Summary
The term “cavernous hemangioma” has been used to describe vascular anomalies with histology featuring dilated vascular spaces, vessel walls consisting mainly of fibrous stromal bands lined by a layer of flattened endothelial cells, and an irregular outer rim of interrupted smooth muscle cells. Hepatic hemangiomas (HHs) and cutaneous venous malformations (VMs) share this histologic pattern, and we examined lesions in both tissues to identify genetic drivers. Paired whole-exome sequencing (WES) of lesional tissue and normal liver in HH subjects revealed a recurrent GJA4 c.121G>T (p.Gly41Cys) somatic mutation in four of five unrelated individuals, and targeted sequencing in paired tissue from 9 additional HH individuals identified the same mutation in 8. In cutaneous lesions, paired targeted sequencing in 5 VMs and normal epidermis found the same GJA4 c.121G>T (p.Gly41Cys) somatic mutation in three. GJA4 encodes gap junction protein alpha 4, also called connexin 37 (Cx37), and the p.Gly41Cys mutation falls within the first transmembrane domain at a residue highly conserved among vertebrates. We interrogated the impact of the Cx37 mutant via lentiviral transduction of primary human endothelial cells. We found that the mutant induced changes in cell morphology and activated serum/glucocorticoid-regulated kinase 1 (SGK1), a serine/threonine kinase known to regulate cell proliferation and apoptosis, via non-canonical activation. Treatment with spironolactone, an inhibitor of angiogenesis, suppressed mutant SGK1 activation and reversed changes in cell morphology. These findings identify a recurrent somatic GJA4 c.121G>T mutation as a driver of hepatic and cutaneous VMs, revealing a new pathway for vascular anomalies, with spironolactone a potential pathogenesis-based therapy.
The authors find that a recurrent somatic GJA4 mutation causes vascular malformations of the liver and skin. Studies in endothelial cells show that mutant expression alters cell morphology and leads to non-canonical activation of SGK1. Both are reversed when cells are exposed to spironolactone, revealing a potential pathogenesis-based therapy.
Introduction
Drivers of vascular overgrowth have been revealed by genetic investigation of lesions in the skin and other tissues, and these findings have informed understanding of vascular biology. Included among these discoveries are mutations in vascular tumors including BRAF (MIM: 194757), VEGFR2 (MIM: 191306), VEGFR3 (MIM: 136352), HRAS (MIM: 190020), KRAS (MIM: 190070), NRAS (MIM: 164790), GNAQ (MIM: 600998), GNA11 (MIM: 139313), and GNA14 (MIM: 604397), which center on RAS-MEK-ERK signaling.1, 2, 3, 4, 5, 6, 7, 8, 9, 10 Somatic activating mutations in PIK3CA (MIM: 171834) and in TEK (MIM: 600221) are also known to drive some cutaneous venous malformations (VMs) (MIM: 600195).11,12 In a recent study, 14 of 39 HH lesions (36%) were found via targeted sequencing to harbor canonical KRAS (G12D, G12S, G13D, or G13S) or BRAF (V600M) mutations, but many cases in the skin and liver remain mutation-unknown, and we sought to identify other genetic causes.13
Hepatic hemangiomas (HHs) have a reported incidence in autopsy series ranging from 1%–7%.14 They are thought to derive from mesenchyme and may occur as solitary or multiple lesions. They are five times more common in women than in men and are commonly diagnosed between ages 30 and 50 years.15 Lesions <10 cm in size are usually asymptomatic and are typically found incidentally, while larger lesions of >10 cm can cause abdominal pain due to distension of the liver capsule, a sensation of abdominal fullness, coagulopathy, Budd-Chiari syndrome (MIM: 600880), or signs of gastric outlet or biliary obstruction.16 A rare, but serious, complication of these lesions is spontaneous rupture, which can lead to intralesional or intra-abdominal hemorrhage, anemia, circulatory shock, and death. In individuals with current or prior malignancy at other sites, HHs are often removed to exclude the diagnosis of liver metastases. Current treatments for HH include interferon alpha, corticosteroid therapy, embolization, and surgery. Surgery is the most definitive treatment for HHs, but post-surgical complications occur in about 25% of adults, with intraoperative bleeding secondary to thrombocytopenia one potential serious complication.16,17
Cutaneous VMs share histologic features with HH, and these lesions can be present at birth or can appear spontaneously in adults and grow over time. Cutaneous VMs can be large, disfiguring, and painful and can undergo localized intravascular coagulopathy. Cutaneous VMs can be found concurrent with visceral vascular anomalies, including HH.18
Syndromes featuring multi-system vascular anomalies include von Hippel Lindau syndrome (VHLS; MIM: 193300) and tuberous sclerosis complex due to mutations in TSC1 (MIM: 191100) and TSC2 (MIM: 613254). VHLS is a rare cancer syndrome caused by mutations in the VHL (MIM: 608537) tumor suppressor gene, which is characterized by the presence of cysts and vascular lesions in multiple organs. A conditional heterozygous inactivation of Vhl in mouse hepatocytes leads to VMs of the liver.19 Similarly, mice with only one functional Tsc2 allele develop hepatic vascular lesions.20 Despite these discoveries in humans and mice, our understanding of the full spectrum of genetic determinants of vascular malformation remains incomplete, and we employed paired whole-exome sequencing (WES) to identify, a priori, the genetic drivers of these VMs in the liver and skin.
Material and methods
DNA extraction, sequencing, and bioinformatic analyses
We selected a total of 14 HH cases in individuals with symptoms that necessitated surgery and 18 cutaneous VM individuals for genetic analysis. The study protocol was approved by the Yale Human Investigation Committee. Lesional and unaffected tissues in all cases were cored from paraffin blocks and genomic DNA extracted with the QIAamp DNA FFPE Tissue Kit (QIAGEN).
WES was performed by the Yale Center for Genome Analysis, using barcoded libraries from sheared genomic DNA. We used the Burrows-Wheeler Aligner (BWA-MEM) to align reads to the hg19 human reference sequence (UCSC Genome Browser) and trimmed them to targeted intervals with Picard.21,22 We employed a Perl script to remove PCR duplicates, recalibrated the resulting BAM files according to the Genome Analysis Toolkit (GATK) Best Practices, and called blood and tissue variants with the GATK Haplotype Caller.22 We used MuTect2 to identify all single-nucleotide variants (SNVs), deletions, and insertions and used ANNOVAR to annotate all variants for functional impact.23,24 Data were filtered for damaging (indels, missense, splice site, and nonsense) mutations that (1) had at least three non-reference reads in lesional tissue and zero, one, or two non-reference reads in unaffected tissue; (2) were present on the left and right strands; and (3) were absent in 1000 Genomes (August 2015 release), the National Heart, Lung, and Blood Institute Exome Sequencing Project Exome Variant Server (ESP6500 release), and the Genome Aggregation Database (gnomAD) Browser control set of >125,000 exomes and >15,000 genomes (v.2.1.1). We examined the remaining mutations with the Broad Institute Integrative Genomics Viewer (IGV) to exclude variants resulting from alignment error.25 GJA4 c.121G>T somatic mutations were verified via Sanger sequencing using the following primers: connexin 37 (Cx37)-forward (5′-AGGTCCAGGAGCACTCGAC-3′) and Cx37-reverse (5′-CAGCACCCAGTAGCGGATG-3′).
Mutant plasmid construction and transfection
GJA4 cDNA (Dharmacon) was cloned into pInducer21 (Addgene) using the Gateway Cloning System (Thermo Fisher Scientific) with forward (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGCCACCATGGGT-3′) and reverse (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACATACTGCTT-3′) primers generating the Cx37-wild type (WT) construct. The mutant construct (Cx37-Gly41Cys) was generated from the Cx37-WT pInducer21 vector using the QuikChange Site-Directed Mutagenesis Kit (Agilent). Hemagglutinin (HA)-tagged constructs were cloned from the nontagged constructs by excluding the TAG stop codon at the end of the respective nontagged cDNA sequences. Lentiviral particles were packaged in HEK293T cells using 1.25 μg vector DNA, 1.25 μg packaging plasmid psPAX2 (Addgene), and 0.5 μg envelope expressing plasmid pMD2.G (Addgene) per well in a 6-well plate with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Lentiviral particles were collected 48 and 72 h after transfection, combined, and concentrated using Lenti-X concentrator (TakaraBio) per manufacturer’s instructions. Human umbilical vein endothelial cells (HUVECs) were transduced using Polybrene (Millipore Sigma) at a concentration of 5 μg/mL. After 72 h, transduction efficiency as evaluated via GFP fluorescence was >80% for all lentiviral constructs. Ectopic gene expression was induced with doxycycline at a concentration of 1 μg/mL unless otherwise indicated.
Western blotting
Protein was isolated from passage 5 HUVECs 18 h after induction of gene expression in 1% SDS lysis buffer containing protease and phosphatase inhibitor. 10 or 20 μg of protein was run on a NuPAGE 4%–12% precast Bis-Tris gel (Invitrogen) and transferred onto nitrocellulose membranes using a semi-dry (Bio-Rad) or a wet transfer method. Anti-human antibodies used are as follows: rabbit anti-phospho-ERK p42/p44 (Cell Signaling Technologies [CST] 9106) 1:1,000, mouse anti-ERK (CST 9102) 1:1,000, rabbit anti-Akt (CST 9272) 1:1,000, rabbit anti-phospho-Akt-T308 (CST 4056), rabbit anti-phospho-Akt-S473 (CST 4051), rabbit anti-serum/glucocorticoid-regulated kinase 1 (SGK1) (CST 12103) 1:1,000, rabbit anti-phospho-SGK1-Ser78 (CST 5599) 1:1,000, anti-phospho-SGK1-Thr256 (Thermo Fisher 44-1260G) 1:1,000, rabbit anti-Cx37 (Abcam 181701) 1:3,000, rabbit anti-LC3A/B (CST 4108) 1:1,000, rabbit anti-ULK1 (CST 8054) 1:1,000, rabbit anti-Atg13 (CST 13273) 1:1,000, rabbit anti-Atg101 (CST 13492) 1:1,000, rabbit anti-RB1CC1 (CST 12436) 1:1,000, rabbit anti-HA (CST 3724) 1:1,000, Actin (Sigma Aldrich A1978) 1:4,000. The following secondary antibodies were used: anti-mouse HRP-linked antibody (CST 7076) 1:2,500 or anti-rabbit HRP-linked antibody (CST 7074) 1:2,500. Quantification of western blots was performed using ImageJ (v1.51).
Cell culture
HUVECs from the Yale Vascular Biology and Therapeutics Core were maintained in endothelial cell growth medium (EGM-2; Lonza). They were transduced with lentiviral constructs containing WT (Cx37-WT), mutant (Cx37-Gly41Cys), HA-tagged WT (HA-Cx37-WT), or HA-tagged mutant (HA-Cx37-Gly41Cys) lentiviral constructs. Where applicable, cells were treated with 50 μM spironolactone (Sigma) or 1 μM tamoxifen (Sigma) in DMSO. Control cells were treated with EGM-2 in an identical amount of DMSO as those in the drug treatments.
Results
A recurrent, somatic GJA4 c.121G>T mutation is present in hepatic and cutaneous VMs
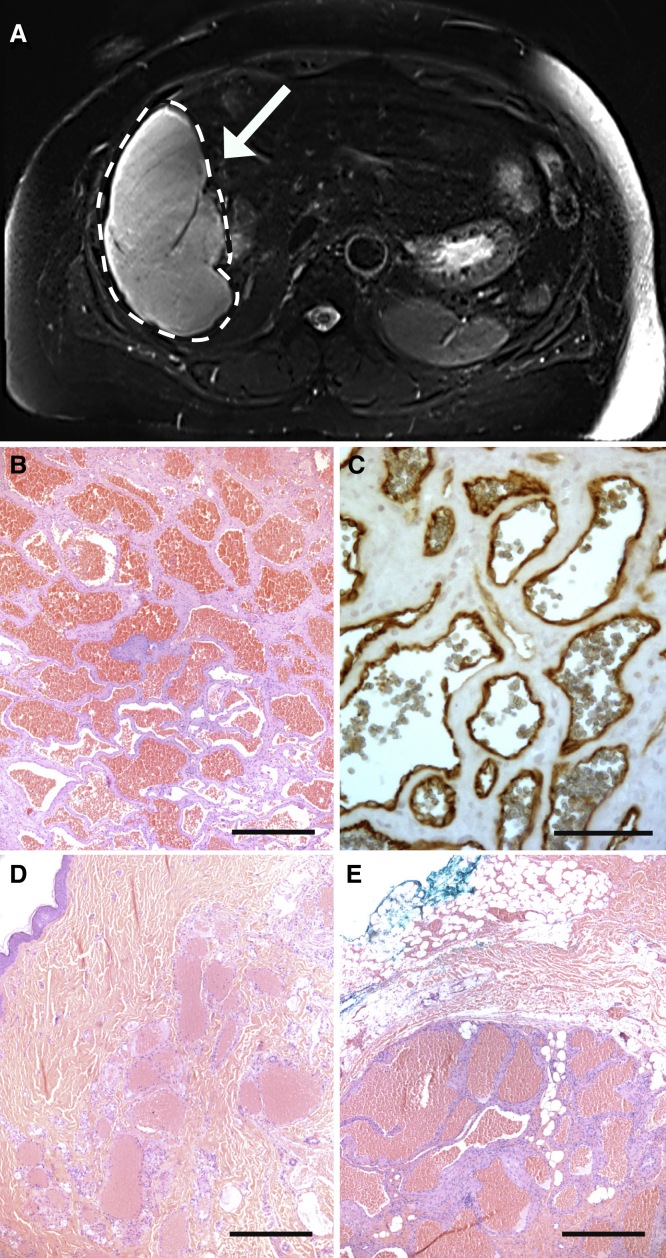
We initially enrolled five unrelated individuals with large, symptomatic HHs in whom diagnosis was confirmed by magnetic resonance imaging showing hyperintense liver masses on T2-weighted sequence (Figure 1; Figure S1). Histologic examination of excised tissue showed dilated vessels lined with CD31-positive endothelial cells (Figure 1). WES identified a recurrent GJA4 (MIM: 121012) (GenBank: NM_002060.3) c.121G>T (p.Gly41Cys) somatic mutation in tissue from 4 out of the 5 HH individuals (80%), confirmed via Sanger sequencing, and absent from 251,000 sequenced alleles in the gnomAD database (Table S1; Figure S2). After verification of somatic variants from Mutect in IGV, the total number of somatic SNVs (sSNVs) observed in each of the four individuals ranged from 11 to 68 (Table S2). Mutations in KRAS or BRAF were not detected, and GJA4 c.121G>T was the only shared mutation among the HH individuals. We also performed targeted sequencing of paired normal and lesional tissue from 9 additional HH individuals (Table 1), finding the GJA4 c.121G>T mutation in 8 of the 9 individuals (89%) (Figure S3). Recognizing shared histology of HH lesions and cutaneous VMs, we assessed cutaneous lesions for this mutation.26,27 Paired Sanger sequencing of normal and affected tissue from 5 subjects with cutaneous VMs with cavernous histology showed the presence of the GJA4 c.121G>T mutation in 3 individuals (60%) (Figures 1D and 1E; Figure S3). The mutation was absent in 12 additional cutaneous VMs that demonstrated other histologic patterns, such as a single, large vascular space lined by endothelial cells or compressed vessels lined by a single layer of endothelial cells dissecting through collagen bundles. Unlike the cavernous lesions associated with the GJA4 c.121G>T mutation, VMs due to TEK mutations are characterized by large vascular spaces with irregular borders lined by endothelial cells, which are in turn bordered by disorganized smooth muscle.11,28 Clinically, VMs due to TEK mutations tend to occur in the subcutis or deeper, whereas cutaneous VMs may occur near the skin surface.11,29
Figure 1.
Shared features of HHs and cutaneous venous malformations
(A) A 16 × 11 × 16 cm vascular mass (white dashed lines and white arrow) in the right liver lobe consistent with HH found in VASC102.
(B) Histology of this lesion demonstrates dilated vessels filled with red blood cells. Scale bar, 500 μm.
(C) Immunohistochemistry (IHC) demonstrating CD31 immunoreactivity in endothelial cells lining vessels confirms that these are vascular lesions. Scale bar, 75 μm.
(D and E) Cutaneous venous malformations show similar histology with dilated vascular spaces and vessel walls consisting mainly of fibrous stromal bands lined by a single layer of flattened endothelial cells. Scale bar, 500 μm.
Table 1.
A somatic mutation in GJA4 causes hepatic hemangiomas and cutaneous venous malformations
| Sample | Age at diagnosis | Sex | Lesion size | Lesion location | Somatic mutation |
|---|---|---|---|---|---|
| VASC101 | 61 | F | 12.0 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC102 | 47 | F | 11.1 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC103 | 34 | F | 16.0 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC104 | 55 | F | 12.6 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC105 | 36 | F | 8.4 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC106 | 55 | F | 16.9 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC107 | 44 | M | 12.0 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC108 | 64 | M | 0.8 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC109 | 36 | F | 13.5 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC110 | 50 | M | 1.5 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC111 | 40 | F | 7.3 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC112 | 48 | F | 5.7 cm | hepatic | GJA4 c.121G>T (p.Gly41Cys) |
| VASC113 | 49 | M | 0.5 cm | cutaneous | GJA4 c.121G>T (p.Gly41Cys) |
| VASC114 | 66 | M | 1.4 cm | cutaneous | GJA4 c.121G>T (p.Gly41Cys) |
| VASC115 | 53 | M | 0.5 cm | cutaneous | GJA4 c.121G>T (p.Gly41Cys) |
Lesional and unaffected tissue from 4 HH individuals (VASC101–VASC104) was subjected to paired WES, revealing a recurrent somatic GJA4 (GenBank: NM_002060.3) c.121G>T (p.Gly41Cys) mutation. Variant allele frequencies for these four individuals are listed in Table S1. Eight additional HH individuals (VASC105–VASC112) and three cutaneous venous malformation individuals (VASC113–VASC115) from male and female adults also harbored the somatic GJA4 c.121G>T mutation. Ages are in years. Abbreviations: F, female; M, male.
GJA4 encodes gap junction protein alpha 4, also called Cx37, which is highly expressed in lung, vasculature, and the ovary.30, 31, 32 There are 21 members of the connexin gene family in humans; all encode tetra-transmembrane-spanning proteins with one cytoplasmic and two extracellular loops, which assemble into connexons comprised of six connexin subunits, with homo- or hetero-polymerization possible.33,34 Connexons can form hemichannels on the cell surface or dock with connexons from adjacent cells to form a gap junction channel permeable to ions and macromolecules.35 Connexins have relatively short half-lives (1.5–5 h) compared to other cell membrane proteins and have central roles in intercellular ion flux and intracellular and intercellular signaling and have been implicated in regulation of cell death, proliferation/survival, and differentiation.36, 37, 38 The identified GJA4 c.121G>T (p.Gly41Cys) mutation occurs within the first transmembrane-spanning domain, which is highly conserved among higher-order vertebrates (Figures S4 and S5) and was predicted to be damaging by SIFT, PolyPhen2, and MutationTaster.39, 40, 41
Cx37-Gly41Cys expression induces vacuole formation and inhibits autophagosome processing in HUVECs
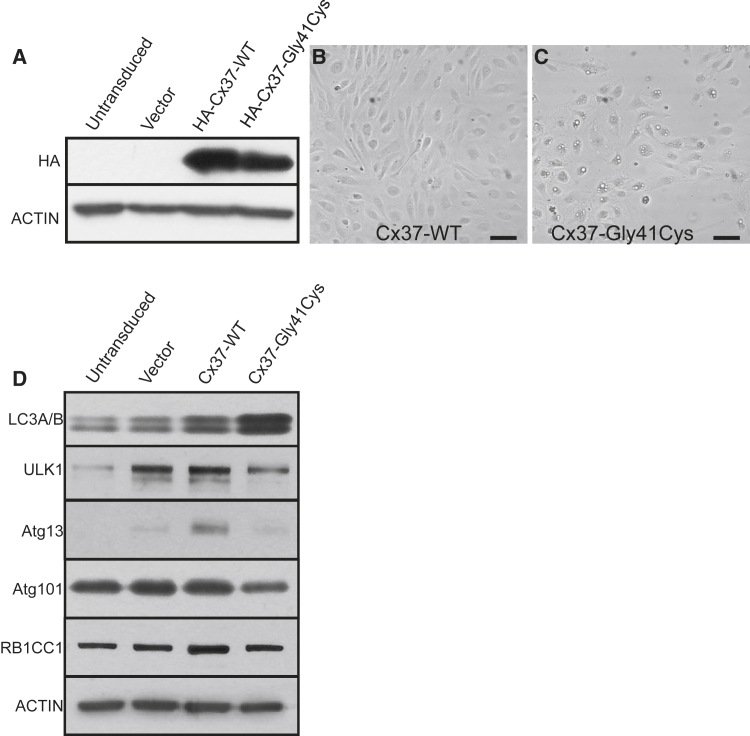
To investigate the pathobiology of the GJA4 c.121G>T (p.Gly41Cys) mutation, we expressed native or HA-tagged Cx37-WT or mutant Cx37 (Cx37-Gly41Cys) in HUVEC cells cloned in tetracycline-regulated lentiviral vectors. These were induced with doxycycline and expression verified via western blotting (Figure 2A; Figure S6). Eight hours after induction of ectopic gene expression, cells expressing Cx37-Gly41Cys exhibited large cytoplasmic vacuoles and spindled morphology, absent in Cx37-WT-expressing cells (Figures 2B and 2C). An oil red O stain for lipids was negative. Via western blotting, we interrogated microtubule-associated proteins MAP1-LC3A and MAP1-LC3B (LC3A/B), which are markers of autophagosome formation, and found increased levels in cells transduced with the Cx37-Gly41Cys construct.42 Quantitation with ImageJ showed a less significant increase in LC3A/B protein levels in cells expressing Cx37-WT compared to those expressing the vector alone (Figure S7). To determine whether the elevated LC3A/B levels were secondary to an upregulation in autophagosome initiation, we blotted for proteins in the autophagy initiation complex comprising ULK1, Atg101, Atg13, and the ULK1 interacting protein RB1CC1, which are negatively regulated by mTOR.43 We found a relative decrease in levels of ULK, Atg101, Atg13, and RB1CC1 in cells expressing Cx37-Gly41Cys compared to cells expressing Cx37-WT (Figure 2). These results suggest that autophagy initiation was downregulated in cells expressing the mutant Cx37 protein. The selective increases in Atg13 and RB1CC1 seen in cells expressing Cx37-WT suggest that the increase in LC3A/B expression levels seen in these cells could derive from an increase in autophagic processing (Figure S7). Altogether, elevated LC3A/B levels and the cytoplasmic vacuoles in cells transduced with Cx37-Gly41Cys suggest a distal blockade in autophagic processing.
Figure 2.
Cx37-Gly41Cys expression induces morphological changes in HUVECs and downregulates expression of proteins comprising the autophagy initiation complex in endothelial cells
(A) Overexpression of HA-tagged Cx37-WT and Cx37-Gly41Cys was verified via western blotting. Actin is blotted as a loading control.
(B) HUVECs expressing Cx37-WT demonstrate a cobblestone morphology. Scale bar, 150 μm.
(C) HUVECs expressing Cx37-Gly41Cys exhibit large cytoplasmic vacuoles and spindled morphology. Scale bar, 150 μm.
(D) Expression of Cx37-Gly41Cys leads to increased levels of the autophagosome markers LC3A/B (LC3A/B). Compared to Cx37-WT, Cx37-Gly41Cys leads to decreased expression of proteins comprising the autophagy initiation complex. Taken together, these data suggest that Cx37-Gly41Cys inhibits autophagosome initiation or processing. Actin is blotted as a loading control.
Cx37-Gly41Cys overexpression does not activate the MAPK pathway or the PI3K-Akt-mTOR axis
Other mutations causing VMs or vascular tumors drive MAPK or PI3K/Akt/mTOR activation, and we interrogated these pathways in HUVECs expressing Cx37-WT and Cx37-Gly41Cys.5,12,44 Western blotting did not show elevation in phospho-ERK1/ERK2 protein levels in cells expressing Cx37-Gly41Cys compared to cells expressing Cx37-WT protein, suggesting that the mutation was not acting through activation of the MAPK pathway (Figure 3A). Recognizing that the PI3K/Akt/mTOR pathway can also contribute to cell survival, growth, and proliferation, we assessed levels of Akt, which mediates effects of the pathway on downstream targets. PI3K activation leads to PDK1-driven phosphorylation of Akt at Thr308, and mTORC2 phosphorylation of Akt at Ser473 is required for full kinase activation.45,46 We found that protein levels of total Akt and Akt-Thr308 were comparable between cells expressing the WT and mutant proteins, whereas Akt-Ser473 levels were decreased in cells expressing Cx37-Gly41Cys compared to cells expressing Cx37-WT protein (Figure 3A). Taken together, these results suggest that the mutant protein does not lead to PI3K- or mTOR-dependent Akt activation.
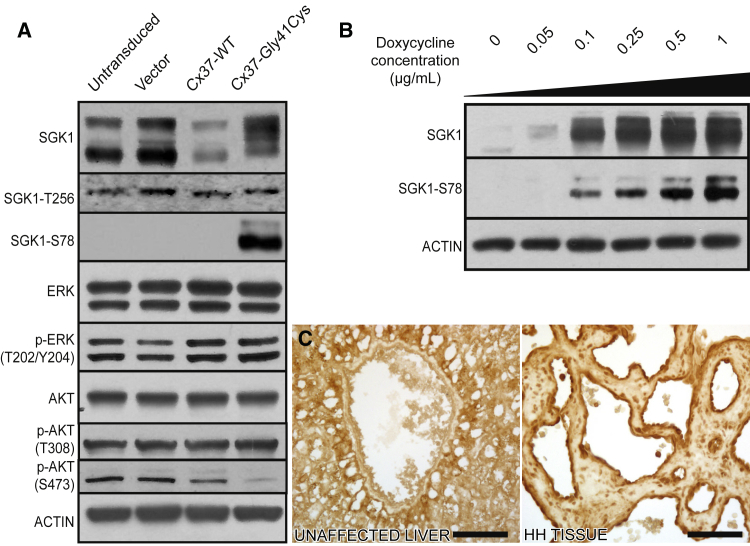
Figure 3.
Cx37-Gly41Cys expression activates SGK1 via a non-canonical pathway
(A) HUVECs transduced with Cx37-G41C express a higher molecular weight phosphorylated form of SGK1, with expression of SGK1-Ser78 seen exclusively in Cx37-Gly41Cys cells. Cx37-Gly41Cys does not alter MAPK expression. Cx37-WT leads to decreased Akt-Ser473 expression, whereas Cx37-Gly41Cys expression results in almost complete abrogation. Actin is blotted as a loading control.
(B) Gradient induction of Cx37-Gly41Cys expression with doxycycline leads to dose-dependent increases in phosphorylation of SGK1 at serine 78 and an increase in total SGK1 expression. In the absence of Cx37-Gly41Cys expression, SGK1 runs as a doublet on SDS-PAGE (lane 1) but migrates as a single band when expression of the mutant protein is induced. These data indicate that Cx37-Gly41Cys leads to SGK1 activation via serine 78 phosphorylation. Actin is blotted as a loading control.
(C) Immunohistochemical staining reveals absence of SGK1-Ser78 in vasculature of normal liver tissue and strong SGK1-Ser78 expression in HH tissue. Scale bars, 75 μm.
Cx37-Gly41Cys upregulates expression of SGK1
Given the absence of MAPK or Akt activation in cells expressing Cx37-Gly41Cys, we examined other downstream mTORC2 targets that have been implicated in cellular overgrowth. One such target is SGK1, a member of the AGC kinase family, which also includes Akt. SGK1 shares 55% identity with Akt, and both proteins are activated by the same kinases. Phosphorylation of Akt at Thr308 and SGK1 at Thr256 by PI3K-directed PDK1 activity leads to full kinase activation.47 SGK1 can also be activated in a PI3K-independent fashion by ERK5 or p38-mediated phosphorylation at serine 78 (SGK1-Ser78).48,49 Western blotting showed similar levels of SGK1-Thr256 in cells expressing the WT and mutant proteins, suggesting that Cx37-Gly41Cys does not cause PI3K-driven canonical activation of SGK1 (Figure 3A). However, SGK1-Ser78 expression was seen only in cells expressing Cx37-Gly41Cys and in a titratable fashion with increasing doxycycline concentration, supporting non-canonical activation of SGK1 (Figure 3B). Immunohistochemical staining confirmed exclusive SGK1-Ser78 expression in HH, but not unaffected tissue (Figure 3C).
Spironolactone, but not tamoxifen, inhibits the effects of Cx37-Gly41Cys expression in HUVECs
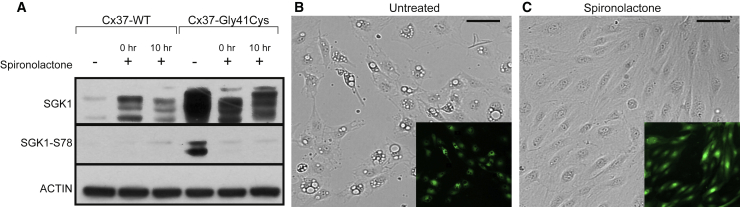
The mineralocorticoid receptor (MR) antagonist spironolactone demonstrates anti-angiogenic properties by inhibiting endothelial cell migration and proliferation.50,51 We therefore evaluated its effect on HUVECs overexpressing Cx37-Gly41Cys. HUVECs expressing Cx37-WT or Cx37-Gly41Cys were exposed to one of three conditions: no spironolactone, spironolactone added at induction of ectopic gene expression (0 h), and spironolactone added 10 h after induction of ectopic gene expression (10 h). The latter condition was selected given that tetracycline-on inducible systems reach maximum gene expression levels at 10 h post-induction.52 Spironolactone was added to the cell culture media at 50 μg/mL, a concentration previously shown to inhibit HUVEC proliferation.51 While western blotting showed strong SGK1-Ser78 expression in Cx37-Gly41Cys cells not exposed to spironolactone, the addition of spironolactone at the time of doxycycline induction or 10 h later completely abrogated SGK1-Ser78 expression, the hallmark of non-canonical activation (Figure 4A). Corroborating these findings, HUVECs expressing Cx37-Gly41Cys that were treated with spironolactone at the time of doxycycline induction did not exhibit vacuoles or other morphological changes compared to untreated cells. Furthermore, spironolactone administration at 10 h after doxycycline induction reversed the vacuolization phenotype in cells expressing Cx37-Gly41Cys within 14 h after treatment (Figures 4B and 4C).
Figure 4.
Spironolactone inhibits and reverses SGK1 non-canonical activation and morphological changes in HUVECS expressing Cx37-Gly41Cys
(A) SGK1 activation via phosphorylation at serine 78 is seen in cells overexpressing Cx37-Gly41Cys in the absence of spironolactone but is not present with addition of spironolactone at the time of induction of Cx37-Gly41Cys expression or 10 h after. Lysates were prepared at 18 h.
(B and C) Cytoplasmic vacuoles are seen in Cx37-Gly41Cys HUVECs that are not treated with spironolactone (B), but addition of spironolactone reverses this phenotype by 14 h after treatment (C). Images were taken at 24 h after induction of Cx37-Gly41Cys expression. Scale bars, 150 μm.
Given the higher prevalence of HHs in women, their pathogenesis has been postulated to involve female sex hormones such as estrogen.53 HUVECs express estrogen receptors, and the effects of estrogen on HUVECs are mediated by estrogen receptor β, activation of which upregulates SGK1 expression and leads to endometrial cell survival.54, 55, 56 To investigate whether blocking estrogen receptor β activity alters the effects of Cx37-Gly41Cys, we treated HUVECs with tamoxifen, a pure antagonist of estrogen receptor β. HUVECs expressing Cx37-WT or Cx37-Gly41Cys were exposed to 1 μmol/mL tamoxifen, which is equivalent to the therapeutic serum concentration in individuals with breast cancer and above the concentrations demonstrated to inhibit estrogen receptor β in endothelial cells.54,57 SGK1-Ser78 levels were unchanged in cells exposed to tamoxifen compared to control cells (Figure S8), showing that non-canonical SGK1 activation is independent of estrogen receptor β activity.
Discussion
Cx37 is expressed in the endothelium of the skin, hepatic artery, hepatic vein, and portal vein, and studies have demonstrated its role in vascular development and cell cycle control.58, 59, 60 Mice lacking both Cx37 and GJA5 (connexin 40) die perinatally after developing vascular abnormalities in lung, intestine, and skin tissue as well as lesions resembling hemangiomas in testis.60 In a mouse model of limb ischemia, Cx37 deficiency was associated with increased angiogenesis, suggesting an anti-proliferative role for Cx37 on vasculature.61 Cx37 also exhibits a growth-suppressive effect on rat insulinoma cells, a property that requires intact pore-forming and C-terminal Cx37 domains.62,63 Nonfunctional channels generated via missense mutations in the Cx37 pore-forming domains coupled with a WT c terminus that localizes to intercellular junctions lack this growth-inhibitory effect. These data suggest that the GJA4 c.121G>T (p.Gly41Cys) mutation, which falls in the first transmembrane domain, may impact the pore function of Cx37 and act, at least in part, via a dominant negative mechanism.64, 65, 66, 67, 68
We show that Cx37-Gly41Cys expression leads to non-canonical activation of SGK1 independent of PI3K. SGK1 activation has been extensively found upregulated in neoplasms, including non-small cell lung cancer, glioblastoma, hepatocellular carcinoma, and breast and colorectal cancer.69, 70, 71, 72, 73, 74 SGK1 is also required for normal vasculogenesis in developing mouse embryos, where it plays a pro-survival role in endothelial and vascular smooth muscle cells.75 SGK1-deficient mouse endothelial cells exhibit diminished migration and tube formation in vitro.76 Furthermore, SGK1 enhances cell-cycle progression in the presence of growth factors or serum.77
Recognizing that SGK1 may undergo canonical or non-canonical activation by phosphorylation at different residues, we examined levels of both phosphorylated forms. The increased levels of SGK1-Ser78, but not SGK1-Thr256, in cells expressing Cx37-Gly41Cys indicate that the identified mutation causes non-canonical SGK1 activation without altering PI3K-directed activation. Alternative activation pathways for AGC kinases are not unique to SGK1, as PI3K-independent Akt activation by ERK4 has also been described.78 The decreased levels of Akt-Ser473 and unchanged levels of Akt-Thr308 in cells expressing the mutant protein provide additional evidence for the absence of canonical PI3K/Akt/mTOR pathway activation as the primary driver of SGK1 activation.
We noted large vesicles in the cytoplasm of cells expressing Cx37-Gly41Cys. The relatively increased levels of the autophagosome markers LC3A/B coupled with the decreased levels of proteins that comprise the autophagy initiation complex suggest the possibility that the large and abundant vacuoles in cells expressing Cx37-Gly41Cys may be an accumulation of stalled autophagosomes. These findings are consistent with evidence shown by others demonstrating that SGK1 inhibits autophagy.79,80 Whether this autophagic block contributes to HH development is unknown. However, autophagy inhibition can lead to increased cell proliferation in vivo, as has been demonstrated in colorectal cancer cells.81 We also find an increase in LC3A/B in cells expressing Cx37-WT coupled with increased levels of proteins comprising the autophagy initiation complex, suggesting an upregulation of autophagic processing. This may be a consequence of Cx37-WT overexpression, as autophagy has a described role in intracellular protein degradation.82,83
Finally, we demonstrate that the MR antagonist spironolactone abrogates Cx37-Gly41Cys-induced SGK1 phosphorylation at serine 78 in HUVECs. Formation of the large cytoplasmic vacuoles seen in cells expressing the mutant protein is also inhibited or reversed by spironolactone treatment (Figure 4). This inhibitory effect persists in cells that are maximally expressing the mutant protein prior to drug administration, suggesting that spironolactone could be an effective therapeutic for established lesions. The modulation of these morphologic and molecular changes by a single agent further suggests that non-canonical SGK1 activation and autophagy inhibition are linked. While the MR agonist aldosterone potently induces SGK1 expression, others have shown that inhibition of HUVEC proliferation by spironolactone is not dependent upon MR blockade.51,84 The mechanism underlying this anti-proliferative effect has not been described. Our results implicate SGK1 in the relevant mechanism, although the point at which spironolactone may be modulating the Cx37-SGK1 axis has yet to be determined.
Our finding of a recurrent somatic GJA4 c.121G>T (p.Gly41Cys) mutation in hepatic and cutaneous VMs in 15 unrelated individuals, combined with evidence that the mutant protein induces non-canonical SGK1 activation, identifies a new pathway for vascular anomalies and links Cx37 to SGK1 regulation. We demonstrate that spironolactone blocks and reverses both non-canonical SGK1 activation and ensuing morphologic changes. While surgery has been considered the most definitive treatment for HHs, a therapeutic targeting the Cx37-SGK1 axis may serve as a treatment alternative for individuals with HHs or cutaneous VMs harboring the Cx37 p.Gly41Cys mutation.
Acknowledgments
We thank the study subjects, their families, and the health care professionals whose participation made this work possible. We thank Jing Zhou, Ronghua Hu, Ruth Halaban, William Chang, Shayan Cheraghlou, and Diana Yanez for technical contributions. This work was supported in part by the National Institutes of Health (NIH R01 AR071491 to K.A.C.), the Jane Danowski Weiss Fellowship and Leon Rosenberg Research Fellowship in Genetics (to N.U.), and UM1 HG006504 (to the Yale Center for Mendelian Genomics).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100028.
Data and code availability
The WES datasets supporting the current study have not been deposited in a public repository because subjects did not consent to data deposition. These data are available from the corresponding author on request. The ClinVar accession numbers for the variant reported in this paper are ClinVar: SCV001479301 and SCV001479325.
Web resources
1000 Genomes Project, https://www.internationalgenome.org/
OMIM, https://www.omim.org/
ImageJ, https://imagej.nih.gov/ij/
UCSC Genome Browser, http://hgdownload.cse.ucsc.edu/downloads.html#human
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, https://evs.gs.washington.edu/EVS/
MutationTaster, http://www.mutationtaster.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Supplemental information
References
- 1.Ayturk U.M., Couto J.A., Hann S., Mulliken J.B., Williams K.L., Huang A.Y., Fishman S.J., Boyd T.K., Kozakewich H.P.W., Bischoff J., et al. Somatic Activating Mutations in GNAQ and GNA11 Are Associated with Congenital Hemangioma. Am. J. Hum. Genet. 2016;98:1271. doi: 10.1016/j.ajhg.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Funk T., Lim Y., Kulungowski A.M., Prok L., Crombleholme T.M., Choate K., Bruckner A.L. Symptomatic Congenital Hemangioma and Congenital Hemangiomatosis Associated With a Somatic Activating Mutation in GNA11. JAMA Dermatol. 2016;152:1015–1020. doi: 10.1001/jamadermatol.2016.2365. [DOI] [PubMed] [Google Scholar]
- 3.Groesser L., Peterhof E., Evert M., Landthaler M., Berneburg M., Hafner C. BRAF and RAS Mutations in Sporadic and Secondary Pyogenic Granuloma. J. Invest. Dermatol. 2016;136:481–486. doi: 10.1038/JID.2015.376. [DOI] [PubMed] [Google Scholar]
- 4.Jinnin M., Medici D., Park L., Limaye N., Liu Y., Boscolo E., Bischoff J., Vikkula M., Boye E., Olsen B.R. Suppressed NFAT-dependent VEGFR1 expression and constitutive VEGFR2 signaling in infantile hemangioma. Nat. Med. 2008;14:1236–1246. doi: 10.1038/nm.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim Y.H., Bacchiocchi A., Qiu J., Straub R., Bruckner A., Bercovitch L., Narayan D., McNiff J., Ko C., Robinson-Bostom L., et al. Yale Center for Mendelian Genomics GNA14 Somatic Mutation Causes Congenital and Sporadic Vascular Tumors by MAPK Activation. Am. J. Hum. Genet. 2016;99:443–450. doi: 10.1016/j.ajhg.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim Y.H., Douglas S.R., Ko C.J., Antaya R.J., McNiff J.M., Zhou J., Choate K.A., Narayan D. Somatic Activating RAS Mutations Cause Vascular Tumors Including Pyogenic Granuloma. J. Invest. Dermatol. 2015;135:1698–1700. doi: 10.1038/jid.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakashima M., Miyajima M., Sugano H., Iimura Y., Kato M., Tsurusaki Y., Miyake N., Saitsu H., Arai H., Matsumoto N. The somatic GNAQ mutation c.548G>A (p.R183Q) is consistently found in Sturge-Weber syndrome. J. Hum. Genet. 2014;59:691–693. doi: 10.1038/jhg.2014.95. [DOI] [PubMed] [Google Scholar]
- 8.Shirley M.D., Tang H., Gallione C.J., Baugher J.D., Frelin L.P., Cohen B., North P.E., Marchuk D.A., Comi A.M., Pevsner J. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter J.W., North P.E., Waner M., Mizeracki A., Blei F., Walker J.W., Reinisch J.F., Marchuk D.A. Somatic mutation of vascular endothelial growth factor receptors in juvenile hemangioma. Genes Chromosomes Cancer. 2002;33:295–303. doi: 10.1002/gcc.10028. [DOI] [PubMed] [Google Scholar]
- 10.Joseph N.M., Brunt E.M., Marginean C., Nalbantoglu I., Snover D.C., Thung S.N., Yeh M.M., Umetsu S.E., Ferrell L.D., Gill R.M. Frequent GNAQ and GNA14 Mutations in Hepatic Small Vessel Neoplasm. Am. J. Surg. Pathol. 2018;42:1201–1207. doi: 10.1097/PAS.0000000000001110. [DOI] [PubMed] [Google Scholar]
- 11.Limaye N., Wouters V., Uebelhoer M., Tuominen M., Wirkkala R., Mulliken J.B., Eklund L., Boon L.M., Vikkula M. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 2009;41:118–124. doi: 10.1038/ng.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castel P., Carmona F.J., Grego-Bessa J., Berger M.F., Viale A., Anderson K.V., Bague S., Scaltriti M., Antonescu C.R., Baselga E., Baselga J. Somatic PIK3CA mutations as a driver of sporadic venous malformations. Sci. Transl. Med. 2016;8:332ra42. doi: 10.1126/scitranslmed.aaf1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janardhan H.P., Meng X., Dresser K., Hutchinson L., Trivedi C.M. KRAS or BRAF mutations cause hepatic vascular cavernomas treatable with MAP2K-MAPK1 inhibition. J. Exp. Med. 2020;217:e20192205. doi: 10.1084/jem.20192205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishak K.G., Rabin L. Benign tumors of the liver. Med. Clin. North Am. 1975;59:995–1013. doi: 10.1016/s0025-7125(16)31998-8. [DOI] [PubMed] [Google Scholar]
- 15.Reddy K.R., Kligerman S., Levi J., Livingstone A., Molina E., Franceschi D., Badalamenti S., Jeffers L., Tzakis A., Schiff E.R. Benign and solid tumors of the liver: relationship to sex, age, size of tumors, and outcome. Am. Surg. 2001;67:173–178. [PubMed] [Google Scholar]
- 16.Yoon S.S., Charny C.K., Fong Y., Jarnagin W.R., Schwartz L.H., Blumgart L.H., DeMatteo R.P. Diagnosis, management, and outcomes of 115 patients with hepatic hemangioma. J. Am. Coll. Surg. 2003;197:392–402. doi: 10.1016/S1072-7515(03)00420-4. [DOI] [PubMed] [Google Scholar]
- 17.Lerner S.M., Hiatt J.R., Salamandra J., Chen P.W., Farmer D.G., Ghobrial R.M., Busuttil R.W. Giant cavernous liver hemangiomas: effect of operative approach on outcome. Arch. Surg. 2004;139:818–821, discussion 821–823. doi: 10.1001/archsurg.139.8.818. [DOI] [PubMed] [Google Scholar]
- 18.Metry D.W., Hawrot A., Altman C., Frieden I.J. Association of solitary, segmental hemangiomas of the skin with visceral hemangiomatosis. Arch. Dermatol. 2004;140:591–596. doi: 10.1001/archderm.140.5.591. [DOI] [PubMed] [Google Scholar]
- 19.Haase V.H., Glickman J.N., Socolovsky M., Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl. Acad. Sci. USA. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T., Minowa O., Kuno J., Mitani H., Hino O., Noda T. Renal carcinogenesis, hepatic hemangiomatosis, and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 1999;59:1206–1211. [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10 33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wassef M., Blei F., Adams D., Alomari A., Baselga E., Berenstein A., Burrows P., Frieden I.J., Garzon M.C., Lopez-Gutierrez J.C., et al. ISSVA Board and Scientific Committee Vascular Anomalies Classification: Recommendations From the International Society for the Study of Vascular Anomalies. Pediatrics. 2015;136:e203–e214. doi: 10.1542/peds.2014-3673. [DOI] [PubMed] [Google Scholar]
- 27.Mulliken J.B., Glowacki J. Hemangiomas and vascular malformations in infants and children: a classification based on endothelial characteristics. Plast. Reconstr. Surg. 1982;69:412–422. doi: 10.1097/00006534-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Shu W., Lin Y., Hua R., Luo Y., He N., Fang L., Tan J., Lu J., Hu Z., Yuan Z. Cutaneomucosal venous malformations are linked to the TIE2 mutation in a large Chinese family. Exp. Dermatol. 2012;21:456–457. doi: 10.1111/j.1600-0625.2012.01492.x. [DOI] [PubMed] [Google Scholar]
- 29.Ye C., Pan L., Huang Y., Ye R., Han A., Li S., Li X., Wang S. Somatic mutations in exon 17 of the TEK gene in vascular tumors and vascular malformations. J. Vasc. Surg. 2011;54:1760–1768. doi: 10.1016/j.jvs.2011.06.098. [DOI] [PubMed] [Google Scholar]
- 30.Gustafsson F., Mikkelsen H.B., Arensbak B., Thuneberg L., Neve S., Jensen L.J., Holstein-Rathlou N.H. Expression of connexin 37, 40 and 43 in rat mesenteric arterioles and resistance arteries. Histochem. Cell Biol. 2003;119:139–148. doi: 10.1007/s00418-002-0493-0. [DOI] [PubMed] [Google Scholar]
- 31.Teilmann S.C. Differential expression and localisation of connexin-37 and connexin-43 in follicles of different stages in the 4-week-old mouse ovary. Mol. Cell. Endocrinol. 2005;234:27–35. doi: 10.1016/j.mce.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Willecke K., Heynkes R., Dahl E., Stutenkemper R., Hennemann H., Jungbluth S., Suchyna T., Nicholson B.J. Mouse connexin37: cloning and functional expression of a gap junction gene highly expressed in lung. J. Cell Biol. 1991;114:1049–1057. doi: 10.1083/jcb.114.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He D.S., Jiang J.X., Taffet S.M., Burt J.M. Formation of heteromeric gap junction channels by connexins 40 and 43 in vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA. 1999;96:6495–6500. doi: 10.1073/pnas.96.11.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Söhl G., Willecke K. Gap junctions and the connexin protein family. Cardiovasc. Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Oshima A. Structure and closure of connexin gap junction channels. FEBS Lett. 2014;588:1230–1237. doi: 10.1016/j.febslet.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 36.Berthoud V.M., Minogue P.J., Laing J.G., Beyer E.C. Pathways for degradation of connexins and gap junctions. Cardiovasc. Res. 2004;62:256–267. doi: 10.1016/j.cardiores.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Naus C.C., Laird D.W. Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer. 2010;10:435–441. doi: 10.1038/nrc2841. [DOI] [PubMed] [Google Scholar]
- 38.Wei C.J., Xu X., Lo C.W. Connexins and cell signaling in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:811–838. doi: 10.1146/annurev.cellbio.19.111301.144309. [DOI] [PubMed] [Google Scholar]
- 39.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Do R., Balick D., Li H., Adzhubei I., Sunyaev S., Reich D. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 2015;47:126–131. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 42.Bampton E.T., Goemans C.G., Niranjan D., Mizushima N., Tolkovsky A.M. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 43.Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uebelhoer M., Nätynki M., Kangas J., Mendola A., Nguyen H.L., Soblet J., Godfraind C., Boon L.M., Eklund L., Limaye N., Vikkula M. Venous malformation-causative TIE2 mutations mediate an AKT-dependent decrease in PDGFB. Hum. Mol. Genet. 2013;22:3438–3448. doi: 10.1093/hmg/ddt198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 46.Bayascas J.R. PDK1: the major transducer of PI 3-kinase actions. Curr. Top. Microbiol. Immunol. 2010;346:9–29. doi: 10.1007/82_2010_43. [DOI] [PubMed] [Google Scholar]
- 47.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 48.Meng F., Yamagiwa Y., Taffetani S., Han J., Patel T. IL-6 activates serum and glucocorticoid kinase via p38alpha mitogen-activated protein kinase pathway. Am. J. Physiol. Cell Physiol. 2005;289:C971–C981. doi: 10.1152/ajpcell.00081.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi M., Tapping R.I., Chao T.H., Lo J.F., King C.C., Yang Y., Lee J.D. BMK1 mediates growth factor-induced cell proliferation through direct cellular activation of serum and glucocorticoid-inducible kinase. J. Biol. Chem. 2001;276:8631–8634. doi: 10.1074/jbc.C000838200. [DOI] [PubMed] [Google Scholar]
- 50.Klauber N., Browne F., Anand-Apte B., D’Amato R.J. New activity of spironolactone. Inhibition of angiogenesis in vitro and in vivo. Circulation. 1996;94:2566–2571. doi: 10.1161/01.cir.94.10.2566. [DOI] [PubMed] [Google Scholar]
- 51.Miternique-Grosse A., Griffon C., Siegel L., Neuville A., Weltin D., Stephan D. Antiangiogenic effects of spironolactone and other potassium-sparing diuretics in human umbilical vein endothelial cells and in fibrin gel chambers implanted in rats. J. Hypertens. 2006;24:2207–2213. doi: 10.1097/01.hjh.0000249698.26983.4e. [DOI] [PubMed] [Google Scholar]
- 52.Heinz N., Hennig K., Loew R. Graded or threshold response of the tet-controlled gene expression: all depends on the concentration of the transactivator. BMC Biotechnol. 2013;13:5. doi: 10.1186/1472-6750-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moser C., Hany A., Spiegel R. Familial giant hemangiomas of the liver. Study of a family and review of the literature. Praxis (Bern 1994) 1998;87:461–468. [PubMed] [Google Scholar]
- 54.Toth B., Scholz C., Saadat G., Geller A., Schulze S., Mylonas I., Friese K., Jeschke U. Estrogen receptor modulators and estrogen receptor beta immunolabelling in human umbilical vein endothelial cells. Acta Histochem. 2009;111:508–519. doi: 10.1016/j.acthis.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Venkov C.D., Rankin A.B., Vaughan D.E. Identification of authentic estrogen receptor in cultured endothelial cells. A potential mechanism for steroid hormone regulation of endothelial function. Circulation. 1996;94:727–733. doi: 10.1161/01.cir.94.4.727. [DOI] [PubMed] [Google Scholar]
- 56.Monsivais D., Dyson M.T., Yin P., Navarro A., Coon J.S.T., Pavone M.E., Bulun S.E. Estrogen receptor β regulates endometriotic cell survival through serum and glucocorticoid-regulated kinase activation. Fertil. Steril. 2016;105:1266–1273. doi: 10.1016/j.fertnstert.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garvin S., Nilsson U.W., Dabrosin C. Effects of oestradiol and tamoxifen on VEGF, soluble VEGFR-1, and VEGFR-2 in breast cancer and endothelial cells. Br. J. Cancer. 2005;93:1005–1010. doi: 10.1038/sj.bjc.6602824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaytor A.T., Martin P.E., Edwards D.H., Griffith T.M. Gap junctional communication underpins EDHF-type relaxations evoked by ACh in the rat hepatic artery. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H2441–H2450. doi: 10.1152/ajpheart.2001.280.6.H2441. [DOI] [PubMed] [Google Scholar]
- 59.Shiojiri N., Niwa T., Sugiyama Y., Koike T. Preferential expression of connexin37 and connexin40 in the endothelium of the portal veins during mouse liver development. Cell Tissue Res. 2006;324:547–552. doi: 10.1007/s00441-006-0165-9. [DOI] [PubMed] [Google Scholar]
- 60.Simon A.M., McWhorter A.R. Vascular abnormalities in mice lacking the endothelial gap junction proteins connexin37 and connexin40. Dev. Biol. 2002;251:206–220. doi: 10.1006/dbio.2002.0826. [DOI] [PubMed] [Google Scholar]
- 61.Fang J.S., Angelov S.N., Simon A.M., Burt J.M. Cx37 deletion enhances vascular growth and facilitates ischemic limb recovery. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1872–H1881. doi: 10.1152/ajpheart.00683.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Good M.E., Nelson T.K., Simon A.M., Burt J.M. A functional channel is necessary for growth suppression by Cx37. J. Cell Sci. 2011;124:2448–2456. doi: 10.1242/jcs.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson T.K., Sorgen P.L., Burt J.M. Carboxy terminus and pore-forming domain properties specific to Cx37 are necessary for Cx37-mediated suppression of insulinoma cell proliferation. Am. J. Physiol. Cell Physiol. 2013;305:C1246–C1256. doi: 10.1152/ajpcell.00159.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J., Scherer S.S., Yum S.W. Dominant Cx26 mutants associated with hearing loss have dominant-negative effects on wild type Cx26. Mol. Cell. Neurosci. 2011;47:71–78. doi: 10.1016/j.mcn.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plantard L., Huber M., Macari F., Meda P., Hohl D. Molecular interaction of connexin 30.3 and connexin 31 suggests a dominant-negative mechanism associated with erythrokeratodermia variabilis. Hum. Mol. Genet. 2003;12:3287–3294. doi: 10.1093/hmg/ddg364. [DOI] [PubMed] [Google Scholar]
- 66.Placantonakis D., Cicirata F., Welsh J.P. A dominant negative mutation of neuronal connexin 36 that blocks intercellular permeability. Brain Res. Mol. Brain Res. 2002;98:15–28. doi: 10.1016/s0169-328x(01)00306-0. [DOI] [PubMed] [Google Scholar]
- 67.Banks E.A., Toloue M.M., Shi Q., Zhou Z.J., Liu J., Nicholson B.J., Jiang J.X. Connexin mutation that causes dominant congenital cataracts inhibits gap junctions, but not hemichannels, in a dominant negative manner. J. Cell Sci. 2009;122:378–388. doi: 10.1242/jcs.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyden L.M., Craiglow B.G., Zhou J., Hu R., Loring E.C., Morel K.D., Lauren C.T., Lifton R.P., Bilguvar K., Paller A.S., Choate K.A. Dominant De Novo Mutations in GJA1 Cause Erythrokeratodermia Variabilis et Progressiva, without Features of Oculodentodigital Dysplasia. J. Invest. Dermatol. 2015;135:1540–1547. doi: 10.1038/jid.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tittarelli A., Guerrero I., Tempio F., Gleisner M.A., Avalos I., Sabanegh S., Ortíz C., Michea L., López M.N., Mendoza-Naranjo A., Salazar-Onfray F. Overexpression of connexin 43 reduces melanoma proliferative and metastatic capacity. Br. J. Cancer. 2015;113:259–267. doi: 10.1038/bjc.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aasen T., Mesnil M., Naus C.C., Lampe P.D., Laird D.W. Gap junctions and cancer: communicating for 50 years. Nat. Rev. Cancer. 2016;16:775–788. doi: 10.1038/nrc.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abbruzzese C., Mattarocci S., Pizzuti L., Mileo A.M., Visca P., Antoniani B., Alessandrini G., Facciolo F., Amato R., D’Antona L., et al. Determination of SGK1 mRNA in non-small cell lung cancer samples underlines high expression in squamous cell carcinomas. J. Exp. Clin. Cancer Res. 2012;31:4. doi: 10.1186/1756-9966-31-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Talarico C., Dattilo V., D’Antona L., Barone A., Amodio N., Belviso S., Musumeci F., Abbruzzese C., Bianco C., Trapasso F., et al. SI113, a SGK1 inhibitor, potentiates the effects of radiotherapy, modulates the response to oxidative stress and induces cytotoxic autophagy in human glioblastoma multiforme cells. Oncotarget. 2016;7:15868–15884. doi: 10.18632/oncotarget.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chung E.J., Sung Y.K., Farooq M., Kim Y., Im S., Tak W.Y., Hwang Y.J., Kim Y.I., Han H.S., Kim J.C., Kim M.K. Gene expression profile analysis in human hepatocellular carcinoma by cDNA microarray. Mol. Cells. 2002;14:382–387. [PubMed] [Google Scholar]
- 74.Liang X., Lan C., Jiao G., Fu W., Long X., An Y., Wang K., Zhou J., Chen T., Li Y., et al. Therapeutic inhibition of SGK1 suppresses colorectal cancer. Exp. Mol. Med. 2017;49:e399. doi: 10.1038/emm.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Catela C., Kratsios P., Hede M., Lang F., Rosenthal N. Serum and glucocorticoid-inducible kinase 1 (SGK1) is necessary for vascular remodeling during angiogenesis. Dev. Dyn. 2010;239:2149–2160. doi: 10.1002/dvdy.22345. [DOI] [PubMed] [Google Scholar]
- 76.Zarrinpashneh E., Poggioli T., Sarathchandra P., Lexow J., Monassier L., Terracciano C., Lang F., Damilano F., Zhou J.Q., Rosenzweig A., et al. Ablation of SGK1 impairs endothelial cell migration and tube formation leading to decreased neo-angiogenesis following myocardial infarction. PLoS ONE. 2013;8:e80268. doi: 10.1371/journal.pone.0080268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buse P., Tran S.H., Luther E., Phu P.T., Aponte G.W., Firestone G.L. Cell cycle and hormonal control of nuclear-cytoplasmic localization of the serum- and glucocorticoid-inducible protein kinase, Sgk, in mammary tumor cells. A novel convergence point of anti-proliferative and proliferative cell signaling pathways. J. Biol. Chem. 1999;274:7253–7263. doi: 10.1074/jbc.274.11.7253. [DOI] [PubMed] [Google Scholar]
- 78.Wang W., Shen T., Dong B., Creighton C.J., Meng Y., Zhou W., Shi Q., Zhou H., Zhang Y., Moore D.D., Yang F. MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J. Clin. Invest. 2019;129:1015–1029. doi: 10.1172/JCI97712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu W., Wang X., Liu Z., Wang Y., Yin B., Yu P., Duan X., Liao Z., Chen Y., Liu C., et al. SGK1 inhibition induces autophagy-dependent apoptosis via the mTOR-Foxo3a pathway. Br. J. Cancer. 2017;117:1139–1153. doi: 10.1038/bjc.2017.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zuleger T., Heinzelbecker J., Takacs Z., Hunter C., Voelkl J., Lang F., Proikas-Cezanne T. SGK1 Inhibits Autophagy in Murine Muscle Tissue. Oxid. Med. Cell. Longev. 2018;2018:4043726. doi: 10.1155/2018/4043726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lauzier A., Normandeau-Guimond J., Vaillancourt-Lavigueur V., Boivin V., Charbonneau M., Rivard N., Scott M.S., Dubois C.M., Jean S. Colorectal cancer cells respond differentially to autophagy inhibition in vivo. Sci. Rep. 2019;9:11316. doi: 10.1038/s41598-019-47659-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 83.Wang X.J., Yu J., Wong S.H., Cheng A.S., Chan F.K., Ng S.S., Cho C.H., Sung J.J., Wu W.K. A novel crosstalk between two major protein degradation systems: regulation of proteasomal activity by autophagy. Autophagy. 2013;9:1500–1508. doi: 10.4161/auto.25573. [DOI] [PubMed] [Google Scholar]
- 84.Chen S.Y., Bhargava A., Mastroberardino L., Meijer O.C., Wang J., Buse P., Firestone G.L., Verrey F., Pearce D. Epithelial sodium channel regulated by aldosterone-induced protein sgk. Proc. Natl. Acad. Sci. USA. 1999;96:2514–2519. doi: 10.1073/pnas.96.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WES datasets supporting the current study have not been deposited in a public repository because subjects did not consent to data deposition. These data are available from the corresponding author on request. The ClinVar accession numbers for the variant reported in this paper are ClinVar: SCV001479301 and SCV001479325.