Abstract
The enzyme NUDT15 efficiently hydrolyzes the active metabolites of thiopurine drugs, which are routinely used for treating cancer and inflammatory diseases. Loss-of-function variants in NUDT15 are strongly associated with thiopurine intolerance, such as leukopenia, and preemptive NUDT15 genotyping has been clinically implemented to personalize thiopurine dosing. However, understanding the molecular consequences of these variants has been difficult, as no structural information was available for NUDT15 proteins encoded by clinically actionable pharmacogenetic variants because of their inherent instability. Recently, the small molecule NUDT15 inhibitor TH1760 has been shown to sensitize cells to thiopurines, through enhanced accumulation of 6-thio-guanine in DNA. Building upon this, we herein report the development of the potent and specific NUDT15 inhibitor, TH7755. TH7755 demonstrates a greatly improved cellular target engagement and 6-thioguanine potentiation compared with TH1760, while showing no cytotoxicity on its own. This potent inhibitor also stabilized NUDT15, enabling analysis by X-ray crystallography. We have determined high-resolution structures of the clinically relevant NUDT15 variants Arg139Cys, Arg139His, Val18Ile, and V18_V19insGlyVal. These structures provide clear insights into the structural basis for the thiopurine intolerance phenotype observed in patients carrying these pharmacogenetic variants. These findings will aid in predicting the effects of new NUDT15 sequence variations yet to be discovered in the clinic.
Keywords: NUDT15, NUDIX, hydrolase, MTH2, inhibitor, crystal structure
Abbreviations: 6-TG, 6-thioguanine; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CETSA, cellular thermal shift assay; DMSO, dimethyl sulfoxide; hNUDT15, human NUDT15; ITDRFCETSA, isothermal dose–response fingerprint CETSA; mNUDT15, mouse NUDT15; Tagg, apparent aggregation temperature; TPMT, thiopurine S-methyltransferase
NUDT15 belongs to the NUDIX (nucleoside diphosphates linked to a moiety x) superfamily of proteins, which are characterized by containing a conserved NUDIX box motif (GX5EX7REVXEEXGU, where U is a hydrophobic residue). These enzymes metabolize a diverse range of substrates including canonical (d)NTPs, oxidized (d)NTPs, nonnucleoside polyphosphates, and capped mRNAs (1). NUDT15 (also known as MTH2) was originally proposed to have a similar function to another NUDIX family member MTH1 (MutT homolog 1), which efficiently hydrolyses oxidized nucleotides, preventing their incorporation into DNA and thereby sanitizing the cellular nucleotide pool (2). However, it has been shown that NUDT15 only displays weak activity toward oxidized nucleotides and the true biological function of the enzyme remains unknown (3, 4). Intriguingly, genome-wide association studies identified NUDT15 mutations (such as Arg139Cys) that are strongly associated with thiopurine intolerance (5, 6). Further studies have conclusively shown that NUDT15 efficiently hydrolyses active metabolites of thiopurine drugs, such as 6-thio-(d)GTP, to their monophosphate forms (4, 7).
Thiopurine drugs are metabolized into cytotoxic guanosine analog metabolites and are commonly used for treating diseases such as acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and inflammatory bowel disease (8, 9). Several different thiopurine drugs are used clinically, such as 6-mercaptopurine, 6-thioguanine (6-TG), and azathioprine. These compounds are ultimately converted to the cellularly active metabolites 6-thio-GTP and 6-thio-dGTP. After 6-thio-dGTP is incorporated into DNA, it is methylated (10, 11, 12), and following a second round of replication, a Me-6-thio-dG:T mismatch is generated, leading to futile DNA repair attempts and ultimately cell death (13, 14). Thiopurine treatment is commonly associated with leukopenia. This is often a result of genetic polymorphism in genes encoding proteins such as thiopurine S-methyltransferase (TPMT), which inactivate thiopurines including 6-mercaptopurine and 6-TG. These variants in TPMT therefore lead to excessive accumulation of thioguanine in the genome, which ultimately causes the severe hematopoietic toxicity observed in patients undergoing thiopurine-based chemotherapy (15, 16, 17, 18, 19). For this reason, it is recommended to adjust the thiopurine dosage according to the results of preemptive TPMT genotyping (20, 21).
Similar to TPMT, loss-of-function variants in NUDT15 are also strongly associated with thiopurine toxicity, because of increased incorporation of 6-thio-dGTP into DNA. It is becoming increasingly clear that thiopurine treatment strategies need to include both NUDT15 and TPMT genotyping to identify patients at-risk of hematopoietic toxicities (20). This is a particularly important consideration for certain racial/ethnic groups as loss-of-function NUDT15 variants are more common in people of Asian or Hispanic descent compared with Caucasians (5). A recent study on a cohort of 270 children diagnosed with ALL and treated with thiopurines identified four mutations in the NUDT15 gene (7), which showed a 74.4% to 100% loss of activity, relative to the WT enzyme. The identified mutations included three single point mutants c.415C>T (Arg139Cys), c.416G>A (Arg139His), c.52G>A (Val18Ile) and a fourth mutant c.36_37insGGAGTC (Val18_Val19insGlyVal), where a microsatellite expansion results in an in-frame insertion of two residues (glycine and valine). The additional residues are inserted into a segment already containing three glycine–valine repeats and the insertion therefore extends this segment by a single Gly-Val repetition.
Based on the activity of NUDT15 toward 6-thio(d)GTP, which reduces cellular thiopurine toxicity, recent studies have also proposed NUDT15 inhibition as a means of improving thiopurine therapy in patients with WT NUDT15. Specifically, the first potent and selective small molecule NUDT15 inhibitor (TH1760) was recently shown to sensitize cells to thiopurines, through the enhanced incorporation of 6-thio-dGTP in DNA (22). This may increase the effectiveness of thiopurine therapy by allowing lower doses of thiopurines to be used while still maintaining good therapeutic effect. In this study, we present the novel small-molecule NUDT15 inhibitor TH7755, which has substantially improved cellular target engagement as well as potentiation of 6-thio-guanine compared to TH1760. We solved the crystal structure of WT NUDT15 in complex with TH7755, which demonstrates that TH7755 adopts a highly similar binding mode to TH1760. Thermal shift analysis indicated that TH7755 significantly increased the stability of NUDT15. No structural information for clinically isolated NUDT15 mutations was previously available because of their inherent instability, which renders them very difficult to crystallize. We utilized the stabilizing effect of TH7755 to determine high-resolution crystal structures of the clinically relevant NUDT15 mutants Arg139Cys, Arg139His, Val18Ile, and V18_V19insGlyVal. Analysis of these structures in relation to the WT enzyme provides clear explanations for the detrimental phenotypic effects observed for these NUDT15 variants.
Results
TH7755 is a potent NUDT15 inhibitor with improved cellular activity
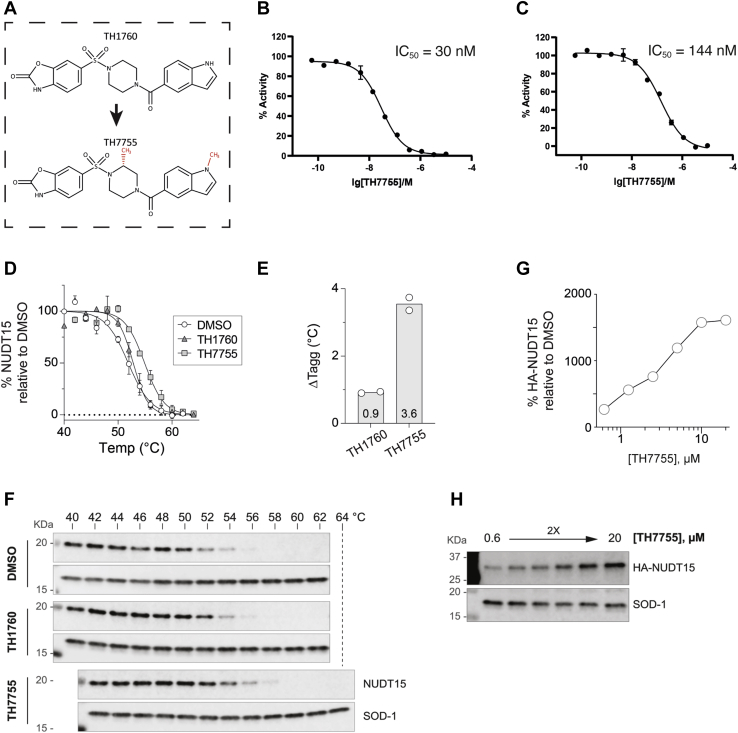
We synthesized the new NUDT15 inhibitor TH7755 based on our previously developed inhibitor TH1760. TH7755 differs from TH1760 in that it contains two additional methyl groups, which are attached to the piperazine ring system and the nitrogen of the indole group (Fig. 1A). The inhibition of human and mouse NUTD15 by TH7755 was determined in vitro using a malachite green–based assay (3, 4). The IC50 values of TH7755 for human and mouse were 30 nM (pIC50 = 7.53 ± 0.03, n = 2) and 144 nM (pIC50 = 6.84 ± 0.02, n = 2), respectively, (Fig. 1, B and C), indicating that the affinity of the inhibitor is higher for the human isoform of the enzyme. The IC50 value of TH7755 for human NUDT15 is very similar to the IC50 value previously reported for TH1760 (25 nM) (22).
Figure 1.
In vitro and in vivo inhibition of NUDT15 by TH7755.A, chemical structures of TH1760 and TH7755, with their differences highlighted in red. B and C, inhibition curves of human (B) and mouse (C) NUDT15 with TH7755. X-axis: [TH7755 concentration], y-axis: percentage activity of NUDT15. In all experiments, formed Pi was determined by addition of malachite green reagent followed by measurement of absorbance at 630 nm. IC50 values were determined using nonlinear regression in GraphPad Prism. The shown IC50 values represent the geometric mean (n = 2). D–F, CETSA of TH7755 and TH1760 in AML HL-60 cells. Intact HL-60 cells were treated with 10 μM TH7755, TH1760, or equal volume of DMSO for 3 h before cells were heated, lysed, and subjected to Western blot to examine soluble NUDT15 and SOD-1 levels. D, The mean signals ± SD of soluble NUDT15 levels are shown, relative to SOD-1 levels and normalized to a DMSO control sample. Two independent experiments are shown. E, Tagg values (°C) calculated from the signals in D, using the Boltzmann sigmoid equation in GraphPad Prism. Two independent experiments are shown. F, corresponding Western blot analysis. G and H, potent engagement of cellular NUDT15 by TH7755 as shown by ITDRFCETSA. Intact HL-60 pInducer20-3xHA-NUDT15 WT cells were induced with doxycycline overnight before being treated with increasing concentrations of TH7755. Cells were heated, lysed, and subjected to Western blot to examine soluble NUDT15 and SOD-1 levels. G, soluble HA-tagged NUDT15 levels, relative to SOD-1 levels and normalized to a DMSO control sample. H, corresponding Western blot analysis. CETSA, cellular thermal shift assay; DMSO, dimethyl sulfoxide; ITDRFCETSA, isothermal dose–response fingerprint CETSA.
To investigate the ability of TH7755 to engage with NUDT15 in cells, we used the cellular thermal shift assay (CETSA) method. Intact AML HL-60 cells were incubated with either TH7755, TH1760, or dimethyl sulfoxide (DMSO) only, after which the cells were heated, lysed, and the thermal stability of cellular NUDT15 was examined by Western blot analysis. At a concentration of 10 μM TH7755 substantially stabilized cellular NUDT15 from heat-induced denaturation and precipitation, as indicated by an increase of 4 °C for the apparent aggregation temperature (Tagg) of NUDT15. Importantly, this significant stabilization strongly suggests TH7755 binds to NUDT15 in the cell (Fig. 1, D–F). In contrast, at the same concentration, the inhibitor TH1760 only increased the Tagg of NUDT15 by 1 °C. The potency of target engagement by TH7755 was further investigated using isothermal dose–response fingerprint CETSA (ITDRFCETSA), where intact HL-60 cells were treated with increasing concentrations of TH7755 before the thermal stability of cellular NUDT15 was determined at a constant screening temperature. TH7755 consistently engaged and thereby stabilized cellular NUDT15, which was observed to start at a concentration of 1 μM and increased with increasing TH7755 concentration (Fig. 1, G and H). Collectively, our CETSA data show that TH7755 engages efficiently with NUDT15 in HL-60 cells and to a larger extent compared with TH1760.
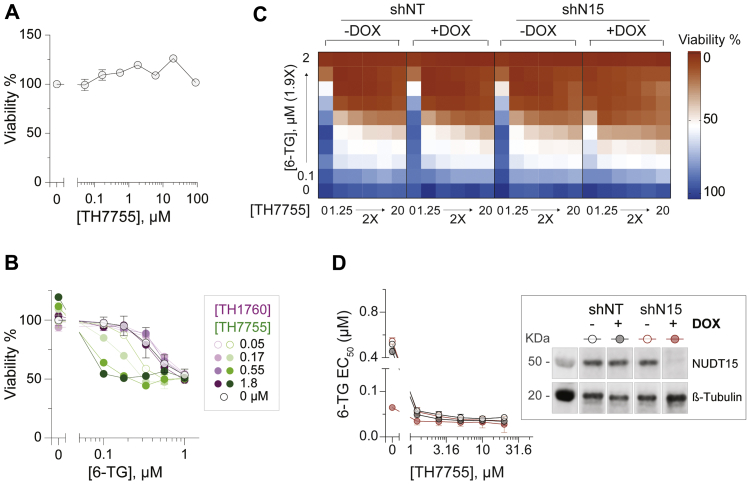
Loss of NUDT15 activity/expression has yet to be associated with an easily quantifiable phenotype. However, inhibition of NUDT15 6-thio-dGTPase activity leads to thiopurine hypersensitivity, which we previously exploited to analyze TH1760 (22). To further characterize TH7755, AML HL-60 cells were treated with a dose–response concentration matrix of thiopurine (6-TG) and TH7755 for 4 days, after which cell viabilities were determined using a resazurin reduction cell viability assay. In agreement with its biochemical inhibitory activity against NUDT15, TH7755 increased the cytotoxicity of 6-TG, as evidenced by a decrease in the 6-TG cytotoxic EC50 (half maximal effective concentration), starting from concentrations as low as 50 nM. Importantly, TH7755 did not demonstrate measurable cytotoxicity when applied alone up to a concentration of 100 μM (Fig. 2, A and B). In agreement with the CETSA data, showing that TH7755 displays an enhanced ability to engage cellular NUDT15 compared with TH1760 (Fig. 1, D–F), TH7755 also displays a far superior in-cell activity as measured by 6-TG potentiation. TH7755 was shown to strongly synergize with 6-TG in NUDT15-proficient AML NB4 cells, which was effectively abrogated upon shRNA-mediated NUDT15 depletion, indicating that TH7755 enhanced 6-TG efficacy through specifically inhibiting NUDT15 (Fig. 2, C and D).
Figure 2.
TH7755 is a cell-active NUDT15 inhibitor as evidenced by thioguanine potentiation.A, cell viability testing of TH7755 in AML HL-60 cells, showing no cytotoxicity up to 100 μM. HL-60 cells were treated with TH7755 for 4 days before cell viabilities were determined using a resazurin viability assay. Mean viabilities ±SD of a single experiment performed in duplicate are shown. B, TH7755 potentiated the antileukemic efficacy of thioguanine (6-TG) in a dose-dependent manner, starting from 0.05 μM. HL-60 cells were treated with a dose–response concentration matrix of 6-TG and NUDT15 inhibitor (TH7755 or TH1760) for 4 days before viabilities were determined. Mean viabilities ± SD of a single experiment performed in duplicate is shown. C and D, NUDT15 depletion abrogated TH7755-induced 6-TG potentiation in AML NB4 cells. NB4 cells expressing doxycycline-inducible shRNA constructs, NUDT15-specific (shN15) or control (shNT), were induced with doxycycline for 3 days before being treated with a dose–response concentration matrix of 6-TG and TH7755. C, mean viabilities of two experiments performed in duplicate are shown. D, Left panel, the cytotoxic EC50 of 6-TG estimated from the mean viabilities determined in C and calculated using a nonlinear curve fitting model with variable slope in GraphPad Prism. Right panel, knockdown efficiency of shN15 shRNA in NB4 cells at 120 h postdoxycycline treatment.
TH7755 is a competitive inhibitor of NUDT15
To investigate the mode of inhibition of TH7755, we produced enzyme saturation curves of NUDT15, with 6-thio-dGTP as substrate, at five different TH7755 concentrations and determined kcat and Km values (Fig. S1, Table S1). We analyzed the dependence of kcat and Km on TH7755 concentration and found that while the Km clearly increased with increasing TH7755 concentration (Fig. S1B), the kcat remained unaltered (Fig. S1C), indicative of a competitive mode of inhibition. The Ki for TH7755 was determined to be 10.2 ± 0.9 nM in agreement with our calculated IC50 value for the compound.
TH7755 recognition by WT NUDT15
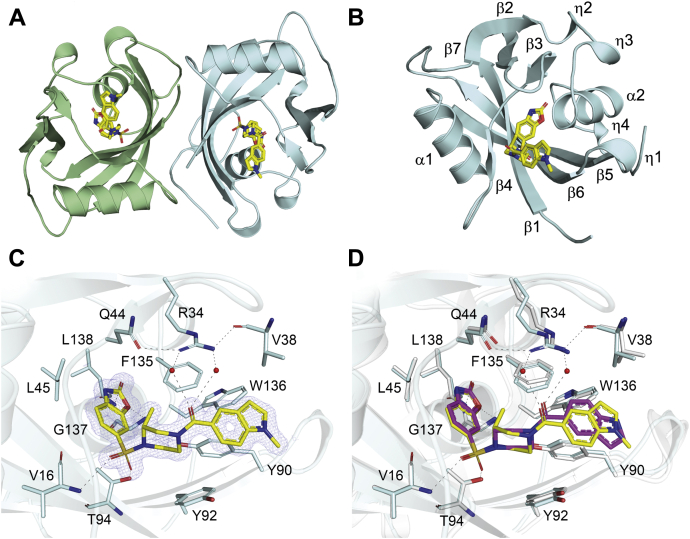
To analyze the inhibitory mechanism of TH7755, we determined the crystal structure of WT NUDT15 in complex with the inhibitor at a resolution of 1.6 Å. The overall structure of NUDT15 is a homodimer (Fig. 3A), consistent with size-exclusion chromatography and small-angle X-ray scattering experiments that show the enzyme is also dimeric in solution (3). The NUDT15 monomer is comprised of two alpha-helices (α1 and α2), seven beta-strands (β1–β7), and four 310 helices (η1–η4) (Fig. 3B). The individual monomers of NUDT15 have a classic NUDIX fold and are very similar to one another, as indicated by the low RMSD value of 0.47 Å for their corresponding Cα-atoms. Each active site contains clear density for a single TH7755 molecule, which is positioned deep within the binding pocket (Fig. 3C). TH7755 is coordinated by extensive hydrophobic and hydrogen bond interactions. The benzoxazolone moiety of TH7755 forms two direct hydrogen bonds with the main chain nitrogen atoms of Gly137 and Leu138 and is further supported by hydrophobic interactions with Leu45 and Phe135. The sulfonamide group of TH7755 is positioned by two interactions involving the main chain nitrogen of Val16 and the main chain oxygen of Thr94. The piperazine ring system of the compound is positioned exclusively by hydrophobic interactions from the residues Phe135, Tyr94, and His49. The piperazine ring has attained a chair conformation with the N-substituents in semi-equatorial orientations. This combined with its absolute R configuration forces the adjacent 2-methyl group on the piperazine into an axial orientation. The amide oxygen of TH7755 engages in water-mediated interactions with Arg34, which itself interacts with the side chain of Gln44 and the main chain oxygen of Val38. Finally, the indole group of the inhibitor is positioned by a perpendicular π-stacking interaction with Tyr90 and is further stabilized by hydrophobic interactions with Val38, Phe135, and Trp136 (Fig. 3C). Comparison of NUDT15-TH7755 with the previously reported NUDT15–TH1760 complex (22) shows that the overall structures superimpose very well, with a low RMSD value of 0.51 Å. There are no significant differences between the two structures with the exception of minor variations in two flexible loop regions. The overall binding mode of TH7755 is highly similar to TH1760, with the surrounding active site residues superimposing well between the structures (Fig. 3D). An interesting point of difference however is the indole ring system, which in TH7755 is flipped 180° relative to the TH1760 structure, likely a result of the additional methyl group present on the indole group of TH7755.
Figure 3.
Crystal structure of the NUDT15 TH7755 complex.A, crystallographic structure of dimeric NUDT15. Individual monomers are shown as ribbon representations, colored cyan and green. The TH7755 ligands are depicted as stick models; C atoms are colored yellow, O atoms red, N atoms blue, and S atoms gold. B, ribbon representation of the NUDT15 monomer in complex with TH7755. The secondary structure elements, alpha-helices (α1-2), beta-strands (β1-7), and 310-helices (η1-4) are labeled. C, hydrogen bond network for TH7755 in the NUDT15 active site. Amino acids involved in ligand coordination are depicted as sticks. Important water molecules are shown as red spheres. Hydrogen bond interactions are shown as dashed lines. The 2Fo − Fc electron density map around TH7755 is contoured at 1.5 σ, and the Fo − Fc electron density map is contoured at 3.0 σ. D, comparison of NUDT15–TH7755 (cyan) with NUDT15 in complex with TH1760 (white, PDB ID: 6T5J). TH1760 is depicted as a stick model; C atoms are colored magenta, O atoms red, N atoms blue, and S atoms gold.
TH7755 substantially increases the thermal stability of NUDT15 variants
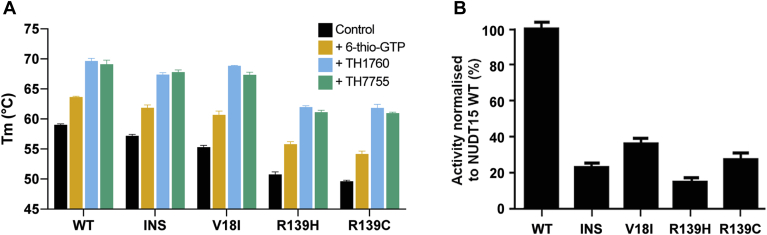
To determine whether inhibitor and substrate binding has a significant stabilizing effect on WT and clinically relevant NUDT15 variants (R139C, R139H, V18I and Val18_Val19insGlyVal), we obtained thermal stability data using the technique differential scanning fluorimetry. In the absence of substrate (6-thio-GTP) or inhibitor (TH7755), all four mutant constructs were less thermally stable compared with WT NUDT15, as indicated by their lower melting temperatures (Tm) (Fig. 4A). The R139C and R139H mutants had larger decreases in Tm compared with WT NUDT15 (−9.4 and −8.2 °C, respectively) compared with the V18I and Val18_Val19insGlyVal constructs (−3.7 and −1.8 °C, respectively). Addition of 6-thio-GTP resulted in a similar stabilization for all of the constructs, with an average increase in Tm of +5 °C (Fig. 4A). TH7755 inhibitor binding had an even more impressive stabilizing effect on the protein, with an average increase in Tm of +11 °C for WT and mutant NUDT15 (Fig. 4A). We also performed thermal stability tests with the previously published inhibitor TH1760, which also significantly stabilizes the enzyme, with an average Tm increase of +11.5 °C for WT and mutant NUDT15 (Fig. 4A).
Figure 4.
Thermal stability and activity analysis of WT NUDT15 and NUDT15 mutants. Differential scanning fluorimetry was used to determine the thermal stability of wildtype NUDT15 (WT) and the mutant NUDT15 constructs R139C, R139H, V18I, and V18_V19insGV (INS). A, melting temperatures (Tm) for each NUDT15 construct in the absence (black) or presence of either 6-Thio-GTP (yellow), TH7755 (green), or TH1760 (blue) are shown. The calculated Tm values represent mean values from three independent experiments ± SD, which were plotted in GraphPad Prism. B, activity of wildtype NUDT15 and NUDT15 mutants in the presence of 25 μM 6-thio-dGTP. Formed Pi was determined by addition of malachite green reagent followed by measurement of the absorbance at 630 nm. The activities of mutant NUDT15 constructs were normalized to the activity of wildtype NUDT15 and plotted in GraphPad Prism. Experiments were performed in triplicate.
NUDT15 mutants display lower enzymatic activity relative to WT
In addition to investigating the thermal stability, we also tested the enzymatic activity of the WT and mutant NUDT15 constructs in vitro. The R139C, R139H, V18I, and Val18_Val19insGlyVal mutants all displayed reduced enzymatic activity relative to WT NUDT15, with a reduction in activity ranging from 65 to 85% (Fig. 4B). The NUDT15 mutant R139H was least active, followed by Val18_Val19insGlyVal, R139C, and lastly, V18I.
Structure determination of clinically relevant NUDT15 mutants
Before this study, no structures of clinically relevant NUDT15 mutants had been determined, as their inherent instability rendered them difficult to crystallize. Previous studies aimed at elucidating the structural mechanisms that affect protein stability therefore had to draw conclusions based on WT NUDT15 structures, instead of using actual mutant NUDT15 structural data. By utilizing the impressive stabilizing effect of TH7755 on NUDT15, we were able to grow high-quality crystals and solve high-resolution crystal structures of the clinically relevant mutants R139C, R139H, V18I, and V18_V19insGV (Fig. 2, A–C). These NUDT15 variant structures are highly structurally similar to WT NUDT15-TH7755, as indicated by the low RMSD values for their corresponding Cα-atoms values of 0.47, 0.42, 0.45, and 0.44 Å, respectively. Comparison of these structures with WT NUDT15 in complex with TH7755 provides clear insights into the structural reasons for their reduced stability.
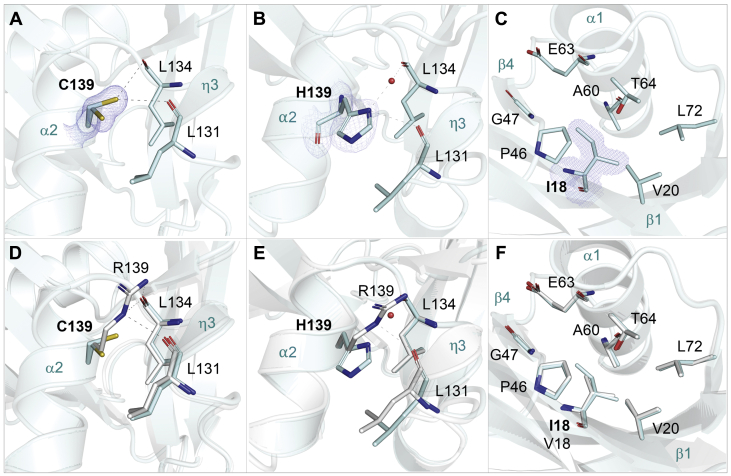
We first solved the structure of the most common pharmacogenetic variant in NUDT15, R139C, to 1.4 Å resolution. The side chain of Cys139 (located in α2) adopts two conformations, one of which interacts with the main chain oxygen atoms of both Leu131 (located in η3) and Leu134 (located in a loop connecting α2 to η3) at distances of 3.2 and 3.4 Å, respectively (Fig. 5A). In our structure of WT NUDT15 in complex with TH7755, Arg139 forms interactions similar to what is observed in the R139C structure. This involves a single interaction with the main chain oxygen of Leu131 (at a distance of 2.9 Å) and two interactions with the main chain oxygen of Leu134 (at distances of 2.9 and 3.2 Å, respectively). Comparison of the R139C mutant structure with WT NUDT15 shows that α2 superimposes very well between the structures. However, in WT NUDT15, the 310-helix η3 is shifted slightly closer to α2, than what is observed in the mutant structure (Fig. 5D).
Figure 5.
Crystal structures of the clinically relevant NUDT15 single-point mutations R139C, R139H, and V18I. The NUDT15 monomer of each mutant structure is shown as a cyan cartoon ribbon representation. The top panels show the hydrogen bond networks of (A) R139C, (B) R139H, and (C) V18I. The 2Fo − Fc electron density maps for the mutated amino acids are contoured at 1.5 σ, and the Fo − Fc electron density maps are contoured at 3.0 σ. Hydrogen bond interactions are shown as dashed lines. Amino acids are depicted as sticks; C atoms are colored cyan, O atoms red, N atoms blue, and S atoms gold. The lower panels (D–F) show superpositions of each NUDT15 mutant (cyan) with wildtype NUDT15 in complex with TH7755 (white). The relevant secondary structure elements, α2, β1, β4, and η3 are also highlighted.
We then determined the structure of another clinically relevant NUDT15 variant at position 139, the R139H, which we solved to 1.6 Å resolution. In contrast to WT and NUDT15 R139C where residue 139 is capable of interactions with both Leu131 and Leu134, the side chain of His139 only interacts with the main chain oxygen of Leu134, at a distance of 3.2 Å (Fig. 5B). As was observed for the R139C structure, comparison of R139H with WT NUDT15 shows α2 to superimpose very well between the structures, while the 310-helix η3 is positioned closer to α2 in the WT enzyme (Fig. 5E).
We then solved the structure of the V18I variant to 1.5 Å resolution. Val18 is close to the dimer interface of NUDT15 and lies on the opposite side of the monomer relative to position 139, which is solvent exposed. The V18 site is located in β1 and is in close proximity to residues from the conserved NUDIX motif, which itself is comprised of a small beta-sheet (β4) linked by a loop region to an alpha-helix (α1). In our V18I mutant structure, the side chain of Ile18 is surrounded by residues from α1 (Thr64, Ala60, and Glu63), β1 (Val20), β4 (Gly47), β5 (Leu72) as well as the loop region connecting β4 and β5 (Pro46) (Fig. 5C). Comparison of the V18I mutant structure with WT NUDT15 shows that the secondary structure in this region superimposes very well between the proteins. However, the additional methyl group present in the side chain of Ile18 slightly disrupts the favorable hydrophobic packing interactions with Pro46, Gly47, and Glu63, which are present in the WT structure with its smaller valine side chain (Fig. 5F).
Finally, we solved the structure of the NUDT15 V18_V19insGV mutant to a resolution of 1.5 Å. The V18_V19insGV mutation leads to an in-frame insertion of the residues glycine and valine into a segment on β1 that already contains three GlyVal repeats, thus extending it by one repetition (Fig. 6, A and B). While the residues Gly15 to Val20 in NUDT15 V18_V19insGV occupy the same position as Gly13 to Val18 in the WT structure, the insertion leads to the preceding residues being out of frame with one another (Fig. 6B). Comparison of the mutant structure with WT NUDT15 shows that β1 superimposes well between the structures. However, as mentioned above, the preceding sequence is out of frame, and the two structures therefore begin to diverge in direction of the N terminus (Fig. 6C). Unfortunately, as the residues at the very beginning of the N termini are disordered in both structures, a full analysis of the structural divergence in this area is not possible. An important difference between the proteins is the residue Val14 in V18_V19insGV NUDT15, which aligns with Pro12 in the WT enzyme (Fig. 6C). Furthermore, as the insertion results in the N-terminal sequences being out of alignment, Pro12 is actually positioned closer the N-terminal end in the NUDT15 V18_V19insGV structure, relative to WT NUDT15.
Figure 6.
Crystal structure of the clinically relevant NUDT15 insertion mutation V18_V19insGlyVal.A, crystal structure the NUDT15 V18_V19insGV dimer in complex with TH7755. Individual monomers are shown as ribbon representations, colored cyan and green. In each monomer, the glycine–valine repeats which include the insertion mutation are shown in orange. B, sequence comparison highlighting the difference between wildtype NUDT15 and NUDT15 V18_V19insGV. The Gly–Val repeats are shown in orange, and the additional Gly–Val repeat present in the mutant sequence is highlighted bold. The surrounding sequence, which is identical in both proteins, is shown in cyan. C, comparison of NUDT15-the NUDT15 V18_V19insGlyVal dimer (cyan and green) with wildtype NUDT15 in complex with TH7755 (white). The 2Fo − Fc electron density map for the Gly–Val repeats (shown as orange sticks) in NUDT15 V18_V19insGlyVal is contoured at 1.5 σ, and the Fo − Fc electron density map is contoured at 3.0 σ. The residues Gly13, Val14, Phe54, and His93 from NUDT15 V18_V19insGlyVal correspond to Arg11, Pro12, Phe52, and His91 in wildtype NUDT15.
Discussion
Thiopurine drugs are routinely used for treating cancer and inflammatory diseases (8, 9). Side-effects such as hematopoietic toxicity are common in patients treated with these drugs, often a consequence of loss of function mutations in genes encoding proteins that metabolize thiopurines such as TPMT (16). In recent years NUDT15 has also been shown to be a major player in thiopurine metabolism. Inactivating mutations in NUDT15 are strongly associated with thiopurine toxicity (19), and thiopurine dosage adjustment is recommended based on NUDT15 genotyping (20, 21). In addition to the role of NUDT15 in thiopurine hypersensitivity, other studies have suggested that thiopurine-based therapies in patients with WT NUDT15, can be improved by NUDT15 inhibition. Recently, we reported that a small molecule NUDT15 inhibitor (TH1760) sensitizes cells to thiopurines (22). A combination treatment might allow for lower thiopurine doses to be administered while still maintaining good therapeutic effect, potentially reducing toxic side-effects during treatment. To obtain a NUDT15 inhibitor with improved activity in cells, we developed the small-molecule NUDT15 inhibitor (TH7755) based on our previously developed inhibitor TH1760. We then utilized the excellent stabilizing properties of TH7755 to solve the structures of several clinically relevant NUDT15 mutants. TH7755 greatly stabilizes the active site of NUDT15, as evidenced by the fact that the WT NUDT15-TH7755 and NUDT15 variant-TH7755 complexes superimpose well in this region (Fig. S2).
The novel NUDT15 inhibitor TH7755 differs from TH1760 in that it contains two additional methyl groups, which are attached to the piperazine ring system and the nitrogen of the indole group. Our assay data indicated that TH7755 was a slightly weaker inhibitor in vitro than TH1760. However, TH7755 actually displayed far superior cellular potency compared with TH1760. As passive cell permeability by small molecules is usually favored by increased lipophilicity (23), the superior cellular effect of TH7755 may be because of its slight increase in lipophilicity (cLogP 2.05) relative to TH1760 (cLogP 1.41). Superposition of our WT NUDT15–TH7755 structure with the previously determined NUDT15–TH1760 complex (22) indicated that the inhibitors have almost identical binding modes. A notable difference, however, is the indole group of TH7755, which is flipped 180° relative to its position in the TH1760 structure. This is likely caused by the additional methyl group attached to the indole nitrogen of TH7755. Comparison of the two structures also provides insights into their slight differences in inhibition in vitro. While there is sufficient space in the binding pocket to accommodate both methyl groups of TH7755, their addition does bring the inhibitor into closer proximity with some ligand coordinating residues than what is observed in the TH1760 structure. First, the methyl group brings in the piperazine ring system into closer range of Phe135, and second, the methyl group bound to the indole nitrogen brings it closer to the main chain oxygen atoms of Glu88 and Asn89. The latter would present an unfavorable environment for a hydrophobic methyl group, which may explain why TH7755 displays slightly weaker NUDT15 inhibition than TH1760, which lacks a methyl at this position.
We also tested the inhibitory potency of TH7755 against mouse NUDT15 (mNUDT15), which is relevant because further development of the inhibitor would likely require testing in mice before human clinical trials. Interestingly, TH7755 was significantly less active against mouse NUDT15 compared with human NUDT15 (hNUDT15). As there is no structure of mouse NUDT15 available, we compared the protein sequences of mNUDT15 and hNUDT15 using Clustal Omega through the EBI webserver (24). Overall, the enzymes are highly similar, sharing 91% amino acid sequence identity. We analyzed the residues required for inhibitor binding in our hNUDT15–TH7755 structure and found two differences between the structures (Fig. S3). These differences are at positions Val38 and Gly137 in hNUDT15, which correspond to Phe37 and Ala136 in mNUDT15. In our human structure, Val38 provides a hydrophobic interaction with the indole group of TH7755, whereas Gly137 hydrogen bonds with the benzoxazolone moiety of the inhibitor through its main chain nitrogen atom. As these amino acid substitutions in mNUDT15 are bulkier and more hydrophobic, we propose this to be a contributing factor for the weaker inhibition of the mouse enzyme.
Before this study, there was no structural information for clinically relevant NUDT15 mutants as their lack of stability rendered them extremely difficult to crystallize. In the current study, we were able to use the impressive stabilizing effect of TH7755 to solve the structures of four clinically isolated NUDT15 variants (R139C, R139H, V18I and V18_V19insGlyVal). In the absence of inhibitor, our thermal stability data showed R139C to be the least stable variant, followed by R139H, V18I and finally Val18_Val19insGlyVal. This trend agrees with previously published thermal stability data by Moriyama et al. (7), who identified these four NUDT15 mutations in a cohort of 270 children diagnosed with ALL and treated with thiopurines. We also tested the enzymatic activity of the mutant NUDT15 constructs, all of which showed clearly lower enzymatic activity relative to the WT enzyme, with values ranging from 65% to 85%. The previously mentioned Moriyama et al. (7) study also showed large decreases in enzyme activity for all four mutants, ranging from 74 to 100% inactivity, relative to WT NUDT15. Interestingly, in that study, the R139C mutant was reported to be completely inactive, which contrasts with a different study by Valerie et al. (4) showing the activity of R139C to be comparable to WT NUDT15. However, it should be noted that the Valerie et al. study did show NUDT15 R139C to be highly unstable, which agrees with our results and those of Moriyama et al. The discrepancies in enzymatic activity may in part result from the choice of buffers used during protein purification and enzymatic assays, which can have a profound effect on protein stability and activity.
Analysis of our NUDT15 mutant structures in relation to the WT enzyme allowed us to assess the underlying structural reasons for their diminished stability. These structures also highlight the importance of analyzing experimentally determined structures when drawing conclusions about the structural effect a mutation has on an enzyme. It was previously proposed on the basis of the WT NUDT15 structure that the R139C mutation might lead to the formation of a disulphide bond between the mutated residue and the neighboring residue Cys140 (3). This would have resulted in the distortion of an α-helix (α2), which could explain the instability of the R139C mutant. In our structure, the sulfhydryl group of Cys139 points away from C140, and no disulphide bond is formed. Instead, the side chain has a dual conformation, one of which interacts with the main chain of Leu131 (η3) and Leu134 (loop connecting α2 to η3), similar to what was observed in our R139H structure, where the side chain of His139 also interacts with the main chain of Leu134 (but not Leu131). In our WT NUDT15 structure, Arg139 also interacts with the main chain oxygen atoms of Leu131 and Leu134; however, the bond distances are shorter than what is observed in the R139C and R139H mutants. This results in η3 being pulled closer to α2 in the WT structure. Clearly, the role of the arginine at this position is to stabilize the position of η3 as well as the loop which connects it to α2. This region of the protein is flexible and solvent exposed, explaining why mutations here are so destabilizing. Based on these structures, we believe that while these mutants are still capable of interacting with Leu131 and Leu134, they are unable to interact with these residues as tightly as Arg139, which has a more electronegative side chain because of its three nitrogen atoms. In addition, the sulfur atom in the side chain of Cys139 is less electronegative than the single nitrogen atom from His139, making it less stabilizing and potentially explaining why the R139C mutant is less thermally stable than R139H.
Analysis of our V18I and Val18_Val19insGlyVal structures also provided insights into their reduced stability and activity compared with WT NUDT15. In the case of the V18I mutant, the additional methyl group in the side chain of Ile18 results in less favorable hydrophobic interactions with Pro46 (loop region connecting β4 and β5), Gly47 (β4), and Glu63 (α1) compared with the WT enzyme with its smaller valine side chain. This could result in slight destabilization of the NUDIX motif, which itself is comprised of a small beta-sheet (β4) linked by a loop region to an alpha-helix (α1). This highly important motif contains residues responsible for the coordination of magnesium and water molecules involved in substrate hydrolysis (1, 3), explaining why the V18I mutation negatively affects both NUDT15 stability and enzyme activity.
With regard to our Val18_Val19insGlyVal structure, the Gly–Val insertion results in the first 12 residues of the protein sequence being out of frame with the WT enzyme, meaning that the two structures begin to physically diverge in direction of the N terminus. A significant difference in the mutant structure is Val14, which aligns with Pro12 in the WT NUDT15. Pro12 (note that both proteins have the same numbering for this residue) is positioned closer to the N-terminal end in the NUDT15 V18_V19insGV structure. The unique structural properties of proline strongly influence the protein main chain geometry and protein stability. Furthermore, in WT NUDT15, Pro12 forms a CH/π interaction with His91 (located on β6), and it is also within range of a hydrophobic interaction with Phe52, which is located in a loop between β4 and α1 from the second monomer. The valine side chain in the mutant structure is not capable of a CH/π interaction with its neighboring histidine residue. Interference with β6 would likely have a destabilizing effect because of its importance for stabilizing the NUDT15 core, whereas introducing strain to β4 would interfere with residues of the NUDIX motif and therefore negatively impact enzyme activity. As mentioned earlier, while the V18I and Val18_Val19insGlyVal proteins are less stable than WT NUDT15, they are still significantly more stable than the R139C and R139H mutants. This may be because of the fact these mutations are located closer to the dimer interface (which stabilizes NUDT15 in solution and these mutations would be unlikely to disrupt it), whereas the variants at position 139 are needed to stabilize a 310-helix (in a flexible solvent exposed region), which is highly sensitive to mutations. However, it should also be noted that the V18I mutation presented here is a relatively conservative mutation and mutations to something much larger (i.e., tryptophan) or significantly charged (i.e., arginine) have previously been shown to be severely destabilizing (21).
In conclusion, we present the development of TH7755, a potent NUDT15 inhibitor with improved cellular potency, which allowed us to determine the structures of four clinically relevant NUDT15 mutants. Analysis of this structural information provided explanations for the detrimental phenotypic effects observed for these variants. TH7755 will provide a valuable tool for future studies of clinically relevant loss-of-function NUDT15 variants and has the potential to be useful in a clinical setting because of its ability to increase the effectiveness of thiopurine therapy.
Experimental procedures
NUDT15 purification
WT NUDT15 was expressed and purified as previously published (3). The NUDT15 mutants R139C, R139H, V18I, and V18_V19insGV were produced, as described by Moriyama et al. (7). Briefly, human NUDT15 cDNA was cloned into a pCold II expression vector with an N-terminal His tag. The NUDT15 mutations analyzed in this study were then introduced into this vector via site-directed mutagenesis. All constructs were expressed in E. coli BL21 cells and purified to homogeneity by immobilized metal affinity chromatography followed by size-exclusion chromatography. Purified proteins were concentrated in sample buffer (10 mM Tris-HCl, 0.1 mM EDTA, 5 mM DTT, 10% (v/v) glycerol pH 8.0) after which the buffer was exchanged to the final storage buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, and 1 mM TCEP). The samples were then flash frozen in liquid nitrogen, and stored at −80 °C.
Crystallization and data collection
All proteins (20 mg/ml) were crystallized via hanging drop vapor diffusion at 21 °C in 0.1 M Tris pH 8.0, 0.2 M sodium acetate, 38% PEG 4000 (WT NUDT15), 0.12 M monosaccharide mix (0.2 M D-Glucose, 0.2 M D-Mannose, 0.2 M D-Galactose, 0.2 M L-Fucose, 0.2 M D-Xylose, 0.2 M N-Acetyl-D-Glucosamine), 0.1 M imidazole/MES pH 6,5, 12.5% MPD, 12.5% PEG 1000, 12.5% PEG 3350 (R139C), 0.1 M MMT pH 5.0, 25.4% PEG 1500 (R139H), 0.12 M monosaccharide mix, 0.1 M imidazole/MES pH 6.5, 20% PEG 500 MME, 20% PEG 2000 (V18I), and 0.09 M SPG pH 5.0, 22.25% PEG 1500, 0.01 M sodium/potassium phosphate, 2.75% 1,2 propandiol, 1.0% glycerol (V18_V19insGV). The R139H crystals were soaked in cryoprotectant solution containing 0.1 M MMT pH 5.0, 30% PEG 1500, 20% ethylene glycol, 2 mM TH7755 for 10 min, and then flash frozen in liquid nitrogen. Diffraction quality crystals appeared after 1 week.
X-ray diffraction data were collected on the BioMAX beamline at MAXIV and the I24 beamline of the Diamond Light Source. The datasets were collected at 100 K at a wavelength of either 0.976 Å (NUDT15 R139C, R139H and V18I), 0.918 Å (WT NUDT15) or 0.968 Å (NUDT15 V18_V19insGV). Single crystals were used for data collection, with the exception of the R139H structure where data from two crystals were collected and later merged.
Structure determination and refinement
All datasets were indexed and integrated using DIALS (v3.2.2) (25), followed by scaling using AIMLESS (26) within the CCP4 suite (27). Structures were solved via molecular replacement with Phaser (28) using the monomer of a WT NUDT15 structure (PDB ID: 5LPG) as the search model. Translation libaration screw parameters were determined using the TLSMD webserver (29). Several rounds of manual model building and refinement were performed using Coot (30), phenix.refine (31) and REFMAC (32) during which waters and ligands were incorporated into the structure. Data processing and refinement statistics are presented in Table 1. The coordinates and structure factors for WT NUDT15-TH7755 and the mutant structures V18I, R139C, R139H, and V18_V19insGV were deposited in the PDB under the codes 7B63, 7B64, 7B65, 7B66, and 7B67, respectively.
Table 1.
Data collection and refinement statistics
| NUDT15 | WT | V18I | R139C | R139H | V18_V19insGV |
|---|---|---|---|---|---|
| Data collection | |||||
| Synchrotron and beamline | MAXIV BioMAX | MAXIV BioMAX | MAXIV BioMAX | MAXIV BioMAX | DLS I24 |
| Space group | P 1 21 1 | C 1 2 1 | P 1 | P 1 | P 1 21 1 |
| Cell dimensions | |||||
| a,b,c (Å) | 63.6, 45.6, 67.0 | 115.5, 34.6, 75,2 | 34.9, 43.8, 60.4 | 41.3, 60.2, 69.3 | 63.4, 45.7, 66.3 |
| α, β, γ (°) | 90, 116, 90 | 90.0, 148.6, 90.0 | 91.6, 106.7, 113.48 | 100.1, 104.1, 90.3 | 90, 115.1, 90 |
| Resolution (Å) | 60.24–1.60 (1.63–1.60)a | 39.22–1.50 (1.53–1.50)a | 35.37–1.60 (1.63–1.60)a | 40.59–1.60 (1.63–1.60)a | 60.07–1.45 (1.47–1.45) |
| Rmeas | 0.064 (0.721)a | 0.119 (0.886)a | 0.059 (0.148)a | 0.060 (0.305)a | 0.086 (1.458)a |
| Rpim | 0.034 (0.385)a | 0.062 (0.472)a | 0.042 (0.105)a | 0.042 (0.216)a | 0.021 (0.362)1 |
| CC1/2 | 0.999 (0.862)a | 0.994 (0.949)a | 0.997 (0.985)a | 0.998 (0.976)a | 1.000 (0.861)a |
| <I/σ(I)> | 15.1 (2.4)a | 8.6 (1.8)a | 16.2 (7.6)a | 11.5 (2.7)a | 24.7 (3.1)a |
| Total observations | 310,958 (15,401)a | 166,790 (8167)a | 140,950 (6.909)a | 289,021 (14,047)a | 1,931,455 (93,528)a |
| Unique observations | 45,919 (2312)a | 25,129 (1235)a | 39,574 (1931)a | 81,339 (3976)a | 61,265 (3026)a |
| Completeness (%) | 100 (100)a | 100 (100)a | 96.6 (95.2)a | 96.8 (95.3)a | 100 (100)a |
| Redundancy | 6.8 (6.7)a | 6.6 (6.6)a | 3.6 (3.6)a | 3.6 (3.5)a | 31.5 (30.9)a |
| Refinement | |||||
| Rwork/Rfree | 0.167/0.196 | 0.180/0.213 | 0.160/0.184 | 0.185/0.220 | 0.167/0.197 |
| B-factors (A2): | |||||
| Protein | 25.5 | 42.3 | 23.1 | 33.1 | 21.4 |
| Solvent | 37.6 | 51.8 | 34.8 | 27.2 | 35.6 |
| Ligand | 20.2 | 39.8 | 24.0 | 39.0 | 17.5 |
| R.m.s.d. bond lengths (Å) | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| R.m.s.d. bond angles (°) | 1.82 | 1.74 | 1.89 | 1.70 | 1.86 |
| Ramachandran statistics: | |||||
| Favored (%) | 98.1 | 96.7 | 98.7 | 97.6 | 98.71 |
| Outliers (%) | 0 | 0 | 0 | 0 | 0.32 |
| PDB ID | 7B63 | 7B64 | 7B65 | 7B66 | 7B67 |
Values in parentheses are for the highest-resolution shell.
Thermal stability tests for WT NUDT15 and NUDT15 mutants
The stability of WT NUDT15 and the R139C, R139H, V18I, and V18_V19insGV variants was examined using differential scanning fluorimetry (33). Each assay condition contained 10 μM protein in sample buffer (20 mM HEPES, 300 mM NaCl, 10% glycerol, 5 mM MgCl2 pH 7.5) with 10X SYPRO Orange dye (Thermo Fisher Scientific), and either 1 mM 6-thio-GTP, 0.1 mM TH7755, or a buffer only control. Experiments were performed using a StepOne real time PCR machine (Applied Biosystems) where the temperature was increased from 25 to 95 °C at a rate of 1 °C/min. The melting temperatures (Tm) were then determined in the Applied Biosystems Protein Thermal Shift Software using the Boltzmann model. Data were collected from three separate experiments and plotted using GraphPad Prism software.
Cellular thermal shift assay
CETSA and ITDRFCETSA were performed as described previously (22, 34). HL-60 cells were treated with putative NUDT15 inhibitors or a DMSO only control for 3 h at 37 °C and 5% CO2 in a humidified incubator, after which cells were harvested, washed by PBS, and then resuspended in TBS supplemented with protease inhibitors. Cell suspensions were then heated at indicated temperatures for 3 min in a Veriti 96-well Thermal Cycler (ABI), followed by cell lysis with three freeze-thaw cycles. Cell lysates were then clarified by centrifugation and prepared for SDS-PAGE and Western blot analysis. For ITDRFCETSA HL-60 cells were transduced with a pInducer20 vector containing 3XHA-tagged WT NUDT15. Cells were induced with 1 μg/ml doxycycline overnight before compound exposure, followed by heating and lysis in the same manner as in the CETSA experiments. Proteins were separated by SDS-PAGE using 4 to 15% Mini-PROTEAN TGX gels and transferred to nitrocellulose membranes. Membranes were then incubated with Odyssey Blocking Buffer (LI-COR) and probed with primary antibodies (anti-NUDT15 antibody, cat. no. sc-84533, Santa Cruz; anti-SOD1 antibody, cat. no. sc-11407, Santa Cruz) followed by species-appropriate secondary antibodies. Protein bands were visualized with an Odyssey Fc Imager, and signal intensities were analyzed using Image Studio Software (Li-Cor Biosciences).
Cell viability assays
Cell viabilities were assayed using the resazurin reduction cell viability assay, as previously described (22). Briefly, AML cell lines HL-60 or NB4 were seeded in 384-well assay plates at 50,000/ml and then treated with various concentrations of 6-TG and/or NUDT15 inhibitors. For NB4 cells conditionally expressing NUDT15-specific or control shRNAs (22), cells were induced with doxycycline for 3 days before seeding. Four days posttreatment, resazurin sodium salt (10 μg/ml) was added to cells for 4 to 6 h, before resazurin reduction was assessed via fluorescence intensity at 544/590 nm (Ex/Em) using a HidexSense plate reader (Hidex). Medium-only and cell-only wells served as negative (0% cell viability) and positive (100% cell viability) controls, respectively. Cytotoxicity EC50 values were estimated by fitting cell viabilities via a nonlinear curve fitting model with variable slope in GraphPad Prism.
Synthesis of TH7755
(R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one (6) was synthesized according to Scheme 1, which is shown in Fig. S4.
Step1: Synthesis of (R)-tert-butyl 3-methyl-4-((2-oxo-2,3-dihydrobenzo[d]oxazol-6-yl)sulfonyl)piperazine-1-carboxylate (3).
A solution of 2-oxo-2,3-dihydrobenzo[d]oxazole-6-sulfonyl chloride (1) (1.0 g; 4.28 mmol) and triethylamine (0.90 ml, 5.42 mmol) in dichloromethane (50 ml) was treated with tert-butyl piperazine-1-carboxylate (2) (0.86 g, 4.28 mmol) and stirred at room temperature overnight. The mixture was diluted with dichloromethane (10 ml) and washed with a saturated solution of sodium bicarbonate (15 ml). The organic phase was dried over anhydrous magnesium sulphate then evaporated to afford the product (3) as a colorless solid which was used without further purification.
Step 2: Synthesis of (R)-3-methyl-4-((2-oxo-2,3-dihydrobenzo[d]oxazol-6-yl)sulfonyl)piperazine hydrochloride (4).
A solution of (R)-tert-butyl 3-methyl-4-((2-oxo-2,3-dihydrobenzo[d]oxazol-6-yl)sulfonyl)piperazine-1-carboxylate (3) in dioxane (10 ml) was treated with a solution of 4 M HCl in dioxane (10 ml) and stirred at room temperature overnight. The mixture was filtered, and the colorless solid was washed with a little dioxane, then dried overnight. m/z 298 (M + H)+. The hydrochloride salt (4) was used directly.
Step 3: Synthesis of (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one (6).
A solution of (R)-3-methyl-4-((2-oxo-2,3-dihydrobenzo[d]oxazol-6-yl)sulfonyl)piperazine hydrochloride (4) (1.40 g; 4.20 mmol), 1-methyl-1H-indole-5-carboxylic acid (5) (0.74 g; 4.20 mmol), and triethylamine (1.76 ml, 12.6 mmol) in dichloromethane (50 ml) was treated with propylphosphonic anhydride (T3P) (1.50 ml; 5.04 mmol) and stirred at room temperature for 48 h. The reaction mixture was washed with saturated aqueous sodium bicarbonate (10 ml), then 1 M aqueous hydrochloric acid (10 ml). The organic extract was dried over anhydrous magnesium sulphate then evaporated to give the crude product, which was purified by column chromatography over silica (DCM-MeOH; 0–6%) to give the title compound (6) (280 mg; 15%). 1H NMR (400 MHz, DMSO) δ 7.73 (d, J = 1.6 Hz, 1H), 7.64 (dd, J = 8.2, 1.8 Hz, 1H), 7.57 (s, 1H), 7.46 (d, J = 8.5 Hz, 1H), 7.41 (d, J = 3.1 Hz, 1H), 7.27 (d, J = 8.2 Hz, 1H), 7.14 (dd, J = 8.5, 1.4 Hz, 1H), 6.48 (dd, J = 3.1, 0.8 Hz, 1H), 4.18 to 3.96 (m, 2H), 3.81 (s, 3H), 3.64 (d, J = 13.1 Hz, 1H), 3.22 to 3.12 (m, 1H), 3.12 to 2.98 (m, 1H), 2.98 to 2.84 (m, 1H), 0.95 (s, 3H). One of the piperazine CH’s is obscured by DMSO. m/z 455 [M + H]+.
Determination of IC50 values of human and mouse NUDT15 with TH7755
Activity of human and mouse NUDT15 (4.75 nM) was determined at 22 °C in assay buffer (Tris-acetate pH 8.0, 40 mM NaCl, 10 mM Magnesium-acetate, 1 mM DTT, 0.005% Tween20, 0.2 U/ml PPase) with dGTP (100 μM) as substrate in the presence of serially diluted TH7755 resulting in final concentrations ranging from 0 to 10 μM. Duplicate samples were used for each inhibitor concentration. Pi formed from the reaction product PPi after hydrolysis by PPase was determined by addition of a malachite green–based reagent (35) followed by measurement of the absorbance at 630 nm using a Hidex plate reader. Controls without NUDT15 were included on the plate to determine the background signal. Background signal was subtracted, and % activity was then calculated. IC50 values were determined by fitting the equation for “log inhibitor versus response-variable slope (four parameters)” to the inhibition data using nonlinear regression in GraphPad Prism. Each experiment was performed twice.
Comparison of activity of WT NUDT15 and NUDT15 mutants
Activity of NUDT15WT (20 nM) and NUDT15 mutants (20 nM) was assayed with 25 μM 6-thio-dGTP in assay buffer (Tris-acetate pH 7.5, 40 mM NaCl, 10 mM magnesium-acetate, 1 mM DTT, 0.2 U/ml PPase). After incubation with shaking at 22 °C for 20 min, the Pi produced from the reaction product PPi was detected using a malachite green reagent (35) followed by measurement of the absorbance at 630 nm. Activities were assayed in triplicate. Controls lacking NUDT15 enzyme were included on the plate, and the background signal was subtracted. Activities were normalized to the activity of NUDT15WT and plotted in GraphPad Prism.
Determination of Ki value and mode of inhibition
Initial rates of NUDT15 (2 nM) were determined in assay buffer (0.1 M Tris acetate pH 8.0, 40 mM NaCl, 10 mM magnesium acetate, 1 mM DTT, 0.005% Tween20) over 6-thio-dGTP concentrations ranging from 0 to 133 μM in the absence or presence of 15, 45, 135, and 405 nM TH7755. Each data point was performed in duplicate, and the experiment was performed twice. PPi produced from 6-thio-dGTP hydrolysis was detected using PPi Light Inorganic Pyrophosphate Assay (Lonza), and the signal was converted to PPi concentration using a PPi standard curve. Initial rates were plotted against 6-thio-dGTP concentration, and the Michaelis-Menten equation was fitted to the initial rates using GraphPad Prism, and the kcat and Km values of NUDT15 at the different inhibitor concentrations were determined. To determine the inhibition constant (Ki), the model for competitive inhibition was fitted to the initial rates determined at different 6-thio-dGTP and TH7755 concentrations using GraphPad Prism.
Data availability
The protein structures presented in this article have been deposited in the Protein Data Bank (PDB) under the accession codes 7B63, 7B64, 7B65, 7B66, and 7B67. All remaining data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no competing financial interests.
Acknowledgments
We thank MAXIV Laboratory (Sweden, proposal MX20190233), Diamond Light Source (United Kingdom, proposal mx21625), and their beamline scientists from the BioMAX and I24 beamlines for their support in data collection. Protein production of WT NUDT15 was facilitated by the Protein Science Facility at Karolinska Institutet/SciLifeLab.
Author contributions
P. S., T. H., and J. Y. conceived the project and supervised the study. D. R., J. D., E. S., and P. S. designed, performed, and analyzed structural biology experiments. S. M. Z. designed, performed, and analyzed all biological experiments. A.-S. J., D. R., and E. S. designed, performed, and analyzed biochemical and biophysical experiments. T. M. and R. N. purified and analyzed proteins. T. K., A. T., O. W., M. D., E. H., and M. S. designed, performed, and analyzed medicinal chemistry experiments. T. H., S. G. R., and P. S. funding acquisition. P. S., T. H., U. W., E. H., and J. Y. supervised the study. E. S., D. R., and P. S. prepared the manuscript. All authors discussed results and approved the manuscript.
Funding and additional information
This project was supported by the Swedish Research Council (P. S. 2018-03406, T. H. 2015-00162, S. G. R. 2018-02114), the European Research Council (TAROX-695376, T. H.), Swedish Cancer Society (201287 PjF, P. S.; CAN2018/0658, T. H.; 19-0056-JI, S. G. R). The Knut and Alice Wallenberg Foundation (KAW2014.0273, T. H.; P. S.), the Swedish Children’s Cancer Foundation (PR2018-0095, T. H.; PR2019-0014, S. G. R.), the Swedish Pain Relief Foundation (SSF/01-05, T. H.), the Crafoord Foundation (P. S.), the Torsten and Ragnar Söderberg Foundation (T. H.), and the Felix Mindus contribution to Leukemia Research (2019-02004 and 2020-02573, S. M. Z.).
Edited by Wolfgang Peti
Contributor Information
Thomas Helleday, Email: thomas.helleday@scilifelab.se.
Pål Stenmark, Email: stenmark@dbb.su.se.
Supporting information
References
- 1.Bessman M.J., Frick D.N., O'Handley S.F. The MutT proteins or “nudix” hydrolases, a family of versatile, widely distributed,“housecleaning” enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 2.Gad H., Koolmeister T., Jemth A.-S., Eshtad S., Jacques S.A., Ström C.E., Svensson L.M., Schultz N., Lundbäck T., Einarsdottir B.O. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 3.Carter M., Jemth A.-S., Hagenkort A., Page B.D., Gustafsson R., Griese J.J., Gad H., Valerie N.C., Desroses M., Boström J. Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nat. Commun. 2015;6:7871. doi: 10.1038/ncomms8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valerie N.C., Hagenkort A., Page B.D., Masuyer G., Rehling D., Carter M., Bevc L., Herr P., Homan E., Sheppard N.G., Stenmark P., Jemth A.S., Helleday T. NUDT15 hydrolyzes 6-thio-DeoxyGTP to mediate the anticancer efficacy of 6-thioguanine. Cancer Res. 2016;76:5501–5511. doi: 10.1158/0008-5472.CAN-16-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang S.K., Hong M., Baek J., Choi H., Zhao W., Jung Y., Haritunians T., Ye B.D., Kim K.J., Park S.H., Park S.K., Yang D.H., Dubinsky M., Lee I., McGovern D.P. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat. Genet. 2014;46:1017–1020. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J.J., Landier W., Yang W., Liu C., Hageman L., Cheng C., Pei D., Chen Y., Crews K.R., Kornegay N., Wong F.L., Evans W.E., Pui C.H., Bhatia S., Relling M.V. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. 2015;33:1235–1242. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriyama T., Nishii R., Perez-Andreu V., Yang W., Klussmann F.A., Zhao X., Lin T.N., Hoshitsuki K., Nersting J., Kihira K., Hofmann U., Komada Y., Kato M., McCorkle R., Li L. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 2016;48:367–373. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford K., Shih D.Q. Optimizing 6-mercaptopurine and azathioprine therapy in the management of inflammatory bowel disease. World J. Gastroenterol. 2011;17:4166–4173. doi: 10.3748/wjg.v17.i37.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmiegelow K., Nielsen S.N., Frandsen T.L., Nersting J. Mercaptopurine/methotrexate maintenance therapy of childhood acute lymphoblastic leukemia: Clinical facts and fiction. J. Pediatr. Hematol. Oncol. 2014;36:503–517. doi: 10.1097/MPH.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson J.A., Carpenter J.W., Rose L.M., Adamson D.J. Mechanisms of action of 6-thioguanine, 6-mercaptopurine, and 8-azaguanine. Cancer Res. 1975;35:2872–2878. [PubMed] [Google Scholar]
- 11.Ling Y.H., Nelson J.A., Cheng Y.C., Anderson R.S., Beattie K.L. 2'-Deoxy-6-thioguanosine 5'-triphosphate as a substrate for purified human DNA polymerases and calf thymus terminal deoxynucleotidyltransferase in vitro. Mol. Pharmacol. 1991;40:508–514. [PubMed] [Google Scholar]
- 12.Warren D.J., Andersen A., Slordal L. Quantitation of 6-thioguanine residues in peripheral blood leukocyte DNA obtained from patients receiving 6-mercaptopurine-based maintenance therapy. Cancer Res. 1995;55:1670–1674. [PubMed] [Google Scholar]
- 13.Swann P.F., Waters T.R., Moulton D.C., Xu Y.Z., Zheng Q., Edwards M., Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 14.Li G.M. The role of mismatch repair in DNA damage-induced apoptosis. Oncol. Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 15.Lennard L., Lilleyman J.S., Van Loon J., Weinshilboum R.M. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 16.Relling M.V., Hancock M.L., Rivera G.K., Sandlund J.T., Ribeiro R.C., Krynetski E.Y., Pui C.H., Evans W.E. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J. Natl. Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 17.Regueiro M., Mardini H. Determination of thiopurine methyltransferase genotype or phenotype optimizes initial dosing of azathioprine for the treatment of Crohn's disease. J. Clin. Gastroenterol. 2002;35:240–244. doi: 10.1097/00004836-200209000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Gardiner S.J., Gearry R.B., Begg E.J., Zhang M., Barclay M.L. Thiopurine dose in intermediate and normal metabolizers of thiopurine methyltransferase may differ three-fold. Clin. Gastroenterol. Hepatol. 2008;6:654–660. doi: 10.1016/j.cgh.2008.02.032. quiz 604. [DOI] [PubMed] [Google Scholar]
- 19.Fotoohi A.K., Coulthard S.A., Albertioni F. Thiopurines: Factors influencing toxicity and response. Biochem. Pharmacol. 2010;79:1211–1220. doi: 10.1016/j.bcp.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Relling M.V., Schwab M., Whirl-Carrillo M., Suarez-Kurtz G., Pui C.H., Stein C.M., Moyer A.M., Evans W.E., Klein T.E., Antillon-Klussmann F.G., Caudle K.E., Kato M., Yeoh A.E.J., Schmiegelow K., Yang J.J. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin. Pharmacol. Ther. 2019;105:1095–1105. doi: 10.1002/cpt.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suiter C.C., Moriyama T., Matreyek K.A., Yang W., Scaletti E.R., Nishii R., Yang W., Hoshitsuki K., Singh M., Trehan A., Parish C., Smith C., Li L., Bhojwani D., Yuen L.Y.P. Massively parallel variant characterization identifies NUDT15 alleles associated with thiopurine toxicity. Proc. Natl. Acad. Sci. U. S. A. 2020;117:5394–5401. doi: 10.1073/pnas.1915680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S.M., Desroses M., Hagenkort A., Valerie N.C.K., Rehling D., Carter M., Wallner O., Koolmeister T., Throup A., Jemth A.S., Almlof I., Loseva O., Lundback T., Axelsson H., Regmi S. Development of a chemical probe against NUDT15. Nat. Chem. Biol. 2020;16:1120–1128. doi: 10.1038/s41589-020-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R., Qin X., Kong F., Chen P., Pan G. Improving cellular uptake of therapeutic entities through interaction with components of cell membrane. Drug Deliv. 2019;26:328–342. doi: 10.1080/10717544.2019.1582730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chojnacki S., Cowley A., Lee J., Foix A., Lopez R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 2017;45:W550–W553. doi: 10.1093/nar/gkx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beilsten-Edmands J., Winter G., Gildea R., Parkhurst J., Waterman D., Evans G. Scaling diffraction data in the DIALS software package: Algorithms and new approaches for multi-crystal scaling. Acta Crystallogr. D Struct. Biol. 2020;76:385–399. doi: 10.1107/S2059798320003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mccoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Painter J., Merritt E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovalevskiy O., Nicholls R.A., Long F., Carlon A., Murshudov G.N. Overview of refinement procedures within REFMAC5: Utilizing data from different sources. Acta Crystallogr. D Struct. Biol. 2018;74:215–227. doi: 10.1107/S2059798318000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 34.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 35.Baykov A.A., Evtushenko O.A., Avaeva S.M. A malachite green procedure for ortho-phosphate determination and its use in alkaline phosphatase-based enzyme-immunoassay. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein structures presented in this article have been deposited in the Protein Data Bank (PDB) under the accession codes 7B63, 7B64, 7B65, 7B66, and 7B67. All remaining data are contained within the article.