Abstract
Purpose
To identify the disease-causing genes of Chinese Han women with idiopathic premature ovarian insufficiency (POI).
Methods
Seventy-four Chinese Han women with idiopathic POI were collected to analyze the genetic etiology. Triplet repeat-primed polymerase chain reaction (TP-PCR) was performed to screen the FMR1 (CGG)n premutation, and then 60 POI-related genes were sequenced by targeted next-generation sequencing (NGS) in POI patients with normal FMR1.
Results
A total of one patient (1/74) with FMR1 premutation was identified. Targeted NGS revealed that 15.07% (11/73) patients had pathogenic or likely pathogenic variants of Mendelian genes (FOXL2, EIF2B2, CYP17A1, CLPP, MCM9, GDF9, MSH5, ERCC6, POLG). Ten novel variants in six Mendelian genes were identified, such as CLPP c.355A>C (p.I119L) and c.688A>C (p.M230L), MCM9 c.1157C>T (p.T386M) and c.1291A>G (p.M431V), GDF9 c. 238C>T (p.Q80X), MSH5 c.604G>C (p.G202R) and c.2063T>C (p.I688T), ERCC6 c.C1769C>T (p.P590L), POLG c.2832G>C (p.E944D), and c.2821A>G (p.I941V).
Conclusion
This study suggested targeted NGS was an efficient etiologic test for idiopathic POI patients without FMR1 premutation and enriched the variant spectrum of POI-related genes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-021-02083-7.
Keywords: Premature ovarian insufficiency, POI, Next-generation sequencing, NGS, Variant
Introduction
Premature ovarian insufficiency (POI), also known as premature ovarian failure (POF), refers to women premature amenorrhea before the age of 40, elevated levels of follicle-stimulating hormone (FSH) and decreased levels of estrogen in serum, and different degrees of hypoestrogenic symptoms, such as hot flashes, sweating, sleeplessness, and decreased libido [1]. The etiology of POI is complex, and the etiological diagnosis of POI is unclear in most patients. It is generally believed that the causes include genetic, immune, infectious, and iatrogenic factors. Genetic factors have been considered as important pathogenic factors of POI, for 10–15% of patients with a family history [2].
Although there is no effective etiological treatment for POI at present, it is of great significance for patients and their family members to make a clear etiological diagnosis: (1) to clarify the etiology and relieve doubts; (2) in families with a clear genetic diagnosis, other members can make pre-symptomatic diagnosis, which is conducive to family planning; (3) as a reference indication for fertility preservation of delayed childbearing age. In addition, defining the patient’s genetic diagnosis is important for understanding the molecular biological basis of ovarian and follicular development and possible gene-targeted therapy in the future [3].
Ovarian development and physiology are exquisitely regulated by hundreds of genes and multiple signaling pathways, which results in low positive rate and limited diagnostic significance of variant screening for single gene. At present, the positive rate of single gene variant analysis is not only very low (usually 1 ~ 2% or less), but also cannot be repeated in different races and different studies [4–7]. Although chromosomal analysis and FMR1 premutation testing are recommended in all women with non-iatrogenic POI, autosomal genetic testing is not indicated in women with POI [1], for the genetic causes of POI are highly heterogeneous. Sanger sequencing is the gold standard for gene sequence analysis of monogenic diseases, but it is time consuming and laborious. Recent study suggested array comparative genomic hybridization (aCGH) or specific next-generation sequencing (NGS) panels should be considered to identify chromosomal deletions/duplications under karyotype resolution or other pathogenic variants in specific genes associated with POI [8]. However, the pathogenicity analysis of the copy number variants (CNVs) found in POI patients by arrays was difficult, and most of them had no definite conclusions because of lack of pedigree samples and small sample size [4, 9, 10]. NSG has overcome the shortcomings of Sanger sequencing and improved the genetic diagnostic rate and efficiency of monogenic disease with high genetic heterogeneity.
In this study, we used FMR1 triplet repeat-primed polymerase chain reaction (TP-PCR) to detect FMR1 (CGG)n premutation in POI patients, and then targeted NGS in POI patients with normal FMR1 (CGG)n repeats was performed to screen POI-related genes, in order to define genetic diagnosis of some patients.
Materials and methods
Patients
The study was approved by Institutional Review Board of the First Affiliated Hospital of Nanjing Medical University. Seventy-four Chinese Han POI patients were recruited in our study. All of the patients had at least 4 months of amenorrhea before the age of 40 years, high basal follicle stimulation hormone (FSH) levels (>25 IU/ml, twice, at least a month apart), and normal female karyotype with the resolution of 400–550 bands. Women having a background of anticancer treatment, ovarian or pelvic surgery, or abnormal karyotype were excluded from the study. All of the patients were referred to our reproductive center for fertility counselling. One hundred healthy fertile women with normal menstrual cycle (age ≥ 38 years old, AMH ≥ 0.5 ng/ml) were selected as controls. With informed consent, peripheral blood was taken, and genomic DNA was extracted using DNA extraction kit (Tiangen, China).
FMR1 (CGG)n–expanded mutation screening
We utilized commercial FMR1 triplet repeat-primed polymerase chain reaction (TP-PCR) reagents (Microread, China) to identify the FMR1 (CGG)n premutation in POI patients. The principle of the method is the same as previously reported [11]. Capillary electrophoresis was performed on Applied Biosystems 3730 DNA analyzer (Applied Biosystems, USA), and data were analyzed by GeneMapper software (Applied Biosystems, USA). Patients with normal FMR1 (CGG)n repeats were submitted to the following targeted NGS.
Targeted NGS and bioinformatics analysis
The PubMed and OMIM databases were used to search peer-reviewed publications using the following terms: “premature ovarian insufficiency,” “primary ovarian insufficiency,” “premature ovarian failure,” “ovarian dysgenesis,” and “hypergonadotropic hypogonadism.” Relevant articles in the English language, published until January 2017, were critically discussed. Sixty POI-related genes (see in Table 1), including known Mendelian genes and suspected candidate genes, were selected in the NGS panel. The suspected candidate genes indicate that animal or functional studies suggest that it is associated with ovarian dysfunction and supported by a small number of POI mutation screening studies, but lack of family studies and repeated validation. FMR1 was not included in the panel because FMR1 (CGG)n–expanded mutation could not be detected by NGS. Some POI-related genes, such as AARS2(OMIM: 612035) and PPM2(OMIM: 601785), which caused syndromic POI with severe nervous system phenotype were not selected as well, since these patients were referred to Neurology Department rather than Reproductive Medicine Department.
Table 1.
Targeted POI genes list
| Genes | No. OMIM | Syndromic/nonsyndromic | Mode of inheritance | OMIM phenotype |
|---|---|---|---|---|
| FSHR | 136435 | Nonsyndromic | AR | Ovarian dysgenesis 1 |
| PSMC3IP | 608665 | Nonsyndromic | AR | Ovarian dysgenesis 3 |
| MCM9 | 610098 | Nonsyndromic | AR | Ovarian dysgenesis 4 |
| SOHLH1 | 610224 | Nonsyndromic | AR | Ovarian dysgenesis 5 |
| DIAPH2 | 300108 | Nonsyndromic | XLD | Premature ovarian failure 2A? |
| POF1B | 300603 | Nonsyndromic | XLR | Premature ovarian failure 2B? |
| BMP15 | 300247 | Nonsyndromic | XLD |
Premature ovarian failure 4 Ovarian dysgenesis 2 |
| NOBOX | 610934 | Nonsyndromic | AD | Premature ovarian failure 5 |
| FIGLA | 608697 | Nonsyndromic | AD | Premature ovarian failure 6 |
| NR5A1 | 184757 | Nonsyndromic | AD | Premature ovarian failure 7 |
| STAG3 | 608489 | Nonsyndromic | AR | Premature ovarian failure 8 |
| HFM1 | 615684 | Nonsyndromic | AR | Premature ovarian failure 9 |
| MCM8 | 608187 | Nonsyndromic | AR | Premature ovarian failure 10 |
| ERCC6 | 609413 | Nonsyndromic | AD | Premature ovarian failure 11 |
| PGBD3 | N/A | Nonsyndromic | AD | Premature ovarian failure 11 |
| SYCE1 | 611486 | Nonsyndromic | AR | Premature ovarian failure 12 |
| MSH5 | 603382 | Nonsyndromic | AR | Premature ovarian failure 13 |
| GDF9 | 601918 | Nonsyndromic | AR/AD | Premature ovarian failure 14 |
| ADAMTS19 | 607513 | Nonsyndromic | AD? | / |
| AMHR2 | 600956 | Nonsyndromic | AD? | / |
| BMPR2 | 600799 | Nonsyndromic | AD ? | / |
| CDKN1B | 600778 | Nonsyndromic | AD ? | / |
| CITED2 | 602937 | Nonsyndromic | AD ? | / |
| COL4A6 | 303631 | Nonsyndromic | XL? | / |
| CPEB1 | 607342 | Nonsyndromic | AD ? | / |
| DACH2 | 300608 | Nonsyndromic | AD ? | / |
| DAZL | 601486 | Nonsyndromic | AR? | / |
| DMC1 | 602721 | Nonsyndromic | AR? | / |
| DNAH6 | 603336 | Nonsyndromic | AD ? | / |
| EIF4ENIF1 | 607445 | Nonsyndromic | AD | / |
| FOXO3 | 602681 | Nonsyndromic | AD ? | / |
| FOXO1 | 136533 | Nonsyndromic | AD ? | / |
| HS6ST1 | 604846 | Nonsyndromic | AD ? | / |
| INHA | 147380 | Nonsyndromic | AD ? | / |
| LHCGR | 152790 | Nonsyndromic | AR | / |
| NLRP5 | 609658 | Nonsyndromic | AD ? | / |
| NANOS3 | 608229 | Nonsyndromic | AD/AR | / |
| NXF5 | 300319 | Nonsyndromic | XL? | / |
| PGRMC1 | 300435 | Nonsyndromic | XL? | / |
| POU5F1 | 164177 | Nonsyndromic | AD ? | / |
| SALL4 | 607343 | Nonsyndromic | AD ? | / |
| SOHLH2 | 616066 | Nonsyndromic | AD ? | / |
| TGFBR3 | 600742 | Nonsyndromic | AD ? | / |
| XPNPEP2 | 300145 | Nonsyndromic | XL? | / |
| FOXL2 | 605597 | Syndromic | AD |
Blepharophimosis, epicanthus inversus, and ptosis, type 1 Premature ovarian failure 3 |
| CYP17A1 | 609300 | Syndromic | AR | 17-alpha-hydroxylase/17,20-lyase deficiency |
| POLG | 174763 | Syndromic | AD/AR | Progressive external ophthalmoplegia |
| AIRE | 607358 | Syndromic | AR | Autoimmune polyendocrinopathy syndrome, type I |
| BLM | 604610 | Syndromic | AR | Bloom syndrome |
| HSD17B4 | 601860 | Syndromic | AR | Perrault syndrome 1 |
| LARS2 | 604544 | Syndromic | AR | Perrault syndrome 2 |
| CLPP | 601119 | Syndromic | AR | Perrault syndrome 3 |
| HARS2 | 600783 | Syndromic | AR | Perrault syndrome 4 |
| C10orf2 | 606075 | Syndromic | AR | Perrault syndrome 5 |
| EIF2B2 | 606454 | Syndromic | AR | Ovarioleukodystrophy |
| EIF2B4 | 606687 | Syndromic | AR | Ovarioleukodystrophy |
| EIF2B5 | 603945 | Syndromic | AR | Ovarioleukodystrophy |
| LMNA | 150330 | Syndromic | AD | Malouf syndrome 1A |
| NOG | 602991 | Syndromic | AD | Symphalangism, proximal, 1A |
| WRN | 604611 | Syndromic | AR | Werner syndrome |
AD autosomal dominant inheritance, AR autosomal recessive inheritance, XLD X-linked dominant inheritance, XLR X-linked recessive inheritance; a question mark indicates that it is candidate gene, and the mode of inheritance is not sure
We used Ion AmpliSeq Designer (https://www.ampliseq.com/browse.action, supplied by ThermoFisher Scientific) to design the amplicon primers. A total of 1021 pairs amplicons in 2 pools were designed to coverage the coding regions of targeted genes; the amplicon range was 125 ~ 375 bp; the panel size was 288.97kb with 99.28% coverage. Detailed coverage information for each gene is outlined in Supplemental Table 1. The amplicon primers were synthesized by ThermoFisher Scientific. Sequencing library was prepared by Ion AmpliSeq Library Kit 2.0 (ThermoFisher Scientific, USA). Prepared library was sequenced on MiseqDX (Illumina, USA) by MiseqDx Universal Kit V3 SBS (Illumina, USA). Experimental operation was according to the standard protocol supplied by Illumina.
Raw data obtained from Illumina MiseqDX and FastQC (version 0.11.5, Babraham Institute, UK) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used for checking data quality. Low quality and adaptor contamination were filtered using Trimmomatic software (version 0.35, RWTH Aachen University, Germany) (http://www.usadellab.org/cms/?page=trimmomatic). The high-quality filtered data were further aligned with the human reference genome (hg19) using BWA software (version 0.7.15-rll40) (http://bio-bwa.sourceforge.net/), after which BAM files were obtained. Variants were identified using samtools software (version 1.5) (http://www.htslib.org/). The variants were then filtered on the basis of read depth and mapping quality using GATK software (version 3.8) (https://software.broadinstitute.org/gatk/. The filtered variations were annotated using annovar software (version 20170716) (http://annovar.openbioinformatics.org/en/latest/). Annotated variations were further reviewed manually. Variations that meet the following criteria will be retained: (1) The variant was annotated in the refGene or ensGene database to the exon region or splice site region; (2) The variant was non-synonymous; (3) If the variant is homozygous, the variant frequency is less than 0.05 in the 1000 Genomes database or ExAc database; (4) If the variant is heterozygous, the variant frequency is less than 0.001 in the 1000 Genomes or ExAc database.
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://siftdna.org/www/Extended_SIFT_chr_coords_submit.html) and Mutation Taster (http://www.mutationtaster.org/) bioinformatics tools were used for assessing potentially damaging effects. Variants classification of Mendelian genes was analyzed according to ACMG/AMP guidelines [12].
Sanger sequencing
PCR was used to amplify the exons from relevant patients and their family members to confirm potentially deleterious candidate variants obtained from targeted NGS assays, and novel mutations of Mendelian genes were furtherly screened in local controls by Sanger sequencing. The primers designed by Primer Premier 5.0 (Premier Biosoft, USA) were seen in Supplemental Table 2. PCR production was purified using shrimp alkaline phosphatase and exonuclease I, following the standard manufacturer’s instructions. Sanger sequencing was performed using a capillary sequencer (Applied Biosystems, USA). Sequencing data were analyzed using Lasergene software (DNAStar, USA) and Chromas software (Technelysium, AU).
Haplotype identification
When two suspected pathogenic variants of autosomal recessive Mendelian genes were found, and the parents’ samples of patients cannot be obtained, we used 10× genomics platform or nanopore sequencing to construct haplotype to confirm whether the two variants were compound heterozygous state.
High molecular weight (HMW) gDNA from blood was extracted and purified using MagAttract HMW Kit (Qiagen, Germany). Genome libraries were constructed using Chromium Genome HT Library & Gel Bead Kit v2 (10× Genomics, USA) according to the manufacturer’s instructions. Libraries sequencing was performed on Hiseq X Ten (Illumina, USA). Analysis of sequencing data used softwares Long Ranger and Loupe Genome Browser (10× Genomics, USA). The pipeline refers to the previously published literature [13].
DNA was extracted from blood using FlexiGene DNA Kit (Qiagen, Germany). Long-range PCR was performed to amplify the targeted genes using Ex Taq kit (TakaRa, Japan). Amplified DNA fragments were sequenced on Nanopore MinION system (Oxford Nanopore, UK). Library preparation and sequencing were carried out according to the manufacturer’s protocol. The raw sequencing output as FAST5 files were converted to FASTQ format using the MINKNOW. We used minimap2 to align the long reads to the human reference genome (hg19). Single nucleotide variants (SNVs) were called by Medaka. Medaka’s pipeline phased the recovered variants using Whatshap to distinguish paternal and maternal haplotypes. Then, medaka consensus was run for each haplotype to obtain improved variant calls. Finally, SNVs were merged and phased from each haplotype.
Results
Patients’ phenotypes
The clinical information of the POI patients with identified variants, including menarche years, amenorrhea years, reproductive history, height, weight, ultrasonography of ovaries and uterus, positive signs, special medical record, and positive family history, was listed in Table 2.
Table 2.
Clinical characteristics of POI patients with known or suspected pathogenic variants
| Sample | Gene | Age (y) | Menarche (y) | Amenorrhea (y) | Reproductive history | Height (cm) | Weight (kg) | Pelvic ultrasound | FSH (IU/L) | Positive signs/special medical record/positive family history | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ovaries | AFC | Uterus | ||||||||||
| FS0005 | SALL4 | 29 | 17 | 21 | No | NA | NA | Invisible | / | Normal | 48 ~ 53 | / |
| FS0011 | COL4A6 | 23 | 12 | 17 | No | NA | NA | Atrophy | 0 | Small | 32 ~ 42 | She had thyroid cancer at 20 years and accepted surgical treatment. |
| FS0018 | ERCC6 | 35 | 15 | 29 | AM:1 | 163 | 62.5 | Atrophy | 0 | Normal | 35 ~ 57 | / |
| FS0028 | DNAH6 | 25 | / | PA | No | 168 | 56 | Atrophy | 0 | Small | 22 ~ 33 | She had breast dysplasia, axillary, and pubic hair scarcity. |
| FS0032 | MSH5 | 39 | 13 | 35 | No | NA | NA | Atrophy | 0 | Normal | 14 ~ 25 | Her elder sister had similar manifestation. |
| FS0033 | DNAH6 | 33 | 13 | 36 | AM:1 | 170 | 55 | Atrophy | 0 | Normal | 20 ~ 58 | / |
| FS0048 | AMHR2 | 25 | 14 | 20 | No | 161 | 60 | Atrophy | 0 | Normal | 52 ~ 94 | She had tetramine poisoning at 3 years old. |
| FS0054 | MCM9 | 31 | 13 | 28 | No | 168 | 60 | Atrophy | 0 | Normal | 83 ~ 111 | / |
| FS0066 | DNAH6 | 29 | 14 | 26 | No | 159 | 60 | Atrophy | 0 | Small | 19 ~ 71 | / |
| FS0074 | DNAH6 | 27 | 11 | 21 | No | 168 | 63 | Atrophy | 0 | Small | 78 ~ 83 | / |
| FS0084 | POLG | 29 | 15 | 27 | No | 158 | 60 | Atrophy | 0 | Normal | 18 ~ 50 | / |
| FS0099 | GDF9 | 26 | 18 | 18 | No | 160 | 57.5 | Invisible | / | Small | 33 ~ 74 |
She had breast dysplasia, axillary and pubic hair scarcity; she was adopted, her biological parents were consanguineous marriage, and her two sisters had similar manifestation. |
| FS0100 | COL4A6 | 24 | 12 | 24 | No | 161 | 47.5 | Atrophy | 1-2 | Normal | 35 ~ 38 | / |
| FS0107 | CYP17A1 | 27 | / | PA | No | 170 | 58 | Invisible | / | PU | 80 ~ 100 | She had breast dysplasia, no axillary hair and pubic hair, pudendal immature type, hypokalemia, hypertension, increased progesterone and decreased androgen; one of her elder sisters had similar manifestation. |
| FS0117 | CLPP | 28 | / | PA | No | 166 | 60.5 | Atrophy | 0 | Small | 99 ~ 108 | She had breast dysplasia, axillary and pubic hair scarcity, fingers and toes malformation. |
| M2070 | FOXL2 | 28 | 15 | 28 | No | NA | NA | Normal | PLC | Normal | 16 ~ 26 | She had narrow palpebral fissure; intermittent ovarian cyst (φ > 30mm) was observed sometimes; controlled ovarian stimulation was performed but failed. AMH is 0.59 ng/ml. |
| FS0133 | SALL4 | 31 | 17 | 20 | No | 173 | 67.5 | Invisible | / | Small | 20 ~ 37 | She had breast dysplasia, axillary and pubic hair scarcity; her sister had irregular menstruation. |
| FS0134 | EIF2B2 | 34 | / | PA | No | 166 | 61 | Atrophy | 0 | Normal | 25 ~ 48 | Shen had breast dysplasia, axillary and pubic hair scarcity, and mild lesions on cranial MRI; she had cataract in both eyes and accepted surgical treatment at the age of 26; her parents were consanguineous marriage. |
| FS0140 | POLG | 28 | 13 | 26 | No | 158 | 51 | Atrophy | 0 | Normal | 58 ~ 59 | Her mother had amenorrhea at 40 years old. |
| FS0147 | FMR1 | 37 | 15 | 32 |
AM:2 SM:4 |
165 | 63 | Atrophy | 0 | Normal | 22 ~ 50 | Two of her uncles and one female cousin had mental retardation. |
| FS0158 | FOXL2 | 27 | 16 | 16 | No | 166 | 54 | Normal | PLC | Normal | 25 ~ 61 | She had narrow palpebral fissure. |
PA primary amenorrhea, AM artificial miscarriage, SM spontaneous miscarriage, AFC antral follicle count, PU primordial uterus, PLC polycystic ovary morphology (PCOM)–like changes
FMR1 (CGG)n–expanded mutation screening results
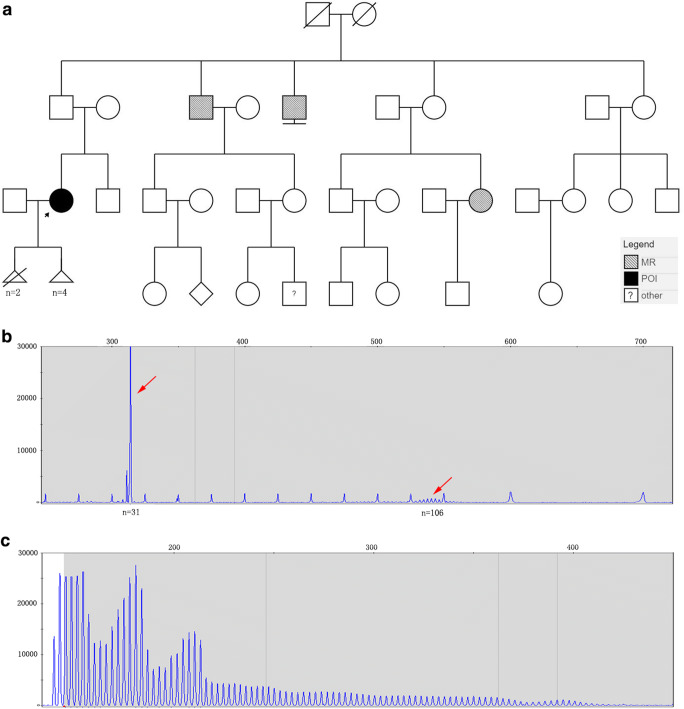
One patient with FMR1 (CGG)n premutation was identified in the 74 POI patients (see in Fig. 1), and the FMR1 (CGG)n repeats numbers of each patient were seen in Supplementary Table 3. The FMR1 (CGG)n repeats number of Sample FS0147 was 31/106, which belonged to premutation (55 ≤ n < 200). She had secondary amenorrhea and a family history of mental retardation (see in Table 2 and Fig. 1).
Fig. 1.
FS00147 pedigree and the FMR1 (CGG)n repeats number detection. a FS00147 pedigree. The black arrow indicates the proband (FS0147). POI refers to premature ovarian insufficiency, MR refers to mental retardation, and other refers to unknown phenotype. b Capillary electrophoresis of full-length amplified product of FMR1 (CGG) n. The red arrow indicates that the two (CGG) n fragments repeats number are 31 and 106. c Capillary electrophoresis of FMR1 (CGG) n repeats TP-PCR products, showing characteristic slope peaks
Targeted NGS results and in silico analysis
Targeted NGS revealed that 20 patients had known or suspected pathogenic variants (See in Table 3). Among these patients, a total of 11 patients had pathogenic or likely pathogenic variants of Mendelian genes (FOXL2, EIF2B2, CYP17A1, CLPP, MCM9, GDF9, MSH5, ERCC6, POLG) with a positive rate of 15.07% (11/73). Ten novel variants in six Mendelian genes were identified, such as CLPP c.355A>C (p.I119L) and c.688A>C (p.M230L), MCM9 c.1157C>T (p.T386M) and c.1291A>G (p.M431V), GDF9 c. 238C>T (p.Q80X), MSH5 c.604G>C (p.G202R) and c.2063T>C (p.I688T), ERCC6 c.C1769C>T(p.P590L), and POLG c.2832G>C (p.E944D) and c.2821A>G (p.I941V). Moreover, suspected pathogenic variants of POI candidate genes (DNAH6, SALL4, CLO4A6, AMHR2) were found in 9 patients (See in Table 3). The totally positive rate was 27.40% (20/73). All these variants were no recording or rare frequency (<0.0001) in the 1000 Genomes Database (http://www.internationalgenome.org/) and the Exome Aggregation Consortium (http://exac.broadinstitute.org/), and they were predicted to be detrimental. All the variants of known Mendelian genes were classified to be pathogenic or likely pathogenic according to ACMG/AMP guidelines (see in Table 3).
Table 3.
Candidate variants identified by targeted NGS and bioinformatics analysis
| Samples | Gene_RefSeq_muation | Het/Hom | Depth | Freq_1KG | Freq_ExAC | PoPh-2 | SIFT | MuTa | Origin | ACMG Classification |
|---|---|---|---|---|---|---|---|---|---|---|
| FS0005 | SALL4:NM_020436.4: c.1252C>T, (p.R418C) | het | 106:78 | / | 8.25E-06 | PRD | T | DC | NA | / |
| FS0011 | COL4A6:NM_001847.3: c.2852G>A, (p.G951E) | het | 296:298 | / | / | PRD | D | DC | NA | / |
| FS0018 | ERCC6:NM_000124.3: c.1769C>T, (p.P590L) | het | 580:329 | / | / | PRD | D | DC | Pat | PM1 PM2 PP3 PP4 (LP) |
| FS0028 | DNAH6:NM_001370.1: c.11689G>A, (p.V3897I) | het | 447:468 | / | 8.91E-05 | PRD | D | DC | NA | / |
| FS0032 | MSH5:NM_025259.5: c.604G>C, (p.G202R) | het | 256:309 | / | / | PRD | D | NA | Pat/Mat* | PM2 PM3 PP2 PP3 PP4 (LP) |
| MSH5:NM_025259.5: c.2063T>C, (p.I688T) | het | 745:779 | / | 1.68E-05 | POD | D | NA | Pat/Mat* | PM1 PM2 PP2 PP3 PP4 (LP) | |
| FS0033 | DNAH6:NM_001370.1: c.5045C>G, (p.P1682R) | het | 252:195 | / | / | PRD | D | DC | NA | / |
| FS0048 | AMHR2:NM_020547.3: c.56C>G, (p.P19R) | het | 302:268 | / | / | PRD | T | DC | NA | / |
| FS0054 | MCM9:NM_017696.2: c.1291A>G, (p.M431V) | het | 223:225 | / | / | PRD | D | DC | Mat | PM1 PM2 PM3 PP4 (LP) |
| MCM9:NM_017696.2: c.1157C>T, (p.T386M) | het | 126:121 | / | / | PRD | D | DC | Pat | PM1 PM2 PM3 PP4 (LP) | |
| FS0066 | DNAH6:NM_001370.1: c.8104A>G, (p.T2702A) | het | 501:536 | / | / | POD | D | DC | NA | / |
| FS0074 | DNAH6:NM_001370.1: c.10942A>G, (p.N3648D) | het | 79:67 | / | / | POD | T | DC | NA | / |
| FS0084 | POLG:NM_002693.2: c.2832G>C, (p.E944D) | het | 1102:1041 | / | / | PRD | T | DC | NA | PM1 PM2 PP3 PP4 (LP) |
| FS0099 | GDF9:NM_005260.5: c.238C>T, (p.Q80X) | hom | 3:818 | / | 8.0E-06 | NA | D | DC | NA | PVS1 PM2 PM3 PP3 PP4 (P) |
| FS0100 | COL4A6:NM_001847.3: c.2653G>A, (p.G885R) | het | 708:703 | / | / | PRD | D | DC | NA | / |
| FS0107 | CYP17A1:NM_000102.3: c.1459_1467del, (p.487_489del) | het | 328:321 | / | 1.291E-05 | NA | NA | DC | Mat | PS3 PM2 PM3 PM4 PP1 PP3 PP4 (P) |
| CYP17A1:NM_000102.3: c.985_987delinsAA, (p.Y329fs) | het | 259:271 | / | 4.947E-05 | NA | NA | DC | Pat | PVS1 PS3 PM2 PM3 PP1 PP3 PP4 (P) | |
| FS0117 | CLPP:NM_006012.2: c.355A>C, (p.I119L) | het | 301:248 | / | / | PRD | D | DC | Not Mat# | PM1 PM2 PP3 PP4 (LP) |
| CLPP:NM_006012.2: c.688A>C, (p.M230L) | het | 177:179 | / | / | BN | T | DC | Mat# | PM1 PM2 PM3 PP4 (LP) | |
| M2070 | FOXL2:NM_023067.3: c.273C>A, (p.Y91X) | het | 36:27 | / | 2.0E-05 | NA | D | DC | NA | PVS1 PS1 PM2 PM4 PM6 PP3 PP4 (P) |
| FS0133 | SALL4:NM_020436.4: c.541G>A, (p.V181M) | Het | 336:180 | / | 4.0E-04 | PRD | D | DC | Not Mat | / |
| FS0134 | EIF2B2:NM_014239.3: c.254T>A, (p.V85E) | Hom | 10:2319 | / | 8.276E-05 | PRD | D | DC | Mat&Pat | PS1 PS3 PM2 PP3 PP4 PP5 (P) |
| FS0140 | POLG:NM_002693.2: c.2821A>G, (p.I941V) | Het | 1145:1123 | / | / | PRD | D | DC | NA | PM1 PM2 PP3 PP4 (LP) |
| FS0158 | FOXL2:NM_023067.3: c.804dupC, (p.G269fs) | Het | 44:50 | / | / | NA | NA | DC | De novo | PVS1 PS1 PS2 PM2 PM4 PP3 PP4 (P) |
All the variants were validated by Sanger sequencing. Het heterozygosity, Hom homozygosity, Freq_1KG frequency data of East Asians from the 1000. Genomes Database (http://www.internationalgenome.org/); ExAC the Exome Aggregation Consortium (http://exac.broadinstitute.org/); “/” indicates no records. PoPh-2 PolyPhen-2 variant prediction software, PRD probable damaging, POD possibly damaging, BN benign. SIFT SIFT variant prediction software, T tolerated, D damaging. MuTa Mutation Taster variant prediction software, DC disease causing. Origin obtained by direct sequencing parents’ samples, Mat maternal origin, Pat paternal origin, NA not available. ACMG Classification standards and guidelines for the interpretation of sequence variants by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology, P pathogenic, LP likely pathogenic
*c.604G>C and c.2063T>C of MSH5 in FS0032 were compound heterozygous confirmed by 10× Genomics, predicted to be biparental origin
#c.355A>C and c.688A>C of CLPP in FS0117 were compound heterozygous confirmed by nanopore sequencing
Sanger sequencing of candidate variants and origin analysis
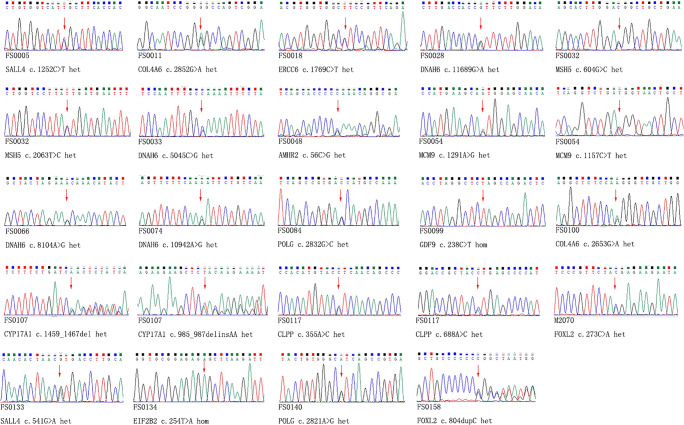
All the candidate variants identified by targeted NGS were validated by Sanger Sequencing, and the pedigree samples sequencing was performed if the samples were available (see in Fig. 2, Table 3 and Supplemental Figure 1 ~ 13). Each variant validation in their pedigrees and species conservation analysis were seen in Supplemental Figure 1 ~ 13, respectively. All of the novel variants of known Mendelian genes (CLPP c.355A>C (p.I119L) and c.688A>C (p.M230L), MCM9 c.1157C>T (p.T386M) and c.1291A>G (p.M431V), GDF9 c. 238C>T (p.Q80X), MSH5 c.604G>C (p.G202R) and c.2063T>C (p.I688T), ERCC6 c.C1769C>T (p.P590L), and POLG c.2832G>C (p.E944D) and c.2821A>G (p.I941V)) were not found in local 100 controls.
Fig. 2.
Sanger sequencing validation
In the cases with no available parents’ samples, MSH5 c.604G>C (p.G202R) and c.2063T>C (p.I688T) in patient FS0032 were compound heterozygous, confirmed by 10×genomics platform (See in Supplementary Table 4). Nanopore sequencing verified that CLPP c.355A>C (p.I119L) and c.688A>C (p.M230L) in sample FS0117 were compound heterozygous state (See in Supplementary Table 5).
Discussion
FMR1 premutation is the most commonly known pathogenic mutation to cause POI. The risk of POI in carriers with FMR1 premutation is 16 ~ 25% [14]. The positive rate of FMR1 premutation in Caucasian POI is about 3.3 ~ 6.7%, and the positive rate is only 0.5% in Chinese POI [15]. In this study, 74 POI patients were screened for FMR1 premutation before targeted NGS, and only one patient (FS0147) with FMR1 (CGG) n premutation (n = 31/106) was found. Several members with mental retardation were observed in this pedigree, which supported the feature of FMR1-related disorder. Therefore, the proportion of FMR1 premutation in Chinese Han POI patients in the present study was 1.35% (1/74), which was obviously lower than that in Caucasian POI patients as reported previously.
In 2007, NGS began to be used in molecular diagnosis of genetic diseases [16, 17]. It was characterized by fast turnaround time and high-throughput data, which has brought revolutionary impact on traditional molecular genetic testing and greatly accelerated the speed of molecular basis identification of monogenic diseases. Targeted NGS can rapidly screen multiple genes at the same time and has been applied to many diseases, such as hereditary non-syndromic deafness [18] and hypertrophic cardiomyopathy [19]. In 2015, Fonseca et al used targeted NGS to screen 70 POI-related genes in 12 POI patients, and found that there were suspected pathogenic variants in 3 patients [20]. In 2016, Bouilly et al. [21] performed NGS with a panel of 19 POI-related genes in 100 POI patients, and at least one rare protein-altered gene variant was found in 19 patients. Among them, seven patients carried two heterozygous pathogenic variants from different genes, suggesting that POI was not a purely monogenic disorder and points to a role of digenicity. Targeted NGS is more suitable for clinical application than exome sequencing, for its lower cost of library preparation and sequencing, better coverage of targeted genes, and simpler data analysis. In regarding to highly suspected hereditary POI patients, when targeted NGS result was negative, exome sequencing could be performed to identify novel disease-causing genes.
In our study, targeted NGS revealed that 11 patients had pathogenic or likely pathogenic variants of Mendelian POI genes (FOXL2, EIF2B2, CYP17A1, CLPP, MCM9, GDF9, MSH5, ERCC6, POLG) with a positive rate of 15.07% (11/73). In addition, suspected pathogenic variants of POI candidate genes (DNAH6, SALL4, CLO4A6, AMHR2) were found in 9 patients with a totally positive rate of 27.4% (20/73). Our results supported that NGS could be an efficient technical means to detect the molecular genetic causes of POI, and it can obtain a relatively high diagnostic rate in FMR1 mutation–negative cases, compared with the extremely low positive rate of previous single gene screening.
For the Mendelian POI genes, variants of c.273C>A (p.Y91X) and c.804dupC (p.G269fs) in FOXL2, c.254T>A (p.V85E) in EIF2B2, c.1459_1467del (p.487_489del), and c.985_987delinsAA (p.Y329fs) in CYP17A1 detected in this study had been reported previously [22–25]. Heterozygous variant of FOXL2 can result in blepharophimosis, ptosis, and epicanthus inversus (BPES), which can be classified into type I (with POI) and type II (without POI). It was interesting to note that pelvic ultrasonography suggested that both ovaries had polycystic ovary morphology (PCOM)–like changes in our two patients with FOXL2 variants. One of the patients (M2070) underwent twice cycles of controlled ovulation stimulations in our hospital, and the first cycle had no dominant follicle growth, and the second cycle had only one dominant follicle growing to mature size, but no eggs were obtained after repeatedly rinsed 14 times. Her AMH was 0.59 ng/ml and significantly low in regarding to her age. So, she does not have resistant ovarian syndrome (ROS). Furthermore, FSHR and LHCGR genes, known disease-causing genes of ROS, were included in our screening panel, and no variants were found. The formation mechanism of ovarian PCOM-like image in these patients remains unknown and needs to be further studied. Leukoencephalopathy with vanishing white matter (VWM) is an autosomal recessive hereditary disease, caused by mutation in five subunit coding genes of eukaryotic initiation factor 2B (EIF2B). The first symptoms of adult onset can be neurological symptoms (68.8%), mental and behavioral disorders (12.5%), and POI (12.5%) [26]. VWM has typical lesions of white matter, which is similar to cerebrospinal fluid (CSF)–like signals (low signal in T1W, high signal in T2W, reduced FLAIR signal) in cranial magnetic resonance imaging (MRI). Cranial MRI examination in our patients (FS0134) indicated multiple abnormal signals in bilateral craniocerebral coronal area, low signal in T1W, high signal in T2W, and FLAIR. There was no typical CSF-like low signal in FLAIR, which may be due to the early stage of the lesion. In this case, the pre-symptomatic diagnosis of the patient was confirmed by targeted NGS. It was of great significance to reduce and delay the risk of disease deterioration by suggesting to avoid inducing factors, such as infection, fever, slight head injury, childbirth, extreme fright, sunbathing, and epilepsy [26]. 17α-Hydroxylase deficiency (17OHD) is a rare autosomal recessive hereditary disease, which is caused by variant of CYP17A1 gene and accounting for less than 1% of congenital adrenal hyperplasia (CAH) [25, 27]. Typical symptoms of 17OHD were hypertension, hypokalemia, and sexual dysplasia. After definite genetic diagnosis, our patient (FS0107) was treated with dexamethasone, and abnormal blood pressure and blood potassium were quickly controlled.
Ten novel variants in six Mendelian genes were identified, such as CLPP c.355A>C (p.I119L) and c.688A>C (p.M230L); MCM9 c.1157C>T (p.T386M) and c.1291A>G (p.M431V); GDF9 c. 238C>T (p.Q80X), MSH5 c.604G>C (p.G202R), and c.2063T>C (p.I688T); and ERCC6 c.C1769C>T(p.P590L), POLG c.2832G>C (p.E944D), and c.2821A>G (p.I941V).
Caseinolytic mitochondrial matrix peptidase proteolytic subunit (CLPP) is a component of a mitochondrial ATP-dependent proteolytic complex [28]. CLPP gene variant leads to autosomal recessive hereditary type 3 Perrault syndrome (PRLTS3), which is characterized by sensorineural deafness and female POI with clinical and genetic heterogeneity [29]. Although most patients have bilateral sensorineural deafness, the severity of deafness can be moderate to severe deafness with or without language disorders. The age of onset of deafness reported in the literature varies from 10 months to 31 years old [29] and there are also cases of unilateral deafness reported [30]. Some patients have Marfan-like skeletal abnormalities as well [30]. In this study, the patient (FS0117) had two heterozygous variants of CLPP c.355A>C (p.I119L) and c.688A>C (p.M230L), both of which were novel variants and predicted to be likely pathogenic by bioinformatics analysis. Sanger sequencing validated that the variant of c.688A>C (p.M230L) was inherited from her mother with normal phenotype, and another variant c.355A>C (p.I119L) was not inherited from her mother. They were compound heterozygous mutations confirmed by nanopore sequencing, which was consistent with autosomal recessive heritance. There was a reported pathogenic variant p.Y229D, closely nearby p.M230L, and functional analysis demonstrated that protease activity was significantly reduced [31]. In our case, there was no obvious bilateral deafness, probably due to phenotypic variation or not yet reaching the age of onset. In addition, the patient had abnormal skeletal changes of the index fingers of both hands and the fourth toes of both feet, which were not reported before (See in supplementary figure 4).
Minichromosome maintenance complex component 9 (MCM9) gene encodes a minichromosome maintenance protein consisting of 1141 amino acids, which is related to homologous recombination of chromosomes and repair of double-stranded DNA breaks. Wood-Trageser MA et al. firstly identified homozygous variants of MCM9 (c.1732+2T>C and c.394C>T (p.Arg132X), respectively) as POI pathogenic gene in two kindreds of Turkish Kurdish families by homozygosity mapping and exome sequencing [32], and the patients presented with primary amenorrhea and short stature. The meiosis of germ cells and somatic DNA repair were more sensitive to MCM9 deficiency, which may be the main cause of POI and short stature in female patients [32]. Fauchereau F et al. [33] reported that two POI sisters in an Arab family had a homozygous pathogenic variant of MCM9 c.1483G>T(p.E495X), and the sisters were primary amenorrhea with normal height, suggesting that short stature was not the characteristic manifestation of MCM9 variant. The patient (FS0054) found in this study had compound heterozygous variants of MCM9 c. 1157C>T (p. T386M) and c.1291A>G (p. M431V), which were inherited from parents with normal phenotype respectively and predicted to be likely pathogenic variants by bioinformatics analysis. Unlike the phenotypes reported in previous studies, the patient in our study was secondary amenorrhea with normal height.
Growth differentiation factor 9 (GDF9) belongs to the superfamily of TGF-β, and it plays an important role in ovarian and follicular development [34]. GDF9 knockout female mice (−/−) were infertility, while GDF9 (−/−) male mice and GDF9 (+/−) female mice had fertility and normal phenotype [35, 36]. In humans, previously several GDF9 heterozygous variants had been reported to be associated with POI, such as c.199A>C and c.646G>A[37]; c.557C>A[38] and c.307C>T[39], and the patients generally presented with secondary amenorrhea. In 2018, Franca MM et al. firstly reported a homozygous variant of GDF9 c.783delC (p.S262Hfs*2) in a Brazilian POI woman with primary amenorrhea. Her parents were consanguineous marriage, and her mother was carrier of heterozygous variant with normal phenotype. All of above suggest that POI caused by GDF9 variant may be autosomal dominant inheritance or autosomal recessive inheritance. In our study, we reported that a POI patient (FS0099) with secondary amenorrhea had a homozygous variant of GDF9 c.238C>T (p.Q80X), which resulted in protein truncation and loss of TGF-β-like domain (located at 316 to 454 amino acids) in GDF9 protein. Our case suggested again that POI caused by GDF9 variant could be autosomal recessive inheritance. The proband’s biological parents were consanguineous marriage, and her two sisters had similar phenotype.
MutS homologue 5 (MSH5) is a member of MutS family, which is related to DNA mismatch repair (MMR). MSH4-MSH5 dimer plays an important role in homologous recombination repair of DNA double strand breaks (DSBs). The paternal heterozygous variant of MSH5 p.P29S in two sisters with POI had been reported to be associated with POI [40]. Guo et al. reported that the homozygous variant of MSH5 c.1459G>T (p.D487Y) in two POI sisters with secondary amenorrhea and the variant could cause ovarian atrophy and no oocyte development in mice [41]. Genotype-phenotype relationship in the family was co-segregation, and carriers of heterozygous variant had no abnormal phenotype, suggesting POI caused by MSH5 was autosomal recessive inheritance. In a POI family (FS0032) reported in this study, two of the four sisters in the family suffered from POI, presenting as secondary amenorrhea. Both sisters were found to have heterozygous variants of MSH5 c.604G>C (p.G202R) and c.2063T>C (p.I688T), both of which were novel and predicted to be likely pathogenic variants by bioinformatics analysis. Although parents’ samples were not available, 10× Genomics detection was performed to confirm the two variants were trans, consistent with autosomal recessive model.
Excision repair cross-complementing, group 6 (ERCC6) is part of the nucleotide excision repair (NER) pathway, a complex system that eliminates a broad spectrum of structural DNA lesions [42]. In a Chinese family in which 4 women over 2 generations experienced secondary amenorrhea, Qin et al. [43] performed exome sequencing and identified a heterozygous variant of ERCC6 p.G746D that segregated with disease in the family. Analysis of ERCC6 in 432 sporadic Chinese POI patients revealed 2 women with heterozygous variants in ERCC6: a nonsense variant (p.E215X) and a missense variant (p.V1056I). Authors noted that POI had not been reported in any of the families of patients with Cockayne syndrome (CSB), which is an autosomal recessive hereditary disease caused by ERCC6. The authors proposed that the novel sporadic variants may act in a dominant-negative fashion. In this study, we found a POI patient (FS0018) with a heterozygous variant of ERCC6 c. C1769C>T(p.P590L). The novel variant originated from her father with normal phenotype, and did not observed in her mother and sister with normal phenotype. Genotype-phenotype relationship was co-segregated in the family, which supported the female-limited autosomal dominant inheritance model.
The DNA polymerase gamma (POLG) protein is composed of a C-terminal polymerase (pol) domain and an amino-terminal exonuclease (exo) domain. The exo domain increases the fidelity of mitochondrial DNA replication by conferring a proofreading activity to the enzyme [44]. POLG is the only enzyme responsible for mitochondrial 16.6 KB DNA replication. It consists of a catalytically active POLG subunit and its affiliated subunit POLG2. POLG variant can lead to a variety of mitochondrial-related diseases, such as progressive external ophthalmoplegia (PEO), Alpers syndrome, and ataxia [45]. Mature oocytes have the highest content of mitochondrial DNA in human cells [46]. The content of mitochondrial DNA in human oocytes increases, and the fertilization rate and embryo formation rate increase [47]. Therefore, the maturation of oocytes may be more sensitive to the defect of POLG activity. Previous literature reported that POI caused by POLG variants were syndromic POI and accompanied by PEO [48, 49]. In this study, two novel heterozygous variants of POLG were found in two (FS0084 and FS0140) of 73 POI patients, respectively. They were c.2832G>C(p.E944D) and c.2821A>G(p.I941V), which were all located in the POLG protein polymerase domain (pol) and speculated to be likely pathogenic by bioinformatics analysis. It was worth noting that PEO did not occur in both patients. The possible reasons were that our patients are 28 ~ 29 years old, not reaching the age of onset of PEO, which varies from 10 to 54 years old according to the literature reported [49]; it may also be caused by different genotype and genetic backgrounds.
In addition to the above variants in known Mendelian genes, we also found that 9 patients had heterozygous missense variants in 4 POI candidate genes, such as c.11689G>A (p.V3897I), c.5045C>G (p.P1682R), c.8104A>G (p.T2702A), and c.10942A > G (p.N3648D) in DNAH6; c.1252C>T (p.R418C) and c.541G>A (p.V181M) in SALL4; c.2852G>A (p.G951E) and c.2653G>A (p.G885R) in COL4A6; and c.56C>G (p.P19R) in AMHR2. These genes were only reported in functional studies and individual case reports [50–54]. Further studies are needed to prove whether they cause Mendelian hereditary POI.
The disadvantages of the current study are listed below: (1) Amplicon capture did not cover 100% of the coding sequence of the targeted genes, although the gap was extremely small; (2) Some newly discovered POI-related genes were not included in the targeted panel, such as FANCM (POF15) [55], ERAL1 (PRLTS6) [56], NUP107 (OD6) [57], MRPS22 (OD7) [58], XRCC2 [59], BNC1 [60], and BRCA2 [61]; (3) The novel suspected pathogenic variants found in this study lack functional studies to further confirm their pathogenicity; (4) Some of the pedigree’s samples were not available for genotype-phenotype co-segregation study.
In conclusion, our study showed a relatively high diagnostic rate in FMR1 mutation–negative cases, and indicated targeted NGS was an efficient etiological test for POI patients without FMR1 premutation. Meanwhile, this study enriched the variant spectrum of POI-related genes.
Supplementary information
(DOCX 35 kb)
(DOCX 15285 kb)
Author contribution
JS, JL, and FD conceived and designed the study. JS, YG, and XM collected samples. DY, FS, JX, and XS performed the experiments. JS, DW, and YC analyzed the data. JS wrote the manuscript. JL and FD revised the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFC1000200, 2017YFC1001602) and the National Natural Science Foundation of China (81471429, 81730041) and Natural Science Foundation of Jiangsu Province (BK20201488).
Data availability
The data that supports the findings of this study are available in the supplementary material of this article.
Code availability
The version of each software applications was indicated in the text.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiayin Liu, Email: jyliu_nj@126.com.
Feiyang Diao, Email: phenix_y@163.com.
References
- 1.POI. tESoHRaEEGGo. Webber L, Davies M, Anderson R, Bartlett J, Braat D, et al. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31(5):926–937. doi: 10.1093/humrep/dew027. [DOI] [PubMed] [Google Scholar]
- 2.De Vos M, Devroey P, Fauser BC. Primary ovarian insufficiency. Lancet. 2010;376(9744):911–921. doi: 10.1016/S0140-6736(10)60355-8. [DOI] [PubMed] [Google Scholar]
- 3.Atabiekov I, Hobeika E, Sheikh U, El Andaloussi A, Al-Hendy A. The role of gene therapy in premature ovarian insufficiency management. Biomedicines. 2018;6(4). 10.3390/biomedicines6040102. [DOI] [PMC free article] [PubMed]
- 4.Qin Y, Jiao X, Simpson JL, Chen ZJ. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21(6):787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen N, Fedewa S, Chen AY. Epidemiology and demographics of the head and neck cancer population. Oral Maxillofac Surg Clin North Am. 2018;30(4):381–395. doi: 10.1016/j.coms.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Jiao X, Ke H, Qin Y, Chen ZJ. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol Metab. 2018;29(11):795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Jiao X, Zhang H, Ke H, Zhang J, Cheng L, Liu Y, Qin Y, Chen ZJ. Premature ovarian insufficiency: phenotypic characterization within different etiologies. J Clin Endocrinol Metab. 2017;102(7):2281–2290. doi: 10.1210/jc.2016-3960. [DOI] [PubMed] [Google Scholar]
- 8.Barros F, Carvalho F, Barros A, Dória S. Premature ovarian insufficiency: clinical orientations for genetic testing and genetic counseling. Porto Biomed J. 2020;5(3):e62. doi: 10.1097/j.pbj.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao C, Fu F, Yang X, Sun YM, Li DZ. Analysis of Chinese women with primary ovarian insufficiency by high resolution array-comparative genomic hybridization. Chin Med J. 2011;124(11):1739–1742. [PubMed] [Google Scholar]
- 10.Bestetti I, Castronovo C, Sironi A, Caslini C, Sala C, Rossetti R, Crippa M, Ferrari I, Pistocchi A, Toniolo D, Persani L, Marozzi A, Finelli P. High-resolution array-CGH analysis on 46,XX patients affected by early onset primary ovarian insufficiency discloses new genes involved in ovarian function. Hum Reprod. 2019;34(3):574–583. doi: 10.1093/humrep/dey389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyon E, Laver T, Yu P, Jama M, Young K, Zoccoli M, Marlowe N. A simple, high-throughput assay for Fragile X expanded alleles using triple repeat primed PCR and capillary electrophoresis. J Mol Diagn. 2010;12(4):505–511. doi: 10.2353/jmoldx.2010.090229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34(3):303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan AK, Marcus M, Epstein MP, Allen EG, Anido AE, Paquin JJ, Yadav-Shah M, Sherman SL. Association of FMR1 repeat size with ovarian dysfunction. Hum Reprod. 2005;20(2):402–412. doi: 10.1093/humrep/deh635. [DOI] [PubMed] [Google Scholar]
- 15.Guo T, Qin Y, Jiao X, Li G, Simpson JL, Chen ZJ. FMR1 premutation is an uncommon explanation for premature ovarian failure in Han Chinese. PLoS One. 2014;9(7):e103316. doi: 10.1371/journal.pone.0103316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39(12):1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 17.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 18.Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Huang J, Zhao J, Chen C, Wang H, Ding H, Wang D, Wang D. Rapid molecular genetic diagnosis of hypertrophic cardiomyopathy by semiconductor sequencing. J Transl Med. 2014;12:173. doi: 10.1186/1479-5876-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonseca DJ, Patino LC, Suarez YC, de Jesus Rodriguez A, Mateus HE, Jimenez KM, et al. Next generation sequencing in women affected by nonsyndromic premature ovarian failure displays new potential causative genes and mutations. Fertil Steril. 2015;104(1):154–162. doi: 10.1016/j.fertnstert.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Bouilly J, Beau I, Barraud S, Bernard V, Azibi K, Fagart J, Fèvre A, Todeschini AL, Veitia RA, Beldjord C, Delemer B, Dodé C, Young J, Binart N. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J Clin Endocrinol Metab. 2016;101(12):4541–4550. doi: 10.1210/jc.2016-2152. [DOI] [PubMed] [Google Scholar]
- 22.Beysen D, De Paepe A, De Baere E. FOXL2 mutations and genomic rearrangements in BPES. Hum Mutat. 2009;30(2):158–169. doi: 10.1002/humu.20807. [DOI] [PubMed] [Google Scholar]
- 23.Nallathambi J, Neethirajan G, Usha K, Jitendra J, De Baere E, Sundaresan P. FOXL2 mutations in Indian families with blepharophimosis-ptosis-epicanthus inversus syndrome. J Genet. 2007;86(2):165–168. doi: 10.1007/s12041-007-0021-z. [DOI] [PubMed] [Google Scholar]
- 24.Matsukawa T, Wang X, Liu R, Wortham NC, Onuki Y, Kubota A, Hida A, Kowa H, Fukuda Y, Ishiura H, Mitsui J, Takahashi Y, Aoki S, Takizawa S, Shimizu J, Goto J, Proud CG, Tsuji S. Adult-onset leukoencephalopathies with vanishing white matter with novel missense mutations in EIF2B2, EIF2B3, and EIF2B5. Neurogenetics. 2011;12(3):259–261. doi: 10.1007/s10048-011-0284-7. [DOI] [PubMed] [Google Scholar]
- 25.Marsh CA, Auchus RJ. Fertility in patients with genetic deficiencies of cytochrome P450c17 (CYP17A1): combined 17-hydroxylase/17,20-lyase deficiency and isolated 17,20-lyase deficiency. Fertil Steril. 2014;101(2):317–322. doi: 10.1016/j.fertnstert.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Labauge P, Horzinski L, Ayrignac X, Blanc P, Vukusic S, Rodriguez D, Mauguiere F, Peter L, Goizet C, Bouhour F, Denier C, Confavreux C, Obadia M, Blanc F, Seze J, Fogli A, Boespflug-Tanguy O. Natural history of adult-onset eIF2B-related disorders: a multi-centric survey of 16 cases. Brain. 2009;132(Pt 8):2161–2169. doi: 10.1093/brain/awp171. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho LC, Brito VN, Martin RM, Zamboni AM, Gomes LG, Inacio M, et al. Clinical, hormonal, ovarian, and genetic aspects of 46,XX patients with congenital adrenal hyperplasia due to CYP17A1 defects. Fertil Steril. 2016;105(6):1612–1619. doi: 10.1016/j.fertnstert.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Jenkinson EM, Rehman AU, Walsh T, Clayton-Smith J, Lee K, Morell RJ, Drummond MC, Khan SN, Naeem MA, Rauf B, Billington N, Schultz JM, Urquhart JE, Lee MK, Berry A, Hanley NA, Mehta S, Cilliers D, Clayton PE, Kingston H, Smith MJ, Warner TT, University of Washington Center for Mendelian Genomics. Black GC, Trump D, Davis JR, Ahmad W, Leal SM, Riazuddin S, King MC, Friedman TB, Newman WG. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet. 2013;92(4):605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marlin S, Lacombe D, Jonard L, Leboulanger N, Bonneau D, Goizet C, Billette de Villemeur T, Cabrol S, Houang M, Moatti L, Feldmann D, Denoyelle F. Perrault syndrome: report of four new cases, review and exclusion of candidate genes. Am J Med Genet A. 2008;146A(5):661–664. doi: 10.1002/ajmg.a.32180. [DOI] [PubMed] [Google Scholar]
- 30.Kim MJ, Kim SJ, Kim J, Chae H, Kim M, Kim Y. Genotype and phenotype heterogeneity in perrault syndrome. J Pediatr Adolesc Gynecol. 2013;26(1):e25–e27. doi: 10.1016/j.jpag.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Brodie EJ, Zhan H, Saiyed T, Truscott KN, Dougan DA. Perrault syndrome type 3 caused by diverse molecular defects in CLPP. Sci Rep. 2018;8(1):12862. doi: 10.1038/s41598-018-30311-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood-Trageser MA, Gurbuz F, Yatsenko SA, Jeffries EP, Kotan LD, Surti U, Ketterer DM, Matic J, Chipkin J, Jiang H, Trakselis MA, Topaloglu AK, Rajkovic A. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet. 2014;95(6):754–762. doi: 10.1016/j.ajhg.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fauchereau F, Shalev S, Chervinsky E, Beck-Fruchter R, Legois B, Fellous M, Caburet S, Veitia RA. A non-sense MCM9 mutation in a familial case of primary ovarian insufficiency. Clin Genet. 2016;89(5):603–607. doi: 10.1111/cge.12736. [DOI] [PubMed] [Google Scholar]
- 34.Elvin JA, Yan C, Matzuk MM. Oocyte-expressed TGF-beta superfamily members in female fertility. Mol Cell Endocrinol. 2000;159(1-2):1–5. doi: 10.1016/S0303-7207(99)00185-9. [DOI] [PubMed] [Google Scholar]
- 35.Carabatsos MJ, Elvin J, Matzuk MM, Albertini DF. Characterization of oocyte and follicle development in growth differentiation factor-9-deficient mice. Dev Biol. 1998;204(2):373–384. doi: 10.1006/dbio.1998.9087. [DOI] [PubMed] [Google Scholar]
- 36.Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383(6600):531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- 37.Dixit H, Rao LK, Padmalatha V, Kanakavalli M, Deenadayal M, Gupta N, Chakravarty B, Singh L. Mutational screening of the coding region of growth differentiation factor 9 gene in Indian women with ovarian failure. Menopause. 2005;12(6):749–754. doi: 10.1097/01.gme.0000184424.96437.7a. [DOI] [PubMed] [Google Scholar]
- 38.Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, Bourcigaux N, Jacquesson L, Bouchard P, Frydman R, Dewailly D, Reyss AĆ, Jeffery L, Bachelot A, Massin N, Fellous M, Veitia RA. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;154(5):739–744. doi: 10.1530/eje.1.02135. [DOI] [PubMed] [Google Scholar]
- 39.Kovanci E, Rohozinski J, Simpson JL, Heard MJ, Bishop CE, Carson SA. Growth differentiating factor-9 mutations may be associated with premature ovarian failure. Fertil Steril. 2007;87(1):143–146. doi: 10.1016/j.fertnstert.2006.05.079. [DOI] [PubMed] [Google Scholar]
- 40.Mandon-Pepin B, Touraine P, Kuttenn F, Derbois C, Rouxel A, Matsuda F, et al. Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur J Endocrinol. 2008;158(1):107–115. doi: 10.1530/EJE-07-0400. [DOI] [PubMed] [Google Scholar]
- 41.Guo T, Zhao S, Zhao S, Chen M, Li G, Jiao X, Wang Z, Zhao Y, Qin Y, Gao F, Chen ZJ. Mutations in MSH5 in primary ovarian insufficiency. Hum Mol Genet. 2017;26(8):1452–1457. doi: 10.1093/hmg/ddx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71(6):939–953. doi: 10.1016/0092-8674(92)90390-X. [DOI] [PubMed] [Google Scholar]
- 43.Qin Y, Guo T, Li G, Tang TS, Zhao S, Jiao X, Gong J, Gao F, Guo C, Simpson JL, Chen ZJ. CSB-PGBD3 Mutations cause premature ovarian failure. PLoS Genet. 2015;11(7):e1005419. doi: 10.1371/journal.pgen.1005419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi GP, Zeviani M. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol. 2002;52(2):211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- 45.Cohen BH, Naviaux RK. The clinical diagnosis of POLG disease and other mitochondrial DNA depletion disorders. Methods. 2010;51(4):364–373. doi: 10.1016/j.ymeth.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 46.Steuerwald N, Barritt JA, Adler R, Malter H, Schimmel T, Cohen J, Brenner CA. Quantification of mtDNA in single oocytes, polar bodies and subcellular components by real-time rapid cycle fluorescence monitored PCR. Zygote. 2000;8(3):209–215. doi: 10.1017/S0967199400001003. [DOI] [PubMed] [Google Scholar]
- 47.Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2006;85(3):584–591. doi: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 48.Pagnamenta AT, Taanman JW, Wilson CJ, Anderson NE, Marotta R, Duncan AJ, Glindzicz MB, Taylor RW, Laskowski A, Thorburn DR, Rahman S. Dominant inheritance of premature ovarian failure associated with mutant mitochondrial DNA polymerase gamma. Hum Reprod. 2006;21(10):2467–2473. doi: 10.1093/humrep/del076. [DOI] [PubMed] [Google Scholar]
- 49.Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors A, Rautakorpi I, Peltonen L, Majamaa K, Somer H, Suomalainen A. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364(9437):875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 50.Ledig S, Ropke A, Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev. 2010;4(4-5):225–232. doi: 10.1159/000314958. [DOI] [PubMed] [Google Scholar]
- 51.Norling A, Hirschberg AL, Rodriguez-Wallberg KA, Iwarsson E, Wedell A, Barbaro M. Identification of a duplication within the GDF9 gene and novel candidate genes for primary ovarian insufficiency (POI) by a customized high-resolution array comparative genomic hybridization platform. Hum Reprod. 2014;29(8):1818–1827. doi: 10.1093/humrep/deu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B, Li L, Ni F, Song J, Wang J, Mu Y, Ma X, Cao Y. Mutational analysis of SAL-Like 4 (SALL4) in Han Chinese women with premature ovarian failure. Mol Hum Reprod. 2009;15(9):557–562. doi: 10.1093/molehr/gap046. [DOI] [PubMed] [Google Scholar]
- 53.Nishimura-Tadaki A, Wada T, Bano G, Gough K, Warner J, Kosho T, Ando N, Hamanoue H, Sakakibara H, Nishimura G, Tsurusaki Y, Doi H, Miyake N, Wakui K, Saitsu H, Fukushima Y, Hirahara F, Matsumoto N. Breakpoint determination of X;autosome balanced translocations in four patients with premature ovarian failure. J Hum Genet. 2011;56(2):156–160. doi: 10.1038/jhg.2010.155. [DOI] [PubMed] [Google Scholar]
- 54.Qin C, Yuan Z, Yao J, Zhu W, Wu W, Xie J. AMH and AMHR2 genetic variants in Chinese women with primary ovarian insufficiency and normal age at natural menopause. Reprod BioMed Online. 2014;29(3):311–318. doi: 10.1016/j.rbmo.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Fouquet B, Pawlikowska P, Caburet S, Guigon C, Makinen M, Tanner L, et al. A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency. Elife. 2017;6. 10.7554/eLife.30490. [DOI] [PMC free article] [PubMed]
- 56.Demain LA, Urquhart JE, O’Sullivan J, Williams SG, Bhaskar SS, Jenkinson EM, et al. Expanding the genotypic spectrum of Perrault syndrome. Clin Genet. 2017;91(2):302–312. doi: 10.1111/cge.12776. [DOI] [PubMed] [Google Scholar]
- 57.Weinberg-Shukron A, Renbaum P, Kalifa R, Zeligson S, Ben-Neriah Z, Dreifuss A, Abu-Rayyan A, Maatuk N, Fardian N, Rekler D, Kanaan M, Samson AO, Levy-Lahad E, Gerlitz O, Zangen D. A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J Clin Invest. 2015;125(11):4295–4304. doi: 10.1172/JCI83553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen A, Tiosano D, Guran T, Baris HN, Bayram Y, Mory A, Shapiro-Kulnane L, Hodges CA, Akdemir ZC, Turan S, Jhangiani SN, van den Akker F, Hoppel CL, Salz HK, Lupski JR, Buchner DA. Mutations in the mitochondrial ribosomal protein MRPS22 lead to primary ovarian insufficiency. Hum Mol Genet. 2018;27(11):1913–1926. doi: 10.1093/hmg/ddy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang YX, Li HY, He WB, Tu C, Du J, Li W, et al. XRCC2 mutation causes premature ovarian insufficiency as well as non-obstructive azoospermia in humans. Clin Genet. 2018;95:442–443. doi: 10.1111/cge.13475. [DOI] [PubMed] [Google Scholar]
- 60.Zhang D, Liu Y, Zhang Z, Lv P, Liu Y, Li J, Wu Y, Zhang R, Huang Y, Xu G, Qian Y, Qian Y, Chen S, Xu C, Shen J, Zhu L, Chen K, Zhu B, Ye X, Mao Y, Bo X, Zhou C, Wang T, Chen D, Yang W, Tan Y, Song Y, Zhou D, Sheng J, Gao H, Zhu Y, Li M, Wu L, He L, Huang H. Basonuclin 1 deficiency is a cause of primary ovarian insufficiency. Hum Mol Genet. 2018;27(21):3787–3800. doi: 10.1093/hmg/ddy261. [DOI] [PubMed] [Google Scholar]
- 61.Qin Y, Zhang F, Chen ZJ. BRCA2 in Ovarian Development and Function. N Engl J Med. 2019;380(11):1086–1087. doi: 10.1056/NEJMc1813800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 35 kb)
(DOCX 15285 kb)
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.
The version of each software applications was indicated in the text.