Abstract
Purpose
Intrauterine growth restriction (IUGR) is a fetal growth complication that can be caused by ineffective nutrient transfer from the mother to the fetus via the placenta. Abnormal placental development and function have been correlated with abnormal expression of imprinted genes, which are regulated by epigenetic modifications at imprinting control regions (ICRs). In this study, we analyzed the expression of imprinted genes known to be involved in fetal growth and epigenetic regulators involved in DNA methylation, as well as DNA methylation at the KvDMR1 imprinting control region and global levels of DNA hydroxymethylation, in IUGR cases.
Methods
Expression levels of imprinted genes and epigenetic regulators were analyzed in term placental samples from 21 IUGR cases and 9 non-IUGR (control) samples, by RT-qPCR. Additionally, KvDMR1 methylation was analyzed by bisulfite sequencing and combined bisulfite restriction analysis (COBRA) techniques. Moreover, global DNA methylation and hydroxymethylation levels were also measured.
Results
We observed increased expression of PHLDA2, CDKN1C, and PEG10 imprinted genes and of DNMT1, DNMT3A, DNMT3B, and TET3 epigenetic regulators in IUGR placentas. No differences in methylation levels at the KvDMR1 were observed between the IUGR and control groups; similarly, no differences in global DNA methylation and hydromethylation were detected.
Conclusion
Our study shows that deregulation of epigenetic mechanisms, namely increased expression of imprinted genes and epigenetic regulators, might be associated with IUGR etiology. Therefore, this study adds knowledge to the molecular mechanisms underlying IUGR, which may contribute to novel prediction tools and future therapeutic options for the management of IUGR pregnancies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-020-02047-3.
Keywords: Genomic imprinting, DNA methylation, DNA hydroxymethylation, Intrauterine growth restriction, Placenta
Introduction
Intrauterine growth restriction (IUGR) is a clinical complication that occurs during pregnancy and is characterized by the inability of the fetus to achieve its genetic growth potential, being the second leading cause of perinatal mortality [1]. The placenta is a fetal-maternal endocrine organ that enables the exchange of nutrients, oxygen, and water [2], secrets hormones and growth factors, and safeguards the fetus from the mother’s immune system [3]. Most IUGR cases are caused by placental insufficiency, which is characterized by alterations in nutrient and oxygen delivery to the placenta and transference of those components across the placenta, influencing the regulation of growth processes [4].
In placental mammals, genomic imprinting plays an important role in fetal development and placentation [5]. Genomic imprinting is an epigenetic mechanism in which genes are mono-allelically expressed, from only one of the parental alleles. It is thought that it first occurred 210–310 million years ago after the divergence of monotremes from marsupials and placental mammals, being evolutionarily linked to placentation [6]. The placenta exhibits enriched expression of many imprinted genes, for example PHLDA2, H19, IFG2, MEST, ZFAT, PLAGL1, AIM1, MEG3, RTL1, PEG10, and DLK1, and the majority of these have an important role in placental function and fetal development [1, 7]. The importance of imprinted genes during placental and embryonic development is supported by the parental-conflict theory that postulates that expression of paternally expressed genes contributes to increasing the allocation of nutrients during gestation, while maternally expressed genes limit the allocation of nutrients to the fetus [8]. Therefore, genomic imprinting influences maternal resources allocation and, in turn, fetal development [6]. Additionally, imprinted genes are associated with placenta physiology, controlling early developmental processes that establish the organ (e.g., Peg10), modulate labyrinth size and the surface area for exchange (Igf2 and Grb10), and vascular branching density (Aquaporin) in mice [9]. Deregulation of epigenetic mechanisms such as genomic imprinting may cause placental dysfunction, culminating in IUGR [6]. Imprinted genes expression is regulated by epigenetic modifications at imprinting control regions (ICR), which include, but are not restricted to, DNA methylation [10]. DNA methylation consists in the incorporation of a methyl group, originating from the S-adenosylmethionine donor (SAM), in cytosines located in CpG dinucleotides, resulting in a 5-methylcytosine (5-mC). This is achieved by DNA methyltransferases (DNMTs). DNA hydroxymethylation, in which pre-existing 5-mC is oxidized into 5-hydroxymethylcytosine (5-hmC) by ten eleven translocation (TETs) family of enzymes, is an epigenetic intermediate involved in the process of active DNA demethylation [11].
The human chromosome 11p15.5 contains an imprinted gene cluster with genes involved in fetal growth. The ICR1 at the 11p15.5 cluster is generally methylated on the paternal allele and regulates the paternally expressed IGF2 (insulin-like growth factor 2) and the maternally expressed H19 imprinted gene (Fig. S1). IGF2 is a major fetal growth hormone in mammals, promoting cellular growth and proliferation. H19 is a noncoding transcript and appears to play an important role in early development. The dysregulation of ICR1 is associated with abnormal fetal growth [12, 13]. Pleckstrin homology like domain, family A, member 2 (PHLDA2), cyclin-dependent kinase inhibitor 1C (CDKN1C), and potassium voltage-gated channel subfamily Q member 1 (KCNQ1) are imprinted genes regulated by a second ICR at 11p15.5 - ICR2 or KvDMR1. On the paternal unmethylated allele, transcription of the non-coding RNA KCNQ1OT1 is one of the mechanisms that results in silencing of the flanking imprinted genes. On the maternal chromosome, the ICR2 is methylated and the anti-sense transcript KCNQ1OT1 is not transcribed, resulting in the expression of the flanking genes [14] (Fig. S1). PHLDA2 is a maternally expressed gene associated with intracellular trafficking, cell signaling, and membrane-cytoskeletal interactions [15]. Additionally, in mouse, Phlda2 was associated with cell cycle inhibition resulting in suppression of trophoblast cell proliferation [16]. CDKN1C is a maternally expressed gene that negatively regulates cell proliferation, inhibiting G1 cyclin/cyclin-dependent kinase complexes [17]. KCNQ1 is a maternally expressed gene that codes a protein with function of a voltage-gated potassium channel and functions as the promoter for the KCNQ1OT1, a long non-coding RNA (lncRNA). KCNQ1 is associated with Silver-Russell syndrome, a restriction growth syndrome, and contains the KvDMR1 within intron 10 [18].
Imprinted genes located on human chromosome 7 are also associated with fetal growth, namely two paternally expressed genes, the mesoderm-specific transcript (MEST/PEG1) and the paternally expressed gene 10 (PEG10) [19]. Maternal uniparental disomy (mUPD) of chromosome 7 in humans is associated with intrauterine and postnatal growth restriction [20]. MEST encodes an α/β hydrolase fold family enzyme that is thought to be involved in angiogenesis in human trophoblast and decidua [21]. PEG10 is a retrotransposon-derived gene associated with trophoblast proliferation [22].
Given the importance of imprinted genes in placental function, we here hypothesized that deregulation of imprinted genes, as well as epigenetic regulators and epigenetic modifications (DNA methylation and hydroxymethylation), could be associated with IUGR. Hence, we evaluated the expression of seven imprinted genes, known to be involved in fetal growth (PHLDA2, CDKN1C, H19, KCNQ1, IGF2, PEG10, and MEST/PEG1) and six epigenetic regulators (DNMT1, DNMT3A, DNMT3B, TET1, TET2, and TET3), in term placentas from IUGR pregnancies. Additionally, epigenetic modifications were also evaluated, such as DNA methylation at the KvDMR1 and, global 5-mC and 5-hmC levels in both control and IUGR placentas.
Material and methods
Placental samples collection
Placental tissue samples were collected from 9 control and 21 IUGR pregnancies, with gestational ages between 35 and 41 weeks (Table 1), in collaboration with the Gynecology and Obstetric Department of Centro Hospitalar de São João (CHSJ), Porto. The IUGR was classified by an obstetrician and the criteria included biometrical parameters of the fetus below percentile 10 for gestational age, fetal anatomy, and placenta evaluation [23, 24]. Only cases with normal karyotype were used in the present study. This study was approved by the Health Ethics Committee of Hospital São João/Faculty of Medicine of Porto, and informed consent was signed and obtained from the patients. Placental samples were collected next to the insertion of the umbilical cord and by the same obstetricians to avoid placenta variability [25, 26].
Table 1.
Samples characteristics according to presence or absence of IUGR
| Sample characteristics | Normal (n = 9) | IUGR (n = 21) | p |
|---|---|---|---|
| Maternal age at birth (years) | 33.78 (4.24) | 31.30 (5.75) | 0.258a |
| Maternal smoking (%) | 11% | 15% | 0.779b |
| Gestational age (weeks) | 38.54 (1.26) | 37.12 (1.10) | 0.005a |
| Birth weight (g) | 3091 (323) | 1997 (416) | < 0.001a |
| Fetal sex |
Female: 67% Male: 33% |
Female: 65% Male: 35% |
0.445b |
Data are expressed as mean (SD) and percentage
IUGR intrauterine grown restriction
at test, bX2 test
After collection, the anonymized placental samples were processed in approximately 5-mm3 segments and stored at − 80 °C in RNA later (Ambion), until RNA and DNA extraction.
RNA and DNA extraction from IUGR and controls placentas
For RNA and DNA extraction, placental tissue was homogenized in a Triple-Pure™ zirconium beads tube (Bertin Techonologies) by a Minilys homogenizer (Bertin Techonologies) with 1 mL of TRIzol reagent (Thermo Fisher Scientific), according to manufacturers’ instructions. Finally, DNA and RNA purity and quantification were determined in a NanoDrop 2000 UV-vis Spectrophotometer (Nanodrop Technologies).
Expression analysis of imprinted genes and epigenetic regulators by quantitative real-time PCR
For cDNA synthesis, 1 μg of total RNA was treated with DNaseI (Thermo Fisher Scientific) and reverse transcription was performed using 1 μg of DNase-treated total RNA and 4 μL of qScriptTM cDNA SuperMix (Quanta Biosciences). The samples were incubated at 25 °C for 5 min, followed by 42 °C for 30 min and 85 °C for 5 min.
Transcript levels of seven imprinted genes (PHLDA2, CDKN1C, H19, KCNQ1, IGF2, PEG10 and MEST/PEG1), DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) genes, and ten eleven translocation (TET1, TET2, and TET3) genes were measured by quantitative real-time PCR (RT-qPCR)on a StepOnePlus™ Real-Time PCR System (Life Technologies Corporation) using 5x HOT FIREPol® EvaGreen® qPCR Supermix (Solis Biodyne) and gene-specific primers, according to manufacturers’ instructions. Gene expression was normalized using two reference genes, RPLP0 (ribosomal protein, large, P0) and TBP (TATA box binding protein) [27]. Primers were selected and designed to be exon-spanning and amplify only the cDNA (primer sequences are available in Table S1). RT-qPCR was performed for each gene and sample, in duplicate, and a negative control for each gene was included to detect any contamination. For each gene, we concomitantly tested normal and IUGR placental samples on the same plate to exclude the effects of intra-plate variation.
The raw data obtained after RT-qPCR was introduced in qbasePlus (BioGazelle), which calculate the relative expression levels. This software converts the quantification cycle values (Cq) to calibrated normalized relative quantities (CNRQ) [28].
Sodium bisulfite conversion and KvDMR1 amplification
Isolated DNA (1 μg) was treated with EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s instructions. This procedure allows the conversion of unmethylated cytosines to uracil with 5-mC remaining unchanged.
Modified DNA was subjected to PCR amplification of the KvDMR1 with HotStarTaq enzyme (Qiagen) and specific primers (Supplementary table 1) [29]. KvDMR1 is an ICR located at 11p15.5 (NCBI Reference Sequence: NC_000011.10; nucleotides 2,608,328-2,699,994), within a CpG island in intron 10 of KCNQ1 that overlaps with the promoter region of KCNQ1 overlapping transcript 1 (KCNQ1OT1) ncRNA, which regulates some of the imprinted genes included in this study. After amplification, the resulting PCR fragment contains 359 base pairs (bp) and 24 CpGs (Genbank Accession Number U90095; nucleotides 66492-66850). The amplification was performed in a 50 μL reaction mix containing 1X buffer with 1.5 mM MgCl2 (Qiagen), 0.12 μM of each dNTP (Invitrogen), 0.5 μM of each KvDMR1 primer (Metabion) and 1.5 U HotStarTaq enzyme (5 U/μL; Qiagen), and 2 μL of the bisulfite modified DNA. The PCR conditions were as follows: initial activation of the HotStarTaq for 15 min at 95 °C, followed by 40 cycles of 1 min denaturation at 94 °C, annealing for 1 min at 60 °C and extension for 1 min at 72 °C, and a final 20 min extension step at 72 °C.
Combined bisulfite restriction analysis of a KvDMR1 CpG and global DNA methylation
Combined bisulfite restriction analysis (COBRA) is a quantitative technique to determine DNA methylation levels at specific gene loci [30]. Briefly, COBRA takes advantage of a restriction site for one restriction enzyme—we utilized HpyCH4IV enzyme—that is modified after bisulfite treatment and allows determination of the methylation level at this specific CpG. First, bisulfite-treated DNA was used for COBRA analysis of KvDMR1 methylation and global DNA methylation by LINE1 analysis [31]. After amplification, PCR products were used for restriction digestion for 15 min at 37 °C with the restriction enzyme HpyCH4IV (New England Biolabs), following an inactivation step at 65 °C for 20 min. This enzyme cleaves only originally methylated DNA as it recognizes and cleaves 5′-ACGT-3′ (5′-A↓CGT-3′; 3′-TGC↑A-5′). This enzyme site selection allows the study of the 13th CpG present in the KvDMR1 fragment, previously amplified. COBRA analysis was run in triplicate for KvDMR1 and duplicate for LINE1, in each sample.
The stained gel was visualized and scanned in a ChemiDoc™ XRS+ System (Bio-Rad Laboratories). Acquired images were processed using Image Lab software (Bio-Rad).
Methylation quantity was calculated using the uncleaved band (~ 359 bp) representing the unmethylated portion and the average intensity of the digested bands (~ 202 bp and ~ 157 bp) which represent the methylated portion. Firstly, the intensity of each band is calculated comparing to the uncut control (which is set to 1). Then, the relative quantity of each band is normalized, in which the relative value of each band is calculated considering the value previously calculated as 1 (band relative quantity normalized = band relative quantity/sum of the uncut band with the mean value of the two cut bands). Finally, the value obtained for the uncleaved fragments represents the value for unmethylated CpG and the mean value of the two cleaved fragments is the value for methylated CpG.
Bisulfite sequencing of KvDMR1
Bisulfite sequencing allows the analysis of methylation at each individual CpG site of the 24 CpGs included in the PCR fragment. The amplified KvDMR1 fragment was purified using Agencourt® AMPure XP (Beckman Coulter Krefeld), according to the manufacturer’s instructions. For this analysis, 18 samples were selected, 9 control and 9 IUGR samples. The selection of IUGR samples considered the highest values of CDKN1C and PHLDA2 expression and fetal sex. The samples were balanced for males and females to avoid differences in methylation due to fetal sex—3 males and 6 females in each group [32, 33].
Adenine residues were added to the PCR fragments after purification for a more efficient ligation to the TOPO TA vector (Invitrogen, Thermo Scientific). The next step was the cloning of purified PCR products with the TOPO TA cloning kit (Invitrogen), using pCR™II-TOPO® (Invitrogen) and 1 μL of the KvDMR1 fragment. After ligation, transformation and cloning were performed using NZY5α chemically competent Escherichia coli cells (NZYTech). Colony PCR was performed with 5 U/μL Taq DNA polymerase (Thermo Scientific). The conditions were as follows: 94° for 10 min, followed by 35 cycles at 94 °C for 45 s, 50 °C for 45 s and 72 °C for 1 min, finally 10 min at 72 °C. After purification with Agencourt® AMPure® XP (Beckman Coulter Krefeld), around 15 independent clones containing the fragment of interest of each sample were sequenced using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc) and were analyzed in an ABI PRISM® 3500 Genetic Analyzer.
The bisulfite sequencing data were analyzed in BiQAnalyzer software, which aligns sequencing results with a reference sequence and determines the methylation status and levels of each CpG [34]. Only clones with more than 90% of non-CpG cytosines converted were included.
Global DNA hydroxymethylation levels
The 5-hmC content of the DNA samples (100 ng) was measured, in duplicate for each sample, using QUEST 5-hmCTM DNA ELISA kit (Zymo research), following manufacturer’s instructions. This procedure allowed the detection and quantification of 5-hmC using anti-5hmC polyclonal antibody and anti-DNA HRP antibody. The color development was measured after 30–60 min (Abs 405), on a 10-min interval, on an ELISA plate reader (Sunrise). This procedure allowed the detection and quantification of 5-hmC (in percentage), comparing the absorbance of each sample to controls and using a standard curve. The standard curve was obtained using 5 controls, in duplicate (control A 0%, control B 0.03%, control C 0.12%, control D 0.23%, control E 0.55%).
Statistical analysis
The Mann-Whitney U test was used to compare the expression of genes between the two groups (IUGR vs control) and the gene expression analysis method was 2-∆∆Cq by Livak and Schmittgen[35]. For gene expression analysis, the definitive outliers for each gene and group were identified in the Statistical Package for Social Sciences (SPSS, IBM) software v24.0 and excluded from the calculations (for DNMT3A analysis, two controls were excluded and for TET1 and TET3 one IUGR sample).
For KvDMR1 analysis, the normalized CpG methylation values, obtained by COBRA, from each group were compared using the Mann-Whitney U test in SPSS. The methylated CpG values after bisulfite sequencing were calculated by dividing the number of observed methylated CpGs by the total number of CpGs analyzed. The mean values and the individual CpG values of each group were compared and analyzed using the t test in SPSS.
Additionally, the linear regressions based in ANOVA between gene expression and normalized KvDMR1 CpG methylation (obtained with COBRA), fetal percentile and normalized KvDMR1 CpG methylation (obtained with COBRA), genes expression and fetal percentile, and lastly, fetal percentile and normalized KvDMR1 CpG methylation (obtained with COBRA) were performed using SPSS. The same was also evaluated with the relative KvDMR1 values obtained with BS-sequencing.
For the global hydroxymethylation study data, the percentage of 5-hmC for each sample was calculated according to manufacturers’ instructions, using the slope and y-intercept values obtained from the standard curve of the known controls. The comparison between groups was evaluated using t test in SPSS.
The results of all tests were considered significant when the p value was below 0.05.
Results
Increased expression of imprinted genes and epigenetic regulators in IUGR term placentas
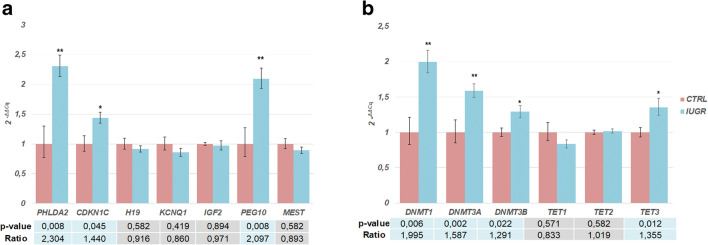
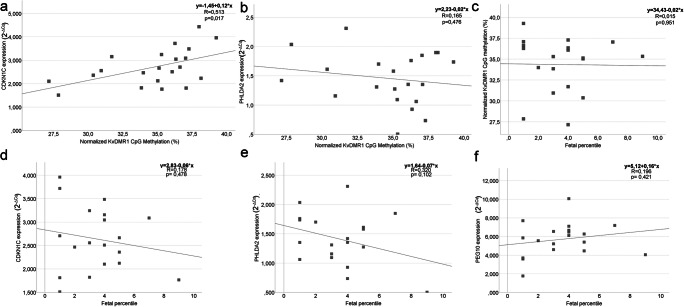
We observed an increased expression in IUGR placentas of three imprinted genes, two maternally expressed -PHLDA2, CDKN1C-, and one paternally expressed- PEG10 (Fig. 1). Additionally, four epigenetic regulators were also more highly transcribed, three involved in DNA methylation- DNMT1, DNMT3A, DNMT3B - and one in the removal of methylation marks -TET3 (Fig. 1). Analysis of correlation, by linear regression, between the fetal percentile and imprinted genes expression, in IUGR, showed no significant correlation (Fig. 2d–f).
Fig. 1.
Relative gene expression in placental samples with IUGR (n = 21) and control placental samples (n = 9). a Relative expression of imprinting genes (PHLDA2, CDKN1C, KCNQ1, IGF2, H19, MEST, and PEG10). The expression of PHLDA2, CDKN1C, and PEG10 is increased in IUGR. b Relative expression of epigenetic regulators (DNMT1, DNMT3A, DNMT3B, TET1, TET2, and TET3). The expression of DNMT1, DNMT3A, DNMT3B, and TET3 is increased in IUGR. The bars represent the mean expression for each gene with respectively error bar ( 2-ΔΔCq ± SEM) * represent the p value < 0.05; ** represent the p value < 0.01. Mann-Whitney U test. CTRL, control samples; IUGR, intrauterine growth restriction samples
Fig. 2.
Linear regression between gene expression and relative KvDMR1 CpG methylation, fetal percentile and relative KvDMR1 CpG methylation, genes expression and fetal percentile or fetal percentile, and relative KvDMR1 CpG methylation. a CDKN1C expression (2-ΔCq) values and KvDMR1 CpG methylation of placental samples with IUGR, observing a moderate linear correlation. b PHLDA2 expression (2-ΔCq) values and KvDMR1 CpG methylation of placental samples with IUGR. c Fetal percentile and KvDMR1 CpG methylation of placental samples with IUGR. d CDKN1C expression (2-ΔCq) values and fetal percentile of placental samples with IUGR. e PHLDA2 expression (2-ΔCq) values and fetal percentile of placental samples with IUGR. f- PEG10 expression (2-ΔCq) values and fetal percentile of placental samples with IUGR. The dots represent each IUGR sample of this study
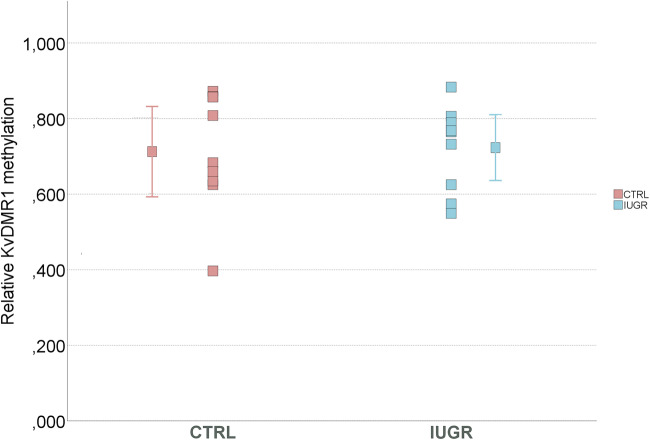
Methylation at KvDMR1 is unaltered in IUGR placentas
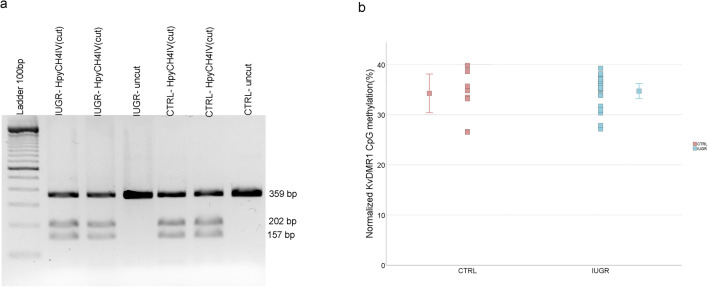
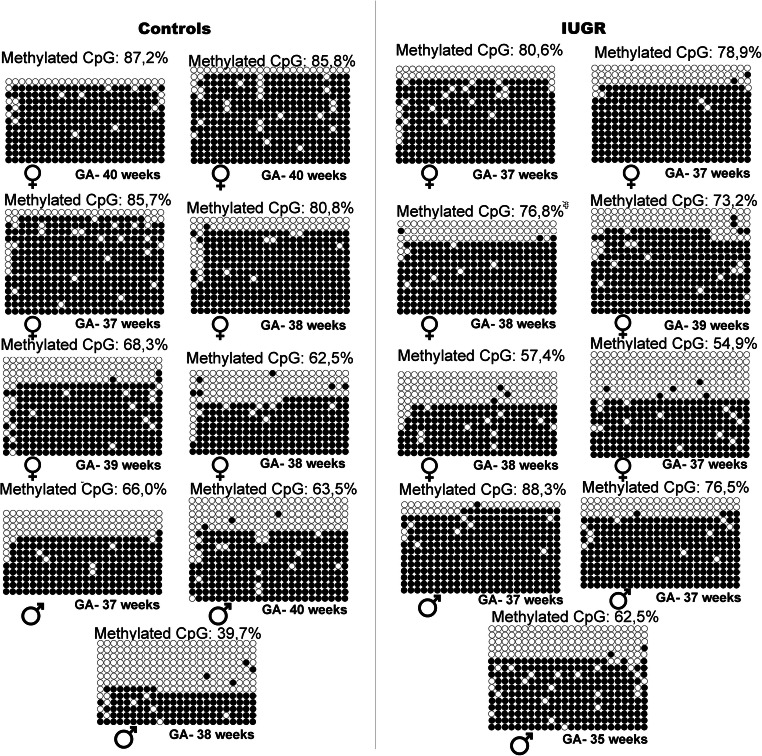
Since altered expression of imprinted genes controlled by KvDMR1 was observed, namely for PHLDA2 and CDKN1C (Fig. 1a and S1), we analyzed methylation at this ICR using combined bisulfite restriction analysis (COBRA) and bisulfite sequencing. Our results, using both methods, showed no differences between control and IUGR groups, in KvDMR1 methylation levels (Figs. 3, 4, and 5). Also, analysis of each CpG individually again did not show differences between the groups (Fig. S2), and the same is true for the CpG13 which was the CpG analyzed by COBRA.
Fig. 3.
KvDMR1 methylation analysis in IUGR placental samples comparing with control samples, using COBRA. a Example of COBRA analysis of KvDMR1 in IUGR placenta and in normal placenta (CTRL). b Comparison of normalized KvDMR1 methylation, using COBRA. No significant difference was observed between the IUGR cases and non-IUGR (CTRL) cases using Mann-Whitney U test. The dots represent the methylation for each sample and the mean methylation for each group. CTRL, control samples; IUGR, intrauterine growth restriction samples
Fig. 4.
Methylation profiles of 24 CpGs KvDMR1 of IUGR cases and non-IUGR (CTRL) cases. The circles represent the CpGs: methylated (black) and non-methylated (non-colored). Each sample has the KvDMR1 methylation percentage, the corresponding fetal sex and each line that represents one clone. CTRL, control samples; IUGR, intrauterine growth restriction samples
Fig. 5.
KvDMR1 methylation analysis in 9 IUGR placental samples comparing with 9 control samples, using BS-sequencing. The mean methylation for each group with respectively error bar. No significant difference was observed between the IUGR cases and non-IUGR (CTRL) cases, using t test. CTRL, control samples; IUGR, intrauterine growth restriction samples
Comparing gene expression and KvDMR1 methylation levels (by COBRA analysis) in IUGR samples, we observed a moderate linear association between CDKN1C expression and KvDMR1 CpG methylation (Fig. 2a; p = 0.017; ANOVA) but not with PHLDA2 expression (Fig. 2b) or with fetal percentile (Fig. 2c).
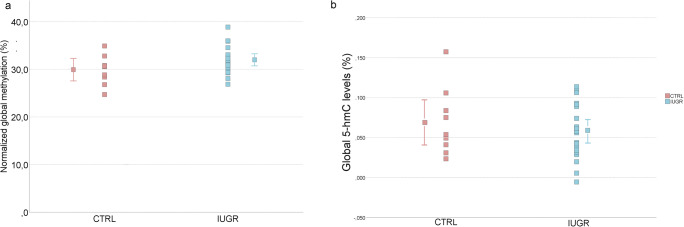
Global 5-mC and 5-hmC levels are similar in IUGR and control placentas
Due to the increased expression of DNMTs in IUGR placentas, we analyzed global DNA methylation levels by LINE1 retrotransposon methylation [31] in IUGR placentas and control placentas (p = 0.082; t test) (Fig. 6a). Since we also observed an increased expression of TET3 in IUGR cases, we analyzed global DNA hydroxymethylation levels using an ELISA-based technique. The IUGR samples exhibited a mean global DNA hydroxymethylation level of 0.058% and the control samples of 0.069% (p = 0,451; t test) (Fig. 6b).
Fig. 6.
a Global DNA methylation by LINE1 analysis in IUGR and control samples. The mean global methylation for each group with respectively error bar. No significant difference was observed between the IUGR cases and non-IUGR (CTRL) cases, using t test. b Comparison of the mean percentage of global 5-hmC between the two groups (IUGR vs. controls). The mean global hydroxymethylation for each group with respectively error bar. No significant difference was observed between the IUGR cases and non-IUGR (CTRL) cases, using t test. CTRL, control samples; IUGR, intrauterine growth restriction samples
Discussion
Placental imprinted genes may be involved in IUGR etiology, as it is well documented that they play an important role in fetal and placental development [36]. Piedrahita proposed that, in IUGR, imprinted genes may be grouped into two categories: genes that participate in enhancing IUGR or reducing fetal growth (negative effectors) and genes that, through a compensatory mechanism, sense that the fetus is at risk and act to increase fetal growth (positive effectors) [37]. We here observed increased expression of two genes that restrict placental and fetal growth (PHLDA2 and CDKN1C) and one gene (PEG10) that enhances fetal growth, therefore potentially acting in a compensatory manner. Overexpression of PHLDA2 and CDKN1C in the human placenta and the association with IUGR was previously described, both in single pregnancies [38–44] and in the placental shares of the smaller fetus in cases of selective IUGR [45, 46]. PHLDA2 is highly expressed in the placenta and encodes a protein with a Pleckstrin-homology domain, where phosphatidylinositol lipids can bind, and it is associated with the inhibition of cell proliferation, migration, and invasion, leading to growth restriction [47]. CDKN1C is a gene that encodes the p57Kip2 protein- CIP/Kip family- which bind and inhibit cyclin/cyclin-dependent kinase complexes, inhibiting the cell cycle [17]. Taking this into account, it is suggested that these two genes may have an important role in IUGR.
The conflict theory for explaining the occurrence of genomic imprinting advocates that paternally expressed genes promote fetal growth [8]. However, we observed overexpression of PEG10, a paternally expressed gene, in placentas from pregnancies with IUGR. Other studies also observed a PEG10 overexpression in IUGR, suggesting that this gene is acting in a compensatory manner to increase the growth of IUGR fetus [37, 40]. PEG10 and MEST paternally expressed genes are located on human chromosome 7. PEG10 and the contiguous gene SGCE are regulated by an ICR located on 7q21.3 that is methylated in the maternal allele. The MEST ICR is also maternally methylated and is located on human chromosome 7q32 [48].
Several studies evaluated methylation at the KvDMR1 or ICR2 that regulate PHLDA2 and CDKN1C imprinted genes in placental samples from IUGR [12, 13, 38, 39, 49, 50]. However, as in our study, others studies did not find changes in methylation at the KvDRM1, suggesting that other epigenetic modifications or methylation at other regions might be contributing to the observed changes in imprinted gene expression [12, 13, 38, 39, 49, 50]. Indeed, Ishida and collaborators observed that the promoter dysregulation altered the expression of PHLDA2 gene, altering human fetal growth [51]. In other types of models like human osteosarcomas, the PHLDA2 expression is regulated by promoter methylation [52] and in goat, Wang and collaborators observed that methylation of PHLDA2 promoter leads to PHLDA2 inhibition in the placenta [53]. Nevertheless, the moderate linear correlation that we observed between CDKN1C imprinted gene and KvDMR1 CpG methylation supports the association between gene expression and KvDMR1 methylation. In humans, it has also been shown that ICR2 deletion in the paternal allele leads to the silencing of KCNQ1OT1 and activation of CDKN1C and PHLDA2, causing IUGR [54].
On the other hand, DNA hydroxymethylation in human placentas from IUGR cases is less explored [55–57]. In our study, we observed increased expression of TET3, one of the dioxygenases involved in oxidation of 5-mC into 5-hmC, in IUGR cases; however, no significant differences in global DNA hydroxymethylation levels were observed. In the placenta, it was shown that the enzymes TET have a role in the specialized trophoblast cells differentiation and regulation [58]. Although increased expression of TET3 and DNMTs was observed in IUGR samples, no significant changes in global levels of methylation and hydroxymethylation occurred, suggesting that regional or gene-specific changes might be the target of this increased expression. It is also plausible that increased transcript levels might be not be reflected into higher protein levels or enzyme activity.
Interestingly, it was shown that TET1 is the principal enzyme responsible for 5-mC oxidation into 5-hmC, while TET2 and TET3 enzymes promote the removal of 5-hmC in the oxidative cascade [59]. In a previous study by our group, we also observed a higher expression of TET2 and TET3 with no significant changes in DNA hydroxymethylation global levels in placental samples from idiopathic spontaneous abortion [49]. The higher expression of TET3 may lead to a faster 5-hmC turnover, by conversion into 5-formylcytosine (5-fC) and 5-carboxycytosine (5-caC); however, we need to take in account that placenta has different cell types and that those cells may have different expression levels that we do not detect, supporting the need to do single-cell analysis. In fact, a study has shown that cell columns, extravillous trophoblast, and syncytiotrophoblast expressed TET1-3 whereas only TET3 was expressed in villus cytotrophoblast cells in first trimester and term human placentas [60].
Nevertheless, analysis of 5-hmC levels specifically at the affected imprinted genes would be of importance to assess whether 5-hmC levels at imprinted genes are altered in IUGR placentas. Indeed, it has been shown that 5-hmC is present in many imprinted genes, both in the human brain and placenta, and it is enriched in the transcribed allele, suggesting 5-hmC is positively associated with transcription at imprinted loci [61]. Furthermore, it has been shown that 5-hmC levels are lower in placenta than brain [62, 63], which positively correlates with transcription in actively transcribed genes [64]. Moreover, the levels of 5-mC at KvDMR and IGF2 DMR0 and 5-hmC at H19 gene body were shown to positively correlate with size at birth [56]. We here confirmed the presence of 5-hmC in placental samples with similar levels to the ones reported by other studies (0.06%) [62, 63].
As IUGR is a multifactorial pathology [4], the dysregulation of growth-related genes, such as PHLDA2 and CDKN1C, may be only one of the factors involved in this pathology. Several other genes involved in different pathways—inflammatory, cardiovascular and metabolic genes—and their altered methylation status in the placenta may be associated with uterine growth [65, 66]. In this study, we showed that PHLDA2 and CDKN1C genes could serve as potential biomarkers for human IUGR. In future, studies will be interesting to investigate which cell types mostly contribute to the observed changes in genes expression and analyze methylation and hydroxymethylation in a genome-wide manner to explore gene-specific epigenetic changes.
Supplementary Information
(DOCX 458 kb)
(XLSX 80 kb)
Acknowledgments
We thank the obstetricians at the Department of Gynecology/Obstetrics of Hospital São João, Porto, Portugal, for collaboration with sample and patients’ data collection. We are also thankful to members of the Genetics Department of FMUP for technical help and insightful discussions.
Abbreviations
- 5-hmC
5-Hydroxymethylcytosine
- 5-mC
5-Methylcytosine
- DNMT
DNA methyltransferase
- ES cells
Embryonic stem cells
- ICR
Imprinting control region
- IUGR
Intrauterine growth restriction
- TET
Ten eleven translocation
Author contributions
C.C. performed the experiments, analyzed the results, and wrote the first draft of the manuscript; S.V. performed experimental work and revised the manuscript; C.R. (MD, Obstetrician) collected placental samples and patients’ data; C.J.M supervised experimental work, analyzed the data, and wrote the final manuscript; S.D. designed the study, analyzed the data, and wrote the final manuscript. All authors revised and approved the final version of the manuscript.
Funding
This work was partly funded by the Portuguese Foundation for Science and Technology (FCT) – UID/BIM/04293/2013 and FCT Investigator award to CJM (IF/00047/2012; CEECIND/00371/2017). C.C. is a recipient of a PhD studentship by FCT (SFRH/BD/141855/2018). S.V. is a recipient of a PhD studentship by FCT (SFRH/BD/147440/2019).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Patient consent
All patients provided a signed informed consent for this study.
Ethics approval
This study was approved by the Health Ethics Committee of Hospital São João/Faculty of Medicine of Porto (N 70/17).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
C. Joana Marques, Email: cmarques@med.up.pt.
Sofia Dória, Email: sdoria@med.up.pt.
References
- 1.Rhon-Calderon EA, Vrooman LA, Riesche L, Bartolomei MS. The effects of assisted reproductive technologies on genomic imprinting in the placenta. Placenta. 2019;84:37–43. doi: 10.1016/j.placenta.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Gude NM, Roberts CT, Kalionis B, King RG. Growth and function of the normal human placenta. Thromb Res. 2004;114:397–407. doi: 10.1016/j.thromres.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol (New York, NY : 1989) 2009;62:78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkins KL, Devaskar SU. Intrauterine growth restriction. Fanaroff Martin's Neonatal-Perinatal Med. 2015;2-Volume Set:227–235. doi: 10.1016/B978-1-4557-5617-9.00016-6. [DOI] [Google Scholar]
- 5.Bressan FF, De Bem THC, Perecin F, Lopes FLL, Ambrosio CE, Meirelles FVV, et al. Unearthing the roles of imprinted genes in the placenta. Placenta. 2009;30:823–834. doi: 10.1016/j.placenta.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Monk D. Genomic imprinting in the human placenta. Am J Obstet Gynecol. 2015;213(4, Supplement):S152–SS62. doi: 10.1016/j.ajog.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 7.Pilvar D, Reiman M, Pilvar A, Laan M. Parent-of-origin-specific allelic expression in the human placenta is limited to established imprinted loci and it is stably maintained across pregnancy. Clin Epigenetics. 2019;11(1):94. doi: 10.1186/s13148-019-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore T. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 9.Cassidy FC, Charalambous M. Genomic imprinting, growth and maternal–fetal interactions. J Exp Biol. 2018;221(Suppl 1):jeb164517. doi: 10.1242/jeb.164517. [DOI] [PubMed] [Google Scholar]
- 10.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 11.Santiago M, Antunes C, Guedes M, Sousa N, Marques CJ. TET enzymes and DNA hydroxymethylation in neural development and function—how critical are they? Genomics. 2014;104(5):334–340. doi: 10.1016/j.ygeno.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Bourque DKK, Avila L, Peñaherrera M, von Dadelszen P, Robinson WPP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Cordeiro A, Neto AP, Carvalho F, Ramalho C, Dória S. Relevance of genomic imprinting in intrauterine human growth expression of CDKN1C, H19, IGF2, KCNQ1 and PHLDA2 imprinted genes. J Assist Reprod Genet. 2014;31:1361–1368. doi: 10.1007/s10815-014-0278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiesa N, De Crescenzo A, Mishra K, Perone L, Carella M, Palumbo O, et al. The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet. 2012;21:10–25. doi: 10.1093/hmg/ddr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen AB, Tunster SJ, John RM. The significance of elevated placental PHLDA2 in human growth restricted pregnancies. Placenta. 2014;35:528–532. doi: 10.1016/j.placenta.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Jin F, Qiao C, Luan N, Shang T. The expression of the imprinted gene pleckstrin homology-like domain family A member 2 in placental tissues of preeclampsia and its effects on the proliferation, migration and invasion of trophoblast cells JEG-3. Clin Exp Pharmacol Physiol. 2015;42(11):1142–1151. doi: 10.1111/1440-1681.12468. [DOI] [PubMed] [Google Scholar]
- 17.Lee MH, Reynisdottir I, Massague J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9(6):639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 18.Jacob K, Robinson W, Lefebvre L. Beckwith–Wiedemann and Silver–Russell syndromes: opposite developmental imbalances in imprinted regulators of placental function and embryonic growth. Clin Genet. 2013;84(4):326–334. doi: 10.1111/cge.12143. [DOI] [PubMed] [Google Scholar]
- 19.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6(7):e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotzot D, Schmitt S, Bernasconi F, Robinson WP, Lurie IW, Ilyina H, Méhes K, Hamel BCJ, Otten BJ, Hergersberg M, Werder E, Schoenle E, Schinzel A. Uniparental disomy 7 in Silver—Russell syndrome and primordial growth retardation. Hum Mol Genet. 1995;4(4):583–587. doi: 10.1093/hmg/4.4.583. [DOI] [PubMed] [Google Scholar]
- 21.Mayer W, Hemberger M, Frank HG, Grümmer R, Winterhager E, Kaufmann P, et al. Expression of the imprinted genes MEST/Mest in human and murine placenta suggests a role in angiogenesis. Dev Dyn. 2000;217(1):1–10. doi: 10.1002/(sici)1097-0177(200001)217:1<1::Aid-dvdy1>3.0.Co;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Sun M, Liu J, Tong C, Meng T. Silencing of paternally expressed gene 10 inhibits trophoblast proliferation and invasion. PLoS One. 2015;10(12):e0144845. doi: 10.1371/journal.pone.0144845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carraca T, Ferraz T, Machado L, Bessa Monteiro S, Rodrigues T, Montengro N. Restrição de crescimento fetal- Rastreio, diagnóstico e orientação clínica. In: Lidel, editor. Protocolos de medicina materno-fetal. 3ª. Lisboa: Lidel; 2008. pp. 133–134. [Google Scholar]
- 24.Figueras F, Gratacós E. Update on the diagnosis and classification of fetal growth restriction and proposal of a stage-based management protocol. Fetal Diagn Ther. 2014;36(2):86–98. doi: 10.1159/000357592. [DOI] [PubMed] [Google Scholar]
- 25.Pidoux G, Gerbaud P, Laurendeau I, Guibourdenche J, Bertin G, Vidaud M, Evain-Brion D, Frendo JL. Large variability of trophoblast gene expression within and between human normal term placentae. Placenta. 2004;25(5):469–473. doi: 10.1016/j.placenta.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Wyatt SM, Kraus FT, Roh CR, Elchalal U, Nelson DM, Sadovsky Y. The correlation between sampling site and gene expression in the term human placenta. Placenta. 2005;26(5):372–379. doi: 10.1016/j.placenta.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Marques CJ, Joao Pinho M, Carvalho F, Bieche I, Barros A, Sousa M. DNA methylation imprinting marks and DNA methyltransferase expression in human spermatogenic cell stages. Epigenetics. 2011;6(11):1354–1361. doi: 10.4161/epi.6.11.17993. [DOI] [PubMed] [Google Scholar]
- 28.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoueiry R, Ibala-Romdhane S, Al-Khtib M, Blachere T, Lornage J, Guerin JF, et al. Abnormal methylation of KCNQ1OT1 and differential methylation of H19 imprinting control regions in human ICSI embryos. Zygote. 2013;21(2):129–138. doi: 10.1017/s0967199411000694. [DOI] [PubMed] [Google Scholar]
- 30.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14(2):67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 32.Gong S, Johnson MD, Dopierala J, Gaccioli F, Sovio U, Constância M, Smith GCS, Charnock-Jones DS. Genome-wide oxidative bisulfite sequencing identifies sex-specific methylation differences in the human placenta. Epigenetics. 2018;13(3):228–239. doi: 10.1080/15592294.2018.1429857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin E, Smeester L, Bommarito PA, Grace MR, Boggess K, Kuban K, Karagas MR, Marsit CJ, O’Shea TM, Fry RC. Sexual epigenetic dimorphism in the human placenta: implications for susceptibility during the prenatal period. Epigenomics. 2017;9(3):267–278. doi: 10.2217/epi-2016-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Ishida M, Moore GE. The role of imprinted genes in humans. Mol Asp Med. 2013;34:826–840. doi: 10.1016/j.mam.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Piedrahita JA. The role of imprinted genes in fetal growth abnormalities. Birth Defects Res A, Clin Molec Teratol. 2011;91:682–692. doi: 10.1002/bdra.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, Weksberg R, Thaker HM, Tycko B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27:540–549. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 39.López-Abad M, Iglesias-Platas I, Monk D. Epigenetic characterization of CDKN1C in placenta samples from non-syndromic intrauterine growth restriction. Front Genet. 2016;7:62. doi: 10.3389/fgene.2016.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diplas AI, Lambertini L, Lee M-J, Sperling R, Lee YL, Wetmur J, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4:235–240. doi: 10.4161/epi.9019. [DOI] [PubMed] [Google Scholar]
- 41.Janssen AB, Tunster SJ, Heazell AEP, John RM. Placental PHLDA2 expression is increased in cases of fetal growth restriction following reduced fetal movements. BMC Med Genet. 2016;17:17. doi: 10.1186/s12881-016-0279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apostolidou S, Abu-Amero S, O’Donoghue K, Frost J, Olafsdottir O, Chavele KM, Whittaker JC, Loughna P, Stanier P, Moore GE. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J Mol Med. 2007;85:379–387. doi: 10.1007/s00109-006-0131-8. [DOI] [PubMed] [Google Scholar]
- 43.Chen XJ, Chen F, Lv PP, Zhang D, Ding GL, Hu XL, Feng C, Sheng JZ, Huang HF. Maternal high estradiol exposure alters CDKN1C and IGF2 expression in human placenta. Placenta. 2018;61:72–79. doi: 10.1016/j.placenta.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Moore GGE, Ishida M, Demetriou C, Al-Olabi L, Leon LJ, Thomas AC, et al. The role and interaction of imprinted genes in human fetal growth. Philos Trans R Soc Lond Ser B Biol Sci. 2015;370:20140074. doi: 10.1098/rstb.2014.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gou C, Liu X, Shi X, Chai H, He ZM, Huang X, Fang Q. Placental expressions of CDKN1C and KCNQ1OT1 in monozygotic twins with selective intrauterine growth restriction. Twin Res Hum Genet. 2017;20(5):389–394. doi: 10.1017/thg.2017.41. [DOI] [PubMed] [Google Scholar]
- 46.Shi X, He Z, Gao Y, Luo Y, Gou C, Fang Q. Placental expression of PHLDA2 in selective intrauterine growth restriction in monozygotic twins. Placenta. 2014;35(6):428–430. doi: 10.1016/j.placenta.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Jin F, Qiao C, Luan N, Li H. Lentivirus-mediated PHLDA2 overexpression inhibits trophoblast proliferation, migration and invasion, and induces apoptosis. Int J Mol Med. 2016;42016:949–957. doi: 10.3892/ijmm.2016.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hannula-Jouppi K, Muurinen M, Lipsanen-Nyman M, Reinius LE, Ezer S, Greco D, Kere J. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. Epigenetics. 2014;9(3):351–365. doi: 10.4161/epi.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasconcelos S, Ramalho C, Marques CJ, Doria S. Altered expression of epigenetic regulators and imprinted genes in human placenta and fetal tissues from second trimester spontaneous pregnancy losses. Epigenetics. 2019;14:1–11. doi: 10.1080/15592294.2019.1634988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miranda Furtado CL, Salomão KB, Verruma CG, Paulino Leite SB, Lopes Rios ÁF, Bialecka M, Moustakas I, Mei H, de Paz CCP, Duarte G, Chuva de Sousa Lopes SM, Ramos ES. Variation in DNA methylation in the KvDMR1 (ICR2) region in first-trimester human pregnancies. Fertil Steril. 2019;111(6):1186–1193. doi: 10.1016/j.fertnstert.2019.01.036. [DOI] [PubMed] [Google Scholar]
- 51.Ishida M, Monk D, Duncan Andrew J, Abu-Amero S, Chong J, Ring Susan M, et al. Maternal inheritance of a promoter variant in the imprinted PHLDA2 gene significantly increases birth weight. Am J Hum Genet. 2012;90(4):715–719. doi: 10.1016/j.ajhg.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Meng G, Guo QN. Changes in genomic imprinting and gene expression associated with transformation in a model of human osteosarcoma. Exp Mol Pathol. 2008;84(3):234–239. doi: 10.1016/j.yexmp.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Wang R, Su L, Yu S, Ma X, Jiang C, Yu Y. Inhibition of PHLDA2 transcription by DNA methylation and YY1 in goat placenta. Gene. 2020;739:144512. doi: 10.1016/j.gene.2020.144512. [DOI] [PubMed] [Google Scholar]
- 54.De Crescenzo A, Sparago A, Cerrato F, Palumbo O, Carella M, Miceli M, et al. Paternal deletion of the 11p15.5 centromeric-imprinting control region is associated with alteration of imprinted gene expression and recurrent severe intrauterine growth restriction. J Med Genet. 2013;50:99–103. doi: 10.1136/jmedgenet-2012-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He Z, Lu H, Luo H, Gao F, Wang T, Gao Y, Fang Q, Wang J. The promoter methylomes of monochorionic twin placentas reveal intrauterine growth restriction-specific variations in the methylation patterns. Sci Rep. 2016;6:20181. doi: 10.1038/srep20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piyasena C, Reynolds RM, Khulan B, Seckl JR, Menon G, Drake AJ. Placental 5-methylcytosine and 5-hydroxymethylcytosine patterns associate with size at birth. Epigenetics. 2015;10:692–697. doi: 10.1080/15592294.2015.1062963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Zheng D, Fang Q, Zhong M. Aberrant hydroxymethylation of ANGPTL4 is associated with selective intrauterine growth restriction in monochorionic twin pregnancies. Epigenetics. 2020;15:1–13. doi: 10.1080/15592294.2020.1737355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Logan PC, Mitchell MD, Lobie PE. DNA methyltransferases and TETs in the regulation of differentiation and invasiveness of extra-villous trophoblasts. Front Genet. 2013;4:265. doi: 10.3389/fgene.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi J-H, Robertson KD. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15(6):R81. doi: 10.1186/gb-2014-15-6-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rakoczy J, Padmanabhan N, Krzak AM, Kieckbusch J, Cindrova-Davies T, Watson ED. Dynamic expression of TET1, TET2, and TET3 dioxygenases in mouse and human placentas throughout gestation. Placenta. 2017;59:46–56. doi: 10.1016/j.placenta.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 61.Hernandez Mora JR, Sanchez-Delgado M, Petazzi P, Moran S, Esteller M, Iglesias-Platas I, Monk D. Profiling of oxBS-450 K 5-hydroxymethylcytosine in human placenta and brain reveals enrichment at imprinted loci. Epigenetics. 2018;13(2):182–191. doi: 10.1080/15592294.2017.1344803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726–870725. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22(3):467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green BB, Houseman EA, Johnson KC, Guerin DJ, Armstrong DA, Christensen BC, Marsit CJ. Hydroxymethylation is uniquely distributed within term placenta, and is associated with gene expression. FASEB J. 2016;30(8):2874–2884. doi: 10.1096/fj.201600310R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chabrun F, Huetz N, Dieu X, Rousseau G, Bouzillé G, Chao de la Barca JM, et al. Data-mining approach on transcriptomics and methylomics placental analysis highlights genes in fetal growth restriction. Front Genet. 2019. 10.3389/fgene.2019.01292. [DOI] [PMC free article] [PubMed]
- 66.O'Callaghan JL, Clifton VL, Prentis P, Ewing A, Miller YD, Pelzer ES. Modulation of Placental Gene Expression in Small-for-Gestational-Age Infants. Genes (Basel). 2020;11(1):80. 10.3390/genes11010080. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 458 kb)
(XLSX 80 kb)