Abstract
Purpose
To identify the pathogenic mutation in PMFBP1 leading to acephalic spermatozoa syndrome.
Methods
Sanger sequencing was used to screen for mutations in the known pathogenic genes SUN5 and PMFBP1 in a patient with acephalic spermatozoa syndrome. Western blotting and immunofluorescence were used to detect the expression and localization of PMFBP1 in sperm. At the same time, a PMFBP1 mutant was constructed, and the expression level of PMFBP1 protein was further verified by in vitro experiments.
Results
We identified a novel homozygous PMFBP1 missense mutation, c.301A>C (p.T101P), in an infertile male from a consanguineous family. Our results showed that the expression of PMFBP1 mutant protein was decreased obviously in sperm of the patient.
Conclusion
Our results showed that the novel homozygous missense mutation of PMFBP1 may be a cause of acephalic spermatozoa syndrome, which provided a basis for genetic counseling for the patient.
Keywords: Acephalic spermatozoa syndrome, PMFBP1, Missense mutation, Sanger sequencing
Introduction
Infertility is a growing reproductive concern worldwide, affecting about 15% of couples. Approximately half of the pathogenic factors are from males, so male infertility is attractingincreasing attention [1, 2]. Male infertility is a multifactorial disease with genetic factors accounting for the etiology of about 15% of infertile men [3]. Teratozoospermia, which comes in many forms, is one of the important reasons for male infertility, such as the rare and severe acephalic spermatozoa syndrome (ASS) [4–10]. Almost no normal sperm are observed during ejaculation in patients with acephalic spermatozoa syndrome, whose sperm morphology in semen includes a large number of headless sperm tails (pinhead sperm), a small number of tailless sperm heads, and sperm with an abnormal head-tail junction. The spermatozoa with abnormal head-tail junctions show a normal-shaped nucleus, an acrosome and midpiece, but the midpiece is not in a linear alignment with the sperm axis [6].
Familial aggregation studies suggest that genetic factors play an important role in acephalic spermatozoa syndrome. To date, there have been reported cases of acephalic spermatozoa syndrome related to mutations in PMFBP1, SUN5, TSGA10, BRDT, HOOK1, and DNAH6 [11–21]. According to previous studies, mutations in SUN5 and PMFBP1 are major causes of ASS in approximately three-quarters of patients [11–15, 18]. However, these mutations do not account for all cases of ASS, whichrequires more studies to elucidate its pathogenesis. Despite the presence of abnormal spermatozoa, there have been cases of successful progeny from patients with ASS attributed to the use of intracytoplasmic sperm injection [11, 13, 15].
Our previous work identified multiple mutations in SUN5 and PMFBP1 as important causes of ASS [11–15, 18]. Hence, in this study, we aimed to screen novel mutations of these genes in a patient with acephalic spermatozoa syndrome from a consanguineous family, where a novel homozygous missense mutation (NM_031293.2, c.301A>C; p.T101P) in PMFBP1 was identified. Both western blotting and immunofluorescence analysis showed the expression level of PMFBP1 was consequently decreased. Our findings thus further confirmed that PMFBP1 may be one of the main pathogenic genes of acephalic spermatozoa syndrome.
Materials and methods
Patient information
A 30-year-old infertile man from family IV-3, whose parents were cousins, was recruited to the Reproductive Center of the First Affiliated Hospital of Anhui Medical University in Aug. 2018. The patient was diagnosed with ASS by performing semen analysis at least twice and Papanicolaou staining. In addition, the patient’s bilateral testicular size, spermatic vein, and prostate were examined by color ultrasound.
The study was approved by the Ethics Committee of Anhui Medical University, and written consent was obtained from the participant.
Semen analysis
According to the Guidelines of the World Health Organization, the patient is instructed to undergo a routine semen examination at the same reproductive center at least twice. The following parameters were observed: semen volume, pH, sperm concentration, sperm motility, and proportion of headless sperm. Sperm morphology was evaluated by Papanicolaou staining.
Sanger sequencing of PMFBP1 and SUN5
The blood samples of patients were collected and preserved in anticoagulant tubes containing EDTA and stored in a − 80 °C refrigerator. Genomic DNA was extracted from the peripheral blood of the proband using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer’s protocol. The coding regions of PMFBP1 and SUN5 genes were amplified as described in previous studies [11, 13]. The PCR products were detected using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Finally, Sanger sequencing was performed to verify whether PMFBP1 and SUN5 contained any mutations, which was predicted using in silico tools.
Plasmid construction
The full-length cDNA of PMFBP1 with different restriction sites of EcoRI or BamHI at both ends (General Biosystems (Anhui) Co., Ltd., Anhui, China) was synthesized by Tongyong Biotechnology Company. The recombinant plasmid was inserted into pCMV-3×FLAG (Sigma) vectors by subcloning. PMFBP1 with c.301A>C mutation plasmid was constructed using the Mut Express II Fast Mutagenesis kit (Vazyme Biotech Co., Ltd., Nanjing, China).
Transfection
HEK293T cells (human embryonic kidney cells) were cultured in Dulbecco’s modified Eagle growth medium (DMEM, Invitrogen) containing 10% (vol/vol) fetal bovine serum (FBS, Hyclone), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C with 5% CO2. Subsequently, FLAG-PMFBP1 (wild-type/WT) and PMFBP1 mutant (p.T101P) expression plasmids were transfected into the cells respectively. The final concentration of each plasmid after transfection was 0.01 μg/μl. After 5 h, the medium was changed with fresh medium containing MG132 (a ubiquitination protease inhibitor), and the cells were harvested after 24 h.
Western blotting
The fresh semen was washed three times with PBS by centrifugation (6000 rpm, 1 min). The supernatant was discarded, and the precipitation was dissolved in 1 mM loading buffer at 100 °C for 10 min. The proteins were separated using SDS-PAGE and were electroblotted onto a nitrocellulose membrane at 100 V for 1.5 h at 4 °C. The membrane was then blocked with 5% BSA solution for 1 h at room temperature before it was incubated overnight at 4 °C with primary antibodies (anti-PMFBP1, 1:1000 dilution; or anti-α-tubulin, 1:1000 dilution; Sigma). The secondary antibodies (goat anti-rabbit IgG or goat anti-mouse IgG, 1:5000 dilution; ProteinTech Group, Hubei, China) were used to detect antigen content.
Immunofluorescence analysis
Immunofluorescence analysis of sperm was performed as previously described [10, 12]. HeLa cells were cultured in DMEM containing 10% FBS, and the corresponding plasmids were transfected into HeLa cells on coverslips in 24-well plates with Lipofectamine 2000 (Invitrogen). After 24 h, the transfected cells were fixed with 2% formaldehyde in PBS for 5 min followed by staining the nuclei of HeLa cells with DAPI (4′, 6-diamidino-2-phenylindole). The images were collected using a confocal laser scanning microscope.
Results
Clinical findings
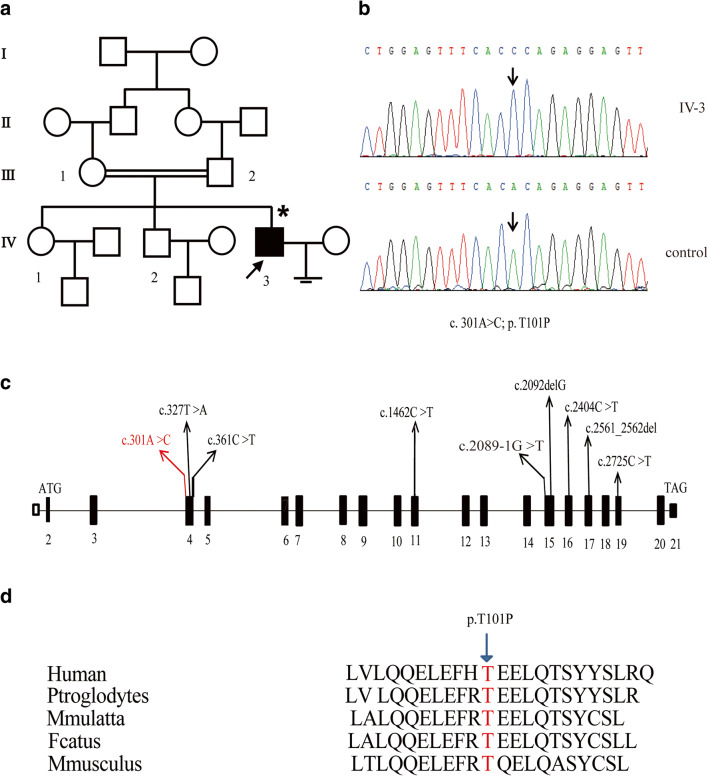
The proband presented with primary infertility over 3 years. The patient was diagnosed with ASS based on the routine semen analysis, which showed complete teratozoospermia with severe oligoasthenozoospermia (Table 1). The karyotype was 46; XY and Y chromosome microdeletions analysis showed no obvious abnormalities. Color ultrasound showed normal size of bilateral testis, and no obvious varicose veins and abnormalities in the prostate. Meanwhile, drugs, environment, poisons, diet, and other possible causes leading to infertility were excluded. The patient had an older sister and brother (Fig. 1a, IV-1, IV-2) who got married and had sons, respectively. The pedigree analysis showed an autosomal recessive inheritance pattern (Fig. 1a).
Table 1.
Semen parameters of the patient with ASS
| Volume (ml) |
pH | Concentration (×106 / ml) |
Motility B+C (%)* | Abnormal spermatozoa | |
|---|---|---|---|---|---|
| 3.1 | 7.4 | 5.6 | 8.4 | 100% |
Abnormal head-tail junction 0.3% Decaudated 0.7% Acephalic 99% |
*Motility A (%)=0
Fig. 1.
a, b Pedigree of a family with inherited PMFBP1 mutation. The individual representing a filled black box was Sanger sequenced. The proband (IV-3) has a homozygous mutation in PMFBP1. c The black boxes represent the coding region and the white box indicates noncoding exon 1, while introns are black bars; the novel missense mutation in this study and other reported mutations in PMFBP1 are represented by red wire and black wires, respectively. d Cross-species comparisons support that PMFBP1 is highly conserved and the detected PMFBP1 mutation in IV-3 is located in a highly conserved region
Identification of the PMFBP1 mutation c.301A>C (p.T101P) (NM_031293.2)
To determine the pathogenic factor of ASS in the patient, peripheral blood DNA was extracted. According to previous reports, PMFBP1 and SUN5 are the main pathogenic genes of ASS. Therefore, we performed Sanger sequencing on the SUN5 and PMFBP1 coding regions of the proband. No mutation was found in SUN5, and a novel homozygous missense mutation in PMFBP1, c.301A>C (p.T101P), was identified in the patient (Fig. 1b). Here, we summarized the known data on PMFBP1 mutations that caused acephalic spermatozoa syndrome (Fig. 1c). Moreover, the amino acid mutation Thr-101/T101 was highly conserved in different species (Fig. 1d).
Effect of the novel PMFBP1 mutation predicted using in silico tools
SIFT, Polyphen2, and Mutation Taster were used to predict the effect of c.301A>C mutation on the function of PMFBP1, which was predicted to be a disease-causing mutation. The mutation was not found in SNPs, genome aggregation database (gnomAD) and exome aggregation consortium database (ExAC).
Expression and localization of PMFBP1 mutant
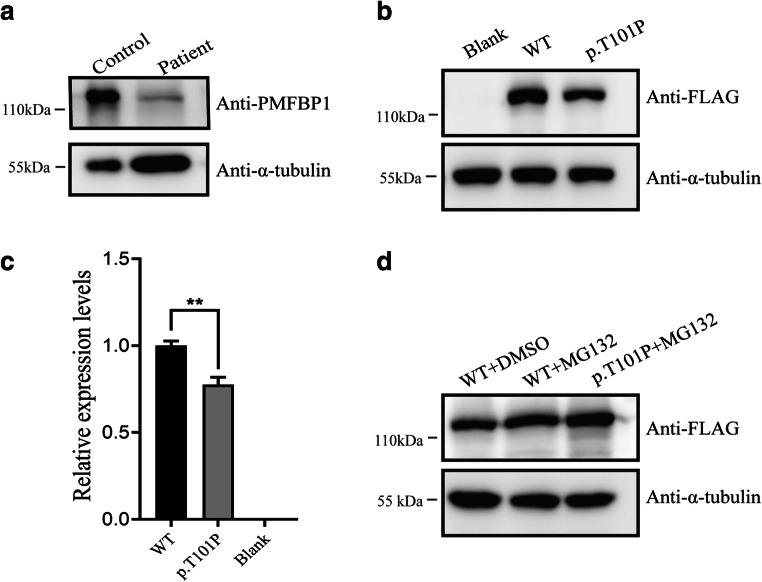
To evaluate the impact of missense mutations on PMFBP1, PMFBP1 expression level and localization in sperm of the patient were analyzed by western blot and immunofluorescence. Our results showed that there was a specific 117-kDa band in sperm samples from control and the patient, but the intensity of band in the patient’s sperm decreased significantly (Fig. 2a). In vitro experiments also confirmed the effect of this mutation on PMFBP1 (Fig. 2b and c). The proteasome inhibitor MG132 treatment could restore the expression level of PMFBP1 mutant (Fig. 2d).
Fig. 2.
Protein expression of PMFBP1 in sperm and HEK293T cells. a Western blotting was performed to test the expression level of PMFBP1 in the normal and patient sperm. b The influence of the c.301A>C mutation on PMFBP1 protein expression levels in HEK293T cells transfected with wild-type and mutant plasmids of PMFBP1 was tested with Western blotting. c The relative expression levels of wild-type and mutant-PMFBP1 (p.T101P) proteins in b represented by bar charts. Information is indicated as the mean and SEM. **P < 0.05. d MG132 was added to the culture medium for 12 h before the cells were collected. The expression levels of wide-type and mutant-PMFBP1 (p.T101P) were measured by western blotting. DMSO treatment alone was used as the control
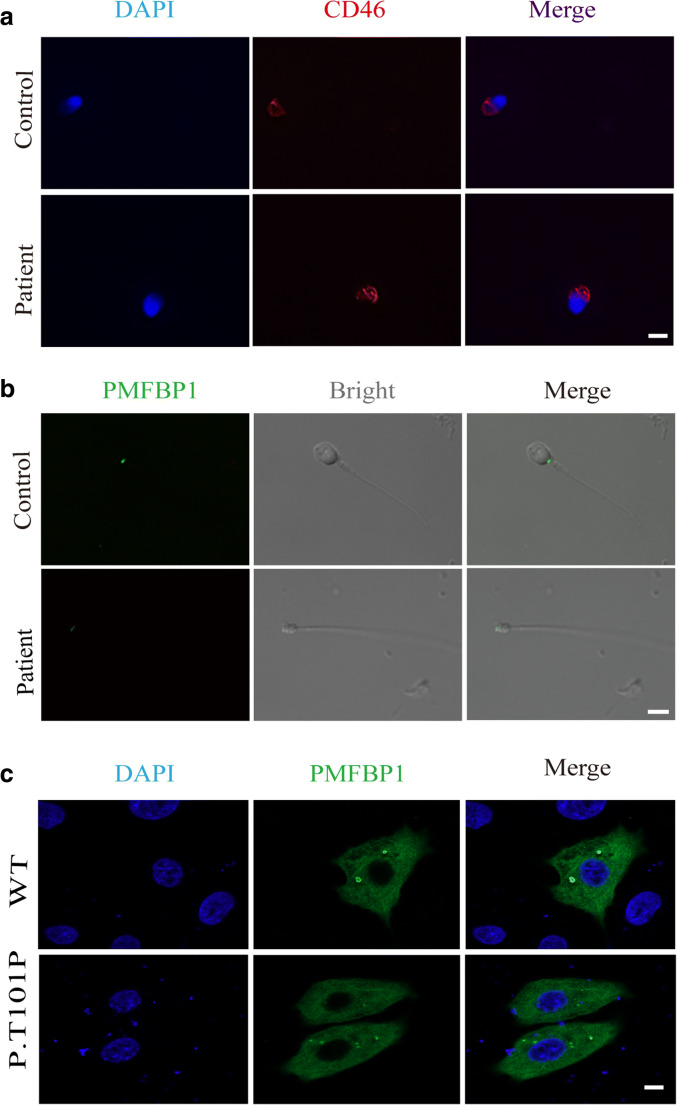
The localization of PMFBP1 was determined by immunofluorescence (Fig. 3), we used 4′, 6-diamidino-2-phenylindole (DAPI), PMFBP1 mouse antibody, and CD46 antibody to stain control and patient’s sperm. The results suggested that nuclear membrane, nucleus, and acrosome were unaffected regardless of mutation (Fig. 3a). In both the two samples, PMFBP1 was detected at the head-tail junction of spermatozoa. However, in the spermatozoa of patient, the fluorescence intensity of PMFBP1 was obviously decreased (Fig. 3b). Furthermore, the immunofluorescence analysis was also performed using HeLa cells, and data showed that the p.T101P mutation did not change the localization of PMFBP1 in the cytoplasm (Fig. 3c).
Fig. 3.
Immunofluorescence analysis and acrosomal staining in sperm of patient and control. a The acrosome and nucleus of the sperm are stained with CD46 antibody (red) and DAPI (blue), respectively. b The localization and expression level of PMFBP1 in sperm from the patient and control were demonstrated by immunofluorescence staining. c Representative images of HeLa cells transfected with FLAG-PMFBP1 and mutant plasmids and stained with anti-FLAG antibody. Bar: 10 μm
Discussion
PMFBP1 is a testis-specific protein located at the head-tail junction of sperm, and forms a “sandwich” structure with SUN5 and SPATA6 proteins. SUN5 is a nuclear membrane protein and SPATA6 is a structural protein, so it is speculated that PMFBP1 may be a scaffold protein connecting the coupling device and the sperm nuclear membrane [11, 12, 22, 23]. In a study of mutations in PMFBP1 leading to acephalic spermatozoa, an independent team found that SUN5 and TSGA10 were upstream and downstream of PMFBP1 respectively. According to previous studies, there is no significant difference in semen parameters when compared to those of patients with acephalic spermatozoa caused by PMFBP1, SUN5, BRDT, and TSGA10 [11–19]. However, another team reported that two patients with ASS, caused by mutations in PMFBP1, not only had sperm with rounded or amorphous heads upon ejaculation but also had lower sperm concentrations, indicating that the phenotype of acephalic spermatozoa caused by PMFBP1 mutation is more severe [12].
It has been established that PMFBP1 and SUN5 mutations are mainly responsible for ASS. Previous reports on PMFBP1 include three compound heterozygous mutations, three homozygous nonsense mutations, and one homozygous frameshift mutation [11, 12, 18]. Because ASS is severe and rare, there are no clear epidemiological statistics on the incidence of this disease. The identification of these mutations in humans represents a major step forward in the study of the disease. Even so, some cases cannot be explained by known mutations. Therefore, more studies are needed to elucidate the pathogenesis of ASS.
In our study, we identified a novel homozygous missense mutation c.301A>C (p.T101P) in exon 4 of PMFBP1 in a patient with ASS. In the present study, the mutant PMFBP1 could be detected in the patient’s sperm but the expression level drops significantly, which was consistent with the in vitro experimental data. To further verify whether the mutant PMFBP1 was degraded by the ubiquitination pathway, the cells transfected with PMFBP1 mutant plasmids were treated or not with MG132. Western blotting analysis showed that MG132 could improve the expression level of PMFBP1 mutant, which suggested that MG132 may block the degradation of the mutant PMFBP1. Furthermore, our hypothesis verified that the degradation of the mutant PMFBP1 was mediated by the ubiquitination pathway [24–26]. In this study, PMFBP1 was expressed in the sperm of the patient, but the expression level of the mutant protein was significantly lower than that of the normal control. The mutation did not change the location of PMFBP, but the immunofluorescence signal was attenuated. These results also verified that the novel missense mutation of PMFBP1 identified in this study may be a pathogenic mutation leading to ASS.
In conclusion, we identified a novel homozygous missense PMFBP1 mutation that could cause ASS. Altogether, these results suggested that PMFBP1 mutations appeared to be a “hotspot” of causative factors underlying patients suffering from ASS. Therefore, identification and characterization of more novel mutations of PMFBP1 would undoubtedly provide more approaches for diagnosis, genetic counseling, and selection of assisted reproductive treatment for patients with the disease.
Funding
This work was supported by grants from National Natural Science Foundation of China (82071701 to F. Z, 81792641 to F. W), Natural Science Foundation of Anhui Province (1908085 J28 to F. Z), Key R&D program of Anhui Province (201904a07020050 to F. Z), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT310002), and Scientific Research Foundation of the Institute for Translational Medicine of Anhui Province (SRFITMAP 2017zhyx29 and ZHYX2020A001) supported this study.
Compliance with ethical standards
This study was approved by the ethics committee of Anhui Medical University. Written informed consent was obtained from each participant.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mengmeng Lu, Shuai Kong, Mingfei Xiang and Yu Wang contributed equally to this work.
Contributor Information
Fengsong Wang, Email: fengsongw@ahmu.edu.cn.
Yunxia Cao, Email: caoyunxia6@126.com.
Fuxi Zhu, Email: fxzhu@ahmu.edu.cn.
References
- 1.Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update. 2015;21(4):411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15:369–384. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- 4.Perotti ME, Giarola A, Gioria M. Ultrastructural study of the decapitated sperm defect in an infertile man. J Reprod Fertil. 1981;63(2):543–549. doi: 10.1530/jrf.0.0630543. [DOI] [PubMed] [Google Scholar]
- 5.Baccetti B, Selmi MG, Soldani P. Morphogenesis of ‘decapitated’ spermatozoa in a man. J Reprod Fertil. 1984;70(2):395–397. doi: 10.1530/jrf.0.0700395. [DOI] [PubMed] [Google Scholar]
- 6.Chemes HE, Puigdomenech ET, Carizza C, Olmedo SB, Zanchetti F, Hermes R. Acephalic spermatozoa and abnormal development of the head-neck attachment: a human syndrome of genetic origin. Hum Reprod. 1999;14(7):1811–1818. doi: 10.1093/humrep/14.7.1811. [DOI] [PubMed] [Google Scholar]
- 7.Toyama Y, Iwamoto T, Yajima M, Baba K, Yuasa S. Decapitated and decaudated spermatozoa in man, and pathogenesis based on the ultrastructure. Int J Androl. 2000;23(2):109–115. doi: 10.1046/j.1365-2605.2000.t01-1-00217.x. [DOI] [PubMed] [Google Scholar]
- 8.Chemes HE, Carizza C, Scarinci F, Brugo S, Neuspiller N, Schwarsztein L. Lack of a head in human spermatozoa from sterile patients: a syndrome associated with impaired fertilization. Fertil Steril. 1987;47(2):310–316. doi: 10.1016/S0015-0282(16)50011-9. [DOI] [PubMed] [Google Scholar]
- 9.Baccetti B, Burrini AG, Collodel G, Magnano AR, Piomboni P, Renieri T, Sensini C. Morphogenesis of the decapitated and decaudated sperm defect in two brothers. Gamete Res. 1989;23(2):181–188. doi: 10.1002/mrd.1120230205. [DOI] [PubMed] [Google Scholar]
- 10.Porcu G, Mercier G, Boyer P, Achard V, Banet J, Vasserot M, et al. Pregnancies after ICSI using sperm with abnormal head-tail junction from two brothers: case report. Hum Reprod. 2003;18(3):562–567. doi: 10.1093/humrep/deg121. [DOI] [PubMed] [Google Scholar]
- 11.Zhu F, Liu C, Wang F, Yang X, Zhang J, Wu H, Zhang Z, He X, Zhang Z, Zhou P, Wei Z, Shang Y, Wang L, Zhang R, Ouyang YC, Sun QY, Cao Y, Li W. Mutations in PMFBP1 cause acephalic spermatozoa syndrome. Am J Hum Genet. 2018;103(2):188–199. doi: 10.1016/j.ajhg.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sha YW, Wang X, Xu X, Ding L, Liu WS, Li P, Su ZY, Chen J, Mei LB, Zheng LK, Wang HL, Kong SB, You M, Wu JF. Biallelic mutations in PMFBP1 cause acephalic spermatozoa. Clin Genet. 2019;95(2):277–286. doi: 10.1111/cge.13461. [DOI] [PubMed] [Google Scholar]
- 13.Zhu F, Wang F, Yang X, Zhang J, Wu H, Zhang Z, Zhang Z, He X, Zhou P, Wei Z, Gecz J, Cao Y. Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am J Hum Genet. 2016;99(4):942–949. doi: 10.1016/j.ajhg.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sha YW, Xu X, Ji ZY, Lin SB, Wang X, Qiu PP, Zhou Y, Mei LB, Su ZY, Li L, Li P. Genetic contribution of SUN5 mutations to acephalic spermatozoa in Fujian China. Gene. 2018;647:221–225. doi: 10.1016/j.gene.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 15.Fang J, Zhang J, Zhu F, Yang X, Cui Y, Liu J. Patients with acephalic spermatozoa syndrome linked to SUN5 mutations have a favorable pregnancy outcome from ICSI. Hum Reprod. 2018;33(3):372–377. doi: 10.1093/humrep/dex382. [DOI] [PubMed] [Google Scholar]
- 16.Sha YW, Sha YK, Ji ZY, Mei LB, Ding L, Zhang Q, Qiu PP, Lin SB, Wang X, Li P, Xu X, Li L. TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin Genet. 2018;93(4):776–783. doi: 10.1111/cge.13140. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Wei X, Sha Y, Li N, Yan X, Cheng L, Qiao D, Zhou W, Wu R, Liu Q, Li Y. Loss-of-function mutation in TSGA10 causes acephalic spermatozoa phenotype in human. Mol Genet Genomic Med. 2020;8(7):e1284. doi: 10.1002/mgg3.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Wang N, Zhang H, Yin S, Dai H, Lin G, Li W. Novel mutations in PMFBP1, TSGA10 and SUN5: expanding the spectrum of mutations that may cause acephalic spermatozoa. Clin Genet. 2020;97(6):938–939. doi: 10.1111/cge.13747. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Sha Y, Wang X, Li P, Wang J, Kee K, Wang B. Whole-exome sequencing identified a homozygous BRDT mutation in a patient with acephalic spermatozoa. Oncotarget. 2017;8(12):19914–19922. doi: 10.18632/oncotarget.15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Zhu Y, Zhu Z, Zhi E, Lu K, Wang X, Liu F, Li Z, Xia W. Detection of heterozygous mutation in hook microtubule-tethering protein 1 in three patients with decapitated and decaudated spermatozoa syndrome. J Med Genet. 2018;55(3):150–157. doi: 10.1136/jmedgenet-2016-104404. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Sha YW, Xu X, Mei LB, Qiu PP, Ji ZY, et al. DNAH6 is a novel candidate gene associated with sperm head anomaly. Andrologia. 2018;50. [DOI] [PubMed]
- 22.Yuan S, Stratton CJ, Bao J, Zheng H, Bhetwal BP, Yanagimachi R, Yan W. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc Natl Acad Sci U S A. 2015;112(5):E430–E439. doi: 10.1073/pnas.1424648112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang Y, Zhu F, Wang L, Ouyang YC, Dong MZ, Liu C, et al. Essential role for SUN5 in anchoring sperm head to the tail. Elife. 2017;6. [DOI] [PMC free article] [PubMed]
- 24.Zimmerman S, Sutovsky P. The sperm proteasome during sperm capacitation and fertilization. J Reprod Immunol. 2009;83(1–2):19–25. doi: 10.1016/j.jri.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Lorès P, Coutton C, El Khouri E, Stouvenel L, Givelet M, Thomas L, et al. Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum Mol Genet. 2018;27(7):1196–1211. doi: 10.1093/hmg/ddy034. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Li Y, Wang X, Xing R, Liu K, Gan Q, Tang C, Gao Z, Jian Y, Luo S, Guo W, Yang C. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J Cell Biol. 2017;216(5):1301–1320. doi: 10.1083/jcb.201608039. [DOI] [PMC free article] [PubMed] [Google Scholar]