Abstract
Nucleic acids have not been widely considered as an optimal material for drug delivery. Indeed, unmodified nucleic acids are enzymatically unstable, too hydrophilic for cell uptake and payload encapsulation, and may cause unintended biological responses such as immune system activation and prolongation of the blood coagulation pathway. Recently, however, three major areas of development surrounding nucleic acids have made it worthwhile to reconsider their role for drug delivery. These areas include DNA/RNA nanotechnology, multivalent nucleic acid nanostructures, and nucleic acid aptamers, which, respectively, provide the ability to engineer nanostructures with unparalleled levels of structural control, completely reverse certain biological properties of linear/cyclic nucleic acids, and enable antibody-level targeting using an all-nucleic acid construct. These advances, together with nucleic acids’ ability to respond to various stimuli (engineered or natural), have led to a rapidly increasing number of drug delivery systems with potential for spatiotemporally controlled drug release. In this review, we discuss recent progress in nucleic acid-based drug delivery strategies, their potential, unique use cases, and risks that must be overcome or avoided.
Introduction
Since the conceptualization of the “magic bullet”, i.e. therapeutic agents that cure diseases without harming the body itself, the delivery of the therapeutic to the target tissue has been recognized as a major means to improve the therapeutic windows and ultimately the health quality and lifespan of the patient [1,2]. A drug delivery system (DDS) alters the intrinsic physiochemical and biological identity of the drug, and can lead to entirely different pharmacokinetic and biodistribution profiles of the loaded cargo [3,4]. An ideal DDS should be able to bind with drugs with tunable loading and remain stable before reaching the target of interest, where a spatiotemporally controlled process unloads the therapeutic [5,6]. Meanwhile, the DDS itself should exhibit low toxicity and non-immunogenicity, and lack long-term adverse effects on the human body [7]. To date, a variety of materials spanning synthetic polymers, lipids, inorganic nanoparticles, engineered microparticles, hydrogels, biomacromolecules, and live/deactivated microorganisms have been explored as the major component for a DDS [8–11].
Nucleic acid, a highly hydrophilic and negatively charged natural biopolymer, has been relatively unnoticed as a material for DDS. Instead, nucleic acids are consistently regarded as a troublesome therapeutic cargo, requiring an advanced DDS to facilitate their delivery. Indeed, unmodified nucleic acids are hopelessly incapable of entering cells and are subject to rapid nuclease cleavage and renal/hepatic clearance [12]. Typically, a particular intracellular localization (e.g. cytosol or nucleus) is often required prior to any mechanism of action, be it gene expression knockdown, mRNA splicing alteration, transcriptional and epigenetic regulation, and genome editing [12]. In addition, certain nucleic acid motifs can elicit a strong activation of the innate immune system even at low concentrations, e.g. certain RNA sequences (e.g. 5’-UGUGU-3’) and DNA sequences containing unmethylated cytosine-phosphate-guanosine (CpG) motifs [13,14]. In fact, these motifs are being explored as potent vaccine adjuvants [15,16]. Given these limitations, nucleic acids in the past have been mainly developed as drugs for rare diseases originating from the liver [17], or in tissues that can be treated by local injection, such as the eye or the spinal cord [18].
With the notion that efficient delivery of nucleic acids necessitates a DDS being firmly established by an overwhelming number of research articles, the idea of using nucleic acids themselves as a DDS had been reduced to the sideline. Interestingly, as research on DNA nanotechnology and other nucleic acid structures thrived in the past decade, new capabilities and unusual physiochemical/biological properties of nucleic acid structures have emerged, which are driving a fresh round of interest toward utilizing nucleic acids as an alternative DDS for certain use cases. This review focuses on the design criteria and application of nucleic acid-based DDSs with an emphasis on their unique benefits and certain limitations. To narrow the scope, only structures that consist mainly or entirely of nucleic acids with no additional carriers are discussed. A variety of payloads are discussed in this review, which include small molecule drugs, biologics, and model drugs such as nanoparticles and fluorescent dyes. With the recent surge of nucleic acid-based DDSs that are able to tackle difficult challenges such as in vivo delivery and tissue-specific activation of protein biologics, it should be safe to assert that nucleic acids are a highly useful class of DDS material, and their properties, potential, and limitations should offer room for much deeper explorations for years to come.
DNA Nanotechnology – From Interesting Structures to Versatile Drug Carriers
The precise nature of the Watson-Crick base pairing opens the possibility to build complex nanostructures by design. Mechanistically, DNA nanostructures are mainly achieved by tile-based assembly, scaffolded DNA origami, and DNA wireframes. The tile-based strategy derives from the immobile Holliday junction originally designed by the Seeman group in 1982 [19], which consists of two DNA duplexes with a crossover (Figure 1). By increasing the number of crossovers to include double [20], triple [21], and paranemic crossovers [22,23], more rigid tiles have been generated and further assembled into 1D through 3D structures [24–29] (Figure 1b,c). DNA origami was first demonstrated by Rothemund in 2006 [30], in which a long single-stranded genomic DNA (scaffold) was folded into different 2D shapes with the assistance of multiple short DNA sequences (staples). By stacking multiple origamis together, different 3D structures (e.g. mololith, square nut, railed bridge, slotted cross) can be formed with near arbitrary control of the morphology [31,32]. A structure of particular interest to the DDS community involves 3D hollow cages enabled by welding the edges of 2D sheets, which can be used to encapsulate macromolecules (Figure 1e) [33,34]. In terms of particle size, tile-based structures are generally smaller (tens of nm) compared with the origami (hundreds of nm). Yin and coworkers recently combined the tile- and scaffold-based approaches by employing single-stranded tiles (SSTs) as building blocks (Figure 1f), which broke the size limit in the traditional origami method posed by the total length of the scaffold DNA [35,36]. A third DNA assembly approach involves DNA wireframe structures, which are assembled from a small number of short DNA strands (Figure 1g,h). Using this strategy, complex structures, such as tetrahedra, prismatic cages, and nanotubes can be prepared with high yields. [37–40] With tremendous structural advances, DNA nanostructures are being increasingly considered for applications such as drug delivery. DNA nanotechnology offers several unique advantages, such as well-defined particle architecture, internal structure, size, composition, stimuli-responsiveness, and lack of intrinsic toxicity.
Figure 1.
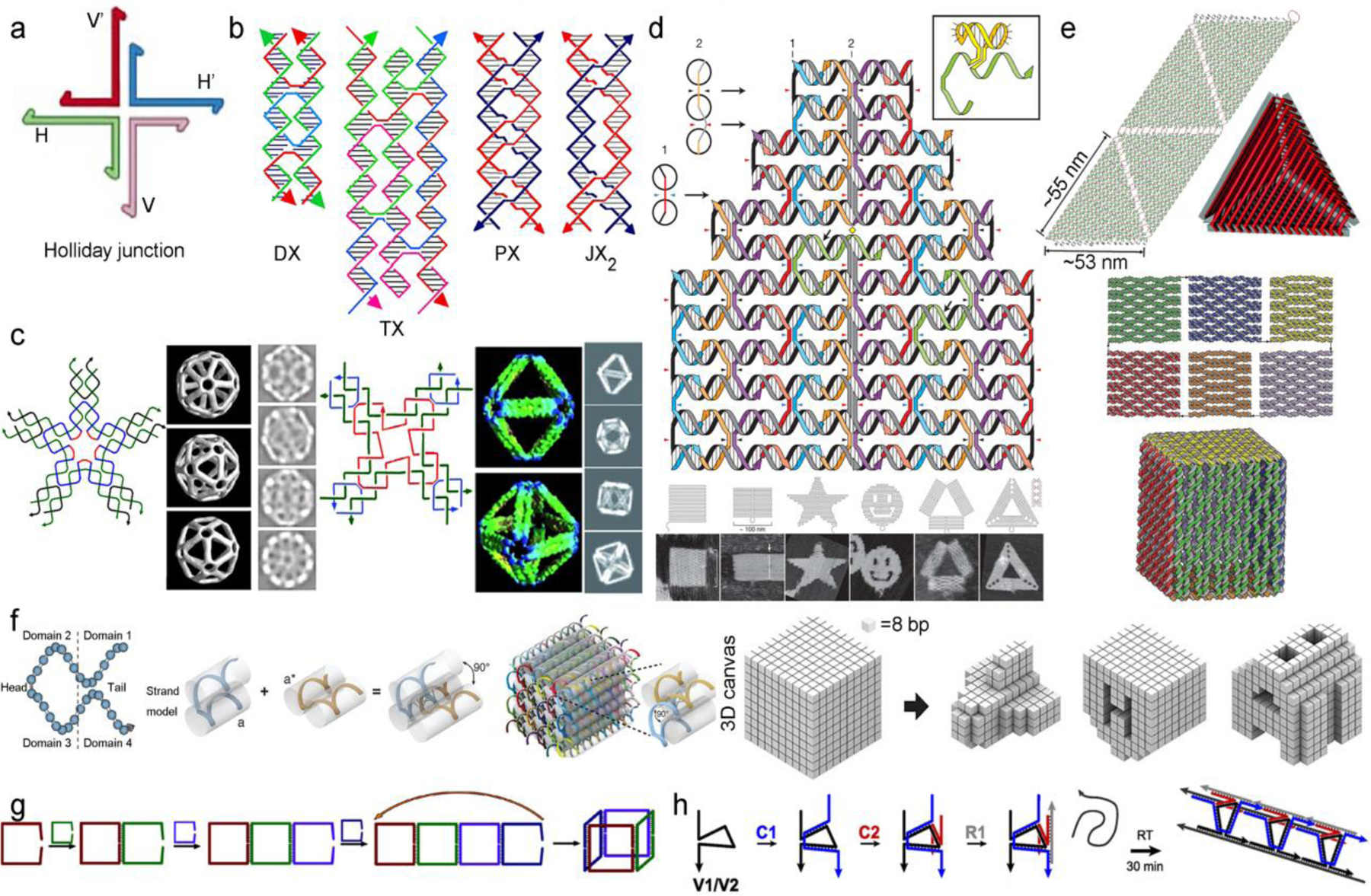
“Bottom-up” and “top-down” design of DNA nanostructures. (a) Holliday junction in DNA nanotechnology. Reproduced with permission from ref 19. Copyright 1982 Elsevier. (b) Different types of crossovers. Reprinted with permission from ref 23. Copyright 2019 American Chemical Society. (c) DNA octahedron and icosahedron assembled from four- and five-point star tiles. Reproduced with permissions from refs 28 and 29. Copyright 2008 National Academy of Sciences and 2010 Wiley-VCH. (d) DNA origami and examples of assembled 2D shapes. Reproduced with permission from ref 30. Copyright 2006 Nature Publishing Group. (e) 3D Hollow tetrahedron and cube, assembled by welding of 2D sheets. Reproduced with permissions from refs 33 and 34. Copyright 2009 Nature Publishing Group and 2009 American Chemical Society. (f) single-stranded tile (SST) design and assembled 3D structures. Reproduced with permission from ref 35. Copyright 2012 American Association for the Advancement of Science. DNA wireframe structures: cubes (g) and nanotubes (h). Reprinted with permission from ref 37 and 38. Copyright 2012, 2013 American Chemical Society.
While nucleic acids generally are safe, it should be noted that the component sequences must be carefully considered in order to minimize unwanted activation of the immune system. Among the mechanisms responsible for inducing immune activity are the toll-like receptors (TLR), which recognize pathogen associated molecular patterns (PAMPs, e.g. lipopolysaccharide, flagellin, or unmethylated CpG-DNA). Viral nucleic acids, such as the genomic phage DNA M13mp18 (often used in DNA origamis), can be recognized by multiple TLRs, notably TLR9 (recognizes CpG-DNA) [41], TLR7 and 8 (recognize ssRNA) [42,43], and TLR3 (recognizes dsRNA) [44]. These nucleic acid-sensing TLRs have the potential to promote the production of type I interferon, among other cytokines. While a limited level of the phage DNA/RNA can persistently stimulate low levels of immune responses without causing any overt symptoms, when used as a DDS, it is conceivable that the concentration of the PAMPs associated with the virome/staples can be too high to ignore. Given that a significant amount of cancer models adopt immunodeficient experimental animals, the need for an analysis of immune system activation is heightened for DNA-based DDSs.
DNA nanostructures are typically sufficiently large to avoid renal clearance (provided enzymatic degradation takes longer compared with the desired pharmacokinetics in plasma) and can engage with enhanced permeability and retention (EPR) effect associated with malignant tumor growth [45]. It is important to note that the EPR effect is often amplified in mice models vs. human, and may vary among different tumor models [46]. One example involves the delivery of the small anticancer drug, doxorubicin (Dox). Dox can intercalate with GC-rich regions of duplex DNA and block the activity of topo isomerase 2, which reduces the growth of tumor cells [47,48]. The intercalation between Dox and DNA duplex is reversible especially under low pH conditions, making DNA nanostructures a unique carrier for this small molecule drug. Ding et al adopted several DNA origamis constructed from the phage DNA (M13mp18) as hosts for Dox [49], including a triangular and a tubular origami (Fig 2a). Both origamis showed enhanced cytotoxicity against a drug-resistant human breast adenocarcinoma cancer cell (MCF 7) as well as improved cellular uptake. The authors also investigated the origami in vivo for the co-delivery of Dox and the p53 tumor suppressor gene [50,51]. The resulting Dox-loaded structures exhibited pronounced antitumor activity in MCF-7R xenografts but little systemic toxicity and immune stimulation in BALB/c mice. Interestingly, a shape effect was also observed: the triangular DNA origami exhibited higher tumor localization and antitumor activity compared with tubular and square shapes, which underline the potential to use shape and size to influence biological responses. However, the mechanism of the shape effect must first be well-understood, and care must be taken in the interpretation of florescence studies from DNA nanostructures. For example, the fluorescent signal may arise from degradation products of the DNA, and the rate of which can conceivably be shape-dependent [52]. In the Ding design, the release of Dox was triggered by the acidic environment in the tumor region. Högberg et al explored the possibility of tuning Dox release kinetics by adjusting the degrees of global twist in the DNA double-helix structure, which allows for different levels of relaxation in the DNA double helix. A twisted nanotube design (12 bp/turn) showed 33% more Dox loading and exhibited an extended period of Dox release than conventional straight design (10.5 bp/turn) in vitro (Fig 2b) [53].
Figure 2.
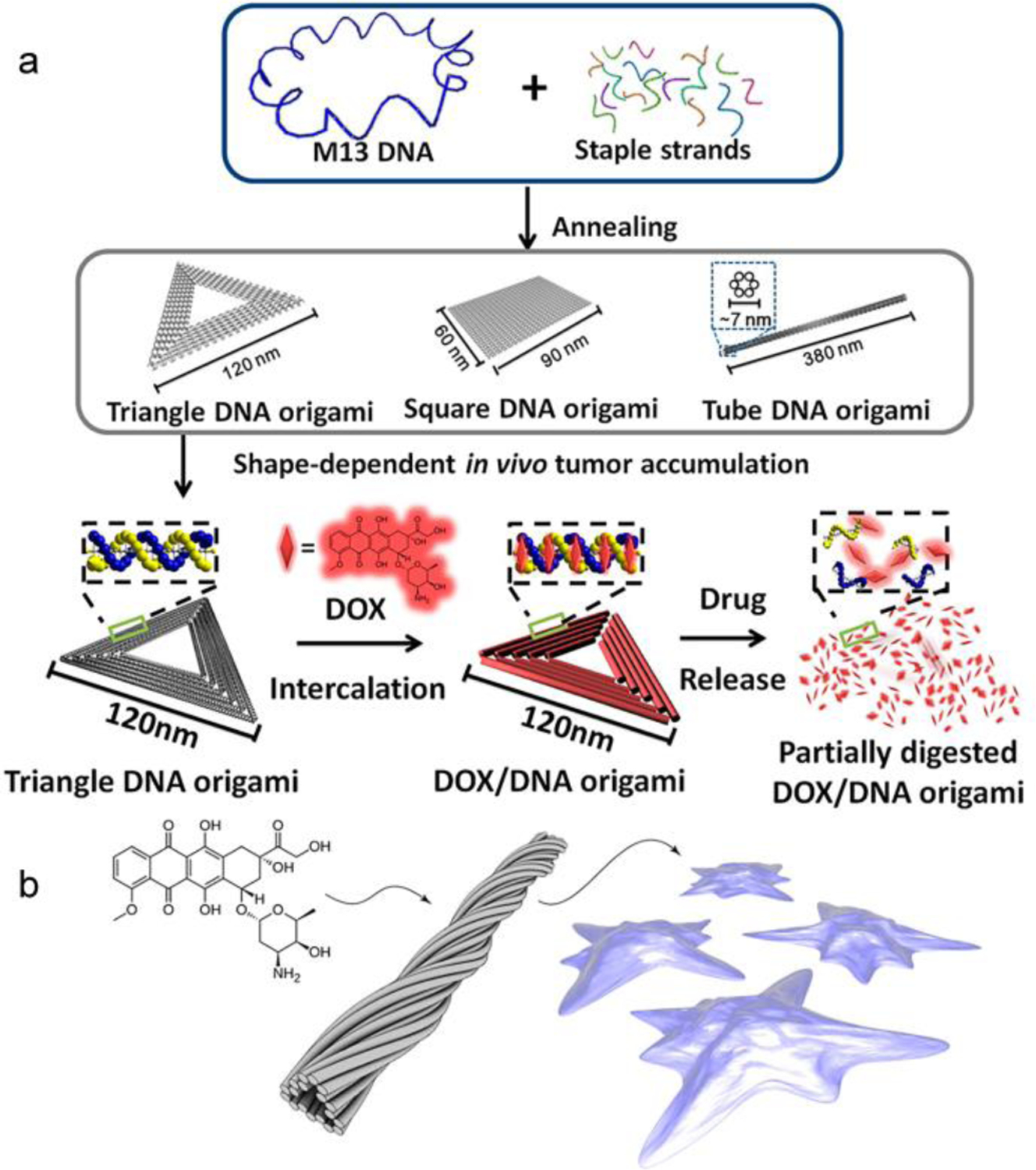
Delivery of small molecule drugs with DNA nanostructures. (a) Intercalation of Dox into various DNA nanostructures and subsequent release. Reproduced with permission from ref 50. Copyright 2014 American Chemical Society. (b). Loading of Dox into a twisted tubular DNA nanostructure. Reproduced with permission from ref 53. Copyright 2012 American Chemical Society.
In addition to small molecule drugs, therapeutic oligonucleotides are another type of payload that have been formulated with DNA nanostructure-based DDS. In this case, the combined DDS and the therapeutic payload are entirely comprised of nucleic acids, which make them a class of self-delivering nucleic acid nanoparticles [54]. This strategy is particularly attractive because no high ζ-potential or surfactant-like co-carriers, such as polyethyleneimine (PEI) and lipofectamine, are involved, which often enhance the delivery the nucleic acid payload at the cost of unacceptable levels of carrier-induced toxicity, blood incompatibility, immunogenicity, among other adverse effects [55–58]. By hybridization with pre-assembled DNA nanostructures, therapeutic oligonucleotides (antisense, siRNA, miRNA, etc) can be effectively delivered into target cells or tissues. For example, Anderson et al reported a tetrahedron DNA nanostructure for the delivery of an siRNA against the firefly luciferase [59]. Each edge of the tetrahedron contains an overhang that was used to bind with siRNA or cancer-targeting ligands such as folate or peptides, thus allowing as many as six siRNA strands to be delivered per particle (Fig 3a). Gene knockdown results suggest that at least three folates per tetrahedron are required to achieve optimal delivery, and that a proper spatial orientation of the ligands is essential. Similarly, Leong et al have designed a DNA Shuriken-like structure for miRNA delivery [60]. In this design, miRNA strands were hybridized with a pre-assembled DNA star, with each assembly carrying three molecules of the tumor suppressive miRNA-145 (Figure 3b). The DNA nanostructure exhibited improved nuclease stability due to the steric hinderance and elevated levels of cellular uptake. The efficacy of the Shurikens was demonstrated in a 3D spheroid tumor model based on DLD-1 cells, showing significant (~30%) shrinkage of the spheroids after two days of treatment. These studies epitomize the unique advantage of DNA nanostructures as a DDS: an unmatched level of control of the particle size, shape, and number/orientation of ligands presented on its surface. This level of control is highly difficult if not impossible for any other kind of nanoparticles.
Figure 3.
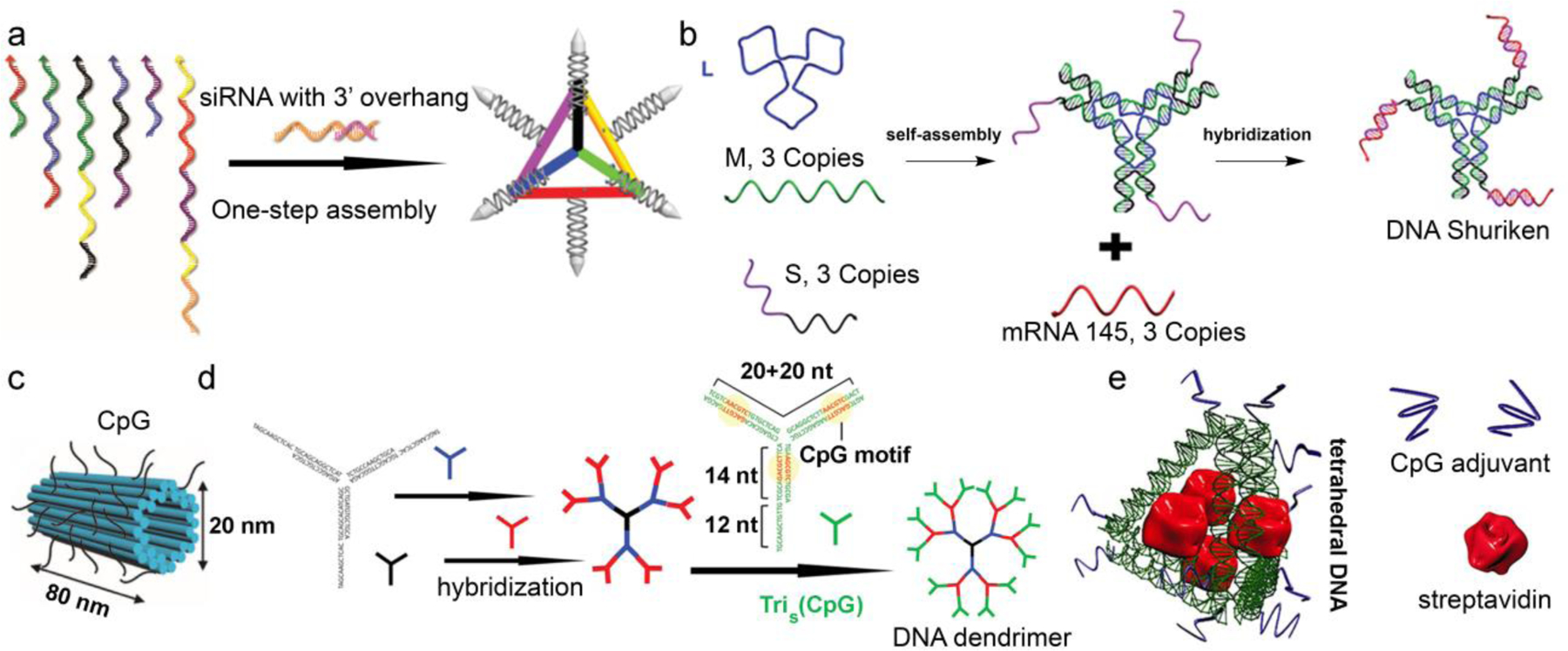
DNA nanostructure-based DDS for the delivery of oligonucleotides. (a) DNA tetrahedron nanoparticles for siRNA delivery. Reproduced with permission from ref 59. Copyright 20012 Nature Publishing Group. (b) Protection and delivery of miRNA via Shuriken-like DNA structures. Reproduced from ref. 60 with permission from the Royal Society of Chemistry. (c) DNA tube- and (d) dendrimer-based delivery of CpG DNA strands. Reproduced with permissions from refs 63 and 64. Copyright 2011 and 2015 American Chemical Society. (e) DNA-scaff olded adjuvant-antigen vaccine complex. Reproduced with permission from ref. 66. Copyright 2012 American Chemical Society.
Another type of therapeutic oligonucleotides involves PAMP motifs that stimulate the immune system, such as unmethylated CpG-rich sequences, which are frequently found in microbial genomes but rarely in vertebrate genomes. Immune system stimulation is a critical component for cancer immunotherapy and infectious disease treatment [61]. DNA nanostructures enable the delivery of CpG-based adjuvants in a fully nucleic acid-based construct by improving cell uptake and promoting enzymatic stability. For instance, Fan et al reported a DNA tetrahedron incorporating CpG-bearing sequences at the four vertices [62]. The resulting DNA tetrahedron showed improved nuclease stability and uptake by RAW264.7 cells, and triggered a strong immune response as evidenced by the secretion of high levels of pro-inflammatory cytokines. The general strategy, i.e. using a DNA nanostructure to present a multitude of CpG-bearing strands, was also adopted by Liedl and Nishikawa in the study of origami tube- and DNA dendrimer-functionalized adjuvants (Figure 3c,d), respectively [63,64]. Such DNA nanostructures appear to be particularly suitable for modulating immunity due to their preferential uptake by immune cells (e.g. macrophages) and their biodistribution in the lymph node [65]. A second significant advantage involves the design flexibility of the DNA nanostructure, which can be tailored to encase protein antigens within the particles. These structures resemble an inversed virus, i.e. a nucleic acid cage with embedded proteins. For example, Yan et al reported a DNA tetrahedron incorporating CpG strands at the vertices and a model antigen, streptavidin (STV), in the cavity of the particle (Figure 3e) [66]. These structures induced high levels of anti-STV antibody in mice, which persisted even after 70 days without apparent immunity against the nanostructure itself. Overall, with the potential to utilize both the surface and the core of the DNA particle to engage with therapeutic payloads, the DNA nanostructure enhances the adjuvant’s activity and allows for the simultaneous delivery of the antigen, completely bypassing the need for a complex co-carrier. Thus, by cleverly choosing the payload and mechanism for disease treatment, the intrinsic immunological difficulties of certain DNA nanostructures can be bypassed and even transformed into a useful feature. One particular area worth looking into is to use the GC-rich fragments to co-deliver Dox to the tumor microenvironment. The combined activation of the immune system and drug-induced tumor cell lysis may lead to enhanced cancer immunotherapy [67].
As the field of DNA nanotechnology advances beyond structural curiosity to include application-driven designs that take advantage of the intrinsic properties of the nucleic acid material as well as the programmability of structure and function, one can expect an increasing number of precisely engineered DNA nanostructures that meet the delivery criteria of a wide variety of therapeutic payloads.
DNA Amphiphiles for DNA/Drug Co-delivery
DNA amphiphiles consist of a hydrophilic DNA block covalently linked to a hydrophobic block. In the aqueous phase, DNA amphiphiles self-assemble to form various micellular nanostructures in which the DNA makes up the outer corona while the hydrophobic block collapse to form the inner core [68, 69]. The DNA micelle can be considered as a form of spherical nucleic acids (SNAs) [70]. SNAs consist of a densely packed layer of DNA strands standing vertically on a central, solid core, which originally was a gold nanoparticle but has since been expanded to include silica, quantum dots, liposomes, polymers, or even empty cavity.[71–76] SNAs exhibit several highly unusual properties that are absent for linear or cyclic nucleic acids, such as rapid endocytosis, enhanced binding affinity with a complementary sequence, and improved stability against nucleases [77]. These properties have been shown to stem from the dense packing and orientation of the DNA strands, irrespective of the core composition.
The unique properties of the SNA have inspired a number of attempts to leverage the SNA core as a depot for therapeutic payloads. One such attempt involves the self-assembly of DNA amphiphiles containing a hydrophobic drug segment. The self-assembly in an aqueous media results in micellar structures that are structurally and functionally analogous to the SNA, but with a core loaded with the desired therapeutics. These self-delivering nanostructures significantly improve the solubility of the hydrophobic drug while promoting the cellular uptake of the DNA due to formation of a dense DNA shell, turning physiochemical disadvantages of both the drug (hydrophobic, insoluble) and the DNA (hydrophilic, unstable) into useful properties. In order for the amphiphiles to robustly form micelles, there must be sufficient hydrophobic driving force from the drug segment to overcome the entropic penalty and the charge-repulsion of the DNA associated with micelle formation. In addition, a drug release mechanism is required if the drugs are covalently linked. Our group recently reported the delivery of camptothecin (CPT) using a DNA-CPT amphiphilic conjugate (Figure 4a, [78]). Three CPT molecules were connected to a phenol-based self-immolative linker before being tethered to a DNA strand. The linker is capped by a light-responsive 2-nitrobenzyl ether moiety. Self-assembly of the conjugate resulted in spherical or cylindrical micelles, depending on the length of the DNA. Regardless of the morphology, the micelles exhibited improved stability against DNase I degradation and enhanced cellular uptake, properties linked to the SNA architecture. The toxicity of the CPT is masked due to being covalently conjugated to the linker. However, upon brief light-irradiation, a series of electronic elimination reactions result in the release of unmodified drug and the full restoration of its cytotoxicity. The general design was taken further in a follow-on study, where paclitaxel (PTX) was polymerized and subsequently conjugated to an antisense DNA strand to form a diblock conjugate, DNA-block-poly(PTX) [79]. With an average of ten PTX molecules per polymer, SNA-like micelles were robustly formed, which enhances cellular uptake of the DNA by more than 100-fold compared with free DNA. Upon endocytosis, native PTX was released from a bioreductive, self-immolative cleavable linker, inducing potent cytotoxicity comparable to that of free PTX. In addition, the antisense shell of the micelle, which targets the antiapoptotic B-cell lymphoma 2 (Bcl2) family proteins, exhibited potent activity at nM concentrations. These studies nicely demonstrate that, by rationally combining nucleic acids and drug molecules, it is possible to co-deliver both components to cells and eliminate the need for complex delivery vehicles.
Figure 4.
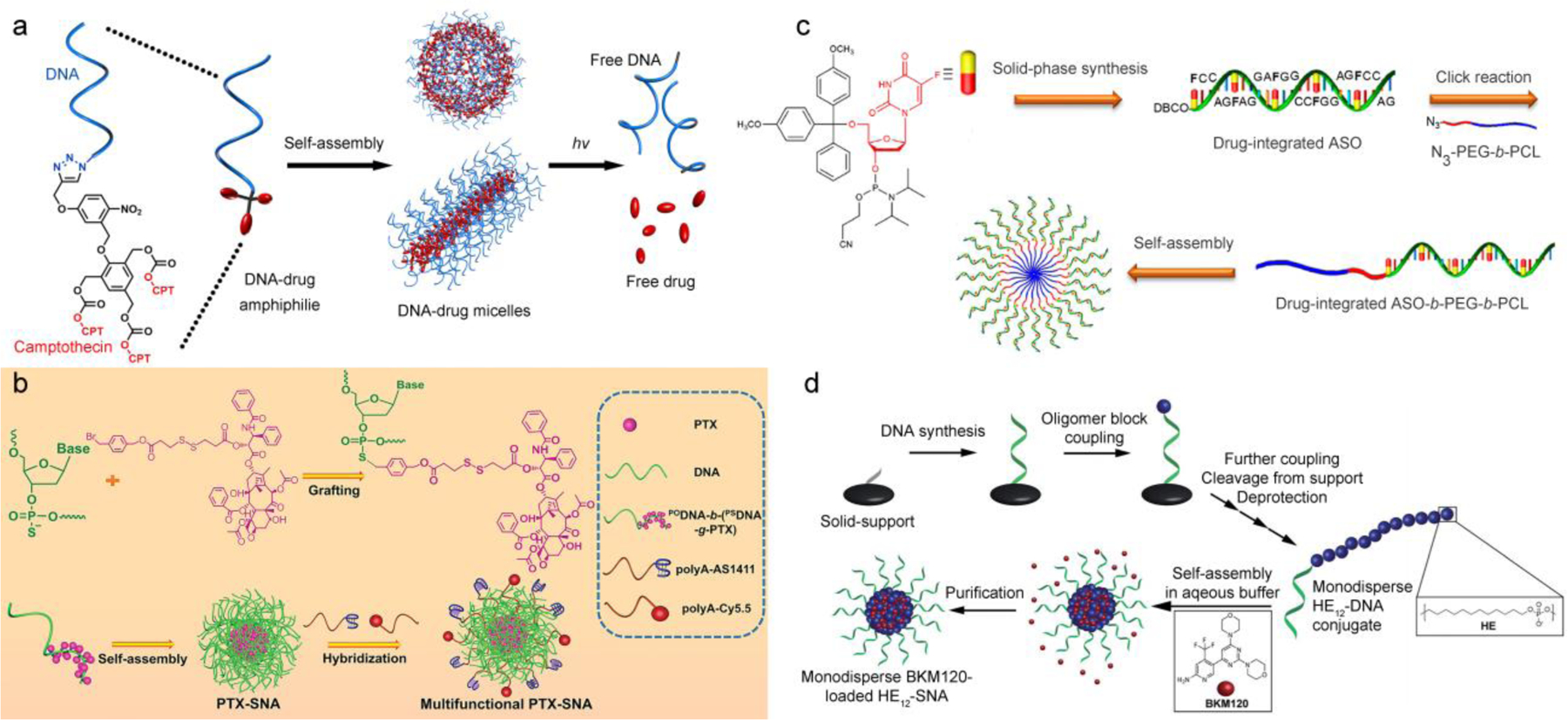
Self-assembled, multivalent DNA nanostructures for drug-DNA co-delivery. (a) DNA-camptothecin nanostructures assembled from photolabile DNA-drug amphiphiles. Reproduced with permission from ref. 78. Copyright 2015 American Chemical Society. (b) SNA-like DNA micelles based on DNA-b-(PSDNA-g-drug) amphiphiles for bioreductively triggered delivery of paclitaxel. Reproduced with permission from ref. 80. Copyright 2019 Wiley-VCH. (c) The concept of chemogene - drug-modified DNA sequences. Reproduced with permission from ref. 81. Copyright 2019 American Chemical Society. (d) Monodisperse DNA amphiphiles for the loading and delivery of BKM120. Reproduced from ref. 83 with permission from the Royal Society of Chemistry.
Instead of relying upon polymer chemistry or small molecule organic synthesis to construct the DNA amphiphile, it is also possible to utilize the automated solid-phase syntheses to accomplish similar syntheses. Zhang et al have recently developed an approach to conjugate several PTX via alkylation of phosphorothioate oligonucleotides (PSDNA) (Figure 4b) [80]. Different from natural DNA with phosphodiester linkages (PODNA), the non-bridging sulfur atoms associated with the PSDNA can serve as sites for nucleophilic reactions with appropriate alkylating agents as described by McLaughlin and Fidanza [81]. In this design, a diblock DNA strand containing both PSDNA and PODNA blocks were alkylated with a PTX-containing bromide prodrug. The alkylation was selective (on the PSDNA sulfur) and quantitative, resulting in an amphiphilic PODNA-b-(PSDNA-g-PTX) conjugate. The conjugates further assembled into SNA-like micellar platform, onto which cell-targeting aptamers and fluorescent reporters were added. These PTX-SNAs were shown to exhibit active tumor targeting, enhanced tumor inhibition, and reversal of drug resistance both in vitro and in vivo. The same group also took advantage of solid-phase synthesis to incorporate floxuridine, a nucleotide analog antitumor drug, into a DNA sequence that is the hydrophilic segment of a DNA-b-poly(ethylene glycol)-b-polycaprolactone triblock amphiphile. Termed “chemogene”, the amphiphile assembled to form SNA-type structures, which enabled the down-regulation of Bcl-2, reversion of chemoresistance, and excellent antitumor activity in vivo upon intravenous injection (Figure 4c) [82]. The advantages of solid-phase methods used by these studies include convenience in the synthesis, better quality control measures, and the lack of polydispersity in the final product.
The Sleiman group demonstrated yet another possibility to take advantage of solid-phase synthesis in the construction of DNA-based micelles for DDSs (Figure 4d) [83]. The micelles were assembled from an amphiphile containing a 19-mer DNA segment and a hydrophobic, 12 hexaethylene (HE12) segment, which was incorporated during the automated solid-phase DNA synthesis using phosphoramidite chemistry. The accurate control in the structure of the amphiphile resulted in extremely well-defined particles. Not surprisingly, the HE12-SNAs exhibited SNA-like properties such as enhanced nuclease stability and cellular uptake. Different from the DNA-drug amphiphilic structures, the drug component, Buparlisib (BKM120, an anticancer drug for the treatment of chronic lymphocytic leukemia), was non-covalently loaded into the core of the micelles via hydrophobic interactions, which is the more traditional means for drug loading. In vivo studies demonstrated that the BKM120-loaded HE12-SNAs sufficiently maintained the efficacy of BKM120 with increased blood circulation times and minimum immune system stimulation. Likely due to the EPR effect, these nanoparticles accumulate at the site of HCT116 colon xenografts over time following both intraperitoneal and intravenous injections. Importantly, the BKM120-loaded DNA particles were not observed to leak drug through the blood-brain barrier (BBB), which is the root cause for major side effects in patients.
In summary, nucleic acid amphiphiles offer a convenient strategy to co-deliver small molecule drugs and nucleic acid payloads simultaneously. Their combination offers benefits such as precisely defined drug loading, carrier-free delivery, improved drug solubility, enhanced DNA stability against nucleases, a new pharmacokinetic profile, among others. Currently this type of “carrier-free” DDS is mainly being considered for cancer chemotherapy, but it is plausible that such structures may be extended to include immunotherapy and other disease areas where SNAs are thought to be effective, such as brain and skin malignancies.
Aptamer-Drug Conjugates for Targeted Delivery
Enriching the therapeutic payloads at targeted disease tissues vs. healthy ones has been the major goal for DDS development [84,85]. In this segment, we discuss how aptamer-drug conjugates (ApDCs) change the game of targeted delivery. To limit the scope, studies involving aptamers serving as targeting ligands in complex DDSs, such as liposomal and polymeric formulations, are not covered.
Nucleic acid aptamers are affinity binders that consist of single-stranded oligonucleotides generated under defined conditions through repeated rounds of in vitro selection, a process known as SELEX (Systematic Evolution of Ligands by EXponential enrichment) [86]. In this process, initial nucleic acid binders against specific molecular targets are separated from a large randomized oligonucleotide library and then amplified by polymerase chain reaction (PCR). High-affinity candidates are further enriched through additional rounds of selection and eventually sequenced. The aptamers fold into unique three-dimensional shapes, which allow them to bind to their targets through non-covalent interactions. Recently, a cell-SELEX process was developed, which uses whole cells or tissues as a mixture of complex targets [87]. To eliminate false positive binders that are not specific to the target cell or tissue, a negative selection step using off-target cell lines is introduced. Cell-SELEX requires no prior knowledge of the targets nor the cells, and targets of interest are screened in their native cellular status [88]. For example, a DNA aptamer XQ-2 was selected by cell-SELEX after 15 rounds of enrichment from a randomized library. XQ-2 can bind to a membrane protein of PL45 cells with a nanomolar Kd. Truncation and chemical modification of XQ-2 leads to the discovery of XQ-2d, an aptamer targeting pancreatic ductal adenocarcinoma that can be used for in vivo imaging and tissue recognition [89]. The use of SELEX and cell-SELEX can thus enable targeted delivery of drugs to different tissues using nucleic acids.
Compared with antibodies, aptamers offer significant benefit in their ability to be chemically engineered, modified, and synthesized in large scales [90]. Thus, various mechanisms of drug loading can be utilized to achieve ApDCs. For example, the secondary structure of an aptamer and/or a chimeric strand can be exploited as drug loading sites by strand hybridization or intercalation [91–93]. Rossi et al reported a pancreatic cancer RNA aptamer (P19) that was loaded with either monomethyl auristatin E (MMAE) or maytansine 1 (DM1) by hybridization via a sticky end [90]. These two antimitotic agents failed as drugs themselves due to lack of sufficient tumor specificity and unacceptable systemic toxicity, and were instead investigated in antibody-drug conjugates [94]. The authors used a truncated version of P19 aptamer (tP19) and engineered the 27-mer ligand with a sticky sequence at the 5’ end to allow for hybridization with a sequence bearing MMAE or DM1. The ApDCs were quickly recognized and internalized by PANC-1 cells, leading to dose-dependent inhibition of cell proliferation. Additionally, a low cytotoxicity level was observed for non-targeted cell lines, suggesting that the aptamers can be an effective alternative to antibodies to achieve targeted delivery. In addition, planar and aromatic molecules such as Dox and 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)porphine (TMPyP4) can be noncovalently loaded into GC-rich motifs and other secondary structures by intercalation (vide supra) [92,93]. A nucleolin-targeting, G-quadruplex forming AS1411 aptamer has been reported to take advantage of such supramolecular interaction to specifically deliver the photosensitizer TMPyP4 to cancer cells [94]. The complex, carrying six intercalated TMPyP4 molecules, resulted in higher drug accumulation and stronger photodamage in nucleolin-overexpressing breast cells (MCF7) vs. normal epithelium cells (M10).
A second mechanism for drug loading is through direct covalent conjugation. The phosphodiester backbone and nucleobases can both be used as potential drug loading sites. Upon delivery, the dormant prodrug must be released from the conjugate in their active form. A number of covalent ApDCs with these characteristics have recently been reported [95–99]. Drugs that are compatible with solid-phase oligonucleotide synthesis may be directly incorporated into the aptamer by phosphoramidite chemistry. For instance, Tan et al reported a fluorouracil (5-FU)-containing phosphoramidite that was compatible with automated synthesis of ApDCs (Figure 5a) [95]. Starting with 3-amino-1,2-propanediol, the authors synthesized an O-nitrobenzyl-linked prodrug phosphoramidite in six steps with reasonable yields. The prodrug was incorporated into a PTK7-targeting sgc8 aptamer at predesigned sites with tunable loading ratios. The target recognition ability of the sgc8 aptamer was not compromised since drug moieties were positioned at the 5’ end of the sequence. In vitro study confirmed that the ApDC can specifically target PTK7-overexpressing HCT116 colon cancer cells with UV-controlled cytotoxicity, and that such effects were not present with a non-target cell line (Ramos lymphoma cells). In addition to the phosphate backbone, the termini of the aptamer sequence are also popular locations for conjugating the therapeutic payload [97,98]. Zhang et al have developed a water-soluble AS1411-PTX conjugate for selective delivery of PTX to tumor targets [97]. The hydroxyl group at 2’ position of PTX, which is essential for its antimitotic activity, was used as an anchor point for tethering with the 3’ end of AS1411 through amidation. The PTX prodrug remained inactive during systemic circulation. Upon cell uptake, a cathepsin B-cleavable dipeptide linker that connects two components is severed, releasing active PTX and restoring its cytotoxicity. As a result, enhanced tumor-suppressive efficacy and reduced off-target cytotoxicity were observed. A third option for drug conjugation involves modification of the nucleobases. Although rarely reported, the nucleobases are intriguing handles because the natural exocyclic aromatic amine groups can serve as anchors for covalent drug loading. For instance, Tan has shown that Dox can form crosslinks with the 2-amine of deoxyguanosines to generate a nuclease-resistant DNA-Dox adduct using formaldehyde as the crosslinking agent [99]. The reaction resulted in several Dox molecules being attached to the sgc8 aptamer sequence via heat-labile methylene bridges. Interestingly, the conjugation did not result in the compromise of the target recognition capability for the aptamer, but did elevate nuclease stability presumably due to steric hindrance associated with Dox. Drug release from the adduct was shown to be gradual under physiological temperature. In a tumor xenograft model, the sgc8-Dox adduct significantly inhibited tumor growth while reducing the risk for cardiomyopathy, a critical detrimental side effect of Dox. This strategy is potentially powerful and general because it enables facile synthesis of ApDCs without prior aptamer modification, although it may not be guaranteed that all aptamers modified this way will retain their binding affinity.
Figure 5.
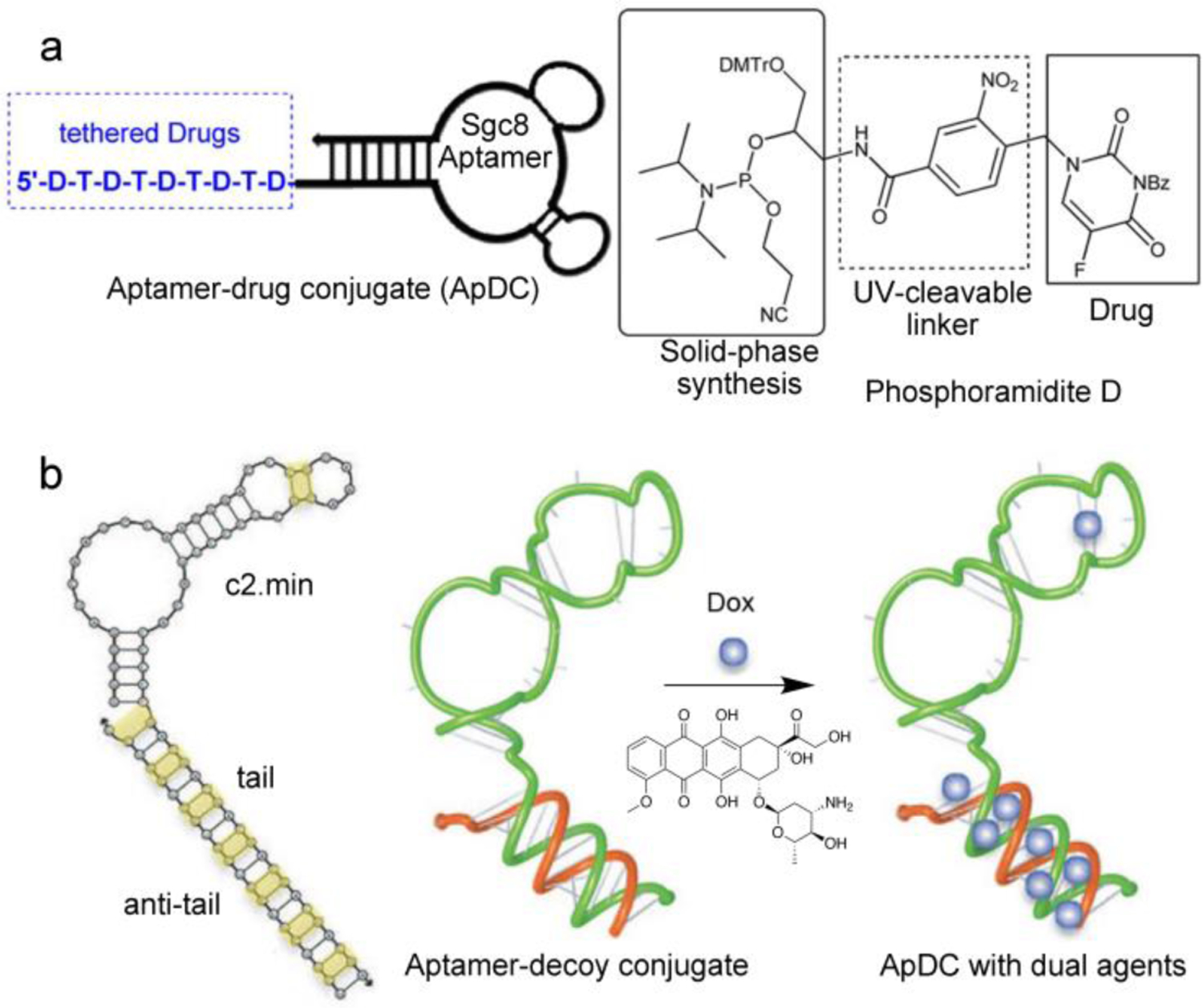
Aptamer-based DDSs. (a) Using an uncanonical phosphoramidite, 5-FU is covalently incorporated into the sgc8 aptamer sequence during solid-phase synthesis. Reproduced with permission from ref. 95. Copyright 2014 American Chemical Society. (b) An RNA aptamer bearing an NF-κB decoy double-stranded oligonucleotide, serving also as intercalation sites to load Dox. Note that yellow highlights indicate possible Dox binding sites. Reproduced with permission from ref. 103. Copyright 2015 American Society of Gene & Cell Therapy.
If one considers a therapeutic oligonucleotide as the payload, then its aptamer conjugate can be regarded as a self-deliverable form of oligonucleotide due to the identical chemical nature of the two components. The aptamer-oligonucleotide chimera can be readily manufactured by solid-phase synthesis without complicated modifications and purification, which represents a significant translational advantage. Importantly, drug release upon delivery becomes non-essential, because the therapeutic portion may retain functionality in the conjugate form, and thus stable chimeras are sometimes preferred over cleavable versions as therapeutics. To date, a variety of oligonucleotide cargos including antisense oligonucleotide (ASO), miRNA, and siRNA have been successfully delivered by aptamers [100–103]. For instance, Esposito et al have developed an aptamer-siRNA chimera for the treatment of glioblastoma (GBM) [100]. GBM is an aggressive Grade IV primary brain tumor. Transcription factors such as signal transducer and activator of transcription-3 (STAT3) have been reported as key regulators in GBM malignancy and proliferation. Oligonucleotides can serve as potent STAT3 antagonists, but their clinical adoption remains difficult due to the lack of tissue selectivity and low cell penetration. Gint4.T is a truncated nuclease-resistant RNA aptamer obtained by cell-SELEX using U87MG GBM cells. The aptamer sequence was elongated with four spacers and a 17-nt sticky handle at the 3’-end so that a STAT3-specific siRNA antagonist can be incorporated by hybridization. As a result, the Gint4.T-siRNA chimera can recognize and bind with platelet-derived growth factor receptor β (PDGFRβ)-expressing GBM cells, leading to STAT3 downregulation and tumor suppression in a xenograft GBM model.
The multitude of mechanisms for covalent/non-covalent loading of drug species onto aptamers makes it possible to simultaneously incorporate different forms of drugs for combination therapy against cancer. Acquired chemoresistance after repeated exposure is an unavoidable issue that has long plagued the efficacy of chemotherapeutics. Because oligonucleotides interfere with cellular events at the genetic level, chemoresistance may be overcome by synergistically combining a gene regulator with a cytotoxic drug. The chemoresistance of Dox, for example, is related to the activation of the nuclear transcription factor κB (NF-κB). Signore et al demonstrated that, with the help of an ApDC targeting an anti-transferrin receptor coupled with a NF-κB decoy oligonucleotide, cell-killing efficiency of intercalated Dox against pancreatic tumor cells can be significantly improved (Figure 5b) [103]. The conjugate consists of two oligonucleotides: an anti-human transferrin receptor (hTfR) RNA aptamer c2.min bearing a (CGA)7 DNA tail at the 3’-end, and an NF-κB decoy double-stranded oligonucleotide tethered with an anti-tail (TCG)7 sequence via a disulfide linker. The complementary, CG-rich tails serve as the linkage between the two oligonucleotides as well as a depot for Dox through intercalation. Through a series of in vitro assessments, selective targeting and anticancer synergy of the decoy- and Dox-loaded ApDC in hTfR-overexpressing MIA PaCa-2 cells were confirmed.
Overall, these studies highlight the design flexibility of nucleic acid aptamers as both a targeting moiety and a drug carrier. While challenges associated with ApDCs remain, which include premature enzymatic damage, undesirable plasma pharmacokinetics, and limited bioavailability due to clearance by the kidney and the immune system [94], given the benign chemical composition of nucleic acids, the powerful SELEX and cell-SELEX processes, and the diversity of mechanisms for drug loading and release, ApDCs remain one of the most appealing and competitive forms of nucleic acid-based DDS for a variety of therapeutics.
DNA-Controlled Drug Release
While certain long-lasting pharmaceuticals are more beneficial when continuously delivered by the DDS for prolonged duration of efficacy, other drugs, such as anticancer drugs and short-lived biologics, are often preferred to be burst-released to maximize their therapeutic response in living systems [105,106]. Therefore, a main purpose of the DDS is to ensure that its cargos are released in a spatiotemporally controlled manner [107]. DNA complexes inherently possess trigger-responsive features owing to the strict base-pairing properties, which are sensitive to environmental factors. This part will survey recently published literature that emphasize the application of nucleic acids for controlled drug release. Release mechanisms that are not directly related to nucleic acids (e.g. chemically cleavable linkers) and composite delivery systems (e.g. DNA-containing hydrogel, DNA-based gatekeepers used in porous nanoparticles, etc.) are excluded here.
Temperature is a factor that directly interferes with nucleic acid hybridization. The thermostability of the delivery system can be programmed by engineering the sequence and length of the target helices [108]. For example, Knudsen et al have demonstrated temperature-controlled encapsulation and release of the enzyme horseradish peroxidase (HRP) using a three-dimensional DNA nanocage (Figure 6a) [109]. This covalently closed DNA cage, which is a truncated octahedron, is assembled from eight oligonucleotides after annealing and ligation. One corner of the cage is modified with a palindromic DNA sequence that can form a hairpin structure. The cage can attain either an open conformation at 37 °C for encapsulation/release of HRP, or a closed conformation at 4 °C to trap the enzyme. Interestingly, entrapped HRP is still catalytically active inside the DNA cage and can convert substrate molecules that penetrate the apertures of the DNA lattice. To achieve temperature-controlled release in vivo, local hyperthermia (~40 °C) is generally required, which can be triggered photothermally using hybrid materials such as gold nanorods. The use of pristine DNA delivery vehicles for temperature-controlled release is rare in the literature. One reason for such limited adoption involves the broad melting transition for DNA duplexes, which can occur over a ~20 °C range. Sharper transitions (~2 °C) have been observed for certain types of DNA structures such as the SNA due to cooperativity in the melting process, which point to their potential application as thermoresponsive DDSs [110].
Figure 6.
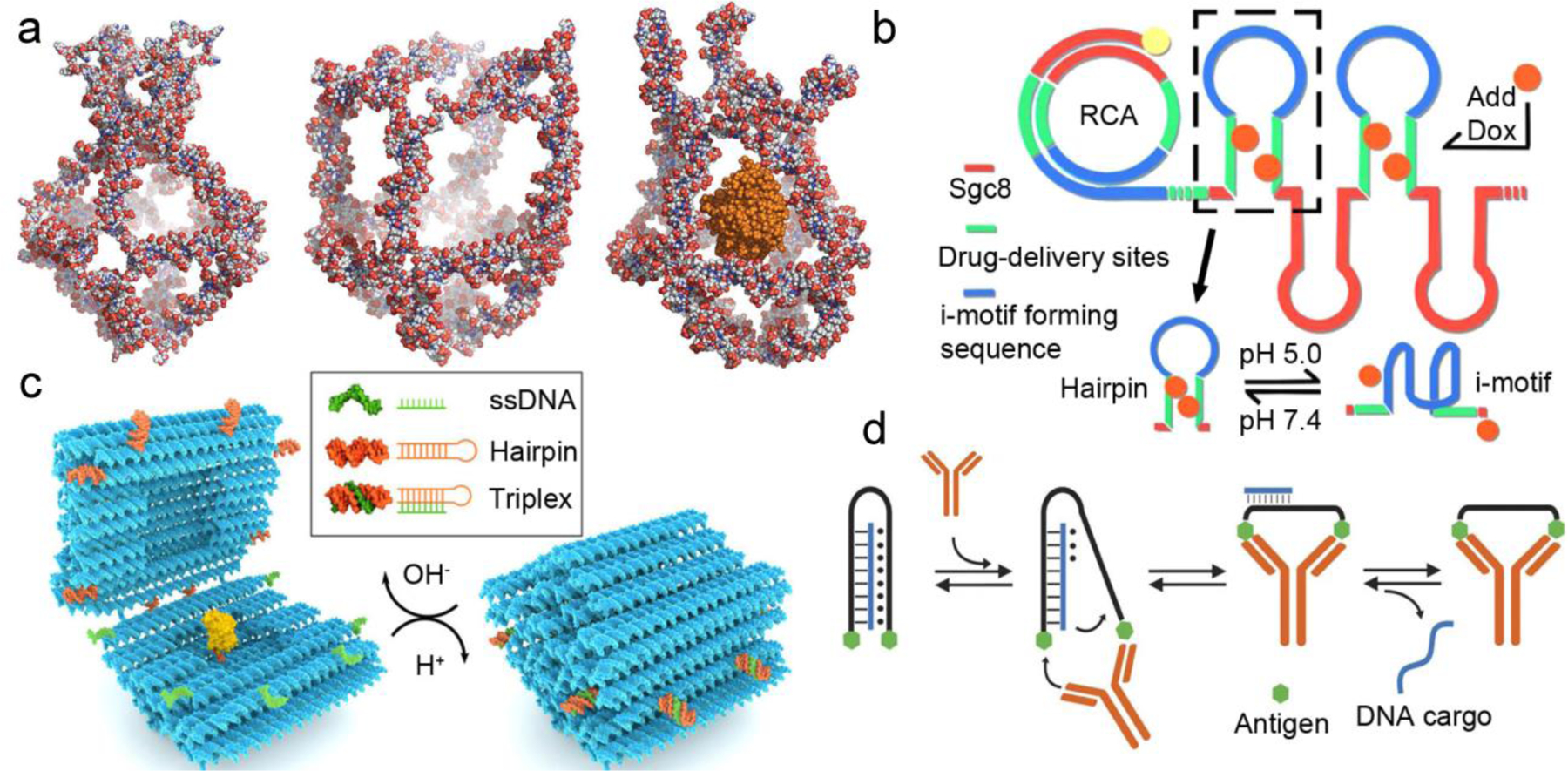
Examples of DNA-mediated drug release. (a) Atomistic models of a DNA nanocage: closed at 4 °C (left), extended at 37 °C (middle), and re-closed cage at 4 °C with HRP (orange) encapsulated. Reproduced with permission from ref. 109. Copyright 2013 American Chemical Society. (b) pH-responsive DNA nanoparticles with i-motif triggers. Intercalated drug is released upon acidification due to formation of i-motifs at low pH. Reproduced with permission from ref. 118. Copyright 2018 American Chemical Society. (c) Action of a pH-responsive, suitcase-like DNA origami. The DNA latch, which is comprised of a hairpin (orange) and a ssDNA (green), forms a triplex DNA staple at low pH, closing the carrier. At higher pH, the triplex latches are unlocked, opening the suitcase and displaying its protein cargo (yellow). Reproduced with permission from ref. 120. Copyright 2019 American Chemical Society. (d) An antibody-responsive DNA-triplex clamp. A clamp strand (black) labeled with two antigens (green hexagons) forms a triplex with a cargo ssDNA (blue). The recognition of an antibody of the two antigens causes a conformational change that ultimately leads to the release of the ssDNA cargo. Reproduced with permission from ref. 124. Copyright 2017 Springer Nature.
Light is another form of external stimulus for drug release. While base pairing generally cannot be directly tuned with light, photoswitchable moieties such as azobenzene can be involved in the design of DNA nanostructures to tune their photostability. The planar trans-azobenzene can stabilize dsDNA via intercalation while the non-planar cis-azobenzene destabilizes the duplex due to steric hindrance [111]. To utilize the opposite stabilities, Sugiyama et al developed an azobenzene-functionalized DNA origami for photoresponsive payload manipulation [112]. The square bipyramidal origami was assembled by mixing and annealing M13 mp18 ssDNA with staple strands. Three out of the four base edges of the capsule were modified with azobenzene-containing staples. Once UV irradiation is applied, the formation of cis-azobenzene disrupts hybridization along three edges of the four linking the two pyramids, allowing them to open with only one edge still attached. This reversible open/close mechanism makes it possible to encapsulate oligonucleotide-functionalized gold nanoparticles following visible light irradiation (closure). UV irradiation and strand displacement can then be applied to open the cage and fully release the cargos from the cavity. While gold nanoparticles were tested as model payloads, one can easily imagine the use of such a system for the delivery of macromolecular therapeutics. In addition to high molecular weight cargos, the delivery of small molecule drugs can also benefit from light-triggered release originating from interrupted hybridization. Using lipid-mediated self-assembly, Famulok et al designed a nanocarrier containing an aptamer against lipidated hepatocyte growth factor receptor (cMet) coupled to a GC-rich DNA hairpin motif with four 2′,6′-dimethyl trans-azobenzene moieties sporadically incorporated into the phosphate backbone [113]. Dox was loaded into the GC-rich domains. After photoisomerization, the non-planar cis-azobenzene was able to induce hairpin destabilization and subsequent Dox release.
Solution acidity/basicity has been extensively studied in the context of drug delivery, because pH gradients exist between different tissues and cell compartments, and are also associated with tumor microenvironments. DNA i-motif and triplex are two uncanonical DNA secondary structures that can respond to pH changes [114,115]. The i-motif is a four-stranded structure with a cytosine-rich sequence. The cytosine and protonated cytosine can form an intercalated and anti-parallel tetramer structure through Hoogsteen base pairing C:C+ interactions. Similarly, the protonated cytosine can also be present in DNA triplexes, which are triple-stranded complexes with the third strand binding to the dsDNA through Hoogsteen base pairing C+:G-C interactions. The hemi-protonated nature of the N3 nitrogens in cytosine makes these folded structures stable at weakly acidic pH (5–6). Thus, the folding/unfolding status of these scaffolds can be coupled to drug release to attain pH-responsiveness [116–120]. For example, the i-motif has been reported as a trigger to release the anti-inflammatory glucocorticoid dexamethasone from DNA nanotubes [116]. The six-helix DNA nanotube was assembled using the single-stranded tile method, and three of the tiles were extended with the i-motif sequence, which was initially hybridized to a drug-conjugated complementary sequence. These polyanionic nanotubes were quickly localized in the endosomes of MH-S macrophages, and the acidic endosomal environment triggered the formation of i-motif, allowing the dexamethasone-containing strand to be released. This pH-responsive handle was demonstrated in tissue resident macrophages from mouse cremaster muscle. Using the same general approach, Tian et al applied rolling circle amplification (RCA) to produce a long ssDNA containing repeating segments of the sgc8 aptamer for cancer cell targeting and segments of an i-motif-forming hairpin sequence for Dox loading and pH-triggered release (Figure 6b) [118]. At physiological pH, the i-motif-forming sequence in the loop of the hairpin remains unfolded, allowing Dox to be stably intercalated in the stem region of the closed hairpin. Under the acidic tumor environment, the formation of the i-motif forces a conformational change of the hairpin, leading to Dox release. Aside from interfering with drug loading sites, another mechanism for i-motif- and/or triplex-triggered drug release involves their use as smart staples to alter the conformation of the DDS. For example, it is possible to tune the open/closed status of a DNA nanocontainer using these motifs to encapsulate/expose/release nano-sized cargos such as proteins and metal nanoparticles [119,120]. Linko et al have developed a DNA “nano-suitcase” that responds to environmental pH (Figure 6c) [120]. The suitcase consists of two halves linked by four ssDNA hinges. Eight pairs of triplex DNA staples, each consisting of one hairpin duplex and one ssDNA, are distributed at the interface between the halves. Under acidic pH, triplex formation results in the closing of the nanocapsule. Higher pH conditions destabilize the triplex staples and trigger the opening of the origami suitcase. The full cycle of cargo loading, encapsulation, and re-exposure was demonstrated at physiologically relevant pH conditions using Förster resonance energy transfer (FRET) and transmission electron microscopy (TEM), and the shielded enzyme remained fully functional after loading and encapsulation.
Due to the plethora of biomolecules that interact natively with nucleic acids, it is potentially a very powerful strategy to use disease-related biomarkers and molecules present in specific tissues and cells to trigger drug release in a spatiotemporally controlled manner. A range of biomacromolecules such as endogenous nucleic acids [121,122], proteins [123], and antibodies [124] have been reported to induce drug release, mainly by reconfiguring or degrading the DNA-based carrier. For instance, Miyoshi et al have developed a G-quadruplex-based DDS that responds to the epidermal growth factor receptor (EGFR) mRNA [121]. The delivery vehicle consists of a G-quadruplex, a G-quadruplex ligand (a telomerase inhibitor), and a long loop that is complementary to the EGFR mRNA. In the absence of EGFR mRNA, the assembled G-quadruplex structure can capture copper(II) anionic phthalocyanine, and the telomerase inhibitor remains inactive. When the complex recognizes its target, the hybridization between the long loop and EGFR mRNA destabilizes the G-quadruplex carrier and releases the active drug as a result. Recently, the tumor-associated protein, nucleolin, was reported as a molecular trigger to alter the conformation of a tube-like “nanorobot”, expose encapsulated thrombin, and induce blood coagulation specifically in tumor tissues [123]. The nanorobot was assembled from a rectangular DNA origami sheet using the M13 bacteriophage genome DNA and predesigned staple strands. Thrombin was anchored on the surface of the DNA sheet by capture strands. Using several fastener strands, the tubular nanorobot was non-covalently closed along the edge of the DNA sheet. Upon intravenous injection, the DNA nanorobots accumulated at tumor-associated blood vessels owing to attached anti-nucleolin aptamers (AS1411). Binding of nucleolin and the fastener strands unwound the nanotube to form nanosheets, which caused the exposure of thrombin and intravascular thrombosis. The resulting tumor necrosis caused tumor growth inhibition in mouse models. Similar to the specific interaction between nucleolin and the fastener strands, antibodies can be generated for specific interactions with an antigen. By tethering antigens to a DNA duplex or triplex structure, it is possible to induce a conformational change of the DNA structure when antibodies recognize the antigen. Ricci et al demonstrated this principle by developing an oligonucleotide-releasing DNA nanomachine [124]. The nanomachine consists of a long ssDNA that forms a clamp-like DNA triplex structure with its specific 12-nt ssDNA cargo via both Watson-Crick and Hoogsteen interactions. The sequence was rationally designed so that the triplex structure is stable while the Watson-Crick duplex between the cargo and core strand denatures at physiological temperature. To introduce antibody-responsiveness into the complex, the two ends of the core strand were conjugated with antigens. The recognition between an antibody and the two antigens causes a conformational change that disrupts the less stable Hoogsteen base pairing, leading to the release of the clamped strand (Figure 6d). By using three orthogonal antigens (digoxigenin, dinitrophenol, and p17 peptide that is recognized by an anti-HIV antibody), the authors showed that the antibody-powered DNA nanomachine can release its DNA cargo in a rapid and highly specific manner.
Notably, with DNA nanotechnology and DNA logic gates, it is possible to use one or more recognition processes to generate a highly specific, or even computed, form of drug release. For instance, a sequence (endogenous or exogenous) can serve as an input through toehold-mediated strand displacement (TMSD), which renders them robust and predictable release triggers [125]. Such triggers were adopted by the “lock-key” mechanism in box-shaped DNA origamis [33,126]. The nanocarrier changes its conformation when the “key” is present, which induces TMSD and subsequently exposes the payload. Similarly, drug release can also be triggered by TMSD-induced carrier disassembly. The Sleiman group have used this strategy to construct several DNA nanostructures that can release a variety of cargos spanning small molecule drugs, siRNA, and gold nanoparticles [127–129]. A notable example involves a DNA nanocube capable of capturing DNA amphiphiles and releasing guest molecules by TMSD [127]. The structure consists of a cube-like nanoscaffold; at each of the eight corners, a dendritic alkyl-DNA amphiphile was attached by hybridization. While typical amphiphile assembly leads to core-shell micellar structures, in this case, the amphiphiles are positioned inside of the DNA cube, making the overall structure equivalent to a cube-shaped micelle. The hydrophobic core formed inside the cube served as a reservoir for molecules such as Nile Red and tyrosine kinase inhibitor Dasatinib by hydrophobic host-guest interactions. TMSD with a target strand caused the disassembly of cube micelle and subsequently the release of the loaded drug molecules. Because the displacement junction responds to its target in a highly specific manner, autonomous logic-gated computing devices can be designed by embedding multiple responsive junctions. DNA nanorobots with AND-gated logic, for example, can only release their cargos after recognizing multiple targets [130,131]. Church et al developed a cell-targeting DNA device that can perform AND-gated release of payloads such as gold nanoparticles and antibody fragments [131]. The hexagonal-shaped barrel carrier consists of two origami domains that are permanently linked at one edge by single-stranded scaffold hinges and are noncovalently tethered at the opposite edge by DNA aptamer-based staples (Figure 7). The staple is comprised of an antigen-binding aptamer and a partially complementary strand. In the absence of the antigen, the lock duplex can fasten the device and encapsulate the payloads. When the antigen is introduced, the aptamer-antigen interaction overpowers the duplex formation, opening the staple. By using two distinct aptamer-based locks, the AND-gated nanorobot can only expose its payloads and trigger cell signaling when both lock duplexes are displaced and opened by their specific cell-surface protein targets.
Figure 7.
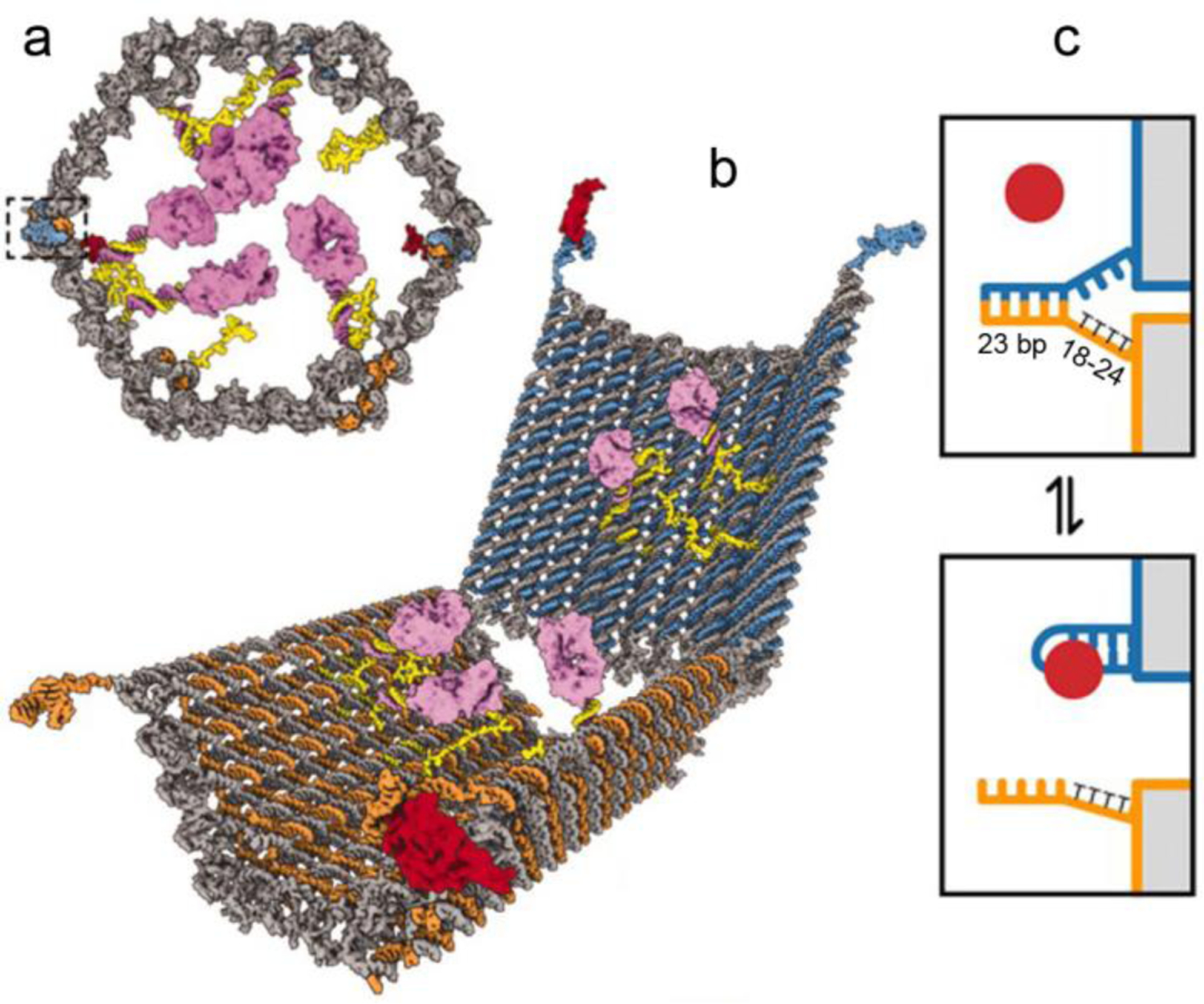
Antigen-responsive DNA nanorobot for AND-gated drug release. (a) Front view of the closed nanorobot loaded with a protein payload (purple). Two DNA-aptamers fasten and close the barrel-shaped carrier (boxed region). (b) Opened nanorobot after protein displacement of aptamer locks. (c) Mechanism of cargo release. The lock duplex is destabilized when its antigen key (red) is present. The lock duplex consists of a DNA aptamer (blue) and a partially complementary strand (orange) with multiple thymine (T) spacers. Reproduced with permission from ref. 131. Copyright 2012 American Association for the Advancement of Science.
Overall, these studies highlight how DNA hybridization can be regulated via different mechanisms and chemical species, natural or exogenous, which lead to conformational or other structural changes (e.g. strand displacement or complete disintegration) of the delivery entity and the release/exposure of the payloads. The built-in chemical and biological responsiveness of DNA makes them remarkable agents for controlled drug loading and release.
Conclusion and Outlook
To summarize, the role that nucleic acids play in therapeutics has been redefined in the last decade. Originally a substance known for information storage and therapeutic potential, nucleic acids have emerged as a game changer in drug delivery in many aspects, spanning traditional delivery approaches, targeted delivery, and smart logic gated devices. DNA nanotechnology, for example, offers simple yet powerful design tools for the construction of intricate nanocarriers for drug delivery that are not replicable in structure and function by any other techniques. DNA amphiphiles can form self-deliverable nanoparticles that simultaneously transport payloads with opposing physical properties into cells without a complex co-carrier. Aptamers impart antibody-level targeting capabilities to associated drugs using an all-nucleic acid composition. Last but not least, the programmability of base pairing and its sensitivity to physiochemical and biological cues render nucleic acids highly versatile in the spatiotemporal control of drug release.
Despite these potential, the inherent biopharmaceutical difficulties of nucleic acids should not be overlooked. Serum stability, for example, may hinder the development of systemically administered nucleic acid-based DDSs. Although chemical enhancements such as modified sugars, unnatural internucleotidic linkages, and modified bases have been reported, chemically modified nucleic acids may introduce additional issues such as compromised binding affinity and liver/cardiovascular toxicity. As a result, nucleic acid-based DDSs in general still fall behind application-leading material classes such as polymeric particles and liposomes. In addition, scrutiny on the sequences used by assembled architectures may be required to reduce unwanted immunogenicity arising from PAMP motifs [132]. Thus, future studies should either address these challenges, or circumvent them by identifying unique use cases where nucleic acid instability and side effects associated with unwanted nucleic acid-protein interactions are insignificant. Oftentimes, pristine nucleic acid carriers are not as favorable as composite DDSs for meeting certain criteria, which has been carefully discussed by several recent reviews [23,133,134]. Overall, it is foreseeable that nucleic acids will continue to play an important role in the design of modern DDSs. With the rapidly expanding scientific community for nucleic acids-based structures and their biological applications, we anticipate that state-of-the-art drug carriers comprised of nucleic acids will soon make a real clinical impact.
Acknowledgements
KZ acknowledges support by the National Institutes of Health (the National Institute of General Medical Sciences Award Number 1R01GM121612) and the National Science Foundation (CAREER Award Number 1453255). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation
References
- [1].Liu D, Yang F, Xiong F, Gu N, The smart drug delivery system and its clinical potential, Theranostics 6 (2016) 1306–1323. 10.7150/thno.14858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anselmo AC, Mitragotri S, An overview of clinical and commercial impact of drug delivery systems, J. Control. Release 190 (2014) 15–28. 10.1016/j.jconrel.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Habtemariam S, Shin HS, Nano based drug delivery systems: recent developments and future prospects, J. Nanobiotechnology 16 (2018) 71. 10.1186/s12951-018-0392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Singh R, Lillard JW, Nanoparticle-based targeted drug delivery, Exp. Mol. Pathol 86 (2009) 215–223. 10.1016/j.yexmp.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Utreja P, Jain S, Tiwary AK, Novel drug delivery systems for sustained and targeted delivery of anti-cancer drugs: Current status and future prospects, Curr. Drug Deliv 7 (2010) 152–161. 10.2174/156720110791011783 [DOI] [PubMed] [Google Scholar]
- [6].Alvarez-Lorenzo C, Concheiro A, Smart drug delivery systems: from fundamentals to the clinic, Chem. Commun 50 (2014) 7743–7765. 10.1039/c4cc01429d [DOI] [PubMed] [Google Scholar]
- [7].Hossen S, Hossain MK, Basher MK, Mia MNH, Rahman MT, Uddin MJ, Smart nanocarrier-based drug delivery systems for cancer therapy and toxicity studies: A review, J. Adv. Res 15 (2018) 1–18. 10.1016/j.jare.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Senapati S, Mahanta AK, Kumar S, Maiti P, Controlled drug delivery vehicles for cancer treatment and their performance, Signal Transduct. Target. Ther 3 (2018) 7. 10.1038/s41392-017-0004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wilczewska AZ, Niemirowicz K, Markiewicz KH, Car H, Nanoparticles as drug delivery systems, Pharmacol. Rep 64 (2012) 1020–1037. 10.1016/S1734-1140(12)70901-5 [DOI] [PubMed] [Google Scholar]
- [10].Verma D, Gulati N, Kaul S, Mukherjee S, Nagaich U, Protein based nanostructures for drug delivery, J. Pharm 2018 (2018) 9285854. 10.1155/2018/9285854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Claesen J, Fischbach MA, Synthetic microbes as drug delivery systems, ACS Synth. Biol 4 (2015) 358–364. 10.1021/sb500258b [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Juliano RL, The delivery of therapeutic oligonucleotides, Nucleic Acids Res 44 (2016) 6518–6548. 10.1093/nar/gkw236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Marques JT, Williams BRG, Activation of the mammalian immune system by siRNAs, Nat. Biotechnol 23 (2005) 1399–1405. 10.1038/nbt1161 [DOI] [PubMed] [Google Scholar]
- [14].Krieg AM, CpG motifs in bacterial DNA and their immune effects, Annu. Rev. Immunol 20 (2002) 709–760. 10.1146/annurev.immunol.20.100301.064842 [DOI] [PubMed] [Google Scholar]
- [15].Bode C, Zhao G, Steinhagen F, Kinjo T, Klinman DM, CpG DNA as a vaccine adjuvant, Expert Rev. Vaccines 10 (2011) 499–511. 10.1586/erv.10.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meng Z, Lu M, RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front Immunol 8 (2017) 331. 10.3389/fimmu.2017.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sehgal A, Vaishnaw A, Fitzgerald K, Liver as a target for oligonucleotide therapeutics, J. Hepatol 59 (2013) 1354–1359. 10.1016/j.jhep.2013.05.045 [DOI] [PubMed] [Google Scholar]
- [18].Shen X, Corey DR, Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs, Nucleic Acids Res 46 (2018) 1584–1600. 10.1093/nar/gkx1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Seeman NC, Nucleic acid junctions and lattices, J. Theor. Biol 99 (1982) 237–247. 10.1016/0022-5193(82)90002-9 [DOI] [PubMed] [Google Scholar]
- [20].Fu TJ, Seeman NC, DNA double-crossover molecules, Biochemistry 32 (1993) 3211–3220. 10.1021/bi00064a003 [DOI] [PubMed] [Google Scholar]
- [21].LaBean TH, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC, Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes, J. Am. Chem. Soc 122 (2000) 1848–1860. 10.1021/ja993393e [DOI] [Google Scholar]
- [22].Gu H, Chao J, Xiao S-J, Seeman NC, A proximity-based programmable DNA nanoscale assembly line, Nature 465 (2010) 202–205. 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hu Q, Li H, Wang L, Gu H, Fan C, DNA nanotechnology-enabled drug delivery systems, Chem. Rev 119 (2019) 6459–6506. 10.1021/acs.chemrev.7b00663 [DOI] [PubMed] [Google Scholar]
- [24].Seeman NC, DNA in a material world, Nature 421 (2003) 427–431. 10.1038/nature01406 [DOI] [PubMed] [Google Scholar]
- [25].Ding B, Sha R, Seeman NC, Pseudohexagonal 2D DNA crystals from double crossover cohesion, J. Am. Chem. Soc 126 (2004) 10230–10231. 10.1021/ja047486u [DOI] [PubMed] [Google Scholar]
- [26].Park SH, Yin P, Liu Y, Reif JH, LaBean TH, Yan H, Programmable DNA self-assemblies for nanoscale organization of ligands and proteins, Nano Lett 5 (2005) 729–733. 10.1021/nl050175c [DOI] [PubMed] [Google Scholar]
- [27].Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH, DNA-templated self-assembly of protein arrays and highly conductive nanowires, Science 301 (2003) 1882–1884. 10.1126/science.1089389 [DOI] [PubMed] [Google Scholar]
- [28].Zhang C, Su M, He Y, Zhao X, Fang P, Ribbe AE, Jiang W, Mao C, Conformational flexibility facilitates self-assembly of complex DNA nanostructures, Proc. Natl. Acad. Sci. U.S.A 105 (2008) 10665–10669. 10.1073/pnas.0803841105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He Y, Su M, Fang P, Zhang C, Ribbe AE, Jiang W, Mao C, On the chirality of self-assembled DNA octahedra, Angew. Chem. Int. Ed 49 (2010) 748–751. 10.1002/anie.200904513 [DOI] [PubMed] [Google Scholar]
- [30].Rothemund PWK, Folding DNA to create nanoscale shapes and patterns, Nature 440 (2006) 297–302. 10.1038/nature04586 [DOI] [PubMed] [Google Scholar]
- [31].Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM, Self-assembly of DNA into nanoscale three-dimensional shapes, Nature 459 (2009) 414–418. 10.1038/nature08016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H, Multilayer DNA origami packed on a square lattice, J. Am. Chem. Soc 131 (2009) 15903–15908. 10.1021/ja906381y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J, Self-assembly of a nanoscale DNA box with a controllable lid, Nature 459 (2009) 73–76. 10.1038/nature07971 [DOI] [PubMed] [Google Scholar]
- [34].Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H, Scaffolded DNA origami of a DNA tetrahedron molecular container, Nano Lett 9 (2009) 2445–2447. 10.1021/nl901165f [DOI] [PubMed] [Google Scholar]
- [35].Ke Y, Ong LL, Shih WM, Yin P, Three-dimensional structures self-assembled from DNA bricks, Science 338 (2012) 1177–1183. 10.1126/science.1227268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wei B, Dai M, Yin P, Complex shapes self-assembled from single-stranded DNA tiles, Nature 485 (2012) 623–626. 10.1038/nature11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hamblin GD, Hariri AA, Carneiro KMM, Lau KL, Cosa G, Sleiman HF, Simple Design for DNA Nanotubes from a Minimal Set of Unmodified Strands: Rapid, Room-Temperature Assembly and Readily Tunable Structure, ACS Nano 7 (2013) 3022–3028. 10.1021/nn4006329. [DOI] [PubMed] [Google Scholar]
- [38].McLaughlin CK, Hamblin GD, Hänni KD, Conway JW, Nayak MK, Carneiro KMM, Bazzi HS, Sleiman HF, Three-Dimensional Organization of Block Copolymers on “DNA-Minimal” Scaffolds, J. Am. Chem. Soc 134 (2012) 4280–4286. 10.1021/ja210313p. [DOI] [PubMed] [Google Scholar]
- [39].Zhang F, Jiang S, Wu S, Li Y, Mao C, Liu Y, Yan H, Complex wireframe DNA origami nanostructures with multi-arm junction vertices, Nature Nanotechnology 10 (2015) 779–784. 10.1038/nnano.2015.162. [DOI] [PubMed] [Google Scholar]
- [40].Jun H, Shepherd TR, Zhang K, Bricker WP, Li S, Chiu W, Bathe M, Automated Sequence Design of 3D Polyhedral Wireframe DNA Origami with Honeycomb Edges, ACS Nano 13 (2019) 2083–2093. 10.1021/acsnano.8b08671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, Lee H, Scuto A, Liu Y, Yang C, Deng J, Soifer HS, Raubitschek A, Forman S, Rossi JJ, Pardoll DM, Jove R, Yu H, In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses, Nat. Biotechnol 27 (2009) 925–932. 10.1038/nbt.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Diebold SS, Kaisho T, Hemmi H, Akira S, e Sousa CR, Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA, Science 303 (2004) 1529–1531. 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- [43].Triantafilou K, Vakakis E, Orthopoulos G, Ahmed MAE, Schumann C, Lepper PM, Triantafilou M, TLR8 and TLR7 are involved in the host’s immune response to human parechovirus 1, Eur. J. Immunol 35 (2005) 2416–2423. 10.1002/eji.200526149 [DOI] [PubMed] [Google Scholar]
- [44].Matsumoto M, Seya T, TLR3: Interferon induction by double-stranded RNA including poly(I:C), Adv. Drug Deliv. Rev 60 (2008) 805–812. 10.1016/j.addr.2007.11.005 [DOI] [PubMed] [Google Scholar]
- [45].Greish K, Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting, Methods Mol. Biol 624 (2010) 25–37. 10.1007/978-1-60761-609-2_3 [DOI] [PubMed] [Google Scholar]
- [46].Fang J, Nakamura H, Maeda H, The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect, Advanced Drug Delivery Reviews 63 (2011) 136–151. 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- [47].Carvalho C, Santos RX, Cardoso S, Correia S, Oliveira PJ, Santos MS, Moreira PI, Doxorubicin: The good, the bad and the ugly effect, Curr. Med. Chem 16 (2009) 3267–3285. 10.2174/092986709788803312 [DOI] [PubMed] [Google Scholar]
- [48].Yang F, Teves SS, Kemp CJ, Henikoff S, Doxorubicin DNA torsion, and chromatin dynamics, Biochim. Biophys. Acta – Rev. Cancer 1845 (2014) 84–89. 10.1016/j.bbcan.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jiang Q, Song C, Nangreave J, Liu X, Lin L, Qiu D, Wang Z-G, Zou G, Liang X, Yan H, Ding B, DNA origami as a carrier for circumvention of drug resistance, J. Am. Chem. Soc 134 (2012) 13396–13403. 10.1021/ja304263n [DOI] [PubMed] [Google Scholar]
- [50].Zhang Q, Jiang Q, Li N, Dai L, Liu Q, Song L, Wang J, Li Y, Tian J, Ding B, Du Y, DNA origami as an in vivo drug delivery vehicle for cancer therapy, ACS Nano 8 (2014) 6633–6643. 10.1021/nn502058j [DOI] [PubMed] [Google Scholar]
- [51].Liu J, Song L, Liu S, Jiang Q, Liu Q, Li N, Wang Z-G, Ding B, A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy, Nano Lett 18 (2018) 3328–3334. 10.1021/acs.nanolett.7b04812 [DOI] [PubMed] [Google Scholar]
- [52].Lacroix A, Vengut-Climent E, de Rochambeau D, Sleiman HF, Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale, ACS Cent. Sci 5 (2019) 882–891. 10.1021/acscentsci.9b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhao Y-X, Shaw A, Zeng X, Benson E, Nyström AM, Högberg B, DNA origami delivery system for cancer therapy with tunable release properties, ACS Nano 6 (2012) 8684–8691. 10.1021/nn3022662 [DOI] [PubMed] [Google Scholar]
- [54].Gudipati S, Zhang K, Rouge JL, Towards self-transfecting nucleic acid nanostructures for gene regulation, Trends Biotechnol 37 (2019) 983–994. 10.1016/j.tibtech.2019.01.008 [DOI] [PubMed] [Google Scholar]
- [55].Lächelt U, Wagner E, Nucleic acid therapeutics using polyplexes: A journey of 50 years (and beyond), Chem. Rev 115 (2015) 11043–11078. 10.1021/cr5006793 [DOI] [PubMed] [Google Scholar]
- [56].Zhang K, Fang H, Shen G, Taylor J-SA, Wooley KL, Well-defined cationic shell crosslinked nanoparticles for efficient delivery of DNA or peptide nucleic acids, Proc. Am. Thorac. Soc 6 (2009) 450–457. 10.1513/pats.200902-010AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Oh SS, Lee BF, Leibfarth FA, Eisenstein M, Robb MJ, Lynd NA, Hawker CJ, Soh HT, Synthetic aptamer-polymer hybrid constructs for programmed drug delivery into specific target cells, J. Am. Chem. Soc 136 (2014) 15010–15015. 10.1021/ja5079464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lv H, Zhang S, Wang B, Cui S, Yan J, Toxicity of cationic lipids and cationic polymers in gene delivery, J. Control. Release 114 (2006) 100–109. 10.1016/j.jconrel.2006.04.014 [DOI] [PubMed] [Google Scholar]
- [59].Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG, Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nat. Nanotechnol 7 (2012) 389–393. 10.1038/nnano.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Qian H, Tay CY, Setyawati MI, Chia SL, Lee DS, Leong DT, Protecting microRNAs from RNase degradation with steric DNA nanostructures, Chem. Sci 8 (2017) 1062–1067. 10.1039/C6SC01829G [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Krieg AM, Yi A-K, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM, CpG motifs in bacterial DNA trigger direct B-cell activation, Nature 374 (1995) 546–549. 10.1038/374546a0 [DOI] [PubMed] [Google Scholar]
- [62].Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C, Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides, ACS Nano 5 (2011) 8783–8789. 10.1021/nn202774x [DOI] [PubMed] [Google Scholar]
- [63].Schüller VJ, Heidegger S, Sandholzer N, Nickels PC, Suhartha NA, Endres S, Bourquin C, Liedl T, Cellular immunostimulation by CpG-sequence-coated DNA origami structures, ACS Nano 5 (2011) 9696–9702. 10.1021/nn203161y [DOI] [PubMed] [Google Scholar]
- [64].Mohri K, Kusuki E, Ohtsuki S, Takahashi N, Endo M, Hidaka K, Sugiyama H, Takahashi Y, Takakura Y, Nishikawa M, Self-assembling DNA dendrimer for effective delivery of immunostimulatory CpG DNA to immune cells, Biomacromolecules 16 (2015) 1095–1101. 10.1021/bm501731f [DOI] [PubMed] [Google Scholar]
- [65].Radovic-Moreno AF, Chernyak N, Mader CC, Nallagatla S, Kang RS, Hao L, Walker DA, Halo TL, Merkel TJ, Rische CH, Anantatmula S, Burkhart M, Mirkin CA, Gryaznov SM, Immunomodulatory spherical nucleic acids, Proc. Natl. Acad. Sci. U.S.A 112 (2015) 3892–3897. 10.1073/pnas.1502850112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H, Chang Y, A DNA nanostructure platform for directed assembly of synthetic vaccines, Nano Lett 12 (2012) 4254–4259. 10.1021/nl301877k [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nishikawa M, Mizuno Y, Mohri K, Matsuoka N, Rattanakiat S, Takahashi Y, Funabashi H, Luo D, Takakura Y, Biodegradable CpG DNA hydrogels for sustained delivery of doxorubicin and immunostimulatory signals in tumor-bearing mice, Biomaterials 32 (2011) 488–494. 10.1016/j.biomaterials.2010.09.013. [DOI] [PubMed] [Google Scholar]
- [68].Kwak M, Herrmann A, Nucleic acid amphiphiles: synthesis and self-assembled nanostructures, Chem. Soc. Rev 40 (2011) 5745–5755. 10.1039/C1CS15138J. [DOI] [PubMed] [Google Scholar]
- [69].Jia F, Lu X, Tan X, Zhang K, Facile synthesis of nucleic acid–polymer amphiphiles and their self-assembly, Chem. Commun 51 (2015) 7843–7846. 10.1039/C5CC01934F. [DOI] [PubMed] [Google Scholar]
- [70].Zakrewsky M, Kumar S, Mitragotri S, Nucleic acid delivery into skin for the treatment of skin disease: Proofs-of-concept, potential impact, and remaining challenges, J. Control. Release 219 (2015) 445–456. 10.1016/j.jconrel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Popat A, Liu J, Qing (Max) Lu G, Zhang Qiao S, A pH-responsive drug delivery system based on chitosan coated mesoporous silica nanoparticles, J. Mater. Chem 22 (2012) 11173–11178. 10.1039/C2JM30501A [DOI] [Google Scholar]
- [72].Tian H, Chen J, Chen X, Nanoparticles for gene delivery, Small 9 (2013) 2034–2044. 10.1002/smll.201202485 [DOI] [PubMed] [Google Scholar]
- [73].Wu Y, Chen W, Meng F, Wang Z, Cheng R, Deng C, Liu H, Zhong Z, Core-crosslinked pH-sensitive degradable micelles: A promising approach to resolve the extracellular stability versus intracellular drug release dilemma, J. Control. Release 164 (2012) 338–345. 10.1016/j.jconrel.2012.07.011 [DOI] [PubMed] [Google Scholar]
- [74].Park K, Controlled drug delivery systems: Past forward and future back, J. Control. Release 190 (2014) 3–8. 10.1016/j.jconrel.2014.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Chen W, Zhong P, Meng F, Cheng R, Deng C, Feijen J, Zhong Z, Redox and pH-responsive degradable micelles for dually activated intracellular anticancer drug release, J. Control. Release 169 (2013) 171–179. 10.1016/j.jconrel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- [76].Cutler JI, Zhang K, Zheng D, Auyeung E, Prigodich AE, Mirkin CA, Polyvalent nucleic acid nanostructures, J. Am. Chem. Soc 133 (2011) 9254–9257. 10.1021/ja203375n [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cutler JI, Auyeung E, Mirkin CA, Spherical Nucleic Acids, J. Am. Chem. Soc 134 (2012) 1376–1391. 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- [78].Tan X, Li BB, Lu X, Jia F, Santori C, Menon P, Li H, Zhang B, Zhao JJ, Zhang K, Light-triggered, self-immolative nucleic acid-drug nanostructures, J. Am. Chem. Soc 137 (2015) 6112–6115. 10.1021/jacs.5b00795 [DOI] [PubMed] [Google Scholar]
- [79].Tan X, Lu X, Jia F, Liu X, Sun Y, Logan JK, Zhang K, Blurring the role of oligonucleotides: Spherical nucleic acids as a drug delivery vehicle, J. Am. Chem. Soc 138 (2016) 10834–10837. 10.1021/jacs.6b07554 [DOI] [PubMed] [Google Scholar]
- [80].Guo Y, Zhang J, Ding F, Pan G, Li J, Feng J, Zhu X, Zhang C, Stressing the role of DNA as a drug carrier: Synthesis of DNA–drug conjugates through grafting chemotherapeutics onto phosphorothioate oligonucleotides, Adv. Mater 31 (2019) 1807533. 10.1002/adma.201807533 [DOI] [PubMed] [Google Scholar]
- [81].Fidanza JA, McLaughlin LW, Introduction of reporter groups at specific sites in DNA containing phosphorothioate diesters, J. Am. Chem. Soc 111 (1989) 9117–9119. 10.1021/ja00207a028 [DOI] [PubMed] [Google Scholar]
- [82].Mou Q, Ma Y, Ding F, Gao X, Yan D, Zhu X, Zhang C, Two-in-one chemogene assembled from drug-integrated antisense oligonucleotides to reverse chemoresistance, J. Am. Chem. Soc 141 (2019) 6955–6966. 10.1021/jacs.8b13875 [DOI] [PubMed] [Google Scholar]
- [83].Bousmail D, Amrein L, Fakhoury JJ, Fakih HH, Hsu JCC, Panasci L, Sleiman HF, Precision spherical nucleic acids for delivery of anticancer drugs, Chem. Sci 8 (2017) 6218–6229. 10.1039/C7SC01619K [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bae YH, Park K, Targeted drug delivery to tumors: Myths, reality and possibility, J. Control. Release 153 (2011) 198–205. 10.1016/j.jconrel.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Rosenblum D, Joshi N, Tao W, Karp JM, Peer D, Progress and challenges towards targeted delivery of cancer therapeutics, Nat. Commun 9 (2018) 1410. 10.1038/s41467-018-03705-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Blind M, Blank M, Aptamer selection technology and recent advances, Mol. Ther. Nucleic Acids 4 (2015) e223. 10.1038/mtna.2014.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Morris KN, Jensen KB, Julin CM, Weil M, Gold L, High affinity ligands from in vitro selection: complex targets, Proc. Natl. Acad. Sci. U.S.A 95 (1998) 2902–2907. 10.1073/pnas.95.6.2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Guo KT, Paul A, Schichor C, Ziemer G, Wendel HP, CELL-SELEX: Novel perspectives of aptamer-based therapeutics, Int. J. Mol. Sci 9 (2008) 668–678. 10.3390/ijms9040668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X, Liu Q, Champanhac C, Teng IT, Ye M, Tan W, DNA aptamer selected against pancreatic ductal adenocarcinoma for in vivo imaging and clinical tissue recognition, Theranostics 5 (2015) 985–994. 10.7150/thno.11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kanwar JR, Roy K, Maremanda NG, Subramanian K, Veedu RN, Bawa R, Kanwar RK, Nucleic acid-based aptamers: Applications, development and clinical trials, Curr. Med. Chem 22 (2015) 2539–2557. 10.2174/0929867322666150227144909 [DOI] [PubMed] [Google Scholar]
- [91].Yoon S, Huang KW, Reebye V, Spalding D, Przytycka TM, Wang Y, Swiderski P, Li L, Armstrong B, Reccia I, Zacharoulis D, Dimas K, Kusano T, Shively J, Habib N, Rossi JJ, Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth, Mol. Ther. Nucleic Acids 6 (2017) 80–88. 10.1016/j.omtn.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wen J, Tao W, Hao S, Iyer SP, Zu Y, A unique aptamer-drug conjugate for targeted therapy of multiple myeloma, Leukemia 30 (2016) 987–991. 10.1038/leu.2015.216 [DOI] [PubMed] [Google Scholar]
- [93].Shieh YA, Yang SJ, Wei MF, Shieh MJ, Aptamer-based tumor-targeted drug delivery for photodynamic therapy, ACS Nano 4 (2010) 1433–1442. 10.1021/nn901374b [DOI] [PubMed] [Google Scholar]
- [94].Dan N, Setua S, Kashyap VK, Khan S, Jaggi M, Yallapu MM, Chauhan SC, Antibody-drug conjugates for cancer therapy: Chemistry to clinical implications, Pharmaceuticals 11 (2018) E32. 10.3390/ph11020032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing FL, Tan W, Automated modular synthesis of aptamer–drug conjugates for targeted drug delivery, J. Am. Chem. Soc 136 (2014) 2731–2734. 10.1021/ja4117395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Park JY, Cho YL, Chae JR, Moon SH, Cho WG, Choi YJ, Lee SJ, Kang WJ. Gemcitabine-incorporated G-quadruplex aptamer for targeted drug delivery into pancreas cancer, Mol. Ther. Nucleic Acids 12 (2018) 543–553. 10.1016/j.omtn.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Li F, Lu J, Liu J, Liang C, Wang M, Wang L, Li D, Yao H, Zhang Q, Wen J, Zhang Z, Li J, Lv Q, He X, Guo B, Guan D, Yu Y, Dang L, Wu X, Li Y, Chen G, Jiang F, Sun S, Zhang B, Lu A, Zhang G, A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer, Nat. Commun 8 (2017) 1390. 10.1038/s41467-017-01565-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Niu W, Chen X, Tan W, Veige AS, N-heterocyclic carbene-gold(I) complexes conjugated to a leukemia-specific DNA aptamer for targeted drug delivery, Angew. Chem. Int. Ed 55 (2016) 8889–8893. 10.1002/anie.201602702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhu G, Cansiz S, You M, Qiu L, Han D, Zhang L, Mei L, Fu T, Chen Z, Tan W, Nuclease-resistant synthetic drug-DNA adducts: Programmable drug-DNA conjugation for targeted anticancer drug delivery, NPG Asia Mater 7 (2015) e169. 10.1038/am.2015.19 [DOI] [Google Scholar]
- [100].Esposito CL, Nuzzo S, Catuogno S, Romano S, de Nigris F, de Franciscis V, STAT3 gene silencing by aptamer-siRNA chimera as selective therapeutic for glioblastoma, Mol. Ther. Nucleic Acids 10 (2018) 398–411. 10.1016/j.omtn.2017.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Hong S, Sun N, Liu M, Wang J, Pei R, Building a chimera of aptamer–antisense oligonucleotide for silencing galectin-1 gene, RSC Adv 6 (2016) 112445–112450. 10.1039/C6RA21250F [DOI] [Google Scholar]
- [102].Esposito CL, Cerchia L, Catuogno S, De Vita G, Dassie JP, Santamaria G, Swiderski P, Condorelli G, Giangrande PH, de Franciscis V, Multifunctional aptamer-miRNA conjugates for targeted cancer therapy, Mol. Ther 22 (2014) 1151–1163. 10.1038/mt.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Porciani D, Tedeschi L, Marchetti L, Citti L, Piazza V, Beltram F, Signore G, Aptamer-mediated codelivery of doxorubicin and NF-κB decoy enhances chemosensitivity of pancreatic tumor cells, Mol. Ther. Nucleic Acids 4 (2015) E235. 10.1038/mtna.2015.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhu G, Niu G, Chen X, Aptamer–drug conjugates, Bioconjug. Chem 26 (2015) 2186–2197. 10.1021/acs.bioconjchem.5b00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fu Y, Kao WJ, Drug release kinetics and transport mechanisms of non-degradable and degradable polymeric delivery systems, Expert Opin. Drug Deliv 7 (2010) 429–444. 10.1517/17425241003602259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Huang X, Brazel CS, On the importance and mechanisms of burst release in matrix-controlled drug delivery systems, J. Control. Release 73 (2001) 121–136. 10.1016/S0168-3659(01)00248-6 [DOI] [PubMed] [Google Scholar]
- [107].Mura S, Nicolas J, Couvreur P, Stimuli-responsive nanocarriers for drug delivery, Nat. Mater 12 (2013) 991–1003. 10.1038/nmat3776 [DOI] [PubMed] [Google Scholar]
- [108].Yakovchuk P, Protozanova E, Frank-Kamenetskii MD, Base-stacking and base-pairing contributions into thermal stability of the DNA double helix, Nucleic Acids Res 34 (2006) 564–574. 10.1093/nar/gkj454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Juul S, Iacovelli F, Falconi M, Kragh SL, Christensen B, Frøhlich R, Franch O, Kristoffersen EL, Stougaard M, Leong KW, Ho YP, Sørensen ES, Birkedal V, Desideri A, Knudsen BR, Temperature-controlled encapsulation and release of an active enzyme in the cavity of a self-assembled DNA nanocage, ACS Nano 7 (2013) 9724–9734. 10.1021/nn4030543 [DOI] [PubMed] [Google Scholar]
- [110].Lytton-Jean AKR, Mirkin CA, A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes, J. Am. Chem. Soc 127 (2005) 12754–12755. 10.1021/ja052255o [DOI] [PubMed] [Google Scholar]
- [111].Liang X, Asanuma H, Kashida H, Takasu A, Sakamoto T, Kawai G, Komiyama M, NMR study on the photoresponsive DNA tethering an azobenzene. Assignment of the absolute configuration of two diastereomers and structure determination of their duplexes in the trans-form, J. Am. Chem. Soc 125 (2003) 16408–16415. 10.1021/ja037248j [DOI] [PubMed] [Google Scholar]
- [112].Takenaka T, Endo M, Suzuki Y, Yang Y, Emura T, Hidaka K, Kato T, Miyata T, Namba K, Sugiyama H, Photoresponsive DNA nanocapsule having an open/close system for capture and release of nanomaterials, Chem. Eur. J 20 (2014) 14951–14954. 10.1002/chem.201404757 [DOI] [PubMed] [Google Scholar]
- [113].Prusty DK, Adam V, Zadegan RM, Irsen S, Famulok M, Supramolecular aptamer nano-constructs for receptor-mediated targeting and light-triggered release of chemotherapeutics into cancer cells, Nat. Commun 9 (2018) 535. 10.1038/s41467-018-02929-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Abou AH, Garavís M, González C, Damha MJ, i-Motif DNA: structural features and significance to cell biology, Nucleic Acids Res 46 (2018) 8038–8056. 10.1093/nar/gky735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Frank-Kamenetskii MD, Mirkin SM, Triplex DNA structures, Annu. Rev. Biochem 64 (1995) 65–95. 10.1146/annurev.bi.64.070195.000433 [DOI] [PubMed] [Google Scholar]
- [116].Sellner S, Kocabey S, Zhang T, Nekolla K, Hutten S, Krombach F, Liedl T, Rehberg M, Dexamethasone-conjugated DNA nanotubes as anti-inflammatory agents in vivo, Biomaterials 134 (2017) 78–90. 10.1016/j.biomaterials.2017.04.031 [DOI] [PubMed] [Google Scholar]
- [117].Kim SH, Kim KR, Ahn DR, Lee JE, Yang EG, Kim SY, Reversible regulation of enzyme activity by pH-responsive encapsulation in DNA nanocages, ACS Nano 11 (2017) 9352–9359. 10.1021/acsnano.7b04766 [DOI] [PubMed] [Google Scholar]
- [118].Zhao H, Yuan X, Yu J, Huang Y, Shao C, Xiao F, Lin L, Li Y, Tian L, Magnesium-stabilized multifunctional DNA nanoparticles for tumor-targeted and pH-responsive drug delivery, ACS Appl. Mater. Interfaces 10 (2018) 15418–15427. 10.1021/acsami.8b01932 [DOI] [PubMed] [Google Scholar]
- [119].Burns JR, Lamarre B, Pyne ALB, Noble JE, Ryadnov MG, DNA origami inside-out viruses, ACS Synth. Biol 7 (2018) 767–773. 10.1021/acssynbio.7b00278 [DOI] [PubMed] [Google Scholar]
- [120].Ijäs H, Hakaste I, Shen B, Kostiainen MA, Linko V, Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo, ACS Nano 13 (2019) 5959–5967. 10.1021/acsnano.9b01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Yaku H, Murashima T, Miyoshi D, Sugimoto N, A mRNA-responsive G-quadruplex-based drug release system, Sensors 15 (2015) 9388–9403. 10.3390/s150409388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Dentinger PM, Simmons BA, Cruz E, Sprague M, DNA-mediated delivery of lipophilic molecules via hybridization to DNA-based vesicular aggregates, Langmuir 22 (2006) 2935–2937. 10.1021/la053005o [DOI] [PubMed] [Google Scholar]
- [123].Li S, Jiang Q, Liu S, Zhang Y, Tian Y, Song C, Wang J, Zou Y, Anderson GJ, Han J, Chang Y, Liu Y, Zhang C, Chen L, Zhou G, Nie G, Yan H, Ding B, Zhao Y, A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo, Nat. Biotechnol 36 (2018) 258–264. 10.1038/nbt.4071 [DOI] [PubMed] [Google Scholar]
- [124].Ranallo S, Prévost-Tremblay C, Idili A, Vallée-Bélisle A, Ricci F, Antibody-powered nucleic acid release using a DNA-based nanomachine, Nat. Commun 8 (2017) 15150. 10.1038/ncomms15150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhang DY, Winfree E, Control of DNA strand displacement kinetics using toehold exchange, J. Am. Chem. Soc 131 (2009) 17303–17314. 10.1021/ja906987s [DOI] [PubMed] [Google Scholar]
- [126].Grossi G, Jepsen MDE, Kjems J, Andersen ES, Control of enzyme reactions by a reconfigurable DNA nanovault, Nat. Commun 8 (2017) 992. 10.1038/s41467-017-01072-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Edwardson TGW, Carneiro KMM, McLaughlin CK, Serpell CJ, Sleiman HF, Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly, Nat. Chem 5 (2013) 868–875. 10.1038/nchem.1745 [DOI] [PubMed] [Google Scholar]
- [128].Bujold KE, Hsu JCC, Sleiman HF, Optimized DNA “nanosuitcases” for encapsulation and conditional release of siRNA, J. Am. Chem. Soc 138 (2016) 14030–14038. 10.1021/jacs.6b08369 [DOI] [PubMed] [Google Scholar]
- [129].Lo PK, Karam P, Aldaye FA, McLaughlin CK, Hamblin GD, Cosa G, Sleiman HF, Loading and selective release of cargo in DNA nanotubes with longitudinal variation, Nat. Chem 2 (2010) 319–328. 10.1038/nchem.575 [DOI] [PubMed] [Google Scholar]
- [130].Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E, An autonomous molecular computer for logical control of gene expression, Nature 429 (2004) 423–429. 10.1038/nature02551 [DOI] [PubMed] [Google Scholar]
- [131].Douglas SM, Bachelet I, Church GM, A logic-gated nanorobot for targeted transport of molecular payloads, Science 335 (2012) 831–834. 10.1126/science.1214081 [DOI] [PubMed] [Google Scholar]
- [132].Surana S, Shenoy AR, Krishnan Y, Designing DNA nanodevices for compatibility with the immune system of higher organisms, Nat. Nanotechnol 10 (2015) 741–747. 10.1038/nnano.2015.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].de Vries JW, Zhang F, Herrmann A, Drug delivery systems based on nucleic acid nanostructures, J. Control. Release 172 (2013) 467–483. 10.1016/j.jconrel.2013.05.022 [DOI] [PubMed] [Google Scholar]
- [134].Zhou L, Ren J, Qu X, Nucleic acid-templated functional nanocomposites for biomedical applications, Mater. Today 20 (2017) 179–190. 10.1016/j.mattod.2016.09.012 [DOI] [Google Scholar]


