protecting factors, thus enhancing autophagy and promoting the barrier function. On the other hand, CUGBP1, AUF1, and TIAR destabilize and/or inhibit the translation of these mRNAs, thereby downregulating the autophagy and barrier function. The intestinal autophagy and barrier function are also tightly regulated by two groups of lncRNAs: the negative lncRNA H19 and positive lncRNAs uc.173, SPRY4-IT1, and Gata6. HuR interacts with both negative and positive lncRNAs and regulates their binding and biological functions synergistically or antagonistically. Maintenance of autophagy and barrier function is dependent on a dynamic balance between the actions of diverse RBPs and lncRNAs, whereas deregulation of RBPS and lncRNAs contributes to pathologic processes of many human diseases.
Clearly, we have learned and will continue to learn a great deal from studies defining the roles and mechanisms of RBPs and lncRNAs in the intestinal epithelium homeostasis. The critical question is how we can translate these knowledges about RBP/lncRNA-mediated changes into human diseases and potential therapeutic application. We must define the exact mechanisms underlying control of the autophagy and gut barrier function by RBPs and lncRNAs and discover molecular signatures that help to early diagnose the acute gut barrier dysfunction in patients with critical disorders. The exact processes controlling the expression levels of these functional RBPs and lncRNAs in the intestinal epithelium remain largely unknown. A better understanding of the signaling control of epithelial RBPs and lncRNAs, the molecular actions of RBPs and lncRNAs, and the regulation of interactions between RBPs/lncRNAs and their target mRNAs are badly needed. We also need more specific ways to activate or inactivate functions of RBPs and lncRNAs or to stimulate autophagy and gut barrier function. Tissue-specific genetic mouse models will continue to provide important information on the in vivo functions of specific RBPs and lncRNAs in the intestinal epithelium. Although studies using human mucosal tissue samples from patients with various critical illnesses are still limited, they are necessary to establish the important impact of altered RBPs and lncRNAs on disease pathogenesis and devise therapeutic venues. With the rapid advance in our understanding of the biology of RBPs and lncRNAs, effective new therapeutics based on targeting RBPs and/or lncRNAs will one day be available to preserve the gut epithelial integrity in the clinical setting.
Funding Statement
This work was supported by Merit-Review Awards from the US Department of Veterans Affairs (to JYW and JNR) and by NIH Grants DK-57819, DK-61972, and DK-68491 (to JYW). JY Wang is a Senior Research Career Scientist, Biomedical Laboratory Research & Development Service, US Department of Veterans Affairs;National Institute of Diabetes and Digestive and Kidney Diseases ;U.S. Department of Veterans Affairs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Abbreviations
| RBPs | RNA-binding proteins |
| ncRNAs | noncoding RNAs |
| lncRNAs | long ncRNAs |
| IECs | intestinal epithelial cells |
| TJs | tight junctions; AJs, adherens junctions |
| ATGs | autophagy-related genes |
| IBD | inflammatory bowel diseases |
| RRM | RNA recognition motif |
| siRNAs | small interfering RNAs |
| LC3 | light chain 3 |
| TLR2 | Toll-like receptor 2 |
| CUGBP1 | CUG-binding protein 1 |
| AUF1 | AU-binding factor 1 |
| CLP | cecal ligation and puncture |
| miRNA | microRNA |
| CNPY3 | canopy3 |
| UTRs | untranslated regions |
| CRs | coding regions |
| TEER | transepithelial electrical resistance |
| DSS | dextran sulfate sodium |
| I/R | ischemia/reperfusion |
| UCRs | ultraconserved regions |
| circRNAs | circular RNAs. |
References
- 1.Beumer J, Clevers H.. How the gut feels, smells, and talks. Cell. 2017;170:1–17. doi: 10.1016/j.cell.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 3.Nighot P, Ma T. Role of autophagy in the regulation of epithelial cell junctions. Tissue Barriers. 2016;4(3):e1171284. doi: 10.1080/21688370.2016.1171284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 5.Yang H, Rao JN, Wang JY. Posttranscriptional regulation of intestinal epithelial tight junction barrier by RNA-binding proteins and microRNAs. Tissue Barriers. 2014;2(1):e28320. doi: 10.4161/tisb.28320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol. 2021;22:39–53. doi: 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 7.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK,Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27(1):19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 9.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 10.Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, Wang C, Ziel J, Van rooijen N, Nuñez G, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe. 2013;14(2):216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Tang L, Wang B, Sun Q, Zhao P, Li W. The role of autophagy in maintaining intestinal mucosal barrier. J Cell Physiol. 2019;234(11):19406–19419. doi: 10.1002/jcp.28722. [DOI] [PubMed] [Google Scholar]
- 12.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu TX, Chung HK, Xiao L, Piao JJ, Lan S, Jaladanki SK, Turner DJ, Raufman JP, Gorospe M, Wang JY. Long noncoding RNA H19 impairs the intestinal barrier by suppressing autophagy and lowering Paneth and Goblet cell function. Cell Mol Gastroenterol Hepatol. 2020;9(4):611–625. doi: 10.1016/j.jcmgh.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011;39(19):8472–8487. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao L, Wang JY. Posttranscriptional regulation of gene expression in epithelial cells by polyamines. Methods Mol Biol. 2011;720:67–79. [DOI] [PubMed] [Google Scholar]
- 16.Xiao L, Wang JY. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol. 2014;19:46–53. doi: 10.1016/j.coph.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebauer F, Schwarzl T, Valcarcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3):185–198. Nov 24, doi: 10.1038/s41576-020-00302-y. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 18.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3(3):195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 19.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Chatterji P, Rustgi AK. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol Med. 2018;24(5):490–506. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geng H, Bu HF, Liu F, Wu L, Pfeifer K, Chou PM, Wang X, Sun J, Lu L, Pandey A, et al. In Inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology. 2018;155(1):144–155. doi: 10.1053/j.gastro.2018.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8(6):479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36(Database):D137–40. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65(20):3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip Rev RNA. 2020;11(3):e1581. doi: 10.1002/wrna.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shorter J. Phase separation of RNA-binding proteins in physiology and disease: an introduction to the JBC Reviews thematic series. J Biol Chem. 2019;294(18):7113–7114. doi: 10.1074/jbc.REV119.007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdelmohsen K, Pullmann R Jr., Lal A, Kim HH, Galban S, Yang X,Blethrow JD, Walker M, Shubert J, Gillespie D, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giammanco A, Blanc V, Montenegro G, Klos C, Xie Y, Kennedy S,Luo J, Chang SH, Hla T, Nalbantoglu I, et al. Intestinal epithelial HuR modulates distinct pathways of proliferation and apoptosis and attenuates small intestinal and colonic tumor development. Cancer Res. 2014;74(18):5322–5335. doi: 10.1158/0008-5472.CAN-14-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 32.Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R,Rilla K, Akhtar S, Provenzani A, D'Agostino VG, et al. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One. 2013;8(7):e69563. doi: 10.1371/journal.pone.0069563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji E, Kim C, Kang H, Ahn S, Jung M, Hong Y,Tak H, Lee S, Kim W, Lee EK. RNA binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol Cell Biol. 2019;39(6):e00508–18. doi: 10.1128/MCB.00508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li XX, Xiao L, Chung HK, Ma XX, Liu X, Song JL, Jin C, Rao JN, Gorospe M, Wang JY. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 Translation. Mol Cell Biol. 2020;40(6):e00492–19. doi: 10.1128/MCB.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 36.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slowicka K, Serramito-Gomez I, Boada-Romero E, Martens A, Sze M, Petta I, Vikkula H, Rycke RD, Parthoens E, Lippens S, et al. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat Commun. 2019;10(1):1834. doi: 10.1038/s41467-019-09667-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Häsler R, et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018;215(11):2868–2886. doi: 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J,Dewan MZ,, Lieberman SR, Lazrak A, Marinis JM, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med. 2017;214(12):3687–3705. doi: 10.1084/jem.20170558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pott J, Kabat AM, Maloy KJ. Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe. 2018;23(2):191–202 e4. doi: 10.1016/j.chom.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 41.Xiao L, Li XX, Chung HK, Kalakonda S, Cai JZ, Cao S,Chen N, Liu Y, Rao JN, Wang HY, et al. RNA-binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3. Gastroenterology. 2019;157(3):731–743. doi: 10.1053/j.gastro.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Christodoulou-Vafeiadou E, Rao JN, Zou T, Xiao L, Chung HK,Yang H, Gorospe M, Kontoyiannis D, Wang JY. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell. 2014;25(21):3308–3318. doi: 10.1091/mbc.e14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L, Zhuang R, Xiao L, Chung HK, Luo J, Turner DJ, Rao JN, Gorospe M, Wang J-Y. HuR enhances Early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol. 2017;37(7):e00574–16. doi: 10.1128/MCB.00574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B,Leal T, Winter SE, Xavier RJ, Hooper LV. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357(6355):1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delgado MA, Deretic V. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009;16(7):976–983. doi: 10.1038/cdd.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. Embo J. 2008;27(7):1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y,Hao B, Bona R, et al. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1(1):79. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10(5):461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang YZ, Cai JZ, Xiao L, Chung HK, Ma XX, Chen LL,Rao JN, Wang JY. RNA-binding protein HuR regulates translation of vitamin D receptor modulating rapid epithelial restitution after wounding. Am J Physiol Cell Physiol. 2020;319(1):C208–C217. doi: 10.1152/ajpcell.00009.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu R, Zhang Y, Xia Y, Zhang J, Kaser A, Blumberg R, Sun J. Paneth cell alertness to pathogens maintained by vitamin D receptor. Gastroenterology. 2020;Nov 17:S0016-5085(20)35405–6. doi: 10.1053/j/gastro.2020.11.015. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5(4):201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M,Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24(2):85–99. doi: 10.1091/mbc.e12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK,Wu J, Donahue JM, Gorospe M, Wang JY. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell. 2015;26(10):1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim C, Kim W, Lee H, Ji E, Choe YJ, Martindale JL,Akamatsu W, Okano H, Kim HS, Nam SW, et al. The RNA-binding protein HuD regulates autophagosome formation in pancreatic beta cells by promoting autophagy-related gene 5 expression. J Biol Chem. 2014;289(1):112–121. doi: 10.1074/jbc.M113.474700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, Guo M, Li Y, Shen M, Kong D, Shao J, Ding H, Tan S, Chen A, Zhang F, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16(8):1482–1505. doi: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xin W, Zhang M, Yu Y, Li S, Ma C, Zhang J, Jiang Y, Li Y, Zheng X, Zhang L, et al. BCAT1 binds the RNA-binding protein ZNF423 to activate autophagy via the IRE1-XBP-1-RIDD axis in hypoxic PASMCs. Cell Death Dis. 2020;11(9):764. doi: 10.1038/s41419-020-02930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao L, Rao JN, Cao S, Liu L, Chung HK, Zhang Y, Zhang J, Liu Y, Gorospe M, Wang JY. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell. 2016;27(4):617–626. doi: 10.1091/mbc.E15-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh V, Gowda CP, Singh V, Ganapathy AS, Karamchandani DM, Eshelman MA,Yochum GS, Nighot P, Spiegelman VS. The mRNA-binding protein IGF2BP1 maintains intestinal barrier function by up-regulating occludin expression. J Biol Chem. 2020;295(25):8602–8612. doi: 10.1074/jbc.AC120.013646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu TX, Gu BL, Yan JK, Zhu J, Yan WH, Chen J,Qian LX, Cai W. CUGBP1 and HuR regulate E-cadherin translation by altering recruitment of E-cadherin mRNA to processing bodies and modulate epithelial barrier function. Am J Physiol Cell Physiol. 2016;310(1):C54–65. doi: 10.1152/ajpcell.00112.2015. [DOI] [PubMed] [Google Scholar]
- 60.Rao JN, Xiao L, Wang JY. Polyamines in gut epithelial renewal and barrier function. Physiology. 2020;35(5):328–337. doi: 10.1152/physiol.00011.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung HK, Wang SR, Xiao L, Rathor N, Turner DJ, Yang P, Gorospe M, Rao JN, Wang JY. α4 coordinates small intestinal epithelium homeostasis by regulating stability of HuR. Mol Cell Biol. 2018;11:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar A, Priyamvada S, Ge Y, Jayawardena D, Singhal M, Anbazhagan AN, Chatterjee I, Dayal A, Patel M, Zadeh K, et al. A novel role of SLC26A3 in the maintenance of intestinal epithelial barrier integrity. Gastroenterology. 2020;Nov 13:S0016-5085(20)35396–8. doi: 10.1053/j.gastro.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23(1):151–162. doi: 10.1091/mbc.e11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22(17):3055–3069. doi: 10.1091/mbc.e11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28(1):68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White EJ, Matsangos AE, Wilson GM. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA. 2017;8(2):8. doi: 10.1002/wrna.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P,Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3ʹ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30(21):5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26(7):2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol Biol Cell. 2008;19(9):3701–3712. doi: 10.1091/mbc.e08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 71.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN,Gorospe M, Wang JY. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol. 2018;13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, Gorospe M, Wang JY. Long noncoding RNA uc. 173 Promotes Renewal of the Intestinal Mucosa by Inducing Degradation of microRNA 195. Gastroenterol. 2018;154:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao L, Gorospe M, Wang J-Y. Long noncoding RNAs in intestinal epithelium homeostasis. Am J Physiol Cell Physiol. 2019;317(1):C93–C100. doi: 10.1152/ajpcell.00092.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Juan V, Crain C, Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28(5):1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 77.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T,Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 78.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G,Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S,Yang Y, Liu N, Zhao X, Santin AD, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34(23):3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 80.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 81.Ghazal S, McKinnon B, Zhou J, Mueller M, Men Y, Yang L,Mueller M, Flannery C, Huang Y, Taylor HS. H19 lnc RNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med. 2015;7(8):996–1003. doi: 10.15252/emmm.201505245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A,Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2(9):e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY,Xu Y, Gorospe M, Wang JY. H19 long noncoding RNA regulates intestinal epithelial barrier function via MicroRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol. 2016;36(9):1332–1341. doi: 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. [DOI] [PubMed] [Google Scholar]
- 85.Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL,Su W, Chung HK, Gorospe M, Wang JY. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24(19):3038–3046. doi: 10.1091/mbc.e13-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, He L, Ye B, Wang S, Meng S, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;20(10):1134–1144. doi: 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu J,Sun Y. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett. 2014;8(5):1947–1952. doi: 10.3892/ol.2014.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu X, Ye X, Lin H, Feng N, Gao S, Zhang X,Wang Y, Yu H, Deng X, Qian B. Knockdown of long non-coding RNA LCPAT1 inhibits autophagy in lung cancer. Cancer Biol Med. 2018;15(3):228–237. doi: 10.20892/j.2095-3941.2017.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tiessen I, Abildgaard MH, Lubas M, Gylling HM, Steinhauer C, Pietras EJ, Diederichs S, Frankel LB, Lund AH. A high-throughput screen identifies the long non-coding RNA DRAIC as a regulator of autophagy. Oncogene. 2019;38(26):5127–5141. doi: 10.1038/s41388-019-0783-9. [DOI] [PubMed] [Google Scholar]
- 90.Li X, Yang L, Chen LL. The Biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 91.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK,Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 93.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):357. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 94.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5(1):12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 96.Li F, Zhang L, Li W, Deng J, Zheng J, An M,Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo J. 2013;32(7):923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Panda AC, Abdelmohsen K, Martindale JL, Di Germanio C, Yang X, Grammatikakis I, Noh JH, Zhang Y, Lehrmann E, Dudekula DB, et al. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1mRNA translation during myogenesis. Nucleic Acids Res. 2016;44(5):2393–2408. doi: 10.1093/nar/gkw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, et al.. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, Finn SP.. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]


