Abstract
COVID-19 is announced as a global pandemic in 2020. Its mortality and morbidity rate are rapidly increasing, with limited medications. The emergent outbreak of COVID-19 prompted by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) keeps spreading. In this infection, a patient's immune response plays pivotal role in the pathogenesis. This inflammatory factor was shown by its mediators that, in severe cases, reach the cytokine at peaks. Hyperinflammatory state may sparks significant imbalances in transporters and drug metabolic machinery, and subsequent alteration of drug pharmacokinetics may result in unexpected therapeutic response. The present scenario has accounted for the requirement for therapeutic opportunities to relive and overcome this pandemic. Despite the diminishing developments of COVID-19, there is no drug still approved to have significant effects with no side effect on the treatment for COVID-19 patients. Based on the evidence, many antiviral and anti-inflammatory drugs have been authorized by the Food and Drug Administration (FDA) to treat the COVID-19 patients even though not knowing the possible drug-drug interactions (DDI). Remdesivir, favipiravir, and molnupiravir are deemed the most hopeful antiviral agents by improving infected patient’s health. Dexamethasone is the first known steroid medicine that saved the lives of seriously ill patients. Some oligopeptides and proteins have also been using. The current review summarizes medication updates to treat COVID-19 patients in an inflammatory state and their interaction with drug transporters and drug-metabolizing enzymes. It gives an opinion on the potential DDI that may permit the individualization of these drugs, thereby enhancing the safety and efficacy.
Keywords: COVID-19, Drug transporters, CYPs, Remdesivir, Dexamethasone, Molnupiravir
Graphical Abstract
1. Introduction
SARS-CoV-2 was initially recognized and designated as COVID-19 infection [1]. The National Health Commission of China confirmed the first of COVID-19 identification in pneumonia patients in the city of Wuhan in China in December 2019. Initially, pneumonia patients stated the normal respiratory infection, which promptly transformed into acute respiratory syndrome [2]. By the end of December 2020, almost 77 million cases have been reported, with about 1.7 million deaths.
There is a very urgent prerequisite to discovering novel antiviral drugs against COVID-19. Based on the evidence, Food and Drug Administration (FDA) approved some medicines that have already been used in the treatment of SARC-CoV and MERC-COV. In these, remdesivir and favipiravir showed the most promising effect against COVID-19 [3]. The antiretroviral (ARV) drug lopinavir has been used for COVID-19 combined with ritonavir (potent anti-HIV drug). This lopinavir-ritonavir combination showed to be efficient against COVID-19 [4]. Anti-malarial drug hydroxychloroquine has demonstrated more promising results against COVID-19, and it is used in higher frequency. Alone and in combination with many other drugs [5], [6]. Regardless of the antiviral drug, dexamethasone has proved little relief against COVID-19. In the initial clinical trial, it lowered the death by one-third on severe patients that were on a ventilator [7]. Fluvoxamine has also shown the potential in outpatients of COVID-19 infection [8]. Molnupiravir is used to treat the COVID-19 condition. It can block the transmission of SARS-CoV-2 within 24 h [9]. Many drugs and peptide some under clinical trial that have been used for treating COVID-19 [10], has shown in Table 1. Vaccine development is typically a long game, and many more vaccines against COVID-19 are in clinical trials, and few are in the final stage [10]. We will have to wait and see how things play out [11].
Table 1.
Drug-drug interaction potential of therapeutic agents used to treat COVID-19.
| Drug | Drug Transporter | Reference | Metabolism | reference | |
|---|---|---|---|---|---|
| 1 | Remdesivir | $Pgp$OATPB1,IOATPB1,IOATPB3,IBSEP,IMRP4,INTCP | [103] | $CYP2C8,$CYP2D6,$CYP3A4,ICYP3A4 | [103] |
| 2 | Favipiravir | IOAT1,IOAT3 | [116] | $AO,ICYP2C8 | [116] |
| 3 | Ribavirin | $NT,$ENT1 | [290], [291] | Phosphorylation, Deribosylation, Amide hydrolysis | [292] |
| 4 | Interferons | $OAT2 | [173] | $CYP1A2,$UGT2B7,ICYP3A,ICYP2D6 | [173], [174] |
| 5 | Lopinavir | $Pgp,$MRP1,$MRP2,$OATP1A2,$OATP1B1IPgp,IBCRP,IOATP1B1,IOATP1B3,IOATP2B1 | [293], [294], [295] | ICYP3A4 | [293], [294], [295] |
| [296], [297], [298], [299] | |||||
| 6 | Ritonavir | $Pgp,$MRP1,$MRP2,IPgp,IMRP1I, BCRP,IOATP1A2,IOATP2B1,IOATP1B1,IOATP1B3,IOCT1, OCT2 | [295] | $CYP1A2,$CYP2C8,$CYP2C9,$CYP2C19,ICYP3A4,ICYP2D6 | [308], [300] |
| [300] | |||||
| [296], [299], [301], [302], [303], [304], [305], [306], [307] | |||||
| 7 | Chloroquine | $OATP1A2 | [309], [310] | $CYP2C8,$CYP3A4$CYP2D6 | [311], [312] |
| 8 | Hydroxy chloroquine | IPgp,IOATP1A2 | [309], [310], [313] | $CYP2C8,$CYP3A4$CYP2D6 | [311], [312], [314] |
| 9 | Dexamethasone | $Pgp,$MRP2 | [315], [316] | $CYP3A4 | [317] |
| 10 | Umifenovir | Unclear | $CYP3A4 and$FMOs | [148] | |
| 11 | Teicoplanin | Unclear | Unclear Metabolic path | [318] | |
| 12 | Nitazoxanide | unclear | $Deacetylase,$UGT | [319] | |
| 13 | Ivermectin | IPgp,IBCRP,IMRP1,IMRP2,IMRP3 | [320], [321], [322] | ICYP2C9,ICYP2C19ICYP2D6,ICYP3A4 | [323] |
| 14 | Atazanavir | $Pgp,$MRP1,$MRP2,IOATP2B1 | [324], [325], [298], [299], [326], [327] | ICYP3A4,IUDGT | [324] |
| 15 | Azithromycin | $Pgp and$MRP2 | [211], [328] | $CYP3A4 | [210], [329] |
| $OATP | |||||
| 16 | Darunavir | $Pgp,$OATP1A2, OATP1B1,IPgp,IOATP2B1 | [330], [331], [332], [333] | ICYP3A4 | [334], [335] |
| 17 | Ruxolitinib | $OATP1B1, and$OCT1,$NTCP,IPgp,IBCRP | [334], [335] | $CYP1A2,$CYP2B6,$CYP2C9$CYP3A4 | [224], [225] |
| 18 | Baricitinib | $P-gp,$BCRP,$OAT3,$MATE-K | [234], [292] | $CYP3A4 | [292], [336] |
| 19 | Imatinib | $Pgp,$OATP1B3 | [337], [338] | $CYP2C8,$CYP3A4 | [238], [339] |
| 19 | Fluvoxamine | Unclear | $CYP1A2,$CYP2C19,$CYP2D6,$CYP3A4 | [340], [341] | |
| ICYP1A2,ICYP2C19 | [247] | ||||
| 20 | Canabinoids | $Pgp,$BCRP,$MRPs | [342] | $CYPs,$UGT | [342], [343] |
| 21 | Molnupiravir | Unclear | Unclear | ||
| 22 | Itolizumab | Unclear | Unclear | ||
| 23 | Tocilizumab | Unclear | Unclear | ||
| 24 | Meplazumab | Unclear | Unclear | ||
| 25 | Eculizumab | Unclear | Unclear | ||
| 26 | AMY101 | Unclear | Unclear | ||
| 27 | ARDS-003 | Unclear | Unclear | ||
| 28 | LCB1 | Unclear | Unclear |
$-substrate of, I-Inhibitor of, Pgp; P-glycoprotein, OAT; Organic Anion Transporter, OATP; Organic Anion Transporter Protein, BCRP; Breast Cancer Resistance Protein, MRP, Multidrug Resistance Associated Protein, OCT; Organic Cation Transporter, NT; Nucleotide Transporter, ENT; Equilibrative Nucleoside Transporter, NTCP; Sodium/Taurocholate Co-transporting polypeptide, BSEP; Bile Salt Export Pump, CYP; Cytochrome P450, UGT, UDP-Glucuronosyltransferase, AO; Aldehyde Oxidase, FMO; Flavin-containing Monooxygenase
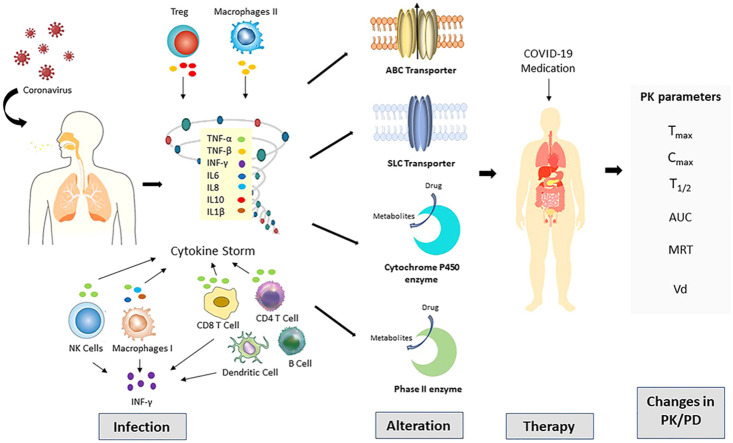
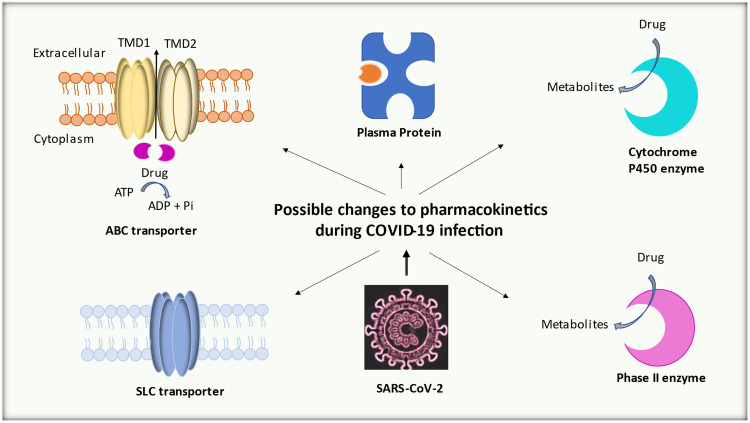
Many pharmacokinetics determinants of used possible COVID-19 medications, including absorption distribution metabolism and elimination, can be modified disease state or during an inflammatory response ( Fig. 1.). Chronic inflammation is produced when an antigen is persisted, and the immune system continuously acts against this antigen. This chronic inflammation is associated with alteration in the level of drug binding proteins [12], [13], [14], in addition, the downregulation of various hepatic and extrahepatic drug metabolizing enzyme [15], [16]. More recently, inflammation-mediated changes in the expression of numerous membranes associated drug transporters have been reported [17]. In the case of co-administration of many drugs, the risk of drug interaction increases, nevertheless in COVID-19, a more complicated disease-drug-drug interaction is anticipated. The SARS-CoV-2 infection has shown an increasing frequency of hospitalization and intensive care unit (ICU) admission [18]. This is considered as a major clinical concern mainly when considering that critically ill patients are more influenced drug interaction that CYPs and efflux pumps are involved in the metabolism and transport respectively of commonly prescribed drugs in ICU [19]. During highly prevalent acute and chronic inflammatory conditions, the alteration in the expression and activity of transporters and DMEs may modify the pharmacokinetics (PK) and pharmacodynamic (PD) properties of therapeutic drugs used in COVID-19 treatment.
Fig. 1.
Alteration in the expression of key transporters (ABC: ATP binding cassette and SLC: Solute carrier) and drug metabolizing enzyme (Cytochrome P450 and Phase II) mediated by acute inflammatory response in SARC-CoV-2 (COVID-19) infection play a role in alteration of drug disposition and pharmacokinetics.
This article will focus on the current state of knowledge and assume PK, disease-drug, and drug-drug interaction of therapeutic drugs used for routine intensive care for COVID-19 patients. Herein, we also reviewed the updated status of the supportive roles of several antivirals, some antibiotics, and some therapeutics peptides that have tested their efficacy in the worldwide treatment of COVID-19.
2. Alteration of drug transporters mechanisms in response to inflammation
Inflammation is associated with various cytokines response. Those are the broad class of small cell-ignaling proteins accountable for keeping homeostasis of the immune system. Proinflammatory cytokines interleukin-1 (IL-1), interleukin-6 (IL-6), and tumor necrosis factor (TNF-α) are mainly responsible for an acute immune response [20]. During infection, these locally producing cytokines can circulate in the bloodstream and create a systemic effect by interacting with cell membrane receptors, transporters on the vascular endothelium, and parenchymal cells of numerous organs. Inflammation and immune reaction represent a significant factor in many acute and chronic diseases involved in changing drug clearance by altering drug transporter's mechanism and drug-metabolizing enzyme activity.
P-glycoprotein (Pgp) is one of the most comprehensively explored ABC transporter that has been studied against the response of inflammations. Several studies suggested the association of IL-6 as direct administration of this cytokine diminished the Pgp mRNA level and protein expression in hepatic cell lines as well as in vivo in mice [21], [22]. In humans, intestinal epithelium showed a decrease in Pgp expression in inflammatory conditions [23]. Caco-2 is a human enterocyte cell line and universal model for permeability experiment was treated with TNF-α resulted in to decrease in Pgp expression and activity [24], [25]. Second, the most comprehensively explored ABC transporter, BCRP (breast cancer resistance protein) was studied in recent years. Like Pgp, BCRP activity and expression were downregulated in primary human hepatocytes after treating with IL-6, while TNF-α treatment led to induction of BCRP [26]. The effect of IFN-γ on primary human hepatocytes significantly decreased the mRNA expression of BCRP [27]. The human brain cell line showed significant decreases in the expression and activity of BCRP after treating IL-β1, IL-6, and TNF-α [28]. It is also believed that proinflammatory cytokines are critical mediators of MRP2. It was recently revealed that IL-β1, IL-6, and TNF-α, all considerably down-regulate the mRNA and protein expression of MRP2 in hepatic cell lines of humans and rats [29], [30]. Overall, alteration in the expression of Pgp, BCRP, MRP2, and several other key transporters is mediated by acute inflammatory response [25], [31], [32], [33], [34], [35]. One study on inflammatory bowel diseases (IBD) patients revealed the effect of inflammation on SLC transporters [36]. SLC transporters (ENT1, ENT2, CNt2, OATP2B1, OATP4A1) mRNA level was dysregulated in IBD patients, which is associated with the inflammation of the tissue and presents an indication about the inflammatory signaling in the regulation of SLC expression [36]. Many transcription factors are also activated during inflammation and play a key role in regulating transporters and drug-metabolizing enzymes [37], [38], [39], [40], [41]. Many excellent reviews have been published on the molecular mechanism of the transporters regulations during inflammation have been described elsewhere [39], [42], [43].
3. Alteration of drug metabolizing enzymes activity in response to inflammation
CYPs are widely involved as the primary contributor to metabolic biotransformation of most drugs [44]. Like drug transporters, regulation of CYPs has also been associated with inflammation in several metabolic and infectious diseases, including viral infection [45]. Inflammatory-induced alteration in hepatic CYPs is caused mainly by cytokines formed during the inflammation process [46]. It has been reported that multiple cytokines may play a part in regulating a single enzyme, whereas a specific cytokine can regulate a subclass of enzymes. The regulation is essential when considering drug interactions because drug pharmacokinetics will ultimately be altered depending on disease type and its released cytokines, as well as the, administered [47], [48]. Many studies have described cytokine-induced CYPs activity alteration in different mammals [49], [50], [51], including humans [52], [53] using hepatocytes as well as in vivo in mice [54], rats [55], and humans [56]. Inflammatory mediators also suppressed the extrahepatic CYPs [57], [58], [59], [60]. Most proinflammatory cytokines are IL-1 [61], [62], [63], IL-6, TNF-α, and IFN-γ that have displayed suppression of CYPs expression and activity [64], [65], [66]. Other cytokines IL-2 and IL-10, also showed the same effect [67], [68], [69]. IL-6 has been identified as the major inflammatory factor that elicits a significant repressive effect on the expression and activity of different CYPs. Human recombinant IL-6 has exhibited concentration dependent inhibition of phenobarbital mediated induction of CYP2B1/2 in rat hepatocytes [70]. It reduced the activity of different CYPs with variable levels [71]. Human recombinant IL-6 treatment markedly suppressed at mRNA level of CYP1A1, CYP1A2, and CYP3A3 in separate human hepatoma cell lines [71]. Chronic inflammatory response, induced by turpentine or bacterial lipopolysaccharide, showed significant suppression in hepatic CYP1A2, CYP2A5, CYP2C1, and CYP3A11 in rats [72], [73]. Several studies have assessed the impact of IL-6 on CYPs, triggered by malignancies [74]. The vital role of IL-6 in cancer-mediated suppression of hepatic CYP3A has been exhibited by mitigating such effects via monoclonal antibodies against IL-6 [75] or IL-6 receptor [76]. Anti-IL-6 antibody intrusion was also tested in IL-6 treated primary human hepatocytes. IL-6 inhibited CYP1A1, CYP1A2, CYP2B6, and CYP3A4 expression and activity. It was also efficient in intervening CYP1A2 and CYP3A4, induced by omeprazole and rifampicin, respectively. The cytokine-induced suppression of CYPs activity is not fully expounded. However, it is believed that strongly suggested that a decrease in the CYPs mRNA strongly recommended a transcriptional mechanism affecting several transcriptional factors [77], [78]. Nuclear factor Kappa B (NF-kB) and the aryl hydrocarbon receptor are the regulatory transcription factor in the inflammatory and immune response, and they regulate the gene expression of many CYPs in human, rats, and mice [77], [78], [79], [80]. For example, pyrrolidine dithiocarbamate is an inhibitor of NF-kB that can the capability to block the inflammatory reduction in CYP1A2 activity [81], [82]. Pregnane X Receptor (PXR) is targeted to many genes, most notably CYP3A4. NF-kB factors regulate PXR, and NF-kB is regulated by inflammatory stimuli results manipulation of Hepatic CYPs expressions [83], [84], [85]. Activation of NF-kB suppressed the glucocorticoid receptors (GR) and constitutive androsterone receptor (CAR) expression. It is associated CYP genes [86]. It has been described that proinflammatory cytokines such as IFN-γ, TNF-α, GM-CSF (granulocyte macrophage colony stimulating factor), MC-SF (macrophage colony stimulating factor), IL-1, IL-6, IL-12 elevated in peripheral blood of COVID-19 patients [2], [87]. IL-6 was found the most important target for anti-cytokine therapy as it is believed to be the key point in the process and its elevation is associated with poor prognosis [88], [89], [90].
It is well recognized that alteration in the expression of a transporter and the activity of DMEs, can lead to alterations in the PK/PD of the prescribed drug; therefore, prescribed medication during inflammation may be an essential to interindividual variability in drug efficacy and toxicity. In the case of co-administration of multiple drugs, the risk of drug interactions is increased. Most prescribed drugs are given below, and their drug interaction potential are shown in Table 1.
4. Agents used to treat COVID-19
4.1. Remdesivir
Remdesivir (GS-5734) is an adenosine triphosphate analog. It was put forth to treat to Ebola and Coronavirus [91]. Remdesivir halts the viral replication by inhibiting the essential replicating enzymes RNA dependent RNA Polymerase. It supports the premature termination of viral transcription by eluding the proofreading activity of exoribonuclease [92]. Remdesivir has broad-spectrum activity against many viruses, including, SARS-CoV, and MERS-CoV [4], [3]. Clinical pharmacokinetics data are still unclear. Moreover, safety data on humans are available online [93]. Remdesivir has already shown the successful inhibition with sub-micromolar concentration in tissue culture experiments against human CoV, and Zoonotic CoV [94], [95]. Similar efficacy was also found in MERS-CoV infected nonhuman primate (rhesus monkey) [96]. It has given the direction for a potential treatment for COVID-19 infected patients [97]. Updated data showed that remdesivir had little or no effect on hospitalized patients with COVID-19, as indicated by overall mortality, initiation of ventilation, and hospital stay duration [98], [99]. Remdesivir plus baricitinib comedication was shown promise for treating COVID-19. This co-medication therapy was superior upon remdesivir alone in lowering recovery and accelerating improvement in clinical status. This combination has been shown fewer serious adverse effects [100]. Using CQ or HCQ with remdesivir may reduce the antiviral activity of remdesivir [101]. Another combination of remdesivir with bericitinib, a substrate of CYP3A4, showed severe side effects despite accelerating clinical status improvement [100].
4.1.1. Drug-drug interaction
Remdesivir demonstrated linear PK. While in healthy people, no significant difference was found in the half-life of remdesivir, but nucleoside metabolite GS-441524 was observed with half-life 24 h [102]. Primary data from healthy human donors confirm that remdesivir is extensively metabolized by CYP2C8, CYP2D6, and CYP3A4 [103]. Clinical studies related to DDI of remdesivir have not been done but mathematical prediction of DDI liability was performed using existing phase I and in-vitro data [104]. The impact of inhibitor/inducer on the PK of remdesivir will be significantly attenuated because of the high to moderate extraction ratio (0.6–0.8) and IV route of administration, if remdesivir had a low hepatic extraction ratio [105]. A strong inducer rifampicin causes the induction of CYP3A and up to 30% decrease of remdesivir exposure is anticipated. Likewise, entire CYP3A inhibition would be supposed to increase 4% level of remdesivir compare to the 20-fold rise in the exposure of midazolam (a probe CYP3A substrate). The use of strong CYP3A inhibitors and moderate CYP3A inducers were permitted phase III clinical program of remdesivir, based on the predictions. Evaluation of the potential for the remdesivir to inhibit DMEs and transporters in vivo, a physiological based pharmacokinetic (PBPK) model that capture the in vivo PK and invitro inhibition potency of remdesivir was developed using SimCYP software. Invitro unbound inhibition constants for BCRP, CYP3A, OATP1B3, and MATE1 were assigned and the alteration in midazolam (CYP3A), rosuvastatin (OATP/BCRP), pravastatin (OATP), and metformin (MATE1) probe exposure (AUC) was anticipated. For these PBPK simulations, the timing of remdesivir administration relative to prob drug was optimized to foresee the maximum DDI, which coarsely related to the administration of remdesivir, so the ending of the infusion occurs at the probe maximum observed plasma concentration. Whereas co-administration of remdesivir is expected to increase probe drug AUC by transporters at therapeutic remdesivir dose with COVID-19. For the treatment of the COVID-19, a 200 mg loading dose along with 100 mg maintenance doses for about 5–10 days, demonstrated steady PK with prior research [104].
4.1.2. Disease-drug interaction
In-vitro data advised that inflammatory response reduces mRNA expression of several CYP450 isoenzymes, including CYP1A2, CYP2B6, CYPC9, CYP2C19, CYP2D6, and CYP3A4 [106] and transporters. Therefore, inflammation could theoretically influence their pharmacokinetics.
4.2. Favipiravir
Favipiravir is an antiviral drug that was used to treat influenza infection in 2014 in Japan [107]. Favipiravir was also used against the Ebola virus, in the absence of a standard cure for Ebola [108]. It was finally accepted for the treatment of Ebola virus infection [109], [110]. Favipiravir also showed the immune response in viral clearance in nonhuman primates [111]. Other studies have also described that the active metabolite of favipiravir (favipiravirribofuranosyl-50-triphosphate) directly halt the transcription by inhibiting the RNA dependent RNA polymerase [112], [113]. The clinical evidence for the efficacy and safety was observed in an open label, nonrandomized control clinical trial [114]. Recently, phase-3 clinical trials for COVID-19 favipiravir with tablet has been initiated [115]. The clearance has granted Appili Therapeutics for evaluating safety and efficacy of favipiravir in the tablets form to control COVID-19 in long-term care services. FDA granted approval to Appili Therapeutics to investigate the broad-spectrum antiviral therapy for favipiravir.
4.2.1. Drug-drug interaction
Favipiravir is also a prodrug that is active after intracellular phosphorylation like remdesivir. It shows hepatic metabolism other than CYPs, mediated by aldehyde oxidase (AO) xanthine oxidase (XO). Asian population must be shown the increased exposure of favipiravir because of genetic polymorphism of AO. Especially variants of aldehyde oxidases are associated with pharmacodynamic outcomes in other drugs that are the substrate of AO. AO inhibition based on DDI has yet to be determined. The DDI based zaleplon and cimetidine is already known. Cimetidine co-administration results in the inhibition on AO catalyzed oxozaleplon development, and caution is included in the Zaleplon level. Possible DDI between favipiravir and these drugs should be thoroughly examined. Hence, potential DDI due to the AO inhibition should not be ignored in the clinical settings. Favipiravir is eliminated as an inactive metabolite (82–92%) through urine. Favipiravir and its metabolite act as a moderate inhibitor of OAT1 and OAT3, resulting in uric acid excretion in the kidney [116]. It is also a week inhibitor of several metabolic enzymes, but its effect is shown on CYP2C8 that must have clinical significance to increase substrate drug exposure [117].
4.3. Lopinavir/ritonavir
Lopinavir (LPV) is a potent anti-HIV drug that is used to treat HIV infection in combination with ritonavir (RTV). Ritonavir inhibits the hepatic drug metabolism of lopinavir to enhance the half-life and activity. Infectious Diseases Society of America (IDSA) advised ritonavir-boosted combination as first-line therapy for HCV patients [118]. LPV/RTV has proven anti-SARS-CoV-2 activity in vitro by preventing the protease in Vero E6 cells [119]. In a comparative study [120], LPV/RTV and ribavirin displayed a risk in SARS-CoV. Furthermore, SARS patients revealed that lopinavir-ritonavir plays a vital role in explaining the clinical consequences [121]. Another comparative study LPV/RTV treatment alone, and combination with IFN enhanced clinical outcomes on some MERS patients. LPV/RTV was found to 40% decrease in the risk of MERS infection [121], [122]. In India, EMR division has advised the dosing program for this drug combination for clinical management of COVID-19 [123]. One randomized open-label clinical trial [124] for LPV/RTV is conducted on patients. The authors did not gain more advantage of LPV/RTV to clinical benefits outside the standard of care, while it was found to have an advantage except secondary endpoints. The efficacy of the LPV/RTV was approved, and future trials will verify the results [124]. The latest study showed that LPV had little or no effect on hospitalized patients with COVID-19, as suggested by overall mortality, initiation of ventilation, and hospital stay duration [98], [99].
4.3.1. Drug-drug interaction
LPV is the substrate or inhibitor of various transporters (P-gp, BCRP, OATP1A2, OATP1B1, OATP1B3 and OATP2B1), and CYP3A4 mediates their metabolism. Ritonavir inhibits the hepatic drug metabolism of lopinavir to enhance the half-life and activity.
4.3.2. Disease-drug interaction
CYP3A activity was approximately 50% lower in HIV-infected patients as compared to healthy volunteers [125]. Later in this study, the effect of inflammation could have been diminished by the presence of booster RTV, which is steadily linked with atazanavir to decrease its clearance. In recent, two short reports specified higher plasma concentration of LPV in severe COVID-19 patients [126], [127], compared to those detected in HIV-patients and concerning inflammation [126]. Since protease inhibitor -based regimens are administered to COVID-19 patients and have a higher potential for interactions then other anti-retroviral drugs, evaluation of DDI is needed to inform proper recommendation of dosing.
4.4. Ribavirin
Ribavirin is a guanosine analog and a broad-spectrum antiviral drug. Combination with IFN, it has been used as a treatment option for hepatitis C infected critically ill patients. It demonstrated lower risk and reduced death in ARDS (Acute respiratory distress syndrome) infection in combination LPV/RTV [120]. Though, in recent in-vitro studies, ribavirin indicated a high effective concentration against COVID-19 [128], [129]. However, ribavirin developed an unexpected adverse effect, which was very harmful to ADRS patients [4].
4.4.1. Drug-drug interaction
Ribavirin is neither substrate nor inhibitor of hepatic or extrahepatic CYPs. Drug-drug interaction is still unclear. It is hydrolyzed and phosphorylated only. It is being used in triple combination with nitazoxanide and hydroxychloroquine.
4.5. Chloroquine, hydroxychloroquine
The two aminoquinolines, chloroquine (CQ) and hydroxychloroquine (HCQ) are used for malaria and rheumatic diseases. They showed the activity against the COVID-19 in Vero E6 cells [130] and recommended it as a primary treatment option for the COVID-19 [131], [132], [133]. CQ and HCQ have weak diprotic features, and they could increase the pH of the endosome during the fusion of the virus to the host cell [134]. Several clinical trials were preceded in China for CQ and HCQ on COVID-19 infected patients. One of them disclosed that promising results in a reduction of the disease progression [135]. One clinical trial was performed in France to find the efficacy of HCQ using at different doses, along with azithromycin on COVID-19 infected patients. The clinical demonstration noticed that the treated rate was considerably higher in HCQ used in combination with azithromycin [136]. Even though this study showed favorable results, extensive clinical data are required to confirm the efficacy and safety of HCQ with azithromycin [137]. Similarly, a postexposure prophylaxis clinical trial (NCT04308668) using an oral dosing regimen has been conducted in the USA. The latest study revealed that HCQ had little or no effect on hospitalized patients with COVID-19, as suggested by overall mortality, initiation of ventilation [98], [99].
4.5.1. Drug-drug interaction
The HCQ elimination half-life is likely > 40 days. The CYP and its isoform could play a vital role in the metabolism of HCQ and CQ. The hepatic metabolism of HCQ is not exactly exemplified. To see the chloroquine data, the effect of CYP3A4/5 CYP2C8 and CYP2D6 were extrapolated. Both change into active metabolites through the dealkylation process by CYP isomers [138], [139]. The single nucleotide polymorphism (SNPs) of CYP2D6 showed on the variable metabolism of HCQ and CQ in SLE [140]. The administration of HCQ for the management of COVID-19 infection was challenged by several side effects among individuals with different CYP genotypes. CYP genotyping, especially for CYP2D6, may help to determine the optimum HCQ dosage in personalized medicine.
4.5.2. Disease-drug interaction
The proinflammatory cytokine IL-6 increases the expression level of CYP2B1 by an epigenetic mechanism. With this higher level of IL-6 in COVID-19 infected patients compared to healthy subjects, there is the possibility that IL-6 interferes with the metabolism of HCQ. The point is highly clinical relevance for drug with a narrow therapeutic index like HCQ and CQ.
4.6. Umifenovir
Umifenovir, is also known as Arbidol, is a broad-spectrum antiviral drug [141]. Umifenovir worked to prevent the fusion of endosome membrane to virus particles [142], [143], [144]. It interacts with the virus hemagglutinin and enhances hemagglutinin stability, thus inhibiting the hemagglutinin transition into a functional state [145]. Umifenovir also disclosed the immunomodulatory and macrophage activation[146]. LPV/RTV and umifenovir were earlier used to treat acute SARC-CoV in clinical practice. The clinical safety and efficacy of the umifenovir monotherapy were analyzed in COVID-19 patients and compared with LPV/RTV therapy. Umifenovir was found better than LVP/RTV for treating COVID-19 [147]. Central Drug Research Institute (CDRI) has acquired the approval for proceeding with the phase III clinical trial of umifenovir. In this randomized, double-blind, placebo-controlled trial, the efficacy, safety, and tolerability will be tested.
4.6.1. Drug-drug interaction
Arbidol was mainly eliminated in the urine; major metabolites as with glucuronide and glucuronide sulfinyl conjugates. A total of 33 metabolites of umifenovir were identified in human plasma, urine, and feces by involving multiple enzymes, including CYP1A1, CYP2C19, CYP2D6, CYP2E1, CYP3A4 CYP3A5, FMO1, FMO3, and FMO5 [148]. After oral administration of arbidol to healthy volunteers, three primary metabolites (M5, M6–1, and M8) were detected in the plasma. The mean elimination half-life of these three metabolites was longer than that of the parent drug. Evaluating the safety and efficacy, M6-1 is essential because of its high exposure and prolonged elimination half-life. In the arbidol metabolism, CYP3A4 was the primary active enzyme followed by FMO3 (responsible for the formation of M6-1)[148]. It indicates possible drug interaction between umifenovir and CYP3A4 inhibitors and inducers. Further investigations are needed to under the importance of M6-1 in the efficacy and safety of umifenovir, because of its high exposure and long half-life (25.0 h).
4.6.2. Disease-drug interaction
There is a possibility to affect the inflammatory cytokine on the DMPK of umifenovir. Because CYP3A is the primary metabolic enzyme involved in the metabolism of umifenovir [148].
4.7. Nitazoxanide
Nitazoxanide is an antiparasitic and broad-spectrum antiviral drug. It has shown potential against SARS-CoV-2 and MERS-CoV in Vero E6 cells. Nitazoxanide prevents viral infection by enhancing the specific host mechanism [149]. The in-vitro activity of nitazoxanide against the SARC-CoV-2 suggested that more clinical data are required to assess the efficacy and safety against COVID-19 [119], [150]. While evaluating the efficacy of nitazoxanide alone and in combination with HQ, it reduced the requirement of insidious ventilator support for COVID-19 patients. Many clinical trials for nitazoxanide are currently proceeding with various doses to treat COVID-19 patients [151]. he FDA has granted Azidus Brasil's approval for nitazoxanide to carry on phase II clinical trial [152].
4.7.1. Drug-drug interaction
Although no drug-drug or disease-drug interaction studies have been performed in vivo, it is believed that no significant interaction would occur when nitazoxanide is co-administered with drugs that either is inducer or inhibitor of CYPs. But it can increase the exposure of valproate and benzodiazepines because of highly competitive protein binding with a low therapeutic window. No significant effect on QTc prolongation has been noticed in COVID-19 patients.
4.8. Ivermectin
Ivermectin is an effective antiparasitic agent that the FDA approved. Ivermectin has shown activity against many viruses [153]. Recently, one in-vitro study revealed that ivermectin strongly impeded the replication of COVID-19 [154]. Its antiviral activity may play a vital role and deliver a potential candidate to treat COVID-19 [155], [156]. Finally, FDA declared a report for self-administration of ivermectin in COVID-19 patients [157]. Ivermectin is broadly offered due to its inclusion on the WHO model list of necessary medicines.
4.8.1. Drug-drug interaction
Several authors assessed the effect of ivermectin in aging patients with different drugs used to control Helminthes. Ivermectin is broadly metabolized to several metabolites by CYP3A4, and it is a substrate for P-gp [158]. Less than 1% ivermectin dose is eliminated in the urine as the parent form. Therefore, a study would require controlling the factor affecting variability in the ivermectin exposure, including administering the dose in the fasted state, and excluding Pgp and CYP3A4 inhibitors which could increase the ivermectin level in the plasma. Since ivermectin is highl.
4.8.2. Disease-drug interaction
The PK of ivermectin in elderly patients has not been described. Ideally, metabolism may lower with age resulting in higher exposure to ivermectin in elderly patients [159]. Drug metabolism and disposition could also be changed by exposure to inflammation.
4.9. Interferons
Interferon (IFN) is a broad-spectrum antiviral agent that inhibits viral replication via interaction with toll-like receptors (TLR) [160]. It established antiviral resistance in cells [161], [162]. Moreover, a member of this family (IFN-λ4) was discovered for viral clearance [163]. IFN-λ was found to be more efficient with a slight increase in inflammation and tissue damage [164], [165] and potentially controlled viral spreading from the nasal epithelium to the upper respiratory tract [166], with efficacy as compared to IFNα-based therapies [167]. IFNα and β displayed activity against the SARS-CoV in-vitro [168], [169]. IFNβ indicated the potential in inhibiting MERS-CoV replication [170], [171]. Mainly type I IFN showed a fast decrease of viral load in mild to moderate COVID-19 patients. In the severe COVID-19 infection, IFN revealed the antiviral response with increased lung cytokine levels, reduced T cell response, and acute clinical relapse [172]. The latest study revealed that IFN had slightly or no effect on hospitalized patients with COVID-19, as suggested by overall mortality, initiation of ventilation [98], [99].
4.9.1. Drug-drug interaction
IFNα is believed to be responsible for the CYP dependent drug interactions interceded by a decrease in CYPs and transporters activities [173], [174].
4.10. Dexamethasone
Dexamethasone is a corticosteroid immunosuppressor. It lessens the ability of B cells to synthesize antibodies [175]. Dexamethasone regulates cytokine’s damaging effects by reducing the level of cytokine [176]. Moreover, dexamethasone prevents macrophages and natural killer cells from clearance secondary nosocomial pathogens [177]. Clinical evidence does not recommend the use of corticosteroids in COVID-19 infection [178]. Even though corticosteroid has been associated with an increase in the viral load, it persisted in the viral load after patients' survival from SARS-CoV [179]. By contrast, a clinical trial proved that dexamethasone saved the life of seriously ill COVID-19 infected patients in the United Kingdom (UK) [7]. UK government stated that dexamethasone was approved as an immediate treatment option for hospitalized patients that were seriously ill and on ventilator [180]. WHO added dexamethasone to the essential medicine list that is readily available at low cost. NIH issued the guideline to recommend dexamethasone as a treatment option for COVID-19 infected patients [181], [182].
4.10.1. Drug-drug interaction
Dexamethasone is a synthetic glucocorticoid, an agonist of nuclear pregnane X receptor (PXR) [183]. It induces the drug-metabolizing enzymes such as CYP3A4 [184], [185] CYP3A11, CYP2B10, and OATP2 [186], [187], [188]. Many variants of the drug-metabolizing enzymes (CYP3A4, CYP3A5, CYP3A7, and GSTT1) and transporters (ABCB1 and MDR1) have been associated with corticosteroid response across multiple disease condition [189]. The metabolic pathway of steroids is complex and genomic determinants with adequate evidence are still unknown for clinical application in COVID-19 infection.
4.10.2. Disease-drug interaction
There is a possibility to affect the inflammatory cytokine on the PK of dexamethasone. Because major inflammatory cytokines inhibit CYPs that primarily involved in the metabolism of dexamethasone.
4.11. Tetracyclines
Tetracyclines are a group of antibiotics. It can be used as a possible treatment option for COVID-19 patients because of its well-known activity to decrease the level of inflammatory cytokines such as IL-1b and IL-6 [190]. Both IL-1b and IL-6 levels were significantly increased during COVID-19 infection [191]. Tetracyclines also revealed that it lessened the inflammatory agent in the circulation and induced programmed cell death [192]. Investigators proposed that tetracyclines must be a better therapeutic option to treat inflammatory disorders [192], [193]. Previously it was documented for the treatment of HIV, west nile virus (WNV), and viral encephalitis diseases [194] and also used for the prevention of septic shock induced by ARDS [195]. It could be selected, as a potential treatment option for COVID-19 infection [196], [197]. In a recent study during the tetracyclines treatment, symptoms resolved in all patients within ten days. Surprisingly, ageusia and anosmia disappeared in the first week of tetracyclines treatment. It appears a promising drug to treat COVID-19 outpatients with mild symptoms [198].
4.11.1. Drug-drug interaction
The drug metabolism of the tetracycline is still unclear. In the clinical studies the antacids have been shown to decrease the bioavailability of tetracycline. Tetracyclines have the ability to form chelates that are frequently insoluble, reducing the absorption in the GI tract [199].
4.12. Teicoplanin
Teicoplanin is an antibiotic and used to treat severe infections caused by gram-positive bacteria. That can inhibit the replication and transcription of the competent virus. It also worked against the MERS and SARS as well [200]. Mechanistic studies revealed that teicoplanin inhibits the activity of the host cell’s cathepsin L and cathepsin B; these proteins are responsible for cleaving the viral glycoprotein, allowing contact receptor-binding domain of its core genome and subsequent release into the cytoplasm of the host cell [201], [202]. Therefore, these studies suggested that teicoplanin could be used as a therapeutic option for treating COVID-19 because SARS-CoV-2 is a cathepsin L-dependent virus. According to Ceccarelli et al., teicoplanin has shown efficacy in COVID-19 infected subjects [203]. Teicoplanin was recommended as a hopeful option for the treatment of COVID-19 even though its safety and efficacy data in humans is still unknown and need to be required. Evidence for drug interaction and path of absorption is still unclear for teicoplanin.
5. Atazanavir
Atazanavir (ATV) is an HIV-1 protease inhibitor (PI) currently suggested as a first-line treatment for naïve HIV-infected patients, as well as switch regimens for patients showing intolerance to other antiretroviral drugs (ARVs) [204]. ATV, alone or in combination with RTV, has shown the potential to inhibit the SARS-CoV-2 replication and pro-inflammatory cytokine production [205], [206]. ATV diminished IL-6 release in COVID-19 infected human primary monocytes. Cellular mortality and cytokine storm-associated intermediaries were lowered after treatment with ATV [206]. The ATV or ATV-RTV has demonstrated a new therapeutic option among clinically approved drugs that should be considered an effective treatment for COVID-19 infected patients.
5.1. Drug-drug interaction
ATV or ATV-RTV potentially inhibits the CYP3A4, UGT1A1, Pgp, BCRP OATP1B1/1B3. ATV is also a week inhibitor of CYP2C8. That have a higher potential for interaction, assessment of DDI is needed to inform dosing recommendations for COVID-19 Patients. Caution should be used when recommending ATV with co-medication that have the potential to increase the QT interval.
5.2. Disease-drug interaction
The effect of inflammation could have been diminished by the presence of booster RTV, which is steadily linked with ATV to decrease its clearance. In recent, two short reports specified higher plasma concentration of LPV in severe COVID-19 patients [126], [127], compared to those detected in HIV-patients and concerning inflammation [126].
6. Azithromycin
Azithromycin (Pfizer, NY USA) was revealed to be active in vitro against Ebola [207]. Moreover, azithromycin is considered to have the ability to prevent severe respiratory tract infection. Orally used azithromycin is distributed to a variety of tissues, particularly the lungs [208]. Azithromycin was used to treat COVID-19 patients in combination with HCQ [209].
6.1. Drug-drug interaction
Azithromycin is a macrolide antibiotic used patents with COVID-19 to cover the risk of secondary infection. It is widely distributed in the body with high tissue affinity. Its elimination half-life between 2 and 4 days. Azithromycin is known to be a P-gp inhibitor and, if co-administered with P-gp substrate (e.g., Digoxin), it has been reported to increase the serum level. It is also a week inhibitor of CYP3A4 [210], OATP1A2 and OATP2B1 [211]. This combination with HCQ was shown a reduction in COVID-19 associated mortality [209]. The risk of possible interactions is not pharmacokinetic, but it interacts pharmacodynamically. While prescribing azithromycin as comedication and showed the effect on QT prolongation. Co-prescription of azithromycin with HCQ increased the QT prolongation, increasing the risk of heart failure and cardiovascular mortality [212].
7. Darunavir
Darunavir is an HIV protease inhibitor approved in combination with cobicistat (pharmaco-enhancer) to treat both naïve and experienced patients infected with HIV-1 [213], [214]. This combination's safety and efficacy profile is already established based on III phase clinical trials [215], [216]. Existing data on the therapeutic effect of HIV protease inhibitors are from thorough in the COVID-19 patients. The inaccessibility of in-vitro activity against SARS-CoV-2, darunavir could not be endorsed to use as the treatment option for COVID-19. The in-vitro activity against SARS-CoV-2, darunavir could not be endorsed to use as the treatment option for COVID-19. Darunavir combined with cobicistat or with RTV should remain exclusively for the treatment of HIV patients [217]. In one study, the darunavir-cobicistat combination was found to be connected with considerable survival gain in critically ill patients of COVID-19 [218]. Darunavir lacks significant evidence for efficacy on COVID-19 patients. Its safety and clinical efficacy data are needed to be required.
7.1. Drug-drug interaction
DRV inhibits CYP3A, CYP2D6 and Pgp. It may interact medication with commonly taken by people, other antiretrovirals and antacids. Approximately 60% of all drugs are primarily metabolized by CYP3A4 and CYP2D6. DRV is a significant drug for DDI.
8. Ruxolitinib
Ruxolitinib is routinely used for the care of myelofibrosis, including polycythemia vera [219], [220]. It is a Janus kinase inhibitor and inhibits the JAK-STAT signaling [221]. Ruxolitinib was investigated against placebo for COVID-19 patients with ARDS. The two randomized phase III clinical trials (NCT04363137 and NCT04377620) evaluated mechanical ventilation requirements for COVID-19 patients with ARDS [222]. Ruxolitinib might be effective against the outcomes of the elevated levels of cytokines in COVID-19 patients [223]. Its safety and efficacy data in humans are still required in critically ill conditions.
8.1. Drug-drug interaction
Ruxolitinib is mainly metabolized by CYP3A4 and CYP2C9 and partially metabolized by CYP1A2 and CYP2B6 [224], [225]. Dual inhibitors of CYP3A4 and CYP29 (like fluconazole) could increase the exposure of ruxolitinib. The magnitude of this interaction was confirmed in healthy subjects based on a physiological-based pharmacokinetic (PBPK) model [226].
8.2. Disease-drug interaction
The inflammatory response reduces the mRNA expression of several CYP450 isozymes including CYP1A2, CYP2B6, CYP2C9 and CYP3A4 [106]. Therefore, inflammation could influence the DMPK properties of ruxolitinib.
9. Baricitinib
Baricitinib is used as a therapeutic option for rheumatoid arthritis. It also a reversible JAK-inhibitor [227], [228], [229]. The JAK-STAT signaling intervenes in the signaling of several cytokines, and interfering with this pathway may be an appealing approach to alter the immunopathology of SARS-CoV-2 [172], [230], [231]. Further, many drugs within this class exhibit antiviral effects, albeit often at supra-therapeutic concentration, by targeting host factors that viruses usurp for cell entry [232], [233]. Baricitinib has the advantage of providing in vitro antiviral activity at concentration achieved with approved dosing [230]. Baricitinib plus remidesivir co-medication was shown promise for treating COVID-19. This co-medication therapy was superior upon remdesivir alone in lowering recovery and accelerating improvement in clinical status. This combination has been shown fewer serious adverse effects [100].
9.1. Drug-drug interaction
Baricitinib is a substrate of OAT3, P-gp, BCRP, multidrug, and toxin extrusion protein (MATE)2K, and it is partially metabolized by CYP3A4. Baricitinib is also an inhibitor of OCT1, but clinical drug-drug interaction is still unclear. Probenecid, an inhibitor of OAT3, increased the exposure of baricitinib and decreased renal clearance in healthy subjects. In vitro, PBPK modeling reproduced the renal clearance and inhibitory effect of probenecid on baricitinib [234].
10. Imatinib
Imatinib is a tyrosine kinase inhibitor used to treat chronic myelogenous leukemia, gastrointestinal stromal tumors. It has been pointed out as an unexplored SARS-CoV-2 infection [235]. One case report on COVID-19 patients exhibited that who received imatinib due to clinical relapse even with dual therapy with HCQ and LPV/RTV [236]. One study was conducted to find the efficacy and safety of imatinib's oral administration combined with the best conventional care (BCC) versus placebo in hospitalized COVID-19 patients [237].
10.1. Drug-drug interaction
Imatinib, is exclusively metabolized by CYP2C8 and CYP3A4 [238]. Imatinib also showed the irreversible mechanism-based inhibition of CYP3A4 [239] and time-dependent autoinhibition of its own CYP3A4 metabolism leading to an essential role for CYP2C8 in the imatinib elimination. Pharmacogenetic polymorphism and drug interaction affect CYP2C8 activity during multiple dosing and may cause mark interindividual variability in the exposure of imatinib.
11. Fluvoxamine
Fluvoxamine is a selective serotonin reuptake inhibitor [240], [241] is used as an antidepressant. It is exceptionally hydrophilic and has fast intracellular uptake [242]. One study was conducted on adult symptomatic COVID-19 patients, and fluvoxamine was given orally to outpatients [8]. However, this study was restricted by a small sample size. This study was triggered by a hypothesis including the influence on the S1R-IRE1 pathway. Cytokine reduction resulting from S1R activation would fit with this recent finding of benefits of the other anti-inflammatory drugs for COVID-19 [243], [244]. The potential advantage of fluvoxamine for outpatient treatment of COVID-19 includes its safety [245], widespread availability, low cost, and oral administration. QT prolongation is not promoted by fluvoxamine like other SSRIs [246]. Nonetheless, fluvoxamine has adverse effects and can cause drug-drug interaction via inhibition of CYP1A2 and CYP2C19 [247].
11.1. Drug-drug interaction
Fluvoxamine half-life is about 30 h. It is known to inhibit CYP1A2. However, it is also a potent inhibitor of CYP2C19. It can slightly affect drugs that are metabolized by CYP3A4. Fluvoxamine is listed in the FDA table as a potent inhibitor for CYP1A2 and CYP2C19 and is commonly used for clinical DDI [248]. PBPK models were developed to characterize the CYP1A2 and CYP2C19 for interaction between fluvoxamine and other drugs [249].
11.2. Tocilizumab
Tocilizumab is a recombinant monoclonal antibody. Tocilizumab is mainly used to treat rheumatoid arthritis. It was conceived as an IL-6 receptor blocker to diminish inflammation. IL-6 drastically increases in patients when COVID-19 infection is produced [191]. That is why tocilizumab was used as a therapeutic option for treating COVID-19. In COVID-19 infected patients, T-lymphocyte and macrophages generate IL-6 to cause the cytokine storm and severe inflammatory responses, mainly in the lungs. Therefore, it is turned into an efficacious therapeutic drug for the treatment of severe COVID-19 infected patients [89], [250]. Tocilizumab exhibited a trend association towards lowered mortality among ICU patients [251]. The Genentech has been given approval by FDA continuing the phase III clinical trial for tocilizumab to assess the safety and efficacy of severe COVID-19 infected patients [252].
11.2.1. Drug-drug interaction
Tocilizumab has no direct inhibitory or inducing effects on CYPs. However, it converses IL-6 induced suppression of CYPs (elevation of IL-6 during inflammation has been revealed to inhibit CYP1A2, CYP2C9, CYP2C19 and CYP3A4 activity, ensuing in higher drug exposure of substrate drugs), which, prior to treatment with tocilizumab, has been regulated to metabolism of drugs. When treatment with tocilizumab is initiated, CYP activity normalizes, thus leading to reduce exposure of drugs which prior to treatment, had been altered to metabolism of therapeutic agents.
11.3. Itolizumab
Itolizumab is a recombinant monoclonal antibody for CD6 (Cluster of Differentiation 6) of IgG1 (Immunoglobulin G1). It is used for the treatment of psoriatic patients [253]. Itolizumab has shown the reduction of inflammatory cytokines, such as IFN-γ, TNF-α, and IL-6 [254], [255], [256]. It could be used as a treatment option for COVID-19 infection [257]. It showed the reduction of IL-6 in critically ill patients [258]. Biocon has acquired consent for itolizumab from the DGCI (Drugs Controller General of India) to treat COVID-19 patients in the emergency state [259]. Cuban regulatory agency has approved for the trial to use of itolizumab for COVID-19. Itolizumab might interrupt the hyperinflammatory cascade and stop COVID-19 morbidity and mortality [260].
11.3.1. Drug-drug interaction
Itolizumab has also no direct inhibitory or inducing effects on CYPs. However, it reverses cytokine induced suppression of CYPs (elevated by IFN-γ, TNF-α, and IL-6 during inflammation has been shown to inhibit various CYPs activity.
11.4. Meplazumab
Meplazumab is a humanized monoclonal antibody for CD147. It efficiently inhibited virus replication in Vero E6 cells [261]. One study has been conducted to ascertain the clinical results of meplazumab and revealed progress in the COVID-19 infected patients [262]. It was previously reported that meplazumab exhibited activity against the Chauge-Strauss syndrome [263]. Phase I clinical trial (NCT04369586) in the healthy volunteers of maplazumab has been achieved for finding the safety, efficacy, tolerability, PK attributes, and dosage regimen for Phase II clinical trial [264]. Phase II clinical trials are going in the USA to find the safety and efficacy of meplazumab injection in COVID-19 infected patients (NCT04275245). This trial will be completed in December 2020 [265]. DDI for maplazumab is still unclear.
11.5. Eculizumab
Eculizumab is a monoclonal antibody for complement C5 protein. It prevents cleavage to C5a and C5b and impedes the creation of the membrane attack complex (MAC) C5b-9 to stop the cell's lysis [266]. Eculizumab was disclosed to be an effective therapeutic option for hematological and neuroinflammatory diseases [266], [267], [268], [269]. Evermore, evidence [270] shows that complement is also a key mediator of lung damage, notably during CoV infection. Therefore, eculizumab might work as an emergency therapy to treat COVID-19 patients associated with ARDS. Some studies [271], [272] supported the eculizumab use as a treatment for severe COVID-19. Studies were performed along with ruxolitinib for confirming the efficacy of eculizumab in severe COVID-19 patients [273], [274]. It has been approved for continuing the clinical trial. DDI for Eculizumab is still unclear.
11.6. AMY101
AMY101 is a highly selective complement C3 inhibitor developed by Amyndas Pharmaceuticals [275], [276], [277]. It is a small-sized cyclic peptide that indicated more promising efficacy in non-human primates [278]. AMY101 has effectively completed the phase I clinical trial with acceptable safety and tolerability, and now it is in phase II clinical trial (NCT04395456) [279], [280]. Some studies [274], [281] have shown the proinflammatory response by activating the complement system (C3) in COVID-19 patients. AMY101 could be a unique therapeutic option to overcome the complement-mediated inflammatory response in COVID-19 patients. The recent clinical study [282], AMY101, revealed the safety and efficacy in patients with severe ARDS due to COVID-19 infection. DDI for AMY101 is still unclear.
11.7. ARDS-003
Cannabinoid (CBD) is also a probable treatment for severe COVID-19 patients [10]. It was designed as an injectable form to treat a serious case of coronavirus “acute respiratory distress syndrome (ARDS).” This syndrome generated a cytokine storm to create inflammation. It will have the advantage of impacting several pro-inflammatory signaling pathways by enhancing the drug's effectiveness to rapidly diminish the release of the cytokines and avert acute outcomes like ARDS. The cannabinoid drug named ARDS-003 has been approved for the phase I clinical trial. It is still being tested by Tetra Bio-Pharma [284]. Firstly, the FDA emphasized that the nonclinical study results were appropriate for starting a study in COVID-19 infected patients [285].
11.7.1. Drug-drug interaction
CBD induces CYP1A2 and it inhibits Pgp. So, this may be significant in patients co-administered a drug metabolized by CYP1A2 and Pgp mediated drug transport. Co-administration of CYP3 inhibitor, ketoconazole approximately AUC was twofold increased, whilst co-administration of CYP3A inducer, rifampicin significantly reduces AUC of CBD. Invitro CBD was observed to be potent inhibitor of CYP2C19. For instance, CBD facilitated inhibition of Clobazam metabolism has been resulted in an up to eight-fold increase clobazam concentration [286]. Conferring to clinician CBD have a capability for drug interactions.
11.8. LCB1
LCB1 revealed as the SARS-CoV-2 neutralizing antibody. It is a computer-designed mini protein that has been synthesized by the researchers of the University of Washington School of Medicine. It binds firmly to SARS-CoV-2 spikes proteins and hinders them from infecting cells. LCB1 appeared to protect the Vero E6 cells from SARS-CoV-2 infection. This synthesized antiviral candidate was conceived to overcome the infection by interfering with the coronavirus mechanism to break into and enter cells. LCB1 is presently being evaluated in rodents [287]. These hyper stable mini-binders provide the starting point for COVID-19 therapeutics [288]. DDI for LCB1 is still unknown and need to be known for such potential drug.
12. Molnupiravir
Molnupiravir (MK448/EIDD-2801) is a prodrug of synthetic nucleoside derivative N4-hydroxy-cytidine and applies its antiviral action via the introduction of copying error during viral RNA replication [289]. Institute for Biomedical Sciences, Georgia State University, developed this antiviral drug. After finding to be active against SARC-CoV-2, MK448/EIDD-2801 was analyzed in the preliminary human study for safety, tolerability, and pharmacokinetics in healthy volunteers in the UK and USA (NCT04392219). On October 19, 2020, Merck started a one-year stage 2/3 trial aimed at hospitalized patients (NCT04575584). MK448/EIDD-2801 can block the transmission of SARS-CoV-2 within 24 h [9]. Plemper’s team repurposed molnupiravir against COVID-19 and applied a ferret model to test the effect of it on containing the spread of the COVID-19 [9]. DDI for molnupiravir needs to be known for better dosing regimen.
13. Conclusions
The present review pointed out the possibilities of risk of drug interaction of mentioned drug in tackling COVID-19. The inflammatory response enforces changes in the expression and activity of transporters and DMEs. Disposition of medicines used to treat COVID-19 infection involves drug metabolism CYPs enzymes and drug transported by ABC and SLC transporters. However, it is already known that ABC and SLC transporters play a central role in the disposition of mostly antiviral drugs and can participate in many drug-drug interactions. Most importantly, the involvement of CYPs in used drugs in COVID-19 infection, drug-drug interaction has been comprehensively known, but some drugs are still unknown. That’s why in COVID-19 conditions, inflammatory responses play a crucial role in disease-drug or drug-drug interactions. Alteration in the transporters and DMEs can lead to changes in the pharmacokinetic parameters of the drug used. Hence, inflammation might play an essential role in drug efficacy and toxicity. The risk of drug interactions should not be prohibited since they are frequently manageable and convenient. LPV/RTV is being used in combination with other drugs. HCQ in combinations with azithromycin and with LPV/RTV has been used in COVID-19 Patients. These medications have been classified as having a risk of developing torsades de points (TdP). Moderate to severe QTc prolongation was observed during these pharmacological treatments [344]. The ATV-RTV has shown new options among clinically approved drugs and should be considered an effective treatment option. Another combination of remdesivir with bericitinib has shown severe side effects even though this combination has shown promise for COVID-19 with accelerating clinical status improvement [100]. The consumption of a single drug may possibly not be more effective but, during co-medication of multiple medications, the risk of drug interaction must be increased. The potential of drug-drug interaction or disease-drug interaction is an important consideration when identifying optimal treatment regimens for individual patients.
Statement of funding source
Authors have not received any funding from any private and government organization.
Author contribution
All authors collected the data, reviewed, and prepared the manuscript and final approval was done by DK for publication.
Declaration of conflicting interests
The authors declared that there are no conflicts of interest
Acknowledgments
Both authors are thankful for UNMC to provide the platform.
References
- 1.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9(4) doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob. Agents Chemother. 2020;64(5) doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitja O., Clotet B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob. Health. 2020;8(5):e639–e640. doi: 10.1016/S2214-109X(20)30114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rismanbaf A. Potential treatments for COVID-19; a narrative literature review. Arch. Acad. Emerg. Med. 2020;8(1) [PMC free article] [PubMed] [Google Scholar]
- 7.Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582(7813):469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- 8.Lenze E.J., Mattar C., Zorumski C.F., Stevens A., Schweiger J., Nicol G.E., Miller J.P., Yang L., Yingling M., Avidan M.S., Reiersen A.M. Fluvoxamine vs placebo and clinical deterioration in outpatients with symptomatic COVID-19: a randomized clinical trial. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trivedi N., Verma A., Kumar D. Possible treatment and strategies for COVID-19: review and assessment. Eur. Rev. Med. Pharmacol. Sci. 2020;24(23):12593–12608. doi: 10.26355/eurrev_202012_24057. [DOI] [PubMed] [Google Scholar]
- 11.Mullard A. COVID-19 vaccine development pipeline gears up. Lancet. 2020;395(10239):1751–1752. doi: 10.1016/S0140-6736(20)31252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Don B.R., Kaysen G. Serum albumin: relationship to inflammation and nutrition. Semin Dial. 2004;17(6):432–437. doi: 10.1111/j.0894-0959.2004.17603.x. [DOI] [PubMed] [Google Scholar]
- 13.Moshage H.J., Janssen J.A., Franssen J.H., Hafkenscheid J.C., Yap S.H. Study of the molecular mechanism of decreased liver synthesis of albumin in inflammation. J. Clin. Investig. 1987;79(6):1635–1641. doi: 10.1172/JCI113000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruot B., Bechereau F., Bayle G., Breuille D., Obled C. The response of liver albumin synthesis to infection in rats varies with the phase of the inflammatory process. Clin. Sci. 2002;102(1):107–114. [PubMed] [Google Scholar]
- 15.Morgan E.T. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin. Pharmacol. Ther. 2009;85(4):434–438. doi: 10.1038/clpt.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan E.T., Goralski K.B., Piquette-Miller M., Renton K.W., Robertson G.R., Chaluvadi M.R., Charles K.A., Clarke S.J., Kacevska M., Liddle C., Richardson T.A., Sharma R., Sinal C.J. Regulation of drug-metabolizing enzymes and transporters in infection, inflammation, and cancer. Drug Metab. Dispos. 2008;36(2):205–216. doi: 10.1124/dmd.107.018747. [DOI] [PubMed] [Google Scholar]
- 17.Cressman A.M., Petrovic V., Piquette-Miller M. Inflammation-mediated changes in drug transporter expression/activity: implications for therapeutic drug response. Expert Rev. Clin. Pharmacol. 2012;5(1):69–89. doi: 10.1586/ecp.11.66. [DOI] [PubMed] [Google Scholar]
- 18.Grasselli G., Pesenti A., Cecconi M. Critical care utilization for the COVID-19 outbreak in lombardy, Italy: early experience and forecast during an emergency response. J. Am. Med. Assoc. 2020;28(323):1545–1546. doi: 10.1001/jama.2020.4031. [DOI] [PubMed] [Google Scholar]
- 19.Mann H.J. Drug-associated disease: cytochrome P450 interactions. Crit. Care Clin. 2006;22(2):329–345. doi: 10.1016/j.ccc.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100(2):446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee G., Piquette-Miller M. Influence of IL-6 on MDR and MRP-mediated multidrug resistance in human hepatoma cells. Can. J. Physiol. Pharmacol. 2001;79(10):876–884. [PubMed] [Google Scholar]
- 22.Sukhai M., Yong A., Kalitsky J., Piquette-Miller M. Inflammation and interleukin-6 mediate reductions in the hepatic expression and transcription of the mdr1a and mdr1b Genes. Mol. Cell Biol. Res. Commun. 2000;4(4):248–256. doi: 10.1006/mcbr.2001.0288. [DOI] [PubMed] [Google Scholar]
- 23.Blokzijl H., Vander Borght S., Bok L.I., Libbrecht L., Geuken M., van den Heuvel F.A., Dijkstra G., Roskams T.A., Moshage H., Jansen P.L., Faber K.N. Decreased P-glycoprotein (P-gp/MDR1) expression in inflamed human intestinal epithelium is independent of PXR protein levels. Inflamm. Bowel Dis. 2007;13(6):710–720. doi: 10.1002/ibd.20088. [DOI] [PubMed] [Google Scholar]
- 24.Belliard A.M., Lacour B., Farinotti R., Leroy C. Effect of tumor necrosis factor-alpha and interferon-gamma on intestinal P-glycoprotein expression, activity, and localization in Caco-2 cells. J. Pharm. Sci. 2004;93(6):1524–1536. doi: 10.1002/jps.20072. [DOI] [PubMed] [Google Scholar]
- 25.Buyse M., Radeva G., Bado A., Farinotti R. Intestinal inflammation induces adaptation of P-glycoprotein expression and activity. Biochem Pharmacol. 2005;69(12):1745–1754. doi: 10.1016/j.bcp.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Le Vee M., Lecureur V., Stieger B., Fardel O. Regulation of drug transporter expression in human hepatocytes exposed to the proinflammatory cytokines tumor necrosis factor-alpha or interleukin-6. Drug Metab. Dispos. 2009;37(3):685–693. doi: 10.1124/dmd.108.023630. [DOI] [PubMed] [Google Scholar]
- 27.Le Vee M., Jouan E., Moreau A., Fardel O. Regulation of drug transporter mRNA expression by interferon-gamma in primary human hepatocytes. Fundam. Clin. Pharmacol. 2011;25(1):99–103. doi: 10.1111/j.1472-8206.2010.00822.x. [DOI] [PubMed] [Google Scholar]
- 28.Poller B., Drewe J., Krahenbuhl S., Huwyler J., Gutmann H. Regulation of BCRP (ABCG2) and P-glycoprotein (ABCB1) by cytokines in a model of the human blood-brain barrier. Cell Mol. Neurobiol. 2010;30(1):63–70. doi: 10.1007/s10571-009-9431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Vee M., Gripon P., Stieger B., Fardel O. Down-regulation of organic anion transporter expression in human hepatocytes exposed to the proinflammatory cytokine interleukin 1beta. Drug Metab. Dispos. 2008;36(2):217–222. doi: 10.1124/dmd.107.016907. [DOI] [PubMed] [Google Scholar]
- 30.Diao L., Li N., Brayman T.G., Hotz K.J., Lai Y. Regulation of MRP2/ABCC2 and BSEP/ABCB11 expression in sandwich cultured human and rat hepatocytes exposed to inflammatory cytokines TNF-{alpha}, IL-6, and IL-1{beta} J. Biol. Chem. 2010;285(41):31185–31192. doi: 10.1074/jbc.M110.107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Herwaarden A.E., Jonker J.W., Wagenaar E., Brinkhuis R.F., Schellens J.H., Beijnen J.H., Schinkel A.H. The breast cancer resistance protein (Bcrp1/Abcg2) restricts exposure to the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Res. 2003;63(19):6447–6452. [PubMed] [Google Scholar]
- 32.Langmann T., Moehle C., Mauerer R., Scharl M., Liebisch G., Zahn A., Stremmel W., Schmitz G. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127(1):26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Mak R.H., Cheung W., Cone R.D., Marks D.L. Mechanisms of disease: cytokine and adipokine signaling in uremic cachexia. Nat. Clin. Pract. Nephrol. 2006;2(9):527–534. doi: 10.1038/ncpneph0273. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki R., Miyamoto S., Yasui Y., Sugie S., Tanaka T. Global gene expression analysis of the mouse colonic mucosa treated with azoxymethane and dextran sodium sulfate. BMC Cancer. 2007;7:84. doi: 10.1186/1471-2407-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Heijden J.W., Dijkmans B.A., Scheper R.J., Jansen G. Drug Insight: resistance to methotrexate and other disease-modifying antirheumatic drugs--from bench to bedside. Nat. Clin. Pract. Rheumatol. 2007;3(1):26–34. doi: 10.1038/ncprheum0380. [DOI] [PubMed] [Google Scholar]
- 36.Wojtal K.A., Eloranta J.J., Hruz P., Gutmann H., Drewe J., Staumann A., Beglinger C., Fried M., Kullak-Ublick G.A., Vavricka S.R. Changes in mRNA expression levels of solute carrier transporters in inflammatory bowel disease patients. Drug Metab. Dispos. 2009;37(9):1871–1877. doi: 10.1124/dmd.109.027367. [DOI] [PubMed] [Google Scholar]
- 37.Ho E.A., Piquette-Miller M. KLF6 and HSF4 transcriptionally regulate multidrug resistance transporters during inflammation. Biochem. Biophys. Res. Commun. 2007;353(3):679–685. doi: 10.1016/j.bbrc.2006.12.090. [DOI] [PubMed] [Google Scholar]
- 38.Kameyama N., Arisawa S., Ueyama J., Kagota S., Shinozuka K., Hattori A., Tatsumi Y., Hayashi H., Takagi K., Wakusawa S. Increase in P-glycoprotein accompanied by activation of protein kinase Calpha and NF-kappaB p65 in the livers of rats with streptozotocin-induced diabetes. Biochim. Biophys. Acta. 2008;1782(5):355–360. doi: 10.1016/j.bbadis.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Teng S., Piquette-Miller M. Regulation of transporters by nuclear hormone receptors: implications during inflammation. Mol. Pharm. 2008;5(1):67–76. doi: 10.1021/mp700102q. [DOI] [PubMed] [Google Scholar]
- 40.Yu C., Argyropoulos G., Zhang Y., Kastin A.J., Hsuchou H., Pan W. Neuroinflammation activates Mdr1b efflux transport through NFkappaB: promoter analysis in BBB endothelia. Cell Physiol. Biochem. 2008;22(5–6):745–756. doi: 10.1159/000185558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan W., Yu C., Hsuchou H., Kastin A.J. The role of cerebral vascular NFkappaB in LPS-induced inflammation: differential regulation of efflux transporter and transporting cytokine receptors. Cell Physiol. Biochem. 2010;25(6):623–630. doi: 10.1159/000315081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kosters A., Karpen S.J. The role of inflammation in cholestasis: clinical and basic aspects. Semin. Liver Dis. 2010;30(2):186–194. doi: 10.1055/s-0030-1253227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirona R.G. Handb Exp Pharmacol. 2011. Molecular mechanisms of drug transporter regulation; pp. 373–402. [DOI] [PubMed] [Google Scholar]
- 44.Harvey R.D., Morgan E.T. Cancer, inflammation, and therapy: effects on cytochrome p450-mediated drug metabolism and implications for novel immunotherapeutic agents. Clin. Pharmacol. Ther. 2014;96(4):449–457. doi: 10.1038/clpt.2014.143. [DOI] [PubMed] [Google Scholar]
- 45.Wu K.C., Lin C.J. The regulation of drug-metabolizing enzymes and membrane transporters by inflammation: evidences in inflammatory diseases and age-related disorders. J. Food Drug Anal. 2019;27(1):48–59. doi: 10.1016/j.jfda.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan E.T. Regulation of cytochromes P450 during inflammation and infection. Drug Metab. Rev. 1997;29(4):1129–1188. doi: 10.3109/03602539709002246. [DOI] [PubMed] [Google Scholar]
- 47.Aitken A.E., Morgan E.T. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab. Dispos. 2007;35(9):1687–1693. doi: 10.1124/dmd.107.015511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muntane-Relat J., Ourlin J.C., Domergue J., Maurel P. Differential effects of cytokines on the inducible expression of CYP1A1, CYP1A2, and CYP3A4 in human hepatocytes in primary culture. Hepatology. 1995;22(4 Pt 1):1143–1153. [PubMed] [Google Scholar]
- 49.Calleja C., Eeckhoutte C., Dacasto M., Larrieu G., Dupuy J., Pineau T., Galtier P. Comparative effects of cytokines on constitutive and inducible expression of the gene encoding for the cytochrome P450 3A6 isoenzyme in cultured rabbit hepatocytes: consequences on progesterone 6beta-hydroxylation. Biochem. Pharmacol. 1998;56(10):1279–1285. doi: 10.1016/s0006-2952(98)00178-6. [DOI] [PubMed] [Google Scholar]
- 50.Tapner M., Liddle C., Goodwin B., George J., Farrell G.C. Interferon gamma down-regulates cytochrome P450 3A genes in primary cultures of well-differentiated rat hepatocytes. Hepatology. 1996;24(2):367–373. doi: 10.1002/hep.510240213. [DOI] [PubMed] [Google Scholar]
- 51.Monshouwer M., Witkamp R.F., Nujmeijer S.M., Van Amsterdam J.G., Van Miert A.S. Suppression of cytochrome P450- and UDP glucuronosyl transferase-dependent enzyme activities by proinflammatory cytokines and possible role of nitric oxide in primary cultures of pig hepatocytes. Toxicol. Appl. Pharmacol. 1996;137(2):237–244. doi: 10.1006/taap.1996.0077. [DOI] [PubMed] [Google Scholar]
- 52.Rubin K., Janefeldt A., Andersson L., Berke Z., Grime K., Andersson T.B. HepaRG cells as human-relevant in vitro model to study the effects of inflammatory stimuli on cytochrome P450 isoenzymes. Drug Metab. Dispos. 2015;43(1):119–125. doi: 10.1124/dmd.114.059246. [DOI] [PubMed] [Google Scholar]
- 53.Abdel-Razzak Z., Loyer P., Fautrel A., Gautier J.C., Corcos L., Turlin B., Beaune P., Guillouzo A. Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Mol. Pharmacol. 1993;44(4):707–715. [PubMed] [Google Scholar]
- 54.Pan J., Xiang Q., Ball S. Use of a novel real-time quantitative reverse transcription-polymerase chain reaction method to study the effects of cytokines on cytochrome P450 mRNA expression in mouse liver. Drug Metab. Dispos. 2000;28(6):709–713. [PubMed] [Google Scholar]
- 55.Morgan E.T. Down-regulation of multiple cytochrome P450 gene products by inflammatory mediators in vivo. Independence from the hypothalamo-pituitary axis. Biochem. Pharmacol. 1993;45(2):415–419. doi: 10.1016/0006-2952(93)90078-b. [DOI] [PubMed] [Google Scholar]
- 56.Frye R.F., Schneider V.M., Frye C.S., Feldman A.M. Plasma levels of TNF-alpha and IL-6 are inversely related to cytochrome P450-dependent drug metabolism in patients with congestive heart failure. J. Card. Fail. 2002;8(5):315–319. doi: 10.1054/jcaf.2002.127773. [DOI] [PubMed] [Google Scholar]
- 57.Liptrott N.J., Penny M., Bray P.G., Sathish J., Khoo S.H., Back D.J., Owen A. The impact of cytokines on the expression of drug transporters, cytochrome P450 enzymes and chemokine receptors in human PBMC. Br. J. Pharmacol. 2009;156(3):497–508. doi: 10.1111/j.1476-5381.2008.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholson T.E., Renton K.W. Role of cytokines in the lipopolysaccharide-evoked depression of cytochrome P450 in the brain and liver. Biochem. Pharmacol. 2001;62(12):1709–1717. doi: 10.1016/s0006-2952(01)00859-0. [DOI] [PubMed] [Google Scholar]
- 59.Bertilsson P.M., Olsson P., Magnusson K.E. Cytokines influence mRNA expression of cytochrome P450 3A4 and MDRI in intestinal cells. J. Pharm. Sci. 2001;90(5):638–646. doi: 10.1002/1520-6017(200105)90:5<638::aid-jps1020>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 60.Nicholson T.E., Renton K.W. Modulation of cytochrome P450 by inflammation in astrocytes. Brain Res. 1999;827(1–2):12–18. doi: 10.1016/s0006-8993(99)01261-5. [DOI] [PubMed] [Google Scholar]
- 61.Dickmann L.J., Patel S.K., Wienkers L.C., Slatter J.G. Effects of interleukin 1beta (IL-1beta) and IL-1beta/interleukin 6 (IL-6) combinations on drug metabolizing enzymes in human hepatocyte culture. Curr. Drug Metab. 2012;13(7):930–937. doi: 10.2174/138920012802138642. [DOI] [PubMed] [Google Scholar]
- 62.Iber H., Chen Q., Cheng P.Y., Morgan E.T. Suppression of CYP2C11 gene transcription by interleukin-1 mediated by NF-kappaB binding at the transcription start site. Arch. Biochem. Biophys. 2000;377(1):187–194. doi: 10.1006/abbi.2000.1772. [DOI] [PubMed] [Google Scholar]
- 63.Parmentier J.H., Kremers P., Ferrari L., Batt A.M., Gielen J.E., Siest G. Repression of cytochrome P450 by cytokines: IL-1 beta counteracts clofibric acid induction of CYP4A in cultured fetal rat hepatocytes. Cell Biol. Toxicol. 1993;9(3):307–313. doi: 10.1007/BF00755608. [DOI] [PubMed] [Google Scholar]
- 64.Bleau A.M., Levitchi M.C., Maurice H., du Souich P. Cytochrome P450 inactivation by serum from humans with a viral infection and serum from rabbits with a turpentine-induced inflammation: the role of cytokines. Br. J. Pharmacol. 2000;130(8):1777–1784. doi: 10.1038/sj.bjp.0703486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donato M.T., Guillen M.I., Jover R., Castell J.V., Gomez-Lechon M.J. Nitric oxide-mediated inhibition of cytochrome P450 by interferon-gamma in human hepatocytes. J. Pharmacol. Exp. Ther. 1997;281(1):484–490. [PubMed] [Google Scholar]
- 66.Nadin L., Butler A.M., Farrell G.C., Murray M. Pretranslational down-regulation of cytochromes P450 2C11 and 3A2 in male rat liver by tumor necrosis factor alpha. Gastroenterology. 1995;109(1):198–205. doi: 10.1016/0016-5085(95)90285-6. [DOI] [PubMed] [Google Scholar]
- 67.Gorski J.C., Hall S.D., Becker P., Affrime M.B., Cutler D.L., Haehner-Daniels B. In vivo effects of interleukin-10 on human cytochrome P450 activity. Clin. Pharmacol. Ther. 2000;67(1):32–43. doi: 10.1067/mcp.2000.103860. [DOI] [PubMed] [Google Scholar]
- 68.Elkahwaji J., Robin M.A., Berson A., Tinel M., Letteron P., Labbe G., Beaune P., Elias D., Rougier P., Escudier B., Duvillard P., Pessayre D. Decrease in hepatic cytochrome P450 after interleukin-2 immunotherapy. Biochem. Pharmacol. 1999;57(8):951–954. doi: 10.1016/s0006-2952(98)00372-4. [DOI] [PubMed] [Google Scholar]
- 69.Tinel M., Robin M.A., Doostzadeh J., Maratrat M., Ballet F., Fardel N., el Kahwaji J., Beaune P., Daujat M., Labbe G., et al. The interleukin-2 receptor down-regulates the expression of cytochrome P450 in cultured rat hepatocytes. Gastroenterology. 1995;109(5):1589–1599. doi: 10.1016/0016-5085(95)90648-7. [DOI] [PubMed] [Google Scholar]
- 70.Williams J.F., Bement W.J., Sinclair J.F., Sinclair P.R. Effect of interleukin 6 on phenobarbital induction of cytochrome P-450IIB in cultured rat hepatocytes. Biochem. Biophys. Res. Commun. 1991;178(3):1049–1055. doi: 10.1016/0006-291x(91)90998-m. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y.L., Florentin I., Batt A.M., Ferrari L., Giroud J.P., Chauvelot-Moachon L. Effects of interleukin-6 on cytochrome P450-dependent mixed-function oxidases in the rat. Biochem. Pharmacol. 1992;44(1):137–148. doi: 10.1016/0006-2952(92)90047-m. [DOI] [PubMed] [Google Scholar]
- 72.Siewert E., Bort R., Kluge R., Heinrich P.C., Castell J., Jover R. Hepatic cytochrome P450 down-regulation during aseptic inflammation in the mouse is interleukin 6 dependent. Hepatology. 2000;32(1):49–55. doi: 10.1053/jhep.2000.8532. [DOI] [PubMed] [Google Scholar]
- 73.Morgan E.T. Suppression of constitutive cytochrome P-450 gene expression in livers of rats undergoing an acute phase response to endotoxin. Mol. Pharmacol. 1989;36(5):699–707. [PubMed] [Google Scholar]
- 74.Robertson G.R., Field J., Goodwin B., Bierach S., Tran M., Lehnert A., Liddle C. Transgenic mouse models of human CYP3A4 gene regulation. Mol. Pharmacol. 2003;64(1):42–50. doi: 10.1124/mol.64.1.42. [DOI] [PubMed] [Google Scholar]
- 75.Kacevska M., Mahns A., Sharma R., Clarke S.J., Robertson G.R., Liddle C. Extra-hepatic cancer represses hepatic drug metabolism via interleukin (IL)-6 signalling. Pharm. Res. 2013;30(9):2270–2278. doi: 10.1007/s11095-013-1042-3. [DOI] [PubMed] [Google Scholar]
- 76.Mimura H., Kobayashi K., Xu L., Hashimoto M., Ejiri Y., Hosoda M., Chiba K. Effects of cytokines on CYP3A4 expression and reversal of the effects by anti-cytokine agents in the three-dimensionally cultured human hepatoma cell line FLC-4. Drug Metab. Pharmacokinet. 2015;30(1):105–110. doi: 10.1016/j.dmpk.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 77.Zordoky B.N., El-Kadi A.O. Role of NF-kappaB in the regulation of cytochrome P450 enzymes. Curr. Drug Metab. 2009;10(2):164–178. doi: 10.2174/138920009787522151. [DOI] [PubMed] [Google Scholar]
- 78.Hayden M.S., West A.P., Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25(51):6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 79.Tian Y., Rabson A.B., Gallo M.A. Ah receptor and NF-kappaB interactions: mechanisms and physiological implications. Chem. Biol. Interact. 2002;141(1–2):97–115. doi: 10.1016/s0009-2797(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 80.Ke S., Rabson A.B., Germino J.F., Gallo M.A., Tian Y. Mechanism of suppression of cytochrome P-450 1A1 expression by tumor necrosis factor-alpha and lipopolysaccharide. J. Biol. Chem. 2001;276(43):39638–39644. doi: 10.1074/jbc.M106286200. [DOI] [PubMed] [Google Scholar]
- 81.Yang H., Sun R., Ma N., Liu Q., Sun X., Zi P., Wang J., Chao K., Yu L. Inhibition of nuclear factor-kappaB signal by pyrrolidine dithiocarbamate alleviates lipopolysaccharide-induced acute lung injury. Oncotarget. 2017;8(29):47296–47304. doi: 10.18632/oncotarget.17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kourylko O., Fradette C., Arcand M., du Souich P. Modulation of CYP1A2 and CYP3A6 catalytic activities by serum from rabbits with a turpentine-induced inflammatory reaction and interleukin 6. Drug Metab. Dispos. 2006;34(1):27–35. doi: 10.1124/dmd.105.006528. [DOI] [PubMed] [Google Scholar]
- 83.Yang J., Hao C., Yang D., Shi D., Song X., Luan X., Hu G., Yan B. Pregnane X receptor is required for interleukin-6-mediated down-regulation of cytochrome P450 3A4 in human hepatocytes. Toxicol. Lett. 2010;197(3):219–226. doi: 10.1016/j.toxlet.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou C., Tabb M.M., Nelson E.L., Grun F., Verma S., Sadatrafiei A., Lin M., Mallick S., Forman B.M., Thummel K.E., Blumberg B. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J. Clin. Investig. 2006;116(8):2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gu X., Ke S., Liu D., Sheng T., Thomas P.E., Rabson A.B., Gallo M.A., Xie W., Tian Y. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J. Biol. Chem. 2006;281(26):17882–17889. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- 86.Assenat E., Gerbal-chaloin S., Maurel P., Vilarem M.J., Pascussi J.M. Is nuclear factor kappa-B the missing link between inflammation, cancer and alteration in hepatic drug metabolism in patients with cancer? Eur. J. Cancer. 2006;42(6):785–792. doi: 10.1016/j.ejca.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 91.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 92.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., Ali R., Coulibaly S., Levine A.C., Grais R., Diaz J., Lane H.C., Muyembe-Tamfum J.J., Group P.W., Sivahera B., Camara M., Kojan R., Walker R., Dighero-Kemp B., Cao H., Mukumbayi P., Mbala-Kingebeni P., Ahuka S., Albert S., Bonnett T., Crozier I., Duvenhage M., Proffitt C., Teitelbaum M., Moench T., Aboulhab J., Barrett K., Cahill K., Cone K., Eckes R., Hensley L., Herpin B., Higgs E., Ledgerwood J., Pierson J., Smolskis M., Sow Y., Tierney J., Sivapalasingam S., Holman W., Gettinger N., Vallee D., Nordwall J., Team P.C.S. A randomized, controlled trial of ebola virus disease therapeutics. N. Engl. J. Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ko W.C., Rolain J.M., Lee N.Y., Chen P.L., Huang C.T., Lee P.I., Hsueh P.R. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.NIH, NIH US National Library of Medicine clinical trial database https://clinicaltrials.gov/ct2/results?cond¼COVID&term¼remdesivir&cntry¼&state¼&city¼&dist¼ Accessed 23 May 2020, 2020.
- 98.Zhang R., Mylonakis E. In inpatients with COVID-19, none of remdesivir, hydroxychloroquine, lopinavir, or interferon beta-1a differed from standard care for in-hospital mortality. Ann. Intern. Med. 2021;174(2):JC17. doi: 10.7326/ACPJ202102160-017. [DOI] [PubMed] [Google Scholar]
- 99.Consortium W.H.O.S.T., Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernandez Garcia C., Kieny M.P., Malekzadeh R., Murthy S., Reddy K.S., Roses Periago M., Hanna P. Abi, Ader F., Al-Bader A.M., Alhasawi A., Allum E., Alotaibi A., Alvarez-Moreno C.A., Appadoo S., Asiri A., Aukrust P., Barratt-Due A., Bellani S., Branca M., Cappel-Porter H.B.C., Cerrato N., Chow T.S., Como N., Eustace J., Garcia P.J., Godbole S., Gotuzzo E., Griskevicius L., Hamra R., Hassan M., Hassany M., Hutton D., Irmansyah I., Jancoriene L., Kirwan J., Kumar S., Lennon P., Lopardo G., Lydon P., Magrini N., Maguire T., Manevska S., Manuel O., McGinty S., Medina M.T., Rubio M.L. Mesa, Miranda-Montoya M.C., Nel J., Nunes E.P., Perola M., Portoles A., Rasmin M.R., Raza A., Rees H., Reges P.P.S., Rogers C.A., Salami K., Salvadori M.I., Sinani N., Sterne J.A.C., Stevanovikj M., Tacconelli E., Tikkinen K.A.O., Trelle S., Zaid H., Rottingen J.A., Swaminathan S. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2021;384(6):497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kalil A.C., Patterson T.F., Mehta A.K., Tomashek K.M., Wolfe C.R., Ghazaryan V., Marconi V.C., Ruiz-Palacios G.M., Hsieh L., Kline S., Tapson V., Iovine N.M., Jain M.K., Sweeney D.A., El Sahly H.M., Branche A.R., Regalado Pineda J., Lye D.C., Sandkovsky U., Luetkemeyer A.F., Cohen S.H., Finberg R.W., Jackson P.E.H., Taiwo B., Paules C.I., Arguinchona H., Goepfert P., Ahuja N., Frank M., Oh M.D., Kim E.S., Tan S.Y., Mularski R.A., Nielsen H., Ponce P.O., Taylor B.S., Larson L., Rouphael N.G., Saklawi Y., Cantos V.D., Ko E.R., Engemann J.J., Amin A.N., Watanabe M., Billings J., Elie M.C., Davey R.T., Burgess T.H., Ferreira J., Green M., Makowski M., Cardoso A., de Bono S., Bonnett T., Proschan M., Deye G.A., Dempsey W., Nayak S.U., Dodd L.E., Beigel J.H. Baricitinib plus remdesivir for hospitalized adults with covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.FDA, FDA EUA Remdesivir Fact Sheet for Health Care Providers. 〈https://www.fda.gov/media/137566/download〉, 2020.
- 102.Humeniuk R., Mathias A., Cao H., Osinusi A., Shen G., Chng E., Ling J., Vu A., German P. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin. Transl. Sci. 2020;13(5):896–906. doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang K. What do we know about remdesivir drug interactions? Clin. Transl. Sci. 2020;13(5):842–844. doi: 10.1111/cts.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Humeniuk R., Mathias A., Kirby B.J., Lutz J.D., Cao H., Osinusi A., Babusis D., Porter D., Wei X., Ling J., Reddy Y.S., German P. Pharmacokinetic, pharmacodynamic, and drug-interaction profile of remdesivir, a SARS-CoV-2 replication inhibitor. Clin. Pharmacokinet. 2021 doi: 10.1007/s40262-021-00984-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kirby B.J., Unadkat J.D. Impact of ignoring extraction ratio when predicting drug-drug interactions, fraction metabolized, and intestinal first-pass contribution. Drug Metab. Dispos. 2010;38(11):1926–1933. doi: 10.1124/dmd.110.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 107.Favipiravir, 〈https://www.drugbank.ca/drugs/DB12466〉 Accessed 26 March 2020, 2020.
- 108.Oestereich L., Ludtke A., Wurr S., Rieger T., Munoz-Fontela C., Gunther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antivir. Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 109.Bai C.Q., Mu J.S., Kargbo D., Song Y.B., Niu W.K., Nie W.M., Kanu A., Liu W.W., Wang Y.P., Dafae F., Yan T., Hu Y., Deng Y.Q., Lu H.J., Yang F., Zhang X.G., Sun Y., Cao Y.X., Su H.X., Sun Y., Liu W.S., Wang C.Y., Qian J., Liu L., Wang H., Tong Y.G., Liu Z.Y., Chen Y.S., Wang H.Q., Kargbo B., Gao G.F., Jiang J.F. Clinical and virological characteristics of ebola virus disease patients treated with favipiravir (T-705)-Sierra Leone, 2014. Clin. Infect. Dis. 2016;63(10):1288–1294. doi: 10.1093/cid/ciw571. [DOI] [PubMed] [Google Scholar]
- 110.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209 doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Madelain V., Mentre F., Baize S., Anglaret X., Laouenan C., Oestereich L., Nguyen T.H.T., Malvy D., Piorkowski G., Graw F., Gunther S., Raoul H., de Lamballerie X., Guedj J. Modeling favipiravir antiviral efficacy against emerging viruses: from animal studies to clinical trials. CPT Pharmacomet. Syst. Pharmacol. 2020;9(5):258–271. doi: 10.1002/psp4.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 113.Delang L., Abdelnabi R., Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antivir. Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 114.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Glenmark begins Phase-3 clinical trials on antiviral drug Favipiravir for COVID-19 patients in India. 〈https://www.thehindu.com/news/national/glenmark-begins-phase-3-clinical-trials-on-antiviral-drug-favipiravir-forcovid-19-patients-in-india/article31563198.ece〉 Accessed 25 May 2020, 2020.
- 116.Mishima E., Anzai N., Miyazaki M., Abe T. Uric acid elevation by favipiravir, an antiviral drug. Tohoku J. Exp. Med. 2020;251(2):87–90. doi: 10.1620/tjem.251.87. [DOI] [PubMed] [Google Scholar]
- 117.Du Y.X., Chen X.P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108(2):242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 118.Lopinavir, Ritonavir. https://www.drugbank.ca/unearth/q?utf8¼%E2%9C%93&query¼lopinavir%2Fritonivir&searcher¼drugs Accessed 02 April 2020(2020).
- 119.Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., Kao R.Y., Poon L.L., Wong C.L., Guan Y., Peiris J.S., Yuen K.Y., Group H.U.S.S. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yao T.T., Qian J.D., Zhu W.Y., Wang Y., Wang G.Q. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J. Med. Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park S.Y., Lee J.S., Son J.S., Ko J.H., Peck K.R., Jung Y., Woo H.J., Joo Y.S., Eom J.S., Shi H. Post-exposure prophylaxis for Middle East respiratory syndrome in healthcare workers. J. Hosp. Infect. 2019;101(1):42–46. doi: 10.1016/j.jhin.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Government of India Ministry of Health & Family Welfare Directorate General of Health Services (EMR Division). Guidelines on Clinical Management of COVID e 19 dated 17th March 2020. 〈https://www.aiims.edu/images/pdf/notice/Guidelines%20Clinical%20Management%20COVID19–23-3–20.pdf,〉, 2020.
- 124.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jetter A., Fatkenheuer G., Frank D., Klaassen T., Seeringer A., Doroshyenko O., Kirchheiner J., Hein W., Schomig E., Fuhr U., Wyen C. Do activities of cytochrome P450 (CYP)3A, CYP2D6 and P-glycoprotein differ between healthy volunteers and HIV-infected patients? Antivir. Ther. 2010;15(7):975–983. doi: 10.3851/IMP1648. [DOI] [PubMed] [Google Scholar]
- 126.Schoergenhofer C., Jilma B., Stimpfl T., Karolyi M., Zoufaly A. Pharmacokinetics of lopinavir and ritonavir in patients hospitalized with coronavirus disease 2019 (COVID-19) Ann. Intern. Med. 2020;173(8):670–672. doi: 10.7326/M20-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gregoire M., Le Turnier P., Gaborit B.J., Veyrac G., Lecomte R., Boutoille D., Canet E., Imbert B.M., Bellouard R., Raffi F. Lopinavir pharmacokinetics in COVID-19 patients. J. Antimicrob. Chemother. 2020;75(9):2702–2704. doi: 10.1093/jac/dkaa195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P., Cai J.P., Chu H., Zhou J., Chen H., Qin C., Yuen K.Y. Treatment with lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Biot C., Daher W., Chavain N., Fandeur T., Khalife J., Dive D., De Clercq E. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J. Med. Chem. 2006;49(9):2845–2849. doi: 10.1021/jm0601856. [DOI] [PubMed] [Google Scholar]
- 132.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 136.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Tissot Dupont H., Honore S., Colson P., Chabriere E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56(1) doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with covid-19. N. Engl. J. Med. 2020;382(25):2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Projean D., Baune B., Farinotti R., Flinois J.P., Beaune P., Taburet A.M., Ducharme J. In vitro metabolism of chloroquine: identification of CYP2C8, CYP3A4, and CYP2D6 as the main isoforms catalyzing N-desethylchloroquine formation. Drug Metab. Dispos. 2003;31(6):748–754. doi: 10.1124/dmd.31.6.748. [DOI] [PubMed] [Google Scholar]
- 139.Furst D.E. Pharmacokinetics of hydroxychloroquine and chloroquine during treatment of rheumatic diseases. Lupus. 1996;5(Suppl 1):S11–S15. [PubMed] [Google Scholar]
- 140.Paniri A., Hosseini M.M., Rasoulinejad A., Akhavan-Niaki H. Molecular effects and retinopathy induced by hydroxychloroquine during SARS-CoV-2 therapy: role of CYP450 isoforms and epigenetic modulations. Eur. J. Pharmacol. 2020;886 doi: 10.1016/j.ejphar.2020.173454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pecheur E.I., Borisevich V., Halfmann P., Morrey J.D., Smee D.F., Prichard M., Mire C.E., Kawaoka Y., Geisbert T.W., Polyak S.J. The synthetic antiviral drug arbidol inhibits globally prevalent pathogenic viruses. J. Virol. 2016;90(6):3086–3092. doi: 10.1128/JVI.02077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Villalain J. Membranotropic effects of arbidol, a broad anti-viral molecule, on phospholipid model membranes. J. Phys. Chem. B. 2010;114(25):8544–8554. doi: 10.1021/jp102619w. [DOI] [PubMed] [Google Scholar]
- 143.Boriskin Y.S., Pecheur E.I., Polyak S.J. Arbidol: a broad-spectrum antiviral that inhibits acute and chronic HCV infection. Virol. J. 2006;3:56. doi: 10.1186/1743-422X-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pecheur E.I., Lavillette D., Alcaras F., Molle J., Boriskin Y.S., Roberts M., Cosset F.L., Polyak S.J. Biochemical mechanism of hepatitis C virus inhibition by the broad-spectrum antiviral arbidol. Biochemistry. 2007;46(20):6050–6059. doi: 10.1021/bi700181j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leneva I.A., Russell R.J., Boriskin Y.S., Hay A.J. Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antivir. Res. 2009;81(2):132–140. doi: 10.1016/j.antiviral.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 146.Liu Q., Xiong H.R., Lu L., Liu Y.Y., Luo F., Hou W., Yang Z.Q. Antiviral and anti-inflammatory activity of arbidol hydrochloride in influenza A (H1N1) virus infection. Acta Pharmacol. Sin. 2013;34(8):1075–1083. doi: 10.1038/aps.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81(1):e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deng P., Zhong D., Yu K., Zhang Y., Wang T., Chen X. Pharmacokinetics, metabolism, and excretion of the antiviral drug arbidol in humans. Antimicrob. Agents Chemother. 2013;57(4):1743–1755. doi: 10.1128/AAC.02282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S., Nambu A., Bavari S., Soloveva V., Sadukhan S., Cassell G.H., Geisbert T.W., Hur S., Goldfeld A.E. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits ebola virus. iScience. 2019;19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pepperrell T., Pilkington V., Owen A., Wang J., Hill A.M. Review of safety and minimum pricing of nitazoxanide for potential treatment of COVID-19. J. Virus Erad. 2020;6(2):52–60. doi: 10.1016/S2055-6640(20)30017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Calderon J.M., Zeron H.M., Padmanabhan S. Treatment with Hydroxychloroquine vs Hydroxychloroquine + Nitazoxanide in COVID-19 patients with risk factors for poor prognosis: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):504. doi: 10.1186/s13063-020-04448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.FDA, 〈https://clinicaltrials.gov/ct2/show/NCT04435314〉, 2020.
- 153.Ketkar H., Yang L., Wormser G.P., Wang P. Lack of efficacy of ivermectin for prevention of a lethal Zika virus infection in a murine system. Diagn. Microbiol. Infect. Dis. 2019;95(1):38–40. doi: 10.1016/j.diagmicrobio.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chaccour C., Hammann F., Ramon-Garcia S., Rabinovich N.R. Ivermectin and COVID-19: keeping rigor in times of urgency. Am. J. Trop. Med. Hyg. 2020;102(f6):1156–1157. doi: 10.4269/ajtmh.20-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Heidary F., Gharebaghi R. Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen. J. Antibiot. 2020;73(9):593–602. doi: 10.1038/s41429-020-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.News, News. FDA approves COVACTA trial for RA drug Actemra in COVID-19 patients. From: 〈https://www.pharmaceuticalbusiness-review.com/news/covacta-trial-actemra-covid-19/〉. (Accessed 28 March 2020), 2020.
- 158.Dunn S.T., Hedges L., Sampson K.E., Lai Y., Mahabir S., Balogh L., Locuson C.W. Pharmacokinetic interaction of the antiparasitic agents ivermectin and spinosad in dogs. Drug Metab. Dispos. 2011;39(5):789–795. doi: 10.1124/dmd.110.034827. [DOI] [PubMed] [Google Scholar]
- 159.Schmith V.D., Zhou J.J., Lohmer L.R.L. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther. 2020;108(4):762–765. doi: 10.1002/cpt.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Uematsu S., Akira S. Toll-like receptors and Type I interferons. J. Biol. Chem. 2007;282(21):15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 161.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 162.Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F.J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K.M. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 163.Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., Chen S., Brand N., Tarway M., Liu L., Sheikh F., Astemborski J., Bonkovsky H.L., Edlin B.R., Howell C.D., Morgan T.R., Thomas D.L., Rehermann B., Donnelly R.P., O’Brien T.R. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45(2):164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Davidson S., McCabe T.M., Crotta S., Gad H.H., Hessel E.M., Beinke S., Hartmann R., Wack A. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO Mol. Med. 2016;8(9):1099–1112. doi: 10.15252/emmm.201606413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Galani I.E., Triantafyllia V., Eleminiadou E.E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., Andreakos E. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46(5):875–890 e6. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 166.Klinkhammer J., Schnepf D., Ye L., Schwaderlapp M., Gad H.H., Hartmann R., Garcin D., Mahlakoiv T., Staeheli P. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018;7 doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Muir A.J., Arora S., Everson G., Flisiak R., George J., Ghalib R., Gordon S.C., Gray T., Greenbloom S., Hassanein T., Hillson J., Horga M.A., Jacobson I.M., Jeffers L., Kowdley K.V., Lawitz E., Lueth S., Rodriguez-Torres M., Rustgi V., Shemanski L., Shiffman M.L., Srinivasan S., Vargas H.E., Vierling J.M., Xu D., Lopez-Talavera J.C., Zeuzem S., E.s. group A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 2014;61(6):1238–1246. doi: 10.1016/j.jhep.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 168.Hensley L.E., Fritz L.E., Jahrling P.B., Karp C.L., Huggins J.W., Geisbert T.W. Interferon-beta 1a and SARS coronavirus replication. Emerg. Infect. Dis. 2004;10(2):317–319. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stroher U., DiCaro A., Li Y., Strong J.E., Aoki F., Plummer F., Jones S.M., Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon- alpha. J. Infect. Dis. 2004;189(7):1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li P.T., Dai J., Mok F.K., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67(6):606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hart B.J., Dyall J., Postnikova E., Zhou H., Kindrachuk J., Johnson R.F., Olinger G.G., Frieman M.B., Holbrook M.R., Jahrling P.B., Hensley L. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014;95(Pt 3):571–577. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jamilloux Y., Henry T., Belot A., Viel S., Fauter M., El Jammal T., Walzer T., Francois B., Seve P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020;19(7) doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen C., Han Y.H., Yang Z., Rodrigues A.D. Effect of interferon-alpha2b on the expression of various drug-metabolizing enzymes and transporters in co-cultures of freshly prepared human primary hepatocytes. Xenobiotica. 2011;41(6):476–485. doi: 10.3109/00498254.2011.560971. [DOI] [PubMed] [Google Scholar]
- 174.Becquemont L., Chazouilleres O., Serfaty L., Poirier J.M., Broly F., Jaillon P., Poupon R., Funck-Brentano C. Effect of interferon alpha-ribavirin bitherapy on cytochrome P450 1A2 and 2D6 and N-acetyltransferase-2 activities in patients with chronic active hepatitis C. Clin. Pharmacol. Ther. 2002;71(6):488–495. doi: 10.1067/mcp.2002.124468. [DOI] [PubMed] [Google Scholar]
- 175.Giles A.J., Hutchinson M.N.D., Sonnemann H.M., Jung J., Fecci P.E., Ratnam N.M., Zhang W., Song H., Bailey R., Davis D., Reid C.M., Park D.M., Gilbert M.R. Dexamethasone-induced immunosuppression: mechanisms and implications for immunotherapy. J. Immunother. Cancer. 2018;6(1):51. doi: 10.1186/s40425-018-0371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen H., Wang F., Zhang P., Zhang Y., Chen Y., Fan X., Cao X., Liu J., Yang Y., Wang B., Lei B., Gu L., Bai J., Wei L., Zhang R., Zhuang Q., Zhang W., Zhao W., He A. Management of cytokine release syndrome related to CAR-T cell therapy. Front. Med. 2019;13(5):610–617. doi: 10.1007/s11684-019-0714-8. [DOI] [PubMed] [Google Scholar]
- 177.Cohn L.A. The influence of corticosteroids on host defense mechanisms. J. Vet. Intern. Med. 1991;5(2):95–104. doi: 10.1111/j.1939-1676.1991.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 178.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee N., Allen Chan K.C., Hui D.S., Ng E.K., Wu A., Chiu R.W., Wong V.W., Chan P.K., Wong K.T., Wong E., Cockram C.S., Tam J.S., Sung J.J., Lo Y.M. Effects of early corticosteroid treatment on plasma SARS-associated Coronavirus RNA concentrations in adult patients. J. Clin. Virol. 2004;31(4):304–309. doi: 10.1016/j.jcv.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., Prudon B., Green C., Felton T., Chadwick D., Rege K., Fegan C., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Montgomery A., Rowan K., Juszczak E., Baillie J.K., Haynes R., Landray M.J. Dexamethasone in hospitalized patients with covid-19 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.M. Bethesda, COVID-19 treatment guidelines: National Institutes of Health, 2020, 2020. [PubMed]
- 182.W.C. , Dexamethasone in the treatment of COVID-19: Implementation and management of supply for treatment in hospitals. London: Medicines and Healthcare Products Regulatory Agency, June 16, 2020, 2020.
- 183.Jiao T., Yao X., Zhao Y., Zhou Y., Gao Y., Fan S., Chen P., Li X., Jiang Y., Yang X., Gonzalez F.J., Huang M., Bi H. Dexamethasone-induced liver enlargement is related to PXR/YAP activation and lipid accumulation but not hepatocyte proliferation. Drug Metab. Dispos. 2020;48(9):830–839. doi: 10.1124/dmd.120.000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Buckley D.B., Klaassen C.D. Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab. Dispos. 2009;37(4):847–856. doi: 10.1124/dmd.108.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cheng X., Maher J., Dieter M.Z., Klaassen C.D. Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab. Dispos. 2005;33(9):1276–1282. doi: 10.1124/dmd.105.003988. [DOI] [PubMed] [Google Scholar]
- 186.Hartley D.P., Dai X., He Y.D., Carlini E.J., Wang B., Huskey S.E., Ulrich R.G., Rushmore T.H., Evers R., Evans D.C. Activators of the rat pregnane X receptor differentially modulate hepatic and intestinal gene expression. Mol. Pharmacol. 2004;65(5):1159–1171. doi: 10.1124/mol.65.5.1159. [DOI] [PubMed] [Google Scholar]
- 187.Wang H., Faucette S.R., Gilbert D., Jolley S.L., Sueyoshi T., Negishi M., LeCluyse E.L. Glucocorticoid receptor enhancement of pregnane X receptor-mediated CYP2B6 regulation in primary human hepatocytes. Drug Metab. Dispos. 2003;31(5):620–630. doi: 10.1124/dmd.31.5.620. [DOI] [PubMed] [Google Scholar]
- 188.Kliewer S.A. The nuclear pregnane X receptor regulates xenobiotic detoxification. J. Nutr. 2003;133(7 Suppl):2444S–2447S. doi: 10.1093/jn/133.7.2444S. [DOI] [PubMed] [Google Scholar]
- 189.Song Q.Q., Xie W.Y., Tang Y.J., Zhang J., Liu J. Genetic variation in the glucocorticoid pathway involved in interindividual differences in the glucocorticoid treatment. Pharmacogenomics. 2017;18(3):293–316. doi: 10.2217/pgs-2016-0151. [DOI] [PubMed] [Google Scholar]
- 190.Henehan M., Montuno M., De Benedetto A. Doxycycline as an anti-inflammatory agent: updates in dermatology. J. Eur. Acad. Dermatol. Venereol. 2017;31(11):1800–1808. doi: 10.1111/jdv.14345. [DOI] [PubMed] [Google Scholar]
- 191.Yoshikawa T., Hill T., Li K., Peters C.J., Tseng C.T. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J. Virol. 2009;83(7):3039–3048. doi: 10.1128/JVI.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sandler C., Ekokoski E., Lindstedt K.A., Vainio P.J., Finel M., Sorsa T., Kovanen P.T., Golub L.M., Eklund K.K. Chemically modified tetracycline (CMT)-3 inhibits histamine release and cytokine production in mast cells: possible involvement of protein kinase C. Inflamm. Res. 2005;54(7):304–312. doi: 10.1007/s00011-005-1358-5. [DOI] [PubMed] [Google Scholar]
- 193.Kritas S.K., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Conti P. Mast cells contribute to coronavirus-induced inflammation: new anti-inflammatory strategy. J. Biol. Regul. Homeost. Agents. 2020;34(1):9–14. doi: 10.23812/20-Editorial-Kritas. [DOI] [PubMed] [Google Scholar]
- 194.Dutta K., Basu A. Use of minocycline in viral infections. Indian J. Med. Res. 2011;133:467–470. [PMC free article] [PubMed] [Google Scholar]
- 195.Griffin M.O., Fricovsky E., Ceballos G., Villarreal F. Tetracyclines: a pleitropic family of compounds with promising therapeutic properties. Review of the literature. Am. J. Physiol. Cell Physiol. 2010;299(3):C539–C548. doi: 10.1152/ajpcell.00047.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bharadwaj S., Lee K.E., Dwivedi V.D., Kang S.G. Computational insights into tetracyclines as inhibitors against SARS-CoV-2 M(pro) via combinatorial molecular simulation calculations. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gautam S.S., Gautam C.S., Garg V.K., Singh H. Combining hydroxychloroquine and minocycline: potential role in moderate to severe COVID-19 infection. Expert Rev. Clin. Pharmacol. 2020;13(11):1183–1190. doi: 10.1080/17512433.2020.1832889. [DOI] [PubMed] [Google Scholar]
- 198.Gironi L.C., Damiani G., Zavattaro E., Pacifico A., Santus P., Pigatto P.D.M., Cremona O., Savoia P. Tetracyclines in COVID-19 patients quarantined at home: literature evidence supporting real-world data from a multicenter observational study targeting inflammatory and infectious dermatoses. Dermatol. Ther. 2021;34(1) doi: 10.1111/dth.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tatro D.S. Tetracycline-antacid interactions. J. Am. Med. Assoc. 1972;220(4):586. [PubMed] [Google Scholar]
- 200.Wang Y., Cui R., Li G., Gao Q., Yuan S., Altmeyer R., Zou G. Teicoplanin inhibits Ebola pseudovirus infection in cell culture. Antivir. Res. 2016;125:1–7. doi: 10.1016/j.antiviral.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Baron S.A., Devaux C., Colson P., Raoult D., Rolain J.M. Teicoplanin: an alternative drug for the treatment of COVID-19? Int. J. Antimicrob. Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., Zhang H. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of ebola virus, middle east respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291(17):9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ceccarelli G., Alessandri F., d’Ettorre G., Borrazzo C., Spagnolello O., Oliva A., Ruberto F., Mastroianni C.M., Pugliese F., Venditti M., C.-S.G.o.S.U. Intensive Care Is teicoplanin a complementary treatment option for COVID-19? The question remains. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.106029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents., Department of Health and Human Services, 2012.
- 205.Rahmani H., Davoudi-Monfared E., Nourian A., Nabiee M., Sadeghi S., Khalili H., Abbasian L., Ghiasvand F., Seifi A., Hasannezhad M., Ghaderkhani S., Mohammadi M., Yekaninejad M.S. Comparing outcomes of hospitalized patients with moderate and severe COVID-19 following treatment with hydroxychloroquine plus atazanavir/ritonavir. Daru. 2020;28(2):625–634. doi: 10.1007/s40199-020-00369-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fintelman-Rodrigues N., Sacramento C.Q., Ribeiro Lima C., Souza da Silva F., Ferreira A.C., Mattos M., de Freitas C.S., Cardoso Soares V., da Silva Gomes Dias S., Temerozo J.R., Miranda M.D., Matos A.R., Bozza F.A., Carels N., Alves C.R., Siqueira M.M., Bozza P.T., Souza T.M.L. Atazanavir, alone or in combination with ritonavir, inhibits SARS-CoV-2 Replication and Proinflammatory Cytokine Production. Antimicrob. Agents Chemother. 2020;64(10) doi: 10.1128/AAC.00825-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2015;1(7):317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 208.Bacharier L.B., Guilbert T.W., Mauger D.T., Boehmer S., Beigelman A., Fitzpatrick A.M., Jackson D.J., Baxi S.N., Benson M., Burnham C.D., Cabana M., Castro M., Chmiel J.F., Covar R., Daines M., Gaffin J.M., Gentile D.A., Holguin F., Israel E., Kelly H.W., Lazarus S.C., Lemanske R.F., Jr., Ly N., Meade K., Morgan W., Moy J., Olin T., Peters S.P., Phipatanakul W., Pongracic J.A., Raissy H.H., Ross K., Sheehan W.J., Sorkness C., Szefler S.J., Teague W.G., Thyne S., Martinez F.D. Early administration of azithromycin and prevention of severe lower respiratory tract illnesses in preschool children with a history of such illnesses: a randomized clinical trial. J. Am. Med. Assoc. 2015;314(19):2034–2044. doi: 10.1001/jama.2015.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Arshad S., Kilgore P., Chaudhry Z.S., Jacobsen G., Wang D.D., Huitsing K., Brar I., Alangaden G.J., Ramesh M.S., McKinnon J.E., O’Neill W., Zervos M., Henry Ford C.-T.F. Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID-19. Int. J. Infect. Dis. 2020;97:396–403. doi: 10.1016/j.ijid.2020.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Westphal J.F. Macrolide - induced clinically relevant drug interactions with cytochrome P-450A (CYP) 3A4: an update focused on clarithromycin, azithromycin and dirithromycin. Br. J. Clin. Pharmacol. 2000;50(4):285–295. doi: 10.1046/j.1365-2125.2000.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lan T., Rao A., Haywood J., Davis C.B., Han C., Garver E., Dawson P.A. Interaction of macrolide antibiotics with intestinally expressed human and rat organic anion-transporting polypeptides. Drug Metab. Dispos. 2009;37(12):2375–2382. doi: 10.1124/dmd.109.028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lane J.C.E., Weaver J., Kostka K., Duarte-Salles T., Abrahao M.T.F., Alghoul H., Alser O., Alshammari T.M., Biedermann P., Banda J.M., Burn E., Casajust P., Conover M.M., Culhane A.C., Davydov A., DuVall S.L., Dymshyts D., Fernandez-Bertolin S., Fister K., Hardin J., Hester L., Hripcsak G., Kaas-Hansen B.S., Kent S., Khosla S., Kolovos S., Lambert C.G., van der Lei J., Lynch K.E., Makadia R., Margulis A.V., Matheny M.E., Mehta P., Morales D.R., Morgan-Stewart H., Mosseveld M., Newby D., Nyberg F., Ostropolets A., Park R.W., Prats-Uribe A., Rao G.A., Reich C., Reps J., Rijnbeek P., Sathappan S.M.K., Schuemie M., Seager S., Sena A.G., Shoaibi A., Spotnitz M., Suchard M.A., Torre C.O., Vizcaya D., Wen H., de Wilde M., Xie J., You S.C., Zhang L., Zhuk O., Ryan P., Prieto-Alhambra D., O.-C.- consortium Risk of hydroxychloroquine alone and in combination with azithromycin in the treatment of rheumatoid arthritis: a multinational, retrospective study. Lancet Rheumatol. 2020;2(11):e698–e711. doi: 10.1016/S2665-9913(20)30276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rezolsta Summary of product characteristics. 〈https://www.ema.europa.eu/en/ documents/product-information/rezolsta-epar-product-information_en.pdf〉., 2020.
- 214.Prezcobix prescribing information. 〈https://www.accessdata.fda.gov/drugsatfda_-docs/label/2016/205395s001lbl.pdf〉, 2020.
- 215.Tashima K., Crofoot G., Tomaka F.L., Kakuda T.N., Brochot A., Van de Casteele T., Opsomer M., Garner W., Margot N., Custodio J.M., Fordyce M.W., Szwarcberg J. Cobicistat-boosted darunavir in HIV-1-infected adults: week 48 results of a Phase IIIb, open-label single-arm trial. AIDS Res Ther. 2014;11:39. doi: 10.1186/1742-6405-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Orkin C., DeJesus E., Khanlou H., Stoehr A., Supparatpinyo K., Lathouwers E., Lefebvre E., Opsomer M., Van de Casteele T., Tomaka F. Final 192-week efficacy and safety of once-daily darunavir/ritonavir compared with lopinavir/ritonavir in HIV-1-infected treatment-naive patients in the ARTEMIS trial. HIV Med. 2013;14(1):49–59. doi: 10.1111/j.1468-1293.2012.01060.x. [DOI] [PubMed] [Google Scholar]
- 217.De Meyer S., Bojkova D., Cinatl J., Van Damme E., Buyck C., Van Loock M., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim E.J., Choi S.H., Park J.S., Kwon Y.S., Lee J., Kim Y., Lee S.Y., Choi E.Y. Use of darunavir-cobicistat as a treatment option for critically Ill patients with SARS-CoV-2 infection. Yonsei Med. J. 2020;61(9):826–830. doi: 10.3349/ymj.2020.61.9.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vannucchi A.M., Kiladjian J.J., Griesshammer M., Masszi T., Durrant S., Passamonti F., Harrison C.N., Pane F., Zachee P., Mesa R., He S., Jones M.M., Garrett W., Li J., Pirron U., Habr D., Verstovsek S. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N. Engl. J. Med. 2015;372(5):426–435. doi: 10.1056/NEJMoa1409002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Verstovsek S., Mesa R.A., Gotlib J., Levy R.S., Gupta V., DiPersio J.F., Catalano J.V., Deininger M., Miller C., Silver R.T., Talpaz M., Winton E.F., Harvey J.H., Jr., Arcasoy M.O., Hexner E., Lyons R.M., Paquette R., Raza A., Vaddi K., Erickson-Viitanen S., Koumenis I.L., Sun W., Sandor V., Kantarjian H.M. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl. J. Med. 2012;366(9):799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Venugopal S., Bar-Natan M., Mascarenhas J.O. JAKs to STATs: a tantalizing therapeutic target in acute myeloid leukemia. Blood Rev. 2020;40 doi: 10.1016/j.blre.2019.100634. [DOI] [PubMed] [Google Scholar]
- 222.El Bairi K., Trapani D., Petrillo A., Le Page C., Zbakh H., Daniele B., Belbaraka R., Curigliano G., Afqir S. Repurposing anticancer drugs for the management of COVID-19. Eur. J. Cancer. 2020;141:40–61. doi: 10.1016/j.ejca.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cao Y., Wei J., Zou L., Jiang T., Wang G., Chen L., Huang L., Meng F., Huang L., Wang N., Zhou X., Luo H., Mao Z., Chen X., Xie J., Liu J., Cheng H., Zhao J., Huang G., Wang W., Zhou J. Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J. Allergy Clin. Immunol. 2020;146(1):137–146 e3. doi: 10.1016/j.jaci.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Febvre-James M., Bruyere A., Le Vee M., Fardel O. The JAK1/2 inhibitor ruxolitinib reverses interleukin-6-mediated suppression of drug-detoxifying proteins in cultured human hepatocytes. Drug Metab. Dispos. 2018;46(2):131–140. doi: 10.1124/dmd.117.078048. [DOI] [PubMed] [Google Scholar]
- 225.Aslanis V., Umehara K., Huth F., Ouatas T., Bharathy S., Butler A.A., Zhou W., Gadbaw B. Multiple administrations of fluconazole increase plasma exposure to ruxolitinib in healthy adult subjects. Cancer Chemother. Pharmacol. 2019;84(4):749–757. doi: 10.1007/s00280-019-03907-1. [DOI] [PubMed] [Google Scholar]
- 226.Umehara K., Huth F., Jin Y., Schiller H., Aslanis V., Heimbach T., He H. Drug-drug interaction (DDI) assessments of ruxolitinib, a dual substrate of CYP3A4 and CYP2C9, using a verified physiologically based pharmacokinetic (PBPK) model to support regulatory submissions. Drug Metab. Pers. Ther. 2019;34(2) doi: 10.1515/dmpt-2018-0042. [DOI] [PubMed] [Google Scholar]
- 227.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh U.K. Across Speciality Collaboration, COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jin M., Tong Q. Rhabdomyolysis as potential late complication associated with COVID-19. Emerg. Infect. Dis. 2020;26(7):1618–1620. doi: 10.3201/eid2607.200445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., Cereda D., Coluccello A., Foti G., Fumagalli R., Iotti G., Latronico N., Lorini L., Merler S., Natalini G., Piatti A., Ranieri M.V., Scandroglio A.M., Storti E., Cecconi M., Pesenti A., C.-L.I. Network Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. J. Am. Med. Assoc. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Richardson P.J., Corbellino M., Stebbing J. Baricitinib for COVID-19: a suitable treatment? - Authors’ reply. Lancet Infect. Dis. 2020;20(9):1013–1014. doi: 10.1016/S1473-3099(20)30270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fragoulis G.E., McInnes I.B., Siebert S. JAK-inhibitors. New players in the field of immune-mediated diseases, beyond rheumatoid arthritis. Rheumatology. 2019;58(Suppl 1):i43–i54. doi: 10.1093/rheumatology/key276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pu S.Y., Xiao F., Schor S., Bekerman E., Zanini F., Barouch-Bentov R., Nagamine C.M., Einav S. Feasibility and biological rationale of repurposing sunitinib and erlotinib for dengue treatment. Antivir. Res. 2018;155:67–75. doi: 10.1016/j.antiviral.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Posada M.M., Cannady E.A., Payne C.D., Zhang X., Bacon J.A., Pak Y.A., Higgins J.W., Shahri N., Hall S.D., Hillgren K.M. Prediction of transporter-mediated drug-drug interactions for baricitinib. Clin. Transl. Sci. 2017;10(6):509–519. doi: 10.1111/cts.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gasmi A., Noor S., Tippairote T., Dadar M., Menzel A., Bjorklund G. Individual risk management strategy and potential therapeutic options for the COVID-19 pandemic. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Morales-Ortega A., Bernal-Bello D., Llarena-Barroso C., Frutos-Perez B., Duarte-Millan M.A., Garcia de Viedma-Garcia V., Farfan-Sedano A.I., Canalejo-Castrillero E., Ruiz-Giardin J.M., Ruiz-Ruiz J., San Martin-Lopez J.V. Imatinib for COVID-19: a case report. Clin. Immunol. 2020;218 doi: 10.1016/j.clim.2020.108518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Emadi A., Chua J.V., Talwani R., Bentzen S.M., Baddley J. Safety and efficacy of imatinib for hospitalized adults with COVID-19: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):897. doi: 10.1186/s13063-020-04819-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Filppula A.M., Neuvonen M., Laitila J., Neuvonen P.J., Backman J.T. Autoinhibition of CYP3A4 leads to important role of CYP2C8 in imatinib metabolism: variability in CYP2C8 activity may alter plasma concentrations and response. Drug Metab. Dispos. 2013;41(1):50–59. doi: 10.1124/dmd.112.048017. [DOI] [PubMed] [Google Scholar]
- 239.Filppula A.M., Laitila J., Neuvonen P.J., Backman J.T. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. Br. J. Pharmacol. 2012;165(8):2787–2798. doi: 10.1111/j.1476-5381.2011.01732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hashimoto K. Activation of sigma-1 receptor chaperone in the treatment of neuropsychiatric diseases and its clinical implication. J. Pharmacol. Sci. 2015;127(1):6–9. doi: 10.1016/j.jphs.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 241.Cobos E.J., Entrena J.M., Nieto F.R., Cendan C.M., Del Pozo E. Pharmacology and therapeutic potential of sigma(1) receptor ligands. Curr. Neuropharmacol. 2008;6(4):344–366. doi: 10.2174/157015908787386113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hallifax D., Houston J.B. Saturable uptake of lipophilic amine drugs into isolated hepatocytes: mechanisms and consequences for quantitative clearance prediction. Drug Metab. Dispos. 2007;35(8):1325–1332. doi: 10.1124/dmd.107.015131. [DOI] [PubMed] [Google Scholar]
- 243.Deftereos S.G., Giannopoulos G., Vrachatis D.A., Siasos G.D., Giotaki S.G., Gargalianos P., Metallidis S., Sianos G., Baltagiannis S., Panagopoulos P., Dolianitis K., Randou E., Syrigos K., Kotanidou A., Koulouris N.G., Milionis H., Sipsas N., Gogos C., Tsoukalas G., Olympios C.D., Tsagalou E., Migdalis I., Gerakari S., Angelidis C., Alexopoulos D., Davlouros P., Hahalis G., Kanonidis I., Katritsis D., Kolettis T., Manolis A.S., Michalis L., Naka K.K., Pyrgakis V.N., Toutouzas K.P., Triposkiadis F., Tsioufis K., Vavouranakis E., Martinez-Dolz L., Reimers B., Stefanini G.G., Cleman M., Goudevenos J., Tsiodras S., Tousoulis D., Iliodromitis E., Mehran R., Dangas G., Stefanadis C., G.-. investigators Effect of colchicine vs standard care on cardiac and inflammatory biomarkers and clinical outcomes in patients hospitalized with coronavirus disease 2019: the GRECCO-19 Randomized Clinical Trial. JAMA Netw. Open. 2020;3(6) doi: 10.1001/jamanetworkopen.2020.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sterne J.A.C., Murthy S., Diaz J.V., Slutsky A.S., Villar J., Angus D.C., Annane D., Azevedo L.C.P., Berwanger O., Cavalcanti A.B., Dequin P.F., Du B., Emberson J., Fisher D., Giraudeau B., Gordon A.C., Granholm A., Green C., Haynes R., Heming N., Higgins J.P.T., Horby P., Juni P., Landray M.J., Le Gouge A., Leclerc M., Lim W.S., Machado F.R., McArthur C., Meziani F., Moller M.H., Perner A., Petersen M.W., Savovic J., Tomazini B., Veiga V.C., Webb S., Marshall J.C. Association between administration of systemic corticosteroids and mortality among critically Ill patients with COVID-19: a meta-analysis. J. Am. Med. Assoc. 2020;324(13):1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Omori I.M., Watanabe N., Nakagawa A., Omori T., Cipriani A., Barbui C., McGuire H., Churchill R., Furukawa T.A., G. Meta-Analysis of New Generation Antidepressants Study Efficacy, tolerability and side-effect profile of fluvoxamine for major depression: meta-analysis. J. Psychopharmacol. 2009;23(5):539–550. doi: 10.1177/0269881108089876. [DOI] [PubMed] [Google Scholar]
- 246.Assimon M.M., Brookhart M.A., Flythe J.E. Comparative cardiac safety of selective serotonin reuptake inhibitors among individuals receiving maintenance hemodialysis. J. Am. Soc. Nephrol. 2019;30(4):611–623. doi: 10.1681/ASN.2018101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Christensen M., Tybring G., Mihara K., Yasui-Furokori N., Carrillo J.A., Ramos S.I., Andersson K., Dahl M.L., Bertilsson L. Low daily 10-mg and 20-mg doses of fluvoxamine inhibit the metabolism of both caffeine (cytochrome P4501A2) and omeprazole (cytochrome P4502C19) Clin. Pharmacol. Ther. 2002;71(3):141–152. doi: 10.1067/mcp.2002.121788. [DOI] [PubMed] [Google Scholar]
- 248.Obach R.S., Ryder T.F. Metabolism of ramelteon in human liver microsomes and correlation with the effect of fluvoxamine on ramelteon pharmacokinetics. Drug Metab. Dispos. 2010;38(8):1381–1391. doi: 10.1124/dmd.110.034009. [DOI] [PubMed] [Google Scholar]
- 249.Kanacher T., Lindauer A., Mezzalana E., Michon I., Veau C., Mantilla J.D.G., Nock V., Fleury A. A physiologically-based pharmacokinetic (PBPK) model network for the prediction of CYP1A2 and CYP2C19 drug-drug-gene interactions with fluvoxamine, omeprazole, S-mephenytoin, moclobemide, tizanidine, mexiletine, ethinylestradiol, and caffeine. Pharmaceutics. 2020;12(12) doi: 10.3390/pharmaceutics12121191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ip A., Berry D.A., Hansen E., Goy A.H., Pecora A.L., Sinclaire B.A., Bednarz U., Marafelias M., Berry S.M., Berry N.S., Mathura S., Sawczuk I.S., Biran N., Go R.C., Sperber S., Piwoz J.A., Balani B., Cicogna C., Sebti R., Zuckerman J., Rose K.M., Tank L., Jacobs L.G., Korcak J., Timmapuri S.L., Underwood J.P., Sugalski G., Barsky C., Varga D.W., Asif A., Landolfi J.C., Goldberg S.L. Hydroxychloroquine and tocilizumab therapy in COVID-19 patients-An observational study. PLoS One. 2020;15(8) doi: 10.1371/journal.pone.0237693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.News, 〈https://www.cancernetwork.com/view/fda-approves-phase-iii-clinical-trial-tocilizumab-covid-19-pneumonia〉, 2020.
- 253.Dogra S., Uprety S., Suresh S.H. Itolizumab, a novel anti-CD6 monoclonal antibody: a safe and efficacious biologic agent for management of psoriasis. Expert Opin. Biol. Ther. 2017;17(3):395–402. doi: 10.1080/14712598.2017.1279601. [DOI] [PubMed] [Google Scholar]
- 254.Aira L.E., Hernandez P., Prada D., Chico A., Gomez J.A., Gonzalez Z., Fuentes K., Viada C., Mazorra Z. Immunological evaluation of rheumatoid arthritis patients treated with itolizumab. MAbs. 2016;8(1):187–195. doi: 10.1080/19420862.2015.1105416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Aira L.E., Lopez-Requena A., Fuentes D., Sanchez L., Perez T., Urquiza A., Bautista H., Falcon L., Hernandez P., Mazorra Z. Immunological and histological evaluation of clinical samples from psoriasis patients treated with anti-CD6 itolizumab. MAbs. 2014;6(3):783–793. doi: 10.4161/mabs.28376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nair P., Melarkode R., Rajkumar D., Montero E. CD6 synergistic co-stimulation promoting proinflammatory response is modulated without interfering with the activated leucocyte cell adhesion molecule interaction. Clin. Exp. Immunol. 2010;162(1):116–130. doi: 10.1111/j.1365-2249.2010.04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Loganathan S., Athalye S.N., Joshi S.R. Itolizumab, an anti-CD6 monoclonal antibody, as a potential treatment for COVID-19 complications. Expert Opin. Biol. Ther. 2020:1–7. doi: 10.1080/14712598.2020.1798399. [DOI] [PubMed] [Google Scholar]
- 258.D. Saavedra, A. Añé Kourí, N. Sánchez, L. Filgueira, J. Betancourt, C. Herrera, L. Manso, E. González, A. Caballero, C. Hidalgo, G. Lorenzo, M. Cepeda Portales, C. Valenzuela, M. Ramos, K. León, Z. Herrera, T. Crombet, An Anti-CD6 Monoclonal Antibody (Itolizumab) Reduces Circulating IL-6 in Severe Covid-19 Elderly Patients, 2020. [DOI] [PMC free article] [PubMed]
- 259.News, 〈https://www.pharmaceutical-technology.com/news/biocon-itolizumab-approval/〉, 2020.
- 260.Caballero A., Filgueira L.M., Betancourt J., Sanchez N., Hidalgo C., Ramirez A., Martinez A., Despaigne R.E., Escalona A., Diaz H., Merino E., Ortega L.M., Castillo U., Ramos M., Saavedra D., Garcia Y., Lorenzo G., Cepeda M., Arencibia M., Cabrera L., Domecq M., Estevez D., Valenzuela C., Lorenzo P., Sanchez L., Mazorra Z., Leon K., Crombet T. Treatment of COVID-19 patients with the anti-CD6 antibody itolizumab. Clin. Transl. Immunol. 2020;9(11) doi: 10.1002/cti2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.W.C. Ke Wang, Yu-Sen Zhou, Jian-Qi Lian, Zheng Zhang, Peng Du, Li Gong, Yang Zhang, Hong-Yong Cui, Jie-Jie Geng, Bin Wang, Xiu-Xuan Sun, Chun-Fu Wang, Xu Yang, Peng Lin, Yong-Qiang Deng, Ding Wei, Xiang-Min Yang, Yu-Meng Zhu, Kui Zhang, Zhao-Hui Zheng, Jin-Lin Miao, Ting Guo, Ying Shi, Jun Zhang, Ling Fu, Qing-Yi Wang, Huijie Bian, Ping Zhu, Zhi-Nan Chen, SARS-CoV-2 invades host cells via a novel route: CD147-spike protein, bioRxiv, 2020.
- 262.Z.-H.Z. Huijie Bian, Ding Wei, Zheng Zhang, Wen-Zhen Kang, Chun-Qiu Hao, Ke Dong, Wen Kang, Jie-Lai Xia, Jin-Lin Miao, Rong-Hua Xie, Bin Wang, Xiu-Xuan Sun, Xiang-Min Yang, Peng Lin, Jie-Jie Geng, Ke Wang, Hong-Yong Cui, Kui Zhang, Xiao-Chun Chen, Hao Tang, Hong Du, Na Yao, Shuang-Shuang Liu, Lin-Na Liu, Zhe Zhang, Zhao-Wei Gao, Gang Nan, Qing-Yi Wang, Jian-Qi Lian, Zhi-Nan Chen, Ping Zhu, Meplazumab treats COVID-19 pneumonia: an open-labelled, concurrent controlled add-on clinical trial., medRxiv, 2020.
- 263.Wechsler M.E., Akuthota P., Jayne D., Khoury P., Klion A., Langford C.A., Merkel P.A., Moosig F., Specks U., Cid M.C., Luqmani R., Brown J., Mallett S., Philipson R., Yancey S.W., Steinfeld J., Weller P.F., Gleich G.J., Team E.M.S. Mepolizumab or placebo for eosinophilic granulomatosis with polyangiitis. N. Engl. J. Med. 2017;376(20):1921–1932. doi: 10.1056/NEJMoa1702079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.NIH, 〈https://clinicaltrials.gov/ct2/show/NCT04369586〉, 2020.
- 265.NIH, 〈https://clinicaltrials.gov/ct2/show/NCT04275245〉, 2020.
- 266.Jodele S., Medvedovic M., Luebbering N., Chen J., Dandoy C.E., Laskin B.L., Davies S.M. Interferon-complement loop in transplant-associated thrombotic microangiopathy. Blood Adv. 2020;4(6):1166–1177. doi: 10.1182/bloodadvances.2020001515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nunius C., Buttner-Herold M., Bertz S., Schiffer M., Buchholz B. Isolated thrombotic microangiopathy of the small intestine in a patient with atypical hemolytic uremic syndrome - a case report. BMC Nephrol. 2020;21(1):104. doi: 10.1186/s12882-020-01766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Olson S.R., Lu E., Sulpizio E., Shatzel J.J., Rueda J.F., DeLoughery T.G. When to stop eculizumab in complement-mediated thrombotic microangiopathies. Am. J. Nephrol. 2018;48(2):96–107. doi: 10.1159/000492033. [DOI] [PubMed] [Google Scholar]
- 269.Roselli F., Karasu E., Volpe C., Huber-Lang M. Medusa’s head: the complement system in traumatic brain and spinal cord injury. J. Neurotrauma. 2018;35(2):226–240. doi: 10.1089/neu.2017.5168. [DOI] [PubMed] [Google Scholar]
- 270.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Diurno F., Numis F.G., Porta G., Cirillo F., Maddaluno S., Ragozzino A., De Negri P., Di Gennaro C., Pagano A., Allegorico E., Bressy L., Bosso G., Ferrara A., Serra C., Montisci A., D’Amico M., Schiano Lo Morello S., Di Costanzo G., Tucci A.G., Marchetti P., Di Vincenzo U., Sorrentino I., Casciotta A., Fusco M., Buonerba C., Berretta M., Ceccarelli M., Nunnari G., Diessa Y., Cicala S., Facchini G. Eculizumab treatment in patients with COVID-19: preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. 2020;24(7):4040–4047. doi: 10.26355/eurrev_202004_20875. [DOI] [PubMed] [Google Scholar]
- 272.NIH, 〈https://clinicaltrials.gov/ct2/show/NCT04288713〉, 2020.
- 273.Giudice V., Pagliano P., Vatrella A., Masullo A., Poto S., Polverino B.M., Gammaldi R., Maglio A., Sellitto C., Vitale C., Serio B., Cuffa B., Borrelli A., Vecchione C., Filippelli A., Selleri C. Combination of ruxolitinib and eculizumab for treatment of severe SARS-CoV-2-related acute respiratory distress syndrome: a controlled study. Front. Pharmacol. 2020;11:857. doi: 10.3389/fphar.2020.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Risitano A.M., Mastellos D.C., Huber-Lang M., Yancopoulou D., Garlanda C., Ciceri F., Lambris J.D. Author correction: complement as a target in COVID-19? Nat. Rev. Immunol. 2020;20(7):448. doi: 10.1038/s41577-020-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mastellos D.C., Yancopoulou D., Kokkinos P., Huber-Lang M., Hajishengallis G., Biglarnia A.R., Lupu F., Nilsson B., Risitano A.M., Ricklin D., Lambris J.D. Compstatin: a C3-targeted complement inhibitor reaching its prime for bedside intervention. Eur. J. Clin. Investig. 2015;45(4):423–440. doi: 10.1111/eci.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Silasi-Mansat R., Zhu H., Georgescu C., Popescu N., Keshari R.S., Peer G., Lupu C., Taylor F.B., Pereira H.A., Kinasewitz G., Lambris J.D., Lupu F. Complement inhibition decreases early fibrogenic events in the lung of septic baboons. J. Cell Mol. Med. 2015;19(11):2549–2563. doi: 10.1111/jcmm.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.van Griensven M., Ricklin D., Denk S., Halbgebauer R., Braun C.K., Schultze A., Hones F., Koutsogiannaki S., Primikyri A., Reis E., Messerer D., Hafner S., Radermacher P., Biglarnia A.R., Resuello R.R.G., Tuplano J.V., Mayer B., Nilsson K., Nilsson B., Lambris J.D., Huber-Lang M. Protective effects of the complement inhibitor compstatin CP40 in hemorrhagic shock. Shock. 2019;51(1):78–87. doi: 10.1097/SHK.0000000000001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zimmerman J.L., Dellinger R.P., Straube R.C., Levin J.L. Phase I trial of the recombinant soluble complement receptor 1 in acute lung injury and acute respiratory distress syndrome. Crit. Care Med. 2000;28(9):3149–3154. doi: 10.1097/00003246-200009000-00004. [DOI] [PubMed] [Google Scholar]
- 279.NIH, 〈https://clinicaltrials.gov/ct2/show/NCT04395456?lead=Amyndas&draw=2&rank=1〉, 2020.
- 280.Reis E.S., Berger N., Wang X., Koutsogiannaki S., Doot R.K., Gumas J.T., Foukas P.G., Resuello R.R.G., Tuplano J.V., Kukis D., Tarantal A.F., Young A.J., Kajikawa T., Soulika A.M., Mastellos D.C., Yancopoulou D., Biglarnia A.R., Huber-Lang M., Hajishengallis G., Nilsson B., Lambris J.D. Safety profile after prolonged C3 inhibition. Clin. Immunol. 2018;197:96–106. doi: 10.1016/j.clim.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mastaglio S., Ruggeri A., Risitano A.M., Angelillo P., Yancopoulou D., Mastellos D.C., Huber-Lang M., Piemontese S., Assanelli A., Garlanda C., Lambris J.D., Ciceri F. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.〈https://www.forbes.com/sites/emilyearlenbaugh/2020/08/20/synthetic-cannabinoid-drug-for-covid-19-approved-for-phase-1-clinical-trials/#314337063329〉, 2020.
- 285.〈https://ir.tetrabiopharma.com/newsroom/press-releases/news-details/2020/FDA-Provides-Positive-Feedback-on-Tetra-Bio-Pharmas-Pre-Investigational-New-Drug-Application-for-ARDS-003-to-Be-Studied-in-COVID-19-Patients-at-Risk-of-Developing-Acute-Respiratory-Distress-Syndrome-ARDS/default.aspx〉, 2020.
- 286.Geffrey A.L., Pollack S.F., Bruno P.L., Thiele E.A. Drug-drug interaction between clobazam and cannabidiol in children with refractory epilepsy. Epilepsia. 2015;56(8):1246–1251. doi: 10.1111/epi.13060. [DOI] [PubMed] [Google Scholar]
- 287.News, 〈https://www.sciencedaily.com/releases/2020/09/200909140314.htm〉, 2020.
- 288.Longxing Cao I.G., Coventry Brian, Case James Brett, Miller Lauren, Kozodoy Lisa, Chen Rita E., Carter Lauren, Walls Alexandra C., Park Young-Jun, Strauch Eva-Maria, Stewart Lance, Diamond Michael S., Veesler David, Baker David. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020:1–10. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Karbanova S., Cerveny L., Jiraskova L., Karahoda R., Ceckova M., Ptackova Z., Staud F. Transport of ribavirin across the rat and human placental barrier: roles of nucleoside and ATP-binding cassette drug efflux transporters. Biochem. Pharmacol. 2019;163:60–70. doi: 10.1016/j.bcp.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 291.Fukuchi Y., Furihata T., Hashizume M., Iikura M., Chiba K. Characterization of ribavirin uptake systems in human hepatocytes. J. Hepatol. 2010;52(4):486–492. doi: 10.1016/j.jhep.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 292.Hodge C., Marra F., Marzolini C., Boyle A., Gibbons S., Siccardi M., Burger D., Back D., Khoo S. Drug interactions: a review of the unseen danger of experimental COVID-19 therapies. J. Antimicrob. Chemother. 2020;75(12):3417–3424. doi: 10.1093/jac/dkaa340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kiser J.J., Gerber J.G., Predhomme J.A., Wolfe P., Flynn D.M., Hoody D.W. Drug/Drug interaction between lopinavir/ritonavir and rosuvastatin in healthy volunteers. J. Acquir Immune Defic. Syndr. 2008;47(5):570–578. doi: 10.1097/QAI.0b013e318160a542. [DOI] [PubMed] [Google Scholar]
- 294.van Waterschoot R.A., ter Heine R., Wagenaar E., van der Kruijssen C.M., Rooswinkel R.W., Huitema A.D., Beijnen J.H., Schinkel A.H. Effects of cytochrome P450 3A (CYP3A) and the drug transporters P-glycoprotein (MDR1/ABCB1) and MRP2 (ABCC2) on the pharmacokinetics of lopinavir. Br. J. Pharmacol. 2010;160(5):1224–1233. doi: 10.1111/j.1476-5381.2010.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Rakhmanina N.Y., Neely M.N., Van Schaik R.H., Gordish-Dressman H.A., Williams K.D., Soldin S.J., van den Anker J.N. CYP3A5, ABCB1, and SLCO1B1 polymorphisms and pharmacokinetics and virologic outcome of lopinavir/ritonavir in HIV-infected children. Ther. Drug Monit. 2011;33(4):417–424. doi: 10.1097/FTD.0b013e318225384f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gupta A., Zhang Y., Unadkat J.D., Mao Q. HIV protease inhibitors are inhibitors but not substrates of the human breast cancer resistance protein (BCRP/ABCG2) J. Pharmacol. Exp. Ther. 2004;310(1):334–341. doi: 10.1124/jpet.104.065342. [DOI] [PubMed] [Google Scholar]
- 297.Janneh O., Jones E., Chandler B., Owen A., Khoo S.H. Inhibition of P-glycoprotein and multidrug resistance-associated proteins modulates the intracellular concentration of lopinavir in cultured CD4 T cells and primary human lymphocytes. J. Antimicrob. Chemother. 2007;60(5):987–993. doi: 10.1093/jac/dkm353. [DOI] [PubMed] [Google Scholar]
- 298.Storch C.H., Theile D., Lindenmaier H., Haefeli W.E., Weiss J. Comparison of the inhibitory activity of anti-HIV drugs on P-glycoprotein. Biochem. Pharmacol. 2007;73(10):1573–1581. doi: 10.1016/j.bcp.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 299.Weiss J., Rose J., Storch C.H., Ketabi-Kiyanvash N., Sauer A., Haefeli W.E., Efferth T. Modulation of human BCRP (ABCG2) activity by anti-HIV drugs. J. Antimicrob. Chemother. 2007;59(2):238–245. doi: 10.1093/jac/dkl474. [DOI] [PubMed] [Google Scholar]
- 300.Marzolini C., Gibbons S., Khoo S., Back D. Cobicistat versus ritonavir boosting and differences in the drug-drug interaction profiles with co-medications. J. Antimicrob. Chemother. 2016;71(7):1755–1758. doi: 10.1093/jac/dkw032. [DOI] [PubMed] [Google Scholar]
- 301.Cvetkovic M., Leake B., Fromm M.F., Wilkinson G.R., Kim R.B. OATP and P-glycoprotein transporters mediate the cellular uptake and excretion of fexofenadine. Drug Metab. Dispos. 1999;27(8):866–871. [PubMed] [Google Scholar]
- 302.Zhang L., Gorset W., Washington C.B., Blaschke T.F., Kroetz D.L., Giacomini K.M. Interactions of HIV protease inhibitors with a human organic cation transporter in a mammalian expression system. Drug Metab. Dispos. 2000;28(3):329–334. [PubMed] [Google Scholar]
- 303.van der Sandt I.C., Vos C.M., Nabulsi L., Blom-Roosemalen M.C., Voorwinden H.H., de Boer A.G., Breimer D.D. Assessment of active transport of HIV protease inhibitors in various cell lines and the in vitro blood--brain barrier. AIDS. 2001;15(4):483–491. doi: 10.1097/00002030-200103090-00007. [DOI] [PubMed] [Google Scholar]
- 304.Huisman M.T., Smit J.W., Crommentuyn K.M., Zelcer N., Wiltshire H.R., Beijnen J.H., Schinkel A.H. Multidrug resistance protein 2 (MRP2) transports HIV protease inhibitors, and transport can be enhanced by other drugs. AIDS. 2002;16(17):2295–2301. doi: 10.1097/00002030-200211220-00009. [DOI] [PubMed] [Google Scholar]
- 305.Perloff M.D., von Moltke L.L., Greenblatt D.J. Fexofenadine transport in Caco-2 cells: inhibition with verapamil and ritonavir. J. Clin. Pharmacol. 2002;42(11):1269–1274. doi: 10.1177/009127002762491370. [DOI] [PubMed] [Google Scholar]
- 306.Tirona R.G., Leake B.F., Wolkoff A.W., Kim R.B. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J. Pharmacol. Exp. Ther. 2003;304(1):223–228. doi: 10.1124/jpet.102.043026. [DOI] [PubMed] [Google Scholar]
- 307.Jung N., Lehmann C., Rubbert A., Knispel M., Hartmann P., van Lunzen J., Stellbrink H.J., Faetkenheuer G., Taubert D. Relevance of the organic cation transporters 1 and 2 for antiretroviral drug therapy in human immunodeficiency virus infection. Drug Metab. Dispos. 2008;36(8):1616–1623. doi: 10.1124/dmd.108.020826. [DOI] [PubMed] [Google Scholar]
- 308.Aungst B.J., Nguyen N.H., Bulgarelli J.P., Oates-Lenz K. The influence of donor and reservoir additives on Caco-2 permeability and secretory transport of HIV protease inhibitors and other lipophilic compounds. Pharm. Res. 2000;17(10):1175–1180. doi: 10.1023/a:1026402410783. [DOI] [PubMed] [Google Scholar]
- 309.Xu C., Zhu L., Chan T., Lu X., Shen W., Madigan M.C., Gillies M.C., Zhou F. Chloroquine and hydroxychloroquine are novel inhibitors of human organic anion transporting polypeptide 1A2. J. Pharm. Sci. 2016;105(2):884–890. doi: 10.1002/jps.24663. [DOI] [PubMed] [Google Scholar]
- 310.Sortica V.A., Lindenau J.D., Cunha M.G., MD O.O., AM R.V., Ribeiro-Dos-Santos A.K., Santos S.E., Guimaraes L.S., Hutz M.H. SLCO1A2, SLCO1B1 and SLCO2B1 polymorphisms influences chloroquine and primaquine treatment in Plasmodium vivax malaria. Pharmacogenomics. 2017;18(15):1393–1400. doi: 10.2217/pgs-2017-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Elewa H., Wilby K.J. A review of pharmacogenetics of antimalarials and associated clinical implications. Eur. J. Drug Metab. Pharmacokinet. 2017;42(5):745–756. doi: 10.1007/s13318-016-0399-1. [DOI] [PubMed] [Google Scholar]
- 312.Babayeva M., Loewy Z. Repurposing drugs for COVID-19: pharmacokinetics and pharmacogenomics of chloroquine and hydroxychloroquine. Pharmgenom. Pers. Med. 2020;13:531–542. doi: 10.2147/PGPM.S275964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Weiss J., Bajraktari-Sylejmani G., Haefeli W.E. Interaction of hydroxychloroquine with pharmacokinetically important drug transporters. Pharmaceutics. 2020;12(10) doi: 10.3390/pharmaceutics12100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lee J.Y., Vinayagamoorthy N., Han K., Kwok S.K., Ju J.H., Park K.S., Jung S.H., Park S.W., Chung Y.J., Park S.H. Association of polymorphisms of cytochrome P450 2D6 with blood hydroxychloroquine levels in patients with systemic lupus erythematosus. Arthritis Rheumatol. 2016;68(1):184–190. doi: 10.1002/art.39402. [DOI] [PubMed] [Google Scholar]
- 315.Courtois A., Payen L., Guillouzo A., Fardel O. Up-regulation of multidrug resistance-associated protein 2 (MRP2) expression in rat hepatocytes by dexamethasone. FEBS Lett. 1999;459(3):381–385. doi: 10.1016/s0014-5793(99)01295-8. [DOI] [PubMed] [Google Scholar]
- 316.Manceau S., Giraud C., Decleves X., Batteux F., Chereau C., Chouzenoux S., Scherrmann J.M., Weill B., Perrot J.Y., Treluyer J.M. Expression and induction by dexamethasone of ABC transporters and nuclear receptors in a human T-lymphocyte cell line. J. Chemother. 2012;24(1):48–55. doi: 10.1179/1120009X12Z.00000000010. [DOI] [PubMed] [Google Scholar]
- 317.Pascussi J.M., Drocourt L., Gerbal-Chaloin S., Fabre J.M., Maurel P., Vilarem M.J. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur. J. Biochem. 2001;268(24):6346–6358. doi: 10.1046/j.0014-2956.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 318.Bernareggi A., Borghi A., Borgonovi M., Cavenaghi L., Ferrari P., Vekey K., Zanol M., Zerilli L.F. Teicoplanin metabolism in humans. Antimicrob. Agents Chemother. 1992;36(8):1744–1749. doi: 10.1128/aac.36.8.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Broekhuysen J., Stockis A., Lins R.L., De Graeve J., Rossignol J.F. Nitazoxanide: pharmacokinetics and metabolism in man. Int J. Clin. Pharmacol. Ther. 2000;38(8):387–394. doi: 10.5414/cpp38387. [DOI] [PubMed] [Google Scholar]
- 320.Lespine A., Dupuy J., Orlowski S., Nagy T., Glavinas H., Krajcsi P., Alvinerie M. Interaction of ivermectin with multidrug resistance proteins (MRP1, 2 and 3) Chem. Biol. Interact. 2006;159(3):169–179. doi: 10.1016/j.cbi.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 321.Jani M., Makai I., Kis E., Szabo P., Nagy T., Krajcsi P., Lespine A. Ivermectin interacts with human ABCG2. J. Pharm. Sci. 2011;100(1):94–97. doi: 10.1002/jps.22262. [DOI] [PubMed] [Google Scholar]
- 322.Houshaymi B., Nasreddine N., Kedees M., Soayfane Z. Oleic acid increases uptake and decreases the P-gp-mediated efflux of the veterinary anthelmintic Ivermectin. Drug Res. 2019;69(3):173–180. doi: 10.1055/a-0662-5741. [DOI] [PubMed] [Google Scholar]
- 323.Neodo A., Schulz J.D., Huwyler J., Keiser J. In vitro and in vivo drug-drug interaction study of the effects of ivermectin and oxantel pamoate on tribendimidine. Antimicrob. Agents Chemother. 2019;63(1) doi: 10.1128/AAC.00762-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Perloff E.S., Duan S.X., Skolnik P.R., Greenblatt D.J., von Moltke L.L. Atazanavir: effects on P-glycoprotein transport and CYP3A metabolism in vitro. Drug Metab. Dispos. 2005;33(6):764–770. doi: 10.1124/dmd.104.002931. [DOI] [PubMed] [Google Scholar]
- 325.Kis O., Zastre J.A., Hoque M.T., Walmsley S.L., Bendayan R. Role of drug efflux and uptake transporters in atazanavir intestinal permeability and drug-drug interactions. Pharm. Res. 2013;30(4):1050–1064. doi: 10.1007/s11095-012-0942-y. [DOI] [PubMed] [Google Scholar]
- 326.Bousquet L., Roucairol C., Hembury A., Nevers M.C., Creminon C., Farinotti R., Mabondzo A. Comparison of ABC transporter modulation by atazanavir in lymphocytes and human brain endothelial cells: ABC transporters are involved in the atazanavir-limited passage across an in vitro human model of the blood-brain barrier. AIDS Res. Hum. Retrovir. 2008;24(9):1147–1154. doi: 10.1089/aid.2007.0022. [DOI] [PubMed] [Google Scholar]
- 327.Zastre J.A., Chan G.N., Ronaldson P.T., Ramaswamy M., Couraud P.O., Romero I.A., Weksler B., Bendayan M., Bendayan R. Up-regulation of P-glycoprotein by HIV protease inhibitors in a human brain microvessel endothelial cell line. J. Neurosci. Res. 2009;87(4):1023–1036. doi: 10.1002/jnr.21898. [DOI] [PubMed] [Google Scholar]
- 328.Sugie M., Asakura E., Zhao Y.L., Torita S., Nadai M., Baba K., Kitaichi K., Takagi K., Takagi K., Hasegawa T. Possible involvement of the drug transporters P glycoprotein and multidrug resistance-associated protein Mrp2 in disposition of azithromycin. Antimicrob. Agents Chemother. 2004;48(3):809–814. doi: 10.1128/AAC.48.3.809-814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Galetin A., Burt H., Gibbons L., Houston J.B. Prediction of time-dependent CYP3A4 drug-drug interactions: impact of enzyme degradation, parallel elimination pathways, and intestinal inhibition. Drug Metab. Dispos. 2006;34(1):166–175. doi: 10.1124/dmd.105.006874. [DOI] [PubMed] [Google Scholar]
- 330.Holmstock N., Mols R., Annaert P., Augustijns P. In situ intestinal perfusion in knockout mice demonstrates inhibition of intestinal p-glycoprotein by ritonavir causing increased darunavir absorption. Drug Metab. Dispos. 2010;38(9):1407–1410. doi: 10.1124/dmd.110.032771. [DOI] [PubMed] [Google Scholar]
- 331.Tong L., Phan T.K., Robinson K.L., Babusis D., Strab R., Bhoopathy S., Hidalgo I.J., Rhodes G.R., Ray A.S. Effects of human immunodeficiency virus protease inhibitors on the intestinal absorption of tenofovir disoproxil fumarate in vitro. Antimicrob. Agents Chemother. 2007;51(10):3498–3504. doi: 10.1128/AAC.00671-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Brown K.C., Paul S., Kashuba A.D. Drug interactions with new and investigational antiretrovirals. Clin. Pharmacokinet. 2009;48(4):211–241. doi: 10.2165/00003088-200948040-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Fujimoto H., Higuchi M., Watanabe H., Koh Y., Ghosh A.K., Mitsuya H., Tanoue N., Hamada A., Saito H. P-glycoprotein mediates efflux transport of darunavir in human intestinal Caco-2 and ABCB1 gene-transfected renal LLC-PK1 cell lines. Biol. Pharm. Bull. 2009;32(9):1588–1593. doi: 10.1248/bpb.32.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Vermeir M., Lachau-Durand S., Mannens G., Cuyckens F., van Hoof B., Raoof A. Absorption, metabolism, and excretion of darunavir, a new protease inhibitor, administered alone and with low-dose ritonavir in healthy subjects. Drug Metab. Dispos. 2009;37(4):809–820. doi: 10.1124/dmd.108.024109. [DOI] [PubMed] [Google Scholar]
- 335.Holmstock N., Gonzalez F.J., Baes M., Annaert P., Augustijns P. PXR/CYP3A4-humanized mice for studying drug-drug interactions involving intestinal P-glycoprotein. Mol. Pharm. 2013;10(3):1056–1062. doi: 10.1021/mp300512r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Takahashi T., Luzum J.A., Nicol M.R., Jacobson P.A. Pharmacogenomics of COVID-19 therapies. NPJ Genom. Med. 2020;5:35. doi: 10.1038/s41525-020-00143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Yamakawa Y., Hamada A., Nakashima R., Yuki M., Hirayama C., Kawaguchi T., Saito H. Association of genetic polymorphisms in the influx transporter SLCO1B3 and the efflux transporter ABCB1 with imatinib pharmacokinetics in patients with chronic myeloid leukemia. Ther. Drug Monit. 2011;33(2):244–250. doi: 10.1097/FTD.0b013e31820beb02. [DOI] [PubMed] [Google Scholar]
- 338.Mlejnek P., Kosztyu P., Dolezel P., Bates S.E., Ruzickova E. Reversal of ABCB1 mediated efflux by imatinib and nilotinib in cells expressing various transporter levels. Chem. Biol. Interact. 2017;273:171–179. doi: 10.1016/j.cbi.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 339.Gschwind H.P., Pfaar U., Waldmeier F., Zollinger M., Sayer C., Zbinden P., Hayes M., Pokorny R., Seiberling M., Ben-Am M., Peng B., Gross G. Metabolism and disposition of imatinib mesylate in healthy volunteers. Drug Metab. Dispos. 2005;33(10):1503–1512. doi: 10.1124/dmd.105.004283. [DOI] [PubMed] [Google Scholar]
- 340.Kashuba A.D., Nafziger A.N., Kearns G.L., Leeder J.S., Gotschall R., Rocci M.L., Jr., Kulawy R.W., Beck D.J., Bertino J.S., Jr. Effect of fluvoxamine therapy on the activities of CYP1A2, CYP2D6, and CYP3A as determined by phenotyping. Clin. Pharmacol. Ther. 1998;64(3):257–268. doi: 10.1016/S0009-9236(98)90174-6. [DOI] [PubMed] [Google Scholar]
- 341.Iga K. Dynamic and static simulations of fluvoxamine-perpetrated drug-drug interactions using multiple cytochrome P450 inhibition modeling, and determination of perpetrator-specific CYP isoform inhibition constants and fractional CYP isoform contributions to victim clearance. J. Pharm. Sci. 2016;105(3):1307–1317. doi: 10.1016/j.xphs.2015.11.044. [DOI] [PubMed] [Google Scholar]
- 342.Alsherbiny M.A., Li C.G. Medicinal cannabis-potential drug interactions. Medicines. 2018;6(1) doi: 10.3390/medicines6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Stout S.M., Cimino N.M. Exogenous cannabinoids as substrates, inhibitors, and inducers of human drug metabolizing enzymes: a systematic review. Drug Metab. Rev. 2014;46(1):86–95. doi: 10.3109/03602532.2013.849268. [DOI] [PubMed] [Google Scholar]
- 344.Padilla S., Telenti G., Guillen L., Garcia J.A., Garcia-Abellan J., Ding C., Mora A., Garcia-Pachon E., Gutierrez F., Masia M., C.O.-E. Group Predictive factors for cardiac conduction abnormalities with hydroxychloroquine-containing combinations for COVID-19. Int. J. Antimicrob. Agents. 2020;56(4) doi: 10.1016/j.ijantimicag.2020.106142. [DOI] [PMC free article] [PubMed] [Google Scholar]