Abstract
Studies of biological function demand probes that can report on processes in real time and in physiological environments. Bioluminescent tools are uniquely suited for this purpose, as they enable sensitive imaging in cells and tissues. Bioluminescent reporters can also be monitored continuously over time without detriment, as excitation light is not required. Rather, light emission derives from luciferase-luciferin reactions. Several engineered luciferases and luciferins have expanded the scope of bioluminescence imaging in recent years. Multicomponent tracking remains challenging, though, due to a lack of streamlined methods to visualize combinations of bioluminescent reporters. Conventional approaches image one luciferase at a time. Consequently, short-term changes in cell growth or gene expression cannot be easily captured. Here we report a strategy for rapid, multiplexed imaging with a wide range of luciferases and luciferins. Sequential addition of orthogonal luciferins, followed by substrate unmixing, enabled facile detection of multiple luciferases in vitro and in vivo. Multicomponent imaging in mice was also achieved on the minutes-to-hours time scale.
Graphical Abstract
INTRODUCTION
Bioluminescence imaging (BLI) is a popular technique for tracing cells and other biological features in heterogeneous environments, including whole animals.1–2 BLI applications rely on genetically encoded luciferase enzymes and luciferin substrates for photon production. Because mammalian tissues do not normally glow, BLI enables sensitive imaging in vivo.3–5 As few as 1–10 cells can be reliably detected using optimized probes in subcutaneous models.6 For these reasons, BLI has long been a go-to technique for monitoring physiological processes in rodents.7–9 More recent advances are further enabling studies in deeper tissues and larger organisms,10–13 including non-human primates.10,14
While ubiquitous, BLI applications in vivo typically track only one cell type or feature at a time.15 A spectrum of bioluminescent probes exists, and some pairs can be color-resolved in rodent models.16–22 Discriminating larger collections is challenging, though, due to the broad emission spectra of bioluminescent reporters and complications from tissue absorption.8 Historically, many applications featuring two BLI reporters combined firefly luciferase (Fluc) and Renilla luciferase (Rluc), two enzymes that use different substrates.23–24 Recent examples have featured Akaluc and Antares or CBG2, additional enzymes that use distinct luciferins.16,25 Multicomponent imaging with these pairs in vivo, though, can require multiple hours, if not days. The bioluminescent substrates are typically administered at saturating doses to maximize photon output,26 and the first probe must clear prior to administering the second. In principle, dozens of other naturally occurring (and orthogonal) luciferases and luciferins could be employed for multiplexed BLI.27–29 In practice, though, most of these enzymes and substrates are not easily applied in vivo owing to suboptimal stabilities, bioavailability, or other parameters.
We aimed to address the need for better probes and practical imaging protocols for multicomponent BLI. Our approach builds on the expanding toolbox of orthogonal bioluminescent reagents.8,15 These probes comprise genetically modified luciferases that are responsive to chemically unique luciferins.12,14,25,30–34 Some of the orthogonal enzymes can be readily discriminated in cells and mouse models based on selective substrate use.17,25,30–32,35–37 Many of these probes are not completely specific (i.e., the luciferins are processed preferentially by one enzyme, but can be turned over to some extent by other enzymes). However, perfect selectivity is not required. The patterns of substrate use (“fingerprints”) can be used to discriminate combinations of luciferases. Indeed, such “fingerprint” analyses are commonly used to differentiate engineered GPCRs and signaling pathways.38–40 Like the examples above, though, traditional applications of the engineered luciferases require long periods of time (hours-to-days) for complete image acquisition.25,31 Dynamic changes in gene expression and cell growth cannot be captured under such conditions. Consequently, bioluminescence has historically been limited to monitoring bulk changes in biological samples. Here we report a method to more rapidly resolve orthogonal bioluminescent probes. The approach is broadly applicable and scalable to multiple reporters, enabling facile multiplexed bioluminescence imaging. We also showcase its utility for monitoring dynamic changes on the minutes time scale.
RESULTS AND DISCUSSION
Identifying Optimal Luciferases
We hypothesized that rapid BLI could be achieved via sequential substrate administration and serial image acquisition. Light outputs would build over time, with each luciferin application resulting in stronger signal. A final processing step41 would unmix the images (Figure 1a). Similar “layering” approaches have vastly expanded fluorescence detection of gene transcripts42 and other cellular features21,43 in recent years. The technique also differs from conventional optical imaging platforms, in that wavelength, lifetime, and other traditional parameters are not factored into the analysis. The probes must simply be substrate resolved (i.e., exhibit some degree of orthogonality) and intensity resolved (Figure 1b). Substrate resolution minimizes cross talk between the probes to provide unique fingerprints. Intensity resolution ensures that signal can be “layered in”: as successively brighter probes are imaged, residual signal from dimmer, earlier images becomes part of the background. If the bioluminescent probes are not intensity resolved, the targets are indistinguishable. The key takeaway from the design is that no time is required for substrate clearance, which is an important consideration for capturing rapid, dynamic events.
Figure 1.
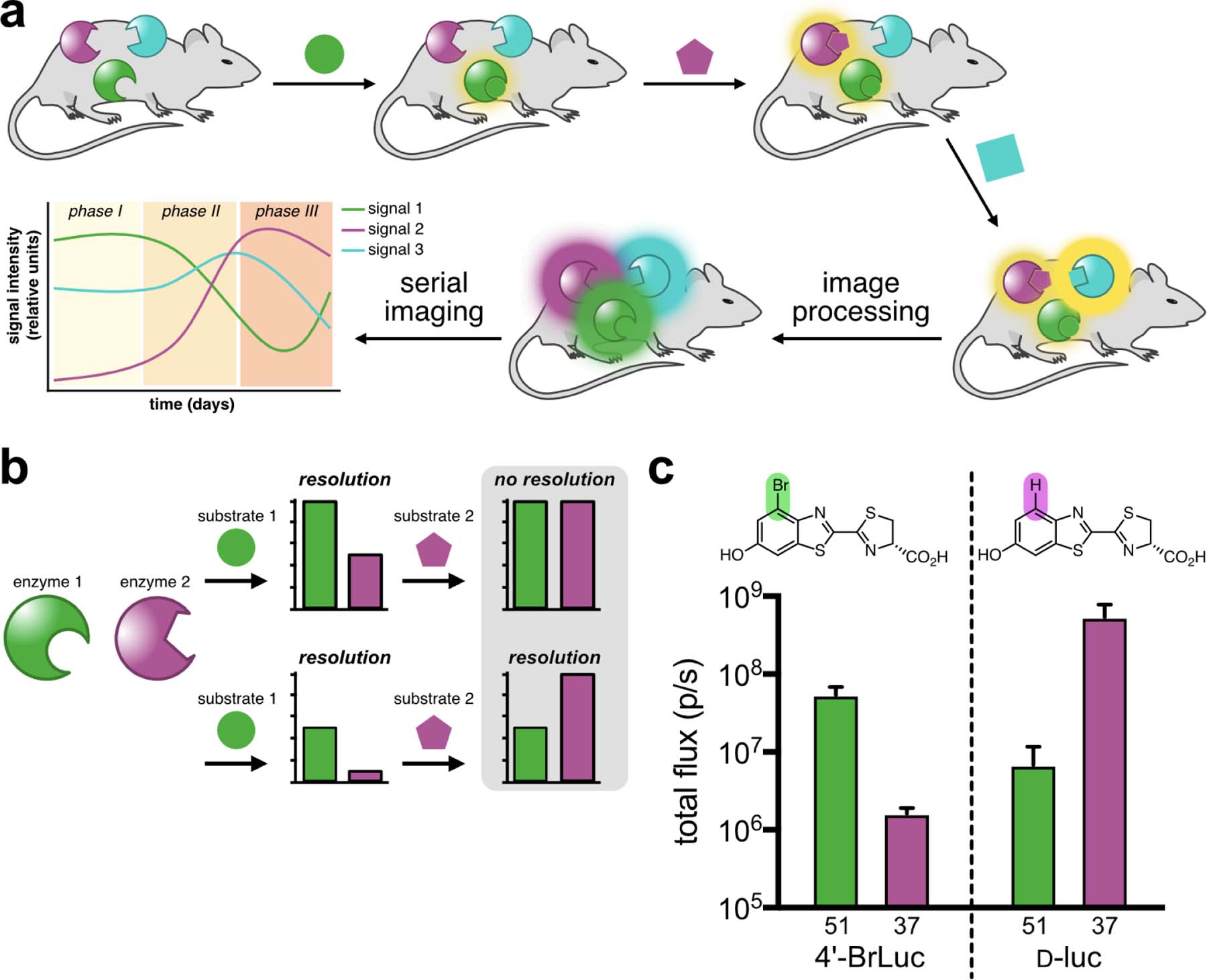
Multicomponent bioluminescence imaging via serial substrate addition and unmixing. (a) Sequential application of orthogonal luciferins (shapes) to illuminate multiple luciferase reporters in vivo. Linear unmixing algorithms can deconvolute substrate signatures, enabling rapid and dynamic readouts of biological processes. (b) Bioluminescent probes must be orthogonal and exhibit differential emission intensities for successful unmixing. When both probes are equally “bright” (top), no resolution is possible. Probes of varying intensity (bottom) can be distinguished when the dimmest probe is administered first. (c) A sample orthogonal bioluminescent pair. Samples expressing mutant 51 or 37 can be resolved using 4’-BrLuc (100 μM) or d-luc (100 μM). Error bars represent the standard error of the mean for n = 3 experiments.
To identify suitable probes for rapid BLI, we focused on two previously reported orthogonal luciferins: 4’-BrLuc and d-luciferin (d-luc, Figure 1c).31,44 These substrates are bright, bioavailable, and accessible in large quantities.45–46 We also previously identified mutant luciferases that could differentiate the analogs.31,47 While orthogonal, these pairs were engineered for maximum brightness and not built with intensity resolution in mind. To identify orthogonal luciferases that exhibited a range of photon outputs, we screened a small panel of mutants known to process C4’-modified analogs (Figure 2a). Screens were performed both in vitro and in vivo (Supplementary Figure 1, Figure 2b). In the latter case, DB7 cells stably expressing mutant luciferases were implanted in mice. 4’-BrLuc and d-luc were administered sequentially. The most orthogonal and intensity resolved pair comprised mutants 51 and 37 (Figure 2b), which we subsequently named Pecan and Cashew, respectively. Bioluminescent signal from 4’-BrLuc/Pecan is lower than that of d-luc/Cashew, meaning that the two pairs are intensity resolved and amenable to rapid sequential imaging (Figure 1c, Figure S1). 4’-BrLuc can be administered first (to illuminate Pecan), followed immediately by d-luc (to illuminate Cashew). Since Cashew signal is brighter than Pecan, the images can be readily unmixed. It is also important to note that while 4’-BrLuc/Pecan signal is reduced relative to d-luc/Fluc, it is still sufficiently intense for applications in vivo.
Figure 2.
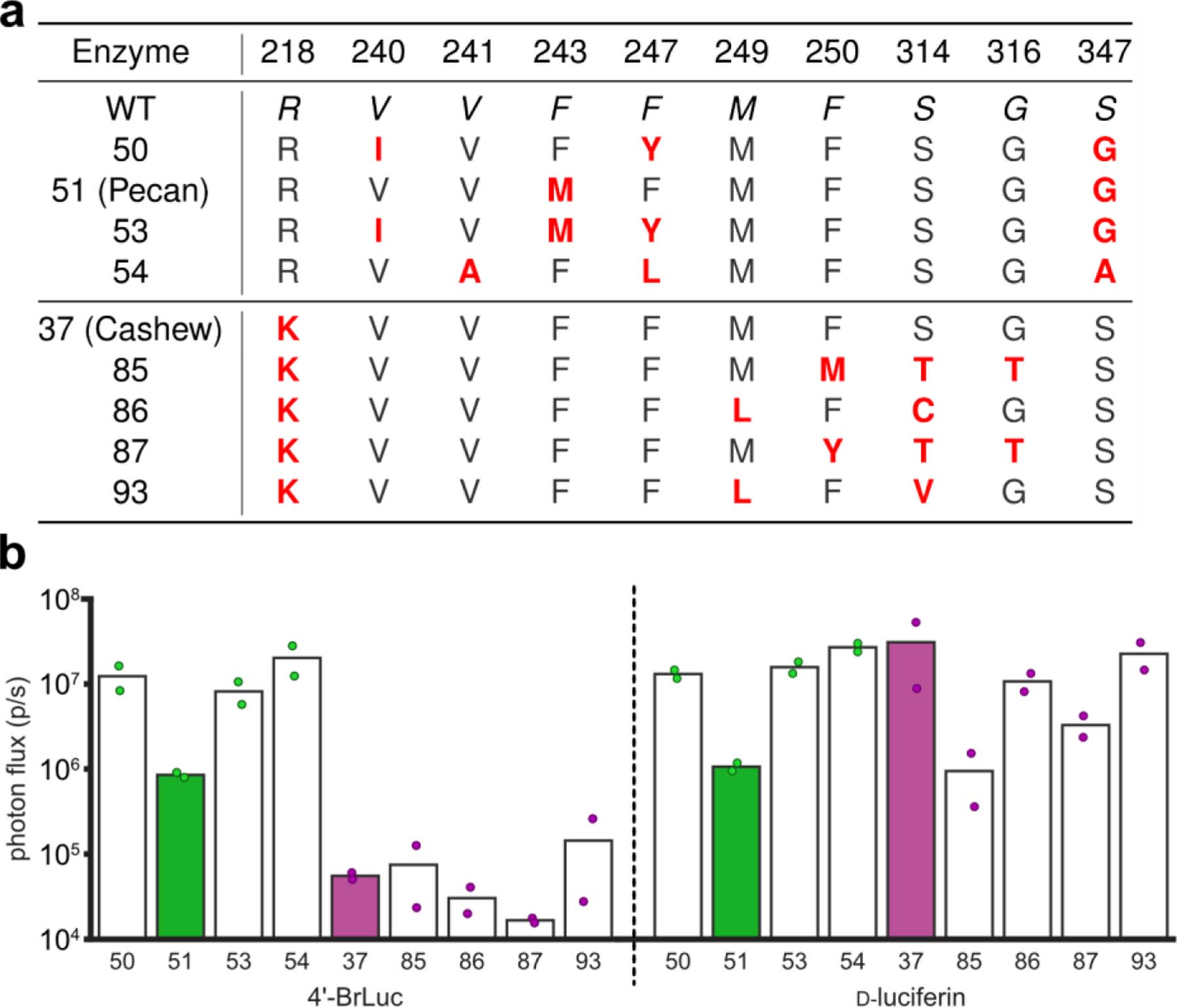
Identifying intensity resolved orthogonal pairs (a) Mutants considered for rapid BLI with 4’-BrLuc and d-luc. Mutants 50, 51 (Pecan), 53, and 54 prefer 4’-BrLuc, while 37 (Cashew), 85, 86, 87, and 93 prefer d-luc.11 (b) Verifying orthogonality and substrate resolution in vivo. DB7 cells expressing different mutant luciferases were implanted into the backs of mice. Sequential application of 4’-BrLuc and d-luc enabled identification of optimal mutant luciferase combinations for multicomponent imaging. Photon flux values from images were quantified and plotted. Mutant luciferases 51 and 37 were identified as optimal candidates for rapid multicomponent BLI.
Resolving Orthogonal Probes via Intensity Resolution and Linear Unmixing
To evaluate the reporters for rapid multicomponent BLI, we performed a series of in vitro experiments. Pecan and Cashew-expressing cells were lysed and distributed across black-well plates. 4’-BrLuc was initially administered to each well, and an image of the plate was acquired (Figure 3a). d-luc was then immediately added to the same wells, and a second image was acquired. Because light output from Cashew is brighter, the Pecan signal fell entirely within the noise of the second image. False colors were assigned to each reporter. The images were then overlaid and a linear unmixing algorithm was employed to determine the relative quantities of each mutant. The measured signal correlated linearly with probe concentration (Figure 3b). Signal outputs from mixed lysate samples were also co-linear with samples comprising a single luciferase, indicating minimal signal crosstalk. Rapid imaging required the dimmest reporter to be visualized first (Supplementary Figure 2). The unmixing algorithm is not necessary when the second target is more abundant than the first.42 When the second target (associated with the “brightest” luciferase) is in low abundance, though, the algorithm is crucial for proper image interpretation. Unmixing ensures that residual signal from the first image is eliminated and doesn’t interfere with the subsequent image (Supplementary Figure 3 and Supplementary Discussion). Since the relative abundance of multiple targets is unknown in a given experiment, the algorithm would be employed in all cases by the end user.
Figure 3.
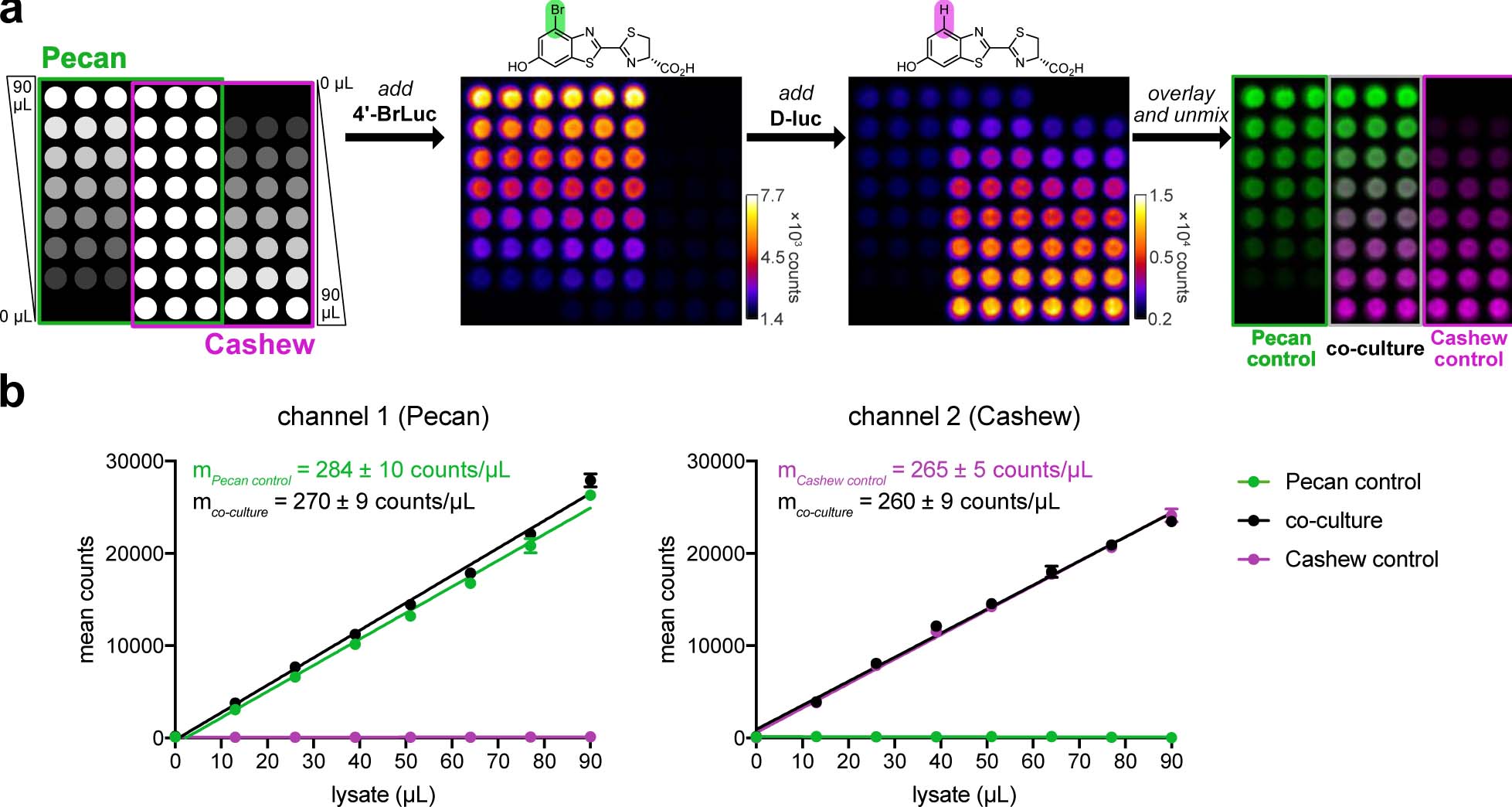
Rapid BLI in vitro. (a) Pecan and Cashew were plated in a gradient fashion (as shown). The samples were treated with 4’-BrLuc (100 μM), followed by D-luc (100 μM). Raw images were acquired after each substrate addition. The substrate-specific signals were unmixed, assigned false colors and overlaid. (b) Quantification of the images from (a), fit via linear regression. In channel 1, R2 values for the Pecan control and co-culture wells are 0.993 and 0.994, respectively. In channel 2, R2 values for Cashew control and co-culture are 0.998 and 0.993, respectively. Error bars represent the standard error of the mean for n = 3 experiments.
These results underscore the notion that perfect substrate selectivity is not required for differentiating multiple probes, broadening the potential impact of the approach. Many dual-component imaging experiments rely on completely different classes of enzymes and substrates that exhibit different bioavailabilities and administration routes.23,25,36 Substrates that are more structurally similar can be easier to work with, but typically exhibit imperfect orthogonality.31,32,45,48–49 The substrate unmixing algorithm takes advantage of these imperfections (in the form of diagnostic fingerprints) and should be able to interface with the dozens of luciferin analogs reported to date. Indeed, the “layering in” approach successfully resolved combinations of d-luciferin analogs and their associated enzymes via rapid luciferin administration (Supplementary Figure 4). Mixtures of Fluc and other commercially available reporters (e.g., NanoLuc and Gaussia luciferase) were readily unmixed following sequential addition of the corresponding substrates (Figure 4). Moreover, combinations of mammalian cells could be visualized in a single imaging session (Supplementary Figure 5). These examples suggest that the unmixing approach is generalizable to multiple luciferase reporters that use unique substrates and exhibit a range of intensities. Additionally, the unmixing approach is applicable to not only well-differentiated bioluminescent tools (e.g., orthogonal insect and marine luciferins), but also structurally similar probes (e.g., d-luciferin analogs). This feature greatly expands the number of luciferase-luciferin pairs that can be imaged in a single experiment, as perfectly orthogonal pairs are not required.
Figure 4.
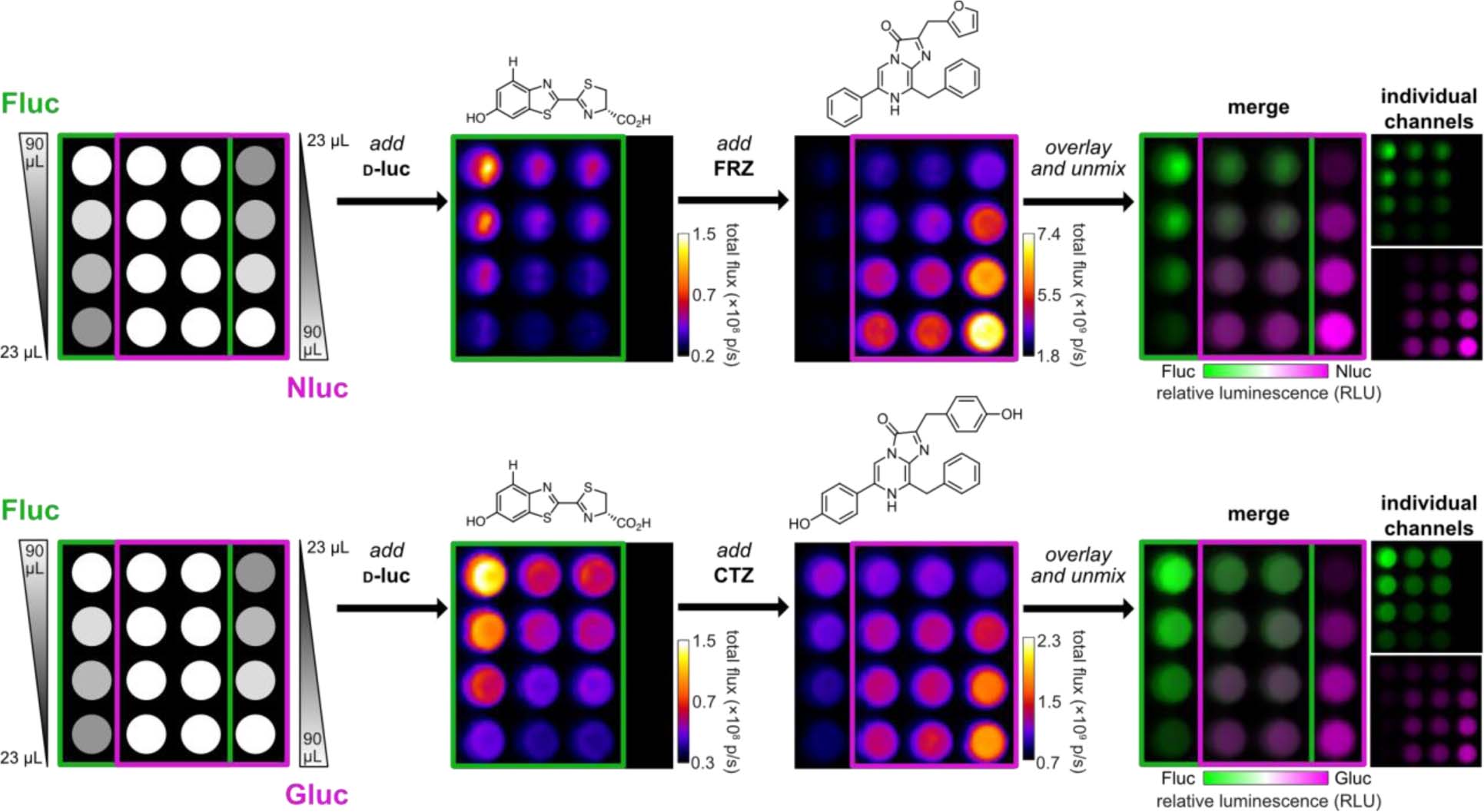
Multiple orthogonal pairs can be rapidly unmixed. Established reporters examined include firefly luciferase (Fluc), NanoLuc (Nluc), and Gaussia luciferase (Gluc). The corresponding luciferins for each reporter are shown. Gradients of the luciferases were plated as shown. The corresponding substrates ([d-luc] = 100 μM, [FRZ] = 1:40 of commercial stock, [CTZ] = 100 μM) were administered, beginning with the dimmest luciferin. Images were acquired after each addition. The raw data were stacked, unmixed, and false colored.
To showcase the utility of rapid unmixing, we performed a head-to-head comparison with conventional bioluminescence imaging. The latter approach entails waiting for signal to clear from one substrate, prior to administering the second. Since saturating doses of luciferins are typically used, the clearance period can be several hours to days. Our approach via sequential substrate application and signal unmixing can dramatically shorten this time frame, enabling improved temporal resolution. As an initial demonstration, mixtures of Pecan- and Cashew-expressing mammalian cells were cultured in varying ratios (Figure 5a). Conventional bioluminescence imaging was performed by adding one luciferin substrate (4’-BrLuc) to illuminate the Pecan-expressing cells. Images were acquired until signal returned to background levels (24 h, bottom row of Figure 5a). The second luciferin (d-luc) was then applied to the cultures to capture Cashew-dependent signal. In total, the two-component imaging study was complete in just over 24 h. By contrast, sequential substrate addition followed by unmixing enabled two-component imaging in only 20 minutes (top row, Figure 5a).
Figure 5.
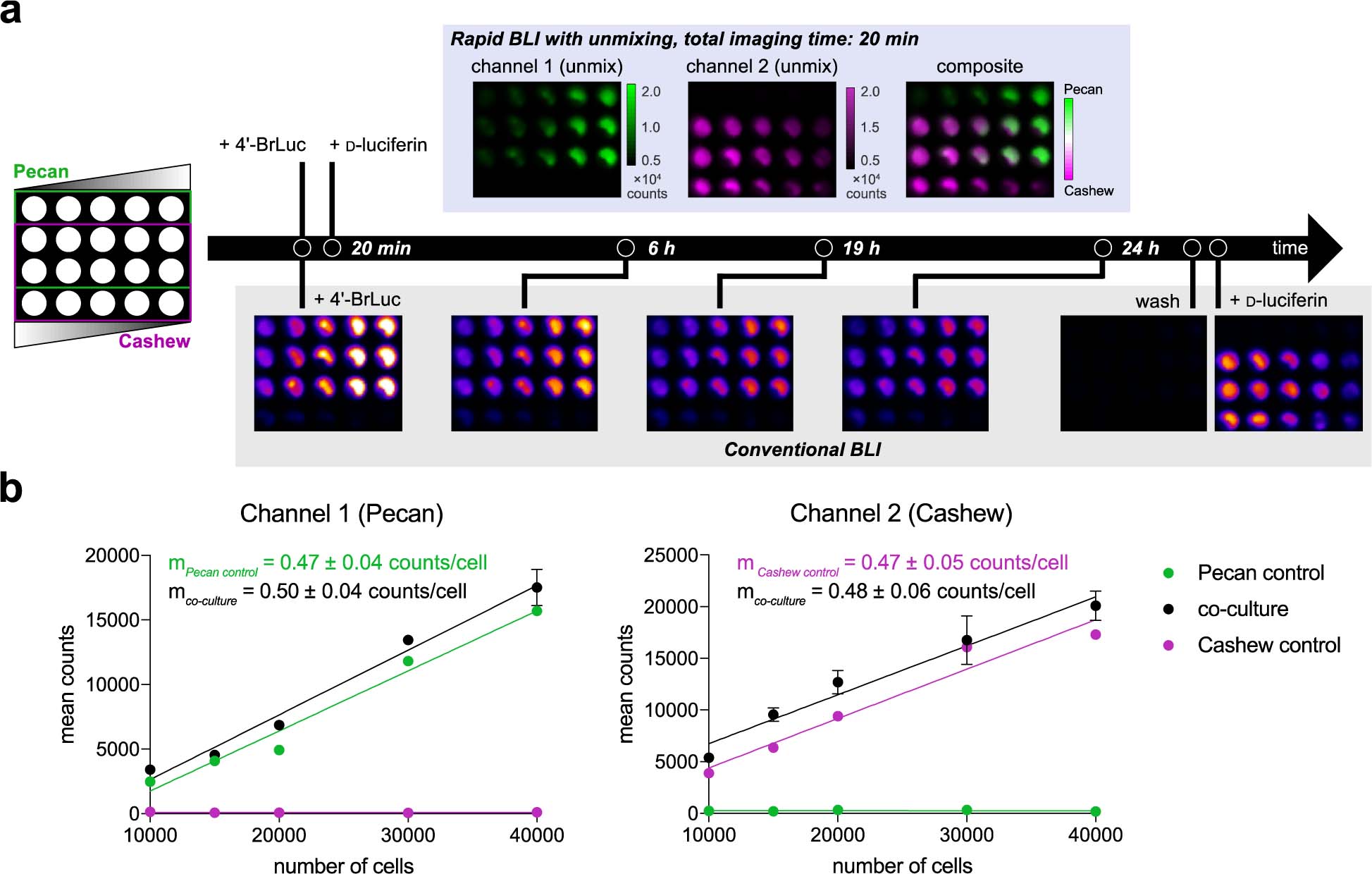
Rapid two-component BLI via substrate unmixing. (a) Cells expressing Pecan or Cashew, were plated in a 96-well plate as shown. Sequential substrate administration (4’-BrLuc, followed by D-luc, top row) and unmixing enabled two-component imaging in only 20 min. Conventional bioluminescence imaging required >24 h to complete (bottom row). Data are representative of n = 3 replicates. (b) Quantified photon outputs for the images in (a), fit via linear regression. In channel 1, R2 values for the Pecan control and co-culture wells are 0.98 and 0.99, respectively. In channel 2, R2 values for the Cashew control and co-culture wells are 0.95 and 0.97, respectively. Error bars represent the standard error of the mean for n = 3 experiments.
We surmised that the unmixing algorithm would also enable rapid imaging in vivo. As an initial demonstration, Pecan- and Cashew-expressing cells were mixed in varying ratios and implanted in mice (106 cells per site, Figure 6a). Upon injection of 4’-BrLuc, Pecan-expressing cells were readily visualized (Figure 6b). Prior to substrate clearance, the brighter luciferin (d-luc) was injected. Signal was then observed from Cashew-expressing cells. Substrate unmixing revealed the expected distributions of Cashew- and Pecan-expressing cells (Figure 6b and Supplementary Figure 6). Notably, the two-component imaging session was complete in ~1 h, a significant improvement from the 6–24 h imaging window common to other substrate-resolved probes.31 The bioavailabilities of the luciferins featured in this experiment are similar, although this is not a requirement for unmixing. While the relative composition of the cell masses can be readily visualized, the unmixing algorithm cannot provide absolute quantification of bioluminescent signals in animals. Rather, the relative amounts of signal are easily discerned and tracked. Samples with a large amount of Pecan will appear “more green than red” in the unmixed images. As the reporter ratios change over time, so too will the unmixed images. These measures are often paramount in optical imaging in vivo, and imaging relative differences is standard practice in most bioluminescent applications.50–52 Similar intensiometric measures are routinely used in imaging with fluorescent sensors.53–55
Figure 6.
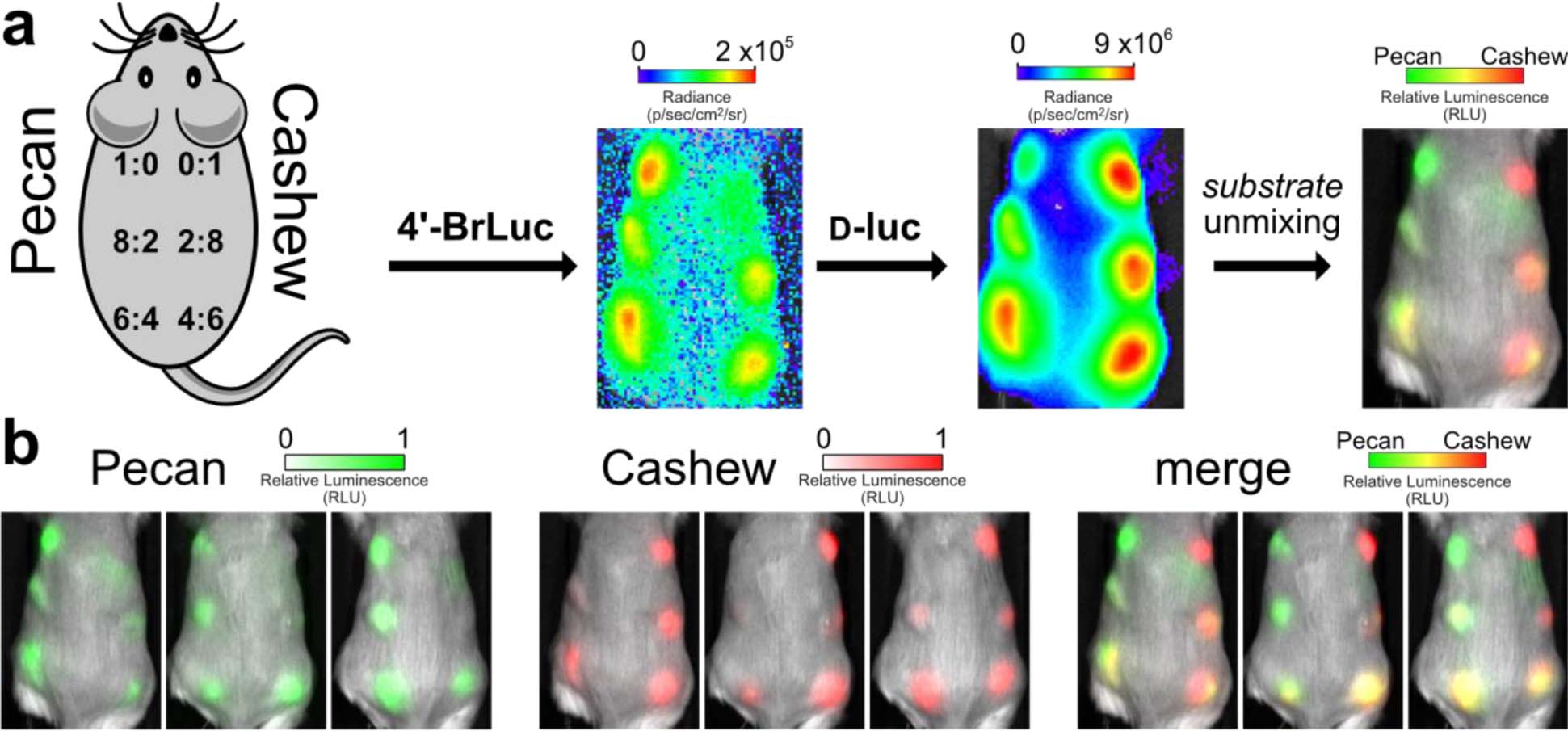
Rapid BLI in vivo. (a) Ratios of Cashew-and Pecan-expressing cells implanted in mice. Orthogonal substrates (65 mM) were administered sequentially via i.p. injection (100 μL). Images were acquired 35 min after each injection. (b) Unmixed channels for each mouse replicate are shown. Color bars indicate normalized luminescence values.
Quantifying the number of cells (or other features) from bioluminescent images is possible, but requires multiple standard curves and external validation. The relevant standard curves are readily generated for in vitro and in cellulo experiments (e.g., see Figures 3 and 5, where bioluminescent outputs correlated with cell number and/or enzyme amount over a broad range.) Such curves are rarely generated for in vivo experiments, though, owing to the complexities involved in standardizing optical signals within heterogeneous tissue.56–57 Thus, applications of bioluminescence imaging in vivo typically involve monitoring relative changes in samples over time. The important takeaway is that the substrate unmixing platform can be immediately employed in conventional applications of bioluminescent probes, where detecting changes over time is often most important.
Generalization of Rapid BLI to Three Probes and Monitoring Cell Function
Having demonstrated the ability to rapidly deconvolute mixtures of two bioluminescent probes, we examined whether the algorithm could be applied to larger collections of luciferase reporters. Triple-component bioluminescence imaging has been historically difficult to achieve, owing to a lack of protocols and methods to distinguish the probes.31,58–59 In one of the only in vivo examples reported, multiple days were required to generate a composite image.35 Signal from one substrate had to clear before another could be administered. We hypothesized that the “layering in” approach could dramatically speed the imaging time, enabling more early events relevant to cell growth to be visualized.
As a model triple component set, we used Cashew and Pecan in combination with Antares, a recently reported marine luciferase variant.60–61 Cashew and Pecan derive from the insect luciferase family, and are thus immediately orthogonal to luciferases (like Antares) that use vastly distinct luciferins (in this case, furimazine).10 Antares also exhibits markedly faster substrate turnover than Cashew, rendering it brighter and intensity resolved from the other two reporters.36 We thus reasoned that the three orthogonal luciferases could be rapidly differentiated by first applying 4’-BrLuc, followed by d-luc, then furimazine to layer in signal from Pecan, Cashew, and Antares, respectively. When the cells were combined and imaged together, the three reporters could be rapidly visualized (<15 min) following sequential substrate addition (Figure 7). Triplet imaging was also readily achieved using other combinations of engineered and native luciferases (Supplementary Figures 7–8).
Figure 7.
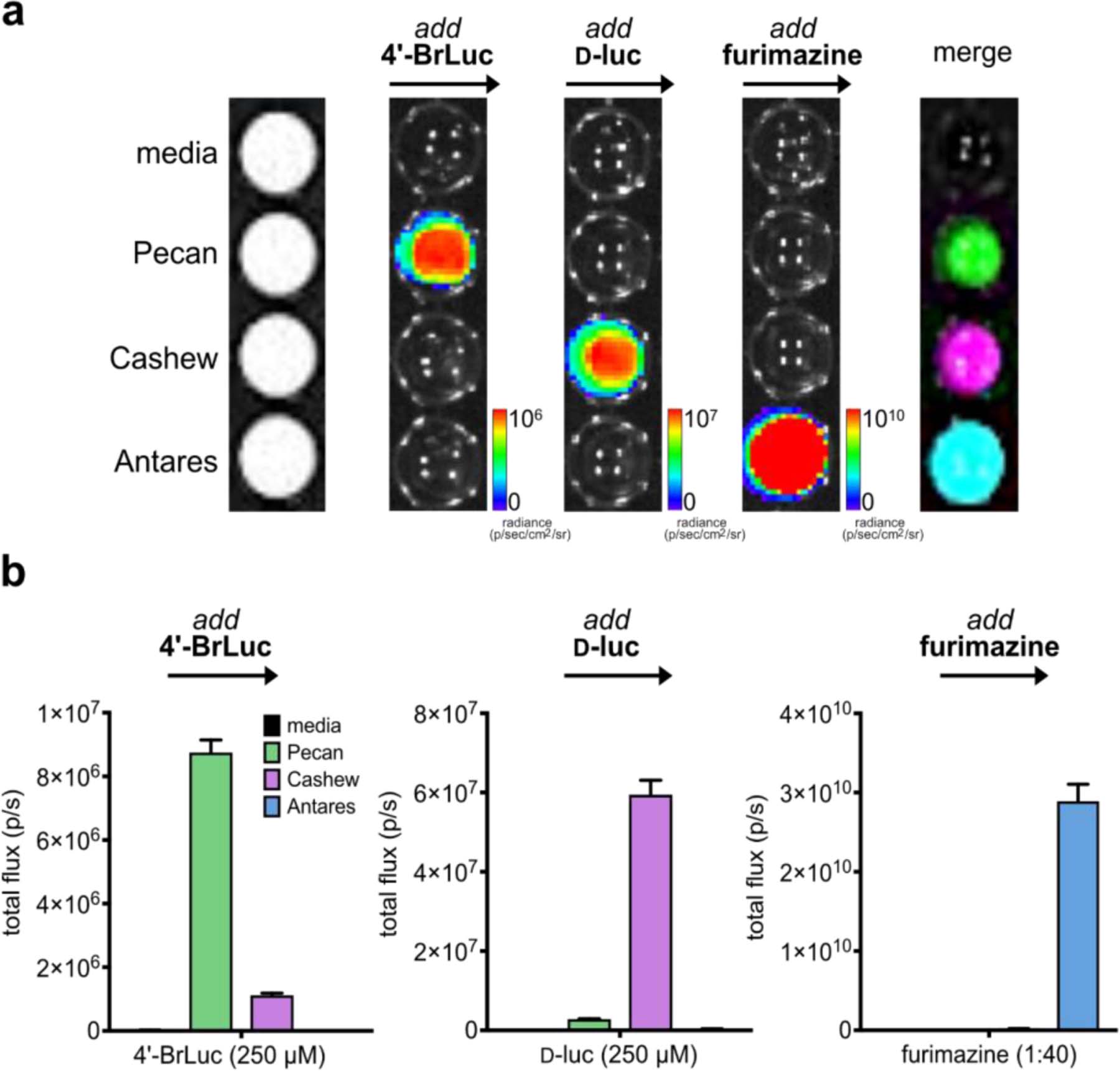
Rapid BLI with three luciferases and luciferins. (a) Cells expressing Pecan, Cashew, Antares, or no luciferase (control) were plated in a 96-well plate. Sequential substrate administration (4’-BrLuc, followed by d-luc, then furimazine) and unmixing enabled three-component imaging. Data are representative of n = 3 replicates. (b) Quantified photon outputs for the images in (a). Error bars represent the standard error of the mean for n = 3 experiments.
CONCLUSIONS AND FUTURE DIRECTIONS
In conclusion, we developed a strategy for rapid multicomponent bioluminescence imaging based on substrate unmixing. This method takes advantage of both substrate and intensity resolution to resolve mixtures of reporters. In this scenario, probe differentiation is less impacted by tissue location, a parameter that has historically hindered efforts to resolve colors in large organisms. We validated the unmixing approach in bacterial lysate, live cells, and mouse models. A variety of bioluminescent reporters were readily resolved, and bioluminescent outputs correlated with cell number and/or enzyme amount over a broad range of concentrations in vitro. The unmixing algorithm further enabled multiple luciferases to be discriminated in vivo using conventional BLI instrumentation.
While the rapid unmixing approach is immediately applicable to experiments with widely available bioluminescent probes, additional questions remain to be addressed. For example, just how much intensity resolution is required for successful unmixing remains unknown. The majority of probe sets tested exhibit >10-fold selectivity between matched enzymes and substrates. It is likely that even less selective probes can be integrated into the approach, though, and expand the number of features that can be imaged in a single setting. “Crosstalk” between more enzyme-substrate pairs could potentially improve the unmixing process, as the molecular signatures become more diagnostic. We also have not yet established the dynamic imaging range that can be achieved with different combinations of reporters. For example, it might not be possible to “see” a particular reporter if it is in low abundance compared to another reporter. The kinetics of light emission and compound transport into tissues must also be fully examined. These parameters could offer additional mechanisms by which to discriminate orthogonal pairs.36,62
The limits of detection will be influenced not only by the inherent photon output (and thus sensitivity) of each luciferase, but also the quantity of each reporter present and its tissue location. The data presented in this manuscript demonstrate that ten-fold changes in relative abundance can be easily discerned. Further theoretical work and experimental data are necessary to provide more definitive thresholds for different tissue types. Additional work is also necessary to streamline the quantification of luciferase reporters in biological samples. We have shown that the substrate unmixing platform can easily monitor relative changes in luciferase levels. Tracking such information is standard practice, as traditional bioluminescence applications rely simply on detecting changes in signal over time. Identifying the absolute quantities of the reporters in unknown mixtures requires external calibration curves. Whether or not such curves must be generated for every independent experiment remains to be determined.
We anticipate that the rapid imaging approach reported here will enable a range of applications, including monitoring multiple cell types and gene expression profiles in vivo. The development of additional intensiometric probes will also increase the number of bioluminescent reporters that can be rapidly imaged in tandem, and work along these lines is ongoing. An expanded toolkit for BLI will enable longstanding questions regarding multicellular interactions to be addressed.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the U.S. National Institutes of Health (R01 GM107630 to J. Prescher). W. Porterfield was supported by the National Science Foundation via the BEST IGERT program (DGE-1144901) and an Allergan Graduate Fellowship. K. Jones. was supported by an institutional Chemical and Structural Biology Training Grant predoctoral fellowship (T32-GM10856). Z. Yao and C. Rathbun were supported by National Science Foundation Graduate Research Fellowships. We thank S. Kasturi for assistance with docking analyses and members of the Prescher laboratory for helpful discussions.
Funding Sources
No competing financial interests have been declared.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Experimental details, spectroscopic data for new compounds, and additional images.
REFERENCES
- 1.Paley MA; Prescher JA Bioluminescence: a versatile technique for imaging cellular and molecular features. MedChemComm 2014, 5, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mezzanotte L; van ‘t Root M; Karatas H; Goun EA; Löwik CWGM In Vivo Molecular Bioluminescence Imaging: New Tools and Applications. Trends Biotechnol. 2017, 35, 640–652. [DOI] [PubMed] [Google Scholar]
- 3.Prescher JA; Contag CH Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol 2010, 14, 80–89. [DOI] [PubMed] [Google Scholar]
- 4.Contag CH; Bachmann MH Advances in In Vivo Bioluminescence Imaging of Gene Expression. Annu. Rev. Biomed. Eng 2002, 4, 235–260. [DOI] [PubMed] [Google Scholar]
- 5.Thorne N; Inglese J; Auld DS Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol 2010, 17, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabinovich BA; Ye Y; Etto T; Chen JQ; Levitsky HI; Overwijk WW; Cooper LJN; Gelovani J; Hwu P Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 14342–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Z; Zhang BS; Prescher JA Advances in bioluminescence imaging: new probes from old recipes. Curr. Opin. Chem. Biol 2018, 45, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathbun CM; Prescher JA Bioluminescent probes for imaging biology beyond the culture dish. Biochemistry 2017, 56, 5178–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller SC; Mofford DM; Adams ST Lessons learned from luminous luciferins and latent luciferases. ACS Chem. Biol 2018, 13, 1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh H-W; Ai H-W Development and applications of bioluminescent and chemiluminescent reporters and biosensors. Annu. Rev. Anal. Chem 2019, 12, 129–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh H-W; Xiong Y; Wu T; Chen M; Ji A; Li X; Ai H-W ATP-Independent Bioluminescent Reporter Variants To Improve in Vivo Imaging. ACS Chem. Biol 2019, 14, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall MP; Woodroofe CC; Wood MG; Que I; van’t Root M; Ridwan Y; Shi C; Kirkland TA; Encell LP; Wood KV; Löwik C; Mezzanotte L Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun 2018, 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaub FX; Reza MS; Flaveny CA; Li W; Musicant AM; Hoxha S; Guo M; Cleveland JL; Amelio AL Fluorophore-NanoLuc BRET Reporters Enable Sensitive In Vivo Optical Imaging and Flow Cytometry for Monitoring Tumorigenesis. Cancer Research 2015, 75, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwano S; Sugiyama M; Hama H; Watakabe A; Hasegawa N; Kuchimaru T; Tanaka KZ; Takahashi M; Ishida Y; Hata J; Shimozono S; Namiki K; Fukano T; Kiyama M; Okano H; Kizaka-Kondoh S; McHugh TJ; Yamamori T; Hioki H; Maki S; Miyawaki A Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 2018, 359, 935–939. [DOI] [PubMed] [Google Scholar]
- 15.Williams SJ; Prescher JA Building biological flashlights: Orthogonal luciferases and luciferins for in vivo imaging. Acc. Chem. Res 2019, 52, 3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zambito G; Hall MP; Wood MG; Gaspar N; Ridwan Y; Stellari FF; Shi C; Kirkland TA; Encell LP; Löwik C; Mezzanotte L ReD-shifted click beetle luciferase mutant expands the multicolor bioluminescent palette for deep tissue imaging. iScience 2021, 24, 101986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stowe CL; Burley TA; Allan H; Vinci M; Kramer-Marek G; Ciobota DM; Parkinson GN; Southworth TL; Agliardi G; Hotblack A; Lythgoe MF; Branchini BR; Kalber TL; Anderson JC; Pule MA Near-infrared dual bioluminescence imaging in mouse models of cancer using infraluciferin. eLife 2019, 8, e45801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markus A; Stefanie V; Cordula S; Amit J; Martin P; Mathias H Quantitative in vivo dual-color bioluminescence imaging in the mouse brain. Neurophotonics 2019, 6, 025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branchini BR; Ablamsky DM; Rosenman JM; Uzasci L; Southworth TL; Zimmer M Synergistic mutations produce blue-shifted bioluminescence in firefly luciferase. Biochemistry 2007, 46, 13847–13855. [DOI] [PubMed] [Google Scholar]
- 20.Mezzanotte L; Que I; Kaijzel E; Branchini B; Roda A; Löwik C Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS ONE 2011, 6, e19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinovink JW; Mezzanotte L; Zambito G; Fransen MF; Cruz LJ; Verbeek JS; Chan A; Ossendorp F; Löwik C A Dual-Color Bioluminescence Reporter Mouse for Simultaneous in vivo Imaging of T Cell Localization and Function. Front. Immunol 2019, 9, 3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarrion-Perdigones A; Chang L; Gonzalez Y; Gallego-Flores T; Young DW; Venken KJT Examining multiple cellular pathways at once using multiplex hextuple luciferase assaying. Nat. Commun 2019, 10, 5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaumik S; Lewis XZ; Gambhir SS Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. J. Biomed. Opt 2004, 9, 578–586. [DOI] [PubMed] [Google Scholar]
- 24.Fan F; Wood KV Bioluminescent assays for high-throughput screening. ASSAY Drug Dev. Techn 2007, 5, 127–136. [DOI] [PubMed] [Google Scholar]
- 25.Su Y; Walker JR; Park Y; Smith TP; Liu LX; Hall MP; Labanieh L; Hurst R; Wang DC; Encell LP; Kim N; Zhang F; Kay MA; Casey KM; Majzner RG; Cochran JR; Mackall CL; Kirkland TA; Lin MZ Novel NanoLuc substrates enable bright two-population bioluminescence imaging in animals. Nat. Methods 2020, 17, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross S; Abraham U; Prior JL; Herzog ED; Piwnica-Worms D Continuous delivery of D-luciferin by implanted micro-osmotic pumps enables true real-time bioluminescence imaging of luciferase activity in vivo. Mol Imaging 2007, 6, 121–130. [PubMed] [Google Scholar]
- 27.Kaskova ZM; Tsarkova AS; Yampolsky IV 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev 2016, 45, 6048–6077. [DOI] [PubMed] [Google Scholar]
- 28.Kotlobay AA; Sarkisyan KS; Mokrushina YA; Marcet-Houben M; Serebrovskaya EO; Markina NM; Somermeyer LG; Gorokhovatsky AY; Vvedensky A; Purtov KV; Petushkov VN; Rodionova NS; Chepurnyh TV; Fakhranurova LI; Guglya EB; Ziganshin R; Tsarkova AS; Kaskova ZM; Shender V; Abakumov M; Abakumova TO; Povolotskaya IS; Eroshkin FM; Zaraisky AG; Mishin AS; Dolgov SV; Mitiouchkina TY; Kopantzev EP; Waldenmaier HE; Oliveira AG; Oba Y; Barsova E; Bogdanova EA; Gabaldón T; Stevani CV; Lukyanov S; Smirnov IV; Gitelson JI; Kondrashov FA; Yampolsky IV Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 12728–12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitiouchkina T; Mishin AS; Somermeyer LG; Markina NM; Chepurnyh TV; Guglya EB; Karataeva TA; Palkina KA; Shakhova ES; Fakhranurova LI; Chekova SV; Tsarkova AS; Golubev YV; Negrebetsky VV; Dolgushin SA; Shalaev PV; Shlykov D; Melnik OA; Shipunova VO; Deyev SM; Bubyrev AI; Pushin AS; Choob VV; Dolgov SV; Kondrashov FA; Yampolsky IV; Sarkisyan KS Plants with genetically encoded autoluminescence. Nat. Biotechnol 2020, 38, 944–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones KA; Porterfield WB; Rathbun CM; McCutcheon DC; Paley MA; Prescher JA Orthogonal luciferase–luciferin pairs for bioluminescence imaging. J. Am. Chem. Soc 2017, 139, 2351–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathbun CM; Porterfield WB; Jones KA; Sagoe MJ; Reyes MR; Hua CT; Prescher JA Parallel screening for rapid identification of orthogonal bioluminescent tools. ACS Cent. Sci 2017, 3, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mofford DM; Reddy GR; Miller SC Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over d-luciferin. J. Am. Chem. Soc 2014, 136, 13277–13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SB; Nishihara R; Citterio D; Suzuki K Fabrication of a new lineage of artificial luciferases from natural luciferase pools. ACS Comb. Sci 2017, 19, 594–599. [DOI] [PubMed] [Google Scholar]
- 34.Yeh H-W; Karmach O; Ji A; Carter D; Martins-Green MM; Ai H.-w. ReD-shifted luciferase–luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 2017, 14, 971–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguire CA; Bovenberg MS; Crommentuijn MH; Niers JM; Kerami M; Teng J; Sena-Esteves M; Badr CE; Tannous BA Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol Ther Nucleic Acids 2013, 2, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stacer AC; Nyati S; Moudgil P; Iyengar R; Luker KE; Rehemtulla A; Luker GD NanoLuc Reporter for Dual Luciferase Imaging in Living Animals. Mol Imaging 2013, 12, 457–469. [PMC free article] [PubMed] [Google Scholar]
- 37.Shah K; Tang Y; Breakefield X; Weissleder R Real-time imaging of TRAIL-induced apoptosis of glioma tumors in vivo. Oncogene 2003, 22, 6865–6872. [DOI] [PubMed] [Google Scholar]
- 38.Masuho I; Ostrovskaya O; Kramer GM; Jones CD; Xie K; Martemyanov KA Distinct profiles of functional discrimination among G proteins determine the actions of G protein–coupled receptors. Science Signaling 2015, 8, ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartfield RM; Schwarz KA; Muldoon JJ; Bagheri N; Leonard JN Multiplexing Engineered Receptors for Multiparametric Evaluation of Environmental Ligands. ACS Synth. Biol 2017, 6, 2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaussabel D; Baldwin N Democratizing systems immunology with modular transcriptional repertoire analyses. Nat. Rev. Immunol 2014, 14, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gammon ST; Leevy WM; Gross S; Gokel GW; Piwnica-Worms D Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal. Chem 2006, 78, 1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eng C-HL; Lawson M; Zhu Q; Dries R; Koulena N; Takei Y; Yun J; Cronin C; Karp C; Yuan G-C; Cai L Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 2019, 568, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo S-M; Veneziano R; Gordonov S; Li L; Danielson E; de Arce KP; Park D; Kulesa AB; Wamhoff E-C; Blainey PC; Boyden ES; Cottrell JR; Bathe M Multiplexed and high-throughput neuronal fluorescence imaging with diffusible probes. Nat. Commun 2019, 10, 4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinhardt RC; Rathbun CM; Krull BT; Yu JM; Yang Y; Nguyen BD; Kwon J; McCutcheon DC; Jones KA; Furche F; Prescher JA Brominated Luciferins Are Versatile Bioluminescent Probes. ChemBioChem 2017, 18, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCutcheon DC; Paley MA; Steinhardt RC; Prescher JA Expedient synthesis of electronically modified luciferins for bioluminescence imaging. J. Am. Chem. Soc 2012, 134, 7604–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCutcheon DC; Porterfield WB; Prescher JA Rapid and scalable assembly of firefly luciferase substrates. Org. Biomol. Chem 2015, 13, 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu MD; Warner EA; Morrissey CE; Fick CW; Wu TS; Ornelas MY; Ochoa GV; Zhang BS; Rathbun CM; Porterfield WB; Prescher JA; Leconte AM Statistical coupling analysis-guided library design for the discovery of mutant luciferases. Biochemistry 2018, 57, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams ST; Mofford DM; Reddy GSKK; Miller SC Firefly luciferase mutants allow substrate-selective bioluminescence imaging in the Mouse Brain. Angew. Chem. Int. Ed 2016, 55, 4943–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang BS; Jones KA; McCutcheon DC; Prescher JA Pyridone luciferins and mutant luciferases for bioluminescence imaging. ChemBioChem 2018, 19, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shachaf CM; Kopelman AM; Arvanitis C; Karlsson Å; Beer S; Mandl S; Bachmann MH; Borowsky AD; Ruebner B; Cardiff RD; Yang Q; Bishop JM; Contag CH; Felsher DW MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004, 431, 1112–1117. [DOI] [PubMed] [Google Scholar]
- 51.Minn AJ; Kang Y; Serganova I; Gupta GP; Giri DD; Doubrovin M; Ponomarev V; Gerald WL; Blasberg R; Massagué J Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest 2005, 115, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luker GD; Bardill JP; Prior JL; Pica CM; Piwnica-Worms D; Leib DA Noninvasive Bioluminescence Imaging of Herpes Simplex Virus Type 1 Infection and Therapy in Living Mice. J. Virol 2002, 76, 12149–12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdelfattah AS; Kawashima T; Singh A; Novak O; Liu H; Shuai Y; Huang Y-C; Campagnola L; Seeman SC; Yu J; Zheng J; Grimm JB; Patel R; Friedrich J; Mensh BD; Paninski L; Macklin JJ; Murphy GJ; Podgorski K; Lin B-J; Chen T-W; Turner GC; Liu Z; Koyama M; Svoboda K; Ahrens MB; Lavis LD; Schreiter ER Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 2019, 365, 699–704. [DOI] [PubMed] [Google Scholar]
- 54.Sanford L; Palmer A Recent Advances in Development of Genetically Encoded Fluorescent Sensors. Methods Enzymol. 2017, 589, 1–49. [DOI] [PubMed] [Google Scholar]
- 55.Chen T-W; Wardill TJ; Sun Y; Pulver SR; Renninger SL; Baohan A; Schreiter ER; Kerr RA; Orger MB; Jayaraman V; Looger LL; Svoboda K; Kim DS Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaincy K; Olivier C; Tamara LT; Heng X; Bradley WR Three-dimensional reconstruction of in vivo bioluminescent sources based on multispectral imaging. J. Biomed. Opt 2007, 12, 1–12. [DOI] [PubMed] [Google Scholar]
- 57.Doyle TC; Burns SM; Contag CH Technoreview: In vivo bioluminescence imaging for integrated studies of infection. Cell. Microbiol 2004, 6, 303–317. [DOI] [PubMed] [Google Scholar]
- 58.Yao Z; Zhang BS; Steinhardt RC; Mills JH; Prescher JA Multicomponent Bioluminescence Imaging with a π-Extended Luciferin. J. Am. Chem. Soc 2020, 142, 14080–14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tannous BA; Kim D-E; Fernandez JL; Weissleder R; Breakefield XO Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther 2005, 11, 435–443. [DOI] [PubMed] [Google Scholar]
- 60.Chu J; Oh Y; Sens A; Ataie N; Dana H; Macklin JJ; Laviv T; Welf ES; Dean KM; Zhang F; Kim BB; Tang CT; Hu M; Baird MA; Davidson MW; Kay MA; Fiolka R; Yasuda R; Kim DS; Ng H-L; Lin MZ A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol 2016, 34, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh Y; Park Y; Cho JH; Wu H; Paulk NK; Liu LX; Kim N; Kay MA; Wu JC; Lin MZ An orange calcium-modulated bioluminescent indicator for non-invasive activity imaging. Nat. Chem. Biol 2019, 15, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeh H-W, Wu T, Chen M, and Ai H-W (2019) Identification of factors complicating bioluminescence imaging, Biochemistry 58, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


