Abstract
Objective:
The unaffected relatives of individuals with non-syndromic orofacial clefts have been shown to exhibit subtle craniofacial differences compared with the general population. Here, we investigate whether these morphological differences extend to the shape of the palate.
Design:
We conducted a geometric morphometric analysis to compare palate shape in the clinically unaffected parents of children with non-syndromic clefting of lip with or without cleft palate (NSCL/P) and adult controls of European, Asian, and African ancestry. We conducted pairwise group comparisons using canonical variates analysis (CVA), and then confirmed and characterized findings of shape differences using Euclidean distance matrix analysis (EDMA).
Results:
Significant differences in palate shape were detected in unaffected mothers (but not fathers) compared to demographically matched controls. The differences in shape were ancestry specific; mothers of Asian-derived and African-derived ancestry showed wider and shorter palates with higher posterior palatal vaults, while mothers of European-derived ancestry showed narrower palates with higher anterior palatal vaults.
Conclusions:
Our findings suggest that altered palate shape is a subclinical phenotypic feature, which may be indicative of elevated orofacial cleft risk. The risk phenotype varied by sex and ancestry, suggesting possible etiologic heterogeneity among demographic groups. Understanding the genetic basis of these informative palate shape traits may reveal new genes and pathways relevant to non-syndromic orofacial clefting.
Keywords: craniofacial morphology, craniofacial biology, palatal morphology, geometric morphometrics, cleft lip and palate
Introduction:
The hard palate, comprised of the horizontally aligned shelves of the maxillae and palatine bones, separates the oral and nasal cavities (Skrzat, et al., 2003; Dursun, et al., 2018). Embryologically, the development of the palate starts by the 6th week of intrauterine life through the fusion of paired palatal shelves from the maxillary processes. Those shelves fuse at the midline as well as with the primary palate anteriorly, thereby forming the hard and soft palates (Mossey, et al., 2009). When this fusion fails, the result is an orofacial cleft (OFC) involving the secondary palate (Mossey, et al., 2009). Such OFCs are most often isolated (i.e., non-syndromic) defects, and can occur along with a cleft of with the lip (CLP) or involve only the secondary palate (CPO). OFCs are among the most common birth defects in humans and are well known to vary in incidence by ancestry, with Asian-derived populations showing the highest rates and African-derived populations showing the lowest (Leslie and Marazita, 2013). Findings from mouse models and human epidemiology show a distinction etiologically and pathogenetically between palatal clefts that occur with cleft lip and those that occur affecting the secondary palate only in isolation (Murray, 2002; Juriloff and Harris, 2008; Mossey, et al., 2009; Watkins, et al., 2014). CLP occurs more commonly in males while CPO also occurs at roughly twice the frequently in females (Mossey, et al., 2009; Leslie and Marazita, 2013). The factors that contribute to the population and sex differences in OFC incidence are still largely unknown.
A long-standing hypothesis states that the shape of certain facial features may be a predisposing factor for OFCs (Ward, et al., 2002). For example, several studies have suggested that a combination of increased upper facial width and reduced midfacial projection is present in the “at-risk” biological relatives of those affected with clefts (Weinberg, et al., 2009; Roosenboom, et al., 2017). In the present study, we study possible morphological differences in the palate, being an integral structure of the midface.
Palate shape varies considerably among modern human populations and between males and females (Maier, et al., 2015; Mustafa, et al., 2019). Studies in mice have suggested that excess palate width may be a predisposing factor for cleft palate (Vergato, et al., 1997). Other evidence from humans suggests that high-arched or vaulted palates may be a risk factor for CP. For example, an elevated rate of high-arched palates has been reported in several genetic syndromes (e.g. Down syndrome, Stickler syndrome, Marfan syndrome) where cleft palate is sometimes a feature (Källén, et al., 1996; Snead and Yates, 1999; Utreja and Evans, 2009). In the current study, we investigate the differences in palate shape between biological parents of individuals affected with orofacial clefting in comparison to unrelated normal controls and explore whether sex and ancestry have an effect on the observed differences. We accomplish this by applying landmark-based morphometrics to 3D maxillary dental models collected on biological mothers and fathers of affected individuals and unrelated controls of African, Asian, and European ancestry.
Materials and Method:
Study Population:
The study sample was comprised of 935 individuals recruited as part of the Pittsburgh Orofacial Cleft study based at the University of Pittsburgh Center for Craniofacial and Dental Genetics. All participants were at least 18 years of age and screened for a personal and family history of oral and craniofacial malformations and prior trauma or surgery involving the palatal region. The parent group included clinically unaffected biological fathers and mothers of individuals with non-syndromic cleft lip with or without cleft palate (NSCL/P). Parents of children with CPO were not included due to inadequate sample numbers. The control group included unrelated individuals with no prior history of oral and craniofacial malformations or disorders. The father group comprised 49 individuals (mean age = 38.6 ± 10.6), while the male control group comprised 356 individuals (mean age = 34.1 ± 15.1). The mother group comprised 92 individuals (mean age = 33.2 ± 7.6), while the female control group comprised 438 individuals (mean age = 36.1 ± 15.1). Individuals were excluded if they presented with torus palatinus, missing canines, or missing first molars, as these features interfered with data collection.
Individuals derived from three ancestral groups were represented: 473 US-Whites of European-derived ancestry recruited from Pittsburgh and Lancaster, Pennsylvania (18 fathers and 157 male controls; 26 mothers and 272 female controls), 338 individuals of African-derived ancestry (13 fathers and 157 male controls; 30 mothers and 138 female controls) and 124 individuals of East Asian-derived ancestry (18 fathers and 42 male controls; 36 mothers and 28 female controls). Individuals of African-derived ancestry were recruited from Nigeria (N = 318), Colombia (N = 13) and Puerto Rico (N = 7). Individuals of East Asian-derived ancestry were recruited from the Philippines (N = 88) and the US (N = 36). The ancestral groups were self-reported by participants.
Data acquisition and Phenotype Capture:
Individuals provided their written informed consent prior to participation in the study. The protocol was approved by the Institutional Review Board (IRB) of the University of Pittsburgh and locally at each of the recruitment sites. Demographic, dental, medical, and social history data were recorded through in-person interviews. Maxillary impressions were taken by standard hydrocolloid impression materials and poured into plaster casts. The casts were then laser-scanned (3Shape, Copenhagen, Denmark). The scanned models were cleaned and processed using the 3Shape Orthoanalyzer software and stored as 3D meshes.
Morphometric Data Collection:
Landmarking was performed using the 3dMD Vultus software (3dMD Inc., Atlanta, Georgia, USA) directly on the 3D digital models of the maxillary arch. Seven landmarks were placed, located at the tip of the incisive papilla (IP), the deepest points of the gingival crevice (or cementoenamel junction) at the right and left canines (CR & CL) and the deepest points of the gingival crevice at the right and left first molars (6R & 6L), the midline between canines (CM), and the midline between first molars (MM). The midline landmarks were placed in a viewing orientation that is perpendicular to the anterior incisal/occlusal plane (Figure 1). The observer was blinded to sex, ancestral group, and parent-control status during landmarking. For intra-observer validation, a subset of 30 casts were landmarked twice, separated by at least 24 hours, yielding intraclass correlation coefficients for coordinate values in each of the three axes (x,y,z) ranging from 0.871 to 0.998, indicating low error.
Figure 1.
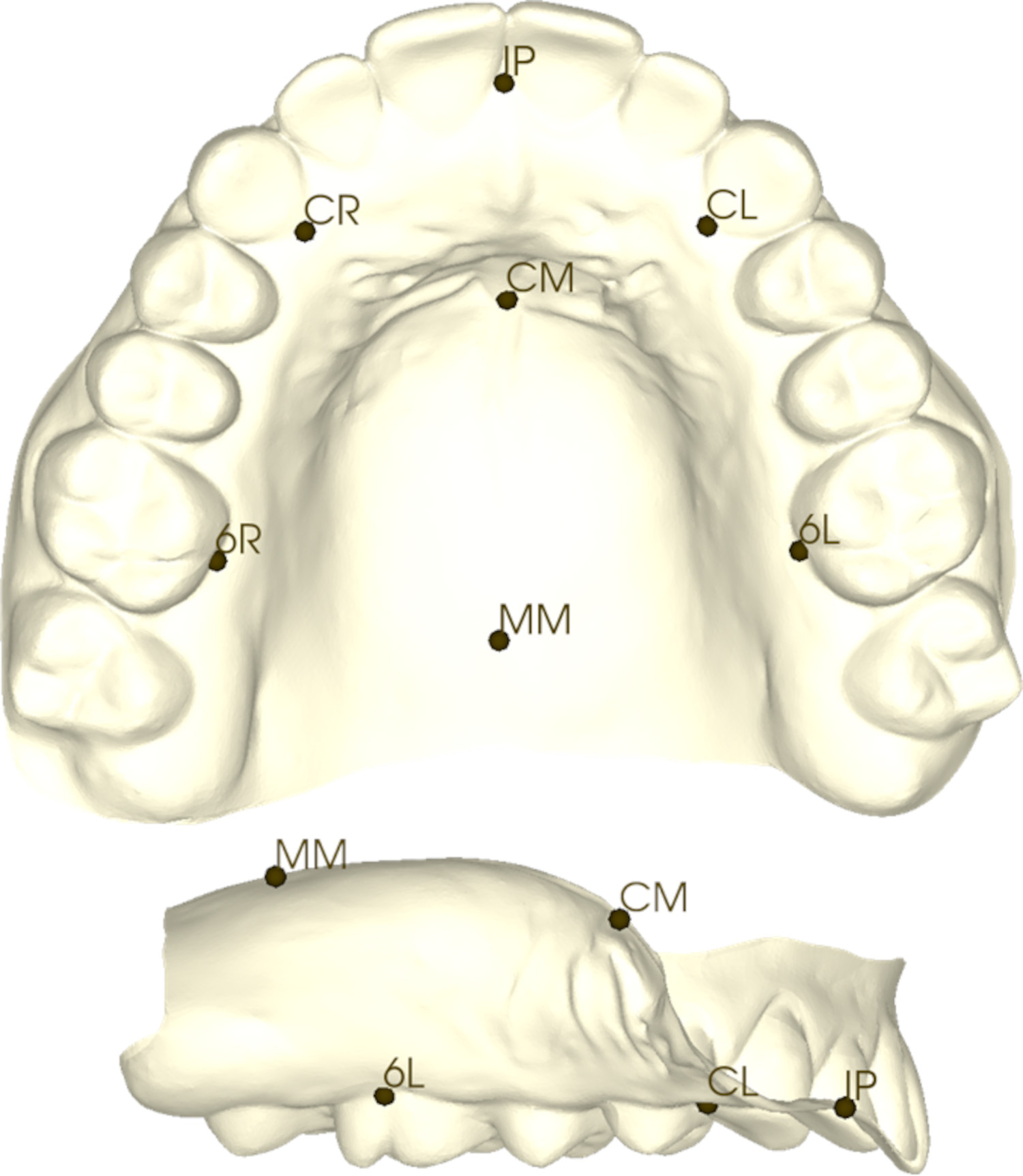
Landmarks on a dental cast in axial and sagittal views. IP = Incisive papilla. CR = Right canine. CL = Left canine. 6R = Right maxillary first molar. 6L = Left Maxillary First Molar. CM = Midline at canines. MM = Midline at first molars.
Statistical Analysis & Visualization:
Landmark configurations were subjected to Generalized Least-squares Procrustes Superimposition. This involved translation, rotation, and scaling into a common orientation. Geometric morphometric analysis was performed on the Procrustes-aligned configurations with two approaches designed to uncover shape differences. First, differences in mean palate shape between mothers and female controls were analyzed using canonical variates analysis (CVA). Similarly, CVA was then used to compare fathers to male controls. These parent-control comparisons were also carried out separately for each of the three ancestral groups. For all tests, p-values were determined based on permutation testing (10000 rounds) of the Procrustes and Mahalanobis distances generated from CVA. Morphometric tests and subsequent visualizations were generated with MorphoJ, R packages geomorph and Morpho, and 3D Slicer (Klingenberg, 2011; Fedorov, et al., 2012; Schlager, 2017; Adams, et al., 2020). The threshold for statistical significance was set at p ≤ 0.05.
Second, to better characterize which landmarks were driving the significant shape differences detected by CVA, Euclidean distance matrix analysis (EDMA) was applied to the Procrustes-aligned landmark configurations (winEDMA, Kansas City) (Cole, 2003). This was done by testing group differences in all possible inter-landmark distances (n=21) through bootstrap resampling (1000 rounds) and generating confidence intervals with an alpha level of 0.1 based on the recommendation by the developers of EDMA (Lele and Richtsmeier, 2001).
Results:
In terms of overall size (i.e., centroid size), fathers had smaller palates than male controls (p = 0.0081). In contrast, mothers had larger palates than female controls (p = 0.0089). However, the effect of allometry (defined as size-related differences shape) was minimal, contributing between 2–3% of the variation in shape.
Canonical Variates Analysis:
No differences in palate shape were detected when comparing fathers to male controls; this was the case with all ancestries combined and within each ancestry separately (Table 1). In contrast, we observed significant differences when comparing mothers to female controls, both in the ancestries-combined sample as well as within each ancestry (Table 1; Figure 2). The nature of the observed morphological differences between mothers and female controls were ancestry specific. In Asian-derived and African-derived individuals, mothers had shorter anteroposterior and wider mediolateral dimensions. Asian-derived and African-derived mothers also had higher posterior palatal vaults in comparison to demographically matched female controls (Figure 2). On the other hand, European-derived mothers had more constricted mediolateral dimensions and more anteriorly located vaults with higher anterior palatal vaults in comparison to demographically matched controls (Figure 2). These group differences were further confirmed by repeating the above analyses after statistically adjusting for the potential confounding effects of scale (centroid size) and age on shape.
Table 1.
Results of canonical variates analysis for testing palatal shape differences between parents and controls.1
| Contrast | Procrustes Distance (p-value) | Mahalanobis Distance (p-value) | |
|---|---|---|---|
| Fathers vs. Male Controls | Ancestries Combined | 0.0081 (0.8915) | 0.2047 (0.9862) |
| Asian | 0.0163 (0.8958) | 0.52 (0.9198) | |
| African | 0.0188 (0.6825) | 0.6671 (0.7115) | |
| European | 0.0159 (0.7183) | 0.8066 (0.2399) | |
| Mothers vs. Female Controls | Ancestries Combined | 0.0157 (0.0352)* | 0.6439 (0.0001)* |
| Asian | 0.0346 (0.0186)* | 1.4662 (<.0001)* | |
| African | 0.0254 (0.0287)* | 0.8957 (0.0093)* | |
| European | 0.025 (0.1062) | 1.0671 (0.0005)* |
p-values based on permutation testing (10000 resamples).
p-values achieving statistical significance (p < 0.05).
Figure 2.
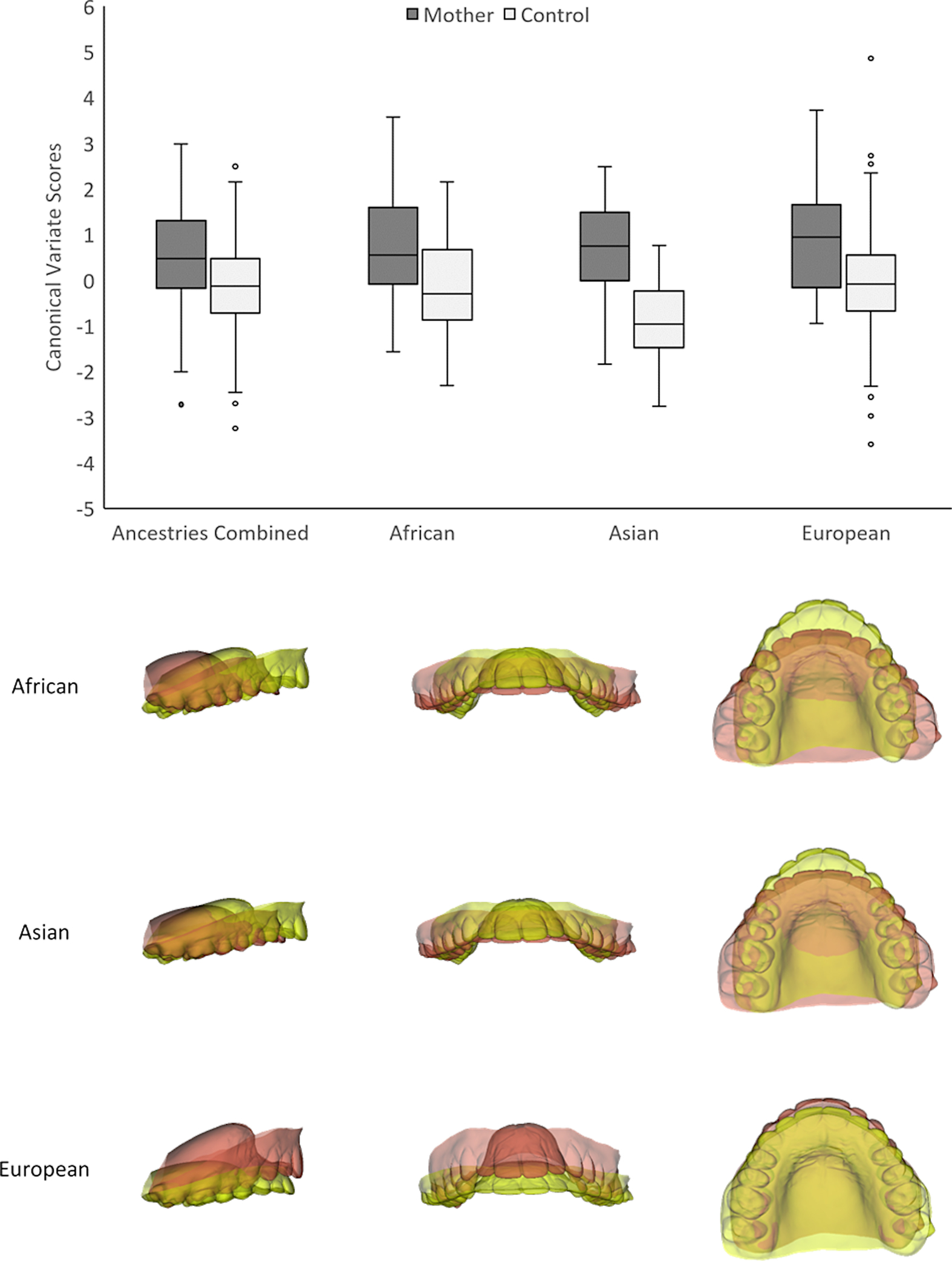
Canonical variates analysis (CVA) of group differences between mothers and female controls. Canonical variate effects are shown in lateral, frontal and axial views. Red = Positive canonical variate effect. Yellow = Negative canonical variate effect. Warp scale factor = 5.
Euclidean Distance Matrix Analysis:
EDMA confirmed the shape differences found by CVA. In Asian-derived and African-derived females, the shortened anteroposterior dimension in mothers was due to a localized retrusion occurring at the anterior palate while the increased palatal vault was occurring more posteriorly (Figure 3). In European-derived females, the constricted mediolateral dimension in mothers was due to medial shifting of the molar landmarks (Figure 3).
Figure 3.
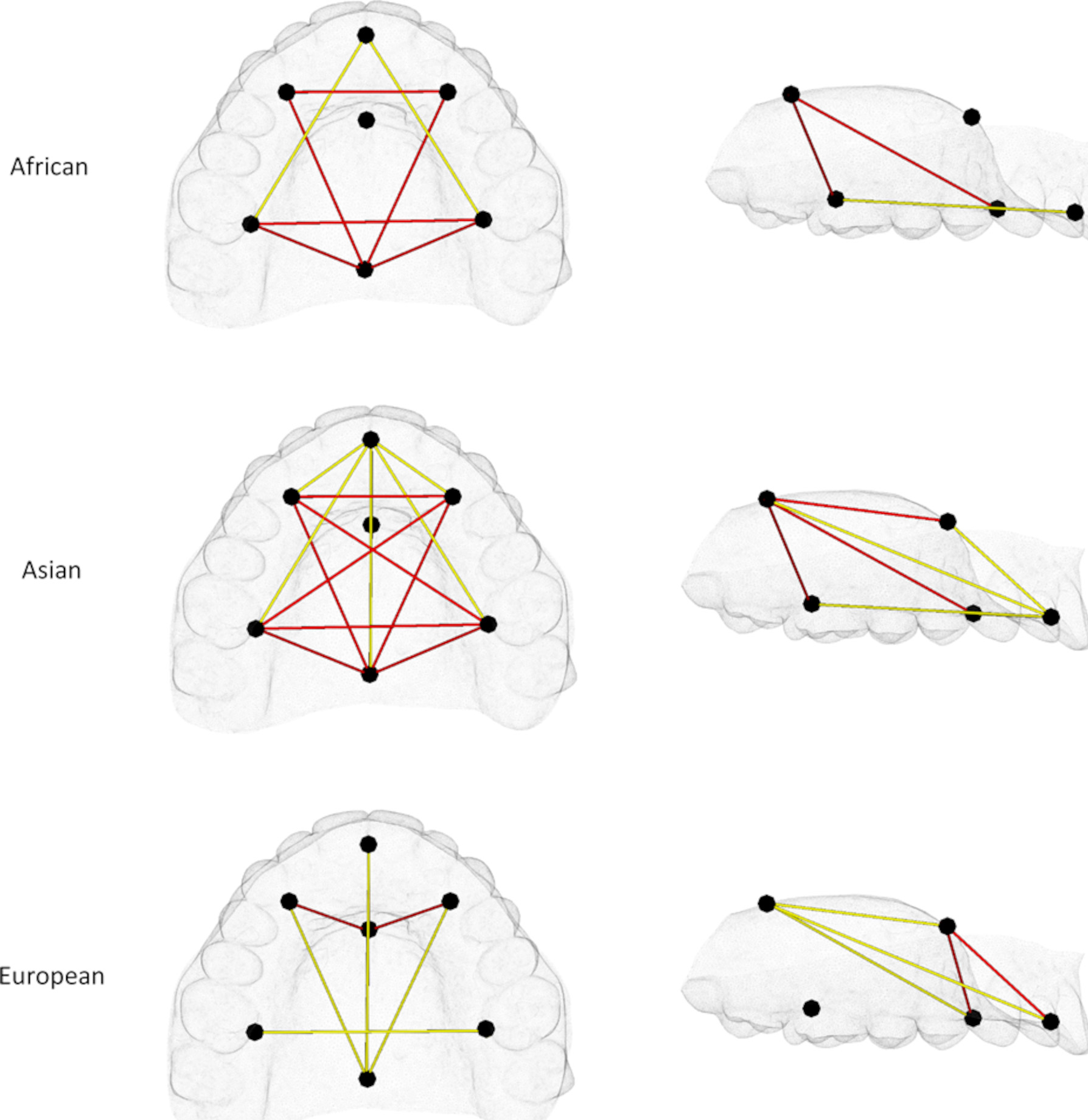
Euclidean distance matrix analysis (EDMA) results of pairwise comparisons between mothers and female controls shown in axial and sagittal views. Red = Distances significantly greater in mothers than controls. Yellow = Distances significantly smaller in mothers than controls.
Discussion:
In this study, we characterize differences in palate shape between the unaffected biological parents of individuals with NSCL/P and demographically matched controls in individuals derived from three ancestral groups with epidemiologically different baseline orofacial cleft risks. We found that fathers had smaller-sized palates than male controls and mothers had larger-sized palates than female controls; however, these size differences did not explain considerable variation in shape. Palate shape differences were sex-specific and limited to unaffected mothers. This may simply reflect a shape differences that is smaller than to be detected given male sample, which was roughly half the size of the female sample. Alternatively, altered shape of the hard palate may indeed be a unique risk factor for females, particularly given the higher rate of CPO in this group. Some have suggested that sex differences in palate development are apparent in the embryo, affecting the timing of palatal shelf elevation (Burdi and Silvey, 1969; Burg, et al., 2016). Further studies with larger samples of males will be needed to sort out these competing explanations.
Among unaffected mothers, the pattern of shape differences varied, depending on ancestry. This is potentially relevant because of the known differences in orofacial clefting incidence across ethnic groups. Orofacial clefting in general and NSCL/P in particular has been reported to be most common in those of Asian ancestry, intermediate in individuals of European ancestry, and least common in those of African ancestry (Leslie and Marazita, 2013). However, Burg et al. reported the prevalence of CPO being highest among two subpopulations of European ancestry (Burg, et al., 2016). In this study, African-derived and Asian-derived mothers showed reduced anteroposterior and wider mediolateral dimensions with higher posterior palatal vaults compared to their demographically matched controls. European-derived mothers, on the other hand, showed a different pattern, displaying narrower mediolateral palatal dimensions posteriorly and higher anterior palatal vaults when compared with their demographically matched controls. A possible explanation for the observed phenotypic heterogeneity is that it reflects population-specific genetic risk factors and/or background loci constituting the genetic architecture of the palatal morphology (Leslie, et al., 2016). Genetic studies of orofacial clefting have revealed population-specific associations (Leslie, et al., 2016); however, some of these findings may have more to do with differences in allele frequencies among ancestral groups, which can affect statistical power. Larger studies of multiple ethnic groups will be needed to map the genetic factors associated with palate shape across diverse human populations.
Earlier studies comparing the facial morphology of unaffected relatives from families with a history of orofacial clefting to controls provide additional supporting evidence that altered palate shape may be indicative of an increased genetic liability to orofacial clefting. Similar to our findings, these subclinical risk phenotypes were often shown to be sex specific. A limitation of these studies, however, is that they were limited to only one ancestral group, which precludes investigating ancestry-specific patterns of morphological differences affecting parents. Beginning in the mid-20th century, the first studies reported the increased presence of discrete morphological defects in the nasal cavity and/or palate of at-risk family members (Fukuhara and Saito, 1962; Fukuhara and Saito, 1963; Niswander, 1968; Weinberg, 2007). A similar study investigated palatal morphology from dental casts, yielding observable, yet not statistically significant, higher arched and narrower palates (Mills, et al., 1968). Later quantitative 2D cephalometric and 3D photogrammetric studies reported differences in the maxillofacial region of unaffected relatives (parents and siblings) indicative of altered palate shape; reductions in vertical nasomaxillary dimensions, increased midfacial breadth, and more concave facial profiles have been reported in the literature (Weinberg, et al., 2006). These previously described facial differences correspond well with the shorter and wider palate shape that we observed in mothers of Asian-derived and African-derived ancestry, but less so in mothers of European-derived ancestry. Regarding palatal vault height, all three groups of mothers showed evidence of increases, but the location varied from posterior (Asian-derived and African-derived ancestry) to anterior (European-derived ancestry). It is difficult to compare our vault height results with previous reports, however, because these reports did not provide detailed assessment of the vault topography.
In conclusion, the current study presented a nuanced assessment of morphological differences in palate shape in the unaffected biological parents of individuals with NSCL/P. The observed findings were found to be sex-specific, occurring among mothers but not fathers. Moreover, the findings in mothers were ancestry-specific, where mothers of African-derived and Asian-derived ancestry showed a distinct pattern of shape difference compared with mothers of European-derived ancestry. These findings will benefit from replication on larger and more well-balanced samples and the use of more advanced morphometric approaches that go beyond descriptions based on a small set of sparsely distributed landmarks. A potential limitation of the current study is the fact that the ancestral groups described here were self-reported by study participants and not confirmed by formal genetic testing, which may reveal possible factors affecting palate shape that are due to the ancestral make-up of each individual. Eventual genetic studies on these palatal traits may yield new insights on possible orofacial clefting risk factors.
Acknowledgements:
This study was funded by the following grants from the NIH/NIDCR: R01-DE016148, R00-DE022378, R01-DE015667, R00-DE024571, S21-MD001830 and U54-MD007587. The funder played no role in the design, analysis, or interpretation of this work. The authors have no conflicts of interest to declare.
References
- Adams D, Collyer M, JKaliontzopoulou A. Geomorph: Software for geometric morphometric analyses. R package version 3.2.1 2020 [Google Scholar]
- Burdi AR, Silvey RG. Sexual differences in closure of the human palatal shelves. Cleft Palate J. January 1969;6:1–7. [PubMed] [Google Scholar]
- Burg ML, Chai Y, Yao CA, Magee W, Figueiredo JC. Epidemiology, Etiology, and Treatment of Isolated Cleft Palate. Front Physiol. 2016;7:67. doi: 10.3389/fphys.2016.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TM. WinEDMA: Software for Euclidean distance matrix analysis . Version 1.0.1 2003; [Google Scholar]
- Dursun A, Öztürk K, Albay S. Development of Hard and Soft Palate During the Fetal Period and Hard Palate Asymmetry. J Craniofac Surg. November 2018;29(8):2358–2362. doi: 10.1097/SCS.0000000000005016 [DOI] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. November 2012;30(9):1323–41. doi: 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara T, Saito S. Genetic consideration on the dysplasia of the nasopalatal segments as a “Formes Frustes” radiologically found in parents of cleft children: a preliminary report. Jpn J Hum Genet. 1962;(7):3. [Google Scholar]
- Fukuhara T, Saito S. Possible carrier status of hereditary cleft palate with cleft lip: report of cases. Bull Tokyo Med Dent Univ. 1963;(10):12. [Google Scholar]
- Juriloff DM, Harris MJ. Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. February 2008;82(2):63–77. doi: 10.1002/bdra.20430 [DOI] [PubMed] [Google Scholar]
- Källén B, Mastroiacovo P, Robert E. Major congenital malformations in Down syndrome. Am J Med Genet. October 1996;65(2):160–6. doi: [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour. March 2011;11(2):353–7. doi: 10.1111/j.1755-0998.2010.02924.x [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. An invariant approach to statistical analysis of shapes : interdisciplinary statistics. Chapman & Hall/CRC interdisciplinary statistics series. Chapman & Hall/CRC; 2001:308 p. [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. July 2016;25(13):2862–2872. doi: 10.1093/hmg/ddw104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Marazita ML. Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet. November 2013;163C(4):246–58. doi: 10.1002/ajmg.c.31381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier CA, Zhang K, Manhein MH, Li X. Palate Shape and Depth: A Shape-Matching and Machine Learning Method for Estimating Ancestry from Human Skeletal Remains. J Forensic Sci. September 2015;60(5):1129–34. doi: 10.1111/1556-4029.12812 [DOI] [PubMed] [Google Scholar]
- Mills LF, Niswander JD, Mazaheri M, Brunelle JA. Minor oral and facial defects in relatives of oral cleft patients. Angle Orthod. July 1968;38(3):199–204. doi: 10.1043/0003-3219(1968)0382.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. November 2009;374(9703):1773–85. doi: 10.1016/S0140-6736(09)60695-4 [DOI] [PubMed] [Google Scholar]
- Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. April 2002;61(4):248–56. [DOI] [PubMed] [Google Scholar]
- Mustafa AG, Tashtoush AA, Alshboul OA, Allouh MZ, Altarifi AA. Morphometric Study of the Hard Palate and Its Relevance to Dental and Forensic Sciences. Int J Dent. 2019;2019:1687345. doi: 10.1155/2019/1687345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswander JD. Laminographic x-ray studies in families with cleft lip and cleft palate. Arch Oral Biol. August 1968;13(8):1019–22. doi: 10.1016/0003-9969(68)90017-4 [DOI] [PubMed] [Google Scholar]
- Roosenboom J, Indencleef K, Hens G, Peeters H, Christensen K, Marazita ML, Claes P, Leslie EJ, Weinberg SM. Testing the face shape hypothesis in twins discordant for nonsyndromic orofacial clefting. Am J Med Genet A. November 2017;173(11):2886–2892. doi: 10.1002/ajmg.a.38471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager S. Morpho and Rvcg – Shape Analysis in R. In: Zheng G, Li S, Szekely G, eds. Statistical Shape and Deformation Analysis. Academic Press; 2017:217–256. [Google Scholar]
- Skrzat J, Holiat D, Walocha J. A morphometrical study of the human palatine sutures. Folia Morphol (Warsz). May 2003;62(2):123–7. [PubMed] [Google Scholar]
- Snead MP, Yates JR. Clinical and Molecular genetics of Stickler syndrome. J Med Genet. May 1999;36(5):353–9. [PMC free article] [PubMed] [Google Scholar]
- Utreja A, Evans CA. Marfan syndrome-an orthodontic perspective. Angle Orthod. March 2009;79(2):394–400. doi: 10.2319/112707-558.1 [DOI] [PubMed] [Google Scholar]
- Vergato LA, Doerfler RJ, Mooney MP, Siegel MI. Mouse palatal width growth rates as an “at risk” factor in the development of cleft palate induced by hypervitaminosis A. J Craniofac Genet Dev Biol. 1997 Oct-Dec 1997;17(4):204–10. [PubMed] [Google Scholar]
- Ward R, Moore E, Hartsfield J. Morphometric characteristics of subjects with oral facial clefts and their relatives. In: Wyszynski D, ed. Cleft lip and palate: From origin to treatment. Oxford University Press.; 2002:66–86. [Google Scholar]
- Watkins SE, Meyer RE, Strauss RP, Aylsworth AS. Classification, epidemiology, and genetics of orofacial clefts. Clin Plast Surg. April 2014;41(2):149–63. doi: 10.1016/j.cps.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Weinberg S Three-dimensional morphometric analysis of the craniofacial complex in the Unaffected relatives of individuals with nonsydnromic orofacial clefts. University of Pittsburgh; 2007. [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Maher BS, Marazita ML. Parental craniofacial morphology in cleft lip with or without cleft palate as determined by cephalometry: a meta-analysis. Orthod Craniofac Res. February 2006;9(1):18–30. doi: 10.1111/j.1601-6343.2006.00339.x [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Naidoo SD, Bardi KM, Brandon CA, Neiswanger K, Resick JM, Martin RA, Marazita ML. Face shape of unaffected parents with cleft affected offspring: combining three-dimensional surface imaging and geometric morphometrics. Orthod Craniofac Res. November 2009;12(4):271–81. doi: 10.1111/j.1601-6343.2009.01462.x [DOI] [PMC free article] [PubMed] [Google Scholar]


