Summary
Appropriate balance of T helper-17 (Th17) and regulatory T (Treg) cells maintains immune tolerance and host defense. Disruption of Th17-Treg cell balance is implicated in a number of immune-mediated diseases, many of which display dysregulation of the insulin-like growth factor (IGF) system. Here, we show that, among effector T cell subsets, Th17 and Treg cells selectively expressed multiple components of the IGF system. Signaling through IGF receptor (IGF1R) activated the AKT-mTOR pathway, increased aerobic glycolysis, favored Th17 cell differentiation over that of Treg cells, and promoted a heightened pro-inflammatory gene expression signature. Group 3 innate lymphoid cells (ILC3s), but not ILC1s or ILC2s, were similarly responsive to IGF signaling. Mice with deficiency of IGF1R targeted to T cells failed to fully develop disease in the EAE model of multiple sclerosis. Thus, the IGF system represents a previously unappreciated pathway by which type 3 immunity is modulated and immune-mediated pathogenesis controlled.
eTOC
The insulin-like growth factor (IGF) system is a well-known regulator of cell growth, differentiation, metabolism and function. DiToro and colleagues demonstrate that insulin like growth factors support inflammatory responses by modulating these processes in Th17 and Treg cells, and in ILC3s.
Introduction
T helper 17 (Th17) and regulatory T (Treg) cells exist in dynamic equilibrium at physiological barrier sites, including the gastrointestinal, respiratory and urogenital tracts, and skin. Tolerance to self-antigens and antigens of the microbiota depends on the suppressive function of Treg cells, while Th17 cells mediate pro-inflammatory responses to invading pathogens (Weaver and Hatton, 2009). Disruption of the Treg–Th17 cell equilibrium is implicated in many autoimmune or immune-mediated diseases, including, but not limited to, type-1 diabetes (T1D), inflammatory boewl disease (IBD), rheumatoid arthritis (RA), and multiple sclerosis (MS). Due to the physiological and pathophysiological relevance of this relationship, factors that modulate the balance of these cell populations can serve as promising drug targets in a variety of diseases.
There is overlap in the programming of Th17 and Treg cells such that their antagonistic roles in immune regulation are developmentally linked. Both Th17 and Treg cells have a shared requirement for transforming growth factor-β (TGFβ) (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006) and T cells activated in the presence of TGFβ often transiently co-express both Foxp3 and RORγt transcription factors. Developmental divergence of these cells is guided by the competing influence of antagonistic signaling pathways. Activation of the PI3K-AKT-mTOR pathway by co-stimulatory signals and sequential STAT3 signaling induced by IL-6 and IL-23 drive Th17 cell differentiation, while STAT5 signaling and low AKT -mTOR or high TGFβ favor Treg cells.
Known for critical roles in development, growth, metabolism, and homeostasis, the insulin-like growth factor (IGF) system drives proliferation, differentiation, and suppresses apoptosis in many cell types by modulating signaling pathways relevant to Th17-Treg balance. The primary signaling receptor, insulin-like growth factor-1 receptor (IGF1R), is a transmembrane tyrosine kinase receptor that binds insulin-like growth factors 1 and 2 (IGF1 and IGF2) and signals through the PI3K-AKT-mTOR and RAS-RAF-MEK-ERK pathways. Expression of IGF1R has also been shown to modulate the phosphorylation of STAT transcription factors, including STAT3 and STAT5. IGF2R is a non-signaling receptor that binds, endocytoses and degrades IGF2 and leukemia inhibitory factor (LIF). IGF2R also cleaves and activates membrane-bound TGFβ (Godár et al., 1999).
IGF1 and IGF2 are highly conserved, 7kDa proteins similar to pro-insulin in sequence and structure. IGF ligands are bound by a family of seven binding proteins, including IGFbp4, which prevent interaction with surface receptors. Cleavage by a family of proteases, each specific for a single or limited set of binding proteins, releases IGF ligands to allow receptor binding. Tissue-specific expression of binding proteins and proteases creates a system in which the function of IGF ligands is highly compartmentalized (Firth and Baxter, 2002; Mohan and Baylink, 2002). Transient induction of both binding proteins and their proteases, often in response to the same signals, further confers temporal control. When coupled with local IGF production, the result is a set of exquisitely regulated and specific mechanisms by which the systemic and local amounts of free biologically active IGF ligands are regulated (Allard and Duan, 2018).
Early studies of lymphocyte biology support a role for the IGF system in T cell development and function (Clark, 1997). Produced by bone marrow stem cells (Abboud et al., 1991), IGF ligands are important drivers of erythropoiesis (Miyagawa et al., 2000), granulopoiesis (Merchav et al., 1988), and lymphopoiesis (Clark et al., 1993; Kecha et al., 2000; Kooijman et al., 1997; 1995; Landreth et al., 1992). IGF1R is expressed on developing and naïve CD4+ T cells (Kooijman et al., 1992). Receptor expression rises following activation but declines on most cells after the peak of proliferation (Johnson et al., 1992; Kozak et al., 1987; Schillaci et al., 1998; Segretin et al., 2003). Exogenous IGF ligands modestly increase T cell numbers in an IGF1R-dependent manner via suppression of apoptosis and possible induction of cell cycle progression (Schillaci et al., 1995; 1994; Yang et al., 2002). Prior attempts to identify a role for IGF ligands in CD4+ T cell differentiation and function pre-dated the discovery of Th17 and Treg cells and produced conflicting results (Smith, 2010).
Microarray studies published by this lab and others indicate that naïve CD4+ T cells activated under Th17 cell conditions retain expression of IGF1R following activation (Lee et al., 2009; Muranski et al., 2011; Purwar et al., 2012; Wei et al., 2009). Furthermore, altered expression of components of the IGF system is seen in multiple pathologies involving a Th17-Treg cell disequilibrium, including T1D (Michaux et al., 2015; Smith, 2010), CD and UC (Lawrance et al., 2001), RA (Pritchard et al., 2004) and MS (Chesik et al., 2006; Lanzillo et al., 2011). In some cases, dysregulation of the IGF system has been directly tied to altered CD4+ T cell function. Some patients with these conditions develop activating autoantibodies against IGF1R, and these antibodies have been shown to suppress apoptosis in CD4+ T cells (Douglas et al., 2007; Pritchard et al., 2004).
In view of the relevance of the signaling pathways modulated by the IGF system to Th17 and Treg cell differentiation, and data demonstrating dysregulation of the IGF system in Th17-mediated diseases, we hypothesized that the IGF system specifically modulates the balance of Th17 and Treg cells. Our results indicate that IGF signaling promotes the differentiation of Th17 cells while simultaneously suppressing development of Treg cells and modulates development of immune-mediated inflammation in experimental autoimmune encephalomyelitis (EAE). Thus, the IGF system represents another key signaling pathway that governs Th17-Treg cell balance. Furthermore, we provide evidence that type 3 innate lymphoid cells (ILC3s) are also affected by IGF signaling, implicating a broader role for the IGF system in regulating type 3 immunity.
Results
Th17 and Treg cells preferentially express multiple components of the IGF system
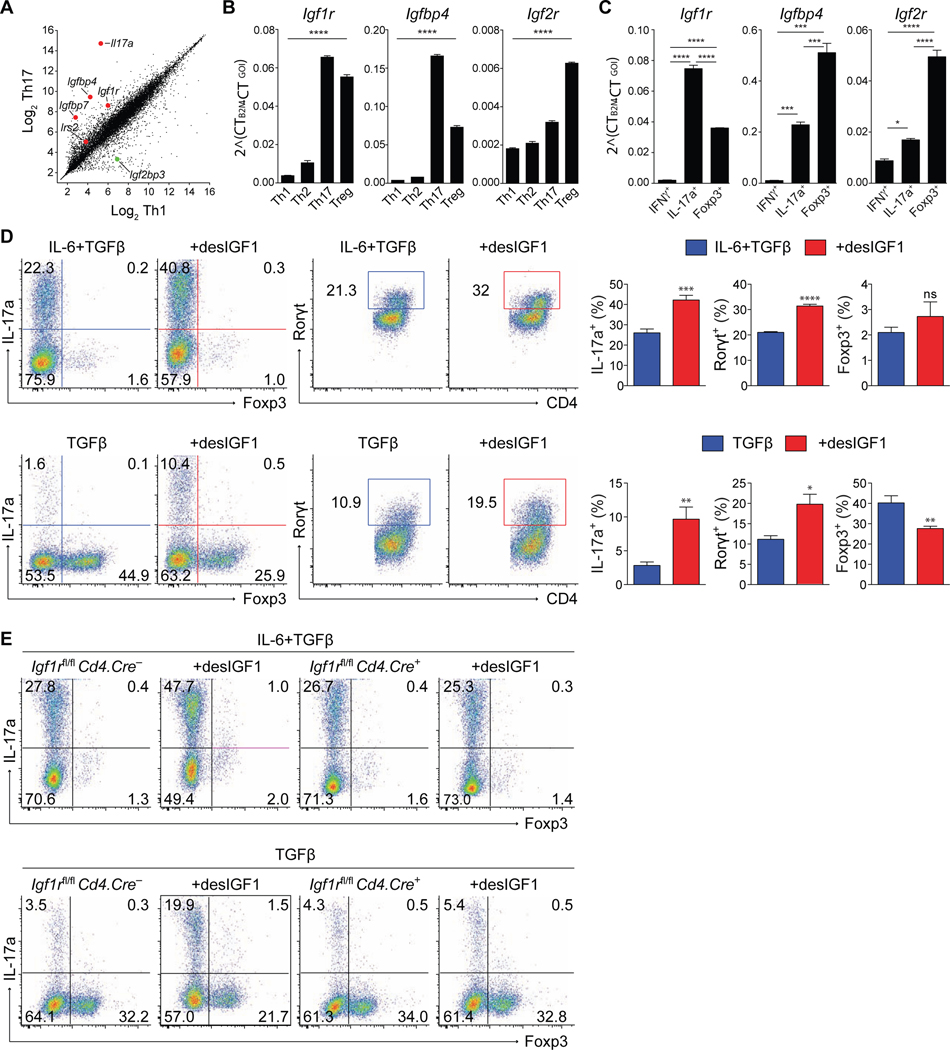
In gene expression studies that compared the Th1 and Th17 cell developmental programs, components of the insulin-like growth factor system were among the genes most differentially expressed (Fig. 1A). Igf1r, Igfbp4, Igfbp7, and Irs2 were enriched in Th17 cells, while Igf2bp3, which suppresses production of IGF2, was enriched in Th1 cells. Published datasets indicate expression of IGF-related genes is also elevated in Treg cells (Feuerer et al., 2010; Fontenot et al., 2005; Gavin et al., 2007). To confirm and extend these findings, naïve CD4+ T cells polarized under Th1, Th2, Th17 and Treg cell conditions were analyzed for expression of IGF-related genes by quantitative PCR (qPCR) (Fig. 1B). Igf1r, Igfbp4 and Igf2r were selectively upregulated in Th17 and Treg cells relative to Th1 and Th2 cells.
Figure 1. Signaling through IGF1R reciprocally regulates Th17 and Treg cell differentiation in-vitro.
(A) Scatterplot of gene expression microarray analysis of RNA from Th1 and Th17 cell conditions. Each data point represents the average of all probes above background for a given gene. (B-C) Sorted naive CD4+ T cells were stimulated in-vitro for 5 days in Th1, Th2, Th17 or Treg cell conditions. (B) qPCR analysis of Igf1R, Igfbp4 and Igf2r mRNA isolated from sorted total WT cells. (C) qPCR analysis of Igf1R, Igfbp4 and Igf2r mRNA (bottom) isolated from sorted reporter-positive IFNγ.Thy1.1-IL-17a.hCD2-Foxp3.GFP cells. (top). Experiments performed 3 times. Data from one representative experiment shown. (D-E) Sorted naive CD4+ T cells from WT (D) or IFGR1-deficient (E) mice were stimulated in-vitro for 5 days in Th17 or Treg cell conditions with or without desIGF1 and subjected to flow cytometric analysis for IL-17a, Foxp3, and RORγt. (D) Dot plots (left panels) and bar plots (right panels) are concatenated from three replicates in a single experiment. Data representative of 3–5 separate experiments. (E) Flow cytometry plots of IL-17a and Foxp3 expression from Igf1rfl/fl Cd4.Cre– and Cd4.Cre+ cells, depicting cell-number controlled concatenated data of three replicates from one of three similar experiment. Student’s t-tests were used to assess significance. See also Supplementary Figs. S1 and S2.
Because polarized effector cells are comprised of subpopulations with differing maturation characteristics, we employed reporter mice to more critically examine the expression of IGF-related genes. An IL-17a reporter mouse that expresses human CD2 (hCD2) under control of the Il17a gene locus (IL-17a.hCD2; Supplementary Figs. S1A,B) was generated and crossed with Foxp3.GFP (Fontenot et al., 2005) and Ifnγ.Thy1.1 (Harrington et al., 2008) reporter mice to generate IL-17a.hCD2-Ifnγ.Thy1.1-Foxp3.GFP triple reporter mice (Supplementary Fig. S1C). Naïve CD4+ T cells isolated from these mice were cultured under Th1, Th17 and Treg cell polarizing conditions, and mRNA isolated from sorted reporter-positive cells was analyzed by qPCR (Fig. 1C and Supplementary Fig. S1C). Consistent with our prior analysis, Igf1r, Igfbp4 and Igf2r were preferentially upregulated in Th17 and Treg cells, with Igf2r highest in Treg cells. In accordance with published data (Wei et al., 2009), we found that Th17 and Treg cells isolated from the spleen, mesenteric lymph nodes and colon of IL-17a.hCD2-Foxp3.GFP mice expressed Igf1r at or above the amount seen in splenic naïve CD4+ T cells (Supplementary Fig. S1D). Expression increased in naïve cells shortly following activation in Th17 conditions (Supplementary Fig. S1E). Flow cytometric analysis of surface IGF1R substantiated the relative mRNA expression patterns seen in naïve CD4+ T cells and in Th1, Th2, Th17 and iTreg cells (Supplementary Fig. S1F,G). Thus, genes encoding the activating receptor, IGF1R, or the regulatory binding protein and receptor, IGFbp4 and IGF2R, respectively, were specifically enriched in mature Th17 and Treg cells relative to Th1 and Th2 cells, with greater expression of the regulatory components by Treg cells. These results implicate functional regulation of Th17 and Treg cells by insulin-like growth factors.
IGFs modulate Th17-Treg balance in an IGF1R-dependent manner
To determine if IGF signaling influences T cell differentiation, naive CD4+ T cells were activated under various differentiation conditions, with or without exogenous IGF ligands. While both IGF1 and IGF2 demonstrated functional activity (Supplementary Fig. S2A, and data not shown), we used desIGF1, a natural ligand variant with reduced affinity for IGF binding proteins (IGFbps) and IGF2R, to avoid the potentially confounding effects of T cell-expressed IGFbp4 and IGF2R. Addition of desIGF1 increased expression of IL-17a and RORγt in both Th17 and Treg cells, while repressing expression of Foxp3 in Treg cells (Figs. 1D and Supplementary Fig. 2B). Moreover, Th17 and Treg cells appeared uniquely responsive, as IGF ligands failed to influence the phenotype or viability of Th1 or Th2 cells (Supplementary Figs. S2C-F).
To extend these findings, the role of IGF1R was examined using mice with targeted deletion of the Igf1r gene in T cells, generated by crossing Igf1rfl/fl mice (Klöting et al., 2008) with Cd4.Cre mice (Cd4Cre/+.Igf1rfl/fl, or Igf1rΔCD4 mice). Naïve CD4+ T cells isolated from Igf1rΔCD4 mice and activated in Th1, Th2, Th17 or Treg cell polarizing conditions did not demonstrate defects in lineage-specific cytokine or transcription factor expression compared to WT controls, however they failed to respond to IGF ligands (Figs. 1E, and data not shown). Collectively, these data indicate that IGF ligands are either not present or are effectively sequestered by IGFbps in culture, and that exogenous IGF ligands modulate the balance of Th17 and Treg cell differentiation in an IGF1R-dependent manner.
IGF1R modulates AKT-mTOR and STAT3 signaling in Th17 and Treg cells
The kinase mTOR integrates multiple environmental cues, including insulin, growth factors, and nutrient availability, to regulate cellular metabolism, proliferation and survival (Guertin and Sabatini, 2007). Downstream of the kinase AKT, mTOR forms the core of mTORC1 and mTORC2, distinct signaling complexes with differential functions and regulation (Kim et al., 2002; Sarbassov et al., 2004). Activation of mTOR is essential for effector T cell development, and directly suppresses induction of Foxp3 (Delgoffe and Powell, 2009). mTORC1 promotes Th1 and Th17 cell development, whereas Th2 cell development depends on mTORC2 (Delgoffe et al., 2011; Lee et al., 2010). Th17 cell development further requires the transcription factor hypoxia-inducible factor 1-alpha (HIF1α). HIF1α mediates the transition from oxidative phosphorylation to aerobic glycolysis, and is itself regulated by mTORC1 (Land and Tee, 2007). In view of the reported AKT-dependent activation of mTOR and HIF1α downstream of IGF1R signaling (Dudek et al., 1997; Fukuda, 2002), we examined this pathway in CD4+ T cells treated with IGF ligands.
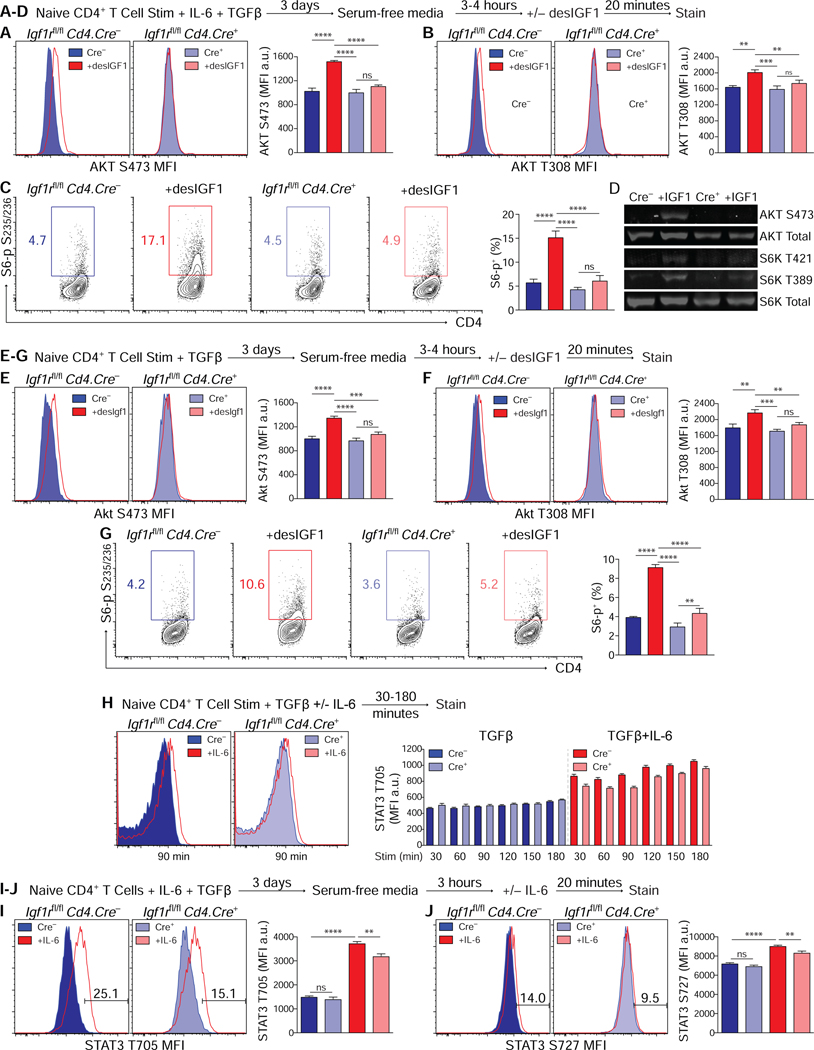
Naïve control and Igf1rΔCD4 CD4+ T cells sorted from littermate mice were activated in Th17 or Treg cell polarizing conditions for 3 days. Recovered cells were serum-starved for 4–5 hours then re-stimulated with desIGF1 and analyzed for phosphorylation of AKT and p70 S6 kinase, a ribosomal protein kinase whose activity is dependent on mTOR (Chung et al., 1992). desIGF1 induced IGF1R-dependent phosphorylation of serine residue 473 (S473) and tyrosine residue 308 (T308) of AKT, as well as serines 235/236 of p70 S6 kinase (S235/236) in both Th17 cells (Figs. 2A-C) and Treg cells ( Figs.2E-G). IGF1R-dependent phosphorylation of AKT and p70 S6 kinase in Th17 cells was confirmed by western-blot analysis (Fig. 2D). desIGF1 induced modest phosphorylation of p70 S6 kinase in T cells stimulated in Th1 or Th2 conditions, suggesting some degree of signaling, but it failed to appreciably activate the AKT (Supplementary Fig. S3), supporting lineage-restricted effects of IGF ligands on the Th17 and Treg pathways.
Figure 2. IGF1R signaling activates AKT-mTOR in Th17 and Treg cells.
(A-G) Sorted naive CD4+ T cells were activated for 3 days in Th17 or Treg cell conditions, washed and serum-starved for 4–5 hours, then re-stimulated with desIGF1 for 15 minutes. (A-C) Flow cytometric analysis of phosphorylation of AKT S473 (A) AKT T308 (B) and S6 ribosomal protein (C), Th17 cell conditions. (D) Immunoblot analysis of phosphorylation of AKT and S6-kinase, Th17 cell conditions. (E-G) Flow cytometric analysis of AKT S473 (E) AKT T308 (F) and S6 ribosomal protein (G), Treg cell conditions. Data for A-G are representative of 2 (D) to 3 (A-C, E-G) experiments.. (H) Splenocytes were stimulated with anti-CD3 and TGFβ with and without IL-6 for the indicated time. Phosphorylation of STAT3 T705 was analyzed by flow cytometry. (I-J) Sorted naive CD4+ T cells were activated for 3 days in-vitro in Th17 cell conditions, washed and serum-starved for 4–5 hours, then re-stimulated with desIGF1 for 15 minutes. Phosphorylation of STAT3 T705 (I) and S727 (J) were analyzed by flow cytometry. Data for I-J are representative of 3 experiments. Data were analyzed using two-way ANOVAs with Tukey’s post-hoc multiple comparisons test. See also Supplementary Fig. S3.
STAT3 is essential for normal development and function of a variety of vertebrate tissues (Takeda et al., 1997; Vogel et al., 2015). Activation of STAT3 by Janus kinases, first in response to IL-6 and later through the actions of IL-23, is required for induction and maturation of Th17 cells. Phosphorylation of STAT3 tyrosine 705 (T705) induces dimerization, nuclear translocation and DNA binding (Leonard and Lin, 2000). Transcriptional activity is regulated by phosphorylation of serine 727 (S727) (Wen et al., 1995). IGF1R modulates phosphorylation of multiple STAT proteins, including STAT3, across a variety of tissues (Himpe and Kooijman, 2009). This effect can be IGF-dependent or -independent (Gan et al., 2010; Zong, 2000). In the latter case, modulation of STAT phosphorylation appears dependent on delayed access to protein phosphatases resulting from a physical interaction between IGF1R, janus kinases and surface receptors (Gan et al., 2013; Huang, 2004). To examine the influence of IGF1R on IL-6-dependent STAT3 phosphorylation, naïve control and IGF1rΔCD4 CD4+ T cells sorted from littermate mice were activated in the presence of TGFβ with and without IL-6 for 30–180 minutes and STAT3 T705 phosphorylation of was assessed. In contrast to cells stimulated with TGFβ alone, Igf1rΔCD4 cells stimulated with TGFβ plus IL-6 had reduced STAT3 phosphorylation compared to controls (Fig. 2H). Next, we determined if IGF1R and IGF ligands affected IL-6-mediated STAT3 phosphorylation in Th17 cells (Figs. 2I,J). Th17 cells derived from control and Igf1rΔCD4 naïve T cells were serum-starved prior to restimulation with IL-6. STAT3 phosphorylation at both T705 and S727 was reduced in cells lacking IGF1R (Figs. 2I,J), and was unaltered by addition of desIGF1 (data not shown). Together, these data indicate that expression of IGF1R is required for optimal STAT3 phosphorylation by IL-6—independently of IGF-induced signaling—both during induction of Th17 cell development and following re-exposure to cytokine.
T cell deficiency of IGF1R protects from autoimmune CNS inflammation
Based on the foregoing studies, we postulated that the IGF system is important in modulating Th17-Treg cell balance, with implications for immune homeostasis and Th17 cell-mediated disease. Given the importance of the IGF system in lymphopoiesis and thymic development, we determined if deletion of Igf1r in T cells influenced their development at steady-state (Supplementary Fig. S4). No alterations in the percent or total number of examined T cell populations were found, suggesting that IGF1R signaling in T cells is dispensable for their normal development and maintenance.
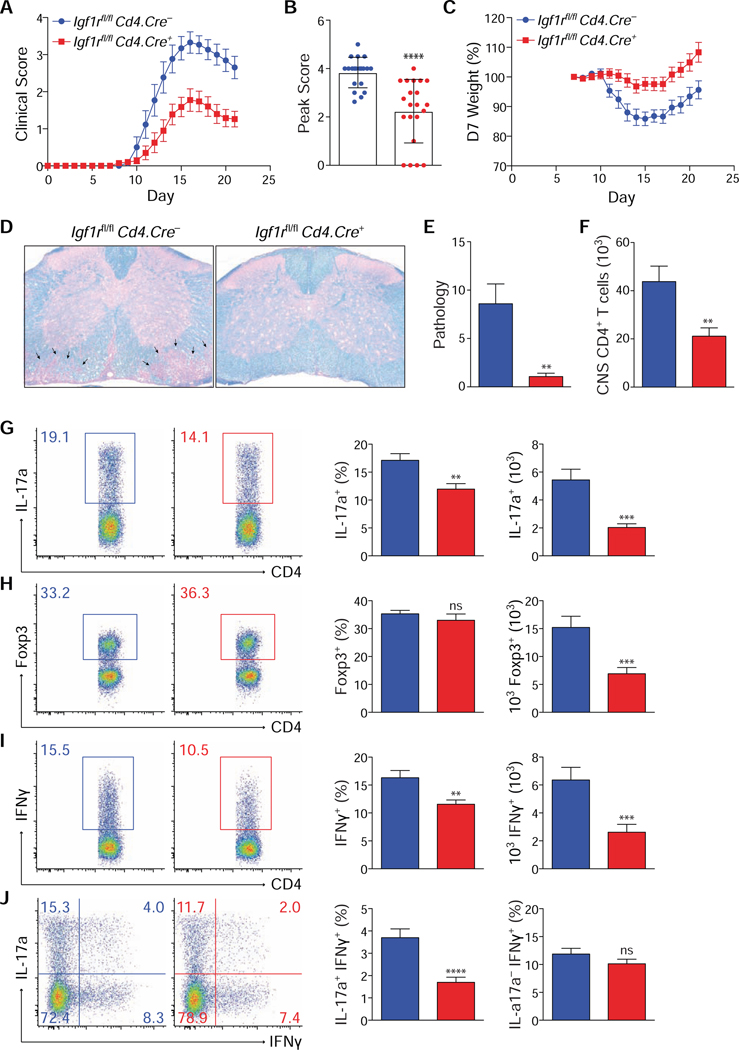
Multiple lines of evidence suggest that IGF ligands are present in the CNS during active inflammation in the EAE model of multiple sclerosis (MS) and affect disease severity, although the mechanisms are not well defined (Liu et al., 1994; Lovett-Racke et al., 1998; Spath et al., 2017). To test if expression of IGF1R directly influences Th17 versus Treg cell differentiation under inflammatory conditions in vivo, Igf1rΔCD4 and control littermate mice were immunized with MOG peptide to induce EAE (Adelmann et al., 1995). Mice lacking expression of IGF1R on T cells were protected from disease, demonstrating reduced weight loss, peak and final clinical scores, and histopathology (Figs. 3A-E).
Figure 3. Deletion of IGF1r in T cells protects against EAE.
(A-J) Gender-matched Igf1rfl/fl Cd4.Cre– and Cd4.Cre+ littermates were immunized with MOG peptide to induce EAE. After 21 days, mice were sacrificed and CNS CD4+ T cells were analyzed by flow cytometry. Mice were scored (A,B) and weighed (C) daily following induction of EAE. (B) Average peak clinical score. (D) Representative histology of Luxol fast blue-stained sections of spinal cord from one of two independent experiments. Arrows highlight areas of inflammation and demyelination in the white-matter tracts. 400x original magnification. (E) Average overall histopathologic score of spinal cord sections. (F) Average number of total CD4+ T cells isolated from CNS tissue. (G-J) Flow cytometric analysis of IL-17a (G+J), Foxp3 (H) and IFNγ (I+J) expression in CD4+ T cells isolated from CNS. Quantifications depict mean and SEM of all samples from 3 separate experiments (WT n=16, cKO n=20), except for histopathological scoring (2 experiments: Igf1rfl/fl Cd4.Cre– n=11; Cd4.Cre+ n=12). Flow cytometry plots depict cell-number controlled concatenated data from one of at least three experiments. Peak clinical scores were analyzed using the Mann-Whitney test. For all other data, student’s t-tests were used to assess significance. See also Supplementary Fig. S4.
Importantly, there was marked reduction in both the percent and total number of CD4+ T cells expressing IL-17a and IFNγ in the CNS (Figs. 3F-J). The reduction in IFNγ was primarily due to the reduction in IL-17a+IFNγ+ cells, as no change in IFNγ expression was seen among IL-17a– cells. The total number of Foxp3+ Treg cells was substantially diminished, though it was unclear if this effect was due to IGF1R-dependent cell-intrinsic factors or reduced disease severity. There was no difference in the percent or total number of corresponding populations in the spleen (data not shown). This could indicate that the source of ligand was restricted to the CNS, but it is possible that potential defects in MOG-specific cells in the spleen were masked by pre-existing non-specific endogenous populations. Taken together, these data indicate that T cell–intrinsic IGF1R signaling is required for the accumulation of pathogenic Th17 cells in the CNS during EAE and full induction of disease.
IGF ligands alter T cell fate by modulating transcription factor expression and suppressing apoptosis
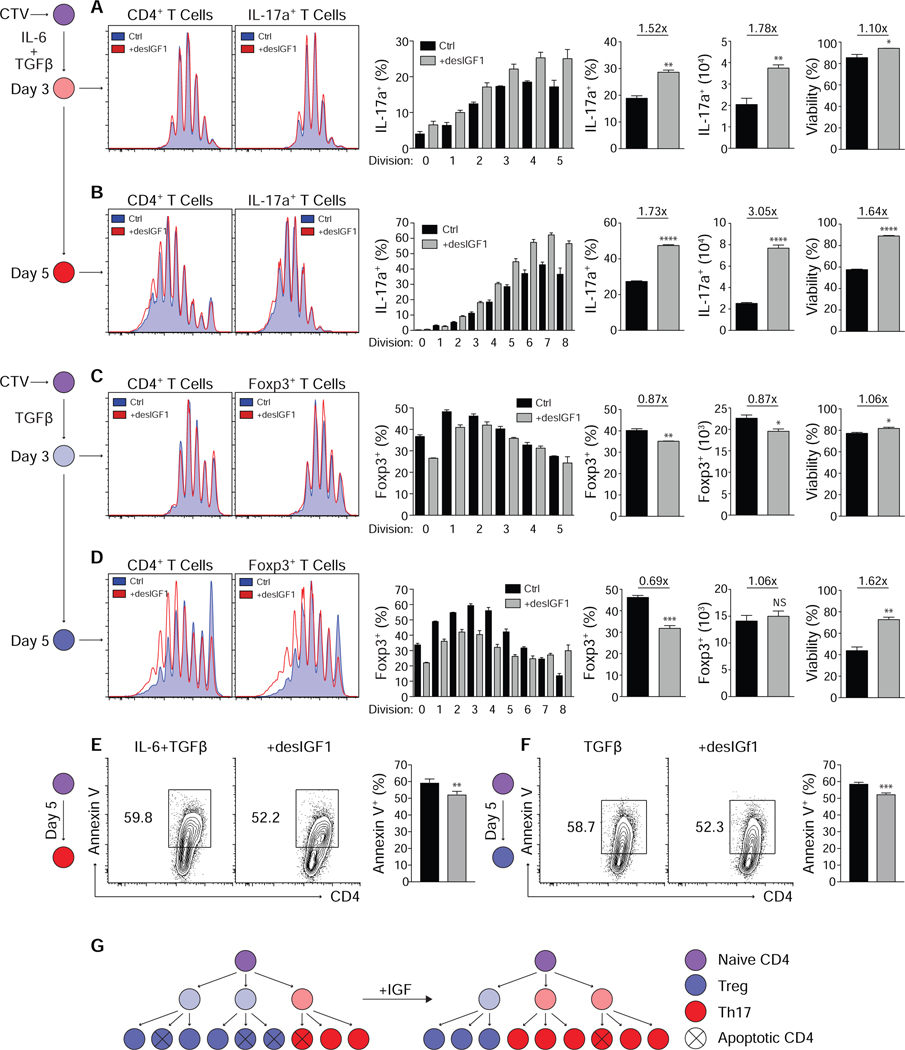
While IGF signaling altered the balance of Th17 and Treg cells and disease severity, the mechanistic basis did not necessarily require alterations in T cell differentiation programs, and may have been multifactorial. Two general mechanisms were considered: those affecting the magnitude of the pro-inflammatory CD4+ T cell response and those affecting its quality. To identify mechanisms by which IGF1R signaling affected the magnitude of the pro-inflammatory response, naïve WT CD4+ T cells were activated under Th1, Th2, Th17 and Treg cell conditions, with or without desIGF1 for 3–5 days. Transcription factor and cytokine expression, as well as cell division, viability and cell numbers were quantitated (Figs. 4A-D and Supplementary Fig. S5). Exogenous IGF failed to influence Th1 or Th2 cell development, proliferation, or viability (Supplementary Fig. S5), in accord with our previous findings. In contrast, IGF signaling increased the percent and number of IL-17a+ cells on day 3 of differentiation without influencing the total cell number or cell divisions (Fig. 4A). The number of cell divisions remained unaltered at day 5, but the percent and total number of IL-17a+ cells was increased (Fig. 4B). The fold-change in number of IL-17a+ cells was greater than the fold-change in percent of cells expressing IL-17a, suggesting that the result was not entirely due to enhanced Th17 cell programming. Absent changes in the numbers of cell divisions, this indicated enhanced cell viability, which was substantiated (Fig. 4B,E). The combined magnitude of increased cell viability and IL-17a expression fully explained the total change in IL-17a+ cell numbers. Thus, IGF signaling enhanced both Th17 cell programming and survival.
Figure 4. Signaling through IGF1R suppresses induction of Foxp3, enhances Th17 cell development and suppresses apoptosis in mature Th17 and Treg cells.
(A-D) Sorted naive CD4+ T cells were labeled with CellTrace Violet and stimulated in Th17 (A+B) or Treg (C+D) cell conditions with or without 20ng/mL desIGF1 for 3 (A+C) or 5 (B+D) days then assessed for expression of IL-17a and Foxp3 by flow cytometry. Flow plots are gated on total live (left) or IL-17a+ and Foxp3+ (right) cells. The left-most bar graphs indicate the percent IL-17a+ (A+B) or Foxp3+ (C+D) within each Cell Trace Violet peak. Bar graphs to the right indicate the percent and total number of IL-17a+ or Foxp3+ cells as well as the percent of cells staining negative for a dead-cell marker (far right). (E+F) Sorted naive CD4+ T cells were stimulated in Th17 (E) or Treg (F) cell conditions for 5 days. Cell viability and surface Annexin V were assessed by flow cytometry. Flow cytometry plots are gated on total live CD4+ cells and depict cell-number controlled concatenated data of three replicates from one representative experiment. (G) Graphic depicting the clonal expansion of naive CD4+ T cells following activation with and without exogenous desIGF1. Data from one representative experiment of 3 is shown. Student’s t-tests were used to assess significance. See also Supplementary Fig. S5.
Under Treg cell polarizing conditions, addition of desIGF1 suppressed the induction of Foxp3 before the initiation of cell division, and enhanced expression of IL-17a subsequent to cell division. At day 3, this led to a decrease in magnitude of both the percent and total number of Foxp3+ cells, and a concomitant and equivalent increase in the percent and total number of IL-17a+ cells—without influencing live cell numbers or number of cell divisions (Fig. 4C). By day 5, however, while IGF ligands suppressed the induction of Foxp3, they also appeared to stabilize its expression after multiple rounds of division (Fig. 4D). Furthermore, there was either no reduction or a small increase in the total number of Foxp3+ cells at day 5, despite the reduction in the percent of cells expressing Foxp3. As before, IGF signaling substantially enhanced cell viability via suppression of apoptosis (Fig. 4F), and the change in total Treg cell numbers was attributable to changes in Foxp3 expression and cell viability.
Collectively, these data support a model in which IGF signaling acts to bias cell fate early by reciprocally suppressing the programming of Treg cells and enhancing programming of Th17 cells, while also acting to enhance mature Th17 and Treg cell numbers by suppressing apoptosis (Fig. 4G). Thus, the net effect on Th17 cell numbers is consistently positive, while the net effect on Treg cell numbers is dependent on the specific balance of opposing genetic and survival signals.
Effects of IGF1R signaling on Th17–Treg cell fate are independent of altered proliferation or migration
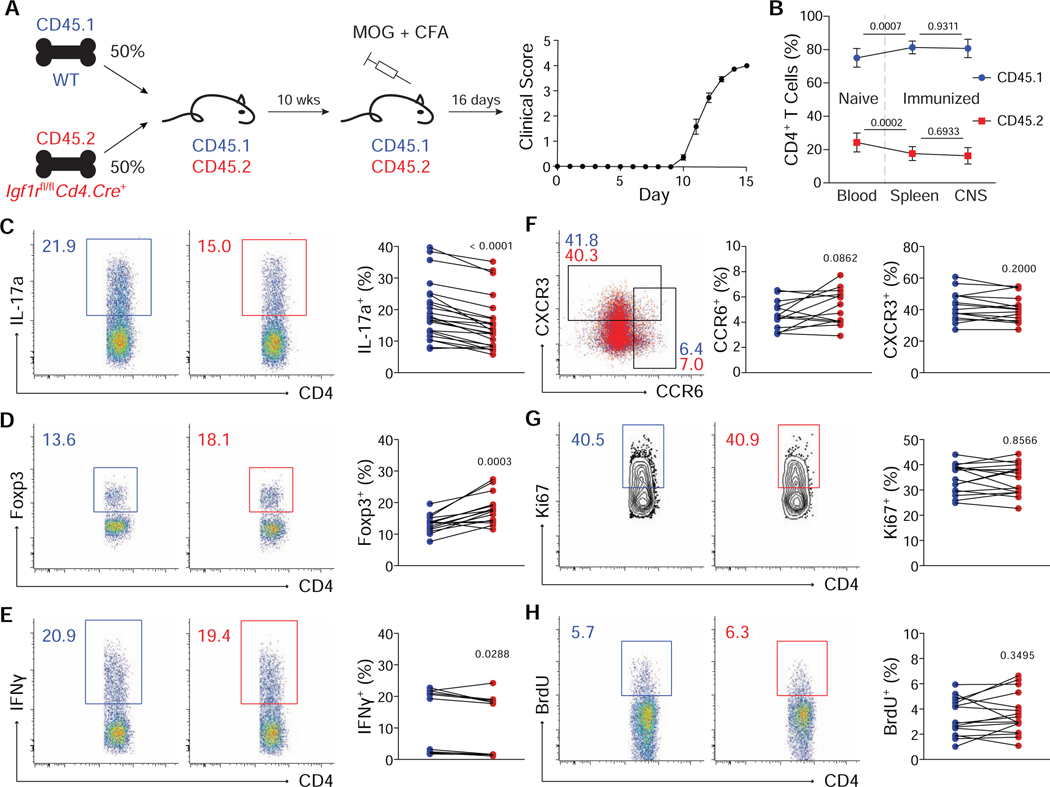
The foregoing studies pointed to combined effects of IGF signaling on both altered developmental programming and enhanced cell survival in favoring Th17 over Treg cell differentiation. To extend these studies in vivo, where effects on cell migration by IGF signaling might also be contributory (Guvakova, 2007), mixed bone marrow chimeras were generated to enable study of IGF1R-deficient T cells in the context of CNS inflammation driven by WT T cells (Fig. 5). Lethally irradiated WT CD45.1 mice were reconstituted with a 1:1 mix of CD45.1 WT and CD45.2 Igf1rΔCD4 bone marrow, then immunized with MOG peptide to induce EAE after a 10-week delay (Fig. 5A). Analysis of peripheral blood lymphocytes after reconstitution but prior to induction of disease revealed that WT T cells preferentially filled the CD4+ (Fig. 5B) and CD8+ (data not shown) T cell compartments, consistent with previous studies showing that IGF signaling accelerates T cell expansion in the context of lymphopenia (Jardieu et al., 1994). Reconstitution of Tcrb–/– mice with a 1:1 mix of WT and Igf1rΔCD4 CD4+ T cells yielded similar results (data not shown). Importantly, there was no difference in the percent of CD4+ T cells that expressed CD45.2 in the spleen or CNS following immunization, indicating that cellular trafficking was unimpaired.
Figure 5. Deletion of Igf1r biases T cell fate in-vivo without altering proliferation or the expression of relevant homing markers.
(A) Age and gender matched CD45.1 mice were lethally irradiated and reconstituted with a 1:1 mix of CD45.1 WT and CD45.2 Igf1rfl/fl Cd4.Cre+ bone marrow. EAE was induced a minimum of 10 weeks following reconstitution. (B) Percent of total CD4+ T cells expressing CD45.1 (blue) or CD45.2 (red) isolated from blood prior to immunization, or spleen and CNS following immunization (n=20). Data for each genotype were analyzed using one-way ANOVA with Tukey’s post hoc comparisons then displayed together on a single plot. (C-E) Flow cytometric analysis of IL-17a (B, n=25), Foxp3 (C, n=15) and IFNγ (D, n=15) expression in CNS CD4+ T cells. (F) Flow cytometric analysis of CXCR3 and CCR6 expression on CNS CD4+ T cells (n=15). (G-H) Flow cytometric analysis of Ki67 (G) and BrdU (H) staining in CNS CD4+ T cells (n=15). Mice were injected with BrdU 18 hours prior to sacrifice. Paired t-tests were used to assess significance for C-H. All flow cytometry plots depict cell-number controlled concatenated data from one representative experiment. Experiments performed 3 times.
Igf1rΔCD4 CD4+ T cells isolated from the CNS at the peak of disease demonstrated a defect in IL-17a production and enhanced expression of Foxp3, suggesting a cell-intrinsic bias in fate outcomes and indicating that, in the context of normal CNS inflammation, deletion of IGF1R enhances Foxp3 expression (Figs. 5C,D). There was no difference in the production of IFNγ. Expression of CXCR3 and CCR6, chemokine receptors important for CD4+ T cell migration into the CNS, was also unchanged (Figs. 5E,F). Similarly, there was no difference in the expression of Ki67 or incorporation of BrdU in the spleen (not shown) or CNS (Figs. 5G,H). These data establish that deletion of IGF1R restricts accumulation of Th17 cells in the inflamed CNS by biasing cell fate decisions and negatively impacting survival without appreciably altering cellular trafficking or proliferation.
IGF signaling alters the transcriptional profile, pathogenicity and metabolism of Th17 cells
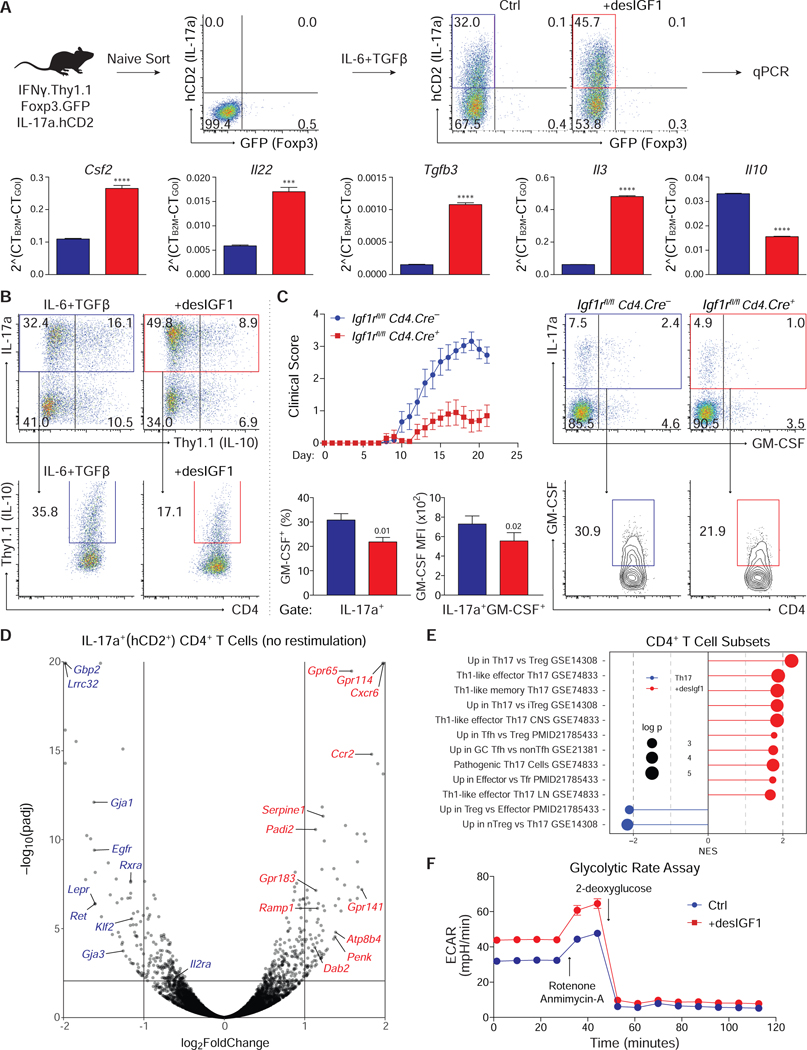
The previous experiments examined mechanisms by which IGF ligands modulate the magnitude of the Th17 and Treg cell responses. However, the relatively modest reduction in IL-17a expression in Igf1rΔCD4 mice during EAE seemed insufficient to explain the defect in disease severity, suggesting instead that IGF ligands may also modulate the quality of the T cell response by affecting Th17 cell pathogenicity. To explore this, expression analysis of selected genes was performed on Th17 cells derived from IL-17a.hCD2-Foxp3.GFP reporter mice in the presence or absence of desIGF1 (Fig. 6A). IGF signaling increased expression of pro-inflammatory cytokine genes Csf2, Il22, Tgfb3, and Il3, among others, while reducing transcripts of the anti-inflammatory cytokine Il10, which was confirmed using an IL-10-Thy1.1 reporter (Fig. 6B) and intracellular staining (Supplementary Fig. S6A). The reduction in IL-10 expression was specific to IL-17a+ cells, as there was no change in Thy1.1 expression among IL-17a– cells. Together, these data indicate that IGF1R signaling alters the expression of at least some genes related to Th17 cell-mediated pathogenicity.
Figure 6. Insulin-like growth factors alter the transcriptional, translational and metabolic identity of Th17 cells.
(A) Sorted naive CD4+ T cells from IFNγ.Thy1.1-IL-17a.hCD2-Foxp3.GFP mice were stimulated in Th17 cell conditions with or without 20ng/mL desIGF1. On day 5, cells were re-stimulated for 3 hours with PMA and Ionomycin, and RNA was isolated from sorted IL-17a (hCD2)+ Foxp3 (GFP)– CD4+ T cells. mRNA was analyzed by qPCR. (B) Sorted naive IL-10.Thy1.1 CD4+ T cells were stimulated in Th17 cell conditions without (blue) and with (red) desIGF1 for 5 days. Expression of Thy1.1 was assessed by flow cytometry. Total live CD4+ T cells (top) and IL17a+CD4+ T cells (bottom) are shown. Data for A and B are representative of two experiments each and were analyzed using a student’s t-test. (C) Gender-matched IGF1rfl/fl Cd4.Cre– and Cd4.Cre+ littermates were immunized with MOG peptide to induce EAE. After 21 days, mice were sacrificed and CNS CD4+ T cells were analyzed by flow cytometry for expression of IL17a and GM-CSF. Flow plots depict cell-number controlled concatenated data from one representative experiment. Total (top) and IL17a+ (bottom) CNS CD4+ T cells are shown. Bar graphs depict percent of IL17a+ cells expressing GM-CSF+ (left) and the MFI of GM-CSF among IL17a+GM-CSF+ cells (right). Experiment performed 3 times (WT n=15 KO n=16). (D-F) Sorted naive CD4+ T cells from IFNγ.Thy1.1-IL-17a.hCD2-Foxp3.GFP mice were stimulated in Th17 cell conditions with or without 20ng/mL desIGF1 for 5 days. RNA-seq was performed on RNA isolated from sorted IL-17a (hCD2)+ Foxp3 (GFP)– CD4+ T cells without re-stimulation. (D) Volcano plot displaying all data points satisfying threshold criteria. (E) Gene Set Enrichment Analysis (GSEA) using gene sets generated from publicly available transcriptomic datasets. Gene lists were analyzed for enrichment of curated query gene sets. A positive Normalized Enrichment Scores (NES) indicate the gene sets are enriched in desIGF1-treated cells (red) or control cells (blue). Adjusted p-values are indicated by diameter (see key, and extended methods for details). (F) Extracellular acidification rate (ECAR) trace of glycolytic rate assay (GRA) performed on Th17 cells generated as in D without (blue) and with (red) desIGF1. Measurements were performed prior to (basal) and following sequential addition of rotenone and antimycin-A (R+A; 1/10 μM), and 2-deoxyglucose (2-DG; 50 mM). Data are representative of three independent experiments. See also Supplementary Fig. S6.
To determine if IGF1R signaling altered pathogenicity of Th17 cells in vivo, Igf1rΔCD4 and control littermates were immunized with MOG peptide to induce EAE. T cells recovered from Igf1rΔCD4 mice exhibited a defect in production of GM-CSF, a key pathogenic cytokine in EAE (Codarri et al., 2011; Ponomarev et al., 2007) (Fig. 6C). The reduced MFI of GM-CSF among IL-17a+GM-CSF+ cells indicated that per-cell expression of GM-CSF was also diminished. Together, these results confirm that, in addition to impacting their number, IGF signaling altered the functional characteristics of Th17 cells in vivo.
In light of these results, we further explored the effect of IGF signaling on Th17 cell gene expression. Comparative transcriptional profiling performed on IL-17a.hCD2+-Foxp3.GFP–Th17 cells generated with or without desIGF1 revealed that IGF signaling affected the expression of multiple genes relevant to Th17 and Treg cell biology (Figs. 6D). Transcripts of genes important for Th17 cell differentiation and function were predominantly upregulated by IGF signaling, whereas untreated cells were enriched for genes relevant to Treg cell differentiation and function. Moreover, gene-set enrichment analysis (GSEA) using publicly available data (Gaublomme et al., 2015; Linterman et al., 2011; Wei et al., 2009; Yusuf et al., 2010) showed that desIGF1-treated Th17 cells were enriched for genes upregulated in Th17 and T follicular helper (Tfh) cells in comparison to Treg cells, and in non-Tfh effectors relative to T follicular regulatory (Tfr) cells (Fig. 6E).
Notably, Th17 cells treated with IGF were enriched for genes expressed by Th17 cell-derived Th1-like cells found in the CNS following induction of EAE (Hirota et al., 2011) (Fig. 6E). Accordingly, GSEA analysis showed that IGF induced expression of genes upregulated in T-bet–sufficient cells, while untreated cells were enriched for genes upregulated in T-bet–deficient cells (Supplementary Fig. S6B). Moreover, we found that IGF-treated Th17 cells were enriched for genes dependent on the expression of Hif1α, IRF4, BATF, STAT3, RORγt (Ciofani et al., 2012) and IL23R (Schiering et al., 2014), and genes repressed by Fosl2 (Ciofani et al., 2012). Thus, IGF signaling augments the core transcriptional program required for Th17 cell differentiation and promotes Th17 transition to pathogenic Th1-like cells (Harbour et al., 2015; Hirota et al., 2011; Lee et al., 2009; Wang et al., 2011); IGF signaling enhances a gene expression signature associated with the T-bet–dependent programming of pathogenic Th17 cells while suppressing genes commonly associated with Treg cells. Importantly, expression of Igf1R and Igfbp4 in Th17 cells is independent of IL-6, TGFβ, IL-1β or IL-23 (Supplementary Fig. S6C). Thus, IGF signaling biases the differentiation of developing Th17 cells away from a Treg cell transcriptional program and towards a more pathogenic Th17 program independently of IL-1β and Il-23.
Insulin and IGFs 1 and 2 can each bind and activate the insulin receptor (INSR) and IGF1R homodimers as well as INSR/IGF1R heterodimers (Clark, 1997; LeRoith et al., 1995; Siddle et al., 1994). IGFs can drive INSR-dependent and -independent effects on cellular metabolism (Kineman et al., 2018), and there is substantial overlap in the transcriptional effects of insulin and IGFs (Cai et al., 2017). We therefore explored the impact of IGFs on metabolic processes in Th17 cells. GSEA showed that many of the most enriched gene sets describe cellular metabolic processes (Supplementary Fig. S6D,E). Moreover, Hk2, the gene encoding hexokinase II, was contained in all gene sets enriched in IGF-treated cells. Induced by AKT-mTOR downstream of both IGF and insulin signaling, HKII catalyzes the phosphorylation of glucose, the rate-limiting step of glycolysis, and is the predominant hexokinase isoform in insulin-sensitive tissues (Roberts and Miyamoto, 2015).
To directly assess the influence of IGFs on Th17 cell metabolism, Th17 cells induced with or without desIGF1 were assayed for extra-cellular acidification rate (ECAR) and oxygen consumption rate (OCR), markers for glycolysis and oxidative phosphorylation, respectively. Th17 cells induced in with desIGF exhibited enhanced basal ECAR (Fig. 6F and Supplementary Fig. S6F), which persisted following inhibition of mitochondrial function with rotenone and antimycin A, but collapsed following inhibition of glycolysis with 2-deoxyglucose. IGF-induced Th17 cells also exhibited enhanced basal OCR (Supplementary Fig. S6G,H). This persisted following inhibition of ATP synthesis with oligomycin, and increased after addition of FCCP, which decouples the electron transport chain (ETC) from the membrane potential, thus maximizing flow through the ETC. Together, these data indicate that IGF signaling in developing Th17 cells enhances expression of HKII, increases glycolytic rates, and may further enhance mitochondrial function, consistent with the induction of enhanced aerobic glycolysis downstream of AKT-mTOR.
IGF signaling enhances the effector function of type 3 ILCs
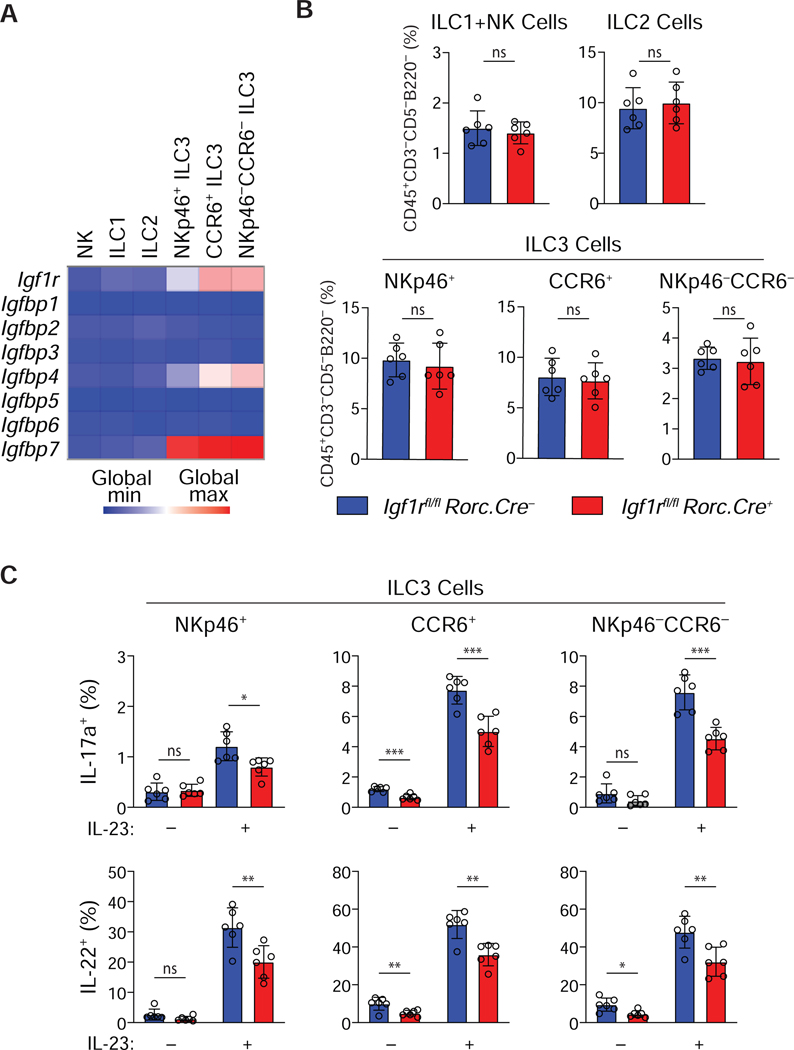
In view of the selective modulation of Th17, but not Th1 or Th2, cell responses by IGF signaling, we examined the possible role of the IGF pathway in regulating innate lymphoid cell subsets, speculating that there might be a more generalized role for the IGF system in promoting type 3 immune responses. Indeed, comparative transcriptional profiling of NK cells, Th1-like ILC1s, Th2-like ILC2s and Th17-like ILC3s from the Immgen dataset revealed that, analogous to their restricted expression by Th17 cells (Fig. 1), IgfF1r, Igfbp4 and Igfbp7 were preferentially expressed by each subset of ILC3s, but not ILC1s or ILC2s (Fig. 7A). This suggested that, like Th17 cells, ILC3s might also be IGF-responsive targets.
Figure 7. Insulin-like growth factors influence the function of Th17-like ILC3s.
(A) Microarray data showing Igf1r and Igfbp transcript expression in ILCs from small intestine lamina propria. (B) ILC frequencies in Igf1rfl/fl and RorcCre/+.Igf1rfl/fl small intestine lamina propria (n=6). (C) IL-17a and IL-22 production by ILC3s isolated from small intestine lamina propria after stimulation with 10 ng/ml IL-23 in vitro for 3 h (n=6). Graphs depict mean +/– SD. ns: not significant, *p≤0.05, **p≤0.01, ***p≤0.001.
To determine if ILC3 function was modulated by IGF signaling, mice with targeted deletion of the Igf1r gene in ILC3s were generated by crossing Igf1rfl/fl mice with mice expressing Cre recombinase under control of the Rorc promoter (RorcCre/+.Igf1rfl/fl; Igf1rΔRORγt) (Eberl and Littman, 2004), and the frequency and function of ILC3s isolated from the intestines were assessed. As with Th17 cells in Igf1rΔCD4 mice, Igf1rΔRORγt mice no defect was found at homeostasis in the frequencies of the three subsets of ILC3s in: NKp46+ ILC3s; CCR6+ ILC3s; and NKp46–CCR6– ILC3s (Fig. 7B). However, in absence of IGF1R expression, all three ILC3 subsets demonstrated impaired IL-17a and IL-22 production upon stimulation with IL-23 ex-vivo (Fig. 7C). Thus, IGF signaling appears to be important in the priming of ILC3s for effector cytokine production, suggesting that at least certain aspects of the lineage-specific effects of IGFs on Th17 cells are recapitulated in their ILC counterparts. Collectively, these data indicate that the functional programs of key components of type 3 innate and adaptive immunity are regulated by IGFs.
Discussion
Here we report that the insulin-like growth factor (IGF) system is an important regulator of Th17 versus Treg cell differentiation that contributes to the development of autoimmunity, and we explore mechanisms underlying its regulatory effects. We find that Th17 cells and Treg cells differ from other CD4+ T cell subsets examined in their expression of IGFbp4, a key regulator of IGF bioavailability, and IGF1R, the major signaling receptor for IGF ligands. Moreover, signaling through IGF1R biases the cell fate decision of naïve CD4+ T cells that share a developmental program dependent on TGFβ: It enhances activation of the AKT-mTOR pathway to suppress Treg cell development and potentiates Th17 cell development and further augments Th17 numbers via suppression of apoptosis. IGF ligands alter the programming of Th17 cells to enhance expression of genes known to be important in promoting the pathogenicity of Th17 cells, while suppressing the expression of anti-inflammatory genes. Expression of IGF1R in T cells is required for full development of pathogenic Th17-mediated CNS inflammation in EAE. We also find that, in parallel with its selective effects on Th17 cells with which they share a requirement for the transcription factor RORγt, the IGF system plays a role in modulating the function of each of the three ILC3 subsets, but not ILC1s or ILC2s. This suggests an important role for IGF regulation of both innate and adaptive components of type 3 immunity.
With specific regard to CNS inflammation, the IGF system has established links. Reactive astrocytes in EAE have been shown to produce IGF1 (Liu et al., 1994), and constitutive expression of GM-CSF by CD4+ T cells increases expression of IGF1 in CNS microglia and infiltrating macrophages during EAE (Spath et al., 2017). Mice administered IGF1 in complex with IGFbp3 during EAE demonstrate enhanced disease severity (Lovett-Racke et al., 1998). And among the genes we found enriched in Th17 cells activated by IGF signaling, several have already been tied to CNS inflammation, and are among genetic variants associated with MS (International Multiple Sclerosis Genetics Consortium et al., 2011). In addition to observations that astrocytes, activated microglia and infiltrating macrophages secrete IGF ligands in the CNS during EAE, Papp-a, the enzyme responsible for cleavage of IGFbp4 and release of free IGF, is expressed by endothelial cells at sites of inflammation in response tumor necrosis factor-α (TNF-α) and IL-1β, and is bound to the surface of macrophages (Conover et al., 2010). In light of these observations, and previous observations suggesting that Th17 cells do not attain a fully pathogenic gene expression signature until entering the inflamed CNS (Hirota et al., 2011), we propose a model whereby Th17 and Treg CD4+ T cells primed in secondary lymphoid tissues express IGF1R and are competent to secrete IGFbp4 during transit into inflamed tissues. There, they encounter macrophages that present antigen, express Papp-a and secrete IGF ligands. Local re-exposure to antigen in the face of enhanced IGF1R-dependent STAT3 and AKT-mTOR signaling may reinforce—or alter—metabolic and transcriptional programming towards a mature, pathogenic Th17 state that drives inflammatory disease.
Multiple aspects of this model remain untested. First, the studies herein do not directly address the source of IGF receptor ligand. Paracrine production of IGFs by CNS-resident myeloid cells is one plausible source, but it remains unclear if production by these cells is directly relevant to T cell biology. Furthermore, there may be alternative sources of ligand. T cells may encounter IGF ligands upon initial activation and prior to egress from lymphoid tissues. It is also possible that endocrine supplies of IGF1, produced mainly by the liver in response to growth hormone, may influence T cell function during an adaptive immune response. If true, such a finding could have important implications for the study of immune dysregulation seen in a variety of metabolic disorders, including obesity and type 2 diabetes, where serum IGFs can be chronically elevated.
The role of IGFbp4 in regulating the local bioavailability of IGF ligands also remains to be better understood. The majority of studies examining the function of IGF binding proteins indicate that IGFbps are required to localize IGF ligands to specific cell populations. This would suggest that expression of IGFbp4 by Th17 and Treg cells increases the bioavailability of IGF ligands for their own use. However, as IGFbp4 is one of only two secreted IGFbps (the remainder being cell-associated), it remains possible that IGFbp4 plays a non-canonical role in this system. IGFbp4 may also have IGF-independent effects on T cells (Mohan and Baylink, 2002). IGFbp4 has been shown to bind Lrp6 and Frz8 in the absence of IGF ligands, suppressing canonical WNT signaling by preventing interaction with Wnt3a (Zhu et al., 2008). In the presence of IGF ligands, IGFbp4 dissociates from Wnt receptors, allowing resumption of signaling. Notably, Wnt3a is produced by effector T cells, and has been shown to inhibit the suppressive capacity of Treg cells (van Loosdregt et al., 2013)1. Thus, it is possible that production of IGFbp4 acts to preserve the suppressive capacity of Treg cells in environments with low concentrations of IGF ligands. Conversely, in the presence of high concentrations of IGF ligands, IGFbp4 may serve to promote pro-inflammatory responses by dissociating from Lrp6 and Frz8, potentiating Wnt3a-mediated inhibition of Treg cell function, and delivering IGF ligands to maturing Th17 cells. Our model is also based on the supposition that expression and function of Papp-a in inflamed CNS tissue is similar to that seen in other models, though this has not been directly addressed. The role of IGF2R in modulating the influence of IGF ligands remains equally unclear. Elevated expression of IGF2R on Treg cells may serve to oppose the effects of IGF signaling via degradation of IGF2 and via enhanced activation of membrane-bound TGFβ. Evidence for such a role would have implications in a wide variety of inflammatory contexts.
The mechanism by which IGF1R exerts its effects on Th17 and Treg cells may have implications for other aspects of T cell biology. The metabolic regulation of T cell development and function has recently become an area of intensive study (Joseph et al., 2018; Klein Geltink et al., 2018). The mTOR pathway and its ability to induce expression of Hif1α are key regulators of T cell metabolism (Dang et al., 2011; Delgoffe et al., 2009; 2011; Lee et al., 2010; Shi et al., 2011). IGF1R, a well-known regulator of metabolic function, has been shown to activate mTOR and Hif1α in other cell types (Dudek et al., 1997; Fukuda, 2002). We show here that IGF1R enhances activation of mTOR in CD4+ T cells. Our RNA-seq data indicate that exposure to IGF ligands strongly induces expression of Hif1α-dependent genes, including Hk2. Recent data indicates hexokinase II directly interacts with the AKT-mTOR pathway to integrate metabolism, proliferation and survival (Roberts and Miyamoto, 2015), consistent with our findings. Thus, through its effects on STAT3 in conjunction with AKT-mTOR, IGF1R may function as a rheostat for type 3 immune responses by simultaneously modulating transcriptional, translational, metabolic and apoptotic pathways.
The data herein extend our understanding of pathways critical for immune regulation and offer targets for clinical intervention in immune-mediated disease. Immune homeostasis, tolerance, and pro-inflammatory responses to extracellular bacteria and fungi all depend on the appropriate recruitment of Th17 and Treg cells. The importance of this balance is evidenced by the co-optation of multiple evolutionarily ancient and highly conserved systems for its regulation, including, among others, those related to TGFβ, vitamin A and retinoic acid, vitamin D, IL-1β, and insulin-like growth factors. The nearly ubiquitous utilization of IGFs during all stages of life, coupled with their physiological importance, has led to the development of an exquisitely complex system that confers spatiotemporal control of their effects. Rational exploitation of this system to modulate the Th17-Treg balance—and ILC3 function—may allow for enhanced target specificity in a wide array of immune-mediated conditions and proffers a number of potential drug targets.
Caution, however, is warranted. Our results indicate that while IGF ligands can activate AKT and mTOR to repress the Treg cell program early in Treg cell development, after multiple divisions IGF ligands appear to stabilize expression of Foxp3 and/or suppress apoptosis in Foxp3+ cells. This suggests that IGF ligands may positively regulate late Treg cell stability and function in addition to biasing against early Treg cell-inducing differentiation events, reinforcing the notion that a full appreciation of timing as well as location will be required to fully leverage this system to control immune-mediated disease. Indeed, while we find that IGF signaling favors Th17 development at the expense of Treg cell development, others have reported that IGF1R is also required for optimal Treg cell responses (Bilbao et al., 2014; Johannesson et al., 2014; Miyagawa et al., 2017), perhaps reflecting the dual effects that IGFs appear to have on the Treg cell pathway. The ultimate role played by IGF signaling in modulating T cells responses may be quite nuanced and attempts to interfere with their actions may lead to counterintuitive results. In this regard, the findings herein may have implications for current efforts at therapeutic manipulation of the IGF system in human disease. More than 10 different drugs targeting IGF1 or IGF1R have entered or completed clinical trials examining their efficacy in a variety of cancers, based on the premise that the IGF system acts on transformed cells to support tumor growth (Iams and Lovly, 2015; King et al., 2014). To date, nearly all of these trials have failed to demonstrate clinical efficacy, and sustained responses were seen in only a small number of specific tumor types, thymoma being one of them (Rajan et al., 2014). These failures occurred despite impressive successes in pre-clinical murine studies. Reasons proffered for the clinical failures include redundant sources of growth factors, the ability of IGF ligands to signal through IGF1R/INSR heterodimers, and poor patient selection. The majority of pre-clinical studies on which these trials were based, however, were performed in mice lacking T cells. In light of the findings herein, consideration of effects of IGF signaling on Th17-Treg balance, and perhaps ILC3 function, will be needed in therapeutic targeting within the tumor microenvironment going forward, and efforts to manipulate of the IGF system for intervention in Th17-mediated autoimmunity may deserve renewed attention.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Casey Weaver (cweaver@uabmc.edu). Mice and plasmids generated in this study are available upon request via a material transfer agreements (MTA).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice
Igf1r floxed mice on a B6 background were a generous gift from Dr. Jens Brüning (Max Planck Institute for Metabolism Research, Köln)(Klöting et al., 2008). B6LY5.2/Cr (congenic B6 CD45.1) were purchased from Frederick Cancer Center. B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J and B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ (Cd4-Cre) and mice were purchased from The Jackson Laboratory (stock #022071). 10BiT (Tg(Il10-Thy1a)1Weav)(Maynard et al., 2007), IFNγ.Thy1.1 (Ifngtm1(Thy1)Weav)(Harrington et al., 2008), and IL-17a.hCD2 mice were generated and bred at the University of Alabama at Birmingham (UAB) animal facility. Foxp3.GFP, IFNγ IFNγ.Thy1.1, and IL-17a.hCD2 mice were intercrossed to generate IFNγ.Thy1.1-IL-17a.hCD2-Foxp3.GFP triple reporter mice. Animals were bred and maintained under specific pathogen–free conditions in accordance with UAB Institutional Animal Care and Use Committee regulations. All studies were performed on gender-matched mice 5–12 weeks of age. Mice homozygous for the Igf1R floxed allele and heterozygous for the CD4-Cre allele were bred with mice homozygous for the Igf1R floxed allele but negative for CD4-Cre to ensure an even distribution of Cre expression among littermates.
For all experiments involving innate-like lymphocyte cells (ILCs), Igf1rtm2Arge/J mice on a C57BL/6 × 129 mixed genetic background were purchased from Jackson Laboratories, and were crossed to B6 RorcCre/+ mice(Eberl and Littman, 2004) provided by Gerard Eberl. All RorcCre/+Igf1rfl/fl mice and their littermate controls were individually typed to check for spontaneous germline deletion of Igf1r; those that exhibited germline deletion were excluded from experiments. Animals were housed in specific pathogen-free facilities at Washington University in Saint Louis, and studies were conducted in accordance with the Washington University Animals Studies Committee.
EAE
Gender matched littermate mice were immunized with MOG peptide using MOG35–55/CFA Emulsion PTX kits (Hooke Labs EK-2160) according to the manufacturer’s instructions. Clinical scores were assigned according to the following rubric: Partial tail paralysis, 1. Complete tail paralysis, 2. Partial hind limb paralysis, 3. Complete hind limb paralysis, 4. Moribund, 5.
Bone Marrow Chimeras
CD45.1 WT recipient mice received two doses of 500 rads given 3 hours apart. CD45.1 WT and CD45.2 Igf1R cKO bone marrow cells were isolated from donor tibias, fibias, and femurs. Cells were re-suspended at 40e6/mL in RPMI then pooled (1:1 mixture). Each recipient mouse was injected with 200uL of cell suspension (4e6 WT and 4e6 cKO) retro-orbital. After 10 weeks, mice were bled to check reconstitution.
METHOD DETAILS Tissue processing
Mice were sacrificed using isoflurane prior to removal of spleen, lymph nodes, colons or CNS tissues. Secondary lymphoid tissues were disrupted in complete RPMI (RPMI medium containing 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, nonessential amino acids, 50 μM β-mercaptoethanol and 2 mM l-glutamine) using a syringe and a 70μm filter. RBCs were lysed using Ack lysis buffer (Fisher 50983219), and the remaining cells were re-suspended in complete RPMI. Colon tissue was flushed, opened longitudinally, and cut into strips 1 cm in length. Tissue fragments were manually disrupted using scissors then incubated for 30–40 min at 37°C with collagenase D (2 mg/mL, Sigma) and DNase (1 mg/mL, Sigma) in complete RPMI. Cells were then purified by Percoll gradient (40%/75%, 20 minutes, 600g, 25°C) and re-stimulated for 4–5 hours in complete RPMI supplemented with 50ng/mL PMA (Sigma), 750ng/mL Ionomycin (Milipore) and GolgiPlug (BD) before staining. For EAE experiments, mice were perfused with PBS prior to removal of tissue. In some experiments, spinal cord tissue was used for histology, and brain tissue was used for flow cytometric analysis. In other experiments, spinal column and brain tissues were pooled for analysis by flow cytometry. After removal, CNS tissue was manually disrupted in complete RPMI using frosted glass slides, then strained over a 70μm filter. Cells were then purified by Percoll gradient (40%/75%, 20 minutes, 600g, 25°C) and re-stimulated for 4–5 hours in complete RPMI supplemented with 50ng/mL PMA (Sigma), 750ng/mL Ionomycin (Milipore) and GolgiPlug (BD) before staining. Small intestine lamina propria single cell suspensions were generated using methods previously described(Bando et al., 2018). Briefly, small intestines were dissected, cleaned with HBSS (Gibco) buffer supplemented with HEPES (Corning), and Peyer’s patches were removed. Intestines were cut longitudinally, chopped into 3 cm-long segments, and agitated in HBSS buffer containing bovine calf serum (HyClone), HEPES, and EDTA (Corning). Tissues were then rinsed, digested with Collagenase IV (Sigma) under agitation for 40 minutes at 37 °C, and filtered through nylon mesh prior to being subjected to density gradient centrifugation (Percoll, GE Healthcare).
Flow cytometry
Surface staining was performed in PBS with 2% FBS for 15–20 minutes at 4°C. T cells from IFNγ.Thy1.1-IL-17a.hCD2-Foxp3.GFP animals were stained for 5 minutes at 4°C with biotinylated anti-hCD2, then washed and stained with streptavidin-PE-Cy7, anti-Thy1.1 and additional surface markers for 15 minutes at 4°C. Intracellular staining for transcription factors and cytokines was performed using the eBioscience Foxp3 staining kit. For phospho-stat flow cytometry, cells were fixed in 4%PFA in PBS for 10 minutes at 37°C, permeabilized in 90% MeOH for 30 minutes on ice, then stained for 1 hour at room temperature in 0.5% BSA in PBS supplemented with cOmplete Protease Inhibitor Cocktail Tablet (Sigma 11836145001). Washes were also performed using 0.5% BSA in PBS supplemented with protease inhibitors. Absolute numbers of T and B cells were calculated using PKH26 reference beads (Sigma P7458–100mL). All flow cytometry data were acquired on an Attune NxT (Thermo Fisher Scientific), LSRII, or an Aria II (BD Immunocytometry Systems, San Jose, CA) and analyzed with FlowJo software (Tree Star, Eugene, OR).
For all experiments involving ILCs, small intestine lamina propria single cell suspensions were incubated with anti-CD16/32 antibodies for 15 minutes to block Fc receptors. Cells were then stained with primary antibodies for 20 minutes, washed, and stained with fluorescently labeled streptavidin for 20 minutes. Live cells were identified using a Live/Dead Fixable Cell Stain Kit (ThermoFisher Scientific). Transcription factors were stained using the Foxp3 Transcription Factor Staining Buffer set from eBioscience. Within the CD45+CD3–CD5–CD19– live lymphocyte gate, ILC subsets were determined using the following markers: pooled ILC1 and NK cells were identified as GATA3int, RORgt-NK1.1+ cells; ILC2 were GATA3hiRORgt- cells, and ILC3 were GATA3int, RORgt+ cells. Intracellular cytokines were stained using the Cytofix/Cytoperm Plus kit from BD Biosciences. In experiments staining for cytokines, ILC3 were identified as CD45+CD3-CD5-CD19- live lymphocytes that expressed high amounts of Thy1.2 and were intermediate for CD45 expression. In each case, ILC3s were further subsetted by NKp46 and CCR6 expression.
Western Blots
Cells were lysed in RIPA buffer (Milipore 20–188) supplemented with cOmplete Protease Inhibitor Cocktail (Sigma 11836145001), Triton X-100, 1mM NaF, 1nM Na3V04 (sodium vanadate), 1mM PMSF, and 100μM Microcystin-LR for 40 minutes on ice, then briefly ultrasonicated. Total protein concentration was estimated using a Micro BCA Protein Assay kit (Thermofisher 23235). Equal amounts of total protein were then run on poly-acrylamide gels poured in-house.
Staining
Figures 1 + 6 + Supplemental Figure 1
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | PE-Cy7 | Tonbo/BD | RM4–5 |
| Thy1.1 | PE | eBioscience | HIS-51 |
| Thy1.1 | PE | BD | OX-7 |
| hCD2 | Biotin | UAB | OKT11 |
| Streptavidin | PE-Cy7 | BD | NA |
| Live/Dead | 633/775 | ThermoFisher | NA |
Figures 1,4 + Supplemental Figures 1-3
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | PE-Cy7 | Tonbo/BD | RM4–5 |
| IL-17a | PE or APC | eBioscience | ebio17B7 |
| RORγt | PE or APC | eBioscience | AFKJS-9 |
| Foxp3 | FITC | eBioscience | FJK-16S |
| Live/Dead | 633/775 | ThermoFisher | NA |
| Cell Trace Violet | 405/450 | ThermoFisher | NA |
Figure 2 Th17 + Supplemental Figure 3
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | PE-Cy7 | Tonbo/BD | RM4–5 |
| Akt T308 | AF647 | Cell Signaling | C31E5E |
| Akt S473 | AF488 | Cell Signaling | D9E |
| S6 S235/236 | PacBlue | Cell Signaling | D57.2.2E |
| Live/Dead | 633/775 | ThermoFisher | NA |
| Akt Total | NA | Cell Signaling | Polyclonal (9272) |
| Akt S473 | NA | Cell Signaling | Polyclonal (9271) |
| P70 S6K Total | NA | Cell Signaling | Polyclonal (2708) |
| P70 S6K T389 | NA | Cell Signaling | 1A5 (9206) |
| P70 S6K T421 | NA | Cell Signaling | Polyclonal (9204) |
Figures 3 + 5 + 6 + Supplemental Figure 6
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | e450/PE-Cy7 | eBioscience/Tonbo | GK1.5 |
| Foxp3 | FITC | eBioscience | FJK-16S |
| IL-17a | PE or APC | eBioscience | ebio17B7 |
| IL-17a | PerCP-Cy5 | BD | TC11–18H10 |
| IFNγ | PE-Cy7/PerCP-Cy5 | eBioscience/BD | XMG1.2 |
| GM-CSF | PE | eBioscience | MP1–27E9 |
| CXCR3 | PE | eBioscience | CXCR3–173 |
| CCR6 | APC | BioLegend | 292L17 |
| Ki67 | PE | eBioscience | SolA15 |
| Anti-BRDU | APC | eBioscience | BU20A |
| Anti-BRDU | APC | BD | 51–23619L |
| CD45.1 | FITC | BD | A20 |
| CD45.2 | BV570 | BioLegend | 104 |
| CD8α | APC-Cy7 | Tonbo | 53–6.7 |
| CD11b | APC-Cy7 | Tonbo | M1/70 |
| CD11c | APC-Cy7 | Tonbo | N418 |
| B220 | APC-Cy7 | eBioscience | RA3–6B2 |
| NK1.1 | APC-Cy7 | BioLegend | PK136 |
| Live/Dead | 633/775 | ThermoFisher | NA |
Figure 6 + Supplemental Figure 6
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | PE-Cy7 | Tonbo/BD | RM4–5 |
| Foxp3 | FITC or PE | eBioscience | FJK-16S |
| IL-17a | APC | eBioscience | ebio17B7 |
| IL-10 | PE | eBioscience | Jess-16E3 |
| Thy1.1 | PerCP-Cy5 | eBioscience | HIS51 |
| Live/Dead | 633/775 | ThermoFisher | NA |
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | PE-Cy7 | Tonbo/BD | RM4–5 |
| IFNγ | PE or APC | eBioscience | XMG1.2 |
| Tbet | PE or APC | eBioscience | ebio4B10 |
| IL-4 | FITC | eBioscience | BVDG-2462 |
| IL-4 | APC | eBioscience | 11B11 |
| Gata3 | PE | eBioscience | TWAJ |
| Live/Dead | 633/775 | ThermoFisher | NA |
| Cell Trace Violet | 405/450 | ThermoFisher | NA |
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| CD4 | PE | BD | GK1.5 |
| CD4 | PE-Cy7 | Tonbo/BD | RM4–5 |
| CD8α | PE or APC | eBioscience | 53–6.7 |
| CD44 | FITC | eBioscience | IM7 |
| CD62l | APC | eBioscience | MEL-14 |
| CD25 | PE | Tonbo | PC61.5 |
| B220 | PE-Cy7 | eBioscience | RA3–6B2 |
| CD19 | FITC | eBioscience | 1D3 |
| Foxp3 | FITC | eBioscience | FJK-16S |
| IL-17a | APC | eBioscience | ebio17B7 |
| IFNγ | PerCP-Cy5 | eBioscience | XMG1.2 |
| Live/Dead | 633/775 | ThermoFisher | NA |
| Marker | Fluor | Vendor | Clone |
|---|---|---|---|
| GATA3 | AF488 | BD | L50–823 |
| RORγt | PE | eBioscience | AFKJS-9 |
| NKp46 | Biotin | Biolegend | 29A1.4 |
| CD3 | PerCP-Cy5 | Biolegend | 145–2C11 |
| CD5 | PerCP-Cy5 | Biolegend | 53–7.3 |
| CD19 | PerCP-Cy5 | Biolegend | 6D5 |
| NK1.1 | APC | Biolegend | PK136 |
| CD45 | APC-Cy7 | Biolegend | 30-F11 |
| CD196 | BV421 | BD | 140706 |
| IL-17a | AF488 | BD | TC11–18H10 |
| IL-22 | PE | eBioscience | 1HPPWSR |
| CD90.2 | APC | BD | 53–2.1 |
| Streptavidin | PE-Cy7 | eBioscience | NA |
In-vitro T cell activation
CD4+ T cells were magnetically enriched from total splenocytes using negative selection (Miltenyi CD4+ T Cell Isolation Kit 130–104-454) on LS Columns (Miltenyi 130–042-401) then stained for CD4, CD44, CD62L, and CD25. Sorted naïve T cells (CD4+CD44− CD62L+CD25−) were stimulated for 3–5 days in 96-well flat-bottom plates (Corning 3596) in complete RPMI supplemented with 5ug/mL anti-CD3 (UAB) and containing irradiated CD4-depleted feeder cells (100e3 CD4s, 450e3 feeder cells per well). Th1 stimulations were further supplemented with 10ng/mL IL-12 (R&D 419-ML-050) and 10μg/mL anti-IL-4 (UAB 11B11). Th2 stimulations were supplemented with 1000U/mL IL-4 (R&D 404-ML-10) and 10ug/mL anti- IFN (UAB XMG1.2). Th17 stimulations were supplemented with 20ng/mL IL-6 (R&D 406-ML-005), 1.125ng/mL Tgfβ (R&D 240-B-10), 10μg/mL anti-IL-4 and 10ug/mL anti-IFNγ. Treg stimulations were supplemented with 2.5ng/mL TGFβ, 10μg/mL anti-IL-4 and 10ug/mL anti-IFNγ. To generate feeder cells, CD4+ T cells were depleted from total splenocytes using Dyna Beads (Invitrogen 11445D). CD4-depleted splenocytes were then exposed to 3000 rads. Human Igf (CU020), human desIgf1 (DU100), human Igf2 (FU020), and human desIgf2 (MU020) were purchased from Gropep and reconstituted according to manufacturer instructions. Cells were treated with 20ng/mL Igf peptides unless otherwise indicated.
For all experiments involving ILCs, lamina propria single cell suspensions were cultured in complete RPMI-10 (RPMI supplemented with bovine calf serum, HEPES, sodium pyruvate (Corning), L-alanyl-L-glutamine dipeptide (Gibco), nonessential amino acids (Corning), kanamycin sulfate (Gibco), and 2-mercaptoethanol (Sigma)) with 10 ng/ml IL-23 (R&D) for 3 h at 37 °C in the presence of brefeldin A (Golgiplug, BD Biosciences).
Histology
Spinal columns were fixed in 10% formalin (Fisher SF93–4) and paraffin embedded. Slide-mounted sections were stained with PAS/LFB or H&E, masked, and scored at UAB’s Department of Comparative Medicine and Pathology.
Real-time PCR
Total RNA was isolated using RNeasy Isolation Kits according to the manufacturer’s instructions (Qiagen 74004). cDNA was synthesized with the iScript Reverse Transcription Supermix (Bio-Rad 170–8841), and real-time PCR was performed on a Bio-Rad CFX Connect Real-Time System using SsoAdvanced Universal SYBR Supermix (Bio-Rad 172–5275). Reactions were run in triplicate. Gene-of-interest (GOI) CTs were normalized to β2m CTs (2^(CTB2M–CTGOI)).
Extracellular Flux Analysis
Analyses of cellular bioenergetics were performed using the Seahorse XFe96 Extracellular Flux Analyzer (Agilent, Santa Clara, CA). Briefly, FACS purified naïve CD4+ T cells were cultured in Th17 conditions with and without desIgf1 as described elsewhere. After 5 days, cells were plated at 250k/well into a XF96 microplate coated with cell-tak. Cells were adhered to the plate by centrifuging at 1000g for 1 minute with no brake. For the Mitochondrial Stress Test (Agilent, 105015–100), XF-DMEM media (DMEM supplemented with 5.5 mM glucose, 1 mM pyruvate and 4 mM L-Glutamine, pH 7.4 at 37 °C) was added and cells were incubated for 1 hour in a non-CO2 incubator at 37 °C. The MST consisted of parallel measures of basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) followed by sequential injections of oligomycin (1 μg/ml), FCCP (1 μM), antimycin-A (10 μM), and 2-deoxyglucose (50 mM). For the Glycolytic Rate Assay (Agilent, 103344–100), cells prepared as in the MST were added to the microplate and incubated for 1 hour in XF Base Medium supplemented with 2 mM glutamine, 10 mM glucose, 1 mM pyruvate, and 5 mM HEPES in a non-CO2 incubator at 37 °C. Basal ECAR was measured followed by sequential injections of rotenone plus Antimycin-A (R+AA; 1/10 μM), and 2-deoxyglucose (2-DG; 50 mM). The glycolytic rate was measured according to the manufacturer’s instruction.
RNA-sequencing and analysis
For sample preparation and hybridization, total RNA from purified Foxp3(GFP)−, IFNγ(Thy1.1)–, IL-17a(hCD2)+ T cells was isolated with Qiazol and miRNeasy micro kits according to manufacturer’s recommendations (Qiagen 217084 and 79306). Libraries were prepared from 500ng of total RNA using TruSeq Stranded mRNA Library Prep Kit (Illumina RS-122–2103). Samples were processed by the La Jolla Institute (LJI) with an Illumina HiSeq2500 in Rapid Run Mode, using single-end reads with lengths of 50 nucleotides (25–40M reads per condition). Reads were mapped onto the mouse genome build GRCm38 (ENSEMBL.mus_musculus.release-75) using STAR (version 2.5.3) (Dobin et al., 2013; Love et al., 2014; Stephens, 2017). BAM files were sorted using SAMtools (version 0.1.18) (Li et al., 2009; Liberzon et al., 2011; Subramanian et al., 2005), and reads were counted for each gene using HTSeq (version 0.7.2) (Anders et al., 2015). Differential expression analysis was performed using DESeq2 (version 1.18.1)(Love et al., 2014) with R (version 3.4.3). Dispersion shrinkage of fold changes was performed with the ASHR algorithm(Stephens, 2017). The fgsea R package (version 1.4.0) (https://github.com/ctlab/fgsea/) was used for gene set enrichment. Gene sets used include those from the MSigDB(Anders et al., 2015; Liberzon et al., 2011; Subramanian et al., 2005) as well as gene sets that were individually curated from several publicly available transcriptomic datasets (see figures and extended methods for details).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
P values were calculated using two-tailed Student’s t-tests and one-way or two-way ANOVA tests with Tukey’s post-hoc multiple comparisons analysis. A P value of <0.05 was considered significant. See figure legends for details. See extended bioinformatics methods below for details regarding statistical approaches utilized during analysis of RNA-sequencing data.
Pre-processing of RNA-sequencing data
The pre-processing pipeline was operated through the UNIX shell. Each raw fastq file was processed using the default settings of each analysis tool except as specified below. Standard Illumina adaptors were used to trim reads with TrimGalore. The ENCODE options for standard long RNA-seq were utilized for mapping using STAR, as defined in the STAR manual(Dobin et al., 2013). Quality of libraries was assessed by overall mapping rate, and libraries with less than 70% mapping rate were discarded from further analysis.
TrimGalore (version 0.4.5) --illumina
STAR (version2.5.3) --outSAMtype BAM SortedByCoordinate Unsorted --outFilterType BySJout --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverReadLmax 0.04 --alignIntronMin 20 --alignIntronMax 100000 --alignMatesGapMax 100000
HTSeq (version 0.7.2) --stranded=yes -t exon
Statistical analysis of RNA-sequencing data
The R software environment (version 3.4.3) with BioConductor (version 3.6) was used for statistical analysis. The random seed was set to 15144305.
Exploratory analysis, quality control
Gene counts were retained only for those genes recorded in ENSEMBL (ENSEMBL.mus_musculus.release-75) as “protein coding.” Transcripts with very low expression levels across all samples were removed. Cleaned sequencing data were subjected to a battery of exploratory analyses using the R package DESeq2 (version 1.18.1) in order to inform model selection for analysis of differential expression. After variance stabilization using the rlog algorithm(Love et al., 2014), we visually inspected heatmaps, distance matrices, and principal component plots to identify low quality samples, outliers, and unexpected trends relating to batch ID or other important covariates to be used in modeling. No worrisome trends relating to data quality were identified. Additional QC metrics (e.g. Cook’s Distances) were computed and examined as described in the DESeq2 vignette.
Count Plots
MA plots were generated using ggplot2 (http://ggplot2.tidyverse.org/) using output from plotCounts, a function in the DESeq2 package. Counts were normalized by sequencing depth and plotted on the y-axis (log10-scale), and groups are plotted along the x-axis. Mean transcript count for each group was used to generate endpoints for each line plotted; thus the lines visualize the mean difference in normalized expression between pairs of groups.
Differential Gene Expression Analysis
Following the quality control measures described above, we analyzed the influence of Igf-ligands on gene expression using a negative binomial generalized linear model described previously(Love et al., 2014). We compared performance of several competing models with and without terms for IL-6 dosing, sex, and interaction effects between these variables. Ultimately, we selected the model on the basis of reduction in type I error and superior model fit:
Transcript Count ~ β0 + β1(IGF treatment) + β2(IL-6 Dose) + β3(Sex) + β4(IGF treatment * IL-6 Dose)
MA plots
MA plots were generated using ggplot2. Shrunken log2 fold change (y-axis) in gene expression attributable to the variable of interest were plotted against the mean of normalized counts for all the samples to generate MA plots. Ashr were used as shrinkage estimators for visualization of log-fold change data(Love et al., 2014; Stephens, 2017), which was in turn used for assessment of model performance as indicated above. For these plots, as elsewhere, coloration (in red) indicates that the adjusted p-value of an observation was less than alpha, set at 0.05. Color saturation was also based on adjusted p-value. Observations with padj < 0.05 were fully saturated, with saturation decreasing to 0.2 as padj increases to 1. Genes were labeled using the ggrepel package in R (http://github.com/slowkow/ggrepel).
Volcano plot
The volcano plot was generated using ggplot2 (http://ggplot2.tidyverse.org/). Shrunken log2 fold change (x-axis) in gene expression attributable to the variable of interest were plotted against the the -log10(adjusted p-value) in the y-axis. Ashr were used as shrinkage estimators for visualization of log-fold change data(Love et al., 2014; Stephens, 2017). Genes were labeled using the ggrepel package in R (http://github.com/slowkow/ggrepel).
Gene Set Enrichment Analysis (GSEA)
Gene lists were ranked by p-value of Wald test multiplied by sign of fold-change and analyzed for enrichment of curated query gene sets using Fast Gene Set Enrichment Analysis (version 1.4.0) (https://github.com/ctlab/fgsea/) with 1 million permutations. Gene set enrichment p-values of Normalized Enrichment Scores (NES) were corrected with the Benjamini-Hochberg procedure. Significant gene sets and pathways were filtered for those with an adjusted p-value less than 0.01. Some gene sets demonstrated a moderate number of genes also enriched in the non-significant experimental condition. The ratio of the enrichment score from the significant and non-significant conditions for each gene set with an adjusted p-value less than 0.01. The bottom quartile of gene sets ranked by this ratio, representing those with substantial gene enrichment in both conditions, were filtered out.
Transcriptomic datasets generated from a variety of T cell lineages (e.g., Th1, Th2, or Th17), activation states (e.g., cytokine stimulated or unstimulated), genetically altered (e.g., Il23r knock-out or Tbx21 knock-out) and phenotypes (e.g., pathogenic or non-pathogenic) were used to create gene sets used in enrichment analysis. In addition, transcriptomic data generated from cells or tissues treated with IGF-1 or insulin were included, along with cells genetically modified to have the Igf1r or Insr genes removed. Pathways from KEGG, HALLMARK, and GO were also used for pathway analysis(Ashburner et al., 2000; Kanehisa and Goto, 2000; Liberzon et al., 2015; The Gene Ontology Consortium, 2017). When possible, analyzed data from original publications were used to create gene sets, however, when these data were not available, raw transcriptomic data were analyzed using the limma R package(Ritchie et al., 2015) for microarray data, or processed using the pipeline described above for RNA-seq data. When making gene sets from microarrays, genes with a fold change less than log2(1.5) were filtered out. The remaining genes were ranked by p-value, and the most statistically significant 125 genes with a positive and negative fold change were selected to make two separate gene sets. For RNA-seq data, a similar approach was used, except that the most significant 250 genes with a positive and negative fold change were selected to generate two distinct gene sets. Genes sets generated from single-cell RNA-seq data from GSE74833 were created by filtering for genes significantly (adjusted p-value < 0.05) expressed in a particular cell type versus all others.
Dotplots
Dotplots of NES values were generated using ggplot2 (http://ggplot2.tidyverse.org/). Coloration of points was based on the experimental condition that each gene set was significant in. The size of dots is directly proportional to -log10 of the adjusted p-value generated from the enrichment of each gene set for each comparison. The distance of each dot from the center (0) represents the NES score of each gene set.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-Seq data generated during this study has been deposited in the NCBI Gene Expression Omnibus with the accessions GSE114733.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Akt T308 AF647 C31E5E | Cell Signaling | 48646 |
| Akt S473 AF647 D9E | Cell Signaling | 40755 |
| Akt Total 9272 | Cell Signaling | 9272S |
| Akt S473 9271 | Cell Signaling | 9271S |
| Anti-BRDU APC BU20A | eBioscience | 17-5071-42 |
| Anti-BRDU APC 51-23619L | BD | 552598 |
| B220 PE-Cy7 RA3-6B2 | eBioscience | 25-0452-82 |
| B220 APC-Cy7 RA3-6B2 | BioLegend | 103223 |
| CCR6 APC 292L17 | BioLegend | 129813 |
| CCR6 BV421 140706 | BD | 564736 |
| CD11b APC-Cy7 M1/70 | Tonbo | 25–0112 |
| CD11c APC-Cy7 N418 | Tonbo | 25–0114 |
| CD19 PerCP-Cy5 6D5 | BioLegend | 115533 |
| CD25 PE PC61.5 | Tonbo | 50–0251 |
| CD4 PE-Cy7 RM4-5 | Tonbo | 60–0042 |
| CD4 PE-Cy7 RM4-5 | BD | 552775 |
| CD4 PE-Cy7 GK1.5 | Tonbo | 60–0041 |
| CD4 e450 GK1.5 | eBioscience | 48-0041-82 |
| CD4 PE GK1.5 | BD | 557308 |
| CD44 FITC IM7 | eBioscience | 11-0441-82 |
| CD45 APC-Cy7 30-F11 | BioLegend | 103115 |
| CD45.1 FITC A20 | BD | 553775 |
| CD45.2 BV570 104 | BioLegend | 109833 |
| CD62l APC MEL-14 | eBioscience | 17-0621-81 |
| CD8α PE 53–6.7 | eBioscience | 12-0081-82 |
| CD8α APC 53–6.7 | eBioscience | 17-0081-82 |
| CD8α APC-Cy7 53–6.7 | Tonbo | 25–0081 |
| Cell Trace Violet 405/450 | ThermoFisher | C34571 |
| CXCR3 PE CXCR3–173 | eBioscience | 12-1831-82 |
| Fixable Dead Cell Dye Near IR 633/775 | ThermoFisher | L10119 |
| Foxp3 FITC FJK-16S | eBioscience | 11-5773-82 |
| Foxp3 PE FJK-16S | eBioscience | 12–5773-82 |
| Gata3 AF488 L50–823 | BD | 560077 |
| Gata3 PE TWAJ | eBioscience | 12-9966-42 |
| GM-CSF PE MP1–27E9 | eBioscience | 12-7331-82 |
| IL-10 PE Jes5–16E3 | eBioscience | 12-7101-41 |
| IL-17a PE ebio17B7 | eBioscience | 12-7177-81 |
| IL-17a APC ebio17B7 | eBioscience | 17-7177-81 |
| IL-17a AF488 TC11-18H10 | BD | 560220 |
| IL-17a PerCP-Cy5 TC11-18H10 | BD | 560666 |
| IL-22 PE 1H8PWSR | eBioscience | 12-7221-82 |
| IL-4 FITC BVDG-2462 | eBioscience | 11-7042-41 |
| IL-4 APC 11B11 | eBioscience | 17-7041-82 |
| IFNγ PE XMG1.2 | eBioscience | 12-7311-82 |
| IFNγ PE-Cy7 XMG1.2 | eBioscience | 25-7311-41 |
| IFNγ APC XMG1.2 | eBioscience | 17-7311-82 |
| IFNγ PerCP-Cy5 XMG1.2 | BD | 560660 |
| Ki67 PE SolA15 | eBioscience | 12-5698-82 |
| NK1.1 APC PK136 | BioLegend | 108710 |
| NK1.1 APC-Cy7 PK136 | BioLegend | 108724 |
| P70 S6K Total 49D7 | Cell Signaling | 2708S |
| P70 S6K T389 1A5 | Cell Signaling | 9206S |
| P70 S6K T421/S424 9204 | Cell Signaling | 9204S |
| RORγt PE AFKJS-9 | eBioscience | 12-6988-82 |
| RORγt APC AFKJS-9 | eBioscience | 17-6988-82 |
| S6 S235/236 Pacific Blue D57.2.2E | Cell Signaling | 8520S |
| Streptavidin PE-Cy7 | BD | 405206 |
| Tbet PE eBio4B110 | eBioscience | 12-5825-82 |
| Tbet APC eBio4B110 | eBioscience | 17-5825-82 |
| Thy1.1 APC 53–2.1 | BD | 561974 |
| Thy1.1 PerCP-Cy5 HIS51 | eBioscience | 45-0900-82 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| IL-1β | R&D | 401-ML-025 |
| IL-12 | R&D | 419-ML-050 |
| IL-23 | R&D | 1887-ML-010 |
| IL-4 | R&D | 404-ML-10 |
| IL-6 | R&D | 406-ML-005 |
| Tgfβ | R&D | 240-B-10 |
| Igf | Gropep | CU020 |
| Igf2 | Gropep | FU020 |
| desIgf1 | Gropep | DU100 |
| desIgf2 | Gropep | MU020 |
| Golgiplug | BD | |
| Phorbol 12-myristate 13-acetate | Sigma-Aldrich | P185–1MG |
| Ionomycin | EMD Bioscience | 4D7–952 |
| Critical Commercial Assays | ||
| CD4+ T Cell Isolation Kit | Miltenyi | 130-104-454 |
| LS Columns | Miltenyi | 130-042-401 |
| Dyna Beads | Invitrogen | 11445D |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | 1725275 |
| iScript cDNA Synthesis Kit | Bio-Rad | 1708891 |
| MOG35-55/CFA Emulsion PTX | Hooke Labs | EK-2160 |
| Mitochondrial Stress Test | Agilent | 105015–100 |
| Glycolytic Rate Assay | Agilent | 103344–100 |
| Qiazol | Qiagen | 217084 |
| miRNeasy | Qiagen | 79306 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | RS-122-2103 |
| Deposited Data | ||
| RNA-Seq, CD4 T Cells | GEO Database | GSE114733 |
| RNA-Seq, Innate Lymphooid Cells | GEO Database | GSE37448 |
| Experimental Models: Cell Lines | ||
| Mouse primary T lymphocytes | Generated in house | |
| Experimental Models: Organisms/Strains | ||
| Igf1rtm1Jcbr | Kloting et al., 2008 | MGI:3818453 |
| Igf1rtm2Arge/J | Jackson Laboratory | Stock No: 012251 |
| Foxp3tm2Ayr | Fontenot et al., 2005 | MGI:3574964 |
| B6-LY5.2/Cr | Charles River | Strain Code 564 |
| B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J | Jackson Laboratory | Stock #022071 |
| 10BiT Tg(Il10-Thy1a)1Weav | Generated in house | MGI:3767675 |
| IFNγ.Thy1.1 (Ifngtm1(Thy1)Weav) | Generated in house | MGI:3831296 |
| IL-17a.hCD2 | Generated in house | N/A |
| RorcCre/+ | Provided by Gerard Eberl | MGI:3054098 |
| Oligonucleotides | ||
| B2m Forward GGTCTTTCTGGTGCTTGTCT | Generated in house | |
| B2m Reverse TATGTTCGGCTTCCCATTCTC | Generated in house | |
| Csf2 Forward GAAGATATTCGAGCAGGGTCTAC | Generated in house | |
| Csf2 Reverse CTTGTGTTTCACAGTCCGTTTC | Generated in house | |
| Foxp3 Forward CCCAGAGTTCTTCCACAACAT | Generated in house | |
| Foxp3 Reverse TTGAGTGTCCTCTGCCTCT | Generated in house | |
| Gata3 Forward TCTGGAGGAGGAACGCTAAT | Generated in house | |
| Gata3 Reverse GGTCTGGATGCCTTCTTTCTT | Generated in house | |
| Ifng Forward CTCTTCCTCATGGCTGTTTCT | Generated in house | |
| Ifng Reverse TTCTTCCACATCTATGCCACTT | Generated in house | |
| Igf1r Forward ACCATCGATTCGGTGACTTCTGCT | Generated in house | |
| Igf1r Reverse AAGGACAAGGAGACCAAGGCATGA | Generated in house | |
| Igf2r Forward AACATTTGTGTGCCCATCTGAGCG | Generated in house | |
| Igf2r Reverse ATTGGGCCAAGGGACTAAGGTCAA | Generated in house | |
| Igfbp4 Forward TCGGAAATCGAAGCCATCCAGGAA | Generated in house | |
| Igfbp4 Reverse GGGTTGAAGCTGTTGTTGGGATGT | Generated in house | |
| Il3 Forward AGGACCCTCTCTGAGGAATAAG | Generated in house | |
| Il3 Reverse TGTAGGCAGGCAACAGTTAAG | Generated in house | |
| Il10 Forward TTGAATTCCCTGGGTGAGAAG | Generated in house | |
| Il10 Reverse TCCACTGCCTTGCTCTTATTT | Generated in house | |
| Il17a Forward CAAACATGAGTCCAGGGAGAG | Generated in house | |
| Il17a Reverse GCTGAGCTTTGAGGGATGAT | Generated in house | |
| Il22 Forward CGACCAGAACATCCAGAAGAA | Generated in house | |
| Il22 Reverse GAGACATAAACAGCAGGTCCA | Generated in house | |
| Rorgt Ex2–4 Forward GAAGACCCACACCTCACAAA | Generated in house | |
| Rorgt Ex2–4 Reverse CAGGAGTAGGCCACATTACAC | Generated in house | |
| Rorgt Ex10–11 Forward TGCAAGACTCATCGACAAGG | Generated in house | |
| Rorgt Ex10–11 Reverse TCCTTATAGAGTGGAGGGAAGG | Generated in house | |
| Tbx21 Forward AGTTCAACCAGCACCAGAC | Generated in house | |
| Tbx21 Reverse CACATCCACAAACATCCTGTAATG | Generated in house | |
| Tgfb3 Forward CGCTACATAGGTGGCAAGAA | Generated in house | |
| Tgfb3 Reverse CAAGTTGGACTCTCTCCTCAAC | Generated in house | |
| Recombinant DNA | ||
| Mouse IL-17a BAC | CHORI | RP24–305023 |
| pL451 | Liu et al. 2003 | N/A |
| pL253 | Na et al. 2013 | N/A |
| pBSK | Addgene | 67504 |
| Human CD2 Y91D | Wolff et al. 1990 | N/A |
| Software and Algorithms | ||
| STAR (version 2.5.3) | Dobin et al., 2013 | |
| SAMtools (version 0.1.18) | Li et al., 2009 | |
| HTSeq (version 0.7.2) | Anders et al., 2015 | |
| DESeq2 (version 1.18.1) | Love et al., 2014 | |
| R (version 3.4.3) | Htt[s://www.r-project.org | |
| ASHR | Stephens et al., 2017 | |
| fgsea R package (version 1.4.0) | https://github.com/ctlab/fgsea/ | |
| MSigDB | Liberzon et al., 2011 |
Highlights.
IGFs favor Th17 cell differentiation over that of Treg cells
IGF1R augments AKT-mTOR and STAT3 signaling and increases aerobic glycolysis
Signaling through IGF1R increases Th17 numbers and pathogenicity in EAE
ILC3 cell function is similarly affected by insulin-like growth factors
ACKNOWLEDGMENTS
The authors thank Drs. Etty Beneveniste, Laurie Harrington, Lou Justement and Robin Lorenz (UAB), and members of the Weaver laboratory for their helpful comments and suggestions. We gratefully acknowledge Jeremy Day (La Jolla Institute of Allergy and Immunology) for assistance with gene expression studies. We also thank Jon Wright, Brenda Dale and Alanna Smith for expert technical assistance, Connie Rutledge for editorial assistance, and the UAB Center for AIDS Research Flow Cytometry Core and UAB Epitope Recognition and Immunoreagent Core Facility flow cytometric sorting and antibody preparations, respectively. This work was supported by an NIH training grant (T32 AI007051; DD), NIH K99 DK118110 (JKB), NIH R01 grants (CHC, SJF, RDH and CTW) and UAB Institutional Funds (C.T.W.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abboud SL, Bethel CR, and Aron DC (1991). Secretion of insulinlike growth factor I and insulinlike growth factor-binding proteins by murine bone marrow stromal cells. J. Clin. Invest 88, 470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelmann M, Wood J, Benzel I, Fiori P, Lassmann H, Matthieu JM, Gardinier MV, Dornmair K, and Linington C. (1995). The N-terminal domain of the myelin oligodendrocyte glycoprotein (MOG) induces acute demyelinating experimental autoimmune encephalomyelitis in the Lewis rat. J. Neuroimmunol 63, 17–27. [DOI] [PubMed] [Google Scholar]
- Allard JB, and Duan C. (2018). IGF-binding proteins: Why do they exist and why are there so many? Front. Endocrinol 9, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando JK, Gilfillan S, Song C, McDonald KG, Huang SC-C, Newberry RD, Kobayashi Y, Allan DSJ, Carlyle JR, Cella M, et al. (2018). The tumor necrosis factor superfamily member RANKL Suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity 48, 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. [DOI] [PubMed] [Google Scholar]
- Bilbao D, Luciani L, Johannesson B, Piszczek A, and Rosenthal N. (2014). Insulin-like growth factor-1 stimulates regulatory T cells and suppresses autoimmune disease. EMBO Mol. Med 6, 1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Sakaguchi M, Kleinridders A, Gonzalez-Del Pino G, Dreyfuss JM, O’Neill BT, Ramirez AK, Pan H, Winnay JN, Boucher J, et al. (2017). Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat. Commun 8, 14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesik D, De Keyser J, Glazenburg L, and Wilczak N. (2006). Insulin-like growth factor binding proteins: regulation in chronic active plaques in multiple sclerosis and functional analysis of glial cells. Eur. J. Neurosci 24, 1645–1652. [DOI] [PubMed] [Google Scholar]
- Chu YW, Schmitz S, Choudhury B, Telford W, Kapoor V, Garfield S, Howe D, and Gress RE (2008). Exogenous insulin-like growth factor 1 enhances thymopoiesis predominantly through thymic epithelial cell expansion. Blood 112, 2836–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Kuo CJ, Crabtree GR, and Blenis J. (1992). Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69, 1227–1236. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. (1997). The somatogenic hormones and insulin-like growth factor-1: stimulators of lymphopoiesis and immune function. Endocr. Rev 18, 157–179. [DOI] [PubMed] [Google Scholar]
- Clark R, Strasser J, McCabe S, Robbins K, and Jardieu P. (1993). Insulin-like growth factor-1 stimulation of lymphopoiesis. J. Clin. Invest 92, 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L, Gyülvészi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, and Becher B. (2011). RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol 12, 560–567. [DOI] [PubMed] [Google Scholar]
- Conover CA, Mason MA, Bale LK, Harrington SC, Nyegaard M, Oxvig C, and Overgaard MT (2010). Transgenic overexpression of pregnancy-associated plasma protein-A in murine arterial smooth muscle accelerates atherosclerotic lesion development. Am. J. Physiol. Heart Circ. Physiol 299, H284–H291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. (2003). Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748. [DOI] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang H-Y, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen H-R, et al. (2011). Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell 146, 772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, and Powell JD (2009). mTOR: taking cues from the immune microenvironment. Immunology 127, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, and Powell JD (2009). The mTOR kinase differentially regulates effector and regulatory t cell lineage commitment. Immunity 30, 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, and Powell JD (2011). The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol 12, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RS, Gianoukakis AG, Kamat S, and Smith TJ (2007). Aberrant expression of the insulin-like growth factor-1 receptor by T cells from patients with Graves’ disease may carry functional consequences for disease pathogenesis. J. Immunol 178, 3281–3287. [DOI] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, and Greenberg ME (1997). Regulation of neuronal survival by the serine-threonine protein kinase AKT. Science 275, 661–665. [DOI] [PubMed] [Google Scholar]
- Duplomb L. (2003). Soluble Mannose 6-Phosphate/Insulin-Like Growth Factor II (IGF-II) Receptor inhibits interleukin-6-type cytokine-dependent proliferation by neutralization of IGF-II. Endocrinology 144, 5381–5389. [DOI] [PubMed] [Google Scholar]
- Eberl G, and Littman DR (2004). Thymic origin of intestinal αß T cells revealed by fate mapping of RORγt+ cells. Science 305, 248–251. [DOI] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, Boehmer, von, H., Mathis, D., and Benoist, C. (2010). Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc. Natl. Acad. Sci. USA 107, 5919–5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth SM, and Baxter RC (2002). Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev 23, 824–854. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, and Rudensky AY (2005). Regulatory T cell lineage specification by the forkhead transcription factor Foxp3. Immunity 22, 329–341. [DOI] [PubMed] [Google Scholar]
- Fukuda R. (2002). Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J. Biol. Chem 277, 38205–38211. [DOI] [PubMed] [Google Scholar]
- Gan Y, Zhang Y, Buckels A, Paterson AJ, Jiang J, Clemens TL, Zhang ZY, Du K, Chang Y, and Frank SJ (2013). IGF-1R modulation of acute GH-induced STAT5 signaling: Role of protein tyrosine phosphatase activity. Mol. Endocrinol 27, 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Zhang Y, DiGirolamo DJ, Jiang J, Wang X, Cao X, Zinn KR, Carbone DP, Clemens TL, and Frank SJ (2010). Deletion of IGF-I receptor (IGF-IR) in primary osteoblasts reduces gh-induced STAT5 signaling. Mol. Endocrinol 24, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. (2015). Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, and Rudensky AY (2007). Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775. [DOI] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang X-P, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. (2010). Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godár S, Horejsi V, Weidle UH, Binder BR, Hansmann C, and Stockinger H. (1999). M6P/IGFII-receptor complexes urokinase receptor and plasminogen for activation of transforming growth factor-beta1. Eur. J. Immunol 29, 1004–1013. [DOI] [PubMed] [Google Scholar]
- Guertin DA, and Sabatini DM (2007). Defining the role of mTOR in cancer. Cancer Cell 12, 9–22. [DOI] [PubMed] [Google Scholar]
- Guvakova MA (2007). Insulin-like growth factors control cell migration in health and disease. Int. J. Biochem. Cell Biol 39, 890–909. [DOI] [PubMed] [Google Scholar]
- Harbour SN, Maynard CL, Zindl CL, Schoeb TR, and Weaver CT (2015). Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc. Natl. Acad. Sci USA 112, 7061–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Janowski KM, Oliver JR, Zajac AJ, and Weaver CT (2008). Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452, 356–360. [DOI] [PubMed] [Google Scholar]
- Himpe E, and Kooijman R. (2009). Insulin-like growth factor-I receptor signal transduction and the janus kinase/signal transducer and activator of transcription (JAK-STAT) pathway. Biofactors 35, 76–81. [DOI] [PubMed] [Google Scholar]
- Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. (2011). Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol 12, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. (2004). Physical and functional interaction of growth hormone and insulin-like growth factor-i signaling elements. Mol. Endocrinol 18, 1471–1485. [DOI] [PubMed] [Google Scholar]
- Iams WT, and Lovly CM (2015). Molecular pathways: clinical applications and future direction of insulin-like growth factor-1 receptor pathway blockade. Clin. Cancer Res 21, 4270–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, et al. (2011). Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardieu P, Clark R, Mortensen D, and Dorshkind K. (1994). In vivo administration of insulin-like growth factor-I stimulates primary B lymphopoiesis and enhances lymphocyte recovery after bone marrow transplantation. J. Immunol 152, 4320–4327. [PubMed] [Google Scholar]
- Johannesson B, Sattler S, Semenova E, Pastore S, Kennedy-Lydon TM, Sampson RD, Schneider MD, Rosenthal N, and Bilbao D. (2014). Insulin-like growth factor-1 induces regulatory T cell-mediated suppression of allergic contact dermatitis in mice. Dis. Model Mech 7, 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EW, Jones LA, and Kozak RW (1992). Expression and function of insulin-like growth factor receptors on anti-CD3-activated human T lymphocytes. J. Immunol 148, 63–71. [PubMed] [Google Scholar]
- Joseph AM, Monticelli LA, and Sonnenberg GF (2018). Metabolic regulation of innate and adaptive lymphocyte effector responses. Immunol. Rev 286, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucl. Acids Res 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kecha O, Brilot F, Martens H, Franchimont N, Renard C, Greimers R, Defresne MP, Winkler R, and Geenen V. (2000). Involvement of insulin-like growth factors in early T cell development: a study using fetal thymic organ cultures. Endocrinology 141, 1209–1217. [DOI] [PubMed] [Google Scholar]
- Kim D-H, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, and Sabatini DM (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175. [DOI] [PubMed] [Google Scholar]
- Kim EY, and Moudgil KD (2017). Immunomodulation of autoimmune arthritis by pro-inflammatory cytokines. Cytokine 98: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kineman RD, Del Rio-Moreno M, and Sarmento-Cabral A. (2018). 40 YEARS of IGF1: Understanding the tissue-specific roles of IGF1/IGF1R in regulating metabolism using the Cre/loxP system. J. Mol. Endocrinol 61, T187–T198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H, Aleksic T, Haluska P, and Macaulay VM (2014). Can we unlock the potential of IGF-1R inhibition in cancer therapy? Cancer Treat. Rev 40, 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Geltink RI, Kyle RL, and Pearce EL (2018). Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu. Rev. Immunol 36, 461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M, and Hafler DA (2013). The plasticity of human Treg and Th17 cells and its role in autoimmunity. Sem. Immunol 25, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöting N, Koch L, Wunderlich T, Kern M, Ruschke K, Krone W, Brüning JC, and Blüher M. (2008). Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes 57, 2074–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman R, van Buul-Offers SC, Scholtens LE, Reijnen-Gresnigt RG, and Zegers BJ (1997). T and B cell development in pituitary insulin-like growth factor-II transgenic dwarf mice. J. Endocrinol 155, 165–170. [DOI] [PubMed] [Google Scholar]
- Kooijman R, van Buul-Offers SC, Scholtens LE, Schuurman HJ, Van den Brande LJ, and Zegers BJ (1995). T cell development in insulin-like growth factor-II transgenic mice. J. Immunol 154, 5736–5745. [PubMed] [Google Scholar]
- Kooijman R, Willems M, De Haas CJ, Rijkers GT, Schuurmans AL, Van Buul-Offers SC, Heijnen CJ, and Zegers BJ (1992). Expression of type I insulin-like growth factor receptors on human peripheral blood mononuclear cells. Endocrinology 131, 2244–2250. [DOI] [PubMed] [Google Scholar]
- Kozak RW, Haskell JF, Greenstein LA, Rechler MM, Waldmann TA, and Nissley SP (1987). Type I and II insulin-like growth factor receptors on human phytohemagglutinin-activated T lymphocytes. Cell. Immunol 109, 318–331. [DOI] [PubMed] [Google Scholar]
- Land SC, and Tee AR (2007). Hypoxia-inducible factor 1 is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J. Biol. Chem 282, 20534–20543. [DOI] [PubMed] [Google Scholar]
- Landreth KS, Narayanan R, and Dorshkind K. (1992). Insulin-like growth factor-I regulates pro-B cell differentiation. Blood 80, 1207–1212. [PubMed] [Google Scholar]
- Lanzillo R, Di Somma C, Quarantelli M, Ventrella G, Gasperi M, Prinster A, Vacca G, Pivonello C, Orefice G, Colao A, et al. (2011). Insulin-like growth factor (IGF)-I and IGF-binding protein-3 serum levels in relapsing-remitting and secondary progressive multiple sclerosis patients. Eur. J. Neurol 18, 1402–1406. [DOI] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. (2007). Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381. [DOI] [PubMed] [Google Scholar]
- Lawrance IC, Maxwell L, and Doe W. (2001). Inflammation location, but not type, determines the increase in TGF-β1 and IGF-1 expression and collagen deposition in IBD intestine. Inflamm. Bowel Dis. 7, 16–26. [DOI] [PubMed] [Google Scholar]
- Le Roith D, Bondy C, Yakar S, Liu JL, and Butler A. (2001). The somatomedin hypothesis: 2001. Endocr. Rev 22, 53–74. [DOI] [PubMed] [Google Scholar]
- Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, and Boothby M. (2010). Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 Cell Subsets via distinct signaling pathways. Immunity 32, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, and Weaver CT. (2009). Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ, and Lin JX (2000). Cytokine receptor signaling pathways. J. Allerg. Clin. Immunol 105, 877–888. [DOI] [PubMed] [Google Scholar]
- LeRoith D, Werner H, Beitner-Johnson D, and Roberts CT, Jr (1995). Molecular and Cellular aspects of the insulin-like growth factor I receptor. Endocr. Rev 16, 143–163. [DOI] [PubMed] [Google Scholar]
- Li C, Yuan J, Zhu Y-F, Yang X-J, Wang Q, Xu J, He S-T, and Zhang J-A (2016a). Imbalance of Th17/Treg in different subtypes of autoimmune thyroid diseases. Cell. Physiol. Biochem 40, 245–252. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P. (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. (2011). Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med 17, 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yao DL, Bondy CA, Brenner M, Hudson LD, Zhou J, and Webster HD (1994). Astrocytes express insulin-like growth factor-I (IGF-I) and its binding protein, IGFBP-2, during demyelination induced by experimental autoimmune encephalomyelitis. Mol. Cell. Neurosci 5, 418–430. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Racke AE, Bittner P, Cross AH, Carlino JA, and Racke MK (1998). Regulation of experimental autoimmune encephalomyelitis with insulin-like growth factor (IGF-1) and IGF-1/IGF-binding protein-3 complex (IGF-1/IGFBP3). J. Clin. Invest 101, 1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, and Weaver CT (2006). Transforming growth factor-β induces development of the TH17 lineage. Nature 441, 231–234. [DOI] [PubMed] [Google Scholar]
- Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, and Weaver CT (2007). Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3– precursor cells in the absence of interleukin 10. Nat Immunol 8, 931–941. [DOI] [PubMed] [Google Scholar]
- McInnes IB, and Schett G. (2007). Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol 7, 429–442. [DOI] [PubMed] [Google Scholar]
- Merchav S, Tatarsky I, and Hochberg Z. (1988). Enhancement of human granulopoiesis in vitro by biosynthetic insulin-like growth factor I/somatomedin C and human growth hormone. J. Clin. Invest 81, 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux H, Martens H, Jaïdane H, Halouani A, Hober D, and Geenen V. (2015). How does thymus infection by coxsackievirus contribute to the pathogenesis of Type 1 diabetes? Front. Immunol 6, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa I, Nakayamada S, Nakano K, Yamagata K, Sakata K, Yamaoka K, and Tanaka Y. (2017). Induction of regulatory t cells and its regulation with insulin-like growth factor/insulin-like growth factor binding protein-4 by human mesenchymal stem cells. J. Immunol 199, 1616–1625. [DOI] [PubMed] [Google Scholar]
- Miyagawa S, Kobayashi M, Konishi N, Sato T, and Ueda K. (2000). Insulin and insulin-like growth factor I support the proliferation of erythroid progenitor cells in bone marrow through the sharing of receptors. Br. J. Haematol 109, 555–562. [DOI] [PubMed] [Google Scholar]
- Mohan S, and Baylink DJ (2002). IGF-binding proteins are multifunctional and act via IGF-dependent and -independent mechanisms. J. Endocrinol 175, 19–31. [DOI] [PubMed] [Google Scholar]
- Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, et al. (2011). Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 35, 972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba T, Watanabe M, Inoue N, and Iwatani Y. (2009). Increases of the Th1/Th2 cell ratio in severe Hashimoto’s disease and in the proportion of Th17 cells in intractable Graves’ disease. Thyroid 19, 495–501. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Shriver LP, Maresz K, Pedras-Vasconcelos J, Verthelyi D, and Dittel BN (2007). GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J. Immunol 178, 39–48. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Han R, Horst N, Cruikshank WW, and Smith TJ (2003). Immunoglobulin activation of T cell chemoattractant expression in fibroblasts from patients with Graves’ disease is mediated through the insulin-like growth factor I receptor pathway. J. Immunol 170, 6348–6354. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Tsui S, Horst N, Cruikshank WW, and Smith TJ (2004). Synovial fibroblasts from patients with rheumatoid arthritis, like fibroblasts from Graves’ disease, express high levels of IL-16 when treated with Igs against insulin-like growth factor-1 receptor. J. Immunol 173, 3564–3569. [DOI] [PubMed] [Google Scholar]
- Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, et al. (2012). Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med 18, 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Carter CA, Berman A, Cao L, Kelly RJ, Thomas A, Khozin S, Chavez AL, Bergagnini I, Scepura B, et al. (2014). Cixutumumab for patients with recurrent or refractory advanced thymic epithelial tumours: a multicentre, open-label, phase 2 trial. Lancet Oncol. 15, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DJ, and Miyamoto S. (2015). Hexokinase II integrates energy metabolism and cellular protection: AKTing on mitochondria and TORCing to autophagy. Cell Death Differ. 22, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, and Colonna M. (2016). Immune modules shared by innate lymphoid cells and T cells. J. Allerg. Clin. Immunol 138, 1243–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim D-H, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, and Sabatini DM (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol 14, 1296–1302. [DOI] [PubMed] [Google Scholar]
- Schiering C, Krausgruber T, Chomka A, Fröhlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, et al. (2014). The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillaci R, Brocardo MG, and Roldán A. (1995). Co-operative effect between insulin-like growth factor-1 and interleukin-2 on DNA synthesis and interleukin-2 receptor-alpha chain expression in human lymphocytes. Immunol. Cell Biol 73, 340–345. [DOI] [PubMed] [Google Scholar]
- Schillaci R, Brocardo MG, Galeano A, and Roldán A. (1998). Downregulation of insulin-like growth factor-1 receptor (IGF-1R) expression in human T lymphocyte activation. Cell. Immunol 183, 157–161. [DOI] [PubMed] [Google Scholar]
- Schillaci R, Ribaudo CM, Rondinone CM, and Roldán A. (1994). Role of insulin-like growth factor-1 on the kinetics of human lymphocytes stimulation in serum-free culture medium. I Immunol. Cell Biol 72, 300. [DOI] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segretin MIAE, Galeano A, Rold aacute n A, and Schillaci R. (2003). Insulin-like growth factor-1 receptor regulation in activated human T lymphocytes. Horm. Res 59, 276–280. [DOI] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, and Chi H. (2011). HIF1-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med 208, 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle K, Soos MA, Field CE, and Navé BT (1994). Hybrid and Atypical insulin/lnsulin-like growth factor I receptors. Horm. Res. Paediatr 41 (suppl 2), 56–65. [DOI] [PubMed] [Google Scholar]
- Smith TJ (2010). Insulin-like growth factor-I regulation of immune function: A potential therapeutic target in autoimmune diseases? Pharmacol. Rev 62, 199–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spath S, Komuczki J, Hermann M, Pelczar P, Mair F, Schreiner B, and Becher B. (2017). Dysregulation of the cytokine GM-CSF induces spontaneous phagocyte invasion and immunopathology in the central nervous system. Immunity 46, 245–260. [DOI] [PubMed] [Google Scholar]
- Stephens M. (2017). False discovery rates: a new deal. Biostatistics 18, 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, and Akira S. (1997). Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94, 3801–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucl. Acids Res 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno A, Ghosh A, Hung D, Li J, and Jijon H. (2015). Th17 plasticity and its changes associated with inflammatory bowel disease. World J. Gastroenterol 21, 12283–12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J, Fleskens V, Tiemessen MM, Mokry M, van Boxtel R, Meerding J, Pals CEGM, Kurek D, Baert MRM, Delemarre EM, et al. (2013). Canonical Wnt signaling negatively modulates regulatory T cell function. Immunity 39, 298–310. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, and Stockinger B. (2006). TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189. [DOI] [PubMed] [Google Scholar]
- Vogel TP, Milner JD, and Cooper MA (2015). The ying and yang of STAT3 in human disease. J. Clin. Immunol 35, 615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LSK, and Herrath, von M. (2016). CD4 T cell differentiation in type 1 diabetes. Clin. Exp. Immunol 183, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brown J, Gu Z, Garcia CA, Liang R, Alard P, Beurel E, Jope RS, Greenway T, and Martin M. (2011). Convergence of the mammalian target of rapamycin complex 1- and glycogen synthase kinase 3-β-signaling pathways regulates the innate inflammatory response. J. Immunol 186, 5217–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, and Hatton RD (2009). Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nat. Rev. Immunol 9, 883–889. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh T-Y, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, and Darnell JE (1995). Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250. [DOI] [PubMed] [Google Scholar]
- Yang Y, Niu J, and Guo L. (2002). The effects of antisense insulin-like growth factor-I receptor oligonucleotide on human cord blood lymphocytes. J. Mol. Endocrinol 28, 207–212. [DOI] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. (2007). Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, DiToro D, Hansen K, Barnett B, and Crotty S. (2010). Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol 185, 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MMW, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. (2008). TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, Naito AT, Nishi J-I, Ueno H, Umezawa A, et al. (2008). IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature 454, 345–349. [DOI] [PubMed] [Google Scholar]
- Zong CS (2000). Mechanism of STAT3 Activation by Insulin-like growth factor I receptor. J. Biol. Chem 275, 15099–15105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.