Abstract
The extracellular matrix (ECM) is a complex meshwork of insoluble fibrillar proteins and signaling factors interacting together to provide architectural and instructional cues to the surrounding cells. Alterations in ECM organization or composition and excessive ECM deposition have been observed in diseases such as fibrosis, cardiovascular diseases, and cancer. We provide here optimized protocols to solubilize ECM proteins from normal or tumor tissues, digest the proteins into peptides, analyze ECM peptides by mass spectrometry, and interpret the mass spectrometric data. In addition, we present here two novel R-script-based web tools allowing rapid annotation and relative quantification of ECM proteins, peptides, and intensity/abundance in mass spectrometric data output files. We illustrate this protocol with ECMs obtained from two pairs of tissues, which differ in ECM content and cellularity: triple-negative breast cancer and adjacent mammary tissue, and omental metastasis from high-grade serous ovarian cancer and normal omentum. The complete proteomics data set generated in this study has been deposited to the public repository ProteomeXchange with the data set identifier: PXD005554.
Keywords: extracellular matrix, matrisome, collagens, microenvironment, hydroxylation, mass-spectrometry-based proteomics
Graphical Abstract
■ INTRODUCTION
The extracellular matrix (ECM), a complex and dynamic meshwork of cross-linked proteins, is a fundamental component of multicellular organisms.1 The ECM provides architectural, mechanical, and biochemical signals interpreted by cell-surface receptors and orchestrates cellular processes such as adhesion, migration, proliferation, survival, and differentiation.2,3 Alterations in the composition and organization of the ECM cause or accompany the development of diseases including fibroses, cardiovascular diseases, and cancers.4−6
Because of its complexity and insolubility, the ECM has been difficult to analyze biochemically, and, until recently, it was not possible to obtain a detailed understanding of its composition.7 We have previously developed a proteomic pipeline to characterize the composition of in vivo ECMs and have shown that any given tissue or tumor includes well over 100 ECM and ECM-associated proteins and that characteristic differences can be detected between different tissues or tumors.8−10 One critical step in this proteomic approach was the development of a method to enrich ECM proteins from tissues and tumors. To do so, we and others devised decellularization methods that deplete intracellular proteins and permit the enrichment of ECM proteins.8,11−20 A second critical step of the approach was the identification and annotation of the ECM components, or “matrisome”, in large proteomic data sets. To this end, we devised a computational approach and defined the in silico matrisome as the ensemble of genes encoding core ECM proteins (ECM glycoproteins, collagens, and proteoglycans) and ECM-associated proteins (these are proteins structurally resembling ECM proteins, ECM remodeling enzymes, and secreted factors including growth factors).8,21
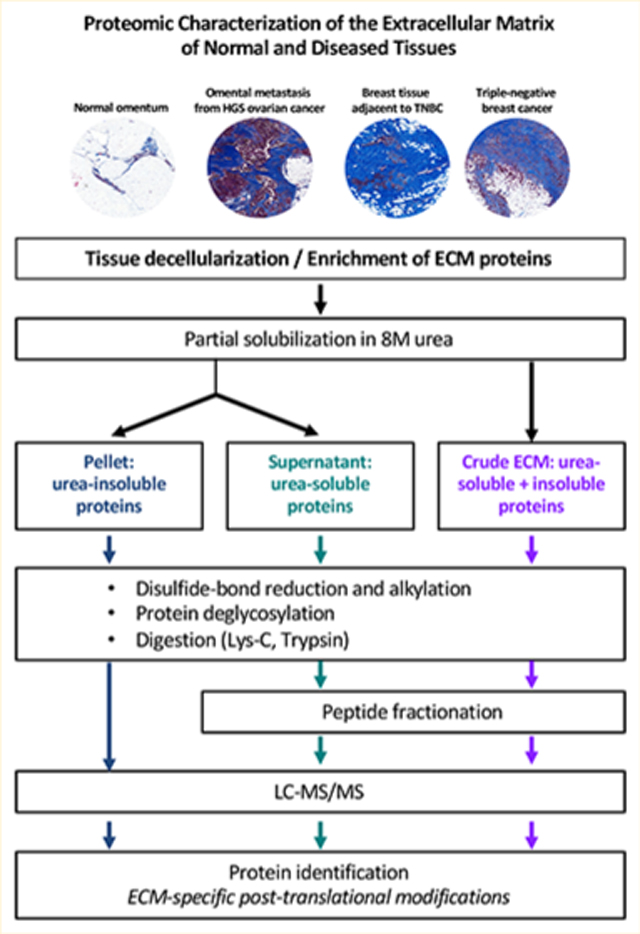
Because the details of the methods applied in the various analyses of ECM proteins using mass-spectrometry-based proteomics in our laboratories and others vary somewhat, we have now explored the impact of variations in the protocols. We provide here updated protocols for (1) digestion of ECM proteins into peptides, (2) analysis of ECM peptides by mass spectrometry, and (3) interpretation of mass spectrometric data. We illustrate the protocols with results obtained on the ECM from two sets of tissues that differ significantly in their ECM content and cellularity: human triple-negative breast cancer samples and adjacent breast tissue, and omental metastases from high-grade serous ovarian cancer (HGSOC) and normal omentum (Figure 1A). The optimized protocols presented here make the comprehensive characterization of the composition of the ECM accessible to a broad range of scientists and should help enhance the identification of disease biomarkers and novel therapeutic targets.
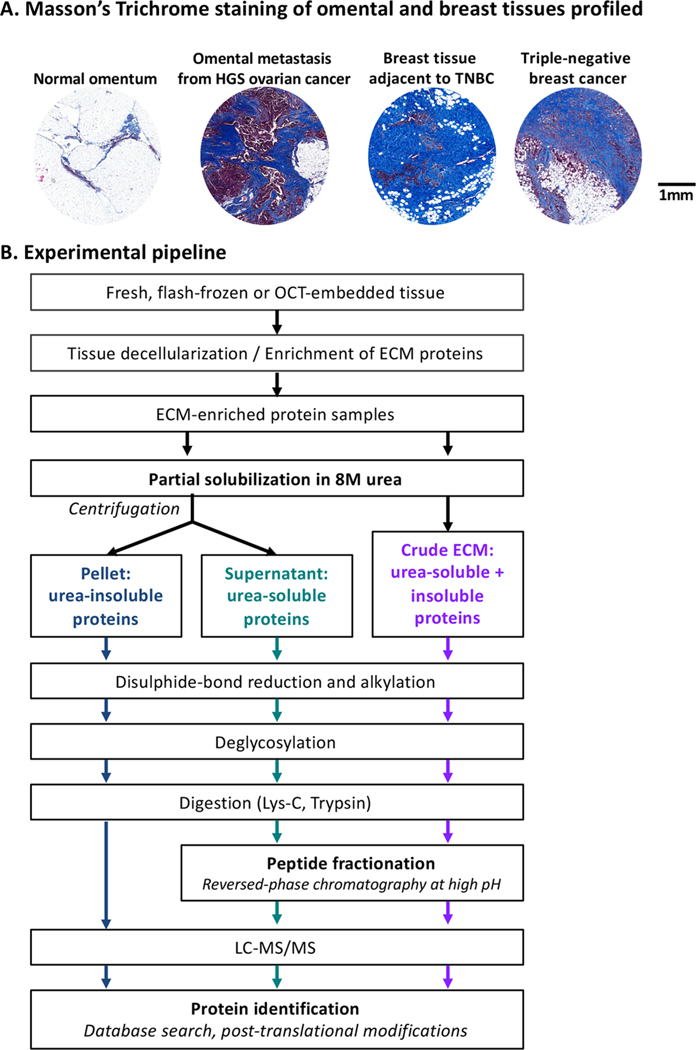
Figure 1.
Experimental pipeline. (A) Masson’s trichrome staining (blue) highlight fibrillar collagen content of the four tissues profiled in this study. (B) Experimental pipeline. A description of the samples analyzed in this study is presented in Supplementary Table S1A.
■ MATERIALS AND METHODS
Samples
Patient samples were kindly donated by women with high-grade serous ovarian cancer (HGSOC; n = 2) or triple-negative breast cancer (TNBC; n = 2) undergoing surgery at Barts Health NHS Trust between 2010 and 2014. Tissues deemed by a pathologist to be surplus to diagnostic and therapeutic requirements were collected together with associated clinical data under the terms of the Barts Gynae Tissue Bank (HTA license number 12199. REC no: 10/H0304/14) and Barts Breast Cancer Tissue Bank (HTA license number 12199. REC no: 15/EE/0192). In both cases TNBC and HGSOC samples are described as containing tumor cells and desmoplasia, while normal or adjacent samples are described as having no obvious tumor cell infiltration or desmoplasia (Supplementary Table S1A). Masson’s trichrome staining was performed according to standard procedures to evaluate the ECM content and cellularity of all four tissue types.
Enrichment for ECM Components
ECM proteins were enriched from frozen whole tissue or tumor sections (20 × 30 μm sections, ∼40−50 mg of tissue), as previously described.15 In brief, tissue sections were resuspended in the cytosolic (“C”) buffer of the CNMCS compartmental protein extraction kit (Millipore) and disrupted by vortexing. The samples were then incubated in a series of buffers to remove sequentially nuclear proteins (buffer “N”), membrane proteins (buffer “M”), and cytoskeletal proteins (buffer “CS”) according to the manufacturer’s instructions and our previous protocol.15 The remaining insoluble pellet is enriched for ECM proteins. Note that the subsequent steps can also be conducted on ECM-enriched samples prepared according to other methods.11−20
ECM Protein Solubilization
ECM-enriched samples were solubilized in 8 M urea, 20 mM HEPES containing Na3VO4 (100 mM), NaF (0.5M), β-glycerol phosphate (1 M), and Na2H2P2O7 (0.25 M). Samples were vortexed for 30 s and held on ice prior to sonication at 50% intensity, 3 times for 15 s, on ice. These resuspended and partially solubilized ECM-enriched samples were either directly digested into peptides as described below (“crude” ECM samples) or were centrifuged at 20 000g for 10 min at 5 °C, and the supernatant was recovered to protein low-bind tubes. This fraction will further be referred to as the urea-soluble fraction. Protein concentration of the urea-soluble fraction was estimated using BCA assay, and 80 μg of protein was digested into peptides as described below. The postcentrifugation urea-insoluble pellet was resuspended in 2 M urea, and proteins were digested as described below.
ECM Protein Digestion into Peptides
Disulfide bonds were reduced by adding 5 mM dithiothreitol to the samples for a 1 h incubation under agitation at room temperature. Free cysteines were alkylated by adding iodoacetamide at a final concentration of 8.3 mM to the samples for a 1 h incubation under agitation at room temperature in the dark. After diluting to 2 M urea, 20 mM HEPES, samples were deglycosylated with 1500 units of PNGaseF (New England BioLabs, Ipswich, MA) by incubation at 37 °C for 2 h under agitation. Samples were pre-digested with 1.6 μg of Lys-C (Pierce) and incubated at 37 °C for 2 h under agitation and digested with bead-immobilized trypsin (40 μL of beads per 250 μg of protein; Pierce) for 16 h at 37 °C under agitation. Samples were acidified with trifluoroacetic acid (TFA, 1% v/v), centrifuged at 2000g for 5 min at 5 °C. Supernatants were transferred to clean microcentrifuge tubes on ice. Glygen TopTips were equilibrated with 100% LC−MS-grade acetonitrile (ACN) and then washed with 99% H2O (+1% ACN, 0.1% TFA) prior to loading the peptide samples. Samples were washed with 99% H2O (+1% ACN, 0.1% TFA), and desalted peptides were eluted with 70/30 ACN/H2O + 0.1% TFA. Desalted peptides were dried and stored at −20 °C.
Liquid Chromatography−Tandem Mass Spectrometry (LC−MS/MS)
Equal volumes of each sample, corresponding to ∼100 ng of material, were initially analyzed via LC−MS/MS, and the total numbers of peptides identified and the sum of the intensities of the precursor ions were used as a normalization metric to determine equivalent peptide amounts. Peptides were separated by reversed-phase HPLC using an EASY-nLC1000 (Thermo Scientific) using a pre-column (made in house, 6 cm of 10 μm C18) and a self-pack 5 μm tip analytical column (12 cm of 5 μm C18, New Objective) over a 140 min gradient before nanoelectrospray using a QExactive mass spectrometer (Thermo). Solvent A was 0.1% formic acid, and solvent B was 80% MeCN/0.1% formic acid. The gradient conditions were 0−10% B (0−5 min), 10−30% B (5−105 min), 30−40% B (105−119 min), 40−60% B (119−124 min), 60−100% B (124−126 min), 100% B (126−136 min), 100−0% B (136−138 min), and 0% B (138−140 min). The mass spectrometer was operated in a data-dependent mode. The parameters for the full MS scan were: resolution of 70 000 across 350−2000 m/z, AGC 3e6, and maximum IT 50 ms. The full MS scan was followed by MS/MS for the top 10 precursor ions in each cycle with a normal collision energy of 28 and dynamic exclusion of 30 s.
Off-Line Fractionation of Peptides by Reversed-Phase Chromatography at High pH
Equivalent peptide amounts were fractionated via high-pH reversed-phase HPLC. Peptides were resuspended in 50 uL of buffer A (10 mM NH4HCO3, pH8) and separated on a 4.6 mm × 250 mm 300Extend-C18, 5 μm column (Agilent) using an 80 min gradient from 1% to 100% buffer B (90% MeCN, 10 mM NH4HCO3, pH8) at a flow rate of 1 mL/min. Fractions were collected over 75 min at 1 min intervals beginning at 5 min after the start of the gradient. The fractions were concatenated into 15 fractions non-contiguously (1 + 16 + 31 + 46 + 61, 2 + 17 + 32 + 47 + 62, etc.).22 The fractions were brought to near dryness in a speed-vac (Thermo Scientific). Each fraction was resuspended in 80 μL of 0.1% formic acid, and 4 μL was analyzed via LC−MS/MS, as described above using the same 140 min gradient.
Protein Identification
Raw mass spectral data files (.raw) were searched using Proteome Discoverer (Thermo Scientific) and Mascot version 2.4.1 (Matrix Science) using the SwissProt Homo sapiens database containing 20 199 entries. Mascot search parameters were: 10 ppm mass tolerance for precursor ions; 15 mmu for fragment-ion mass tolerance; two missed cleavages of trypsin; fixed modification was carbamidomethylation of cysteine; and variable modifications were oxidized methionine, deamidation of asparagine, pyro-glutamic acid modification at N-terminal glutamine, and hydroxylation of lysine and proline (see Results section). Only peptides with a Mascot score ≥25 and an isolation interference ≤30 were included in the data analysis. The average false discovery rate was 0.0074 (ranging from 0.0032 to 0.0144). For each protein, total precursor ion intensity (further referred to as “peptide intensity”) was calculated as the sum of MS1-precursor-ion-peak areas for all identified peptides of the given protein. Confidently identified proteins were further annotated as being part of the extracellular matrix as previously defined8,21 and using the novel tool Matrisome Annotator presented in this publication (see Results section).
The raw mass spectrometry data have been deposited to the ProteomeXchange23 via the PRIDE partner repository24 with the data set identifier: PXD005554.
■ RESULTS
Optimization of Methods To Solubilize and Digest ECM Proteins
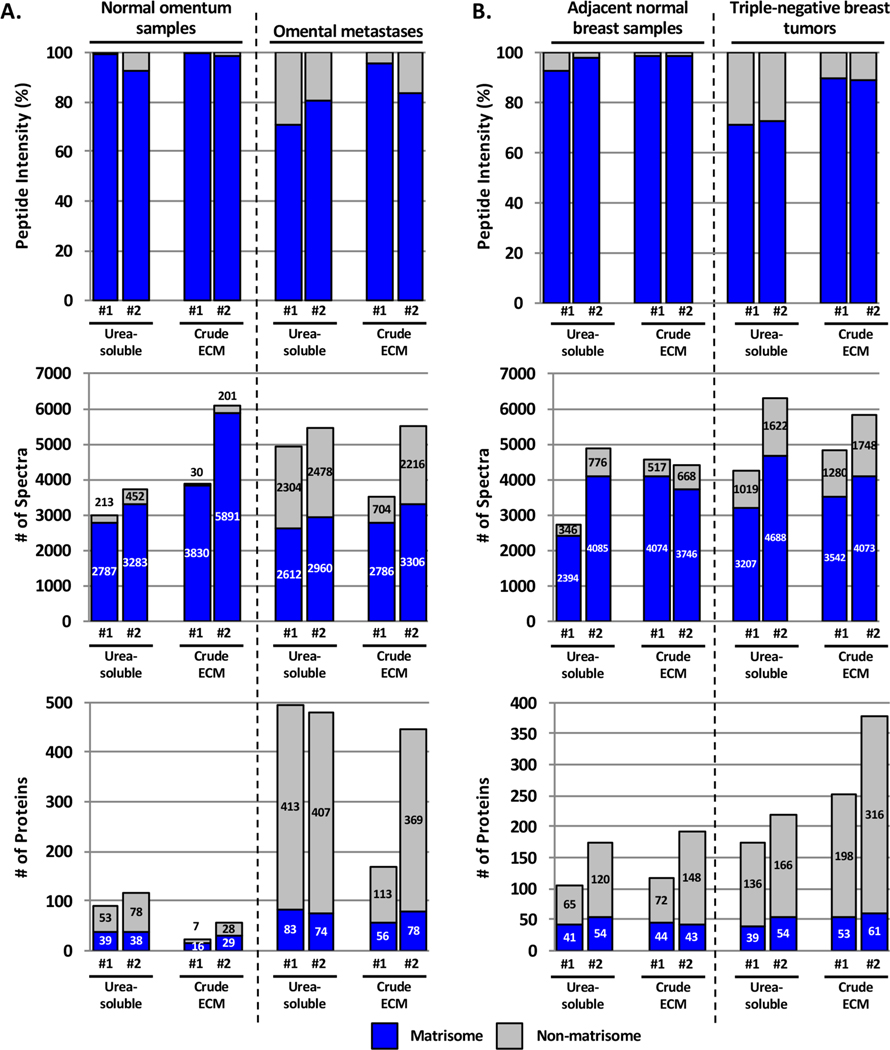
We initially devised a method that consisted of enrichment of tissue samples for ECM proteins, partial solubilization in 8 M urea, and digestion of this “crude” ECM extract into peptides. Using this protocol, the most abundant proteins identified were fibrillar collagens.8 We hypothesized that the abundance of fibrillar collagens could prevent the identification of other ECM or ECM-associated proteins present in lower abundance. To test this, we compared the original method with a method consisting of digesting only the urea-soluble fraction of ECM-enriched samples (Figure 1B). We observed that for the four tissues analyzed (breast cancer and adjacent breast tissue samples and omentum and ovarian-cancer-derived omental metastasis samples; n = 2 for each sample type) the abundance of ECM peptides (Figures 2A,B, upper panels) and the numbers of identified spectra (Figures 2A,B, middle panels) were somewhat greater in most of the crude ECM preparations as compared with the corresponding urea-soluble extracts (Supplementary Table S1B,C). However, the numbers of ECM proteins confidently identified were mostly similar (Figures 2A,B, lower panels, and Supplementary Table S1B,C), although urea-soluble preparations of normal and diseased omentum generally gave a higher number of ECM proteins compared with crude ECM preparations. This might be explained by the low content of fibrillar ECM in this tissue (Figure 1A).
Figure 2.
Comparison of matrisome and non-matrisome proteins identified in urea-soluble versus crude ECM samples. Bar charts represent the distribution of matrisome (blue) versus non-matrisome (gray) proteins in terms of peptide abundance (upper panels), number of spectra (middle panels), and number of proteins (lower panels) in urea-soluble or crude ECM extracts from normal omental tissue and omental metastases from ovarian tumors (A) and normal breast tissues and triple-negative breast tumors (B). Two independent samples were compared for each tissue and methodology. Related to Supplementary Table S1B,C.
We next sought to analyze the composition of the urea-insoluble protein fraction (Supplementary Figure S1A and Supplementary Table S1B) and observed that only four proteins were detected solely in the pellet and not in either urea-soluble or crude ECM preparations (namely, AMBP and LMAN1L, which is, in fact, a transmembrane protein in TNBC-adjacent breast tissue and COL6A5 and MFAP2 in TNBC samples) (Supplementary Figure S1B and Supplementary Table S1B). However, the total number of matrisome proteins detected in the pellet was less than either the crude or urea-soluble preparations. It is possible that with more extensive analysis, including peptide fractionation (see below), these proteins would also be detected in the urea-soluble or crude ECM preparations. Therefore, in general, both crude and urea-soluble extractions result in similar numbers of ECM proteins detected, although one method may be superior to the other depending on the tissue and, in particular, depending on the collagen content of the tissue.
Optimization of Methods To Fractionate ECM-Protein-Derived Peptides
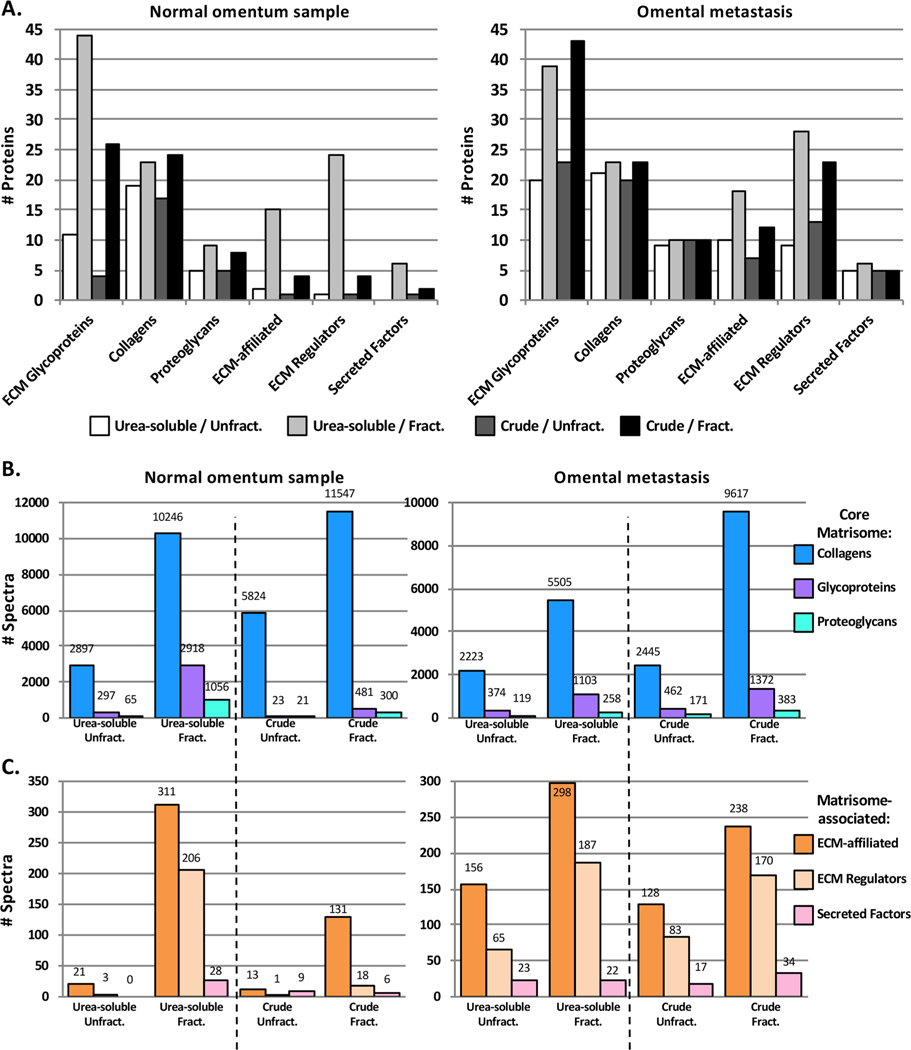
We have previously reported that off-line fractionation of peptide mixtures by off-gel electrophoresis increased by a factor of 3 the number of matrisome peptides confidently identified, which resulted in the identification of twice as many core ECM components and over 10 times the number of matrisome-associated proteins.8 Off-gel electrophoresis is a lengthy process and is now being replaced in proteomic pipelines by fractionation using high-pH liquid chromatography (also termed basic-reversed-phase or bRP-LC), orthogonal to the LC separation in-line with the tandem mass spectrometry. We sought to determine whether fractionation by bRP-LC was beneficial to identify otherwise undetected ECM proteins from both urea-soluble and crude ECM preparations. Independent of the tissue type or the method employed to digest ECM-enriched samples into peptides, fractionation prior to LC−MS/MS resulted in the identification of a greater number of spectra and proteins (Figure 3 and Supplementary Table S1D). In the case of omental samples, the urea-soluble preparation, when coupled to bRP-LC fractionation, consistently resulted in higher numbers of ECM proteins detected when compared with fractionated crude preparations (Figure 3A). This is presumably a consequence of the reduced amount of collagens in the urea-soluble fraction (Supplementary Table S1D). Furthermore, peptide fractionation was particularly beneficial to identify lower-abundance matrisome-associated proteins such as ECM-affiliated proteins and ECM regulators (including ECM cross-linking enzymes or proteases) (Figure 3B,C and Supplementary Table S1D).
Figure 3.
Peptide fractionation increases the number of ECM and ECM-associated proteins identified by mass spectrometry in both urea-soluble and crude ECM samples. (A) Bar charts represent the number of matrisome proteins identified proteins in urea-soluble or ECM samples from normal omentum (left) or omental metastasis from ovarian cancer (right) fractionated or not by basic-reverse phase liquid chromatography prior to LC−MS/MS. (Bar charts represent the number of identified spectra corresponding to core matrisome (B) or matrisome-associated (C) proteins in urea-soluble or ECM samples from normal omentum (left) or omental metastasis from ovarian cancer (right) fractionated or not by basic-reverse phase liquid chromatography prior to LC−MS/MS. Related to Supplementary Table S1C,D.
Importance of Allowing for Proline Hydoxylations for in Silico Protein Identification
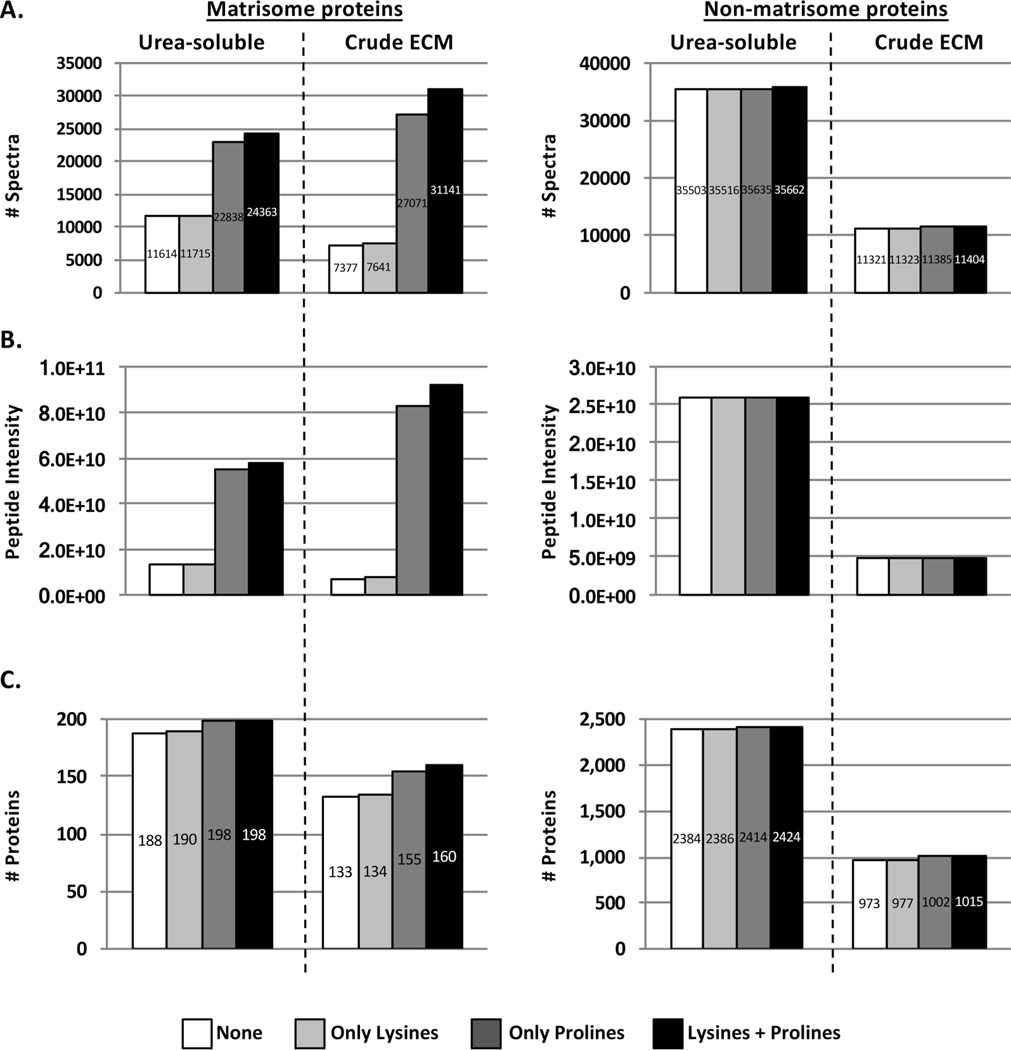
Peptide and subsequent protein identification from tandem mass spectrometry data rely on the comparison of experimental spectral mass values with databases containing theoretical spectral data. It is important that search algorithms include post-translational modifications (PTMs) that alter the mass of amino acids. Hydroxylated prolines and lysines are obligate PTMs of triple-helical collagens (prolines represent 17% of amino acids in collagen I) and other ECM proteins.25−27 However, these PTMs are not routinely included when interrogating databases for protein identification because they increase the search time. To examine the significance of including the dynamic modifications of lysine and proline hydoxylation, we selected a data set and conducted a database search in four different ways: without allowing for lysine or proline hydroxylation, allowing only lysine hydroxylation, allowing only proline hydroxylation, or allowing both lysine and proline hydroxylation. Allowing for proline hydroxylations (and to a lesser extent for lysine hydroxylations) led to a significant increase in the number of identified spectra of matrisome proteins by a factor of 2 (Figure 4A, left panels), as also previously reported by Barallobre-Barreiro and collaborators,28 and thus to a more accurate estimation of the abundance of matrisome proteins (Figure 4B, left panel). Allowing these PTMs also allowed the identification of several additional ECM proteins (Figure 4C, left panel, and Supplementary Table S1E,F). Importantly, allowing for these two PTMs was equally beneficial to the analysis of urea-soluble and crude ECM extracts. Of note, these two PTMs did not significantly increase the number of spectra or unique peptides of non-matrisome proteins (Figure 4A−C, right panels). Therefore, allowing for proline and lysine hydroxlations is beneficial specifically for the detection of ECM proteins, and we recommend they be included.
Figure 4.
Importance of allowing hydroxylation of prolines when searching databases to identify and quantify ECM proteins. Bar charts represent the number of spectra (A), peptide abundance (B), and number of proteins (C) for matrisome (left) and non-matrisome (right) proteins identified by searching the data allowing or not for prolines or lysines hydroxylations as dynamic modifications. Post-fractionation data to conduct this comparative analysis were acquired on urea-soluble or crude ECM extracts from omental metastasis from high-grade-serous ovarian cancer. Related to Supplementary Table S1E,F.
Scripts To Annotate Matrisome Proteins and Calculate Mass-Spectrometric Metrics
To facilitate the annotations of matrisome proteins in large data sets, we developed a script called “Matrisome Annotator”. Providing that a data set contains Entrez or HUGO gene symbols for each entry, the script will return an output file in which each entry will be annotated as being part of the matrisome or not and will be tagged with matrisome division (core matrisome vs matrisome-associated) and category (ECM glycoproteins, collagens, proteoglycans, ECM-affiliated proteins, ECM regulators, or secreted factors). “Matrisome Annotator” can be used to annotate not only proteomic data but also any kind of list of genes/proteins. We also developed a second script, called “Matrisome Analyzer”, that calculates the proportion of ECM content in terms of number of spectra, number of unique peptides, number of proteins and peptide intensity (i.e., protein abundance) in proteomics data set. This script allows rapid evaluation of the abundance of matrisome versus non-matrisome proteins in any given data set input as a delimited text file and exports the calculation in tables and graphs. Both scripts are available as webtools and to download under the Analytical Tools section of the Matrisome Project Web site (http://matrisome.org/).
■ DISCUSSION
We provide here a detailed description of mass-spectrometry-based methods allowing in-depth and comprehensive characterization of the composition of ECM of normal and diseased tissues. The major advantage of these methods is that each step, from the solubilization of ECM proteins to the interpretation of mass spectrometric data, has been optimized to take into account biochemical characteristics of ECM and ECM-associated proteins. For example, we recommend allowing for proline and lysine hydroxylation when searching databases, as this significantly increases ECM protein detection and identification. The preferred method to solubilize ECM-enriched protein samples for digestion to peptides depends on the tissue and its ECM content, and, in some cases, restricting the analysis to urea-soluble ECM proteins permits detection of additional proteins. This is because significant proportions of the collagens remain insoluble. Few proteins are missed by ignoring the urea-insoluble fraction, although, of course, the relative amounts of proteins detected are altered, which compromises semi-quantitative conclusions. Furthermore, the inclusion of a pre-fractionation step by bRP-LC chromatography provides an alternative means, allowing the detection of additional proteins. It is worth noting that high-pH reversed-phase peptide fractionation can be achieved using commercially available spin columns requiring only a bench centrifuge. In addition, any number of fractions can be generated depending on the complexity of the samples and the desired coverage.
Although not discussed in the manuscript, these methods can be coupled to label-based quantitative proteomics,29 which allows quantification of the abundance of proteins and peptides in different samples and also allows multiplexing, thus reducing mass-spectrometer time.
■ CONCLUSIONS
Alterations in the composition and organization of the ECM cause or accompany the development of diseases such as fibrosis, cardiovascular diseases, musculo-skeletal diseases, and cancers.
The application of the proposed methods to profile the composition of the ECM of, for example, pairs of normal versus diseased tissues, treated versus untreated tissues, poorly versus highly metastatic tumors, or cell or organoid cultures can lead to the identification of novel ECM proteins that could serve as prognostic or diagnostic biomarkers or novel therapeutic targets.
Supplementary Material
Supplementary Figure S1: Composition of urea-insoluble protein samples. (PDF)
Supplementary Table S1: Complete mass spectrometry data set. (XLSX)
■ ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute − Tumor Microenvironment Network (U54 CA126515/CA163109), DoD BRCP Innovator Award (BC131410) to R.O.H., the Howard Hughes Medical Institute of which R.O.H. is investigator, and in part by the Cancer Center Support (Core) Grant P30-CA14051 from the National Cancer Institute. O.M.T.P. and F.R.B. were funded by the European Research Council (ERC322566) and Cancer Research UK (A16354). We thank Prof. Louise Jones and Dr. Sally Dreger (Center for Tumor Biology, Barts Cancer Institute) for providing triple-negative breast cancer tissues and Dr. Michelle Lockley (Center for Molecular Oncology, Barts Cancer Institute) for providing HGSOC samples used in the study, Dr. Sam Nichols (Center for Cancer & Inflammation, Barts Cancer Institute), and Karl Clauser (Proteomic Platform, Broad Institute) for helpful discussions and suggestions.
Footnotes
Notes
The authors declare no competing financial interest.
The complete proteomics data set generated in this study has been deposited to the public repository ProteomeXchange with the data set identifier: PXD005554.
■ ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.7b00191.
■ REFERENCES
- (1).Hynes RO; Yamada KM Extracellular Matrix Biology; Cold Spring Harbor Perspectives in Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2012. [Google Scholar]
- (2).Rozario T; DeSimone DW The extracellular matrix In development and morphogenesis: a dynamic view. Dev. Biol 2010, 341 (1), 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bonnans C; Chou J; Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol 2014, 15 (12), 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Pickup MW; Mouw JK; Weaver VM The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15 (12), 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cox TR; Erler JT Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis. Models & Mech 2011, 4 (2), 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bergmeier W; Hynes RO Extracellular Matrix Proteins in Hemostasis and Thrombosis. Cold Spring Harbor Perspect. Biol 2012, 4 (2), a005132−a005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wilson R. The extracellular matrix: an underexplored but important proteome. Expert Rev. Proteomics 2010, 7 (6), 803–806. [DOI] [PubMed] [Google Scholar]
- (8).Naba A; Clauser KR; Hoersch S; Liu H; Carr SA; Hynes RO The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics 2012, 11 (4), M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Naba A; Clauser KR; Lamar JM; Carr SA; Hynes RO Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife 2014, 3, e01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Naba A; Clauser KR; Whittaker CA; Carr SA; Tanabe KK; Hynes RO Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer 2014, 14 (1), 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Barallobre-Barreiro J; Didangelos A; Yin X; Domenech N;́ Mayr MA Sequential Extraction Methodology for Cardiac Extracellular Matrix Prior to Proteomics Analysis. In Heart Proteomics; Vivanco F, Ed.; Humana Press: Totowa, NJ, 2013; Vol. 1005, pp 215–223. [DOI] [PubMed] [Google Scholar]
- (12).Byron A; Humphries JD; Humphries MJ Defining the extracellular matrix using proteomics. Int. J. Exp. Pathol 2013, 94 (2), 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Crapo PM; Gilbert TW; Badylak SF An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32 (12), 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Krasny L; Paul A; Wai P; Howard BA; Natrajan RC; Huang PH Comparative proteomic assessment of matrisome enrichment methodologies. Biochem. J 2016, 473 (21), 3979–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Naba A; Clauser KR; Hynes RO Enrichment of extracellular matrix proteins from tissues and digestion into peptides for mass spectrometry analysis. J. Visualized Exp 2015, 101, e53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Schiller HB; Fernandez IE; Burgstaller G; Schaab C; Scheltema RA; Schwarzmayr T; Strom TM; Eickelberg O; Mann M. Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol. Syst. Biol 2015, 11 (7), 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sato N; Taniguchi T; Goda Y; Kosaka H; Higashino K; Sakai T; Katoh S; Yasui N; Sairyo K; Taniguchi H. Proteomic Analysis of Human Tendon and Ligament: Solubilization and Analysis of Insoluble Extracellular Matrix in Connective Tissues. J. Proteome Res 2016, 15 (12), 4709–4721. [DOI] [PubMed] [Google Scholar]
- (18).Randles MJ; Humphries MJ; Lennon R. Proteomic definitions of basement membrane composition in health and disease. Matrix Biol. 2017, 57–58, 12−28. [DOI] [PubMed] [Google Scholar]
- (19).de Castro Bras LE; Ramirez TA; DeLeon-Pennell KY; Chiao YA; Ma Y; Dai Q; Halade GV; Hakala K; Weintraub ST; Lindsey ML Texas 3-Step decellularization protocol: Looking at the cardiac extracellular matrix. J. Proteomics 2013, 86, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hellewell AL; Rosini S; Adams JC A Rapid, Scalable Method for the Isolation, Functional Study, and Analysis of Cell-derived Extracellular Matrix. J. Visualized Exp 2017, No. 119, e55051−e55051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Naba A; Clauser KR; Ding H; Whittaker CA; Carr SA; Hynes RO The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yang F; Shen Y; Camp DG; Smith RD High pH reversed-phase chromatography with fraction concatenation as an alternative to strong-cation exchange chromatography for two-dimensional proteomic analysis. Expert Rev. Proteomics 2012, 9 (2), 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Vizcaíno JA; Deutsch EW; Wang R; Csordas A; Reisinger F; Ríos D; Dianes JA; Sun Z; Farrah T; Bandeira N; et al. ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nat. Biotechnol 2014, 32 (3), 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Vizcaíno JA; Csordas A; del-Toro N; Dianes JA; Griss J; Lavidas I; Mayer G; Perez-Riverol Y; Reisinger F; Ternent T; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44 (D1), D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gjaltema RAF; Bank RA Molecular insights into prolyl and lysyl hydroxylation of fibrillar collagens in health and disease. Crit. Rev. Biochem. Mol. Biol 2017, 52, 74–95. [DOI] [PubMed] [Google Scholar]
- (26).Myllyharju J. Intracellular Post-Translational Modifications of Collagens. In Collagen; Brinckmann J, Notbohm H, P K, Eds.; Topics in Current Chemistry; Springer: Berlin, 2005; pp 115–147. [Google Scholar]
- (27).Yamauchi M; Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem. 2012, 52, 113–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Barallobre-Barreiro J; Oklu R; Lynch M; Fava M; Baig F; Yin X; Barwari T; Potier DN; Albadawi H; Jahangiri M; et al. Extracellular matrix remodelling in response to venous hypertension: proteomics of human varicose veins. Cardiovasc. Res 2016, 110 (3), 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Naba A; Clauser KR; Mani DR; Carr SA; Hynes RO Quantitative proteomic profiling of the extracellular matrix of pancreatic islets during the angiogenic switch and insulinoma progression. Sci. Rep 2017, 7, 40495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Composition of urea-insoluble protein samples. (PDF)
Supplementary Table S1: Complete mass spectrometry data set. (XLSX)