Abstract
Because regulation of gene expression is heritable and context-dependent, we investigated AD-related gene expression patterns in cell types in blood and brain. Cis-expression quantitative trait locus (eQTL) mapping was performed genome-wide in blood from 5257 Framingham Heart Study (FHS) participants and in brain donated by 475 Religious Orders Study/Memory & Aging Project (ROSMAP) participants. The association of gene expression with genotypes for all cis SNPs within 1 Mb of genes was evaluated using linear regression models for unrelated subjects and linear-mixed models for related subjects. Cell-type-specific eQTL (ct-eQTL) models included an interaction term for the expression of “proxy” genes that discriminate particular cell type. Ct-eQTL analysis identified 11,649 and 2533 additional significant gene-SNP eQTL pairs in brain and blood, respectively, that were not detected in generic eQTL analysis. Of note, 386 unique target eGenes of significant eQTLs shared between blood and brain were enriched in apoptosis and Wnt signaling pathways. Five of these shared genes are established AD loci. The potential importance and relevance to AD of significant results in myeloid cell types is supported by the observation that a large portion of GWS ct-eQTLs map within 1 Mb of established AD loci and 58% (23/40) of the most significant eGenes in these eQTLs have previously been implicated in AD. This study identified cell-type-specific expression patterns for established and potentially novel AD genes, found additional evidence for the role of myeloid cells in AD risk, and discovered potential novel blood and brain AD biomarkers that highlight the importance of cell-type-specific analysis.
Subject terms: Psychiatric disorders, Genomics
Introduction
Recent expression quantitative trait locus (eQTL) analysis studies suggest that changes in gene expression have a role in the pathogenesis of AD1,2. However, regulation of gene expression, as well as many biological functions, has been shown to be context-specific (e.g., tissue and cell types, developmental time point, sex, disease status, and response to treatment or stimulus)3–6. One study of 500 healthy subjects found over-representation of T cell-specific eQTLs in susceptibility alleles for autoimmune disease and AD risk alleles polarized for monocyte-specific eQTL effects7. In addition, disease and trait-associated cis-eQTLs were more cell-type-specific than average cis-eQTLs7. Another study classified 12% of more than 23,000 eQTLs in blood as cell-type-specific4. Large eQTL studies across multiple human tissues have been conducted by the GTEx consortium, with a study on genetic effects on gene expression levels across 44 human tissues collected from the same donors characterizing patterns of tissue specificity recently published8.
Microglia, monocytes, and macrophages share a similar developmental lineage and are all considered to be myeloid cells9. It is believed that a large proportion of AD genetic risk can be explained by genes expressed in myeloid cells and not other cell types10. Several established AD genes are highly expressed in microglia9,11, and a variant in the AD-associated gene CELF1 has been associated with lower expression of SPI1 in monocytes and macrophages10. AD risk alleles have been shown to be enriched in myeloid-specific epigenomic annotations and in active enhancers of monocytes, macrophages, and microglia12, and to be polarized for cis-eQTL effects in monocytes7. These findings suggest that a cell-type-specific analysis in blood and brain tissue may identify novel and more precise AD associations that may help elucidate regulatory networks. In this study, we performed a genome-wide cis ct-eQTL analysis in blood and brain, respectively, then compared eQTLs and cell-type-specific eQTLs (ct-eQTLs) between brain and blood with a focus on genes, loci, and cell types previously implicated in AD risk by genetic approaches.
Materials, subjects and methods
Study cohorts
Framingham Heart Study (FHS)
The FHS is a multigenerational study of health and disease in a prospectively followed community-based and primarily non-Hispanic white sample. Procedures for assessing dementia and determining AD status in this cohort are described elsewhere13. Clinical, demographic, and pedigree information, as well as 1000 Genomes Project Phase 1 imputed SNP genotypes and Affymetrix Human Exon 1.0 ST array gene expression data from whole blood, were obtained from dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v31.p12). Requisite information for this study was available for 5257 participants. Characteristics of these subjects are provided in Supplementary Table S1.
Religious Orders Study (ROS)/Memory and Aging Project (MAP)
ROS enrolled older nuns and priests from across the US, without known dementia for longitudinal clinical analysis and brain donation and MAP enrolled older subjects without dementia from retirement homes who agreed to brain donation at the time of death14 (http://www.eurekaselect.com/99959/article). RNA-sequencing brain gene expression and whole-genome sequencing (WGS) genotype data were obtained from the AMP-AD knowledge portal (https://www.synapse.org/#!Synapse:syn3219045) (https://www.synapse.org/).
Data processing
Generation, initial quality control (QC), and pre-processing procedures of the FHS GWAS and expression data are described elsewhere13. Briefly, the Robust Multichip Average (RMA) method15,16 was used for background adjustment and normalization of gene expression levels and further adjusted for the first principal component of ancestry. ROSMAP gene expression data were log-normalized and adjusted for known and hidden variables detected by surrogate variable analysis (SVA)17 in order to determine which of these variables should be included as covariates in analysis models for eQTL discovery. Additional filtering steps of FHS and ROSMAP GWAS and gene expression data included eliminating subjects with missing data, restricting gene expression data to protein-coding genes, and retaining common variants (MAF ≥ 0.05) with good imputation quality (R2 ≥ 0.3).
Cis-eQTL mapping
Cis-eQTL mapping was performed using a genome-wide design (Supplementary Fig. S1). The association of gene expression with SNP genotypes for all cis SNPs within 1 Mb of protein-coding genes was evaluated using linear-mixed models adjusting for family structure in FHS and linear regression models for unrelated individuals in ROSMAP. In FHS, lmekin function in the R kinship package (version 1.1.3)18 was applied assuming an additive genetic model with covariates for age and sex, and family structure modeled as a random-effects term for kinship—a matrix of kinship coefficients calculated from pedigree structures. The linear model for analysis of FHS can data be expressed as follows:
where Yi is the expression value for gene i, Gj is the genotype dosage for cis SNP j, Aij and Sij are the covariates for age and sex respectively, Uij is the random effect for family structure, and β1, β2, and β3 are regression coefficients.
ROSMAP data were analyzed using the lm function in the base stats package in R (http://www.R-project.org/). The regression model, which included covariates for age, sex, postmortem interval (PMI), study (ROS or MAP), and a term for a surrogate variable (SV1) derived from analysis of high dimensional data, can be expressed as:
where Yi is the expression value for gene i, Gj is the genotype dosage for cis SNP j, Aij, Sij, PMij, S2ij, and SV1ij are the covariates for age, sex, PMI, study, and SV1, respectively, ɛij is the residual error, and the βs are regression coefficients.
Cis ct-eQTL mapping
Models testing associations with cell-type-specific eQTLs (ct-eQTLs) included an interaction term for expression levels of “proxy” genes that represent cell types. Proxy genes representing ten cell types in whole blood4 and five cell types in brain19–21 were incorporated in cell-type-specific models (Supplementary Table S2). These proxy genes for cell types in blood were established previously using BLUEPRINT expression data to validate cell-type-specific expression in each cell-type module4 and the proxy genes for brain cell types have been incorporated in several studies19–21. Cell-type-specific expression analyses in blood of FHS participants were conducted using the following model:
where in each eQTLij pair, Yi is the eQTL expression value for gene i, Gj is the genotype dosage for cis SNP j, P is the proxy gene, P * Gj is the interaction term representing the effect of genotype in a particular cell type, Aij and Sij are covariates for age and sex, respectively, Uij is the random effect for family structure, and βs are regression coefficients. Models with significant interaction terms indicate cell-type-specific eQTLs.
The following model was used to evaluate cell-type-specific expression in the brain in ROSMAP:
where in each eQTLij pair, variables Yi, Gj, P, Aij, Sij, Pij, ɛij, and βs are as described above, and PMij, S2ij, and SV1ij are covariates for PMI, study, and SV1, respectively.
A Bonferroni correction was applied to determine the significance threshold for each analysis (Supplementary Table S3).
We assessed the relevance of the significant findings more directly to AD in two ways. In one approach, AD status was included as a covariate in the eQTL and ct-eQTL analysis models. In addition, the significant eQTLs and ct-eQTLs were evaluated separately in AD cases and controls separately in the ROSMAP brain expression dataset, but not in the FHS blood expression dataset due to the paucity of AD cases (2%) in that sample.
Selection of eQTLs in AD loci and gene-set pathway enrichment analysis
AD loci were determined based on the review of published GWAS and linkage studies of AD and AD-related traits, and this list was augmented with genes that are well recognized as functionally related to AD by experimental approaches (Supplementary Table S4). AD genes identified by GWAS met genome or study-wide significance thresholds and some of these were annotated as the closest gene to an intergenic association signal. eGenes (genes whose expression levels are associated with variation at a particular eSNP) included 88 genes and 80 eSNPs (no SNPs that significantly influence gene expression) which include genome-wide significant “peak” SNPs (i.e., top-ranked SNP within an association signal) for AD. Gene-set enrichment analysis was performed using the PANTHER (Protein ANalysis THrough Evolutionary Relationships) software tool22 to determine if the unique genes in the significant eQTL/ct-eQTL pairs shared by both brain and blood datasets are associated with a specific biological process or molecular function. The significance of the pathways was determined by the Fisher’s Exact test with false discovery rate (FDR) multiple test correction.
Colocalization analyses
Assessment of causal variants shared by adjacent GWAS and eQTL signals was performed using a Bayesian colocalization approach implemented in the R package coloc23. This analysis incorporated SNP summary statistics from a recent large AD GWAS24 and eQTL analyses described above. For the purpose of this study, a peak SNP refers to the most significantly associated AD-SNPs under a particular GWAS signal and a lead eQTL variant is defined as the eSNP showing the strongest association with gene expression. Following recommended guidelines, the variants were deemed to be colocalized by a high posterior probability that a single shared variant is responsible for both signals (PP4 > 0.8)23,25. A lower threshold for statistical significance with a false discovery rate (FDR) < 0.05 for eQTL significant results was applied to maximize detection of colocalized pairs. Regional plots were constructed with LocusZoom26.
Differential expression analysis of potential AD biomarker genes
The 386 distinct eGenes in shared eQTL pairs in significant blood and brain results were further examined for differentially expressed genes (DEG) between AD cases and controls in the AD enriched ROSMAP RNA-Seq dataset. After filtering, 308 of the total 386 genes were tested in the DEG analysis. The differences in expression among the groups were computed using the log2 transformation of the fold-change (log2FC). The differential analysis was performed using a linear model to identify DE genes between AD cases and controls implemented in R package limma (Linear Model for Microarray Data) version 3.32.7 (http://www.R-project.org/). The P values were adjusted for multiple testing to control the False Discovery Rate (FDR), with the gene considered DE when the adjusted P value was ≤0.05.
This study was approved by the Boston University Institutional Review Board.
Results
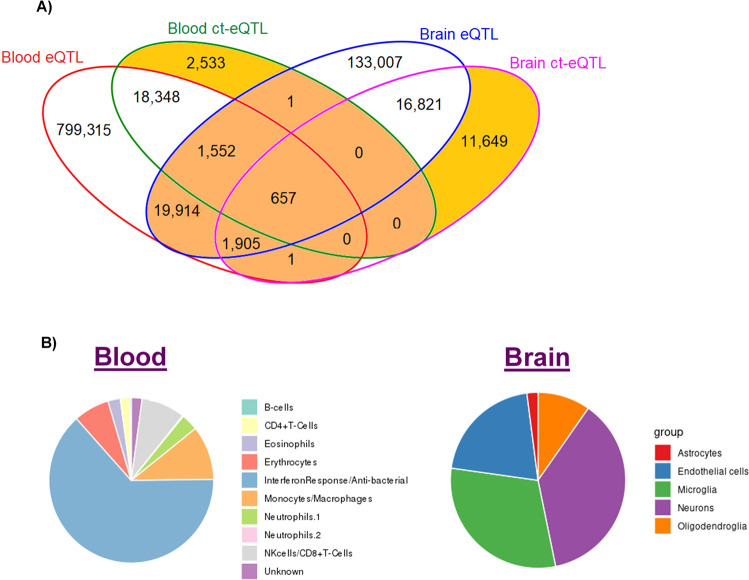
A total of 173,857 eQTLs and 51,098 ct-eQTLs in the brain, and 847,429 eQTLs and 30,405 ct-eQTLs in blood were significant after Bonferroni correction (Supplementary Table S3 and Supplemental Resources). Additional significant gene-SNP eQTLs pairs in the brain (n = 11,649) and blood (n = 2533) were observed in ct-eQTL analysis that were not detected in eQTL analysis (Fig. 1A).
Fig. 1. Significant gene-SNP eQTLs and ct-eQTLs in blood and brain tissue genome-wide.
A Venn diagram shows the number of overlapping eQTLs and ct-eQTLs in blood and brain. Gold color indicates significant eQTLs that are cell-type-specific. Orange color indicates significant eQTLs that are shared between blood and brain. B Cell-type distributions of significant genome-wide ct-eQTL results in blood and brain.
eQTLs and ct-eQTLs common to blood and brain
Of note, 24,028 significant gene-SNP eQTL pairs were shared between blood and brain. The 386 distinct eGenes among these shared eQTL pairs (Supplementary Table S5) are most enriched in the apoptosis signaling (P = 0.023) and Wnt signaling (P = 0.036) pathways (Supplementary Table S6). Five of these eGenes (HLA-DRB5, HLA-DRB1, ECHDC3, CR1, and WWOX) were previously associated with AD24,27. Three eSNPs in eQTLs involving HLA-DRB1/HLA-DRB5 (rs9271058) and ARL17A/LRRC37A2 (rs2732703 and rs113986870, which are near KANSL1 and MAPT) were previously associated with AD risk at the genome-wide significance level24,28 (Table 1).
Table 1.
eQTLs and ct-eQTLs in established AD loci appearing in both blood and brain.
| (A) eQTLs and ct-eQTLS in established AD genes in both blood and brain. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| eGene | Tissue | Cell type | Lead eSNP | Position | MAF | Beta | Std error | P value | Number of total significant eSNPs in gene/cell-type | AD GWAS peaks |
| CR1 | Blood | NA | rs7533408 | 1:207673631 | 0.25 | 0.059 | 0.006 | 3.60E-22 | 169 | NA |
| HLA-DRB5 | Blood | NA | rs9269008 | 6:32436217 | 0.17 | −2.580 | 0.057 | <1.0E-314 | 72 | NA |
| HLA-DRB1 | Blood | NA | rs9271058 | 6:32575406 | 0.14 | −2.950 | 0.028 | <1.0E-314 | 630 | Lead eSNP |
| ECHDC3 | Blood | NA | rs11257290 | 10:11780324 | 0.28 | 0.041 | 0.005 | 2.91E-19 | 115 | NA |
| WWOX | Blood | NA | rs7202722 | 16:78282458 | 0.40 | 0.023 | 0.003 | 2.60E-14 | 45 | NA |
| HLA-DRB5 | Blood | Interferon response(+)/antibacterial (−) | rs9269047 | 6:32438783 | 0.12 | −7.120 | 0.335 | 3.04E-100 | 9 [all (−)] | NA |
| HLA-DRB5 | Blood | Monocytes/macrophages | rs9269047 | 6:32438783 | 0.12 | −11.600 | 1.030 | 2.02E-29 | 1 | NA |
| HLA-DRB5 | Blood | NK cells/CD8 + T cells | rs9269047 | 6:32438783 | 0.12 | −7.660 | 0.994 | 1.30E-14 | 1 | NA |
| HLA-DRB1 | Blood | NK cells/CD8 + T cells | rs9270928 | 6:32572461 | 0.15 | −4.070 | 0.377 | 3.60E-27 | 287 | rs9271058 |
| HLA-DRB1 | Blood | Eosinophils | rs9270994 | 6:32574250 | 0.14 | −2.700 | 0.415 | 7.72E-11 | 42 | NA |
| HLA-DRB1 | Blood | Interferon response (+)/antibacterial (−) | rs9271147 | 6:32577385 | 0.14 | −5.510 | 0.250 | 1.19E-107 | 346 [260 (−)/86 (+)] | rs9271058 |
| HLA-DRB1 | Blood | Monocytes/macrophages | rs9271148 | 6:32577442 | 0.13 | −6.110 | 0.709 | 6.83E-18 | 222 | rs9271058 |
| CR1 | Brain | NA | rs12037841 | 1:207684192 | 0.17 | −0.096 | 0.007 | 9.25E-44 | 64 | rs6656401 |
| HLA-DRB5 | Brain | NA | rs3117116 | 6:32367017 | 0.12 | −2.780 | 0.070 | <1.0E-314 | 10537 | rs9271058, rs9271192 |
| HLA-DRB1 | Brain | NA | rs73399473 | 6:32538959 | 0.26 | −2.050 | 0.058 | 8.78E-272 | 10792 | rs9271058, rs9271192 |
| ECHDC3 | Brain | NA | rs866770710 | 10:11784320 | 0.0002 | −0.252 | 0.018 | 4.61E-44 | 45 | NA |
| WWOX | Brain | NA | rs12933282 | 16:78124987 | 0.45 | −0.133 | 0.017 | 1.13E-15 | 75 | NA |
| HLA-DRB5 | Brain | Microglia | rs67987819 | 6:32497655 | 0.14 | −1.900 | 0.137 | 9.82E-44 | 754 | NA |
| HLA-DRB5 | Brain | Endothelial cells | rs67987819 | 6:32497655 | 0.14 | −2.410 | 0.220 | 6.32E-28 | 343 | NA |
| HLA-DRB1 | Brain | Microglia | rs72847627 | 6:32538512 | 0.28 | −2.130 | 0.125 | 4.15E-65 | 2305 | rs9271058, rs9271192 |
| HLA-DRB1 | Brain | Neurons | rs115480576 | 6:32538570 | 0.26 | −2.210 | 0.153 | 2.72E-47 | 3263 | rs9271058, rs9271192 |
| HLA-DRB1 | Brain | Endothelial cells | rs9269492 | 6:32542924 | 0.30 | −2.250 | 0.243 | 2.06E-20 | 351 | rs9271192 |
| HLA-DRB5 | Brain | Neurons | rs9270035 | 6:32553446 | 0.14 | −2.520 | 0.137 | 1.46E-75 | 2540 | rs9271058, rs9271192 |
| ECHDC3 | Brain | Neurons | rs866770710 | 10:11784320 | 0.0002 | 0.328 | 0.045 | 3.13E-13 | 2 | NA |
| (B) eQTLs and ct-eQTLs involving AD GWAS association peak SNPs in both brain and blood. | ||||||||
|---|---|---|---|---|---|---|---|---|
| eGene | Tissue | Cell type | eSNP + GWAS SNP | Positiona | MAF | Beta | Std error | P value |
| HLA-DRB1 | Blood | NA | rs9271058 | 6:32575406 | 0.27 | −2.950 | 0.028 | <1.0E-314 |
| ARL17A | Blood | NA | rs2732703 | 17:44353222 | 0.21 | 0.147 | 0.023 | 5.95E-11 |
| ARL17A | Blood | NA | rs113986870 | 17:44355683 | 0.09 | 0.166 | 0.025 | 2.30E-11 |
| HLA-DRB1 | Blood | Interferon response (+)/antibacterial (−) | rs9271058 | 6:32575406 | 0.27 | −3.010 | 0.159 | 6.36E-80 |
| HLA-DRB1 | Blood | NK cells/CD8 + T cells | rs9271058 | 6:32575406 | 0.27 | −4.090 | 0.464 | 1.20E-18 |
| HLA-DRB1 | Blood | Monocytes/macrophages | rs9271058 | 6:32575406 | 0.27 | −3.540 | 0.497 | 1.06E-12 |
| HLA-DRB1 | Brain | NA | rs9271058 | 6:32575406 | 0.27 | −1.690 | 0.054 | 1.94E-213 |
| HLA-DRB5 | Brain | NA | rs9271058 | 6:32575406 | 0.27 | −1.770 | 0.081 | 2.28E-106 |
| LRRC37A2 | Brain | NA | rs2732703 | 17:44353222 | 0.21 | 1.370 | 0.053 | 4.13E-150 |
| LRRC37A2 | Brain | NA | rs113986870 | 17:44355683 | 0.09 | 1.260 | 0.068 | 1.98E-76 |
| ARL17A | Brain | NA | rs113986870 | 17:44355683 | 0.09 | −0.326 | 0.047 | 4.96E-12 |
| HLA-DRB1 | Brain | Microglia | rs9271058 | 6:32575406 | 0.27 | −1.400 | 0.111 | 1.80E-36 |
| HLA-DRB1 | Brain | Neurons | rs9271058 | 6:32575406 | 0.27 | −1.650 | 0.135 | 2.37E-34 |
| HLA-DRB5 | Brain | Neurons | rs9271058 | 6:32575406 | 0.27 | −1.550 | 0.201 | 1.24E-14 |
| LRRC37A2 | Brain | Neurons | rs2732703 | 17:44353222 | 0.21 | 1.520 | 0.140 | 1.84E-27 |
| LRRC37A2 | Brain | Microglia | rs2732703 | 17:44353222 | 0.21 | 1.480 | 0.147 | 7.65E-24 |
| LRRC37A2 | Brain | Endothelial cells | rs2732703 | 17:44353222 | 0.21 | 1.750 | 0.233 | 5.88E-14 |
| LRRC37A2 | Brain | Microglia | rs113986870 | 17:44355683 | 0.09 | 1.530 | 0.195 | 4.29E-15 |
| LRRC37A2 | Brain | Neurons | rs113986870 | 17:44355683 | 0.09 | 1.400 | 0.184 | 2.77E-14 |
aPosition according to GRCh37 assembly.
MAF = minor allele frequency of variant in 1000 Genomes Combined European Population; cell-type-specific result rows are in bold.
eQTLs involving CR1, ECHDC3, and WWOX were much more significant in the brain than blood, whereas HLA-DRB5 and HLA-DRB1 were more significant in blood when comparing the effect sizes. ECHDC3 was a significant eGene in blood and brain eQTLs (specifically in neurons). HLA-DRB5 and HLA-DRB1 were the only eGenes ascribed to significant ct-eQTLs in both blood and brain noting that of the ten distinct lead eSNPs, five are unique to each tissue (Table 1). Although the eQTLs involving these genes with the largest effect were observed in blood across multiple cell types, the total number of significant eSNP-eGene combinations was far greater in brain (particularly in microglia and neurons). The only instance in which the lead eSNP is also associated with AD risk at the GWS level was observed in the blood eQTL pair of HLA-DRB1 with eSNP rs9271058 (Table 1). Among the AD-associated SNPs at the GWS level, rs9271058 is a significant eSNP for HLA-DRB1 in both blood and brain cell types (the most significant association by P value was observed in antibacterial cells and microglia) and rs9271192 is a significant ct-eQTL for the gene in multiple brain cell types (Table 1). Both of these SNPs are also eSNPs for HLA-DB5 in the brain in neurons only.
There were 657 gene–SNP eQTL pairs comprising 16 unique eGenes that were significant in blood and brain overall as well as in specific cell types in both blood and brain (Supplementary Table S7). None of these eGenes were observed in significant pathways enriched for AD genes, however, they included AD-associated genes HLA-DRB1 and HLA-DRB5.
eQTLs and ct-eQTLs among previously established AD loci
Slightly more than half (42/80 = 52.5%) of the established AD associations (Supplementary Table S3) are eGene targets for significant eQTLs in blood (Supplementary Table S8). By comparison, only seven established AD loci were eGene targets for significant eQTLs in the brain, among which OARD1 was significant in endothelial cells only (Supplementary Table S8). Many GWS SNPs for AD risk are eSNPs affecting the expression of the nearest gene, which is usually recognized as the causative gene, but several GWS SNPs target other genes (Supplementary Table S9). For example, AD-associated eSNPs rs113986870 and rs2732703 in the MAPT/KANSL1 region target ARL17A in blood, but are paired in seven of eight eQTLs and ct-eQTLs with LRRC37A2 in the brain (Supplementary Table S9). HLA-DRB1 is the only AD gene with a significant ct-eQTL in blood, whereas many AD genes have significant blood eQTLs. In the brain, only four AD loci (CR1, HLA-DRB1/DRB5, IQCK, and MAPT/KANSL1) have significant brain eQTLs of which HLA-DRB1/DRB5 and MAPT/KANSL1 are the only brain ct-eQTLs, noting that all are significant in microglia, neurons, and endothelial cells.
Next, we evaluated whether the most significant eSNPs and SNPs genome-wide significantly associated with AD status (i.e., AD-SNPs) co-localize and thus to identify a single shared variant responsible for both signals (posterior probability of shared signals (PP4) > 0.8). This analysis revealed eight eQTL/ct-eQTL signals that colocalized with seven AD GWAS signals and half of the colocalized signals involved a ct-eQTL (Table 2 and Supplementary Fig. S2). Two different eSNPs for CD2AP, rs4711880 (eQTL P = 1.4 × 10−104) and rs13201473 (NK/CD8 + T cell ct-eQTL P = 1.47 × 10−9), flank CD2AP GWAS SNP rs10948363 which is also the second most significant eQTL (P = 2.32 × 10−104) and the second most significant ct-eQTL in NK cells/CD8 + T cells (P = 2.66 × 10−9). These three SNPs span a 9.0-kb region in intron 2 and are in complete linkage disequilibrium (LD, r2 = 1.0), indicating that any one or more of them could affect the function of target gene CD2AP. Rs6557994 is the most significant eSNP for and located in PTK2B (blood interferon ct-eQTL P = 2.58 × 10−9) and is moderately correlated with the PTK2B GWAS SNP (rs28834970, r2 = 0.78, P = 1.58 × 10−9). Thus, it is not surprising that rs6557994 is also significantly associated with AD risk (P = 8.19 × 10−7). Rs6557994 is also correlated with a GWAS SNP in CLU, located approximately 150 kb from PTK2B, that is not significantly associated with the expression of any gene. Because PTK2B and CLU are independent AD risk loci27, it is possible that this eSNP has an effect on AD pathogenesis through independent pathways (Supplementary Fig. S2). The most significant eSNP in MADD (rs35233100, P = 2.88 × 10−10) was predicted to have functional consequences because it is a stop-gained mutation. This brain eQTL is colocalized (PP4 = 0.95) and weakly correlated with a GWAS SNP (P = 1.91 × 10−5) in CELF1 rs10838725 (r2 = 0.12).
Table 2.
Colocalized AD GWAS/lead eQTL SNP pairs.
| Regiona | AD GWAS Variant | Lead eQTL variant | eQTL type | PP4 | r2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Nearest gene | MAF | P value | eQTL P value | eGene | Cell type | rsID | MAF | eGene | eQTL P value | Cell type | GWAS P value | ||||
| 6:46487762–48487762 | rs10948363 | CD2AP | 0.72 | 1.77E-07 | 2.32E-104 | CD2AP | NA | rs4711880 | 0.23 | CD2AP | 1.36E-104 | NA | 2.57E-07 | Blood eQTL | 0.909 | 1.00 |
| 6:46487762–48487762 | rs10948363 | CD2AP | 0.72 | 1.77E-07 | 2.66E-09 | CD2AP | NK cells/CD8 + T cells | rs13201473 | 0.27 | CD2AP | 1.47E-09 | NK cells/CD8 + T cells | 2.74E-07 | Blood ct-eQTL | 0.917 | 1.00 |
| 8:26195121–28195121 | rs28834970 | PTK2B | 0.63 | 1.58E-09 | 9.15E-09 | PTK2B | Interferon response/antibacterial cells | rs6557994 | 0.41 | PTK2B | 2.58E-09 | Interferon response/antibacterial cells | 8.19E-07 | Blood ct-eQTL | 0.990 | 0.78 |
| 8:26467686–28467686 | rs9331896 | CLU | 0.61 | 3.62E-16 | Not an eSNP | rs6557994 | 0.45 | PTK2B | 2.58E-09 | Interferon response/antibacterial cells | 8.19E-07 | Blood ct-eQTL | 0.990 | 0.00 | ||
| 1:206692049–208692049 | rs6656401 | CR1 | 0.19 | 2.17E-15 | 1.05E-43 | CR1 | NA | rs12037841 | 0.19 | CR1 | 9.25E-44 | NA | 1.77E-15 | Brain eQTL | 0.993 | 1.00 |
| 11:46557871–48557871 | rs10838725 | CELF1 | 0.68 | 1.91E-05 | Not an eSNP | rs35233100 | 0.068 | MADD | 2.88E-10 | NA | 1.25E-03 | Brain eQTL | 0.954 | 0.12 | ||
| 11:58923508–60923508 | rs983392 | MS4A6A | 0.59 | 4.76E-15 | Not an eSNP | rs11230563 | 0.35 | CD6 | 2.31E-113 | NA | 0.48 | Brain eQTL | 0.854 | 0.00 | ||
| 19:44411941–46411941 | rs429358 | APOE | 0.78 | < 1.0E-300 | Not an eSNP | rs74253343 | 0.47 | RELB | 1.9E-14 | Oligodendroglia | 0.23 | Brain ct-eQTL | 0.971 | 0.00 | ||
aMap position within 1 Mb of AD GWAS SNP according to GRCh37 assembly.
MAF minor allele frequency, NA not available, PP4 posterior probability of colocalization, r2 correlation of AD and eQTL variants.
ct-eQTLs genome-wide
Examination of the distribution of the significant ct-eQTL results genome-wide showed that nearly two-thirds of the ct-eQTLs in blood occurred in interferon response/antibacterial cells which are defined as type I interferon viral response cells in upregulated genes and type II interferon antibacterial inflammatory response cells in downregulated genes4, whereas brain ct-eQTLs are highly represented in endothelial cells, neurons, and microglia (Fig. 1B and Supplementary Table S10). Further examination of significant results within myeloid cell lineages (i.e., microglia and monocytes/macrophages) which account for a large proportion of the genetic risk for late-onset AD10 revealed that 3234 or 10.6% of all significant ct-eQTLs in blood were in monocytes/macrophages. This subset includes 128 unique eGenes which are significantly enriched in the AD amyloid secretase pathway (FDR P = 0.013, Supplementary Table S11). A total of 974 or 30.1% of ct-eQTLs including 4 of the 20 most significant eGenes in monocytes/macrophages are located within 1 Mb of established AD loci. One of the eGenes in this top-ranked group (HLA-DRB5) is an established AD gene, and three others that are near established AD loci (DLG2 near PICALM29, C4BPA near CR130, and MYO1E near ADAM10 31) are reasonable AD gene candidates based on evidence using non-genetic approaches (Table 3). Microglia accounted for 15,560 (30.5%) of significant ct-eQTLs in the brain (Supplementary Table S10) which involved 304 unique eGenes. Approximately 52% of significant ct-eQTLs in microglia are located in AD regions including five of the 20 most significant ct-eQTLs in this group (Table 3). One of these five eGenes is an established AD gene (HLA-DRB1) and two others (ALCC32 and WNT333) have been linked to AD in previous studies.
Table 3.
Top-ranked ct-eQTLs in myeloid cell types.
| (A) Monocytes/macrophages | |||||||
|---|---|---|---|---|---|---|---|
| eGene | Lead eSNP | Positiona | MAF | Beta | Std error | P value | Number of significant eSNPs in gene/cell type |
| SLC12A1 | rs8037626 | 15:48606346 | 0.17 | −3.340 | 0.219 | 1.62E-52 | 126 |
| DLG2 | rs75798025 | 11:84018349 | 0.01 | 5.350 | 0.364 | 6.66E-49 | 597 |
| ABCA9 | rs4147976 | 17:66925923 | 0.44 | 0.872 | 0.068 | 1.97E-37 | 48 |
| PTPRG | rs116497321 | 3:62245373 | 0.01 | 2.650 | 0.221 | 3.96E-33 | 10 |
| CLNK | rs5028371 | 4:10452986 | 0.50 | 1.060 | 0.092 | 7.66E-31 | 272 |
| NFXL1 | rs10938499 | 4:47848377 | 0.33 | −1.270 | 0.112 | 8.38E-30 | 73 |
| FCRL5 | rs12760587 | 1:157526021 | 0.23 | 2.140 | 0.19 | 1.99E-29 | 93 |
| HLA-DRB5 | rs9269047 | 6:32438783 | 0.12 | −11.600 | 1.03 | 2.02E-29 | 1 |
| FMOD | NA | 1:203263699 | NA | 2.110 | 0.2 | 5.08E-26 | 42 |
| ABCA6 | rs144031521 | 17:67162715 | 0.01 | 8.620 | 0.833 | 4.27E-25 | 9 |
| INPP5F | rs181735165 | 10:121555618 | 0.02 | 7.150 | 0.701 | 1.99E-24 | 11 |
| RBMS3 | rs192885607 | 3:29612955 | 0.00 | 2.570 | 0.257 | 1.52E-23 | 34 |
| ARHGAP44 | NA | 17:12750576 | NA | 1.760 | 0.177 | 2.69E-23 | 54 |
| C4BPA | rs74148971 | 1:207275799 | 0.07 | −2.300 | 0.234 | 8.44E-23 | 24 |
| DCLK2 | rs114930380 | 4:150954757 | 0.03 | 1.630 | 0.169 | 5.16E-22 | 39 |
| PAM | NA | 5:102153433 | NA | −0.691 | 0.073 | 2.00E-21 | 47 |
| MYO1E | rs146483144 | 15:59422810 | 0.03 | 4.300 | 0.453 | 2.26E-21 | 17 |
| DSP | rs4960328 | 6:7495948 | 0.42 | 0.554 | 0.061 | 9.30E-20 | 6 |
| ROR1 | rs1557596882 | 1:64453767 | 0.01 | 3.570 | 0.393 | 1.05E-19 | 31 |
| CACNB2 | rs117299889 | 10:18404550 | 0.06 | 1.510 | 0.168 | 2.52E-19 | 61 |
| (B) Microglia | |||||||
|---|---|---|---|---|---|---|---|
| eGene | Lead eSNP | Positiona | MAF | Beta | Std error | P value | Number of significant eSNPs in gene/cell type |
| AC142381.1 | rs199931530 | 16:33047273 | 0.45 | −0.401 | 0.012 | 3.89E-233 | 43 |
| MLANA | rs201480524 | 9:68457329 | 0.50 | −0.176 | 0.007 | 4.82E-124 | 18 |
| AC015688.3 | rs62058902 | 17:25303954 | 0.50 | −0.213 | 0.009 | 1.94E-113 | 11 |
| HNRNPCL1 | rs75627772 | 1:13182567 | 0.00 | 0.186 | 0.008 | 4.31E-113 | 8 |
| AL050302.1 | rs3875276 | 21:14472722 | 0.50 | −0.890 | 0.040 | 2.62E-111 | 142 |
| ALLC | rs9808287 | 2:3624799 | 0.11 | 0.833 | 0.044 | 1.41E-79 | 12 |
| FAM21B | NA | 10:47917284 | NA | −2.210 | 0.118 | 2.88E-78 | 22 |
| WNT3 | rs9904865 | 17:44908263 | 0.37 | −1.570 | 0.084 | 3.90E-78 | 1 |
| RPL9 | rs1458255 | 4:39446549 | 0.28 | −2.370 | 0.137 | 4.76E-67 | 37 |
| HLA-DRB1 | rs72847627 | 6:32538512 | 0.32 | −2.130 | 0.125 | 4.15E-65 | 2305 |
| XRCC2 | rs80034602 | 7:152104360 | 0.50 | 2.120 | 0.128 | 1.30E-61 | 5 |
| WI2-3308P17.2 | rs4067785 | 1:120576209 | 0.50 | −0.184 | 0.011 | 1.33E-58 | 9 |
| DEFB121 | rs117541536 | 20:29422202 | 0.49 | −7.420 | 0.460 | 1.56E-58 | 1 |
| GINS1 | rs75374582 | 20:26109209 | 0.50 | −33.200 | 2.060 | 1.95E-58 | 5 |
| EXOSC10 | rs2580511 | 1:121113600 | 0.50 | −4.390 | 0.276 | 5.78E-57 | 5 |
| TRIM49B | rs202086299 | 11:48363026 | 0.50 | 0.205 | 0.013 | 2.16E-54 | 5 |
| TMPRSS9 | rs7248384 | 19:23936403 | 0.48 | −1.410 | 0.093 | 1.83E-52 | 1 |
| LDHC | NA | 11:18432033 | NA | 0.428 | 0.030 | 1.07E-47 | 73 |
| HLA-DOB | rs201194354 | 6:32796857 | NA | 1.190 | 0.084 | 4.39E-46 | 70 |
| DEFB119 | rs78099404 | 20:29617870 | 0.50 | −0.377 | 3.51E-15 | <1.0E-314 | 142 |
aMap position according to GRCh37 assembly.
MAF minor allele frequency, NA not available.
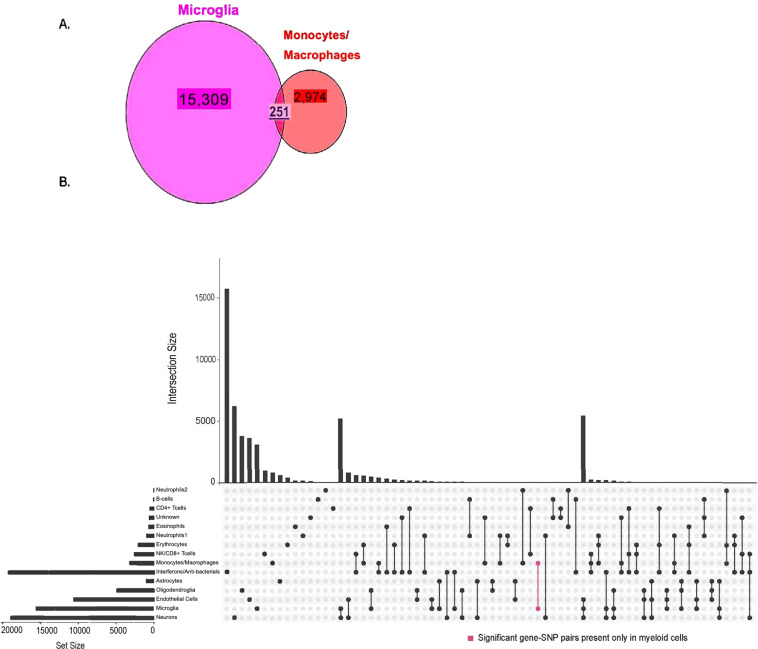
Overlap of eQTLs and ct-eQTLs among myeloid cell types
Considering significant eGene–eSNP pairs in myeloid cell types, 251 pairs including five distinct eGenes (BTNL3, FAM118A, HLA-DOB, HLA-DRB1, and HLA-DRB5) are shared between microglia and monocytes/macrophages (Table 4A and Fig. 2A). Three of these pairs involving eSNPs rs3763355, rs3763354, and rs1183595100 have the same target gene HLA-DOB and occur only in microglia and monocytes/macrophages (Table 4B). Among the significant ct-eQTLs in the brain, the cell types with the largest proportion that were also significant in monocytes/macrophages were microglia (1.6%) and neurons (1.3%) (Table 4). Conversely, among the significant ct-eQTLs in blood, the cell types with the largest proportion that were also significant in microglia were NK/CD + T cells (12.9%) and monocytes/macrophages (7.8%). Among ct-eQTLs which are significant only for one cell-type each in blood and one in the brain, monocytes/macrophages shared three ct-eQTLs with microglia but with no other brain cell types (Fig. 2B and Table 4C). By comparison, microglia shared 63 ct-eQTLs with interferons/antibacterial cells, but with no other blood cell types. The proportions of overlap of ct-eQTLs between blood and brain across ten paired cell types are significantly different (Fisher’s Exact test = 789.8, P = 2.2 × 10−16). The much larger number of ct-eQTLs in microglia that were common with interferons/bacterial cells than monocytes/macrophages may reflect the substantially greater proportion of significant eQTLs in blood involving interferons/antibacterial cells (64%) than monocytes/macrophages (10.6%) (Supplementary Table S10). The only other ct-eQTLs that were unique to a pair of cell types in brain and blood cell type involved neurons paired with neutrophils (n = 3) and with interferons/antibacterial cells (n = 65) (Fig. 2B).
Table 4.
Overlap of ct-eQTLs in myeloid cell types in brain and blood.
| (A) Unique eGenes shared in significantly associated ct-eQTLs in monocytes/macrophages and microglia. Number below each gene represents significant eGene-eSNP eQTL pairs in each gene. | ||||
|---|---|---|---|---|
| BTNL3 | FAM118A | HLA-DOB | HLA-DRB1 | HLA-DRB5 |
| 1 | 43 | 6 | 200 | 1 |
| (B) eSNP-eGene pairs among ct-eQTLs significant in both monocytes/macrophages and microglia. | ||||||||
|---|---|---|---|---|---|---|---|---|
| eGene | eSNP | Position | MAF | Monocytes/macrophages | Microglia | AD GWAS P value23 | ||
| Beta | P value | Beta | P value | |||||
| HLA-DOB | rs3763355 | 6:32786882 | 0.06 | −2.02 | 9.98E-15 | 0.938 | 3.89E-14 | 0.001 |
| HLA-DOB | rs3763354 | 6:32786917 | 0.15 | −1.11 | 1.40E-10 | −0.642 | 2.80E-13 | 0.652 |
| HLA-DOB | rs1183595100 | 6:32768232 | NA | −1.13 | 8.34E-11 | −0.605 | 1.98E-11 | NA |
| (C) Overlap of significant eQTLs in brain and blood with ct-eQTLs in myeloid cell types. | ||||
|---|---|---|---|---|
| Cell types | Monocytes/macrophages | Microglia | ||
| Blood | # ct-eQTLs common to cell-type pair | # ct-eQTLs unique to cell-type pair | # ct-eQTLs common to cell-type pair | # ct-eQTLs unique to cell-type pair |
| Neutrophils1 | 3 (0.3%)a | 0 | ||
| CD4 + T cells | 3 (0.5%) | 0 | ||
| NK/CD8 + T cells | 337 (12.9%) | 0 | ||
| Erythrocytes | 119 (5.6%) | 0 | ||
| Monocytes/macrophages | 251 (7.8%) | 3 | ||
| Unknown | 0 | 0 | ||
| Interferon/antibacterial | 628 (3.3%) | 63 | ||
| Neutrophils2 | 0 | 0 | ||
| B cells | 0 | 0 | ||
| Eosinophils | 38 (5.2%) | 0 | ||
| Brain | ||||
| Endothelial cells | 55 (0.5%) | 0 | ||
| Neurons | 250 (1.3%) | 0 | ||
| Microglia | 251 (1.6%) | 3 | ||
| Astrocytes | 0 | 0 | ||
| Oligodendroglia | 0 | 0 | ||
aNumber in parentheses represent the proportion of ct-eQTLs for each cell type on the left that were also observed in either microglia or monocytes/macrophages.
Fig. 2. Intersection of significant gene-SNP eQTL pairs between cell types in blood and brain tissue.
A Venn diagram showing overlap of ct-eQTL pairs in myeloid cell types (microglia and monocytes/macrophages). B Number of significant eQTLs unique to and that overlap cell types in blood and brain. The bar chart on the left side indicates the number of significant eQTLs involving each cell type and the bar chart above the matrix indicates the number of significant eQTLs that are unique to each cell type and set of cell types. Pink colored bar indicates the number of eQTLs pairs that are unique to microglia and monocytes/macrophages.
Effect of AD status on significant eQTLs and ct-eQTLs
None of the significant eQTLs and ct-eQTLs observed in the brain (Table 1) were influenced by the inclusion of AD status in the analysis models. Stratified analyses revealed that the top findings involving eSNPs that were previously associated with AD at the genome-wide significant level were evident in both AD cases and controls (Supplementary Table 12A). Although most of the findings were more significant in AD cases than controls (noting that the ROSMAP brain sample of AD cases was 44% larger than the control sample), the effect size for most eSNP–eGene pairs was similar. However, patterns among AD cases and controls differed when focusing on the most significant eQTLs and ct-eQTLs in established AD genes. For example, eQTLs observed in undifferentiated brain cells involving CR1 paired with rs6656401 (P = 7.85 × 10−22), in endothelial cells involving HLA-DRB1 paired with rs73399473 (P = 2.5 × 10−10) and HLA-DRB5 paired with rs1064697 (P = 2.18 × 10−14), in microglia involving HLA-DRB1 paired with rs72847627 (P = 4.43 × 10−51), and in neurons involving ECHDC3 paired with rs866770710 (P = 5.79 × 10−13) were significant only in AD cases (Supplementary Table 12B). Other eQTLs observed in multiple cell types involving these same genes (HLA-DRB1: rs111976080, P = 1.68 × 10−25; HLA-DRB5: rs2395517, P = 8.64 × 10−12, rs9271184, P = 5.42 × 10−41, and rs80141235, P = 3.94 × 10−9) were significant only in controls. Several other eQTLs and ct-eQTLs in CR1, HLA-DRB1, and HLA-DRB5 were highly significant in one group but showing a much less significant effect in the opposite direction in the other group.
Among the 386 eGenes that were significant in both blood and brain (Supplementary Table S5), 87 were differentially expressed between AD cases and controls (Supplementary Table S13). This includes WWOX (Padj = 1.02 × 10−4) and LRRC2 (Padj = 2.38 × 10−3) which have been associated with AD risk by GWAS24,34.
Discussion
We identified several novel AD-related eQTLs that highlight the importance of cell-type-dependent context. It is noteworthy that there were more significant ct-eQTLs in the brain (n = 51,098) than blood (n = 30,405) even though the dataset containing expression data from blood (FHS) is several times larger than the brain expression dataset (ROSMAP). This could be due to greater cell-type heterogeneity in the brain, the enrichment of AD cases in the ROSMAP dataset who may show different patterns of gene expression compared to persons without AD, or highly variable gene expression across cell types in the nervous system35. Because expression studies in the brain are often constrained by the small number of specimens compared to studies in other tissues, postmortem changes that may affect gene expression in the brain36, and the growing recognition that AD is a systemic disease37–39, incorporating expression data from multiple tissues can enhance discovery of additional genetic influences on AD risk and pathogenesis.
Although most significant findings were tissue-specific, the 386 distinct eGenes among more than 24,000 significant gene-SNP eQTL pairs that were shared between blood and brain were enriched in the apoptosis signaling pathway which has been suggested to contribute to the underlying pathology associated with AD40,41. Six established AD genes (CR1, ECHDC3, HLA-DRB1, HLA-DRB5, LRRC2, and WWOX24,27,34) were shared eGenes in the brain and blood. They were also involved in eQTLs and ct-eQTLs that showed different patterns of association in cases versus controls (i.e., CR1, HLA-DRB1, HLA-DRB5, and ECHD3) or differentially expressed in AD cases versus controls (i.e., WWOX and LRRC2).
The complement receptor 1 (CR1) gene encodes a transmembrane glycoprotein functioning in the innate immune system by promoting phagocytosis of immune complexes, cellular debris, and Aβ42. CR1 is an eGene for several eSNPs, including AD GWAS peak SNP rs6656401 located within the gene, in brain and blood eQTLs and the effects on CR1 expression are opposite in blood and brain. There are multiple possible explanations for the effect direction differences across tissues. The effect of eSNP rs6656401 on CR1 expression may be developmental, noting that the average age of the FHS subjects (a group with expression data in blood) is more than 30 years younger than the ROSMAP subjects (group with expression data in the brain). The difference between brain and blood may also reflect postmortem changes in the brain that are not indicative of expression in vivo. Alternatively, these effects may be related to AD because few FHS subjects were AD cases at the time of blood draw, whereas 60% of subjects in the ROSMAP sample are AD cases. This idea is supported by the observation of a larger and positive effect of rs6656401 on CR1 expression in AD (β = 0.020) compared to control brains (β = −0.0086). Opposite effect directions of expression in brain and blood from AD patients compared to controls have been observed for several ribosomal genes43. GWS variants located in the region spanning ECHDC3 and USP6NL have previously been associated with AD44. Altered ECHDC3 expression in AD brains45 supports the idea that this gene has a role in AD. Knockout of WWOX in mice leads to aggregation of amyloid-β (Aβ) and Tau, and subsequent cell death46,47. LRRC2 is located in a region including GWS variants that modify the inverse relationship between education attainment and AD34. A recent study showed that the expression of a LRRC2 long noncoding RNA (LCCR2-AS1) is significantly increased in children with autism spectrum disorder compared to children with normal development48.
The human leukocyte antigen (HLA) region is the key susceptibility gene in many immunological diseases and many associations have been reported between neurodegenerative diseases and HLA haplotypes49. In addition, the most widely used marker to examine activated microglia in normal and diseased human brains is HLA-DR and microglia activation increases with the progression of AD50,51. HLA-DRB5 and HLA-DRB1 have been implicated in numerous GWAS studies as significantly associated with AD risk24,27 and appeared frequently among significant results in blood and brain in this study. Rs9271058, which is located approximately 17.8 kb upstream of HLA-DRB1, is significantly associated with AD risk (P = 5.1 × 10−8)24. and when paired with HLA-DRB1 is a significant eQTL and ct-eQTL in multiple cell types in blood and brain including myeloid lineage cells (i.e., monocytes/macrophages and microglia). This eSNP is also a significant eQTL in the brain and specifically in neurons when paired with HLA-DRB5. Rs9271192, which is adjacent to rs9271058 and also significantly associated with AD risk (P = 2.9 × 10−12)27, is a significant eQTL and ct-QTL with multiple cell types in brain but not blood when paired with HLA-DRB5 and HLA-DRB1.
Significant associations for AD have been reported with variants spanning a large portion of the major histocompatibility (MHC) region in HLA class I, II, and III loci49,52,53. While the strongest statistical evidence for association in this region is with variants in HLA-DRB124, fine mapping in this region suggests that a class I haplotype (spanning the HLA-A and HLA-B loci) and a class II haplotype (including variants in HLA-DRB1, HLA-DQA1, and HLA-DQB1) are more precise markers of AD risk. Given the complexity of the MHC region and extensive LD, further work is needed to confirm whether this is a true eQTL or a signal generated from a specific HLA allele or haplotype. Although functional studies may be required to discern which HLA variants have AD relevant consequences and HLA polymorphisms methods would be required to detect differential gene expression between the HLA alleles, our findings support a role for the immune system in AD37,54 and the hypothesis that a large proportion of AD risk can be explained by genes expressed in myeloid cells10.
The potential importance and relevance to AD of eQTLs and ct-eQTLs in myeloid cell types are supported by the observation that a large portion of GWS ct-eQTLs we identified map within 1 Mb of established AD loci, and 58% (12/20 in monocytes/macrophages and 11/20 in microglia) of the most significant eGenes have been previously implicated in AD. DLG2 encodes a synaptic protein whose expression was previously reported as downregulated in an AD proteome and transcriptome network55 and inversely associated with AD Braak stage29. Genome-wide significant associations of AD risk with PTPRG was observed in a family-based GWAS56 and with CLNK in a recent large GWAS for which the evidence was derived almost entirely with a proxy AD phenotype in the UK Biobank57. NFXL1 is a novel putative substrate for BACE1, an important AD therapeutic target58. FCRL5 may interact with the APOE*E2 allele and also modifies AD age of onset59. C4BPA was shown to be consistently downregulated in MCI and AD patients, and the protein encoded by this gene accumulates in Aβ plaques in AD brains30,60. Lower levels of the PAM have been observed in the brains and CSF of AD patients compared to healthy controls61 and MYO1E is expressed by anti-inflammatory disease-associated microglia31. As a calcium channel protein, CACNB2 may affect AD risk by altering calcium levels which could cause mitochondrial damage and then induce apoptosis62,63.
Likewise, several eGenes of top-ranked ct-eQTLs in microglia that are not established AD loci may have a role in the disease. It was shown that copy number variants (CNVs) near HNRNPCL1 overlapped the coding portion of the gene in AD cases but not controls64. A region of epigenetic variation in ALLC was associated with AD neuropathology32. FAM21B, a retromer gene in the endosome-to-Golgi retrieval pathway, was associated with AD in a candidate gene study65. Vacuolar sorting proteins genes in this pathway including SORL1 have been functionally linked to AD through the trafficking of Aβ66. One study demonstrated that WNT3 expression in the hippocampus was increased by exercise and alleviated AD-associated memory loss by increasing neurogenesis33. Expression of RPL9 is downregulated in severe AD67 and significantly differs by sex among persons with the APOE ɛ4 allele68. Significant evidence of association with a TRIM49B SNP was found in a genome-wide pleiotropy GWAS of AD and major depressive disorder (MDD)69.
HLA-DOB, which is one of the five distinct eGenes (BTNL3, FAM118A, HLA-DOB, HLA-DRB1, and HLA-DRB5) for significant ct-eQTLs shared between microglia and monocytes/macrophages, and is the target gene for three eSNPs (rs3763355, rs3763354, and rs1183595100) that were evident only in these myeloid cell types. These eSNPs have similar eQTL p-values in both cell types, but have slightly larger effect sizes in monocytes (Fig. 2). The effect of rs3763355 on expression is in opposite directions in monocytes and microglia which suggests HLA-DOB may be acting in different immune capacitates in AD in blood and brain. Though the functions of the genes BTNL3 and FAM118A are unknown, a BTNL8-BTNL3 deletion has been correlated with TNF and ERK1/AKT pathways, which have an important role in immune regulation inducing inflammation, apoptosis, and proliferation, suggesting the deletion could be correlated to inflammatory disease70. This suggests that the majority of the shared myeloid cell-type genes—the HLA genes and possibly BTNL3—are all immune-related. Ct-eQTLs involving microglia and monocytes/macrophages had a larger proportion of total intersection, an isolated set interaction and a statistically significant overlap (P < 1.0 × 10−314), demonstrating a stronger connection than other brain/blood cell types in this study and thus providing further evidence for the importance of the immune system in AD.
The proportions of significant ct-eQTLs in NK cells/CD8 + T cells, monocytes/macrophages, and eosinophils are comparable to those observed in reference blood tissue (https://www.miltenyibiotec.com/US-en/resources/macs-handbook/human-cells-and-organs/human-cell-sources/blood-human.html#gref), (https://www.stemcell.com/media/files/wallchart/WA10006-Frequencies_Cell_Types_Human_Peripheral_Blood.pdf). Similarly, significant eQTL distributions in endothelial cells, neurons, and glia are consistent with reference brain tissue71. The majority of significant blood eQTLs were type I interferon response cells which cross-regulate with pro-inflammatory cytokines that drive the pathogenesis of autoimmune diseases including AD and certain heart diseases72–74 and the enrichment of interferon ct-eQTLs in this study could possibly be due to the high proportion of subjects these diseases in the FHS dataset. In contrast, the proportion of significant ct-eQTLs among glial cells is much lower in astrocytes and oligodendrocytes and three-fold higher in microglia than in reference brain tissue (https://www.stemcell.com/media/files/wallchart/WA10006-Frequencies_Cell_Types_Human_Peripheral_Blood.pdf). Because many AD risk genes are expressed in myeloid cells including microglia10, the large number of microglia ct-eQTLs is consistent with the high proportion of AD subjects in the ROSMAP dataset.
Several SNPs previously reported to be associated with AD at the GWS level were associated with eGenes that differ from genes ascribed to AD loci and thus may have a role in AD. Karch et al. observed that the expression of PILRB, which is involved in immune response and is the activator receptor to its inhibitory counterpart PILRA, an established AD gene75,76, was highest in microglia11. CNN2, the eGene for eSNP rs4147929 located near the end of ABCA7, significantly alters extracellular Aβ levels in human induced pluripotent stem cell-derived neurons and astrocytes77. Rs4147929 also targeted HMHA1 which plays several roles in the immune system in an HLA-dependent manner78. The eSNP/GWAS SNP rs3740688 located in SPI1 also affects expression of MYBPC3 and MADD. MYBPC3 was recently identified as a target gene for eSNPs located in CELF1 and MS64A6A in a study of eQTLs in blood for GWS AD loci79. MADD is expressed in neurons11, is involved in neuronal cell death in the hippocampus80, and was shown to be a tau toxicity modulator81. Although eSNP rs113986870 in KANSL1 when paired with the nearby eGene LRRC37A2 was a significant brain eQTL and ct-eQTL, LRRC37A2 encodes a leucine-rich repeat protein that is expressed primarily in testis and has no apparent connection to AD. However, rs113986870 also significantly influenced the expression of another gene in this region, ARL17A, that was previously linked to progressive supranuclear palsy by analysis of gene expression and methylation82.
The aim of this study was to identify context-dependent (i.e., cell-type-specific) eQTLs in blood and brain among older individuals including AD cases using a genome-wide approach. Previous studies have evaluated ct-eQTLs using purified cells, but they focused on only one or two cell types7,10. Other studies examined multiple cell types but using expression data generated from individuals who were on average much younger than the FHS and ROSMAP participants4,83,84. With the exception of a recent report by Patrick et al. who applied a deconvolution approach to estimate cell-type proportions from in cortical tissue obtained from ROSMAP participants but did not examine ct-eQTLs85, our study is one of the first to study eQTLs and ct-eQTLs in a sample enriched for AD cases and link these findings to established AD genes and AD risk.
Our study has several noteworthy limitations. The proxy genes individually or collectively may not accurately depict cell-type-specific context. In addition, the comparisons of gene expression in blood and brain may yield false results because they are based on independent groups ascertained from a community-based longitudinal study of health (FHS—blood) and multiple sources for studies of AD (ROSMAP—brain) which were not matched for age, sex, ethnicity and other factors which may affect gene expression. Moreover, the FHS cohort contains many elderly but relatively few AD cases, whereas ~60% of the ROSMAP participants in this autopsy sample are AD cases. Although the dataset for eQTL analysis in blood was much larger than the dataset derived from the brain, the effect sizes associated with many of the eQTLs common to both tissues were similar. Also, findings in the brain may reflect postmortem changes unrelated to disease or cell-type different expression36. Another limitation of our findings is that some cell types are vastly underrepresented compared to others. Because myeloid cell types are represented in only a small proportion of the total cell populations in the brain and blood (generally ~10%), it is difficult to identify myeloid-specific signals12. Despite this limitation, many of the most significant and noteworthy results of this study involved monocytes/macrophages and microglia.
Conclusion
Our observation of cell-type-specific expression patterns for established and potentially novel AD genes, finding of additional evidence for the role of myeloid cells in AD risk, and discovery of potential novel blood and brain AD biomarkers highlight the importance of cell-type-specific analysis. Future studies that use more robust computational approaches such as deconvolution to reliably estimate cell type proportions83–85, compare cell-type-specific differential gene expression among AD cases and controls using single-cell RNA-sequencing or cell count data and conduct functional experiments are needed to validate and extend our findings.
Supplementary information
Acknowledgements
This study was supported by NIH grants RF1-AG057519, 2R01-AG048927 U01-AG058654, P30-AG13846, 3U01-AG032984, U01-AG062602, and U19-AG068753. Framingham brain bank data was supported by grants 75N92019D00031 and HHSN2682015000011. Collection of study data provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, U01AG61358, a grant from the Illinois Department of Public Health, and the Translational Genomics Research Institute.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01373-z.
References
- 1.Rao S, et al. An APOE-independent cis-eSNP on chromosome 19q13.32 influences tau levels and late-onset Alzheimer’s disease risk. Neurobiol. Aging. 2018;66:178.e1–178.e8. doi: 10.1016/j.neurobiolaging.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou F, et al. Gene expression levels as endophenotypes in genome-wide association studies of Alzheimer disease. Neurology. 2010;74:480–486. doi: 10.1212/WNL.0b013e3181d07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonkers IH, Wijmenga C. Context-specific effects of genetic variants associated with autoimmune disease. Hum. Mol. Genet. 2017;26:R185–R192. doi: 10.1093/hmg/ddx254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobbyn A, et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 2018;102:1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhernakova DV, et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 6.Town, T & Weitz, T.M. Microglia in Alzheimer’s disease: it’s all about context. Int J Alzheimer Dis 2012, 314185 (2012) [DOI] [PMC free article] [PubMed]
- 7.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efthymiou, A. G. & Goate, A. M. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener.12, 1–12 (2017). [DOI] [PMC free article] [PubMed]
- 10.Huang K, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 2017;20:1052–1061. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karch, C. M., Ezerskiy, L. A., Bertelsen, S. & Goate, A. M. Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 Loci. PLoS ONE11, e0148717 (2016). [DOI] [PMC free article] [PubMed]
- 12.Novikova G, et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat Commun. 2021;12:1610. doi: 10.1038/s41467-021-21823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Identification of common genetic variants controlling transcript isoform variation in human whole blood. Nat. Genet. 2015;47:345–352. doi: 10.1038/ng.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2013;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the Rush Memory and Aging Project. Curr. Alzheimer. Res. 2012;6:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Therneau, T. The Lmekin Function. https://cran.r-project.org/web/packages/coxme/vignettes/lmekin.pdf (2018).
- 19.Penney J, Ralvenius WT, Tsai L. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry. 2020;25:148–167. doi: 10.1038/s41380-019-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenzie AT, et al. Brain cell type specific gene expression and co-expression network architectures. Sci. Rep. 2018;8:8868. doi: 10.1038/s41598-018-27293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren, Y. et al. TMEM106B haplotypes have distinct gene expression patterns in aged brain. Mol. Neurodegener.13, 1–11 (2018). [DOI] [PMC free article] [PubMed]
- 22.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, et al. Colocalization of GWAS and eQTL signals at loci with multiple signals identifies additional candidate genes for body fat distribution. Hum. Mol. Genet. 2019;28:4161–4172. doi: 10.1093/hmg/ddz263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jun G, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry. 2016;21:108–117. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hondius DC, et al. Profiling the human hippocampal proteome at all pathologic stages of Alzheimer’s disease. Alzheimers Dement. 2016;12:654–668. doi: 10.1016/j.jalz.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Sjöberg AP, Trouw LA, Blom AM. Complement activation and inhibition: a delicate balance. Trends Immunol. 2009;30:83–90. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Rangaraju S, et al. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol. Neurodegener. 2018;13:24. doi: 10.1186/s13024-018-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AR, et al. Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer’s disease. Clin. Epigenet. 2019;11:52. doi: 10.1186/s13148-019-0636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Jung S, Kim K, Kim C. Treadmill exercise ameliorates Alzheimer disease-associated memory loss through the Wnt signaling pathway in the streptozotocin-induced diabetic rats. J. Exerc. Rehabil. 2016;12:276–283. doi: 10.12965/jer.1632678.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghavan NS, Vardarajan B, Mayeux R. Genomic variation in educational attainment modifies Alzheimer disease risk. Neurol. Genet. 2019;11:e310. doi: 10.1212/NXG.0000000000000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blair, J. A. et al. Individual case analysis of postmortem interval time on brain tissue preservation. PLoS ONE11, e0151615 (2016). [DOI] [PMC free article] [PubMed]
- 37.Le Page A, et al. Role of the peripheral innate immune system in the development of Alzheimer’s disease. Exp. Gerontol. 2018;107:59–66. doi: 10.1016/j.exger.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Goldeck D, Witkowski JM, Fülop T, Pawelec G. Peripheral immune signatures in Alzheimer disease. Curr. Alzheimer Res. 2016;13:739–749. doi: 10.2174/1567205013666160222112444. [DOI] [PubMed] [Google Scholar]
- 39.Harris SA, Harris EA. Herpes simplex virus type 1 and other pathogens are key causative factors in sporadic Alzheimer’s disease. J. Alzheimers Dis. 2015;48:319–353. doi: 10.3233/JAD-142853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obulesu M, Lakshmi MJ. Apoptosis in Alzheimer’s disease: an understanding of the physiology, pathology and therapeutic avenues. Neurochem. Res. 2014;39:2301–2312. doi: 10.1007/s11064-014-1454-4. [DOI] [PubMed] [Google Scholar]
- 41.Fu H, et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 2019;22:47–56. doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crehan H, et al. Complement receptor 1 (CR1) and Alzheimer’s disease. Immunobiology. 2012;217:244–250. doi: 10.1016/j.imbio.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 43.Rasmussen L, et al. Differential expression of ribosomal genes in brain and blood of Alzheimer’s disease patients. Curr. Alzheimer Res. 2015;12:984–989. doi: 10.2174/1567205012666151027124017. [DOI] [PubMed] [Google Scholar]
- 44.Jun GR, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 2017;13:727–738. doi: 10.1016/j.jalz.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desikan RS, et al. Polygenic overlap between C-reactive protein, plasma lipids, and Alzheimer disease. Circulation. 2015;131:2061–2069. doi: 10.1161/CIRCULATIONAHA.115.015489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang J-T, Chang N-S. WWOX dysfunction induces sequential aggregation of TRAPPC6AΔ, TIAF1, tau and amyloid β, and causes apoptosis. Cell Death Discov. 2015;1:15003. doi: 10.1038/cddiscovery.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, et al. WWOX phosphorylation, signaling, and role in neurodegeneration. Front. Neurosci. 2018;12:563. doi: 10.3389/fnins.2018.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Honarmand Tamizkar, K. et al. Altered expression of lncRNAs in autism spectrum disorder. Metab. Brain Dis. 10.1007/s11011-021-00681-z (2021). [DOI] [PubMed]
- 49.Steele NZR, et al. Fine-mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: a case–control study. PLoS Med. 2017;14:e1002272. doi: 10.1371/journal.pmed.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang Z, Haroutunian V, Ho L, Purohit D, Pasinetti GM. Microglia activation in the brain as inflammatory biomarker of Alzheimer’s disease neuropathology and clinical dementia. Dis. Markers. 2006;22:95–102. doi: 10.1155/2006/276239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker DG, Lue L. Immune phenotypes of microglia in human neurodegenerative disease: challenges to detecting microglial polarization in human brains. Alzheimers Res. Ther. 2015;7:56. doi: 10.1186/s13195-015-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neill D, et al. Risk for Alzheimer’s disease in older late-onset cases is associated with HLA-DRB1*03. Neurosci. Lett. 1999;275:137–140. doi: 10.1016/S0304-3940(99)00761-2. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann DJ, et al. HLA class I, II & III genes in confirmed late-onset Alzheimer’s disease. Neurobiol. Aging. 2001;22:71–77. doi: 10.1016/S0197-4580(00)00180-9. [DOI] [PubMed] [Google Scholar]
- 54.Lambert J, et al. Implication of the immune system in Alzheimer’s disease: evidence from genome-wide pathway analysis. J. Alzheimers Dis. 2010;20:1107–1118. doi: 10.3233/JAD-2010-100018. [DOI] [PubMed] [Google Scholar]
- 55.Hallock P, Thomas MA. Integrating the Alzheimer’s disease proteome and transcriptome: a comprehensive network model of a complex disease. OMICS. 2012;16:37–49. doi: 10.1089/omi.2011.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herold C, et al. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol. Psychiatry. 2016;21:1608–1612. doi: 10.1038/mp.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson JL, Chambers E, Jayasundera K. Application of a bioinformatics-based approach to identify novel putative in vivo BACE1 substrates. Biomed. Eng. Comput. Biol. 2013;5:1–15. doi: 10.4137/BECB.S8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vélez JI, et al. APOE*E2 allele delays age of onset in PSEN1 E280A Alzheimer’s disease. Mol. Psychiatry. 2016;21:916–924. doi: 10.1038/mp.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song F, et al. Plasma protein profiling of mild cognitive impairment and Alzheimer’s disease using iTRAQ quantitative proteomics. Proteome Sci. 2014;12:5. doi: 10.1186/1477-5956-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wand GS, et al. Alzheimer’s disease: low levels of peptide alpha-amidation activity in brain and CSF. Neurology. 1987;37:1057–1061. doi: 10.1212/WNL.37.6.1057. [DOI] [PubMed] [Google Scholar]
- 62.Liang X, et al. Genomic convergence to identify candidate genes for Alzheimer disease on chromosome 10. Hum. Mutat. 2009;30:463–471. doi: 10.1002/humu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pang C, et al. Identification and analysis of Alzheimer’s candidate genes by an amplitude deviation algorithm. J. Alzheimers Dis. Parkinsonism. 2019;9:460. doi: 10.4172/2161-0460.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swaminathan S, et al. Genomic copy number analysis in Alzheimer’s disease and mild cognitive impairment: an ADNI study. Int J. Alzheimers Dis. 2011;2011:729478. doi: 10.4061/2011/729478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vardarajan BN, et al. Candidate gene study in the endosome-to-Golgi retrieval pathway reveals association of retromer genes with Alzheimer’s disease. Alzheimer’s Dement.: J. Alzheimer’s Assoc. 2010;6:S194. doi: 10.1016/j.jalz.2010.05.618. [DOI] [Google Scholar]
- 66.Rogaeva E, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer’s disease. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong W, et al. Independent component analysis of Alzheimer’s DNA microarray gene expression data. Mol. Neurodegener. 2009;4:5. doi: 10.1186/1750-1326-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu M, Dedhia M, Crusio WE, Delprato A. Sex differences in gene expression patterns associated with the APOE4 allele. F1000Res. 2019;8:387. doi: 10.12688/f1000research.18671.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lutz, M. W., Sprague, D., Barrera, J. & Chiba-Falek, O. Shared genetic etiology underlying Alzheimer’s disease and major depressive disorder. Transl. Psychiatry10, 1–14 (2020). [DOI] [PMC free article] [PubMed]
- 70.Guo Y, Wang AY. Novel immune check-point regulators in tolerance maintenance. Front. Immunol. 2015;6:421. doi: 10.3389/fimmu.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.von Bartheld CS, Bahney J, Herculano-Houzel S. The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J. Comp. Neurol. 2016;524:3865–3895. doi: 10.1002/cne.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dinkins MB, et al. The 5XFAD mouse model of alzheimer’s disease exhibits an age-dependent increase in anti-ceramide IgG and exogenous administration of ceramide further increases anti-ceramide titers and amyloid plaque burden. J. Alzheimers Dis. 2015;46:55–61. doi: 10.3233/JAD-150088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abou-Raya A, Abou-Raya S. Inflammation: a pivotal link between autoimmune diseases and atherosclerosis. Autoimmun. Rev. 2006;5:331–337. doi: 10.1016/j.autrev.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel, T. et al. Whole-exome sequencing of the BDR cohort: evidence to support the role of the PILRA gene in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 44, 506–521 (2017). [DOI] [PMC free article] [PubMed]
- 76.Logue MW, et al. Search for age-related macular degeneration risk variants in Alzheimer disease genes and pathways. Neurobiol. Aging. 2014;35:1510.e7–18. doi: 10.1016/j.neurobiolaging.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Se S, et al. Candidate-based screening via gene modulation in human neurons and astrocytes implicates FERMT2 in Aβ and TAU proteostasis. Hum. Mol. Genet. 2019;28:718–735. doi: 10.1093/hmg/ddy376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Kreuk, B. et al. The human minor histocompatibility antigen1is a RhoGAP. PLoS ONE8, e73962 (2013). [DOI] [PMC free article] [PubMed]
- 79.Katsumata Y, Nelson PT, Estus S, Fardo DW. Translating Alzheimer’s disease–associated polymorphisms into functional candidates: a survey of IGAP genes and SNPs. Neurobiol. Aging. 2019;74:135–146. doi: 10.1016/j.neurobiolaging.2018.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc. Natl Acad. Sci. USA. 2004;101:4210–4215. doi: 10.1073/pnas.0307349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dourlen P, et al. Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology. Mol. Psychiatry. 2017;22:874–883. doi: 10.1038/mp.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allen M, et al. Gene expression, methylation and neuropathology correlations at progressive supranuclear palsy risk loci. Acta Neuropathol. 2016;132:197–211. doi: 10.1007/s00401-016-1576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim-Hellmuth S, et al. Cell type–specific genetic regulation of gene expression across human tissues. Science. 2020;369:eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Donovan MKR, D’Antonio-Chronowska A, D’Antonio M, Frazer KA. Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants. Nat. Commun. 2020;11:955. doi: 10.1038/s41467-020-14561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patrick E, et al. Deconvolving the contributions of cell-type heterogeneity on cortical gene expression. PLoS Comput. Biol. 2020;16:e1008120. doi: 10.1371/journal.pcbi.1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.