Abstract
Medicago sativa ‘Deqin’ is an excellent alfalfa landrace with strong drought and heat resistant which can grow and propagate very well in Deqin, a xerothermic valley of Jinsha River, China. In this study, the complete chloroplast genome of M. sativa ‘Deqin’ was assembled. The complete chloroplast genome of M. sativa ‘Deqin’ represents a typical circular with 125,470 bp in length, containing one inverted repeat (IR) region. Gene prediction revealed 110 genes encoding 76 proteins, 30 transfer RNAs, and four ribosome RNAs. Three genes (rps16, rpl22 and infA) are absent. The overall GC content is 33.9%. The phylogenetic analysis revealed that M. sativa ‘Deqin’ belonged to the IR lacking clade, and was closely related to M. sativa with a 100% bootstrap support.
Keywords: Medicago sativa ‘Deqin’, special landrace, chloroplast genome, phylogenetic analysis
The genus Medicago contains about 87 species (Small 2010) and belongs to tribe Trifolieae, which is nested within the invert repeat lacking clad (IRLC, Cardoso et al. 2015). Medicago sativa L. (alfalfa) is the most important forage crop in the world. Wild type alfalfa distributed in large and continuous area as dominant species in Deqin, xerothermic valley of Jinsha River with annual precipitation of 303.9–660.0 mm, Yunnan, China (Bi et al. 2007; Ma 2010). It can grow and propagate very well with highly digestible and rich in proteins; therefore, it was used as forage by local villagers (Bi et al. 2005; Zhao et al. 2013). In 2010, it was approved by the National Grass Variety Application Committee of China as a landrace, named M. sativa ‘Deqin.’ However, its phylogenetic position in the genus Medicago is still unclear. In this study, we first reported and characterized the complete chloroplast genome of M. sativa ‘Deqin’ (GenBank Accession Number: MN 218692), which will be useful for its further development and utilization, and for the future phylogenetic studies of Medicago.
Fresh leaves of M. sativa ‘Deqin’ were collected from Deqin (Yunnan, China; 28°14′43.1″N, 99°18′17.9″E). Voucher specimen (BYF2004Dq003) was deposited at Herbarium, Kunming Institute of Botany, CAS (KUN). The total genomic DNA was extracted using modified CTAB method (Doyle and Doyle 1987). Reads of the complete chloroplast genome were assembled using CLC Genomic Workbench v10 (CLC Bio., Aarhus, Denmark). All the contigs were checked against the reference genome of M. sativa (MK460489) using BLAST (https://blast.ncbi.nlm.nih.gov/) and aligned contigs were oriented according to the reference genome. The complete chloroplast genomes were then constructed using Geneious v4.8.5 (Biomatters Ltd., Auckland, New Zealand) and was automatically annotated using DOGMA (http://dogma.ccbb.utexas.edu/). To identify the phylogenetic position of M. sativa ‘Deqin’, a maximum-likelihood (ML) tree was conducted by MEGA v7.0 (Kumar et al. 2016) with 1000 bootstrap replicates based on the alignments created by the online program MAFFT (https://mafft.cbrc.jp/alignment/server/index/index.html) using already published complete chloroplast genomes.
The complete chloroplast genome of M. sativa ‘Deqin’ represents a typical circular with 125,470 bp in length, containing only one inverted repeat region. Gene prediction revealed 110 genes encoding 76 proteins, 30tRNAs, and four rRNAs. A total of sixteen genes have one intron, and ycf3 is the only gene with two introns. Three genes (rps16, rpl22, infA) are absent. The overall GC content of M. sativa ‘Deqin’ complete chloroplast genome is 33.9%.
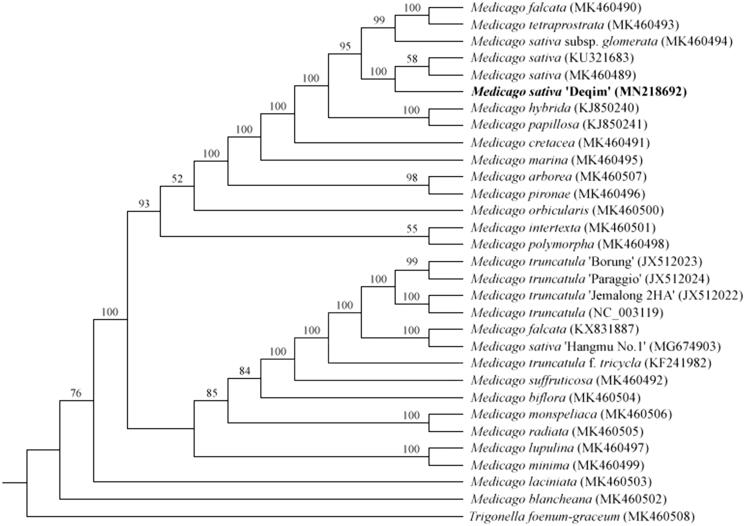
To investigate the phylogenetic position of M. sativa ‘Deqin,’ 29 published complete chloroplast genomes of the genus Medicago were used to construct a phylogeny tree, using Trigonella foenum-graceum (MK460508) in Papilionoideae as the outgroup. The results showed that M. sativa ‘Deqin’ closely related to M. sativa with a 100% bootstrap support (Figure 1), and belonged to the IRLC which was consistent with previous study (Choi et al. 2019). This complete chloroplast genome can be subsequently used for phylogenetic and genetic engineering studies of M. sativa ‘Deqin,’ and would be fundamental to formulate potential development and management strategies for this special landrace.
Figure 1.
Phylogenetic relationship among Medicago species based on the maximum-likelihood (ML) analysis of the complete chloroplast genome sequences. Bootstrap support values (%) are indicated in each node.
Acknowledgements
The authors appreciate the Laboratory of Molecular Biology in the Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences for providing experimental platform.
Funding Statement
This work was supported by the National Natural Science Foundation of China [No. 31660683].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bi YF, Che WG, Gu L.. 2007. Polymorphic characteristics and origin analysis of wild alfalfa population (Medicago sative L.) in Deqin area. Acta Agrestia Sin. 15:306–311. [Google Scholar]
- Bi YF, Li JR, Che WG.. 2005. Studies on nutrition value of less fall dormancy alfalfa. J Yunnan Agric Univ (Natural Science). 20:252–257. [Google Scholar]
- Cardoso D, São-Mateus WMB, da Cruz DT, Zartman CE, Komura DL, Kite G, Prenner G, Wieringa JJ, Clark A, Lewis G.. 2015. Filling in the gaps of the papilionoid legume phylogeny: the enigmatic Amazonian genus Petaladenium is a new branch of the early-diverging Amburaneae clade. Mol Phylogenet Evol. 84:112–124. [DOI] [PubMed] [Google Scholar]
- Choi IS, Jansen R, Ruhlman T.. 2019. Lost and found: return of the inverted repeat in the Legume clade defined by its absence. Genome Biol Evol. 11(4):1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XL. 2010. Study on genetic diversity and evolution on acid and aluminum resistance of the wild and escaped alfalfa germplasm resources in Yunnan [unpublished PhD dissertation]. Kunming: Yunnan Agricultural University. [Google Scholar]
- Small E. 2010. Alfalfa and relatives: evolution and classification of Medicago. Ottawa, Canada: NRC Research Press. [Google Scholar]
- Zhao Y, Bi YF, Che WG.. 2013. Comparison of cytoplasmic proteins variation in Medicago sativa cultivars under heat stress. Chin J Grassland. 35:13–18. [Google Scholar]