Abstract
Colorectal cancer (CRC) is one of the most common and deadly cancers, and the incidence of CRC is on the rise. Due to the lack of early diagnosis method and high metastasis of the disease, the prognosis of CRC remains very poor. Exploring the underlying molecular mechanisms of CRC is very necessary for effective therapy. In this study, we investigated the function of circBANP in CRC. The results showed that circBANP was elevated in both CRC tissues and cells and its level positively correlated with the stage of CRC. Knockdown of circBANP greatly suppressed the epithelial-mesenchymal transition (EMT) process and CRC cell proliferation, migration, and invasion. In addition, knockdown of circBANP inhibited CRC tumor growth and metastasis in vivo. Further, circBANP directly bound to let-7d-5p and regulated CRC development via acting as a let-7d-5p sponge. Let-7d-5p directly targeted HMGA1 and thus circBANP/let-7d-5p regulated Wnt/β-catenin signaling via HMGA1. Collectively, circBANP promotes CRC development and metastasis via acting as a let-7d-5p sponge to regulate HMGA1/Wnt/β-catenin signaling, providing a potential biomarker and therapeutic target for the management of CRC.
Keywords: circBANP, HMGA1, EMT, CRC, let-7d-5p, Wnt/β-catenin
Graphical abstract
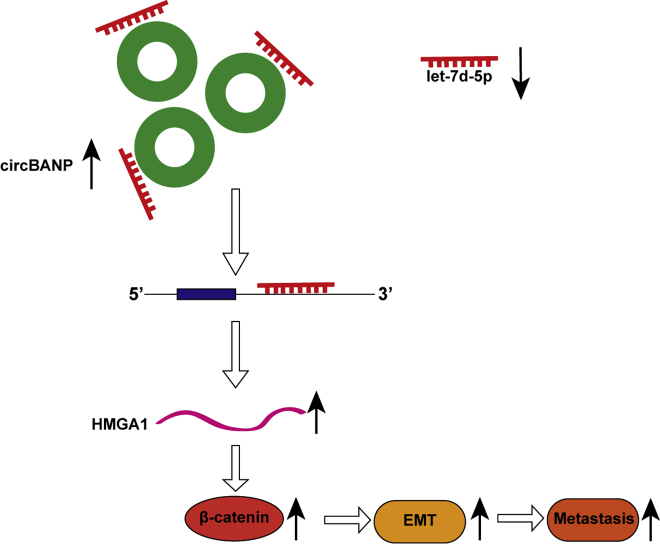
This study reveals that circBANP promotes colorectal cancer development and metastasis via acting as a let-7d-5p sponge to regulate HMGA1/Wnt/β-catenin signaling, providing a potential biomarker and therapeutic target for the management of colorectal cancer.
Introduction
Colorectal cancer (CRC) is one of the most prevalent and aggressive malignancies in the world, ranking as the third deadliest of all cancers and the fourth most commonly diagnosed cancer.1,2 What is worse, the past decades have witnessed a steady increase of CRC incidence. Owing to the difficulty of early diagnosis and its high distal metastasis rate, the prognosis of CRC is very poor and the 5-year survival rate for patients with distant metastasis is only about 14%.3 The pathogenesis of CRC is complex, and three major mechanisms have been proposed: chromosomal instability, microsatellite instability, and CpG island methylator phenotype.4 Many genes and molecules are involved in the development and progression of the disease, such as WNT signaling and transforming growth factor (TGF-β) signaling.5,6 One key mechanism underlying the metastasis of CRC is the epithelial-mesenchymal transition (EMT) process,7,8 which involves the formation of migratory mesenchymal cells from epithelial cells that enables the invasion.9 Nevertheless, the molecular mechanisms of CRC development and EMT process remain largely unknown. Searching for new biomarkers of early CRC diagnosis and understanding the underling mechanisms are very necessary for better outcomes.
Circular RNA (circRNAs) are a novel class of endogenous, non-coding RNAs.10,11 They are covalently closed and thus have very stable structures.10,11 They can regulate gene expression by acting as microRNA (miRNA) or protein decoys. Accumulating evidence indicates that circRNAs have critical roles in diverse processes including physiological processes and pathological processes, particularly in cancers.12 CircRNAs can regulate cell proliferation, early lineage differentiation, pluripotency, and cell migration and invasion by modulating downstream gene expression or protein stability and translation.11 In addition, due to their diversity and stable structure, they could serve as diagnose biomarkers for some diseases. In CRC, many dysregulated circRNAs have been reported.13,14 For instance, circRNA has_circ_0044556, is upregulated in CRC cells, and promotes CRC proliferation and invasion.15 Similarly, circRNA has_circ0007142 contributes to the progression of CRC by targeting miR-103a-2-5p.16 circBANP has been observed to be elevated in CRC cells.14 circBANP is a circRNA derived from exons 5–11 of BANP gene. However, what is the function of circBANP in CRC and what is the mechanism underlying its roles in CRC are not clear. Here, we attempted to study how circBANP is involved in CRC.
MiRNAs are a well-known class of endogenous non-coding RNAs that play key roles in varieties of cellular processes.17,18 It is widely accepted that many circRNAs exert their functions by acting as miRNA sponges.10 Our preliminary bioinformatic analysis suggested that miRNA let-7d-5p might be a target of circBANP. Indeed, let-7d-5p has been implicated in CRC and its level is reduced in CRC tissues although the mechanisms are not well understood.19 Interestingly, with the help of starBase, we found that let-7d-5p could potentially bind high mobility group A1 (HMGA1) mRNA. HMGA1 is an architectural transcription factor that regulate gene expression.20 One of its downstream targets is β-catenin and HMGA1 can positively regulate Wnt/β-catenin signaling,21 which is a very crucial pathway for EMT process.22 Therefore, we hypothesized that circBANP might participate in CRC development and EMT by regulating Wnt/β-catenin signaling via the let-7d-5p/HMGA1 axis.
In the present study, we investigated the role of circBANP/let-7d-5p/HMGA1 axis in CRC. We confirmed that circBANP was elevated in CRC human tissues and cells and that its level correlated with the stage of the cancer. Knockdown of circBANP suppressed EMT process and CRC cell proliferation, migration, and invasion, as well as CRC tumor growth and metastasis in vivo. At the molecular level, we demonstrated that circBANP directly bound let-7d-5p, while let-7d-5p targeted HMGA1 mRNA. circBANP/let-7d-5p regulated Wnt/β-catenin signaling potentially via modulating HMGA1. Our study reveals an essential role of circBANP/let-7d-5p/HMGA1 in CRC and provides insights into the molecular mechanisms of EMT process and the cancer development.
Results
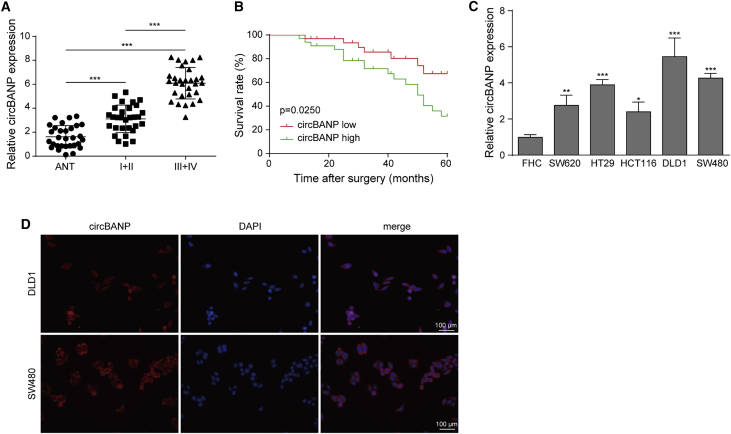
circBANP was elevated in CRC tissues and cells
To investigate the function of circBANP in CRC, we measured its expression level in CRC tissues and cells. We collected human CRC samples from diagnosed patients and divided them into two groups based on their disease stages (I+II, III+IV, TNM staging; Table 1). Compared to normal adjacent non-tumor colon tissues, we found that circBANP level was significantly elevated in CRC tissues and the increase was bigger in patients at advanced stages (III+IV) than in patients at early stages (I+II; Figure 1A). Moreover, we found that the survival rate of patients with a high circBANP level was significantly lower than patients with a low circBANP level (Figure 1B). We also examined circBANP expression in CRC cell lines and observed that circBANP level was much higher in CRC cells than in normal human colon cells (Figure 1C). With an RNA fluorescence in situ hybridization (FISH) experiment, we found that circBANP was primarily located in the cytoplasm (Figure 1D). Together, these results show that circBANP is upregulated in CRC. The level of circBANP was higher in SW480 and DLD1 cells than the rest of the three cell lines, and thus we used these two cell lines for subsequent investigations.
Table 1.
Clinicopathological characteristics of patients with CRC
| Characteristic | Cases of patients (n = 60) |
|---|---|
| Sex | |
| Female | 34 |
| Male | 26 |
| Age (years) | |
| >60 | 24 |
| ≤60 | 36 |
| Histological grade | |
| Well | 11 |
| Moderately | 33 |
| Poorly | 16 |
| Tumor location | |
| Colon | 33 |
| Rectum | 27 |
| Tumor size, cm | |
| >5 | 16 |
| ≤5 | 44 |
| TNM stage | |
| I + II | 30 |
| III + IV | 30 |
| Lymph node metastasis | |
| N0 | 34 |
| N1-2 | 26 |
Figure 1.
circBANP was elevated in CRC tissues and cells
(A) Relative circBANP levels in CRC tissues from patients at various stages. (B) Kaplan-Meier survival curve in patients with high or low circBANP level. (C) Relative circBANP levels in CRC cells compared to normal human colon cells. (D) RNA FISH analysis of subcellular localization of circBANP, scale bar = 100 μm. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
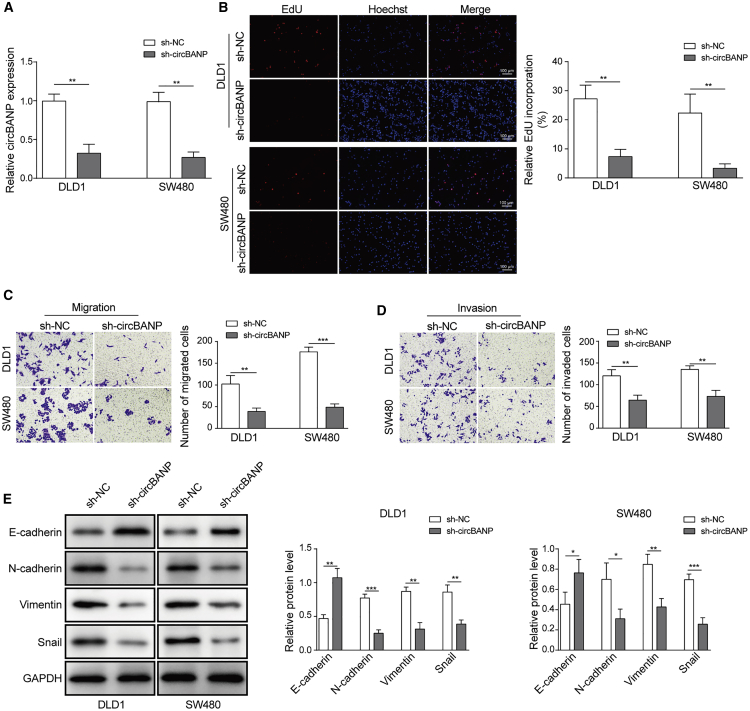
Knockdown of circBANP suppressed EMT and CRC cell proliferation, migration, and invasion
To further study the role of circBANP in CRC development, we used short hairpin RNA (shRNA) to manipulate the level of circBANP in cells and examined ensuing effects on cancer cell proliferation and migration. We confirmed that cells transfected with sh-circBANP had a much lower level of circBANP than cells transfected with sh-negative control (NC; Figure 2A). Using Cell Counting Kit-8 (CCK-8) and 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay, we found that sh-circBANP-transfected CRC cells had a significantly lower cell viability and EdU incorporation rate than sh-NC-transfected cells, indicating that knockdown of circBANP inhibits CRC cell proliferation (Figures S2A and 2B). Using Transwell assay, we showed that knockdown of circBANP greatly diminished the numbers of migration and invasion CRC cells (Figures 2C and 2D). EMT process plays a critical role in the cancer development, and thus we evaluated how circBANP affected EMT process. Using western blot, we found that CRC cells transfected with sh-circBANP had significant lower levels of N-cadherin, Vimentin, and Snail, but a higher level of E-cadherin compared to control sh-NC-transfected cells (Figure 2E), suggesting that knockdown of circBANP suppresses EMT process. Taken together, we demonstrate that inhibition of circBANP represses EMT process and CRC cell proliferation, migration, and invasion.
Figure 2.
Knockdown of circBANP suppressed EMT and CRC cell proliferation, migration, and invasion
(A) Relative circBANP levels in transfected CRC cells. (B) EdU incorporation analysis of cell proliferation in transfected cells, scale bar = 100 μm. (C and D) Transwell assay was utilized to measure the migration and invasion capabilities of transfected CRC cells. (E) Western blot analysis of protein levels of EMT-related proteins including N-cadherin, E-cadherin, Sail, and Vimentin. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
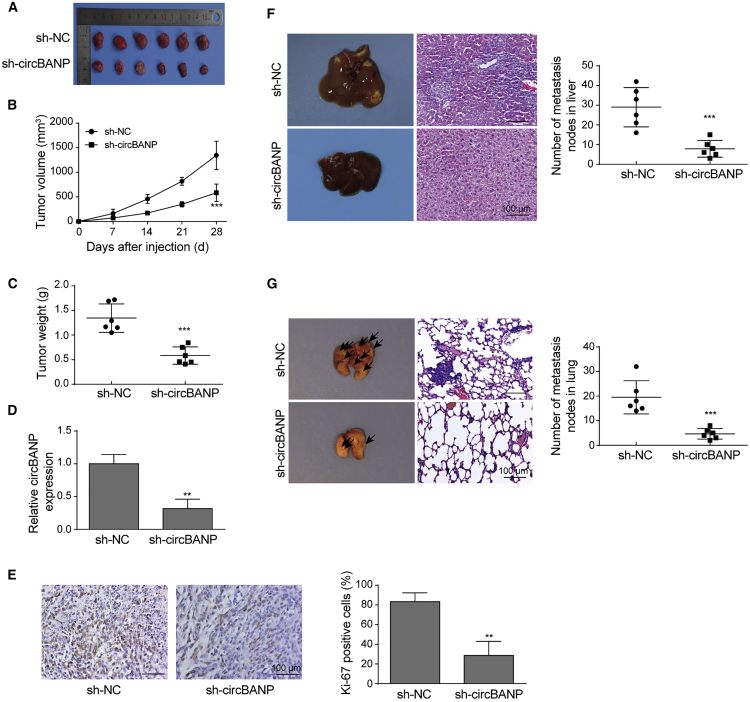
Knockdown of circBANP inhibited CRC tumor growth and metastasis in vivo
We then evaluated the function of circBANP in CRC in vivo by using the nude mouse xenograft model. We infected CRC cells with sh-circBANP or sh-NC lentiviruses for 24 h, subcutaneously injected them into nude mice, and then monitored the tumor growth every 7 days for 4 weeks. We found that the tumor in mice bearing sh-NC-transfected CRC cells progressively increased with time while the tumor volume in mice implanted with sh-circBANP-transfected CRC cells was significantly and consistently smaller (Figures 3A and 3B). At the end of 4 weeks, the tumor weight in sh-circBANP mice was remarkably smaller than in sh-NC mice (Figure 3C). These results indicate that knockdown of circBANP represses CRC tumor growth in animals. We confirmed that tumors transfected with sh-circBANP had a much lower level of circBANP compared to tumors transfected with sh-NC (Figure 3D). With immunohistochemistry (IHC) staining, we observed a significant lower level of Ki-67 in the sh-circBANP group than that in the sh-NC group, suggesting a reduced cell proliferation (Figure 3E). Next, we examined the lung and liver metastasis of CRC by performing hematoxylin & eosin (H&E) staining with the lung and liver tissues. We showed that the numbers of cancer nodules in the liver and lung were significantly smaller in mice bearing sh-circBANP-transfected cells than in mice implanted with sh-NC-transfected cells (Figures 3F and 3G). Taken together, these data reveal that knockdown of circBANP remarkably suppresses CRC tumor growth and metastasis in vivo.
Figure 3.
Knockdown of circBANP inhibited CRC tumor growth and metastasis in vivo
(A) Representative tumor images in mice bearing sh-NC- or sh-circBANP-transfected CRC cells. (B) Tumor volume in each group of mice with time. (C) Tumor weight in each group of mice. (D) Relative circBANP levels in tumors from mice implanted with sh-NC- or sh-circBANP-transfected CRC cells. (E) IHC analysis of Ki-67 level in each group of mice. (F and G) H&E staining to measure the number of tumor nodules in the lung and liver in each group of mice, scale bar = 100 μm. Data were presented as means ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
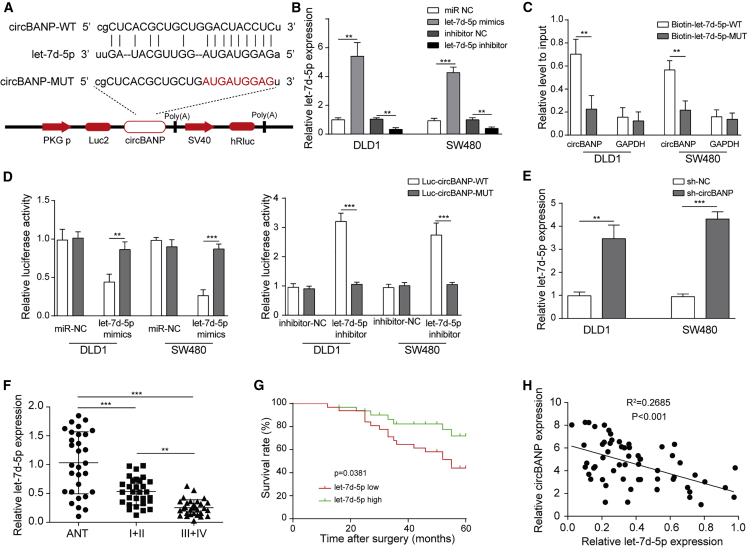
circBANP acted as a let-7d-5p sponge
We next investigated the molecular mechanisms underlying circBANP function. With bioinformatic analysis (starBase: http://starbase.sysu.edu.cn/), we observed some complementary binding sites between circBANP and let-7d-5p (Figure 4A). To directly examine the interaction, we performed an RNA pull-down experiment and dual luciferase assay. First, we confirmed that let-7d-5p mimics greatly increased let-7d-5p level while let-7d-5p inhibitor decreased (Figure 4B). Immunoprecipitation of biotin-labeled let-7d-5p-wild-type (WT) significantly pull down a lot more circBANP than immunoprecipitation of biotin-labeled let-74-5p-mutant (MUT) with the binding sites of circBANP mutated (Figure 4C). Consistently, we observed that let-7d-5p mimics diminished the relative luciferase activity of circBANP-WT but not the activity of circBANP-MUT with the binding sites of let-7d-5p mutated (Figure 4D). In contrast, let-7d-5p inhibitor greatly increased the luciferase activity of circBANP-WT but not circBANP-MUT (Figure 4D). These results strongly prove that circBANP directly binds let-7d-5p. Further, cells transfected with sh-circBANP had higher levels of let-7d-5p than control sh-NC-transfected cells (Figure 4E). Also, let-7d-5p expression was diminished in CRC tissues from the patients, and we detected a significantly negative correlation between circBANP level and let-7d-5p level (Figures 4F and 4H). Further, the survival rate of patients with high let-7d-5p level was higher than patients with low let-7d-5p level (Figure 4G). Therefore, we conclude that circBANP functions as a let-7d-5p sponge in CRC.
Figure 4.
circBANP acted as a let-7d-5p sponge
(A) Complementary binding sites between circBANP and let-7d-5p. (B) Relative let-7d-5p levels in transfected cells. (C) Relative circBANP levels pulled down by immunoprecipitation with specific antibodies. (D) Relative luciferase activity of WT-circBANP and MUT-circBANP in transfected cells. (E) Relative let-7d-5p levels in transfected cells. (F) Relative let-7d-5p levels in CRC tissues. (G) Kaplan-Meier survival curve in patients with high or low let-7d-5p level. (H) Correlation between circBANP level and let-7d-5p level in CRC tissues. Data were presented as means ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
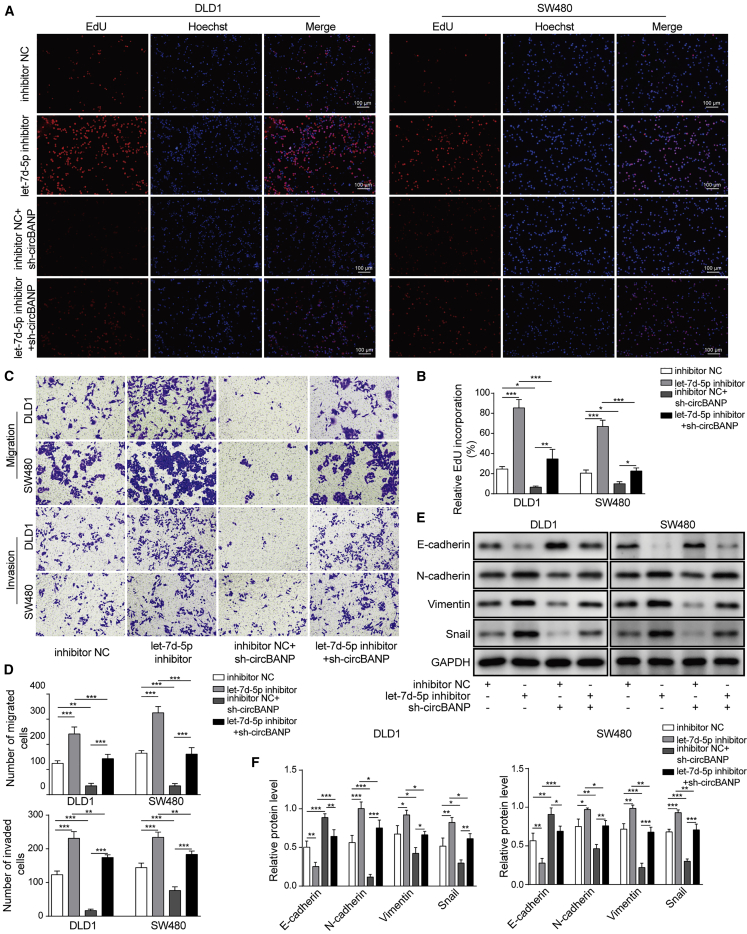
Knockdown of circBANP blocked the effects of let-7d-5p inhibition on EMT process and CRC cell proliferation, migration, and invasion
We then examined the functional role of circBANP/let-7d-5p interaction in CRC. As expected, transfection of let-7d-5p inhibitor diminished let-7d-5p level but increased circBANP level while sh-circBANP decreased circBANP level and upregulated let-7d-5p level. Co-transfection of cells with let-7d-5p inhibitor and sh-circBANP restored the levels of let-7d-5p and circBANP back to baseline (Figures S1A and S1B). With CCK-8 and EdU incorporation assay, we showed that let-7d-5p inhibitor alone drastically increased CRC cell proliferation while knockdown of circBANP with sh-circBANP alone had the opposite effect (Figures S2B; Figures 5A and 5B). Notably, knockdown of circBANP suppressed the increase of cell viability and EdU incorporation induced by let-7d-5p inhibitor (Figure S2B; Figures 5A and 5B). Similarly, using a Transwell assay, we found that let-7d-5p inhibitor alone significantly increased the numbers of migration and invasion CRC cells while overexpression of sh-circBANP decreased (Figures 5C and 5D). Co-transfection of cells with let-7d-5p and sh-circBANP recovered the numbers to baseline (Figures 5C and 5D). Regarding the EMT process, let-7d-5p inhibitor significantly upregulated the levels of N-cadherin, Vimentin, and Snail but downregulated the E-cadherin level while sh-circBANP showed the opposite effects (Figures 5E and 5F). Again, sh-circBANP reversed the suppression effects of let-7d-5p inhibitor on the EMT process (Figures 5E and 5F). Together, the above results demonstrate that inhibition of let-7d-5p enhances EMT and CRC cell proliferation, migration, and invasion while knockdown of circBANP could block those effects.
Figure 5.
Knockdown of circBANP blocked the effects of let-7d-5p inhibition on the EMT process and CRC cell proliferation, migration, and invasion
(A and B) EdU incorporation analysis of cell proliferation in transfected cells, scale bar = 100 μm. (C and D) Transwell assay was utilized to measure the migration and invasion abilities of transfected CRC cells. (E and F) Western blot analysis of protein levels of EMT-related proteins including N-cadherin, E-cadherin, Snail, and Vimentin. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
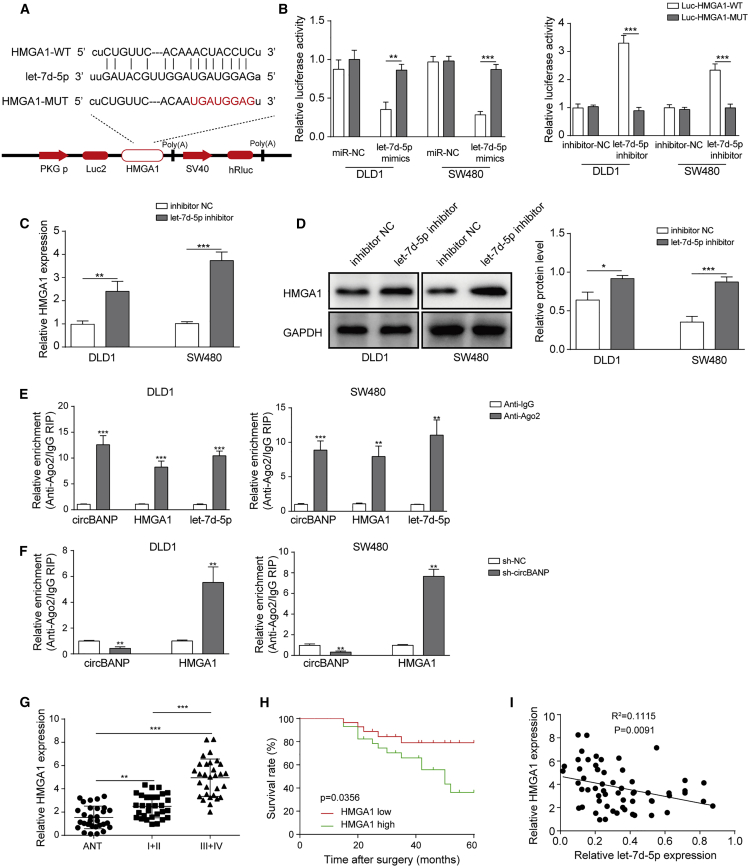
Let-7d-5p targeted HMGA1 in CRC cells
miRNAs usually exert their functions via binding to target mRNAs.17 Through bioinformatic analysis (starBase), we found some complementary binding sites between let-7d-5p and HMGA1(Figure 6A). To directly examine that interaction, we employed dual luciferase assay. Let-7d-5p mimics greatly diminished the relative luciferase activity of HMGA1-WT but not HMGA1-MUT in which the predicted binding sites with let-7d-5p were mutated, while let-7d-5p inhibitor showed the opposite effect (Figure 6B). In addition, cells transfected with let-7d-5p inhibitor had significantly higher levels of HMGA1 mRNA and protein than cells transfected with inhibitor NC (Figures 6C and 6D), suggesting that let-7d-5p negatively regulates HMGA1 expression. We then validated the sponging effect of circBANP. First, an RNA immunoprecipitation (RIP) assay with Ago2 antibody showed that circBANP, HMGA1, and let-7d-5p were mainly enriched to Ago2. Moreover, knockdown of circBANP decreased the enrichment of Ago2 to circBANP, while it increased the enrichment of Ago2 to HMGA1 (Figures 6E and 6F), suggesting that inhibition of circBANP strengthens the let-7d-5p/HMGA1 mRNA binding. In human CRC tissues, we found that HMGA1 mRNA was significantly elevated compared to normal colon tissues and that patients at late stages (III+IV) had higher level of HMGA1 than patients at early stages (I+II; Figure 6G). Consistently, the survival rate of patients with high HMGA1 level was lower than patients with low HMGA1 level (Figure 6H) Moreover, we identified a strong negative correlation between the HMGA1 level and let-7d-5p level in CRC tissues (Figure 6I). Taken together, these results show that let-7d-5p binds HMGA1 mRNA and negatively regulates HMGA1 expression in CRC cells.
Figure 6.
Let-7d-5p targeted HMGA1 in CRC cells
(A) Complementary binding sites between let-7d-5p and HMGA1 mRNA. (B) Relative luciferase activity of HMGA1-WT and HMGA1-MUT in transfected cells. (C and D) Relative HMGA1 mRNA and protein levels in transfected cells. (E and F) Relative circBANP, let-7d-5p, and HMGA1 mRNA levels in transfected cells following Ago2 or immunoglobulin G (IgG) immunoprecipitation. (G) Relative HMGA1 mRNA levels in CRC tissues. (H) Kaplan-Meier survival curve in patients with high or low HMGA1 level. (I) Correlation between HMGA1 mRNA level and let-7d-5p level in CRC tissues. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
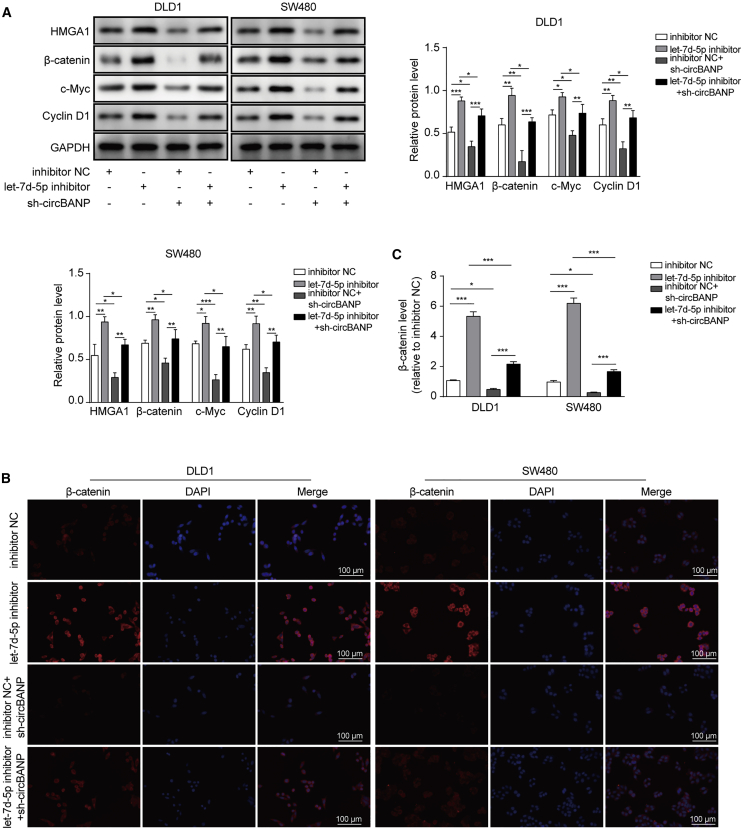
circBANP/let-7d-5p regulated HMGA1-mediated Wnt/β-catenin signaling
It has been shown that HMGA1 modulates Wnt/β-catenin signaling which is crucial for cancer development.21,23 In the end, we examined whether circBANP/let-7d-5p affected Wnt/β-catenin pathway. First, using western blot, we found that let-7d-5p inhibitor increased HMGA1 level, as well as β-catenin, c-Myc, and cyclin D1, while sh-circBANP decreased (Figure 7A). Further, sh-circBANP suppressed the upregulations of HMGA1, β-catenin, c-Myc, cyclin D1 caused by let-7d-5p inhibitor in CRC cells (Figure 7A). Second, with the immunostaining method, we observed similar results. Let-7d-5p inhibitor alone increased the β-catenin signal while sh-circBANP alone decreased (Figures 7B and 7C). Co-transfection of sh-circBANP and let-7d-5p inhibitor recovered the signal to the control level (Figures 7B and 7C). These data indicate that circBANP/let-7d-5p regulates Wnt/β-catenin signaling in CRC cells, most likely through targeting HMGA1.
Figure 7.
circBANP/let-7d-5p regulated HMGA1-mediated Wnt/β-catenin signaling
(A) Western blot analysis of protein levels of HMGA1, β-catenin, c-Myc, and cyclin D1 in transfected cells. (B and C) Immunostaining analysis of β-catenin signaling in transfected cells, scale bar = 100 μm. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
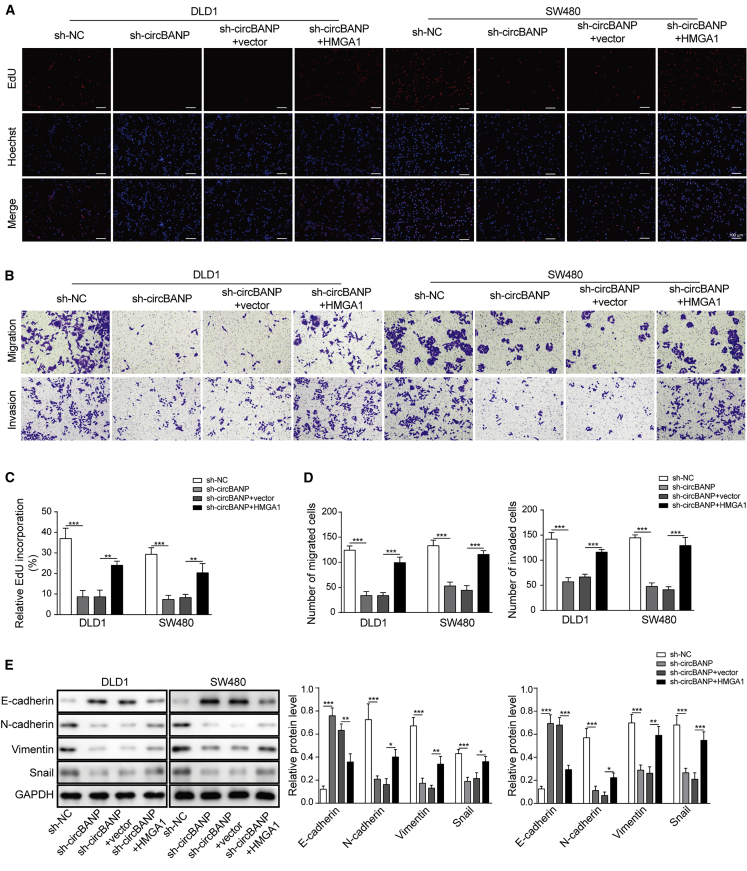
circBANP regulated EMT and CRC cell proliferation, migration, and invasion via disinhibiting HMGA1
To directly study whether circBANP regulated CRC progression via sponging let-7d-5p to disinhibit HMGA1 expression, we examined how HMGA1 overexpression affected the effects of circBANP knockdown. As expected, transfection of sh-circBANP greatly decreased circBANP level, as well as HMGA1 mRNA level, while co-overexpression of HMGA1 recovered HMGA1 level in sh-circBANP-transfected cells (Figure S1C). Consistent with the aforementioned results, sh-circBANP-transfected CRC cells had a significantly lower cell viability and EdU incorporation rate than sh-NC-transfected cells, indicating that knockdown of circBANP suppresses CRC cell proliferation (Figure S2C; Figures 8A and 8C). In contrast, co-overexpression of HMGA1 restored the viability and EdU incorporation rate (Figure S2C; Figures 8A and 8C). With Transwell assay, we found that the decreases in the numbers of migration and invasion of CRC cells caused by sh-circBANP were recovered by co-overexpression of HMGA1 (Figure 8B and 8D). Furthermore, sh-circBANP-induced suppression on EMT process was restored by HMGA1 co-overexpression (Figure 8E). The levels of N-cadherin, Vimentin, Snail, and E-cadherin in sh-circBANP- and HMGA1-transfected cells were comparable to sh-NC-transfected cells (Figure 8E). We therefore conclude that circBANP regulates EMT process and CRC cell proliferation, migration, and invasion by de-repressing HMGA1 expression.
Figure 8.
circBANP regulated EMT and CRC cell proliferation, migration, and invasion via disinhibiting HMGA1
(A and B) EdU incorporation analysis of cell proliferation in transfected cells, scale bar = 100 μm. (C and D) Transwell assay was utilized to measure the migration and invasion capabilities of transfected CRC cells. (E) Western blot analysis of protein levels of EMT-related proteins including N-cadherin, E-cadherin, Sail, and Vimentin. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
As one of the most prevalent and deadly cancers, CRC has been affecting millions of people worldwide, leaving a lot of burden to the world.24,25 Despite tremendous advances in research made in the past, the pathogenesis of CRC is still not well understood and it is very necessary to improve the prognosis of the disease.1 Here, we fully elucidated the function of circBANP in CRC. We showed that circBANP was greatly elevated in CRC and that the elevation was higher in patients at advanced stages. Knockdown of circBANP significantly inhibited the EMT process and CRC cell proliferation, migration, and invasion, as well as tumor growth and metastasis in vivo. Mechanistically, we found that circBANP acted as a let-7d-5p sponge while let-7d-5p targeted HMGA1. circBANP/let-7d-5p regulated Wnt/β-catenin signaling. Together, we demonstrate that circBANP/let-7d-5p/HMGA1 axis plays a critical role in EMT process and CRC development. This elucidation suggests that circBANP level could serve as a diagnosis biomarker for CRC and targeting circBANP/let-7d-50/HMGA1 axis might be an avenue to treat CRC.
Since their discoveries, circRNAs have been indicated to play critical functions in many cellular processes and diseases, particularly in cancers.12,26 In CRC, recent studies have reported many aberrant expressions of circRNAs during the development, such as circACAP2, circITCH, and circDDX17.13,27, 28, 29 circBANP, generated from exons 5–11 of BANP gene, was first identified as an overexpressed circRNA in CRC tissues by a large-scale of microarray, and knockdown of circBANP greatly attenuated CRC cell proliferation.14 Subsequently, circBANP has been shown involved in multiple types of cancers such as lung cancer and gastric cancer.30,31 The exact function of circBANP in CRC remains largely unknown. Here, we first confirmed that circBANP was significantly elevated in CRC tissues and cells. Moreover, we showed that its level correlated with the stage of the disease with higher expression in patients at advanced stages than at early stages. We also provided strong evidence that knockdown of circBANP suppressed EMT process and CRC cell proliferation, migration, and invasion, as well as the tumor growth and metastasis in vivo. Our study, together with previous research, supports the conclusion that circBANP acts as an oncogene in CRC.
It is well acknowledged that most circRNAs function by binding to miRNAs or proteins to regulate gene expression or protein translation.10 After demonstrating the oncogene role of circBANP in CRC, we went to examine the underlying molecular mechanisms. We identified that circBANP directly bound let-7d-5p and negatively regulated its expression. Interestingly, a lower level of let-7d-5p in CRC has been observed before,32 which is consistent with our data and model. Further, we showed that knockdown of circBANP reversed the effects of let-7d-5p inhibitor on the EMT process and CRC cell proliferation, migration, and invasion, indicating that circBANP regulates the EMT process and CRC development by acting as a let-7d-5p sponge. circBANP has been shown to bind some other miRNAs such as miR-503 and let-7a.30,31 Therefore, it is possible that circBANP may exert its function by targeting other miRNAs besides let-7d-5p in CRC. Future studies are necessary to test that hypothesis. In addition, some circRNAs can directly bind with proteins.11,13 It remains to further explore whether any proteins bind with circBANP.
The Wnt/β-catenin signaling is a conserved, classic pathway that is involved in many cellular functions such as cell proliferation, growth, and survival.33,34 Aberrant Wnt/β-catenin activation has been reported in varieties of cancers including breast, lung, and colorectal cancers and greatly contributes to tumor recurrence as well.5,6,23 One key mechanism underlying the role of Wnt/β-catenin in cancers is that Wnt/β-catenin signaling pathway regulates the EMT process to promote the migration and invasion of cancer cells.22,35 In the present study, we showed that circBANP/let-7d-5p regulated Wnt/β-catenin signaling as well. Let-7d-5p directly targeted HMGA1, and HMGA1 has been shown to activate Wnt/β-catenin signaling.21 Consistently, we observed that let-7d-5p inhibitor activated HMGA1/Wnt/β-catenin signaling while knockdown of circBANP inhibited. Therefore, circBANP/let-7d-5p modulates EMT process and CRC metastasis most likely through HMGA1/Wnt/β-catenin pathway (Figure 9). The interaction of let-7d-5p with HMGA1 mRNA has been reported in other types of cancers, such as breast cancer and bladder cancer.36,37 Therefore, let-7d/5p/HMGA1 axis might be a conserved signaling during the cancer development and progression.
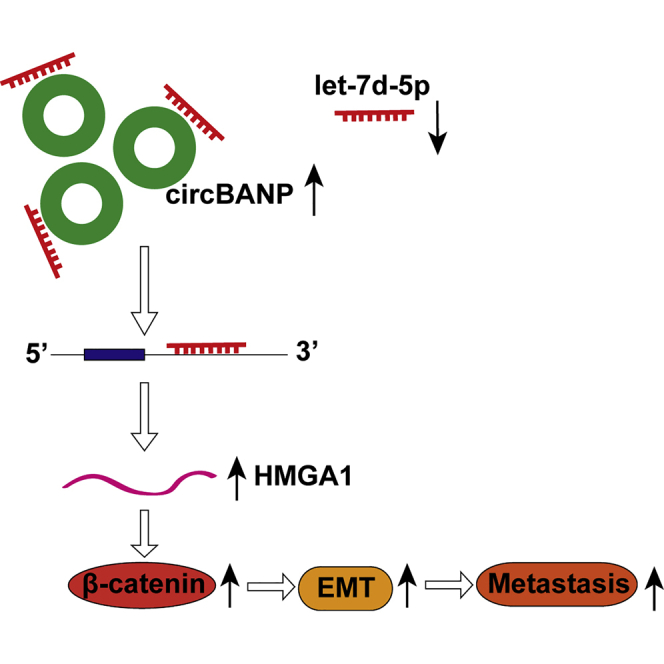
Figure 9.
The schematic cartoon of circBANP in CRC
circBANP promotes CRC development and metastasis via acting as a let-7d-5p sponge to regulate HMGA1/Wnt/β-catenin signaling.
In summary, we provide evidence that circBANP functions as an oncogene in CRC and promotes CRC development by acting as a let-7d-5p sponge to regulate HMGA1/Wnt/β-catenin signaling. Treatments targeting circBANP/let-7d-5p/HMGA1 axis might be useful for CRC therapy.
Materials and methods
Human CRC samples
Human CRC tissues were collected from 60 diagnosed CRC patients during surgical resection from Changzhou Second People’s Hospital Affiliated to Nanjing Medical University. The stages of CRC were diagnosed based on the pathological analysis. The adjacent non-tumor colorectal tissues were collected as control samples. The patients did not receive preoperative treatments before the collection. The procedure and protocol have been reviewed and approved by the ethics committee of our hospital. All patients have consented to the study and signed a written form. All specimens were snap-frozen immediately in liquid nitrogen and stored at −80°C for subsequent experiments.
Cell culture and transfection
Five human CRC cell lines (SW620, SW480, HCT116, HT29, and DLD1) and one normal human colon cell line (FHC) were used in this study and were purchased from ATCC (Manassas, VA, USA). The medium used for cell culture was composed of Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) plus 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin-streptomycin (Gibco). The cells were maintained in the cell culture incubator at 37°C. Lipofectamine 3000 (Invitrogen, St. Louis, MO, USA) was utilized as the reagent for cell transfection. In brief, cells were cultured to ∼70% confluence and then construct was added together with Lipofectamine 3000. Cells were harvested for further analysis 48 h after transfection.
RNA extraction and qRT-PCR
Trizol (Invitrogen) was employed to extract total RNAs from CRC human tissues or cells as the manufacturer’s instructions described. DNaseI was included in the lysis buffer to avoid the contamination of DNA. Commercial kit of cDNA synthesis kits (Thermo Fisher Scientific) was utilized to generate cDNAs through reverse transcription. SYBR Green Master Mix (Invitrogen) was used for the quantitative PCR. Relative expression levels of let-7d-5p or circBANP and mRNAs were normalized to U6 or GAPDH mRNA, respectively, as internal controls. The relative expression level was calculated by 2−ΔΔCt method. The primers listed as follows were from Genepharma (Shanghai, P.R. China):
circBANP forward primer (FP): 5′-TGGCCGATGTGGTTCAGATT-3′,
circBANP reverse primer (RP): 5′-CTGCGGAATTCTCGTGGTGT-3′;
let-7d-5p FP: 5′-GGCGAGAGGTAGTAGGTTGC-3′,
let-7d-5p RP: 5′-CGGCCCAGTGTTCAGACTAC-3′;
HMGA1 FP: 5′-CCTCCAAGCAGGAAAAGGAC-3′,
HMGA1 RP: 5′-CTTCCTGGAGTTGTGGTGGT-3′;
U6 FP: 5′-TGGCGGGTGTATTAAACCAC-3′,
U6 RP: 5′-TTCACGAATTTGCGTGTCATC-3′;
GAPDH FP: 5′-CCAGGTGGTCTCCTCTGA-3′,
GAPDH RP: 5′-GCTGTAGCCAAATCGTTGT-3′.
RNA FISH
Stellaris kit (Biosearch Technologies, Novato, CA, USA) was utilized to carry out the RNA FISH experiment as the manufacturer’s protocol described. Cells were washed with iced cold PBS and then fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature. 0.2% Triton X-100 in PBS was used to permeabilize cells for 10 min at room temperature. Cells were subsequently washed with PBS again and incubated with the probe in hybridization buffer overnight at 37°C in the dark, followed by 3 washes with wash buffer. Cells were mounted in 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium and imaged with confocal microscopy. The following probes used were generated and purchased from Stellaris: Cys-labeled circBANP.
EdU) incorporation assay
EdU is a nucleoside analog of thymidine and can be incorporated into DNA during DNA synthesis. Therefore, cell proliferation could be determined by the EdU incorporation assay. The commercial kit (Invitrogen) was utilized to carry out the EdU incorporation assay as the manufacturer’s protocol described. Briefly, cells were plated in the 96-well plate and grown up to 80% confluence. EdU was added to incubate with cells for 4 h. The residual EdU was washed off by PBS. Cells were fixed with 4% PFA first for 15 min at room temperature and then permeabilized with 0.2% Triton X-100 in PBS for 10 min. Afterward, the cells were blocked with 3% BSA and stained with Apollo 567 and Hoechst working solution for 0.5 h in the dark followed by imaging with the fluorescence microscopy.
Transwell migration and invasion assays
Transfected CRC cells or normal colon cells were seeded in the culture medium with no serum on top of the filter membrane. Full culture medium that contains 10% FBS was put in the lower chamber. After 24 h incubation, the upper filter was discarded. Cells growing on the lower dish were cells migrated there from the filter. They were fixed in 4% PFA first for 15 min at room temperature, and then 0.1% crystal violet was added to stain the cells followed by imaging. To analyze the invasive ability of cells, we pre-coated the upper filter with Matrigel (Corning, Corning, NY, USA) overnight before cells were seeded. Following 24 h growth, cells residing in the lower dish were cells that invaded from the top. They were proceeded for crystal violet staining similarly as described above.
Western blot analysis
Proteins from cultured CRC cells were extracted by utilizing the radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, P.R. China) according to standard protocol. DC Protein Assay Kit (Bio-Rad, P.R. China) was utilized to quantify the protein concentrations. Equal protein from each sample was loaded into SDS-polyacrylamide gels and separated through electrophoresis. Later proteins in the gels were transferred to polyvinylidene fluoride (PVDF) membranes. 3% bovine serum albumin (BSA) was used to block the membranes for 60 min at room temperature and then specific primary antibodies (Abcam, USA) were added to incubate at 4°C overnight. The antibodies were discarded and TBST was utilized to wash the membranes 3 times before incubation with specific secondary antibodies (1:20,000, Abcam) for 1 h at room temperature. Protein band intensities were detected by using the ECL kit (Bio-Rad). Primary antibodies used in the study were: anti-E-cadherin antibody (1:10,000), anti-N-cadherin antibody (1:1,000), anti-Vimentin antibody (1:2,000), anti-Snail (1:1,000), anti-HMGA1 antibody (1:10,000), anti-β-catenin antibody (1:5,000), anti-c-Myc (1:1,000), anti-Cyclin D1 antibody (1:25,000), and anti-GAPDH (1:2,500). The films were scanned, and the intensities were quantified using the ImageJ.
Nude mice experiments
All animal experiments have been reviewed and received approval by the Animal Care and Use Committee of our hospital. Nude mice were purchased from SJA Laboratory Animal (Hunan, P.R. China) and raised in the standard animal facility room. Then nude mice were unilaterally subcutaneously injected with 5 × 106 transfected CRC cells (sh-NC-transfected cells, sh-circBANP-transfected cells) to induce tumors. Tumors were monitored on a daily base for 28 days. Tumor length (L) and width (W) were analyzed to quantify the tumor volume (V): V(mm3) = 0.5 × (W)2 × (L). In the end, tumors were dissected out to measure weight. To measure metastasis, transfected CRC cells (5 × 105; control sh-NC group, sh-circBANP group) were tail injected via the vein. Lung and liver tissues were harvested after 42 days for further analysis (H&E staining and IHC).
IHC assay
The tumor tissues were fixed in 4% PFA at 4°C overnight, washed by PBS, and then embedded in paraffin. The tumor tissues were sliced into 5 μm thick sections and dried overnight at 37°C on glass slides. The dried slices were deparaffinized in xylene first followed by rehydration through a graded concentration of alcohol. 3% hydrogen peroxide was used to quench the sections and 5% BSA was added to block the slices for 1 h at room temperature. Primary antibody (anti-Ki-67; 1:1,000; Invitrogen, USA) was added to incubate with the slices at 4°C overnight. The next day, the antibody was washed off by PBS and secondary antibodies were added to incubate with sections for 1 h at room temperature. Substrates of the Envision system-horseradish peroxidase (HRP) from the kit (Dako REAL Ebision kit; DAKO, Glostrup, Denmark) were added to incubate with stained slices as the manufacturer’s protocol described and the signals were analyzed with a light microscope.
H&E staining
Lung and liver tissues were dissected out and then placed in 4% PFA overnight for fixation at 4°C. The tissues were then embedded in paraffin and then 5 μm thick paraffin sections were cut. Paraffin slices were stained with H&E as the manufacturer’s protocol described.
Biotinylated RNA pull-down assay
The biotinylated RNA pull-down assay was utilized to verify the direct binding between circBANP and let-7d-5p. In brief, the biotinylated WT/MUT let-7d-5p was transfected into CRC cells together with circBANP by using Lipofectamine 3000. 2 days after overexpression, cells were harvested and lysed using lysis buffer. Magnetic beads were added to incubate with cell lysates overnight at 4°C to pull down biotinylated RNAs. The next day, the cell lysates were discarded, and the beads were washed with lysis buffer followed by elution. The elution was subjected to qRT-PCR to determine the expression levels of circBANP.
RIP assay
In order to strengthen the functional validity of sponging effect, RIP assay was performed. In brief, MS2bs-circBANP, MS2bs-circBANPmt, or MS2bs-Rluc and MS2bp-GFP were cotransfected into cells for 48 h. Then, Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) was allowed for RIP. The purified RNA complexes were used for quantification of let-7d-5p level.
For the RIP assay for Ago2, RIP was performed with an anti-Ago2 antibody (Millipore). RNAs were then purified, and the levels of circBANP, HMGA1, and let-7d-5p were measured.
Dual luciferase report assay
The WT sequences or mutated binding sites of let-7d-5p in 3′ untranslated region (UTR) of HMGA1mRNA and the full length of circBANP were cloned into the luciferase report vector (psiCHECK2). Commercial kit (the Phusion Mutagenesis kit, Thermo Fisher Scientific) was utilized to mutate the predicted binding sites as the protocol described. CRC cells were seeded in individual wells of the 24-well plates first overnight and then recombinant constructs were transfected into CRC cells together with let-7d-5p mimics or miR-NC by using the Lipofectamine 3000. 48 h after transfection, the cells were harvested in the Reporter Lysis Buffer from the commercial kit (Promega, P.R. China) and relative luciferase activities were measured.
Immunostaining
Transfected CRC cells were washed with ice-cold PBS and incubated with 4% PFA at room temperature for 15 min for fixation. Fixed cells were permeabilized in 0.1% Triton X-100 for 10 min at room temperature. Cells were washed again with PBS and blocked with 1% BSA in PBS for 1 h at room temperature. Primary antibody rabbit anti-β-catenin (1:500) was added to incubate with cells at 4°C overnight. The next day, antibodies were washed off by PBS and secondary antibody conjugated with tetramethylrhodamine was added to incubate with cells for 2 h at room temperature. The secondary antibody was washed off by PBS and cells were mounted in DAPI-containing mounting medium. Images were taken using the confocal microscope.
Statistical analysis
All experiments were carried out with at least three biological replicates and the data were analyzed in GraphPad Prism 7. Statistical details were calculated by Student t test (two groups) or one-way ANOVA (multiple groups). The difference was considered significant if p <0.05. All experimental data are presented as mean ± standard deviation (SD).
Acknowledgments
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Author contributions
Conceptualization, methodology, software, and writing – original draft preparation, Y.N.; data curation, C.L.; visualization and investigation, W.W.; software and validation, W.G.; conceptualization, writing – reviewing & editing, and writing – original draft preparation, C.Y.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.03.012.
Supplemental information
References
- 1.Rawla P., Sunkara T., Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A., Jemal A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Tariq K., Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol. Med. 2016;13:120–135. doi: 10.28092/j.issn.2095-3941.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng X., Xu X., Chen D., Zhao F., Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed. Pharmacother. 2019;110:473–481. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]
- 7.Grady W.M., Markowitz S.D. The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig. Dis. Sci. 2015;60:762–772. doi: 10.1007/s10620-014-3444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vu T., Datta P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers (Basel) 2017;9:E171. doi: 10.3390/cancers9120171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roche J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers (Basel) 2018;10 doi: 10.3390/cancers10020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhu L.P., He Y.J., Hou J.C., Chen X., Zhou S.Y., Yang S.J., Li J., Zhang H.D., Hu J.H., Zhong S.L. The role of circRNAs in cancers. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170750. BSR20170750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao S., Cong L., Qu R., Liu R., Zhang G., Li Y. Emerging roles of circular RNAs in colorectal cancer. OncoTargets Ther. 2019;12:4765–4777. doi: 10.2147/OTT.S208235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu M., Xu Y., Chen Y., Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed. Pharmacother. 2017;88:138–144. doi: 10.1016/j.biopha.2016.12.097. [DOI] [PubMed] [Google Scholar]
- 15.Jing L., Wu J., Tang X., Ma M., Long F., Tian B., Lin C. Identification of circular RNA hsa_circ_0044556 and its effect on the progression of colorectal cancer. Cancer Cell Int. 2020;20:427. doi: 10.1186/s12935-020-01523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu C.L., Sha X., Wang Y., Li J., Zhang M.Y., Guo Z.Y., Sun S.A., He J.D. Circular RNA hsa_circ_0007142 Is Upregulated and Targets miR-103a-2-5p in Colorectal Cancer. J. Oncol. 2019;2019:9836819. doi: 10.1155/2019/9836819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J., Liu H.L., Tao L., Lin X.Y., Yang Y.D., Tan S.W., Wu B. Let-7d inhibits colorectal cancer cell proliferation through the CST1/p65 pathway. Int. J. Oncol. 2018;53:781–790. doi: 10.3892/ijo.2018.4419. [DOI] [PubMed] [Google Scholar]
- 20.Cleynen I., Van de Ven W.J. The HMGA proteins: a myriad of functions (Review) Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 21.Xing J., Cao G., Fu C. HMGA1 interacts with β-catenin to positively regulate Wnt/β-catenin signaling in colorectal cancer cells. Pathol. Oncol. Res. 2014;20:847–851. doi: 10.1007/s12253-014-9763-0. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y.G., Luo Y., He D.L., Li X., Zhang L.L., Peng T., Li M.C., Lin Y.H. Role of Wnt/beta-catenin signaling pathway in epithelial-mesenchymal transition of human prostate cancer induced by hypoxia-inducible factor-1alpha. Int. J. Urol. 2007;14:1034–1039. doi: 10.1111/j.1442-2042.2007.01866.x. [DOI] [PubMed] [Google Scholar]
- 23.Jung Y.S., Park J.I. Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Exp. Mol. Med. 2020;52:183–191. doi: 10.1038/s12276-020-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 25.GBD 2017 Colorectal Cancer Collaborators The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019;4:913–933. doi: 10.1016/S2468-1253(19)30345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M., Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 2018;11:21. doi: 10.1186/s13045-018-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J.H., Li Y.G., Han Z.P., Zhou J.B., Chen W.M., Lv Y.B., He M.L., Zuo J.D., Zheng L. The CircRNA-ACAP2/Hsa-miR-21-5p/ Tiam1 Regulatory Feedback Circuit Affects the Proliferation, Migration, and Invasion of Colon Cancer SW480 Cells. Cell. Physiol. Biochem. 2018;49:1539–1550. doi: 10.1159/000493457. [DOI] [PubMed] [Google Scholar]
- 28.Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS ONE. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X.N., Wang Z.J., Ye C.X., Zhao B.C., Li Z.L., Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J. Exp. Clin. Cancer Res. 2018;37:325. doi: 10.1186/s13046-018-1006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han J., Zhao G., Ma X., Dong Q., Zhang H., Wang Y., Cui J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem. Biophys. Res. Commun. 2018;503:2429–2435. doi: 10.1016/j.bbrc.2018.06.172. [DOI] [PubMed] [Google Scholar]
- 31.Xun J., Wang C., Yao J., Gao B., Zhang L. CircBANP acts as a sponge of let-7a to promote gastric cancer progression via the FZD5/Wnt/β-catenin pathway. RSC Advances. 2020;10:7221–7231. doi: 10.1039/c9ra09887a. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Chen W., Lin G., Yao Y., Chen J., Shui H., Yang Q., Wang X., Weng X., Sun L., Chen F. MicroRNA hsa-let-7e-5p as a potential prognosis marker for rectal carcinoma with liver metastases. Oncol. Lett. 2018;15:6913–6924. doi: 10.3892/ol.2018.8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lecarpentier Y., Schussler O., Hébert J.L., Vallée A. Multiple Targets of the Canonical WNT/β-Catenin Signaling in Cancers. Front. Oncol. 2019;9:1248. doi: 10.3389/fonc.2019.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu S., Cheriyamundath S., Ben-Ze’ev A. Cell-cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Res. 2018;7:1488. doi: 10.12688/f1000research.15782.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K., Zhang C., Li T., Ding Y., Tu T., Zhou F., Qi W., Chen H., Sun X. Let-7a inhibits growth and migration of breast cancer cells by targeting HMGA1. Int. J. Oncol. 2015;46:2526–2534. doi: 10.3892/ijo.2015.2949. [DOI] [PubMed] [Google Scholar]
- 37.Qin M.M., Chai X., Huang H.B., Feng G., Li X.N., Zhang J., Zheng R., Liu X.C., Pu C. let-7i inhibits proliferation and migration of bladder cancer cells by targeting HMGA1. BMC Urol. 2019;19:53. doi: 10.1186/s12894-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.