Abstract
BACKGROUND & AIMS:
Wilson disease is caused by mutations in the copper transporter ATP7B, with its main pathology attributed to copper-mediated oxidative damage. The limited therapeutic effect of copper chelators and the early occurrence of mitochondrial deficits, however, undermine the prevalence of this mechanism.
METHODS:
We characterized mitochondrial DNA copy number and mutations as well as bioenergetic deficits in blood from patients with WD and in livers of tx-j mice, a mouse model of hepatic copper accumulation. In vitro experiments with hepatocytes treated with CuSO4 were conducted to validate in vivo studies.
RESULTS:
Here, for the first time, we characterized the bioenergetic deficits in Wilson disease as consistent with a mitochondrial DNA depletion-like syndrome. This is evidenced by enriched DNA synthesis/replication pathways in serum metabolomics and decreased mitochondrial DNA copy number in blood of Wilson disease patients as well as decreased mitochondrial DNA copy number, increased citrate synthase activity, and selective Complex IV deficit in livers of the tx-j mouse model of Wilson disease. Tx-j mice treated with the copper chelator penicillamine, methyl donor choline, or both ameliorated mitochondrial DNA damage but further decreased mitochondrial DNA copy number. Experiments with copper-loaded HepG2 cells validated the concept of a direct copper-mitochondrial DNA interaction.
CONCLUSIONS:
This study underlines the relevance of targeting the copper-mitochondrial DNA pool in the treatment of Wilson disease separate from the established copper-induced oxidative stress-mediated damage.
Keywords: copper, mitochondria, bioenergetics, oxidative stress, mitochondrial DNA, penicillamine
Lay summary:
Wilson disease in patients and in mice is characterized by reduced mitochondrial DNA copy number and features of a mitochondrial DNA depletion syndrome. The mitochondrial DNA-copper pool should be a target of anti-copper treatments.
Introduction
Wilson Disease (WD) is a genetic condition characterized by multi-organ copper accumulation due to dysfunction of the ATP-dependent copper-transporting ATPase encoded by the ATP7B gene. Mutations affecting the ATP7B gene are responsible for impaired copper biliary excretion and deficient copper-carrier ceruloplasmin maturation through the trans-Golgi network. Consequently, copper accumulates in the hepatocytes, cytosol, lysosomes, and mitochondria1. The main clinical phenotypes are generally described with hepatic and neurologic involvements; however, within these categories, patients present high phenotypic variability with no genotype-phenotype correlation2.
Mitochondria morphological and functional impairments have been described in patients with WD and in animal models of copper accumulation. While mitochondria require adequate copper levels to maintain efficient Complex IV activity, copper overload is associated with early mitochondria structural changes, including separation of outer and inner membranes, cristae expansions, and giant mitochondria as well as mitochondrial protein crosslinks3–5. In rodents, high dietary copper results in a 40% reduction in copper-containing Complex IV activity6. Impaired mitochondrial oxygen uptake is also associated with copper accumulation in early and late stages of liver disease without a clear age correlation or progression7. Copper-induced mitochondrial dysfunction in mice resulted in reduced serum acylcarnitine levels starting as early as 3 weeks of age, reflecting mitochondrial fatty acid beta-oxidation defects8. Emerging data are showing a diet x genotype interaction in models of hepatic copper accumulation9,10. A high-caloric diet worsened the severity of liver injury in Atp7b−/− compared to Atp7b+/− rats, including mitochondrial morphology, reactive oxygen species (ROS) production, and aspartate aminotransaminase levels10. Removal of mitochondrial and cytosolic copper through methanobactin treatment in Atp7b−/− rats increased the number of hepatocytes with normal mitochondrial membrane potential and improved liver pathology4,10. The early hepatocellular phenotype observed in Atp7b−/− rodent models is characterized by increased mitochondrial copper accumulation associated with progressive alterations of their structure11, a phenocopy of that reported in WD patients and the Jackson Laboratory toxic milk mouse model, C3He-Atp7btx-J/J (tx-j)5,7,12,13.
The current dogma is mitochondrial dysfunction and the main multi-organ and varied clinical manifestations of WD are due to copper-induced oxidative stress-mediated damage of macromolecules5 via a copper-dependent Fenton/Haber-Weiss chemistry. However, several lines of evidence point to an unexplored and overlooked mechanism to explain the mitochondrial dysfunction observed. First, mitochondrial target deficits due to copper overload in patients with WD cannot be solely explained by increased oxidative stress. Decreased aconitase activity was reported in subjects with WD and tx-j mice5,14, similar to the superoxide dismutase mutant Sod2−/− mouse model of increased mitochondrial ROS (mtROS) and oxidative stress15. In this model, a significant accumulation of mitochondrial DNA (mtDNA) oxidative damage is accompanied by tissue-specific decreases in Complexes I, II, aconitase, and citrate synthase (CS) activities, consistent with free radical damage to mtDNA and iron-sulfur cluster-containing enzymes (except CS), respectively. However, in WD rodent models and patients, inconsistent results have been reported for the activities of Complex I and Complexes II-III with Complex IV activity the only consistently reported deficiency5,7,8,14, a deficit not observed in Sod2−/− mice. Moreover, higher CS activity in tx-j mice7 and higher superoxide dismutase activity in Atp7b−/− mice8 have been described, with lower and no activity, respectively, in Sod2−/− mice15. In the Atp7b−/− mouse liver, impairment of respiratory chain complexes was evident only at 47 weeks8, an advanced age suggesting an age x genotype interaction supported by the decline in oxidative phosphorylation (OXPHOS) capacity associated with mtDNA damage accumulation11.
Second, mitochondrial dysfunction is observed before the occurrence of histological abnormalities. Fetal tx-j livers already show altered gene expression and global DNA methylation16, suggesting these early molecular events lead to later liver pathology. Indeed, mitochondria are a likely target in tissues with impaired copper homeostasis as they contain about 30% of cellular copper and accumulate it in a membrane potential-driven manner. Furthermore, the disease progression follows the accumulation of mitochondrial copper and not that of cytosolic/total copper4.
Third, some mitochondrial changes seem to reach various degrees of recovery after long-term copper chelation treatments17; the high-affinity copper-binding methanobactin4 seemed to be more effective at recovering some aspects of mitochondrial morphology and membrane potential by removing excess mitochondrial copper. As the difference among chelators is based on their different binding capacity, e.g. the clinically-used copper chelator penicillamine (PCA, 2.38 × 10−16 M)18 vs. experimental methanobactin (10−21 M)19, we reasoned that copper tightly bound to mitochondrial structures, including mtDNA20, could not only promote mtDNA damage via redox reactions but, more importantly, copper bound specifically to mtDNA could compromise mtDNA replication and transcription20. For instance, cuprizone-treated rats exhibited increased mtDNA synthesis21,22 whereas decreased Complex IV activity and mtDNA content have been reported in a yeast model of Friedreich’s ataxia supplemented with iron23. Of note, mitochondrial ATP binding cassette transporter mutants are characterized by accumulation of mitochondrial iron and depletion of mtDNA24.
Intriguingly, significantly lower levels of critical proteins required for mtDNA replication (TFAM and SSBP) were reported in tx-j mice but with no changes in mtDNA content tested by long-range PCR7. Based on the previous arguments and this observation, we tested the hypothesis that mitochondrial dysfunction reported in patients and animal models of WD is the result of copper-induced toxicity which not only induces oxidative stress-mediated damage but also creates a complex intersection with the sequestration and/or binding of copper to mtDNA.
Methods
Subjects and animals
Humans.
Thirty-seven healthy controls and 61 patients with a diagnosis of WD according to Leipzig criteria were previously described in detail25–27 and included in the metabolomic study; of these, 47 WD were included in the mtDNA study (Fig. S3 and Table S3). Informed written consent was obtained from all subjects and patients, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Institutional Review Board at the University of California, Davis.
Animals.
Tx-j and C3HeB/FeJ (C3H) control mice from the Jackson Laboratory (Bar Harbor, Maine) were bred in-house on the UC Davis campus. All animal husbandry and experimental procedures were approved by the UC Davis Institutional Animal Care and Use Committee and abide by the National Research Council’s Guide for the Care and Use of Laboratory Animals.
For additional details about subjects, animals, assays, and statistical analyses please see Supplementary Information.
Results
Altered mitochondrial metabolic markers in serum of WD patients
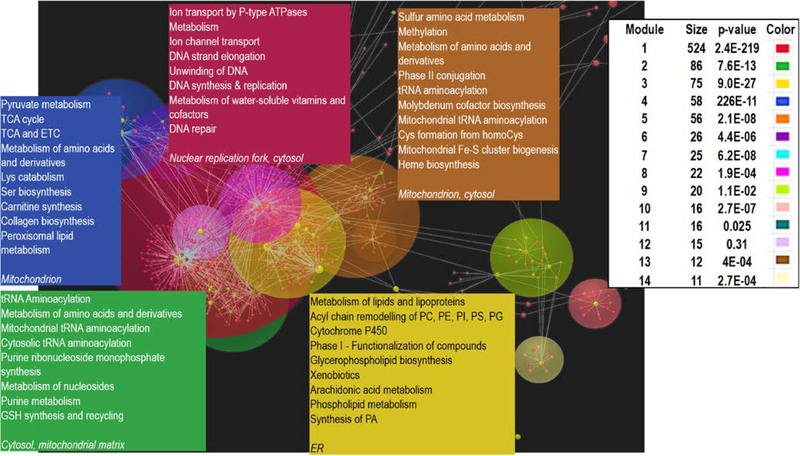
Metabolomic analyses were conducted on 61 WD and 15 healthy control serum samples. A parallel network pathway analysis utilizing the WalkTrap algorithm revealed the presence of 14 modules highlighting multiple interconnected pathways with mitochondrial metabolism and DNA replication (Fig. 1). The most affected pathway (>500 metabolites; module 1 – purple quadrant) included metabolites involved with ion transport by P-type ATPases (such as ATP7B); nuclear DNA strand elongation; and DNA unwinding, synthesis, replication, and repair, indicating an underlying defect in DNA processing. The second most affected pathway (86 metabolites; module 2 – green quadrant) was related to metabolites involved in tRNA aminoacylation, including mitochondrial tRNA aminoacylation. Other pathways included the tricarboxylic acid cycle and the mitochondrial electron transport chain (module 4 – blue quadrant), acyl-chain remodeling (module 3 – yellow quadrant), and iron-sulfur biogenesis and amino acid metabolism with emphasis on sulfur-containing amino acids (module 5 – light brown quadrant).
Figure 1. Network analysis of metabolomic data highlighting.
Metabolites selected via the partial least squares discriminant analysis with a variable importance in projection ≥ 0.8 were analyzed by using the OmicsNet49 platform and subsequently with the WalkTrap algorithm. Each module was then analyzed for pathway over-representation against the Reactome database. This last analysis revealed the presence of 14 modules (shown with size of nodes and significance) highlighting multiple interconnected pathways to copper transport, DNA synthesis and repair, and mitochondria metabolism. Matching the color of the top 5 modules, the top pathways were indicated (plain text) along with their subcellular localization (in italics). More detailed information on each of the modules is found in Dataset S1.
Untargeted metabolomics clearly distinguished WD from healthy subjects in both unsupervised (heat map; Fig. S1a) and supervised (Fig. S1b) analyses. Pathway analysis revealed enrichment of several metabolic pathways in WD vs. control, including bile acid biosynthesis, ammonia and urea metabolism, aspartate metabolism, and the malate-aspartate shuttle, matching the WD phenotype but also intersecting with mitochondrial metabolism (Fig. S1c). Metabolomic analysis revealed a shift of the glucose flow in WD from glycolysis to the pentose phosphate pathway which produces ribose for DNA synthesis and repair (Fig. S2), consistent with metabolic stress. In addition, decreased levels of γ-tocopherol and cysteine along with increased 2-hydroxybutyrate signals are indicative of oxidative stress and detoxification of xenobiotics.
Age- and BMI-independent mitochondrial DNA copy number changes in WD blood
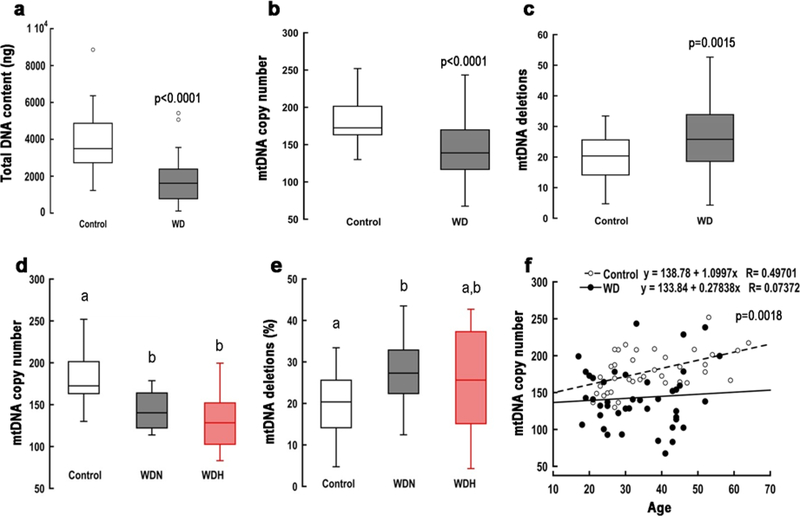
Based on the metabolomic results indicating altered mitochondrial integrity and DNA synthesis in WD, we quantified the levels of circulating total DNA and mtDNA in 37 controls and 47 WD subjects for which samples were available (Fig. S3). Compared to healthy controls, WD samples exhibited lower levels of both total DNA content and mtDNA copy number per cell (Fig. 2a, b). The mtDNA copy number in each group was neither correlated nor associated with the platelet count (Pearson’s r = −0.084; p = 0.584), suggesting it mainly originated from peripheral blood monocytic cells. The odds ratio of having a WD patient with mtDNA copy number lower than the 25th percentile (first quartile) was 5.84 (95% CI 2.19–15.54; Z = 3.53; p = 0.0004). The lower mtDNA copy number in WD was unexpected because increased copies of mtDNA are usually observed under oxidative stress conditions in a likely attempt to maintain wild-type template28. In addition, a previous study reported a lack of hepatic mtDNA depletion by long-range PCR analysis utilizing the tx-j mouse model7. As expected with copper-induced oxidative stress conditions, the diagnosis of WD was associated with more damaged mtDNA as determined by mtDNA deletions (Fig. 2c); however, the differences in mtDNA copy number and deletions were associated with neither neurologic nor hepatic subphenotypes (Fig. 2d, e).
Figure 2. Genomic and mtDNA outcomes in patients with WD.
Box plot representing (a) the amount of total genomic DNA, (b) mitochondrial DNA (mtDNA) copy number per cell, and (c) mtDNA deletions in peripheral blood monocytic cells obtained from healthy controls and Wilson disease (WD) patients (hepatic + neurologic + asymptomatic). The mtDNA was quantified via qPCR using dual-labeled probes. The ratio of mitochondrial ND1 to nuclear PK was taken as the mtDNA copy number per cell. Deletions were quantified as the ratio of CYTB to ND1 as described in the Methods section. The coefficient of variation within assays < 1% and between assays < 5%. All statistical analyses were performed by using a 2-tailed, unpaired Student’s t test given that all data followed a normal distribution. Panels (d) and (e) represent mtDNA copy number and deletions for the WD subphenotypes (neurologic and hepatic, WDN and WDH, respectively). Statistical analysis was performed with one-way ANOVA followed by Bonferroni post-hoc analysis. (f) Correlation between mtDNA copy number and age for each of the diagnostic groups. p-value was obtained via Pearson’s analysis. Healthy subjects n = 16 M/21 F; WDN n = 11 M/7 F; WDH n = 7 M/11 F; WD asymptomatic n = 6 M/5 F.
The mtDNA copy number was inversely correlated with the levels of circulating bilirubin (Pearson’s r = −0.350; p = 0.0197). Given that high bilirubin levels are associated with cholestatic or hepatocellular disease and that an equal number of patients in each subphenotype presented low mtDNA copy number (mtDNA <99% CI; 36% and 46% for hepatic and neurologic groups; Chi-squared test not significant), it is reasonable to infer both groups have an underlying hepatic disease. While mtDNA copy number in blood cells from healthy subjects positively correlated with age, as shown previously29, no significant correlation was observed in WD subjects (Fig. 2f). A weak positive association between BMI and mtDNA deletions was observed in both healthy subjects and patients with WD (Fig. S4a). A marginal but non-significant positive correlation between mtDNA deletions and copy number was observed only in subjects with WD (Fig. S4b). Together, these results suggest additional mechanisms, beside oxidative stress-mediated DNA damage, contribute to increased mtDNA deletions and reduced copy number in WD.
Phenocopy of WD human mitochondrial pathology in a mouse model of WD
Based on the module analysis from Fig. 1, we reasoned the lower mtDNA copy number in WD could be the result of decreased mtDNA replication or decreased mitochondrial mass (biogenesis). To evaluate these possibilities, we utilized the tx-j mouse model of WD, which harbors an Atp7b mutation leading to copper accumulation, compared to wild-type C3H syngenic mice.
The electron paramagnetic resonance (EPR) technique was applied to investigate the amount and nature of copper binding to its ligands in the tx-j mouse model (Fig. S5). No signal was observed at physiological pH in any of the mouse liver samples with field scans ranging from 2500 G to 39000 G. This is consistent with the EPR-silent signal obtained with cuprous-thiolate in either yeast Cu(I)-thionein30 or in rat liver Cu(I)-metallothionein clusters31. Under acidic conditions, copper is released from these complexes and oxidized to Cu(II) by oxygen, generating an EPR signal30. Consistent with this concept, a differential EPR signal was obtained at acidic pH with liver homogenates from tx-j mice (Fig. S5a). The signal was presented roughly as having half-Lorentzian and half-Gaussian broadenings with a g value of 2.16. The signal matched the signal observed with a standard solution of Cu(I)-cysteine at acidic pH (Fig. S5b)32, fitting the spectrum of copper sulfate at acidic pH (3 N HCl). This signal was characterized by a copper-chloride/water complex in fast dynamics and/or coupled by exchange and magnetic dipolar interaction with spectroscopic parameters consistent with data reported for Cu(II)33. The copper concentration in the liver homogenate was calculated and corrected for the dilution resulting in 3.06 mM (179 μg copper/g liver wet weight). This value was about 10-fold the copper-metallothionein binding capacity31 and within the range of values reported previously utilizing atomic absorption to quantify copper in livers from either the same mouse model (100–200 μg copper/g liver wet weight)7,8 or from three WD-affected subjects (312–725 μg/g)14.
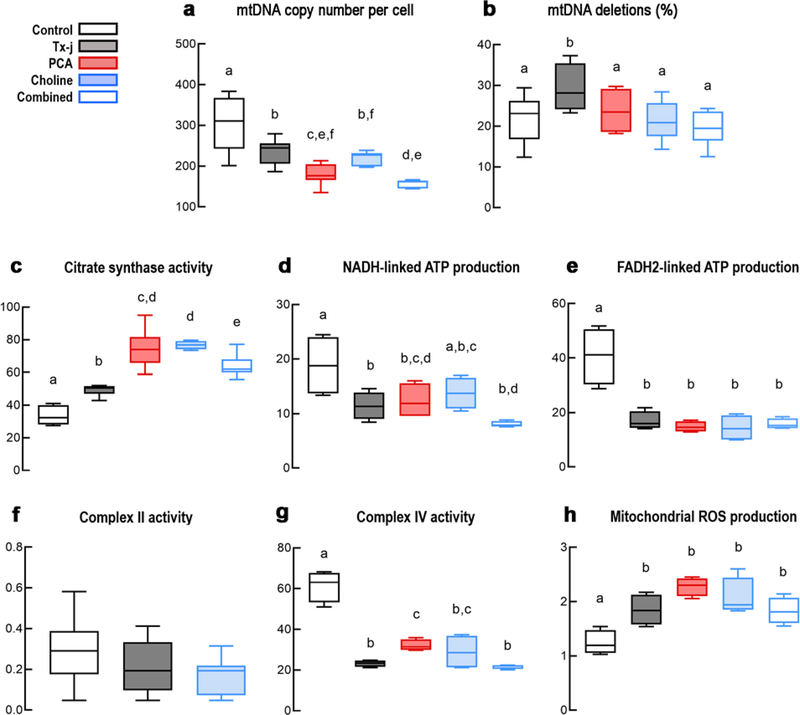
As expected, tx-j mice exhibited significantly decreased mtDNA copy number compared to C3H control (Fig. 3a) and significantly more mtDNA deletions (Fig. 3b). These findings resemble those observed in WD subjects, thereby validating the use of this mouse model for studying WD mitochondrial deficits in humans and the presence of a likely common mechanism in both species leading to similar detrimental mitochondrial outcomes.
Figure 3. Hepatic mtDNA and mitochondrial bioenergetic outcomes in tx-j mice.
The mtDNA copy number (a) and deletions (b) were evaluated in C3HeB/FeJ (C3H) control and C3He-Atp7btx-J/J (tx-j) mouse livers. (c) Citrate synthase activity (CS, as a marker of mitochondrial matrix) was evaluated by spectrophotometry. (d) The NADH-linked and (e) FADH2-linked oxygen uptake was utilized to evaluate hepatic mitochondrial ATP production in C3H control and tx-j mice. (f) Complex II activity was evaluated by a colorimetric method. (g) KCN-sensitive Complex IV activity was evaluated by polarography. Activities were expressed as nmol oxygen (panels d, e, g) or product (panels c, f) x (min x mg protein)−1. (h) Proton leak/mitochondrial ROS at Complex III was evaluated as the oxygen uptake obtained in the presence of an FADH2-linked substrate, rotenone, and antimycin, and normalized by the basal respiration. All data are shown as Tukey’s box plots. Statistical analyses were performed by using ANOVA followed by uncorrected Fisher’s Least Significant Difference post-hoc test. Equal letters on top of each box signify no statistically significant differences among groups (p > 0.05). C3H control n = 10 M/12 F; tx-j n = 11 M/11F; PCA n = 12 M/10 F; Choline n = 8 M/13 F; Combined n = 10 M/12 F.
Mitochondrial deficits are resistant to copper chelation and one-carbon supply
Pharmacologic treatments provided to tx-j mice were designed to test the effects of the copper chelator PCA (from 12 to 24 weeks of age) on reducing copper-induced oxidative damage and choline supplementation (conception to 24 weeks of age) on facilitating mtDNA replication and repair through increased dietary methyl donors (Fig. 3). While all treatments rescued the mtDNA damage as expected, PCA, alone or in combination with choline, decreased the mtDNA copy number further. This exacerbated decrease is likely independent from oxidative stress (PCA-resistant) and could be explained by either an increased mitophagy or by a direct inhibition of mtDNA replication (Fig. 1, module 1). No increased mitophagy was observed as CS activity (a surrogate marker of mitochondrial mass) was elevated in tx-j mice and further increased after all provided treatments (Fig. 3c). The increased CS activity in tx-j mice is consistent with enlargement of existing hepatic mitochondria, accumulation of damaged mitochondria34, or mtDNA depletion as these disorders show often-elevated CS activity35,36.
Based on our previous work and to determine if differentially methylated nuclear genes were associated with mtDNA replication/translation, we re-analyzed existing genome-wide methylome data from the same WD subjects25. Pathway analysis revealed 114 WD differentially methylated genes with essential roles in one-carbon metabolism, mitochondrial network organization, and mitochondrial protein synthesis as well as mtDNA transcription, replication, and stability (Table S1). From this subset of nuclear encoded genes with mitochondrial function, we evaluated the hepatic transcript levels of mitochondrial serine hydroxymethyltransferase (Shmt2) in tx-j mice along with that of its cytosolic counterpart, Shmt1, and other selected genes with differential methylation relevant to mitochondrial function (Fig. S6). SHMT2, the mitochondrial enzyme that catalyzes formation of the essential purine biosynthesis intermediate 5,10-methylenetetrahydrofolate, was selected to discern the effect of mitochondrial translation via folate-dependent tRNA methylation, independent of changes in mtDNA copy number or deletions37. Among the genes whose hepatic transcript levels were altered in tx-j mice compared to C3H controls were Shmt1, Shmt2, and Pgc1a (down-regulated), and Atp5b, Sirt2, and Ndufs6 (up-regulated), with no differences for Ppard. When testing treatment effects on the above-indicated genes, 6 out of 8 showed differential expression with PCA treatment.
Whole genome DNA methylation analyses identified several genes epigenetically altered in WD as possible regulatory candidates for Complex IV activity (COX20, PET100) and mitochondrial DNA replication with some implicated in mtDNA depletion syndromes (Table S2 in bold). However, differential methylation status of these genes did not correspond to expression changes or PCA treatment response in tx-j mice, suggesting a limited impact on mitochondrial biogenesis as well as on deficits in mtDNA replication and bioenergetics observed in WD. Thus, the low mtDNA copy number (Fig. 3a) and the relatively high CS activity (Fig. 3c) seem consistent with a mtDNA depletion-like syndrome in WD characterized by a lower mtDNA replication capacity separate from the copper-induced oxidative stress damage.
As most mtDNA depletion syndromes are associated with impaired activities of complexes with mtDNA-encoded subunits (I, III, IV, and V), we evaluated the hepatic mitochondrial ATP capacity of tx-j mice vs. C3H control with NADH- and FADH2-linked substrates. Segments encompassing Complexes I-III-IV and II-III-IV were decreased in tx-j mice (Fig. 3d, e) with no changes in the activity of the nuclear DNA-encoded Complex II (Fig. 3f), suggesting deficits in mtDNA-encoded components. Consistent with several studies from subjects with WD and mouse models of copper accumulation5,7,8,14, Complex IV activity was significantly decreased (by 60%) in tx-j livers relative to controls (Fig. 3g). The mtROS production from Complex I was not different between C3H control and tx-j (Fig. S7), whereas Complex III-driven ROS levels were significantly higher in tx-j mice (Fig. 3h). None of the treatments significantly improved deficits in ATP production or Complex IV activity (1.4-fold recovery by PCA, but mainly attributed to increased CS activity), or reduced mitochondrial ROS production (Fig. 3 and Fig. S7), again ruling out oxidative stress or shortage in one-carbon supply as the major contributors to the deficits observed in these outcomes. Nuclear DNA-encoded ATP5B, COXIV, and CS protein levels in tx-j mice were mildly affected by the treatments and mtDNA-encoded MTCO1 levels were higher in untreated tx-j mice, not affected by PCA treatment, and lower with choline (Fig. S7); however, none of the treatments were associated with an improvement in Complex IV and/or bioenergetics in general (Fig. 3).
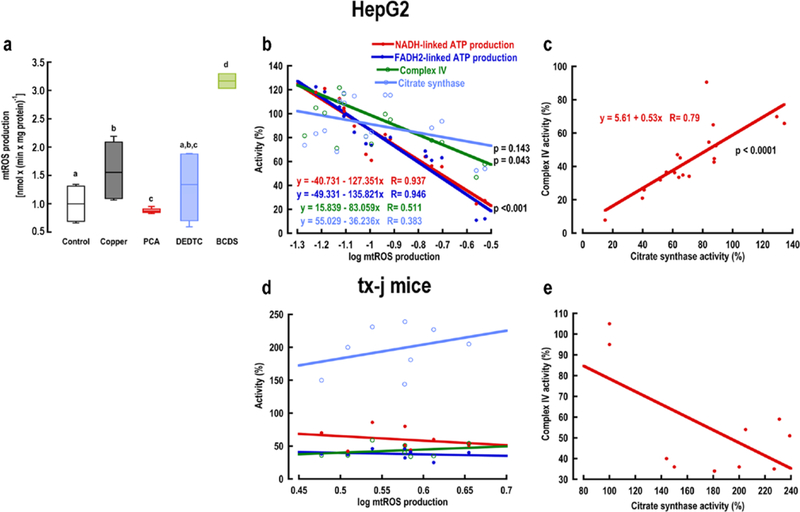
To provide evidence of tx-j bioenergetics differing from a copper-induced oxidative stress injury, we modulated mtROS production by utilizing copper chelators in copper-loaded HepG2 cells. The chelators bathocuproine disulfonate (BCDS), diethyldithiocarbamate (DEDTC), and PCA were selected based on their cell permeability and affinity for copper: cell impermeant BCDS with a stability constant of log β2 at pH 7.4 = 18.7 calculated from β2max 38; cell permeant DEDTC with log β2 at pH 7.4 = 18 18 or log β2 at pH 7.5 = 16.1 39; and cell impermeant PCA40 with log β2 at pH 7.4 = 15.6 18. As predicted, the use of chelators modulated mtROS production with higher rates in copper-treated HepG2 cells compared to controls (Fig. 4a). PCA significantly reduced mtROS production to control values, whereas BCDS increased it significantly when compared to any of the other treatments; no significant changes were recorded for DEDTC. The production of ATP fueled by NADH- and FADH2-linked substrates was significant and inversely correlated with mtROS production (Pearson’s p < 0.001; Fig. 4b). The fitted lines for these outcomes were not significantly different by ANCOVA suggesting mtROS damages a target (Complex III or Complex IV) shared by both of these substrates. Albeit at a lower significance (p = 0.043), the correlation of mtROS with Complex IV activity showed the same trend as the rates of ATP production via Complex I and II, whereas no correlation was observed with the activity of CS. Notably, the rate of mtROS production required to decrease both rates of ATP production by 50% was 0.145 ± 0.05 nmmol x (min x mg protein)−1, whereas to achieve the same effect on Complex IV the rate was 3.4-fold higher, indicating (i) the decrease in ATP production is likely due to mtROS-induced damage of Complex III and (ii) Complex IV is significantly more resistant to oxidative stress than the rest of the electron transport chain. A strong direct correlation (Pearson’s r = 0.79) was obtained between Complex IV and CS activities indicating these outcomes changed coordinately in response to different rates of mtROS production (Fig. 4c). The same type of analyses conducted with the bioenergetic outcomes tested in mouse liver (Fig. 3) showed no correlation with mtROS production under any treatment condition (Fig. 4d, e).
Figure 4. Mitochondrial bioenergetic outcomes in oxidative stress-induced HepG2 cells.
(a) Modulation of mitochondrial reactive oxygen species (mtROS) in copper-loaded HepG2 cells by copper chelators. The rate represents the log of the total mtROS production by Complex I and Complex III. Statistical analysis was performed with one-way ANOVA followed by uncorrected Fisher’s Least Significant Difference test. Equal letters on top of each box signify no statistically significant differences among groups (p > 0.05). Bioenergetic results were expressed as a percentage of average untreated control values for (b, c) HepG2 cells and (d, e) C3He-Atp7btx-J/J (tx-j) mice. Pearson’s coefficients (R) and two-tailed p-values are reported for each correlation. Tx-j mice n = 4 M/4 F. HepG2 cells n = 3 T-75 flasks per treatment group.
Discussion
This study compiles several novel lines of evidence from unbiased metabolomic and mitochondrial bioenergetic screens along with our direct investigation of mtDNA in both WD patients and the tx-j mouse model of WD to reveal unexplored mechanisms underlying WD mitochondrial dysfunction. Until now, detrimental changes in mitochondrial integrity have been attributed to copper-induced oxidative damage via a Fenton/Haber-Weiss chemistry5,14,41,42. However, our findings highlight a novel mechanism independent from and possibly complimentary to epigenetics and oxidative stress-mediated damage at the core of the mitochondrial deficits in WD molecular pathogenesis. Here, we discuss our results in the framework of an altered WD mitochondrial metabolism due to a direct copper-mtDNA interaction with an ensuing mtDNA depletion-like syndrome which contributes to the pathogenesis of WD and adds important new metabolic evidence for the precise nature of WD mitochondrial deficits (Fig. 5).
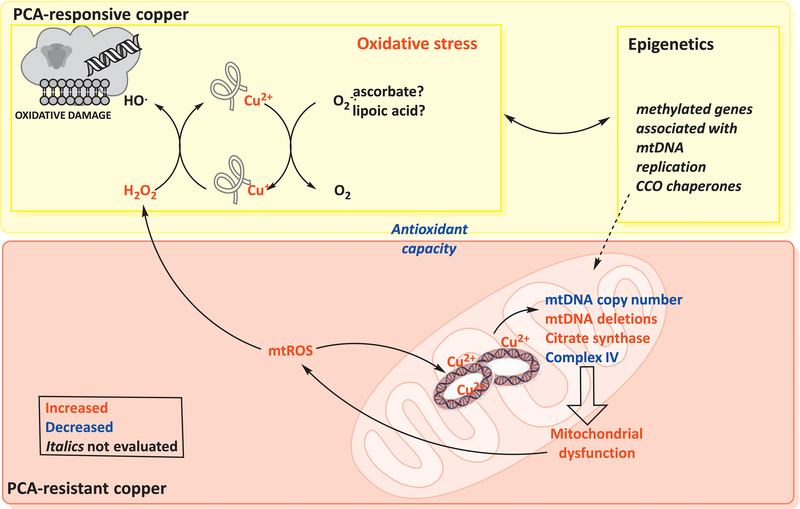
Figure 5. Mitochondria-centered representation of WD pathogenesis.
Altered Wilson disease (WD) mitochondrial metabolism appears to be the downstream effect of a direct copper-mitochondrial DNA (mtDNA) interaction, leading to a mtDNA depletion-like syndrome. Under conditions of copper overload, deletions or damage to the D-loop region of mtDNA could result in halted replication, affecting the overall mitochondrial ATP production capacity. The deficit in ATP7B results in lower copper elimination with subsequent copper accumulation in the cytosol and mitochondria. Upon penicillamine (PCA) treatment, two pools can be functionally defined as PCA-resistant (mitochondrial) and PCA-labile (other than mitochondrial). In the context of the labile pool, copper can form adducts with adequate ligands (nuclear DNA, proteins, glutathione). Intracellular reductants, e.g. glutathione and superoxide anion, can then reduce Cu(II)-adduct to Cu(I)-adduct; in turn, this reduced adduct can catalyze the cleavage of hydrogen peroxide (mainly produced by mitochondria) resulting in the high-reactive species hydroxyl radical. This radical causes cellular damage by initiating lipid peroxidation, DNA strand breaks, and protein crosslinkings, breaks, and aggregates. The increased oxidative stress can trigger epigenetic changes, some of which may have some impact on mitochondrial metabolism. By binding to mtDNA, mitochondrial copper accumulation prevents normal replication and transcription, resulting in Complex IV deficits, lower mtDNA copy number, and increased deletions with higher citrate synthase. The deletions are acquired during the Haber-Weiss reaction with mtDNA-copper adducts, mitochondrial ROS, and mitochondrial reductants20,50; this reaction can also affect other mitochondrial components such as aconitase or initiate lipid peroxidation. As a result, mitochondria are unable to perform fatty acid β-oxidation; heme synthesis becomes impaired (anemia, hemolysis, splenomegaly); and a dysfunctional urea cycle leads to increased ammonia levels, altered neurotransmitter levels, and energy deficits.
The molecular pathology of WD is consistent with that of a mtDNA depletion-like syndrome. This is supported by the enriched nuclear and mtDNA synthesis and translation pathways (metabolomics of WD patients), the lower mtDNA copy number (in tx-j mouse livers and blood of WD patients), and the decreased Complex IV activity with higher CS activity in tx-j mice compared to C3H controls. Together, these results led us to consider copper accumulation may result in lower content of functional mtDNA. As the threshold for true mtDNA depletion is set at values ≤ 30% of controls, and the mtDNA copy number from tx-j mice and WD subjects was about 80% relative to their respective controls, we reasoned the fraction of functional mtDNA in WD falls below that required to sustain a viable respiratory chain. The same scenario is observed in cases of iron overload with loss of mtDNA and in individuals with heteroplasmic deletions and tRNA point mutations of mtDNA11,43, the latter example in concordance with altered mitochondrial tRNA aminoacylation and translation indicated in our pathway analyses (Fig. 1, modules 2 and 5; Table S1).
The mtDNA deletions were increased in tx-j mice and in patients with WD. Our findings are consistent with a higher incidence of large single or multiple mtDNA deletions in livers from patients with WD compared to controls41, although patients were on chelation therapy and assessed only for large mtDNA deletions in this previous study. The mtDNA damage assessed here was responsive to both treatments, copper chelator PCA and methyl donor choline, but this was not mirrored by improvements in mtDNA copy number. The low content of mtDNA, albeit damaged, in subjects with WD as well as tx-j mice and the weak correlation between mtDNA deletions and copy number indicate a small percentage of deletions was acquired during the replication process, suggesting issues with replication itself. Furthermore, mtDNA deletions in WD do not appear to originate from the normal aging process, which leads to cumulative oxidative mtDNA damage, but rather represent an early and persistent feature throughout the disease progression, indicated by the lack of correlation between mtDNA copy number in blood cells and age in patients with WD. Indeed, although our anti-copper treatment observation is limited to animal models, the recovery of mtDNA damage upon PCA treatment would suggest the mtDNA repair capacity is intact. However, the lack of PCA-mediated recovery of bioenergetics indicates bioenergetic deficits are unrelated to mtDNA damage.
The deficits in energy metabolism identified by human serum metabolomics were further elucidated and functionally complemented by detailed studies in the tx-j mouse liver. The increased abundance of 2-hydroxybutyrate, a by-product of cystathionine cleavage in conditions of low glutathione and an early marker of impaired glucose metabolism regulation44, the decreased N-acetylglutamate, and the increased citrulline levels point to a compromised disposal of excess nitrogen resulting from amino acid catabolism. The higher levels of glycerol, glyceric acid, and 1-acylglycerol suggest increased lipolysis, probably to swap damaged fatty acids from membrane pools. The pool size of TCA cycle intermediates (oxoglutarate) and associated amino acids (Glu, Asp, Phe, Tyr) was increased while Gln and Asn were decreased, reflecting an increased glycolytic activity and compromised mitochondrial fatty acid beta-oxidation45.
In tx-j mice, mitochondrial ATP production, especially Complex IV activity, was significantly decreased, whereas mtROS was increased. To date, none of the mitochondrial findings reported in the context of WD were obtained by testing the whole electron transport chain, overlooking the effect of moderate changes in each complex’s specific activity on overall ATP production capacity. In our study, the defect in the bioenergetic capacity was mainly attributed to Complex IV, a deficiency also reported by others in WD patients and rodent models5,7,8,14 and in one study in heterozygous subjects who do not accumulate significant hepatic copper (total, not necessarily mitochondrial)46. The lower ATP capacity in tx-j mouse livers did not improve significantly with any of the treatments. These results preclude the possibility that oxidized cardiolipin, lipid critical for Complex IV activity and a target of ROS47, was at the core of this deficiency as no activity was rescued after 12 weeks of copper chelation with PCA. The specificity of decreased Complex IV activity in WD could be explained by the fact that Complex IV is the only copper-containing complex with two copper-binding sites required for its full activity; the lower mtDNA content may affect Complex IV more than any other complex given that mtDNA-encoded proteins represent 30% of those in Complex IV vs. only 7–16% of other complexes with mtDNA-encoded subunits. Moreover, our data also exclude an effect of mtDNA damage on mtDNA replication and transcription as mtDNA deletions were recovered by all treatments, whereas mitochondrial function was not.
In contrast to the lack of significant improvement in bioenergetics, the expression of most differentially methylated nuclear-encoded genes related to mitochondrial metabolism was modulated by individual treatments in mice. In addition to copper-mtDNA toxicity, it is possible other factors not tested in this study could contribute to WD-associated mitochondrial dysfunction such as the differential methylation of Complex IV chaperone genes (COX20, PET100; Table S2) and LRPPRC. Further studies are warranted to elucidate the epigenetic role of mitochondrial-encoding genes and defective bioenergetics in WD.
The apparent discrepancy with previous studies supporting a major role for copper-mediated oxidative stress damage in WD can be bridged by considering our findings with copper-loaded HepG2 cells in which mtROS production was inversely correlated with some bioenergetic outcomes (NADH-/FADH2-linked ATP production and Complex IV activity), whereas no correlation was identified between the same outcomes in the mouse model.
As mitochondrial copper accumulation has been widely established in WD, a copper-mtDNA adduct is certainly plausible, leading to mitochondrial dysfunction and treatment inefficacy of mitochondrial bioenergetics by lowering the number of “functional” mtDNA copies available for normal replication, transcription, and possibly translation. This is supported by differences between WD bioenergetic deficits and our own model of copper-induced oxidative stress injury along with previous iron- and copper-overload studies. In cuprizone-treated rats, low intracellular copper conditions increase mtDNA content and mtDNA synthesis in association with duplications of D-loop, the origin of mtDNA replication21,22. Therefore, it is likely that, under conditions of copper overload, D-loop deletion/damage or excess copper bound to mtDNA could block replication, affecting the capacity to generate ATP (Fig. 5). Taken together, these findings resemble those reported for sideroblastic anemia in which iron-mtDNA adducts result in mtDNA relaxation (creating a more receptive state for further metal-DNA binding and iron colloid growth on the strands) that is not completely prevented by either nitrogen purging or antioxidants, precluding a major role for ROS48. Rats with long-term copper overload showed lower hepatic oxygen uptake and selectively decreased Complex IV activity, pointing to this oxidase as a key target in a copper toxicity model6. Our model is also in line with the therapeutic effect of methanobactin, a potent bacterial copper-binding protein that significantly reduces copper content in Atp7b−/− rat mitochondria, even at late stages of disease4. Therefore, copper-DNA complexes may serve as effective ligands (with an estimated binding constant as high as 10−19 M), supporting a site-specific mechanism for mtDNA unwinding which leads to conformational changes and precludes normal replication and transcription. In tx-j mice, the PCA-mediated recovery of mtDNA damage, but not replication, provides support to the concept of direct copper-mtDNA interaction as a trigger for mitochondrial dysfunction over that driven by a downstream oxidative damage mechanism. This is further supported by the differences between tx-j mice and copper-loaded HepG2 cells as models of copper-induced toxicity.
In conclusion, our findings overcome some limitations of previous studies and have relevant clinical implications that could modify chelation therapy in WD by determining and monitoring mitochondrial function at the molecular level for chelator efficacy. As with other mitochondrial disorders, it is tempting to propose the number and extent of clinical manifestations increase as more organ systems become compromised with mitochondrial copper overload and the ensuing depletion of “functional” mtDNA. Our study highlights the need for developing more clinically effective copper chelators, e.g. methanobactin4, possibly bioengineered to specifically target subcellular organelles, thereby increasing efficacy while reducing undesired and potentially dangerous side-effects, e.g. not competing with endogenous, normal copper ligands such as SCO2. Clinical assays to determine and monitor recovery of mitochondrial function should be requisite to test whether or not new treatments could reverse the compromised bioenergetic capacity, likely an early hallmark of the disease.
Supplementary Material
Acknowledgments
Grant support: The research was supported by the National Institutes of Health through grant number R01DK104770 (to V.M.) and by S10RR023586 and partly by R21HD086769 (to C.G.).
Abbreviations
- BCDS
bathocuproine disulfonate
- C3H
C3HeB/FeJ
- CS
citrate synthase
- DEDTC
diethyldithiocarbamate
- EPR
electron paramagnetic resonance
- mtDNA
mitochondrial DNA
- mtROS
mitochondrial ROS
- OXPHOS
oxidative phosphorylation
- PCA
penicillamine
- ROS
reactive oxygen species
- tx-j
Jackson Laboratory toxic milk mouse model C3He-Atp7btx-J/J
- WD
Wilson disease
Footnotes
Disclosures: The authors have declared no conflict of interest.
REFERENCES
- 1.Czlonkowska A, Litwin T, Dusek P, et al. Wilson disease. Nat Rev Dis Primers. 2018;4(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferenci P, Stremmel W, Czlonkowska A, et al. Age and Sex but Not ATP7B Genotype Effectively Influence the Clinical Phenotype of Wilson Disease. Hepatology. 2019;69(4):1464–1476. [DOI] [PubMed] [Google Scholar]
- 3.Sternlieb I Fraternal concordance of types of abnormal hepatocellular mitochondria in Wilson’s disease. Hepatology. 1992;16(3):728–732. [DOI] [PubMed] [Google Scholar]
- 4.Lichtmannegger J, Leitzinger C, Wimmer R, et al. Methanobactin reverses acute liver failure in a rat model of Wilson disease. J Clin Invest. 2016;126(7):2721–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zischka H, Lichtmannegger J, Schmitt S, et al. Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease. J Clin Invest. 2011;121(4):1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol RJ, Devereaux MW, O’Brien K, Khandwala RA, Loehr JP. Abnormal hepatic mitochondrial respiration and cytochrome C oxidase activity in rats with long-term copper overload. Gastroenterology. 1993;105(1):178–187. [DOI] [PubMed] [Google Scholar]
- 7.Roberts EA, Robinson BH, Yang S. Mitochondrial structure and function in the untreated Jackson toxic milk (tx-j) mouse, a model for Wilson disease. Mol Genet Metab. 2008;93(1):54–65. [DOI] [PubMed] [Google Scholar]
- 8.Sauer SW, Merle U, Opp S, et al. Severe dysfunction of respiratory chain and cholesterol metabolism in Atp7b(−/−) mice as a model for Wilson disease. Biochim Biophys Acta. 2011;1812(12):1607–1615. [DOI] [PubMed] [Google Scholar]
- 9.Wooton-Kee CR, Robertson M, Zhou Y, et al. Metabolic dysregulation in the Atp7b (−/−) Wilson’s disease mouse model. Proc Natl Acad Sci U S A. 2020;117(4):2076–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Einer C, Leitzinger C, Lichtmannegger J, et al. A High-Calorie Diet Aggravates Mitochondrial Dysfunction and Triggers Severe Liver Damage in Wilson Disease Rats. Cell Mol Gastroenterol Hepatol. 2019;7(3):571–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256(5057):628–632. [DOI] [PubMed] [Google Scholar]
- 12.Sternlieb I Mitochondrial and fatty changes in hepatocytes of patients with Wilson’s disease. Gastroenterology. 1968;55(3):354–367. [PubMed] [Google Scholar]
- 13.Huster D, Finegold MJ, Morgan CT, et al. Consequences of copper accumulation in the livers of the Atp7b−/− (Wilson disease gene) knockout mice. Am J Pathol. 2006;168(2):423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu M, Cooper JM, Butler P, et al. Oxidative-phosphorylation defects in liver of patients with Wilson’s disease. Lancet. 2000;356(9228):469–474. [DOI] [PubMed] [Google Scholar]
- 15.Melov S, Coskun P, Patel M, et al. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc Natl Acad Sci U S A. 1999;96(3):846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medici V, Kieffer DA, Shibata NM, et al. Wilson Disease: Epigenetic effects of choline supplementation on phenotype and clinical course in a mouse model. Epigenetics. 2016;11(11):804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sternlieb I, Feldmann G. Effects of anticopper therapy on hepatocellular mitochondria in patients with Wilson’s disease: an ultrastructural and stereological study. Gastroenterology. 1976;71(3):457–461. [PubMed] [Google Scholar]
- 18.Smirnova J, Kabin E, Jarving I, et al. Copper(I)-binding properties of de-coppering drugs for the treatment of Wilson disease. alpha-Lipoic acid as a potential anti-copper agent. Sci Rep. 2018;8(1):1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semrau JD, Jagadevan S, DiSpirito AA, et al. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ Microbiol. 2013;15(11):3077–3086. [DOI] [PubMed] [Google Scholar]
- 20.Giulivi C, Boveris A, Cadenas E. Hydroxyl radical generation during mitochondrial electron transfer and the formation of 8-hydroxydesoxyguanosine in mitochondrial DNA. Arch Biochem Biophys. 1995;316(2):909–916. [DOI] [PubMed] [Google Scholar]
- 21.Albring M, Radsak K, Thoenes W. Enhanced DNA synthesis in isolated megamitochondria. FEBS Lett. 1973;35(1):4–6. [DOI] [PubMed] [Google Scholar]
- 22.Guerineau M, Guerineau S, Gosse C. Abnormal mitochondrial DNA molecules in megamitochondria from cuprizone-treated rats. Eur J Biochem. 1974;47(2):313–319. [DOI] [PubMed] [Google Scholar]
- 23.Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich’s ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411(2–3):373–377. [DOI] [PubMed] [Google Scholar]
- 24.Kispal G, Steiner H, Court DA, Rolinski B, Lill R. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem. 1996;271(40):24458–24464. [DOI] [PubMed] [Google Scholar]
- 25.Mordaunt CE, Kieffer DA, Shibata NM, et al. Epigenomic signatures in liver and blood of Wilson disease patients include hypermethylation of liver-specific enhancers. Epigenetics Chromatin. 2019;12(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarode GV, Kim K, Kieffer DA, et al. Metabolomics profiles of patients with Wilson disease reveal a distinct metabolic signature. Metabolomics. 2019;15(3):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazi TA, Sarode GV, Czlonkowska A, et al. Dysregulated Choline, Methionine, and Aromatic Amino Acid Metabolism in Patients with Wilson Disease: Exploratory Metabolomic Profiling and Implications for Hepatic and Neurologic Phenotypes. Int J Mol Sci. 2019;20(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giulivi C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu CS, Tsai CS, Kuo CL, et al. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free radical research. 2003;37(12):1307–1317. [DOI] [PubMed] [Google Scholar]
- 30.Rupp H, Cammack R, Hartmann HJ, Weser U. Oxidation-reduction reactions of copper-thiolate centres in Cu-thionein. Biochim Biophys Acta. 1979;578(2):462–475. [DOI] [PubMed] [Google Scholar]
- 31.Shishido N, Nakayama K, Takazawa A, Ohyama T, Nakamura M. Cu-metallothioneins (Cu(I)8-MTs) in LEC rat livers 13 weeks after birth still act as antioxidants. Arch Biochem Biophys. 2001;387(2):216–222. [DOI] [PubMed] [Google Scholar]
- 32.Gala L, Lawson M, Jomova K, et al. EPR spectroscopy of a clinically active (1:2) copper(II)-histidine complex used in the treatment of Menkes disease: a Fourier transform analysis of a fluid CW-EPR spectrum. Molecules. 2014;19(1):980–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Śmigielska HLG, Goslar J, Hoffmann SK Binding of the Trace Elements: Cu(II) and Fe(III) to the Native and Modified Nutritive Potato Starches Studied by EPR. Acta Physica Polonica Series. 2005;108:303–310. [Google Scholar]
- 34.Napoli E, Song G, Panoutsopoulos A, et al. Beyond autophagy: a novel role for autism-linked Wdfy3 in brain mitophagy. Sci Rep. 2018;8(1):11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ducluzeau PH, Lachaux A, Bouvier R, et al. Depletion of mitochondrial DNA associated with infantile cholestasis and progressive liver fibrosis. J Hepatol. 1999;30(1):149–155. [DOI] [PubMed] [Google Scholar]
- 36.Stiles AR, Simon MT, Stover A, et al. Mutations in TFAM, encoding mitochondrial transcription factor A, cause neonatal liver failure associated with mtDNA depletion. Mol Genet Metab. 2016;119(1–2):91–99. [DOI] [PubMed] [Google Scholar]
- 37.Morscher RJ, Ducker GS, Li SH, et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554(7690):128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao Z, Loughlin F, George GN, Howlett GJ, Wedd AG. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J Am Chem Soc. 2004;126(10):3081–3090. [DOI] [PubMed] [Google Scholar]
- 39.Banci L, Bertini I, Ciofi-Baffoni S, et al. Affinity gradients drive copper to cellular destinations. Nature. 2010;465(7298):645–648. [DOI] [PubMed] [Google Scholar]
- 40.Gupte A, Wadhwa S, Mumper RJ. Enhanced intracellular delivery of the reactive oxygen species (ROS)-generating copper chelator D-penicillamine via a novel gelatin--D-penicillamine conjugate. Bioconjug Chem. 2008;19(7):1382–1388. [DOI] [PubMed] [Google Scholar]
- 41.Mansouri A, Gaou I, Fromenty B, et al. Premature oxidative aging of hepatic mitochondrial DNA in Wilson’s disease. Gastroenterology. 1997;113(2):599–605. [DOI] [PubMed] [Google Scholar]
- 42.Sokol RJ, Twedt D, McKim JM Jr., et al. Oxidant injury to hepatic mitochondria in patients with Wilson’s disease and Bedlington terriers with copper toxicosis. Gastroenterology. 1994;107(6):1788–1798. [DOI] [PubMed] [Google Scholar]
- 43.Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–178. [DOI] [PubMed] [Google Scholar]
- 44.Napoli E, Schneider A, Wang JY, et al. Allopregnanolone Treatment Improves Plasma Metabolomic Profile Associated with GABA Metabolism in Fragile X-Associated Tremor/Ataxia Syndrome: a Pilot Study. Mol Neurobiol. 2019;56(5):3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56. [DOI] [PubMed] [Google Scholar]
- 46.Shokeir MH, Shreffler DC. Cytochrome oxidase deficiency in Wilson’s disease: a suggested ceruloplasmin function. Proc Natl Acad Sci U S A. 1969;62(3):867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzagoloff A, MacLennan DH. Studies of the electron-transfer system. LXIV. Role of phospholipid in cytochrome oxidase. Biochim Biophys Acta. 1965;99(3):476–485. [DOI] [PubMed] [Google Scholar]
- 48.Yaffee M, Walter P, Richter C, Muller M. Direct observation of iron-induced conformational changes of mitochondrial DNA by high-resolution field-emission in-lens scanning electron microscopy. Proc Natl Acad Sci U S A. 1996;93(11):5341–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou G, Xia J. Using OmicsNet for Network Integration and 3D Visualization. Curr Protoc Bioinformatics. 2019;65(1):e69. [DOI] [PubMed] [Google Scholar]
- 50.Massa EM, Giulivi C. Alkoxyl and methyl radical formation during cleavage of tert-butyl hydroperoxide by a mitochondrial membrane-bound, redox active copper pool: an EPR study. Free Radic Biol Med. 1993;14(5):559–565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.