ABSTRACT
The SET1 family of enzymes are well known for their involvement in the histone 3 lysine 4 (H3K4) methylation, a conserved trait of euchromatin associated with transcriptional activation. These methyltransferases are distinct, and involved in various biological functions in the cell. Impairment in the function of SET1 family members leads to a number of abnormalities such as skeletal and neurological defects, leukaemogenesis and even lethality. Tremendous progress has been made in understanding the unique biological roles and the mechanism of SET1 enzymes in context with H3K4 methylation/canonical functions. However, in recent years, several studies have indicated the novel role of SET1 family proteins, other than H3K4 methylation, which are equally important for cellular functions. In this review, we focus on these non-canonical function of SET1 family members.
KEYWORDS: MLL, SETD1A, H3K4 histone methyltransferase, non-canonical functions, cell cycle, non-histone protein substrates
Introduction
Transcription is majorly regulated by two epigenetic modifications, namely DNA methylation and histone modifications [1–3]. Acetylation, phosphorylation, methylation and ubiquitylation are some of the important post-translational modifications in histones which regulate the chromatin functions [1,2]. Histone methylation majorly occurs on the side chains of lysines (Ks) and arginines (Rs) [1]. In general, methylation of histones is associated with both transcriptional repression and activation [4–6]. Till now, over 100 specific histone lysine methyltransferases (HKMTs) have been discovered and they bring about methylation of six lysines in histone H3 tail (K4, K9, K23, K27, K36 and K79) and one lysine in histone H4 tail (K20) [3,7,8]. SET1/MLL (mixed lineage leukemia) family of methyltransferases is conserved from yeast to mammals. They catalyse the mono-, di-, or tri-methylation of histone 3 at lysine 4 (H3K4) on the chromatin using their Suppressor of variegation 3–9, Enhancer of Zeste, Trithorax (SET) domain [9–11]. In humans, six H3K4 methyltransferses (HMT) – MLL1 (MLL/KMT2A), MLL2 (KMT2B), MLL3 (KMT2C), MLL4 (KMT2D), SETD1A (KMT2F) and SETD1B (KMT2G) – are required, while in yeast, only one HMT – Set1/complex proteins associated with Set1 (COMPASS) – is capable of catalysing all the three states of methylation [10,12,13].
SET1 family members
The six SET1 family members characterized in mammalian cells, have distinct functions with unique biological roles [13,14]. Based on phylogenetic analysis, human SET1 family can be divided into three pairs, with each pair related to a single Drosophila protein. MLL and MLL2 are closely associated with trithorax(trx) itself, while MLL3 and MLL4 are highly similar to trithorax-related (trr) protein. SETD1A and SETD1B are closest to Drosophila dSet1 protein, which in turn is related to Set1 of S. cerevisiae [15,16].
Mll1, located in the chromosome 11q23.3 encodes for MLL protein. It was first discovered for its involvement in chromosomal translocation observed in children and adults suffering from haematological malignancies including acute lymphoid leukaemia and acute myeloid leukaemia [17–19]. MLL regulates the homeobox (Hox) gene expression, which has been implicated in haematopoiesis and embryonic development [20–22]. The genomic architecture (exon/intron structure) of proteins encoded by Mll1 and Mll2 genes is comparable and most of the tissues in adult human ubiquitously express both these proteins. Mll2 gene, located on human chromosome 19q13.12, is amplified in some solid tumours [23,24]. The cytogenetic location of Mll3 gene is on human chromosome 7q36.1, a region usually deleted in myeloid leukaemia cases whereas Mll4 lies on chromosome 12q13.12 [25,26]. Further, loss-of-function mutations of MLL3 and MLL4 are associated with a wide spectrum of cancers implicating these SET1 family members as tumour suppressors [27–29]. SETD1A and SETD1B proteins are encoded by genes located on human chromosome 16p11.2 and 12q24.31 respectively. Despite being highly homologous proteins, SETD1A and SETD1B exhibit distinct subnuclear distributions, suggesting that each of these proteins is targeted to unique set of genomic sites, performing specific functions [30–32].
The commonality between these SET1 family members is an evolutionarily conserved SET domain, which catalyzes the transfer of methyl moiety from S-adenosylmethionine to the ε-amine on the lysine 4 of histone H3 [9,33]. Members of SET1 family exhibit moderate to weak enzymatic activity. However, their interaction with WDR5, RbBP5, Ash2L and Dpy30 (WRAD) subunits markedly enhances the methyltransferase activity of the SET domain [34–37]. In vitro studies show that in the presence of WRAD, MLL1/2 display mono- and di-methylation activity, MLL3/4 predominantly show mono-methylation activity while SETD1A/B can catalyse all the three states of methylation [37]. Further studies show that not only the interaction between SET domain and HKMT substrates is stabilized by RbBP5-Ash2L heterodimer, but it also enhances the H3K4 methyltransferase activity of SET1 family proteins [38,39]. However, MLL shows a weak interaction with RbBP5-Ash2L heterodimer, and it requires WDR5 as a bridging molecule which interacts with WDR5-interacting (Win) motif in MLL and VDV motif in RbBP5, further stimulating the methyltransferase activity of MLL [38,40,41]. In contrast, WDR5 is dispensable for the regulation of catalytic activity of other SET1 family proteins [38]. Recently, the cryo-electron microscopy (EM) structures of SET1 family protein complex revealed that the variation in the efficiency of methyltransferase activity of these family members is due to their distinct structural organizations of interfaces between WDR5, SET domain and RbBP5 subunits [42,43]. Furthermore, studies showed that the H2B ubiquitination mark on nucleosome, stimulates the catalytic efficiency of all SET1 family members except MLL3 [43,44].
Beyond the shared SET domain and core complex partners, members of this family display unique protein interactions. MLL interacts with tumour suppressors (Menin), cell cycle regulators (E2Fs and HCF), nuclear cyclophilin 33 (Cyp 33), histone deacetylase (HDAC1), histone acetylase (CBP/P300) and chromatin remodelling factors (INI1/SNF5) [45–51]. Menin upregulates the expression of MLL target genes such as Hox genes and its co-regulator (Hoxa9, Meis1), and CDK inhibitors (p27 and p18), which are necessary for MLL-mediated leukaemogenesis [52–55]. Further, MLL and MLL2 interact with Menin/LEDGF sub-complex, which in turn binds H3K36me2 modified chromatin and promote transcriptional activation [56,57]. MLL3 and MLL4 interact with multiple proteins including H3K27me3 demethylase UTX, PTIP, PA1 and regulate transcription of target genes [58–60]. NCOA6/ASC-2 is a unique subunit of both MLL3 and MLL4 in a complex named ASCOM, that acts as a p53 co-activator and thus implicated in tumour suppressor pathway [61,62]. SETD1A and SETD1B interact with unique proteins such as WDR82 and CXXC finger protein 1 (CFP1). WDR82 not only contributes to the stability of the SETD1A/B complexes but also binds to RNA polymerase II (Pol II) through serine 5 phosphorylated C terminal domain, essential for transcription initiation whereas CFP1 binds to unmethylated CpGs and guides deposition of H3K4me3 mark on chromatin, required for the expression of CpG island-associated genes [63–65].
Structure and function
SET1 family members are large (approximately 1707–5537 amino acid) proteins with a 130-amino-acid long C-terminal SET domain (Figure 1). This is followed by a post-SET domain which also participates in the HMT activity [9,33,66]. However, beyond the SET and post-SET domains, different SET1 family members display substantial differences in their protein domain structures. MLL1-4 methyltransferases contain varying numbers of plant homeodomain (PHD) fingers, that mediate interactions with methylated histones and several other proteins [67]. They also have FYRN/FYRC (phenylalanine and tyrosine-rich N-terminal/C-terminal) domain, which is found in many chromatin-associated transcription factors [68]. In addition, MLL and MLL2 proteins possess a series of AT hook motifs that mediate their binding to minor groove of AT-rich DNA region, a CXXC (Cysteine-rich region) domain that binds to unmethylated CG-rich DNA (CpG islands) and an atypical bromodomain, that recognizes the acetylated lysine residues in histone tail (Figure 1) [63,69,70]. The MLL3 and MLL4 proteins contain a unique DNA-binding motif, a high mobility group (HMG) box, associated with DNA-dependent functions [71,72]. MLL and MLL2 are different from the MLL3 and MLL4, as they possess specific recognition sequence, where these proteins are cleaved by threonine aspartase – Taspase 1. After cleavage, the N-terminal and C-terminal fragments self-associate by non-covalent intra molecular interaction and form a complex which confers protein stability and subnuclear localization [73–75]. SETD1A and SETD1B proteins of SET1 family have a RNA recognition motif (RRM) at the N-terminal and an N-SET domain adjacent to the SET domain, required for high levels of H3K4 trimethylation [76–78]. The variation in the domain architecture across the methyltransferases indicate the binding and functional diversity of the SET1 family.
Figure 1.
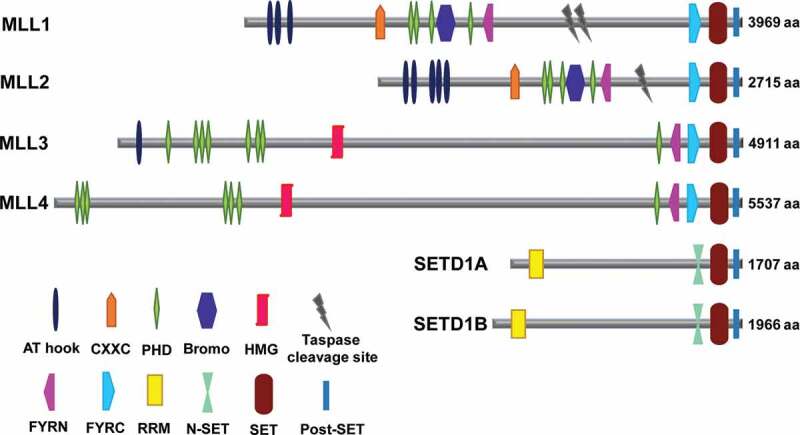
Mammalian SET1 family of methyltransferases. The figure depicts the structure of each family member with variable number of DNA binding motifs and protein-protein interaction domains. All the six methyltransferases contain a SET and post-SET domain at the C-terminal. The number of amino acids (aa) shown, represents the length of each protein. AT hook: AT-rich region, CXXC: Cysteine-rich region, PHD: Plant homeodomain, Bromo: Bromodomain, HMG: High mobility group box, FYRN/C: Phenylalanine and tyrosine-rich region N-terminal/C-terminal, RRM: RNA recognition motif, SET: Suppressor of variegation 3–9, enhancer of zeste and trithorax, N-SET: N-terminal of SET, Post-SET: C-terminal of SET
Members of SET1 family have been implicated as critical epigenetic regulators of development. The complex subunit of this family members, WDR5 interacts with long non-coding RNA (lncRNA) such as HOTTIP and NeST, and maintains H3K4 methylation mark on chromatin for active gene transcription [79–81]. Further, each of these proteins exhibit specialized function in mammalian development. Knockout studies in mouse revealed that SET1 family members display distinct phenotypes, even between highly related gene pairs, suggesting that functions of SET1 proteins are non-redundant [22,30,82–84]. The SET1 family members, MLL and MLL2 are vital for maintaining the pattern of Hox gene expression during embryonic development [22,82]. Mll1 knockout results in embryonic lethality in homozygous null mice, while heterozygous mice display retarded growth, abnormal haematopoiesis and anterio-posterior skeletal defects [22]. Further, deletion of the MLL SET domain in mice exhibit defects in body patterning, suggesting a role for H3K4 methylation in the epigenetic regulation during mammalian developmental process. However, the effects on embryogenesis and haematopoiesis was observed to a lesser extent, suggesting methylation-independent functions of MLL [85,86]. In addition to the Hox genes, MLL also regulates intracellular and extracellular regulators of Wnt signalling [87]. Even though, MLL and MLL2 are closely related proteins, Mll2 gene knockout display early embryonic lethality with widespread evidence of apoptosis [82]. Also, loss of MLL2 affects members of HoxB gene clusters, distinct from those regulated by MLL which primarily affects HoxA and HoxC [52,82,88]. Further, MLL2 knockout in mouse embryonic stem cells (mESCs) leads to rapid methylation of the CpG islands at Magoh2 gene promoter, implying that the MLL2 occupancy protects this promoter region from aberrant methylation and thus, maintains transcriptionally active promoter [89,90].
Regardless of the similarities between MLL3 and MLL4, the null phenotypes in mouse development are dramatically different. MLL3 null mice die at birth with no evident phenotype, while MLL4 knockout embryos die at E10.5 [83,84,91]. MLL3 and MLL4 play crucial role in normal differentiation of haematopoietic stem and progenitor cell during development [92,93]. Further, MLL4 show partial functional redundancy with MLL3 in adipogenesis and myogenesis [83,84]. Studies have shown that MLL3 and MLL4 are the major regulators of mono-methyltransferase activity at enhancers [38,66,84,94,95]. In addition, MLL3 and MLL4 acts as transcriptional coactivator and loss of MLL3/4 leads to disruption of the epigenetic landscape at enhancers, including loss of H3K4me1, H3K27 acetylation and RNA Pol II loading, resulting in reduced enhancer RNA (eRNA) synthesis [84,96,97]. Recently, in mESCs, it has been shown that the H3K4 methyltransferase activities of MLL3 and MLL4 are inessential for transcription of genes in the vicinity of enhancers. Thus, at enhancers, the main function of MLL3 and MLL4 is likely to be independent of H3K4 methylation [96,98].
Though SETD1A and SETD1B are highly similar proteins, they are discretely essential for mouse embryogenesis. Knockout of SETD1A in mouse embryo resulted in gastrulation failure, while SETD1B knockout embryos display severe growth retardation and die at E11.5 [30]. Further, SETD1A is crucial for embryonic stem cell proliferation [30]. Conditional knockout studies in mice revealed that ablation of SETD1A resulted in aberrant B cell and erythroid lineage differentiation while ablation of SETD1B disturbed the homoeostasis of haematopoietic stem and progenitor cells, suggesting that both SETD1A and SETD1B regulate adult haematopoiesis [99–101]. SETD1A maintains the bulk of genomic H3K4me3 in the cell while the in vivo methyltransferase activities of SETD1B are yet to be determined [30]. Recently it has been shown that the catalytic activity of SETD1A is dispensable for embryonic cell proliferation and self-renewal. However, embryonic stem cells are unable to undergo normal differentiation upon loss of SET domain of SETD1A, indicating the importance of H3K4 methylation in this process [102].
Non-canonical roles of SET1 family members
Despite conserved functions of SET1 in the methylation of histones, emerging studies have reported crucial methylation independent role of SET1 family. In vivo studies revealed that MLL can work through non-canonical pathways, evident by the fact that mice bearing homozygous SET-domain deletion survive embryogenesis with mild abnormalities, while MLL-null mice fail to survive past E10.5 [22,86]. Similarly, loss of MLL3 and MLL4, but not H3K4me1, led to significant depletion of enhancer Pol II occupancy with subsequent downregulation of transcription of target genes in mESC knockouts of these proteins [96]. Thus, SET1 family members are likely to have methyltransferase-independent functions. However, most studies concentrate on the functions regulated via H3K4 methylation activities and not much has been explored about the non-canonical functions of these members. Here, in this review we will discuss in detail about the non-histone methyltransferase functions of SET1 family members in various cellular processes (see Table 1).
Table 1.
Non-canonical functions of SET1 family members
| SET1 family member | Non-canonical roles | Loss of function | References |
|---|---|---|---|
| MLL | Spindle organization | Elongated spindles with dense or low microtubule formation/multipolar spindle formation. | [130] |
| Chromosomal alignment and segregation | Delayed chromosomal alignment at metaphase plate. Formation of micronuclei. | [111,130] | |
| Cytokinesis | Formation of binucleated cells. | [111,135] | |
| Cell proliferation | Growth arrest at G1 Phase/defects in S-Phase progression. | [111] | |
| Ubiquitination process (as E3 ligase) | Delayed degradation and increased stability of MLL. | [184] | |
| MLL2 | Cell proliferation | Defects in S-Phase progression. | [111] |
| Ubiquitination process, as E3 ligase | Delayed degradation and increased stability of MLL2. | [184] | |
| MLL3 | Cell proliferation | Defects in S-Phase progression. | [111] |
| Transcriptional coactivator, Pol II loading and eRNA synthesis at enhancers | Reduces Pol II density in adjacent gene-bodies. | [96] | |
| MLL4 | Cell proliferation | Defects in S-Phase progression | [111] |
| Transcriptional coactivator, Pol II loading and eRNA synthesis at enhancers | Reduces Pol II density in adjacent gene-bodies. | [96] | |
| SETD1A | DNA damage response | Supresses DDR genes and induces p53-dependent apoptosis/impaired DDR | [120,123] |
| Chromosomal alignment and segregation | Delayed chromosomal alignment at metaphase plate. Formation of micronuclei. | [111,130] | |
| Cytokinesis | Formation of binucleated cells. | [111] | |
| Non-histone protein methylation | Reduced cellular proliferation in cancer cells. | [168,173] | |
| Cell proliferation/viability | Induced growth arrest | [120] | |
| SETD1B | Metabolic processes | Reduction in HADHA, a mitochondrial trifunction protein and thus leads to accumulation of lipids. | [32] |
| Cell proliferation/ viability | Altered cancer cell survival and growth. | [32] |
SET1 family in cell cycle regulation
The general network topology of eukaryotic cell cycle from yeast to mammals remains conserved, though evolutionary processes have given rise to differences in the regulatory proteins and their functions involved in cell cycle control [103,104]. In yeast, Set1/COMPASS regulates the cell cycle progression and proper mitotic assembly by methylation of H3K4 [105]. Similarly, numerous studies have investigated MLL’s role in cell cycle control (see Box 1) [51,55,106,107].
Box 1.
| Expression of MLL peaks at boundary of G1/S and G2/M phases, necessary to enter the S phase and progression into mitosis, respectively. Two major E3 complexes SCFSkp2 and APCCdc20 ensure MLL’s degradation through the cell cycle, regulating its expression at specific stages as required [107]. Taspase 1-mediated cleavage is another post-translational modification required for the MLL/MLL2 to control cell cycle. MLL is proteolytically cleaved by Taspase 1 to generate a fully functional mature MLLN320/C180 heterodimer [74]. Following cleavage by Taspase 1, MLL/MLL2 target to cyclin promoters to methylate histone H3K4, leading to the activation of genes involved in cell cycle regulation [74]. However, in contrast to this observation, another in vivo study on mouse indicates that MLL-dependent gene activation is independent of Taspase 1-mediated proteolytic cleavage, and rather depends on the intramolecular interaction between the MLLC and MLLN subunits [108]. Upon loss of intra-molecular interaction, FYRN domain is exposed which engenders MLL degradation resulting in loss of its function [108]. MLL and H3K4 methylation, both regulate the cell cycle in a distinct and dynamic manner, and play significant roles in the differential expression of Hox genes (HoxA5, HoxA7 and HoxA10) associated with cell cycle regulation [109]. MLL, normally associates with euchromatin at G1 phase, detaches from condensed mitotic chromatin followed by re-association towards the late telophase. Distinct to this observation, H3K4 trimethylation mark remains associated with chromatin throughout the cell cycle [109]. Contrary to this study, Blobel et.al. reported that MLL occupies chromatin throughout mitosis [110]. They also demonstrated that MLL recruits Menin, Ash2L, RbBP5 to mitotic chromatin and thus, ensures rapid onset of transcriptional activities following completion of mitosis [110]. |
Most of the activities of MLL or other SET1 family members are histone methylation dependent. However, recent researches have shown non-canonical/histone-methylation independent role in cell cycle regulation of this family.
Role in S-phase DNA damage response (DDR)
SET1 family members play an important role in S phase of cell cycle progression. RNAi-mediated knock down of many SET1 family members or WRAD complex components shows growth arrest, which suggests their essential participation in S phase transition [111]. MLL’s transcriptional activity but not methyltransferase activity, is required for the progression of cells into S phase [111]. During S phase, any mishaps with DNA replication triggers a checkpoint signalling cascade to resolve the error. A broad range of cellular functions such as cell-cycle progression, gene expression in S phase, stress response, DNA-replication and repair are controlled by S-phase DDR [112,113]. Under S-phase progression, SET1 family members are actively involved key elements in DDR regulation through conventional pathways (see Box 2).
Box 2.
| The SET1 family has been shown to play critical role in the process of DDR through histone modifications as well as nucleosome mobility and are essential in the process of DNA proof-reading. MLL is identified as a crucial factor involved in the mammalian S-phase check-point. Dysfunction of MLL at this check-point is a key underlying mechanism of MLL leukaemias [114,187]. Normally, MLL expression peaks at G1/S phase and is subsequently degraded by SCFSkp2 proteasomes [107]. However, upon DNA damage, interaction between MLL and SCFSkp2 is disrupted and MLL gets accumulated on the chromatin in response to ATR kinases [187]. The recruitment of CDC45 to the pre-replication complex is an essential step in initiation of DNA replication. To delay the DNA replication, stabilized MLL protein methylates H3K4 at late replication origins and prevents this recruitment of CDC45, and thus regulates S-phase checkpoint [187]. In addition, several members of MLL family regulate G1-to S-phase progression by initiating the transcriptional activation of E2F-target genes by directly or indirectly interacting with core transcription factors of the cell cycle – the E2F proteins [51,74]. Akin to MLL, another SET1 family member – MLL3 – is involved in DDR. Loss of MLL3 leads to decreased expression of genes involved in DDR and homologous recombination (HR)-mediated DNA repair [115]. Suppressed expression of these genes is linked with reduced H3K4me3 levels [115]. Knock down of MLL3 in bladder cancer cells results in low expression of HR repair proteins such as RAD51 which results in genetic instability with increased frequency of micronuclei and chromosomal aberrations. Moreover, in the absence of MLL3, cells heavily rely on the another DNA damage repair responder PARP1/2 [115]. Furthermore, in yeast cells, Set1 and H3K4me3 mark controls the DNA damage process through double strand break (DSB) repair. Set1 recruitment to the site of DSB is based on the presence of chromatin structure remodelling (RSC) complex. Upon loss of Set1 and RSC, the cells display defects in the DSB repair process [116]. In mammals, PTIP associates with MLL3/4 to recruit DSB repair protein MRE11 to stalled replication forks. The recruitment of MRE11 correlates with MLL3/4-mediated H3K4 mono and trimethylation. Authors used Mll4−/- null cells which were deficient in BRCA1, to show that catalytic activity of SET domain helps in the stalled forks degradation [117]. In contrast, SETD1A catalyses the methylation of H3K4 residues and escalates the FANCD2-dependent histone chaperoning/nucleosome mobility, helps in the recruitment and stabilization of RAD51 at the stalled replication forks which in turn protects the forks from degradation [118,119]. |
Interestingly, SETD1A is involved in DDR independent of its histone methylation activity as well (Figure 2)[120]. A novel highly conserved region ‘FLOS1’ (functional location on SETD1A:F1) domain on SETD1A is required for the DDR [120]. RNA seq. analysis from the SETD1A wild-type and SETD1A mutant MLL-AF9 leukaemic cells, show that the absence of SETD1A significantly decreases the DNA repair and Fanconi pathway-associated genes such as Fancd2, Mlh1 and increases p53 target genes. Authors show that FLOS1 domain acts as a cyclin-K-binding site and disruption of this domain suppresses DDR genes while inducing p53-dependent apoptosis. In addition, it was shown that Cyclin K/CDK12 complex is an obligation for SETD1A function, required for leukaemic cell growth. Using the non-invasive cell cycle indicator, Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) [121], they show that Cyclin K is highly expressed in the S phase and knock down of either SETD1A or Cyclin K suppresses cell cycle-dependent expression of FANCD2 [120], required for the repair of DNA inter-strand crosslinks [122].
Figure 2.

SET1 family in cell cycle regulation. The figure represents the function of SET1 family members in different stages of cell cycle progression. S-phase DNA damage response (DDR): in one pathway SETD1A FLOS1 domain interact with Cyclin K to regulate the expression of DDR associated genes and in other pathway SETD1A interact with RAD18 to repair DNA damage. Chromosomal alignment and segregation/Spindle assembly: MLL complex helps in proper chromosomal alignment and segregation, and also in the spindle organization by interacting with kinesin family members, Kinesin 13 (Kif2A, Kif2B, Kif2C), Kinesin1 (Kif5A, Kif5B) and Dynein. Cytokinesis: MLL and WDR5 both localize on the midbody. PRC1 and CYK4/MLKP1 proteins associates with WDR5 and help in its localization to the mid body. FLOS1: functional location on SETD1A: F1, Kif: Kinesin like protein, PRC1: Protein required for cytokinesis, CYK4: a Rho family GTPase activating protein (GAP), MKLP1: Mitotic kinesin-like protein 1
Another recent study also implicates SETD1A in DDR [123]. This report discloses the association of SETD1A methyltransferases to DNA Damage repair protein RAD18. RAD18 is an E3 ubiquitin ligase protein implicated in the repair of UV damaged DNA post-replication [124]. Interestingly, in humans, it is involved specifically in single-strand break repair during S phase [125]. Loss of either SETD1A or RAD18 results in impaired DNA damage repair in HEK293T cells. From the mass spectrometry analysis of nuclear extracts of HEK293T cells, it was found that RAD18 specifically interacts with SETD1A, but not with the SETD1A core complex (RBBP5 and ASH2L). Knockdown of SETD1A or RAD18 using siRNA, revealed their mutual regulation, as they downregulated each other’s expression at mRNA and protein levels, without affecting the global histone H3K4me3 levels. These findings suggest SETD1A together with RAD18 congregate to form a complex which is distinct from the conventional COMPASS complex [123].
Role in M phase
Following S phase, replicated chromosomes separate during mitosis. SET1 family proteins exhibit multiple essential functions in the mitotic control which do not involve histone methyltransferase activity. SET1 family members, such as MLL, are seen to be involved in mitotic entry, spindle assembly organization, spindle assembly check point, chromosomal segregation and cytokinesis (Figure 2).
Knockdown of MLL or SETD1A through siRNA results in high number of binucleated cells and micronuclei in human cell lines [111]. The reconstitution of the full-length MLL expression rescues the cells from mitotic defects signifying MLL’s specific role in cell division [111]. Interestingly, like in other MLL functions, the MLLC subunit is the effector in mitosis also, although the exact role of MLLN subunit has not been worked out as yet. Complementation studies using domain-deletion mutations of TAD or SET domain of MLLC revealed that the mitotic defects observed after MLL knock down were independent of the transcriptional and/or the methyltransferase activity [111]. Curiously, MLL-WRAD interaction, even in the absence of SET domain, is necessary for the proper mitotic progression indicating that in addition to enhancing catalytic activity/stability, WRAD has other functions with MLL [111,126].
Chromosome alignment and segregation
During cell division, any errors in chromosomal alignment or segregation may lead to aneuploidy, yielding non-viable cells or cells susceptible to oncogenic transformation [127,128]. Proper chromosomal alignment is necessary for the error free chromosomal segregation [129]. Depletion of MLL or WDR5 leads to delayed chromosomal alignment at the metaphase plate with extended prophase/metaphase which in turn delays the onset of anaphase [130]. Interestingly, the functional capabilities for chromosomal alignment resides in MLLC subunit, but not via its enzymatic HMT activity. Point mutation in conserved Win motif in MLL could not re-establish proper chromosome alignments [130]. Only MLL- and SETD1A- but not MLL3-depleted cells displayed misaligned chromosomes in metaphase [130]. This was consistent with our previous observations where RNAi of MLL2 or MLL3 did not result in any aberrant mitosis [111]. Remarkably, MLL and SETD1A depletion also leads to formation of micronuclei, implying that in addition to chromosomal alignment, both proteins are critical for proper chromosomal segregation as well [111].
In yeast, Set1 suppresses the abnormal chromosome segregation during mitosis, which is commonly seen in cells carrying a temperature sensitive Ipl1 mutation [131]. The mutation in lysine 4 of histone H3 imitates this abnormal chromosome segregation to some extent, implying that the observed phenotype cannot be attributed solely to H3K4 methylation. Instead, Set1 complex methylates a non-histone substrate – Dam 1 – a kinetochore protein [131]. Equilibrium between methylation of Dam1 by Set1 complex and Ipl1 Aurora kinase-mediated phosphorylation is critical for normal chromosome segregation as well as cell viability [131].
Spindle organization
Mitosis or meiosis both require proper spindle assembly for nuclear division. Bipolar microtubule (MT)-based spindle assembly is pivotal for the accurate and well timed segregation of the chromosomes during mitosis [132]. Spindle, microtubule associated proteins (MAPs) are the key molecules which outline spindle MT length, dynamics, orientation and location, as well as where and when MTs are generated [127,132]. Many reports suggest H3K4 methyltransferases coordinates cell-cycle progression and appropriate spindle-assembly during mitosis and meiosis using their HMT activity (see Box 3) [105,131,133,134]; however non-canonical pathways remain to be explored.
Box 3.
| The CFP1 protein, a conserved member of the SETD1A complex, binds to the chromatin at non-methylated CpG islands [63]. In mouse oocytes, deletion of the CFP1, leads to failure in bivalent metaphase II spindle assembly, which suggests that CFP1 may regulate α-tubulin polymerization in oocytes [134]. H3K4 trimethylation regulation via CFP1 makes the chromatin accessible to transcriptional machinery ensuring timely transcription during oocyte formation [134]. In yeast, Mad2, a spindle assembly check point (SAC) component, binds to the methylated H3K4 and regulates deactivation of the spindle assembly checkpoint [133]. Loss of Set1 or the G951S mutation, which abrogates the ability of Set1 to methylate histone H3K4, shows benomyl resistance phenotype [133]. Benomyl resistance phenotype is characterized by enhanced microtubule stability or enhanced spindle formation as cells continue to grow in the presence of benomyl (which interfere with the mitotic spindle assembly by depolymerizing microtubules and negatively affects the mitotic progression) [105]. Abrogation of H3K4 methylation also gives rise to benomyl resistance and thick mitotic spindle phenotype. Mad2 HORMA domain was revealed as conformation specific reader of H3K4me [133]. Concurring with this study, another group also implicates loss of Set1, and in turn defective methylation of H3K4 in benomyl resistance phenotype in yeast. ΔSet1 mutants show abnormal gene expression during G1/S, accompanied with deferred S-phase entry and mimic benomyl resistance. They suggest that Set1 and H3K4 methylation work in conjunction to regulate the cell-cycle progression and chromosome segregation during mitosis [105] |
Recent study from our lab has found that MLL complex localizes to spindle apparatus during mitosis. Enrichment of MLL to the centrosome and spindle compartments suggests important role in spindle assembly [130,135]. Even though only C subunit seems to have a defined role on spindle apparatus, both the subunits of MLL (MLLN and MLLC) are capable of localizing on the spindles and spindle poles (centrosome) independently throughout mitosis [135]. Similar to MLL, SETD1A also exhibited staining on spindle apparatus [130]. Remarkably, many members of the MLL complex including MLL, WDR5 and Menin have been reported as MAPs previously [55,136–138]. Consistent with a possible functional role, RNAi of MLL or WDR5 led to multiple spindle defects with either elongated spindles with long and dense MT formation, or low MTs, or multipolar spindles, suggesting their crucial roles in spindle assembly [130].
The question then arises: how MLL’s localization on spindles helps in their assembly? Mass spectrometric experiments identified Kif2A, Kif5A and dynein heavy chain (DYNC1H1), as putative binding partners of WDR5 [130]. Kif2A is a member of kinesin 13 family proteins, which act as microtubule depolymerizing enzymes [139] while Kif5A, a member of kinesin1 protein family, is highly processive motor protein, essential for vesicle transport in neuronal cells [140]. DYNC1H1, on the other hand, is an important protein in higher eukaryotes that carries out different functions like maintaining the orientation of MTs and chromosomes at the kinetochores, proper nuclear positioning, endosomal movement specificities and Golgi maintenance [141]. All three motor proteins identified either have closely related family members or occur as large protein complex. In order to test specificity of interaction of MLL complex with these proteins, we tested other proteins from kinesin 13 family, kinesin 1 family and dynein motor complex using HeLa cells lines expressing localization and affinity purification (LAP)-tagged bacterial artificial chromosome transgenes expressed from their indigenous promoters [130,142,143]. We found that Kif2A, Kif2B, Kif2C, Kif5A, Kif5B, Dynein intermediate chain (DYNC1I2) and dynactin subunit p150 (DCTN1), all interacted with MLL complex members,WDR5 and RbBP5, while unrelated motor Kif11 did not show an interaction [130]. Interestingly, few kinesins have been identified as WDR5-interacting protein before but these interactions has not been explored extensively [138,144].
During spindle assembly, both kinesins and dynein are required for spindle pole formation, and chromosomal positioning while kinesins alone take care of establishing spindle bipolarity [145]. Inhibition of kinesin or dynein leads to spindle defects such as monopolar spindles or long spindles, kinetochore misalignments and misorientation or misaligned chromosomal arms [145]. In our study, we specifically focused on Kif2A, as it was highly enriched protein in the WDR5 pull down assays [130]. Knock down of Kif2A leads to monopolar spindle formation and misaligned chromosomes in mammalian cells [146], and defective spindles, misaligned chromosomes, reduced microtubule depolymerization in mouse oocytes [147]. We demonstrated that knockdown of MLL and WDR5 showed elongated spindles and misaligned chromosomes similar to Kif2A knockdown, and reduced Kif2A levels at the spindle poles [130]. All of these findings suggested that MLL complex helps in the localization of Kif2A to the spindles. However, how MLL may get recruited to the spindles is not known. It is possible that dynein or other kinesins helps in the transportation/recruitment of MLL complex to spindles and other tubulin rich structures during the cell division processes.
Interestingly, we found the evolutionarily conserved Win motif, so far identified in histone H3 and SET1 family members [148–152], in the N terminus of Kif2A [130]. Even though Kif2B and Kif2C also interacted with WDR5, this motif was unique to Kif2A and majorly responsible for Kif2A-WDR5 interaction. Curiously, MLL was able to interact with Kif2A independently of WDR5. Nonetheless, a mutation in the Win motif of Kif2A abolishes its spindle localization [130]. The MLL-WDR5-Kif2A is reminiscent of the MLL-WDR5-H3 complex where use of Win motif by multiple partners may ensure stepwise regulation of a temporal recruitment process. While the mechanism of how the Win motif based interactions bring about spindle organization will have to wait for a detailed structural investigation, there is a strong likelihood that such interactions are a crucial component of MLL/WDR5 modus operandi.
Cytokinesis
SET1 family member, MLL, may play an important role in cytokinesis. A previous study, showed that WDR5 resides at the midbody [138]. Knockdown of WDR5 hinders the process of abscission (separation of daughter cells) and therefore increases the prevalence of multi-nucleated cells, which is a hallmark for cytokinesis failure [138]. Similarly, MLL can be detected at the midbody in mammalian cells, and depletion of MLL leads to binucleated cells, most likely due to failed cytokinesis [111,135]. However, how exactly MLL is targeted to the midbody and controls the cytokinesis process is yet to be elucidated. It is possible that MLL associates with midbody regulatory proteins, as it is shown that WDR5 interacts with a number of midbody proteins, including – Protein Regulator of Cytokinesis-1 (PRC1) and centralspindlin complex proteins CYK4 (a Rho family GTPase activating protein (GAP)/MKLP1(Mitotic kinesin-like protein 1) – known midbody-localized microtubule regulators [138,153–156]. PRC1, a conserved non-motor microtubule associated protein, is known to associate with kinesin family member Kif4 [157], which helps in organelle transport and chromosome movement [158]. It has been shown that Kif4 translocates PRC1 to the spindle and helps in the midzone formation and cytokinesis as well as regulates the midzone length during mitosis [136,159]. MKLP1, a member of Kinesin 6 family is required for cytokinesis and spindle polarity [158]. However, WDR5 directly interacts with kinesin family member MKLP1, but not with Kif4 [138]. The mutational analysis showed that the central arginine binding cavity of WDR5 (that interacts with Win motif) appears to be required for this targeting it to the midbody dark zone [138]. It is likely that similar to its interaction with Kif2A, WDR5 utilizes the central arginine binding cavity to interact with proteins involved in cytokinesis.
SET1 family in metabolic processes
H3K4me3 has been associated with modifying the lifespan of various species including yeast, worms and flies [160]. Recent studies have explored the impact of methyltransferases in metabolism [161,162]. Lack of H3K4me3 methyltransferase complex affects the monounsaturated fatty acids (MUFA) metabolism [161]. It is speculated that they might regulate lipid metabolism by methylating non-histone proteins or by affecting the methyl pool utilized by other enzymes [161]. In support of this hypothesis of a non-catalytic role, recent study indicates the involvement of SETD1B/COMPASS in regulating the fat metabolism [32]. Remarkably, mass spectrometric study of purified COMPASS complex revealed majority of SETD1B components in the cytoplasmic fractions of MCF-7 cells [32]. Immunofluorescence re-confirmed the localization of SETD1B mainly in the cytoplasm while SETD1A in the nucleus. Depletion of BOD1, which is a cytoplasmic-specific subunit of SETD1B, dramatically accelerated the degradation of the SETD1B protein, and the loss of SETD1B also destabilizes BOD1, indicating that these proteins need to occur as a complex to maintain their stability even in the cytoplasm. Surprisingly, unlike its correspondent homolog, SETD1A, knock down of SETD1B did not affect the bulk of H3K4 methylation in multiple cell lines [32]. RNA seq. analysis from the MDA-MB-231 cells infected with a lentivirus expressing BOD1 or SETD1B shRNAs shows that depletion of either BOD1 or SETD1B exhibits a 50% overlap in genes regulated by both proteins [32]. SETD1B loss significantly increased the expression of genes involved in cell metabolism such as ADIPOR1, PRKAR2A, COX7C, SDC4 and COQ7 and consequently lead to the accumulation of lipids in human breast cancer cells [32]. Nonetheless, a mitochondrial trifunctional protein (HADHA/B) was copurified with BOD1 in the cytoplasm, which also interacts with SETD1B/COMPASS [32]. Knockdown of SETD1B remarkably reduces the HADHA protein levels, suggesting cytoplasmic SETD1B complex stabilizes the mitochondrial trifunction protein, and thus regulates the metabolic processes in catalytic-domain independent manner. Further mechanistic approach suggested that adiponectin receptor 1 (AdipoR1) signalling may be relevant to SETD1B function, as many SETD1B downstream genes are involved in the AdipoR1 signalling [32].
SET1 family in methylation of non-histone proteins
As described above, SET1 family of proteins specially methylate histone 3 on lysine 4 and critically regulate chromatin packing and gene transcription. However, recent studies have reported the novel role of SET1 family members in catalysing the methylation of proteins other than histones in mammalian cells (Figure 3).
Figure 3.
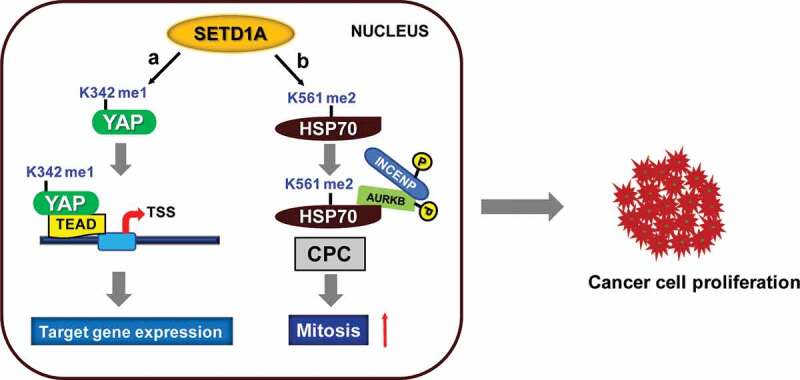
Non-histone protein methylation by SETD1A. The figure illustrates the pathway by which SETD1A regulates cancer cell proliferation by protein methylation. (a) SETD1A mono-methylates YAP (at K342) which interacts with transcriptional factor, TEAD and promotes the expression of target genes. (b) Di-methylation of HSP70 at K561 by SETD1A regulates the cell cycle progression. The di-methylated HSP70 binds to AURKB, which in turn interacts with INCENP, resulting in kinase activation of AURKB and formation of CPC, essential for mitotic progression. YAP: Yes associated protein; K: Lysine; TEAD: TEA domain; TSS: Transcription start site; HSP70: Heat shock protein 70; AURKB: Aurora kinase B; INCENP: Inner centromere protein; CPC: Chromosomal passenger complex
Methylation of non-histone proteins on lysine and arginine residues has emerged as an important post translational modification with wide range of cellular functions. In particular, methylation of lysine residues of non-histone proteins has been demonstrated to play a pivotal role in the regulation of various cellular signalling pathways including WNT, HIPPO, MAPK, JAK-STAT and BMP [163]. However, similar to lysine methylation on histone, the primary function of non-histone lysine methylation is to regulate protein-protein interaction by which it controls various downstream processes including protein stability, subcellular localization and DNA binding. Furthermore, lysine methylation cross talk with other post-translational modifications, adding another level of regulation [164,165].
In yeast, methylation of non-histone protein by SET1 protein complex has already been reported [131]. The yeast ortholog of SETD1A responsible for H3K4 methylation, also di-methylates lysine residue (K233) of kinetochore protein Dam1, a member of DASH complex. Further, Dam1 is also regulated by Aurora kinase Ipl1, which phosphorylates this protein at adjacent serine residues (S232, S234, S235) within ‘SKSS’ motif. This phosphorylation by Aurora kinase Ipl1 is essential for kinetochore formation [166,167]. Interestingly, di-methylation of Dam1 K233 opposes the phosphorylation of flanking serines in ‘SKSS’ motif, thus regulating kinetochore functions of Dam1, and subsequent chromosome segregation during cell division [131]
In humans, SET1 family member, SETD1A has been found to di-methylate HSP70, a ubiquitous molecular chaperone, at lysine 561 (K561) and regulates Aurora kinase B activity in cancer cells. Post-translational modifications of HSP70 such as K561 methylation and C-terminal phosphorylation are known to increase the rate of cancer cell proliferation [168,169]. SETD1A di-methylates HSP70 and regulates its subcellular localization. The di-methylated HSP70 interacts with Aurora kinase B and promotes various types of cancer cell proliferation [168]. Aurora kinase B, as a member of chromosomal passenger complex, utilizes its kinase activity to regulate mitosis [170–172]. Even though, a direct role of Set1/SETD1A in Aurora kinase activity has not been discovered so far, the cross talk between Set1/SETD1A with Aurora kinase, is observed in yeast (Ipl1) and in relation to HSP70 regulation in mammals. These two interactions along with SETD1A’s role in chromosome segregation, a process intimately linked to Aurora kinase B, point towards a more intricate role between the two proteins.
Recently, another study reported the role of SETD1A in the regulation of Yes associated protein (YAP), a key downstream regulator of tumour suppressor pathway (Hippo signalling pathway) [173,174]. YAP is a nucleocytoplasmic shuttling protein, which regulates cell proliferation and critically functions in organ size and development, tissue regeneration and self-renewal of stem cells [175]. Earlier studies have shown that SET7 methylates YAP at K494, retains YAP in the cytoplasm and inhibits its function [176]. However, it has been shown that SETD1A acts as a positive regulator of YAP activity. SETD1A as a multi-subunit protein complex (with WRAD), mediate the mono-methylation of YAP at lysine residue K342. SETD1A-mediated K342 methylation regulates YAP activity by blocking its interaction with CRM1, a nuclear export protein which consequently blocks YAP nuclear export and thus promotes cell proliferation which is required for tumorigenesis [173].
These studies indicate that not only non-histone substrates exist for SET1 family members in mammals but lysine methylation of these non-histone proteins play crucial roles in the regulation of cellular functions, sometimes even independent of transcription. SET1 family member, SETD1A tightly regulates the subcellular localization of proteins associated with proliferation of many types of cancer cells and thus tumorigenesis. Hence, this SET1 family methyltransferase may be used as a potential target for cancer therapy.
SET1 family as E3 Ubiquitin ligase
MLL 1–4 can be distinguished based on the number of PHD fingers. They contain around 23 PHD fingers (MLL 1 and 2 each contain four, MLL 3 contains eight and MLL 4 has seven PHD fingers) possessing a signature C4HC2C/H sequence which is stabilized by two zinc ions (Figure 1)[67]. Though MLL contains 4 PHD fingers, MLL fusion proteins lack all of them [177]. PHD fingers as epigenome readers serve multiple functions such as controlling gene expression via recruitment of multi-protein complexes and DNA binding [178]. Several reports suggest that PHD fingers are involved in ubiquitination process and have E3 ligase activity [179–181], however few studies report contrary [182,183]. Hess group discovered that second PHD (plant homeodomain) finger of SET1 family member, MLL has intrinsic E3 ubiquitin ligase activity [184]. This E3 ligase activity is also conserved in the closest analogue, MLL2 [184]. Endogenous MLL in 293 cells co-precipitates with CDC34, which is known as ubiquitin conjugating enzyme (E2) and regulates cell cycle [185,186]. CDC34, thus, interacts with MLL and by binding to PHD2, expedites the E3 ligase activity [184]. PHD2 finger of MLL also helps in regulating its transcriptional activity. As stated earlier, MLL undergoes a bimodal degradation during cell cycle with cell-cycle specific SCFSkp2 and APCCdc20E3 ligases [107,187]. It is possible that PHD2 finger also helps in degradation of MLL with its E3 ligase activity, as mutant PHD2 MLL displays prolonged degradation and increased stability [184]. Even though, different PHD fingers of MLL can read various states of H3K4 methylation [188], the E3 ligase function may be independent of its histone methylation catalytic activity.
Conclusions
Dynamic role of classical histone lysine methylation in transcriptional gene regulation, maintenance of chromosomal structure and stability is well known. Recent advances in the non-canonical functions of SET1 family of H3K4 methyltransferases have provided new concepts in epigenetic cellular regulations. Here, we have reviewed the roles of SET1 family members in regulating functions related to cell cycle, metabolic processes and non-histone protein methylations. In future, detailed mechanistic studies may fully uncover their functional significances. Although, SET1 family members possess various functional domains such as enzymatic, DNA-binding and protein-interacting domains, majority of research has been limited to SET domain. It is imperative to explore the role of other domains in order to better appreciate the function of these family members in a broader context. Presently all SET1 family proteins are known to be associated with various pathological conditions including skeletal and neurodevelopmental disorders, syndrome like Wiedemann–Steiner and Kabuki, and rearrangements and mutations in many types of cancer. These studies indicate that it is requisite to understand the roles of all SET1 family members beyond H3K4 methylation for improved understanding of their disease pathogenesis.
Acknowledgements
We thank A. Bakshi and R. Joshi for their critical reading, and G.B. Srinivas for editing of the manuscript. JS is recipient of Research Associateship from Department of Biotechnology (DBT), India.
Funding Statement
This work was supported in part by the DBT/Wellcome Trust India Alliance Senior Fellowship to S.T.[IA/S/18/2/503981], and grants by Department of Biotechnology (DBT) [BT/PR13351/BRB/10/1403/2015] and Department of Science and Technology [EMR/2016/000406], Government of India to S.T.
Author contributions
S.T. conceived and designed the content of the review. J.S. and J.G. prepared the figures and the tables. J.S., J.G and S.T. wrote the manuscript.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- [1].Bannister AJ, Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Allfrey VG, Faulkner R, Mirsky AE.. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miller JL, Grant PA. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem. 2013;61:289–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Briggs SD, Bryk M, Strahl BD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in saccharomyces cerevisiae. Genes Dev. 2001;15(24):3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Krogan NJ, Dover J, Khorrami S, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277(13):10753–10755. [DOI] [PubMed] [Google Scholar]
- [6].Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16438–16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sims RJ 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19(11):629–639. [DOI] [PubMed] [Google Scholar]
- [8].Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol. 2019;26(10):880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38(12):621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Malik S, Bhaumik SR. Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. Febs J. 2010;277(8):1805–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taverna SD, Li H, Ruthenburg AJ, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vedadi M, Blazer L, Eram MS, et al. Targeting human SET1/MLL family of proteins. Protein Sci. 2017;26(4):662–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ruthenburg A, Allis C, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. [DOI] [PubMed] [Google Scholar]
- [14].Piunti A, Shilatifard A. Epigenetic balance of gene expression by polycomb and COMPASS families. Science. 2016;352(6290):aad9780. [DOI] [PubMed] [Google Scholar]
- [15].Mohan M, Herz H-M, Smith ER, et al. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31(21):4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Balgobind BV, Raimondi SC, Harbott J, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114(12):2489–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Winters AC, Bernt KM. MLL-rearranged leukemias—an update on science and clinical approaches. Front Pediatr. 2017;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691–700. [DOI] [PubMed] [Google Scholar]
- [20].Hess JL, Yu BD, Li B, et al. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90(5):1799–1806. [PubMed] [Google Scholar]
- [21].Yu BD, Hanson RD, Hess JL, et al. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci U S A. 1998;95(18):10632–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yu BD, Hess JL, Horning SE, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–508. [DOI] [PubMed] [Google Scholar]
- [23].FitzGerald KT, Diaz MO. MLL2: A new mammalian member of the trx/MLL family of genes. Genomics. 1999;59(2):187–192. [DOI] [PubMed] [Google Scholar]
- [24].Huntsman DG, Chin S-F, Muleris M, et al. MLL2, the second human homolog of the Drosophila trithorax gene, maps to 19q13.1 and is amplified in solid tumor cell lines. Oncogene. 1999;18(56):7975–7984. [DOI] [PubMed] [Google Scholar]
- [25].Ruault M, Brun ME, Ventura M, et al. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284(1–2):73–81. [DOI] [PubMed] [Google Scholar]
- [26].Prasad R, Zhadanov AB, Sedkov Y, et al. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to drosophila trithorax. Oncogene. 1997;15(5):549–560. [DOI] [PubMed] [Google Scholar]
- [27].Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2010;331(6016):435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bledau AS, Schmidt K, Neumann K, et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141(5):1022–1035. [DOI] [PubMed] [Google Scholar]
- [31].Lee J-H, Tate CM, You J-S, et al. Identification and characterization of the human Set1B histone H3-lys4methyltransferase complex. J Biol Chem. 2007;282(18):13419–13428. [DOI] [PubMed] [Google Scholar]
- [32].Wang L, Collings CK, Zhao Z, et al. A cytoplasmic COMPASS is necessary for cell survival and triple-negative breast cancer pathogenesis by regulating metabolism. Genes Dev. 2017;31(20):2056–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Qian C, Zhou MM. SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci. 2006;63(23):2755–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dou Y, Milne TA, Ruthenburg AJ, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. [DOI] [PubMed] [Google Scholar]
- [35].Jiang H, Shukla A, Wang X, et al. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144(4):513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Patel A, Dharmarajan V, Vought VE, et al. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284(36):24242–24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shinsky SA, Monteith KE, Viggiano S, et al. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem. 2015;290(10):6361–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li Y, Han J, Zhang Y, et al. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016;530(7591):447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cao F, Chen Y, Cierpicki T, et al. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS One. 2010;5(11):e14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Avdic V, Zhang P, Lanouette S, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19(1):101–108. [DOI] [PubMed] [Google Scholar]
- [41].Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem. 2008;283(47):32158–32161. [DOI] [PubMed] [Google Scholar]
- [42].Xue H, Yao T, Cao M, et al. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature. 2019;573(7774):445–449. [DOI] [PubMed] [Google Scholar]
- [43].Park SH, Ayoub A, Lee Y-T, et al. Cryo-EM structure of the human MLL1 core complex bound to the nucleosome. Nat Commun. 2019;10(1):5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kwon M, Park K, Hyun K, et al. H2B ubiquitylation enhances H3K4 methylation activities of human KMT2 family complexes. Nucleic Acids Res. 2020;48(10):5442–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Caslini C, Yang Z, El-Osta M, et al. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen Y-X, Yan J, Keeshan K, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Nat Acad Sci. 2006;103(4):1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ernst P, Wang J, Huang M, et al. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21(7):2249–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Park S, Osmers U, Raman G, et al. The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression. Biochemistry. 2010;49(31):6576–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rozenblatt-Rosen O, Rozovskaia T, Burakov D, et al. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci U S A. 1998;95(8):4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xia Z-B, Anderson M, Diaz MO, et al. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci U S A. 2003;100(14):8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tyagi S, Chabes AL, Wysocka J, et al. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27(1):107–119. [DOI] [PubMed] [Google Scholar]
- [52].Milne TA, Dou Y, Martin ME, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102(41):14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yokoyama A, Somervaille TCP, Smith KS, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. [DOI] [PubMed] [Google Scholar]
- [54].Zeisig BB, Milne T, García-Cuéllar M-P, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24(2):617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Milne TA, Hughes CM, Lloyd R, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A. 2005;102(3):749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhu L, Li Q, Wong SHK, et al. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 2016;6(7):770–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cho Y-W, Hong T, Hong S, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim J-H, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Patel SR, Kim D, Levitan I, et al. The BRCT-domain containing protein PTIP Links PAX2 to a Histone H3, Lysine 4 methyltransferase complex. Dev Cell. 2007;13(4):580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee S, Kim D-H, Goo YH, et al. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol Endocrinol. 2009;23(5):610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lee J, Kim D-H, Lee S, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci U S A. 2009;106(21):8513–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280(50):41725–41731. [DOI] [PubMed] [Google Scholar]
- [64].Brown DA, Di Cerbo V, Feldmann A, et al. The SET1 complex selects actively transcribed target genes via multivalent interaction with CpG island chromatin. Cell Rep. 2017;20(10):2313–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28(2):609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang Y, Mittal A, Reid J, et al. Evolving catalytic properties of the MLL family SET domain. Structure. 2015;23(10):1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ali M, Hom RA, Blakeslee W, et al. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim Biophys Acta. 2014;1843(2):366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].García-Alai MM, Allen MD, Joerger AC, et al. The structure of the FYR domain of transforming growth factor beta regulator 1. Protein Sci. 2010;19(7):1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Allen MD, Grummitt CG, Hilcenko C, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. Embo J. 2006;25(19):4503–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zeleznik-Le NJ, Harden AM, Rowley JD. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci U S A. 1994;91(22):10610–10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19(8):5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chauhan C, Zraly CB, Parilla M, et al. Histone recognition and nuclear receptor co-activator functions of Drosophila cara mitad, a homolog of the N-terminal portion of mammalian MLL2 and MLL3. Development. 2012;139(11):1997–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hsieh JJ-D, Ernst P, Erdjument-Bromage H, et al. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23(1):186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Takeda S, Chen DY, Westergard TD, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20(17):2397–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yokoyama A, Kitabayashi I, Ayton PM, et al. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100(10):3710–3718. [DOI] [PubMed] [Google Scholar]
- [76].Schlichter A, Cairns BR. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. Embo J. 2005;24(6):1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Trésaugues L, Dehé P-M, Guérois R, et al. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J Mol Biol. 2006;359(5):1170–1181. [DOI] [PubMed] [Google Scholar]
- [78].Wu M, Wang PF, Lee JS, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28(24):7337–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gomez JA, Wapinski O, Yang Y, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152(4):743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Szabó B, Murvai N, Abukhairan R, et al. Disordered regions of mixed lineage leukemia 4 (MLL4) protein are capable of RNA binding. Int J Mol Sci. 2018;19(11):3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Glaser S, Schaft J, Lubitz S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133(8):1423–1432. [DOI] [PubMed] [Google Scholar]
- [83].Lee J, Saha PK, Yang Q-H, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci U S A. 2008;105(49):19229–19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lee J-E, Wang C, Xu S, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2. DOI: 10.7554/eLife.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mishra BP, Zaffuto K, Artinger E, et al. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep. 2014;7(4):1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Terranova R, Agherbi H, Boned A, et al. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103(17):6629–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang P, Lin C, Smith ER, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–1117. [DOI] [PubMed] [Google Scholar]
- [89].Glaser S, Lubitz S, Loveland KL, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ladopoulos V, Hofemeister H, Hoogenkamp M, et al. The histone methyltransferase KMT2B is required for RNA polymerase II association and protection from DNA methylation at the MagohB CpG island promoter. Mol Cell Biol. 2013;33(7):1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Jang Y, Wang C, Zhuang L, et al. H3K4 methyltransferase activity is required for MLL4 protein stability. J Mol Biol. 2017;429(13):2046–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Chen C, Liu Y, Rappaport A, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25(5):652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Santos MA, Faryabi RB, Ergen AV, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514(7520):107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hu D, Gao X, Morgan MA, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33(23):4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Weirich S, Kudithipudi S, Kycia I, et al. Somatic cancer mutations in the MLL3-SET domain alter the catalytic properties of the enzyme. Clin Epigenetics. 2015;7(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dorighi KM, Swigut T, Henriques T, et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell. 2017;66(4):568–576.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lin-Shiao E, Lan Y, Coradin M, et al. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev. 2018;32(2):181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wang C, Lee J-E, Lai B, et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci U S A. 2016;113(42):11871–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Li Y, Schulz VP, Deng C, et al. Setd1a and NURF mediate chromatin dynamics and gene regulation during erythroid lineage commitment and differentiation. Nucleic Acids Res. 2016;44(15):7173–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Schmidt K, Zhang Q, Tasdogan A, et al. The H3K4 methyltransferase Setd1b is essential for hematopoietic stem and progenitor cell homeostasis in mice. Elife. 2018;7. DOI: 10.7554/eLife.27157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tusi BK, Deng C, Salz T, et al. Setd1a regulates progenitor B-cell-to-precursor B-cell development through histone H3 lysine 4 trimethylation and Ig heavy-chain rearrangement. Faseb J. 2015;29(4):1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Sze CC, Cao K, Collings CK, et al. Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 2017;31(17):1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Harashima H, Dissmeyer N, Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013;23(7):345–356. [DOI] [PubMed] [Google Scholar]
- [104].Cross FR, Buchler NE, Skotheim JM. Evolution of networks and sequences in eukaryotic cell cycle control. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Beilharz TH, Harrison PF, Miles DM, et al. Coordination of cell cycle progression and mitotic spindle assembly involves histone H3 lysine 4 methylation by Set1/COMPASS. Genetics. 2017;205(1):185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Xia ZB, Popovic R, Chen J, et al. The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc Natl Acad Sci U S A. 2005;102(39):14028–14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21(19):2385–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yokoyama A, Ficara F, Murphy MJ, et al. MLL becomes functional through intra-molecular interaction not by proteolytic processing. PLoS One. 2013;8(9):e73649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. Febs J. 2009;276(6):1629–1640. [DOI] [PubMed] [Google Scholar]
- [110].Blobel GA, Kadauke S, Wang E, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36(6):970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ali A, Veeranki SN, Tyagi S. A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia. Nucleic Acids Res. 2014;42(12):7611–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. [DOI] [PubMed] [Google Scholar]
- [113].Willis NA, Zhou C, Elia AEH, et al. Identification of S-phase DNA damage-response targets in fission yeast reveals conservation of damage-response networks. Proc Natl Acad Sci U S A. 2016;113(26):E3676–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Tyagi S, Herr W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. Embo J. 2009;28(20):3185–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Rampias T, Karagiannis D, Avgeris M, et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. 2019;20(3):e46821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8):e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ray Chaudhuri A, Callen E, Ding X, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535(7612):382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Begum S, Goula A, Bayley R, et al. On your marks, get SET(D1A): the race to protect stalled replication forks. Mol Cell Oncol. 2018;5(6):e1511209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Higgs MR, Sato K, Reynolds JJ, et al. Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol Cell. 2018;71(1):25–41.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Hoshii T, Cifani P, Feng Z, et al. A non-catalytic function of SETD1A regulates cyclin K and the DNA damage response. Cell. 2018;172(5):1007–1021 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zielke N, Edgar BA. FUCCI sensors: powerful new tools for analysis of cell proliferation. Wiley Interdiscip Rev Dev Biol. 2015;4(5):469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Liang CC, Li Z, Lopez-Martinez D, et al. The FANCD2-FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun. 2016;7:12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Alsulami M, Munawar N, Dillon E, et al. SETD1A methyltransferase is physically and functionally linked to the DNA damage repair protein RAD18. Mol Cell Proteomics. 2019;18(7):1428–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Palle K, Vaziri C. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle. 2011;10(10):1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shiomi N, Mori M, Tsuji H, et al. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2006;35(2):e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ali A, Tyagi S. Diverse roles of WDR5-RbBP5-ASH2L-DPY30 (WRAD) complex in the functions of the SET1 histone methyltransferase family. J Biosci. 2017;42(1):155–159. [DOI] [PubMed] [Google Scholar]
- [127].Petry S. Mechanisms of mitotic spindle assembly. Annu Rev Biochem. 2016;85(1):659–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Potapova T, Gorbsky GJ. The consequences of chromosome segregation errors in mitosis and meiosis. Biology (Basel). 2017;6(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Guo Y, Kim C, Mao Y. New insights into the mechanism for chromosome alignment in metaphase. Int Rev Cell Mol Biol. 2013;303:237–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Ali A, Veeranki SN, Chinchole A, et al. MLL/WDR5 complex regulates Kif2A localization to ensure chromosome congression and proper spindle assembly during mitosis. Dev Cell. 2017;41(6):605–622.e7. [DOI] [PubMed] [Google Scholar]
- [131].Zhang K, Lin W, Latham JA, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122(5):723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18(3):187–201. [DOI] [PubMed] [Google Scholar]
- [133].Schibler A, Koutelou E, Tomida J, et al. Histone H3K4 methylation regulates deactivation of the spindle assembly checkpoint through direct binding of Mad2. Genes Dev. 2016;30(10):1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Yu C, Fan X, Sha -Q-Q, et al. CFP1 regulates histone H3K4 trimethylation and developmental potential in mouse oocytes. Cell Rep. 2017;20(5):1161–1172. [DOI] [PubMed] [Google Scholar]
- [135].Karole AM, Chodisetty S, Ali A, et al. Novel sub-cellular localizations and intra-molecular interactions may define new functions of mixed lineage leukemia protein. Cell Cycle. 2018;17(24):2684–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Hu C-K, Coughlin M, Field C, et al. KIF4 regulates midzone length during cytokinesis. Curr Biol. 2011;21(10):815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Sauer G, Körner R, Hanisch A, et al. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics. 2005;4(1):35–43. [DOI] [PubMed] [Google Scholar]
- [138].Bailey JK, Fields AT, Cheng K, et al. WD repeat-containing protein 5 (WDR5) localizes to the midbody and regulates abscission. J Biol Chem. 2015;290(14):8987–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Ems-McClung SC, Walczak CE. Kinesin-13s in mitosis: key players in the spatial and temporal organization of spindle microtubules. Semin Cell Dev Biol. 2010;21(3):276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Verhey KJ, Kaul N, Soppina V. Kinesin assembly and movement in cells. Annu Rev Biophys. 2011;40:267–288. [DOI] [PubMed] [Google Scholar]
- [141].Schiavo G, Greensmith L, Hafezparast M, et al. Cytoplasmic dynein heavy chain: the servant of many masters. Trends Neurosci. 2013;36(11):641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Maliga Z, Junqueira M, Toyoda Y, et al. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15(3):325–334. [DOI] [PubMed] [Google Scholar]
- [143].Poser I, Sarov M, Hutchins JRA, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5(5):409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].van Nuland R, Smits AH, Pallaki P, et al. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol. 2013;33(10):2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Loughlin R, Riggs B, Heald R. SnapShot: motor proteins in spindle assembly. Cell. 2008;134(3):548–548 e1. [DOI] [PubMed] [Google Scholar]
- [146].Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166(4):473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Yi Z-Y, Ma X-S, Liang Q-X, et al. Kif2a regulates spindle organization and cell cycle progression in meiotic oocytes. Sci Rep. 2016;6(1):38574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Dharmarajan V, Lee J-H, Patel A, et al. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem. 2012;287(33):27275–27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhang P, Lee H, Brunzelle JS, et al. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012;40(9):4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Song -J-J, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283(50):35258–35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Karatas H, Townsend EC, Bernard D, et al. Analysis of the binding of mixed lineage leukemia 1 (MLL1) and histone 3 peptides to WD repeat domain 5 (WDR5) for the design of inhibitors of the MLL1-WDR5 interaction. J Med Chem. 2010;53(14):5179–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Patel A, Vought VE, Dharmarajan V, et al. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem. 2008;283(47):32162–32175. [DOI] [PubMed] [Google Scholar]
- [153].Sasabe M, Boudolf V, De Veylder L, et al. Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc Natl Acad Sci U S A. 2011;108(43):17844–17849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Jantsch-Plunger V, Gönczy P, Romano A, et al. CYK-4: a Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149(7):1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Jiang W, Jimenez G, Wells NJ, et al. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2(6):877–885. [DOI] [PubMed] [Google Scholar]
- [156].Liu X, Li Y, Meng L, et al. Reducing protein regulator of cytokinesis 1 as a prospective therapy for hepatocellular carcinoma. Cell Death Dis. 2018;9(5):534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Subramanian R, Ti S-C, Tan L, et al. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 2013;154(2):377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15(9):467–476. [DOI] [PubMed] [Google Scholar]
- [159].Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci U S A. 2005;102(2):343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Han S, Brunet A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012;22(1):42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Han S, Schroeder EA, Silva-García CG, et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature. 2017;544(7649):185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Parkhitko AA, Jouandin P, Mohr SE, et al. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell. 2019;18(6):e13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Biggar KK, Li SSC. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2014;16(1):5–17. [DOI] [PubMed] [Google Scholar]
- [164].Baron B. The lysine multi-switch: the impact of lysine methylation on transcription factor properties. Biohelikon: Cell Biol. 2014;2:a13. [Google Scholar]
- [165].Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15(2):110–124. [DOI] [PubMed] [Google Scholar]
- [166].Nogales E, Ramey VH. Structure-function insights into the yeast Dam1 kinetochore complex. J Cell Sci. 2009;122(21):3831–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Cheeseman IM, Anderson S, Jwa M, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111(2):163–172. [DOI] [PubMed] [Google Scholar]
- [168].Cho H-S, Shimazu T, Toyokawa G, et al. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat Commun. 2012;3:1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Muller P, Ruckova E, Halada P, et al. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2012;32(25):3101–3110. [DOI] [PubMed] [Google Scholar]
- [170].Sessa F, Mapelli M, Ciferri C, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18(3):379–391. [DOI] [PubMed] [Google Scholar]
- [171].Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol. 1987;105(5):2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Vader G, Medema RH, Lens SMA. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173(6):833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Fang L, Teng H, Wang Y, et al. SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell. 2018;34(1):103–118.e9. [DOI] [PubMed] [Google Scholar]
- [174].Abylkassov R, Xie Y. Role of Yes-associated protein in cancer: an update. Oncol Lett. 2016;12(4):2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zhao B, Li L, Lei Q, et al. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24(9):862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Oudhoff MJ, Freeman S, Couzens A, et al. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev Cell. 2013;26(2):188–194. [DOI] [PubMed] [Google Scholar]
- [177].Milne TA, Kim J, Wang GG, et al. Multiple Interactions Recruit MLL1 and MLL1 Fusion Proteins to the HOXA9 Locus in Leukemogenesis. Mol Cell. 2010;38(6):853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36(7):364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Coscoy L, Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003;13(1):7–12. [DOI] [PubMed] [Google Scholar]
- [180].Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155(7):1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Dul BE, Walworth NC. The plant homeodomain fingers of fission yeast Msc1 exhibit E3 ubiquitin ligase activity. J Biol Chem. 2007;282(25):18397–18406. [DOI] [PubMed] [Google Scholar]
- [182].Scheel H, Hofmann K. No evidence for PHD fingers as ubiquitin ligases. Trends Cell Biol. 2003;13(6):285–287.author reply 287-8. [DOI] [PubMed] [Google Scholar]
- [183].Aravind L, Iyer LM, Koonin EV. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle. 2003;2(2):123–126. [DOI] [PubMed] [Google Scholar]
- [184].Wang J, Muntean AG, Wu L, et al. A subset of mixed lineage leukemia proteins has plant homeodomain (PHD)-mediated E3 ligase activity. J Biol Chem. 2012;287(52):43410–43416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Cocklin R, Heyen J, Larry T, et al. New insight into the role of the Cdc34 ubiquitin-conjugating enzyme in cell cycle regulation via Ace2 and Sic1. Genetics. 2011;187(3):701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Goebl MG, Yochem J, Jentsch S, et al. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241(4871):1331–1335. [DOI] [PubMed] [Google Scholar]
- [187].Liu H, Takeda S, Kumar R, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467(7313):343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Chang P-Y, Hom RA, Musselman CA, et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]


