Abstract
The complete chloroplast (cp) genome sequence of Clematis taeguensis Y.N.Lee (Ranunculaceae) was determined to be 159,534 bp in length, consisting of large (79,326 bp) and small (18,338 bp) single-copy regions and a pair of identical inverted repeats (30,935 bp). The genome contains 92 protein-coding genes, 32 tRNA genes, 8 rRNA genes, and 1 pseudogene (infA). Phylogenetic analysis of 19 taxa inferred from the chloroplast genome showed a relationship with C. taeguensis, which is also recognized as a species endemic to the Korean Peninsula. The complete cp genome sequence of C. taeguensis reported here provides important information for future phylogenetic and evolutionary studies in Ranunculaceae.
Keywords: Chloroplast genome, Clematis taeguensis, phylogenetic relationship, endemic species
The genus Clematis L. (Ranunculaceae) comprises about 300 species with worldwide distribution (Wang and Bartholomew 2001; Wang and Li 2005; Kadota 2006; Kim 2017). To date, 13 species have been reported from South Korea (Kim 2017). Among them, four species, including Clematis taeguensis Y.N.Lee, are known to be endemic to the Korean Peninsula (Lee 1982, 1996, 2006; Wang 2003a, 2003b; Wang and Li 2005; Chung et al. 2017, Park et al. 2021). This species is easily distinguishable from its closely related taxa by the lack of shoot remains in the winter season, hairy abaxial surface and margin of the sepal, and yellow to brown hairs on the style (Ko and Park 2018; Park et al. 2021). In the present study, we reported the complete chloroplast genome of C. taeguensis, and we carried out a phylogenetic analysis to investigate the phylogenetic relationships of C. taeguensis with other Clematis species.
Clematis taeguensis was sampled from Beommul-dong, Daegu-si, South Korea (Voucher specimen: BK Park, Beommuldong-190719-001, N35°49′00.2″ E128°39′16.5″). Fresh leaves were silica-dried for DNA extraction. The collected material was stored at the Herbarium of the Korea National Arboretum (KH). DNA was extracted from the silica-dried leaves using a DNeasy Plant Mini Kit (Qiagen, Seoul, Korea) following the manufacturer’ s protocol. The extracted DNA was detected on agarose gel (2%), and high quality gDNA was sequenced using an Illumina MiSeq platform with a 550 bp insert size. Finally, 8,467,638 reads were obtained. The complete chloroplast genome was assembled with Geneious Prime v2020.2.4 (Biotmatters, Auckland, New Zealand). The cp genome was annotated with the Geseq tool (Tillich et al. 2017) and Geneious Prime v2020.2.4 (Biotmatters, Auckland, New Zealand).
The complete cp genome sequence of C. taeguensis was 159,534 bp long. The obtained genome sequence was deposited in GenBank (GenBank number MW201572.1). It comprised one large single-copy region (LSC, 79,326 bp), one small single-copy region (SSC, 18,338 bp), and a pair of inverted repeats (IRs, 30,935 bp). The overall GC content of the cp genome was 38%, and in the LSC, SSC, and IRs, it was 36.3%, 31.3%, and 42.1%, respectively. The chloroplast genome contained 136 unique genes, including 92 coding genes, 8 rRNA genes, 32 tRNA genes, and one pseudogene (infA). We found that 25 genes were duplicated in the IR regions.
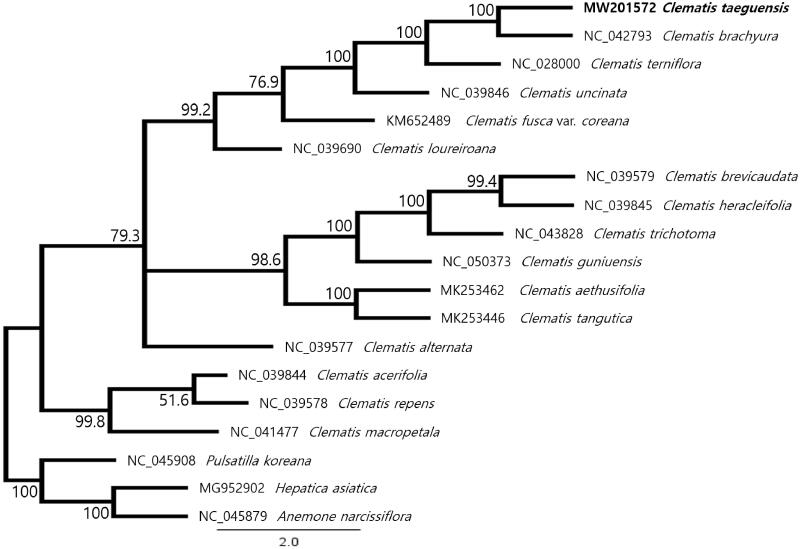
To infer the phylogenetic relationships among Clematis species, the complete cp genome sequences of 19 Clematis species were downloaded from GenBank (Pulsatilla koreana, Hepatica asiatica and Anemone narcissiflora species were used as outgroups). A total of 19 species alignments, including C. taeguensis, were performed using MAFFT v7.450 (Katoh and Standley 2013; Katoh et al. 2002). The maximum likelihood (ML) bootstrap analysis with 1000 replicates was carried out using RAxML v8.2.11 (Stamatakis 2014). Model selection was performed with JModelTest version 2.1.10 (Darriba et al. 2012) to find the best-fit model (GTR + G + I). The phylogenetic tree showed that C. taeguensis was closely related to C. brachyura taxa (Figure 1), and plastome was consistent with gene order and content of C. brachyura. However, morphologically distinguished from C. brachyura because it abaxial and marginal hairs on the sepal, yellowish to brown style hairs, achenes not winged and a persistent style is more than 1.9 cm long (Park et al. 2021). The cp genome sequence of C. taeguensis obtained in this study can provide important information for future phylogenetic and evolutionary studies of Ranunculaceae.
Figure 1.
The maximum likelihood phylogenetic tree of Clematis taeguensis Y.N.Lee and related taxa based on 92 protein-coding genes. The numbers on each node are the bootstrap support values. The species Pulsatilla koreana, Hepatica asiatica, and Anemone narcissiflora were set as outgroups.
Funding Statement
This work was supported by the research project of the Korea National Arboretum under grant KNA-1-1-13, 14-1.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW201572.1
References
- Chung GY, Chang KS, Chung J-M, Choi HJ, Paik W-K, Hyun J-O.. 2017. A checklist of endemic plants on the Korean Peninsula. Korean J Pl Taxon. 47(3):264–288. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y. 2006. Clematis L. In: Iwatsuki K, Boufford DE, Ohba H, editors. Flora of Japan. Vol. IIa. Angiospermae-Dicotyledoneae: Archichlamydeae. Tokyo: Kodansha; p. 298–308. [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS. 2017. Clematis L. In: Flora of Korea editorial committee and National Institute of Biological Resources. Flora of Korea. Vol. 2a. Incheon (Korea): National Institute of Biological Resources; p. 69–76. [Google Scholar]
- Ko SC, Park BK.. 2018. Daegu clematis. In: Oh BU, Oh SH (eds.). Silvics of Korea. Vol. 2. Korea National Arboretum, Pocheon, Republic of Korea. p. ; 103–112. [Google Scholar]
- Lee YN. 1982. New taxa of Korean flora (4). Kor J Bot. 25(4):175–180. [in Korean]. [Google Scholar]
- Lee YN. 1996. Flora of Korea. Seoul, Korea: Kyohak Publishing Co. Ltd. [in Korean]. [Google Scholar]
- Lee YN. 2006. New Flora of Korea. Vol. 1. Seoul, Korea: Kyohaksa. [in Korean]. [Google Scholar]
- Park BK, Ghimire B, Son DC.. 2021. Taxonomic identity of Korean endemic Clematis taeguensis Y. N. Lee (Ranunculales: Ranunculaceae. J Asia-Pac Biodivers. 14(1):70–77. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45 (W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WT, Bartholomew B.. 2001. Clematis L. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 6. St. Louis, MO: Science Press, Beijing and Missouri Botanical Garden Press; p. 333–386. [Google Scholar]
- Wang WT, Li LQ.. 2005. A new system of classification of the genus Clematis (Ranunculaceae). Acta Phytotax Sin. 43(5):431–488. [Google Scholar]
- Wang WT. 2003a. A revision of Clematis sect. Clematis (Ranunculaceae) (continued). Acta Phytotax Sin. 41(2):97–172. [Google Scholar]
- Wang WT. 2003b. A revision of Clematis sect. Clematis (Ranunculaceae). Acta Phytotax Sin. 41(1):1–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW201572.1