ABSTRACT
Human coronaviruses (HCoVs) are associated with a range of respiratory complications. In the last two decades, three major outbreaks have been reported due to HCoVs including the current pandemic. In December 2019, a newly emerged virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in Wuhan city, China. This paper presents a detailed review of the literature and discusses the uncertain spread of coronavirus disease 2019 (COVID-19) using fuzzy set as classical set theory logic to measure uncertainty and vagueness of COVID-19 in China. Our findings show that both infection and death rate touched the peak (normal fuzzy sets) and have shown a decline. The graphs are not convex, which shows that there remains much uncertainty in the spread of COVID-19. Effective vaccines are clearly needed to control and prevent the COVID-19 pandemic.
KEYWORDS: SARS-CoV-2, COVID-19, China, fuzzy set approach, vaccines
Introduction
Human coronaviruses (HCoVs) are characterized as a main group of coronavirus (CoV) which can cause several respiratory infections of different levels of severity, some of the common manifestations of which are pneumonia, bronchitis, and common cold.1 In HCoVs, a very rapid evolution occurs by recombination and genome nucleotide substitution.2 In recent years, the rapid evolution in HCoVs has occurred due to poultry farming and urbanization.3 These factors have facilitated the regular mixing of species and contribute the genome recombination of HCoVs.3 The transmission of these highly pathogenic viruses occurs from animals to humans due to their close contacts. Climate changes are also one of the facilitating factors in the transmission of these pathogenic viruses by disturbing the geographical places of viral carrier animals and insects.4 Seven HCoVs have been identified: HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Among them the first four HCoVs are spread globally in the human population and cause nearly one-third of common cold infections.5 In immunocompromised patients, HCoV-NL63 and HCoV-229E cause light infection. These two CoVs originate in the bats of Africa.−6,7 HCoV-229E is Camelid, an intermediate host.8 HKU1 and HCoV-OC43 have origin of rodents and harmless for humans. Rhinolophus-bat CoV HKU2 is a novel strain which is called SADS coronavirus (SADS-CoV) which can cause swine acute diarrhea-syndrome (SADS) and originates from piglets.9 HCoV-229E and HCoV-OC43 infections accompanied by multiple respiratory and systemic symptoms in the elderly age are frequent, likely causing substantial medical disease burden.10,11 Infections may lead to some serious complications such as neurological problems.12,13 Some recent studies suggest that most RNA viruses are likely to have a much more recent human adaptation and evolution.14,15
The worldwide spread of recently emerged SARS-CoV-2 is very uncertain. The emerging of such pathogenic viruses is a big challenge for medical science to discover the unknown zoonotic source of zoonotic viruses, to develop rapid diagnostic techniques, and to create drugs and vaccines to treat and control thee deadly infectious viruses. Vaccine and drugs against these highly pathogenic viruses have not yet been developed; thus, only symptomatic therapy is used for infected patients.4
The aim of this article is to describe the epidemiology of CoVs, genetics, vaccines, and the current scenario of COVID-19. We also have performed a fuzzy set analysis to measure uncertainty and vagueness of COVID-19 in China.
Genetics of CoVs
CoVs are enveloped RNA viruses, belongs to family Coronaviridae, and order Nidovirale. Based on comparison of whole viral genome sequences, the International Committee for Taxonomy of Viruses (ICTV) has further divided CoVs into four genera, Alpha, Beta, Gamma, and Delta.16,17 CoV can cause infection in humans, swine, and avian. HCoVs belong to Alpha or Beta genera. Alpha CoVs include HCoV-NL63 and HCoV-229E, and Beta CoVs include SARS-CoV, MERS-CoV, HCoV-OC43, and HCoV-HKU1. Under electron microscopy, CoV virions have a spherical, pleomorphic shape, and “club-like” projections are present on the outer surface due to spike proteins.18,19 The virion contains a helical nucleocapsid which protects a positive-sense single-stranded RNA viral genome of 26–32 kilobases.20 CoVs have four structural proteins named spike, envelope, membrane, and nucleocapsid. Moreover, other accessory overlapping open reading frames (ORFs) are also present. Their number and location are different among CoV species.21
According to the sequence databases all HCoVs have a recognized animal origin. SARS-CoV-2, MERS-CoV, SARS-CoV, HCoV-229E, and HCoV-NL63 origin in bats, and HKU1 and HCoV- OC43 in rodents.22,23 The Phylogenetic relationship of CoVs is presented in Figure 1.
Figure 1.
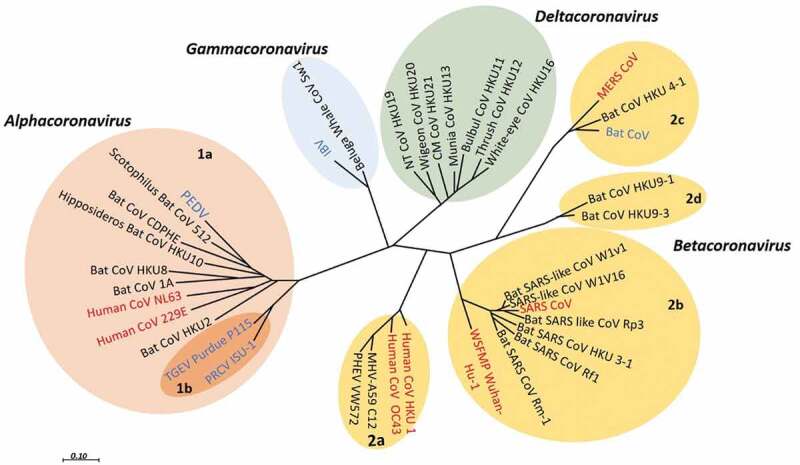
Phylogenetic relationship of CoVs. The phylogenetic tree illustrates the relationship among some HCoVs (red) and animal CoVs (blue) as a reference used in the tree, on the basis of complete genome nucleotide sequences. The viruses are grouped and subgrouped as (prototype shown): Alpha-CoV (pink, subgroup;1a,1b), Beta-CoV (light brown, subgroup; 1a,1b,1 c,1d), Gamma-CoV (light blue), and Delta-CoV (green). This tree is reconstructed with RNA-dependent RNA polymerase-coding region complete sequences of CoVs (with MEGA 7.2 software for maximum likelihood method).24 Porcine enteric diarrhea virus (PEDV); infectious bronchitis virus (IBV); SARS- CoV; transmissible gastroenteritis virus (TGEV) MERS- CoV; Porcine respiratory CoV ISU-1 (PRCV ISU-1); and Wuhan seafood market pneumonia (Wuhan-Hu-1)
SARS-CoV
Due to the recombination of bat SARSr-CoVs, a new virus emerged called SARS-CoV.25 The SARS-CoVs cause epidemics by infecting humans and civets.4,26 The SARS-CoV outbreak was reported in 2002–3 in China and spread to 37 countries. The number of SARS-CoV confirmed cases were 8273 with 775 (9.4%) deaths.4 ACE2 is a receptor binding site of the SARS-CoV.27,28
MERS-CoV
The occurrence of highly pathogenic CoV in the Middle East, called MERS-CoV, focused attention on HCoVs. MERS-CoV is a highly pathogenic CoV discovered in 2012 in the Kingdom of Saudi Arabia.29 MERS-CoV infected people in Saudi Arabia, Jordan, Oman, Qatar, Egypt, Jordan, and the United Arab Emirates. Some of the peoples were also infected in European countries who had a history of visit to the Arabian Peninsula or contact with infected peoples. There is evidence of person-to-person transmission of MERS-CoV from infected persons to healthy persons. However, the transmission of MERS-CoV occurs in those who have very close contact with infected persons, like the close contact of health care staff with infected patients. People who are immunocompromised and have secondary infections are more susceptible to MERS-CoV infection.30
Studies conducted in Egypt and Oman showed that neutralizing antibodies against MERS-CoV are found in dromedary camels “Camelus dromedaries”,31 which shows that the MERS-CoV may have the dromedary camels as intermediate host. A recent study finds that the infected person MERS-CoV RNA matches with that of camels in Qatar.32 MERS-CoV isolation has not been isolated yet from camels, but it seems that the transmission of MERS-CoV is camel to human. According to phylogenetic analysis MERS-CoV shows close similarity with BtCoV-HKU5 and BtCoV-HKU4 which are the known CoVs.33 The reservoir of MERS-CoV may be bat as well as the European hedgehog “Erinaceuseuropaeus” in which a MERS-CoV close relative is found.34
SARS-CoV-2
Pneumonia cases were reported in December 2019 with an unknown etiological agent. The initial cases were epidemiologically related with the fresh Seafood Market located in Wuhan city, Hubei province, China.35 Genome sequence of five patients’s shows 79.5% identical similarity to SARS-CoV. In addition, on the basis of whole genome sequence, the SARS-CoV-2 is 96% identical with bat CoV. The results of the pairwise proteins sequence analysis of seven conserved nonstructural proteins showed that this novel SARS-CoV-2 belongs to the SARSr-CoV species.36 The disease caused by SARS-CoV-2 named as COVID-19.
The SARS-CoV-2 was isolated from the patient’s bronchoalveolar lavage fluid. As with SARS-CoV, SARS-CoV-2 uses the same receptor called ACE2 for cell entry. SARS-CoV-2 contains six ORFs and other accessory genes. The analysis showed that the genes of SARS-CoV-2 show less than 80% nucleotide sequence similarity with SARS-CoV.36
For the classification of SARS-CoV-2, the seven conserved replicase domains of ORF1ab were used, which showed that between SARS-CoV and SARS-CoV-2, the amino acid sequence is 94.6% identical, indicating that SARS-CoV-2 and SARS-CoV belong to the same species. The results of phylogenetic analysis also revealed that SARS-CoV-2 is the closest relative of RaTG13, and it has different ancestry from other SARSr-CoV.36
The previously detected bat CoV from Rhinolophus affine called “BatCoV RaTG13” from Yunnan province China showed maximum sequence similarity to SARS-CoV-2. The results of Simplot analysis showed that RaTG13 genome is very similar to that of SARS-CoV-2, which show 96.2% genome sequence similarity. The gene of receptor spike binding-proteins (S) of SARS-CoV-2 is very different from those of other CoVs, which show <75% nucleotide sequence similarity with the other SARSr-CoVs, while RaTG13 shows 93.1% identity.36
The receptor spike binding-protein gene of RaTG13 and the receptor spike binding-protein gene of SARS-CoV-2 are longer than those of other SARSr-CoVs. The main difference in SARS-CoV-2 compared with SARS-CoV is that SARS-CoV-2 has three short insertions in the N-terminal domain, and 4 out of 5 key residues changes in the receptor-binding motif. The recent finding shows close phylogenetic relationship to BatCoV RaTG13, which provides enough evidence for a bat origin of the recent SARS-CoV-2.36
By electron microscopy, SARS-CoV-2 is of spherical shape with some pleomorphism and 60–140 nm in diameter. The virus particle consists of unique 9–12 nm spikes, which give the virus an appearance like the solar corona.37 The cases of COVID-19 have been reported in essentially all countries/regions.38 Initial COVID-19 cases had a history of travel to China.39 The transmission cycle of SARS-CoV, MERS-CoV, and SARS-CoV-2 is presented in Figure 2.
Figure 2.
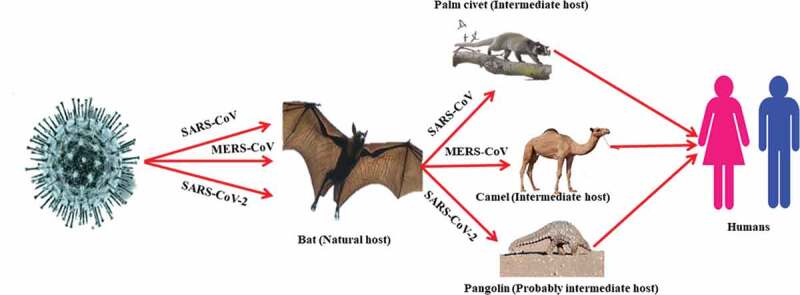
Transmissions cycle of SARS-CoV, MERS-CoV, and SARS-CoV-2
Replication cycle
CoV infection begins when the spike protein S1 domain attaches to the ACE2 receptor. This attachment brings structural changes in the S2 subunit of spike protein, which leads to the mutual membrane fusion of cell plasma membrane and virus resulting in the nucleocapsid entering the cytoplasm. Through ribosomal frameshifting the translation of viral RNA occurs, producing two polyproteins pp1a and pp1ab. Sixteen non-structural proteins (NSPs) are generated from pp1a and pp1ab autoproteolytically processed in the presence of host and viral proteases, and then replicase-polymerase is formed from its assembly. The replicase polymerase is responsible for virus replication, a process by which structural proteins are formed due to replication of genomic RNA and by the transcription and translation of sub-genomic RNA. The viral products assemble in the specific region called ERGIC and form a bud which is a smooth wall vesicle and finally release virus from the cell by a process called exocytosis.40,41 The replication cycle of SARS-CoV-2 is presented in Figure 3.
Figure 3.
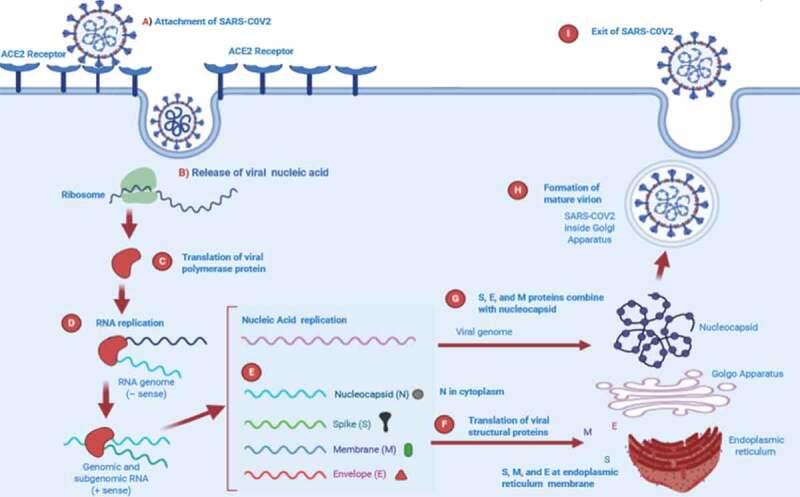
Proposed replication cycle of SARS-CoV-2
COVID-19 spread in China, a fuzzy set approach
The fuzzy set concept was developed by Zadeh in the mid-60’s to account for numerous concepts used in vague and imprecise reasoning, e.g., tall, old, and other similar parameters. It is a very convenient method for the representation of uncertainty. Fuzzy set theory has many applications; it also allows the graduate assessment of membership of element in a set, described with the aid of membership function values in interval [0, 1]. Fuzzy set is capable of measuring uncertainty of certain diseases. Hence it seems quite useful in case of COVID-19, as its spread is very uneven. We can understand it with following example.
Example: Assume that we have the following data which we have extracted from different sources as presented in Table 1.
Now we will use the concept of fuzzy set for which we must find the membership function as shown in Table 2.
By plotting the data we get Figure 4.
If we take only one attribute i.e. we get Figure 5.
If we take another attribute, then we get Figure 6.
Table 1.
Trend of COVID-19 cases in China (01–22-2020 to 04–03-2020)
| Date | COVID-19 daily cases | COVID-19 daily deaths |
|---|---|---|
| 1/22/2020 | 571 | 17 |
| 1/23/2020 | 259 | 8 |
| 1/24/2020 | 457 | 16 |
| 1/25/2020 | 688 | 15 |
| 1/26/2020 | 769 | 24 |
| 1/27/2020 | 1771 | 26 |
| 1/28/2020 | 1459 | 26 |
| 1/29/2020 | 1737 | 38 |
| 1/30/2020 | 1981 | 43 |
| 1/31/2020 | 2099 | 46 |
| 2/1/2020 | 2589 | 45 |
| 2/2/2020 | 2825 | 57 |
| 2/3/2020 | 3235 | 64 |
| 2/4/2020 | 3884 | 65 |
| 2/5/2020 | 3694 | 73 |
| 2/6/2020 | 3143 | 73 |
| 2/7/2020 | 3385 | 86 |
| 2/8/2020 | 2652 | 89 |
| 2/9/2020 | 2973 | 97 |
| 2/10/2020 | 2467 | 108 |
| 2/11/2020 | 2015 | 97 |
| 2/12/2020 | 14108 | 146 |
| 2/13/2020 | 5090 | 121 |
| 2/14/2020 | 2641 | 143 |
| 2/15/2020 | 2008 | 142 |
| 2/16/2020 | 2048 | 105 |
| 2/17/2020 | 1888 | 98 |
| 2/18/2020 | 1749 | 136 |
| 2/19/2020 | 391 | 114 |
| 2/20/2020 | 889 | 118 |
| 2/21/2020 | 823 | 109 |
| 2/22/2020 | 648 | 97 |
| 2/23/2020 | 214 | 150 |
| 2/24/2020 | 508 | 71 |
| 2/25/2020 | 406 | 52 |
| 2/26/2020 | 433 | 29 |
| 2/27/2020 | 327 | 44 |
| 2/28/2020 | 427 | 47 |
| 2/29/2020 | 573 | 35 |
| 3/1/2020 | 202 | 42 |
| 3/2/2020 | 125 | 31 |
| 3/3/2020 | 119 | 38 |
| 3/4/2020 | 139 | 31 |
| 3/5/2020 | 143 | 30 |
| 3/6/2020 | 99 | 28 |
| 3/7/2020 | 44 | 27 |
| 3/8/2020 | 40 | 22 |
| 3/9/2020 | 19 | 17 |
| 3/10/2020 | 24 | 22 |
| 3/11/2020 | 15 | 11 |
| 3/12/2020 | 20 | 7 |
| 3/13/2020 | 11 | 13 |
| 3/14/2020 | 20 | 10 |
| 3/15/2020 | 16 | 14 |
| 3/16/2020 | 21 | 13 |
| 3/17/2020 | 13 | 11 |
| 3/18/2020 | 34 | 8 |
| 3/19/2020 | 39 | 3 |
| 3/20/2020 | 41 | 7 |
| 3/21/2020 | 46 | 6 |
| 3/22/2020 | 39 | 9 |
| 3/23/2020 | 78 | 7 |
| 3/24/2020 | 47 | 4 |
| 3/25/2020 | 67 | 6 |
| 3/26/2020 | 55 | 5 |
| 3/27/2020 | 54 | 3 |
| 3/28/2020 | 45 | 5 |
| 3/29/2020 | 0 | 0 |
| 3/30/2020 | 79 | 5 |
| 3/31/2020 | 36 | 7 |
| 4/1/2020 | 35 | 6 |
| 4/2/2020 | 31 | 4 |
| 4/3/2020 | 19 | 4 |
| Total | 81639 | 3326 |
Table 2.
Membership function
| Date = di | a1 | a2 | a1/am | a2/am |
|---|---|---|---|---|
| 1/22/2020 | 571 | 17 | 0.04 | 0.113 |
| 1/23/2020 | 259 | 8 | 0.02 | 0.053 |
| 1/24/2020 | 457 | 16 | 0.03 | 0.107 |
| 1/25/2020 | 688 | 15 | 0.05 | 0.100 |
| 1/26/2020 | 769 | 24 | 0.05 | 0.160 |
| 1/27/2020 | 1771 | 26 | 0.13 | 0.173 |
| 1/28/2020 | 1459 | 26 | 0.10 | 0.173 |
| 1/29/2020 | 1737 | 38 | 0.12 | 0.253 |
| 1/30/2020 | 1981 | 43 | 0.14 | 0.287 |
| 1/31/2020 | 2099 | 46 | 0.15 | 0.307 |
| 1/2/2020 | 2589 | 45 | 0.18 | 0.300 |
| 2/2/2020 | 2825 | 57 | 0.20 | 0.380 |
| 2/3/2020 | 3235 | 64 | 0.23 | 0.427 |
| 2/4/2020 | 3884 | 65 | 0.28 | 0.433 |
| 2/5/2020 | 3694 | 73 | 0.26 | 0.487 |
| 2/6/2020 | 3143 | 73 | 0.22 | 0.487 |
| 2/7/2020 | 3385 | 86 | 0.24 | 0.573 |
| 2/8/2020 | 2652 | 89 | 0.19 | 0.593 |
| 2/9/2020 | 2973 | 97 | 0.21 | 0.647 |
| 2/10/2020 | 2467 | 108 | 0.17 | 0.720 |
| 2/11/2020 | 2015 | 97 | 0.14 | 0.647 |
| 2/12/2020 | 14108 | 146 | 1.00 | 0.973 |
| 2/13/2020 | 5090 | 121 | 0.36 | 0.807 |
| 2/14/2020 | 2641 | 143 | 0.19 | 0.953 |
| 2/15/2020 | 2008 | 142 | 0.14 | 0.947 |
| 2/16/2020 | 2048 | 105 | 0.15 | 0.700 |
| 2/17/2020 | 1888 | 98 | 0.13 | 0.653 |
| 2/18/2020 | 1749 | 136 | 0.12 | 0.907 |
| 2/19/2020 | 391 | 114 | 0.03 | 0.760 |
| 2/20/2020 | 889 | 118 | 0.06 | 0.787 |
| 2/21/2020 | 823 | 109 | 0.06 | 0.727 |
| 2/22/2020 | 648 | 97 | 0.05 | 0.647 |
| 2/23/2020 | 214 | 150 | 0.02 | 1.000 |
| 2/24/2020 | 508 | 71 | 0.04 | 0.473 |
| 2/25/2020 | 406 | 52 | 0.03 | 0.347 |
| 2/26/2020 | 433 | 29 | 0.03 | 0.193 |
| 2/27/2020 | 327 | 44 | 0.02 | 0.293 |
| 2/28/2020 | 427 | 47 | 0.03 | 0.313 |
| 2/29/2020 | 573 | 35 | 0.04 | 0.233 |
| 3/1/2020 | 202 | 42 | 0.01 | 0.280 |
| 3/2/2020 | 125 | 31 | 0.01 | 0.207 |
| 3/3/2020 | 119 | 38 | 0.01 | 0.253 |
| 3/4/2020 | 139 | 31 | 0.01 | 0.207 |
| 3/5/2020 | 143 | 30 | 0.01 | 0.200 |
| 3/6/2020 | 99 | 28 | 0.01 | 0.187 |
| 3/7/2020 | 44 | 27 | 0.00 | 0.180 |
| 3/8/2020 | 40 | 22 | 0.00 | 0.147 |
| 3/9/2020 | 19 | 17 | 0.00 | 0.113 |
| 3/10/2020 | 24 | 22 | 0.00 | 0.147 |
| 3/11/2020 | 15 | 11 | 0.00 | 0.073 |
| 3/12/2020 | 20 | 7 | 0.00 | 0.047 |
| 3/13/2020 | 11 | 13 | 0.00 | 0.087 |
| 3/14/2020 | 20 | 10 | 0.00 | 0.067 |
| 3/15/2020 | 16 | 14 | 0.00 | 0.093 |
| 3/16/2020 | 21 | 13 | 0.00 | 0.087 |
| 3/17/2020 | 13 | 11 | 0.00 | 0.073 |
| 3/18/2020 | 34 | 8 | 0.00 | 0.053 |
| 3/19/2020 | 39 | 3 | 0.00 | 0.020 |
| 3/20/2020 | 41 | 7 | 0.00 | 0.047 |
| 3/21/2020 | 46 | 6 | 0.00 | 0.040 |
| 3/22/2020 | 39 | 9 | 0.00 | 0.060 |
| 3/23/2020 | 78 | 7 | 0.01 | 0.047 |
| 3/24/2020 | 47 | 4 | 0.00 | 0.027 |
| 3/25/2020 | 67 | 6 | 0.00 | 0.040 |
| 3/26/2020 | 55 | 5 | 0.00 | 0.033 |
| 3/27/2020 | 54 | 3 | 0.00 | 0.020 |
| 3/28/2020 | 45 | 5 | 0.00 | 0.033 |
| 3/29/2020 | 0 | 0 | 0.00 | 0.000 |
| 3/30/2020 | 79 | 5 | 0.01 | 0.033 |
| 3/31/2020 | 36 | 7 | 0.00 | 0.047 |
| 4/1/2020 | 35 | 6 | 0.00 | 0.040 |
| 4/2/2020 | 31 | 4 | 0.00 | 0.027 |
| 4/3/2020 | 19 | 4 | 0.00 | 0.027 |
| Total | 81639 | 3326 |
Figure 4.
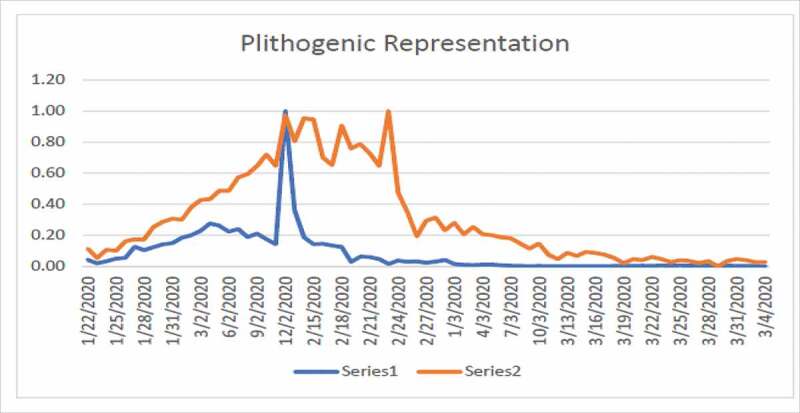
Plithogenic representation of COVID-19 cases in China from 01/22/2020 to 03/04/2020
Figure 5.
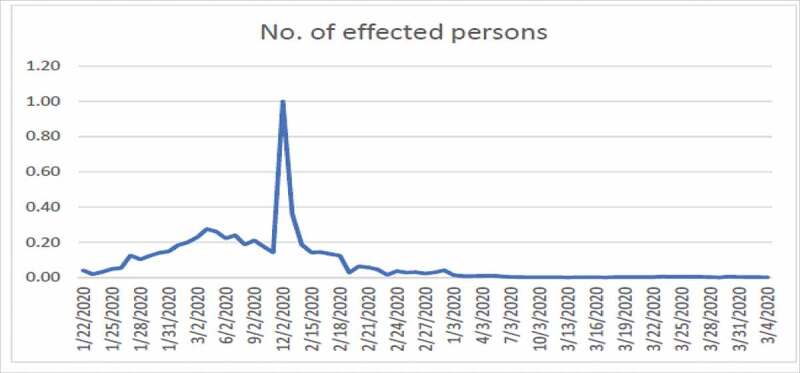
Number of infected person from COVID-19 in China
Figure 6.
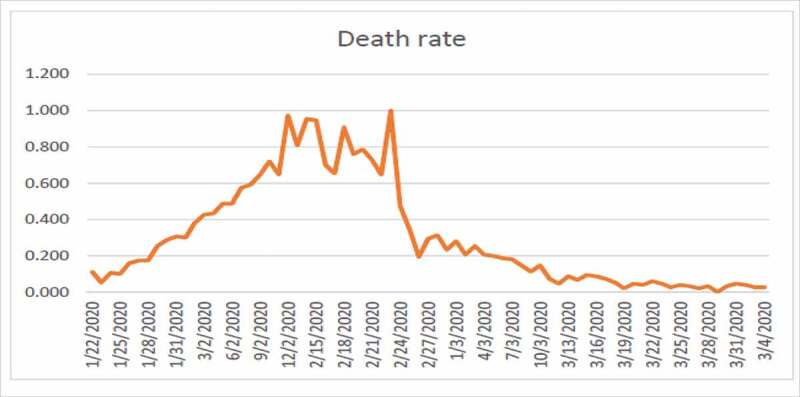
Death rate due to COVID-19 in China
Figure 4 shows high fluctuation in the data hence no one can predict about the new cases in near or far future. Figure 4 shows the combined effects of series 1 and 2. Series 1 indicates the number of affected individuals while series 2 represents mortality rate. From Figure 5, we observe that until 12/02/2020 the infected touched the peak and after that this rate has been decreasing until now which is a positive sign. From Figure 6, we observe that until 24/02/2020 the rate of deaths in China touched the peak and after that this rate keeps decreasing. The graphs are not convex which shows that a lot of uncertainty exists in the spread of COVID-19.
Diagnosis and treatment
Being the novel CoV, many aspects of the SARS-CoV-2 are still unrevealed. Therefore, a fast and accurate diagnostic method is crucial to diagnose the disease on time to prevent its transmission. Currently different methods are being used such as serological testing, chest computed tomography (CT), nucleic acid amplification tests (NAAT), viral sequencing, and viral culture.42 There is no specific antiviral treatment of SARS-CoV-2. Ribavirin with recombinant interferons show limited effects against CoV infection.43 Many researchers have developed different agents after the epidemic of SARS-CoV and MERS-CoV against of their polymerases, entry proteins, and proteases but till now none of them proved to be helpful.44–46 It is proposed that for the treatment of COVID-19 the antibody and plasma of recovering patients can be used.47 In addition, convalescent plasma therapy is an experimental treatment which shows a potential therapeutic effect and low risk in the treatment of severe COVID-19 patients.48 For the treatment of COVID-19 infection, only remdesivir or in combination with interferon beta or chloroquine was found active; this approach has not triggered any significant adverse effects.49–51 To confirm the effects of remdesivir more investigation is necessary. Common therapeutic objectives can be of more significance as CoV shares significant genomic features. The best approach for the treatment of COVID-19 may be therapeutic agents to target the viral nucleic acids, nucleotides, nucleosides, and enzymes which are involved in the transcription and replication of CoV.52
Vaccines
Vaccines are immediately required to control and prevent infrequent viral outbreaks and epidemics which occurred due to newly emerging viruses like the recent outbreak due to SARS-CoV-2. In 2003 the SARS-CoV was controlled, similarly MERS-CoV later as well. The current SARS-CoV-2 pandemic is spreading constantly with a continuous increase in infection and mortality rate. So effective vaccine is required to control SARS-CoV-2. For SARS-CoV, live-attenuated vaccines designed may be assessed for infected individuals as well.49
Many strategies are used for the vaccine development of CoV such as viral vector-based vaccines, inactivated viruses, subunit vaccines, inactivated viruses, recombinant proteins, live-attenuated viruses, and DNA vaccines, and these all were tested in animal models.53,54 Given the conserved RBDs of SARS-CoV and bat SARSr-CoVs, some anti-SARS-CoV strategies in development, such as anti-RBD antibodies or RBD-based vaccines, could be tested against bat SARSr-CoVs. Recent studies demonstrated that anti-SARS-CoV strategies worked against only WIV1 and not SHC014 (rEFs71,88,89). Some strategies are being used to develop vaccine against SARS-CoV and recombinant SARSr-CoV by using the anti-receptor binding domains antibodies or receptor binding domain-based vaccine.43,55,56 The different strategies of vaccine development are presented in Figure 7.
Figure 7.
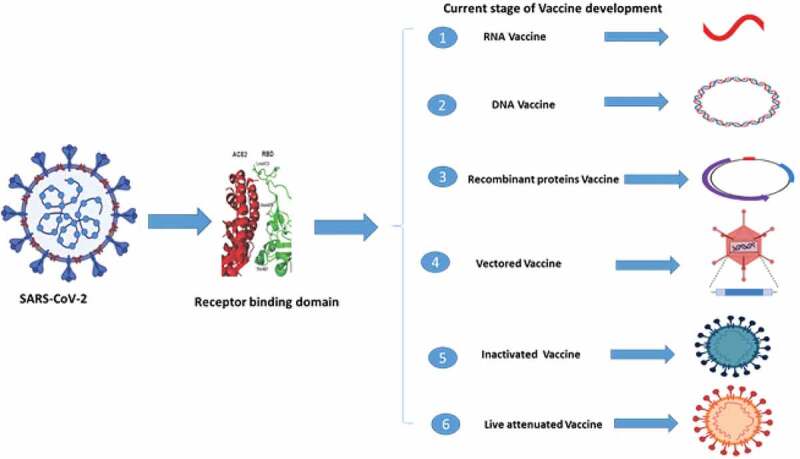
Different strategies of vaccine development against SARS-CoV-2
Moreover, protein cage nanoparticles and rhesus θ-defensin-1 are innate immune-modulators with great anti-SARS-CoV ability.57,58 A protein cage nanoparticle which is prepared for SARS-CoV can be assessed for SARS-CoV-2 based on the SARS-CoV-2 and SARS-CoV higher resemblances and phylogenetic similarity. In the meantime, following the related approaches applied for SARS-CoV, novel protein cage nanoparticles identified for novel CoV can be prepared on an emergency basis. Based on the current situation of COVID-19 pandemic, vaccination approaches based on viral-like particles, recombinant protein, and viral vectors which have been established for MERSS or SARS can be reformed for application against SARS-CoV-2.59
Limitations of study
This study has certain limitations. It discusses the different aspects of COVID-19 with respect to China only, hence data from other regions or countries are not applicable.
Conclusions
Since the start of the COVID-19 pandemic the morbidity and mortality rate keeps increasing with an alarming and uncontrolled conditions worldwide. There has been an imperative need to accelerate the research and development of COVID-19 candidate vaccine (s) and drugs.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at Majmaah University for funding this work. We thank Dr. Kuldeep Dhama (Indian Veterinary Research Institute) for his critical review and valuable suggestions. The authors also acknowledge their respective institutes and universities.
Funding Statement
This work was supported by Deanship of Scientific Research at Majmaah University under project number [RGP-2019-1].
Authors’ contributions
FMK and TA: Study design and wrote the manuscript. TA: Data collection and compilation. MG and WC: Preformed the analysis. TA, MK, and JH: Edited and reviewed the manuscript. All the authors read and approved the final manuscript for publication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Pene F, Merlat A, Vabret A, Rozenberg F, Buzyn A, Dreyfus F, Cariou A, Freymuth F, Lebon P.. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis. 2003;37(7):929–32. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijgen L, Keyaerts E, Moës E, Maes P, Duson G, Van Ranst M.. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human Coronavirus OC43 and 229E. J Clin Microbiol. 2005;43(11):5452–56. doi: 10.1128/JCM.43.11.5452-5456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, McKeever D, Mutua F, Young J, McDermott J, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci U S A. 2013;110(21):8399–404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinese SARS Molecular Epidemiology Consortium . Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–69. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 5.van der Hoek L. Human Coronavirus: what do they cause? Antivir Ther. 2007;12:651–58. [PubMed] [Google Scholar]
- 6.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol. 2012;86(23):12816–25. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S. Surveillance of bat coronavirus in Kenya identifies relatives of human coronavirus NL63 and 229E and their recombination history. J Virol. 2017;91(5):e01953–16. doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman VM, Eckerle I, Memish ZA, Liljander AM, Dijkman R, Jonsdottir H, Juma Ngeiywa KJZ, Kamau E, Younan M, Al Masri M, et al. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci U S A. 2016;113(35):9864–69. doi: 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–58. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh EE, Shin JH, Falsey AR. Clinical impact of human Coronavirus 229E and OC43 infection in diverse adult populations. J Infect Dis. 2013;208(10):1634–42. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorse GJ, O’Connor TZ, Hall SL, Vitale J, Nichol K. Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. J Infect Dis. 2009;199(6):847–57. doi: 10.1086/597122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbour N, Day R, Newcombe J, Talbot PJ. Neuroinvasion by human respiratory Coronavirus. J Virol. 2000;74(19):8913–21. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smuts H. Human coronavirus NL63 infections in infants hospitalised with acute respiratory tract infections in South Africa. Influenza Other Respir Viruses. 2008;2(4):135–38. doi: 10.1111/j.1750-2659.2008.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc Natl Acad Sci USA. 2011;108(1):238–43. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447(7142):279–83. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride R, Fielding BC. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4(11):2902–23. [Published 2012 November7]. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya AE, Snijder EJ, Spaan WJ. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004;78(15):7863–66. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolesnikova L, Slenczka W, Brodt H-R, Klenk H-D, Becker S. Electron microscopy in diagnostics of SARS case. Microsc Microanal. 2003;9(S03):438–39. doi: 10.1017/S1431927603035104. [DOI] [Google Scholar]
- 19.Marsolais G, Berthiaume L, DiFranco E, Marois P. Rapid diagnosis by electron microscopy of avian coronavirus infection. Can J Comp Med. 1971;35:285–88. [PMC free article] [PubMed] [Google Scholar]
- 20.Masters PS. The molecular biology of Coronavirus. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other Coronavirus. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronavirus. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B, Zeng LP, Yang XL, Ge X-Y, Zhang W, Li B, Xie J-Z, Shen X-R, Zhang Y-Z, Wang N, et al. Discovery of a rich gene pool of bat SARS-related Coronavirus provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11):e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song HD, Tu CC, Zhang GW, Wang S-Y, Zheng K, Lei L-C, Chen Q-X, Gao Y-W, Zhou H-Q, Xiang H, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A. 2005;102(7):2430–35. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013;100(1):246–54. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Wong SK, Li F, Kuhn JH, Huang I-C, Choe H, Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. 2006;80(9):4211–19. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia [published correction appears in N Engl J Med. 2013 Jul 25;369(4):394]. N Engl J Med. 2012;367(19):1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 30.Coleman CM, Frieman MB. Coronavirus: important emerging human pathogens. J Virol. 2014;88(10):5209–12. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, Muth D, Raj VS, Vries LSD, Corman VM, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13(10):859–66. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, Godeke G-J, Jonges M, Farag E, Diab A, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14(2):140–45. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus ADME, Haagmans BL, Gorbalenya AE, Snijder EJ, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3(6):e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357(6377):420–22. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wuhan City Health Committee . Wuhan Municipal Health and Health Commission’s briefing on the current pneumonia epidemic situation in our city. 2019. [accessed 8April 2020]. http://wjw.wuhan.gov.cn/front/web/showDetail/2019123108989
- 36.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–73. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coronavirus COVID-19 global cases by the centre for system science and engineering (CSSE) at John Hopkins University. [accessed 9April 2020]. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6
- 39.Ahmad T, Khan M, Khan FM, Hui J. Are we ready for the new fatal coronavirus: scenario of Pakistan? Hum Vaccin Immunother. 2020;16(3):736–38. doi: 10.1080/21645515.2020.1724000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo H, Chen Q, Chen J, Chen K, Shen X, Jiang H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579(12):2623–28. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corman VM, Kallies R, Philipps H, Gopner G, Muller MA, Eckerle I, Brunink S, Drosten C, Drexler JF. Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J Virol. 2014;88(1):717–24. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization . Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Interim guidance; 2020. March 2 [accessed 1July 2020]. https://apps.who.int/iris/bitstream/handle/10665/331329/WHO-COVID-19-laboratory-2020.4-eng.pdf
- 43.Menachery VD, Yount BL Jr, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, et al. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci USA. 2016;113(11):3048–53. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan JF, Chan KH, Kao RY, To KKW, Zheng B-J, Li CPY, Li PTW, Dai J, Mok FKY, Chen H, et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect. 2013;67(6):606–16. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng KW, Cheng SC, Chen WY, Lin M-H, Chuang S-J, Cheng I-H, Sun C-Y, Chou C-Y. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Sun Y, Wu A, Xu S, Pan R, Zeng C, Jin X, Ge X, Shi Z, Ahola T, et al. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J Virol. 2015;89(16):8416–27. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mair-Jenkins J, Saavedra-Campos M, Baillie JK, Cleary P, Khaw F-M, Lim WS, Makki S, Rooney KD, Nguyen-Van-Tam JS, Beck CR, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211(1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020;117(17):9490–96. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–71. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–36. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronavirus - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–47. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging Coronavirus. Nat Rev Microbiol. 2013;11(12):836–48. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronavirus. Nat Rev Microbiol. 2016;14(8):523–34. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng LP, Ge XY, Peng C, Tai W, Jiang S, Du L, Shi Z-L. Cross-neutralization of SARS coronavirus-specific antibodies against bat SARS-like Coronavirus. Sci China Life Sci. 2017;60(12):1399–402. doi: 10.1007/s11427-017-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menachery VD, Yount BL Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge X-Y, Donaldson EF, et al. A SARS-like cluster of circulating bat Coronavirus shows potential for human emergence [published correction appears in Nat Med. 2016 Apr;22(4):446]. Nat Med. 2015;21(12):1508–13. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wohlford-Lenane CL, Meyerholz DK, Perlman S, Zhou H, Tran D, Selsted ME, McCray PB. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J Virol. 2009;83(21):11385–90. doi: 10.1128/JVI.01363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiley JA, Richert LE, Swain SD, Harmsen A, Barnard DL, Randall TD, Jutila M, Douglas T, Broomell C, Young M, et al. Inducible bronchus-associated lymphoid tissue elicited by a protein cage nanoparticle enhances protection in mice against diverse respiratory viruses. PLoS One. 2009;4(9):e7142. doi: 10.1371/journal.pone.0007142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mubarak A, Alturaiki W, Hemida MG. Middle East respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J Immunol Res. 2019;2019:6491738. [Published 2019 April7]. doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]


