Abstract
Background
Human apurinic/apyrimidinic (AP) endonuclease 1 (APE1) plays a critical role in DNA base excision repair (BER) pathway and has been reported to be overexpressed in multiple cancers. Previously, we have shown that histone chaperone FACT complex (Facilitates Chromatin Transcription, a heterodimer of SSRP1 and SPT16 proteins) facilitates the chromatin access and DNA repair function of APE1, and their expression levels are correlated with promoting drug resistance in cancer. FACT inhibitor has been introduced in phase I and II clinical trials for chemosensitization of advanced solid cancers. However, the expression profile and prognostic significance of APE1 and FACT complex in bladder cancer remains largely unknown.
Methods
Retrospectively, 69 bladder cancer samples were retrieved and submitted for immunohistochemical staining of APE1 and SSRP1. Expression profile including cytoplasmic and nuclear staining of APE1 and expression level of SSRP1 was examined and semi-quantified to render a H-score. The prognostic significance of APE1 and SSRP1 was evaluated by Kaplan-Meier survival analysis in our cohort and R2 database.
Results
APE1 expression is elevated in bladder cancer compared to normal adjacent tissues. Compared with low grade tumors, high grade tumors show a shift in the staining pattern including higher intensity and positive cytoplasmic staining. Carcinoma in situ has a similar staining pattern to high grade tumors. APE1 and SSRP1 staining intensity increases as tumor progresses with stage. There is a correlation between APE1 and SSRP1 staining in invasive bladder cancer (Spearman r = 0.5466, p < 0.0001). The increased expression of APE1 and SSRP1 is associated with poor survival in Kaplan-Meier analysis in our cohort and in R2-TCGA bladder cancer database.
Conclusions
The expression levels of APE1 and SSRP1 are significantly elevated in bladder cancer as compared to normal adjacent tissues. APE1 correlates with SSRP1 expression in high grade tumors. Overexpression of APE1 and SSRP1 is associated with poor survival in bladder cancer. This suggests the usage of FACT inhibitor curaxins in muscle invasive bladder cancer to target FACT complex and APE1 to improve chemosensitization after further validation.
Keywords: APE1, SSRP1, Bladder cancer
APE1, SSRP1, bladder cancer
1. Introduction
Bladder cancer is the most common cancer of the urinary tract with approximately 430,000 new cases and 165,000 deaths per year worldwide [1]. Urothelial carcinoma of the bladder comprises two long-recognized disease entities with distinct molecular features and clinical outcome [2]. Low-grade, non-muscle-invasive bladder cancer (NMIBC, pTa/pT1 stage) recurs frequently and requires long-term surveillance, whereas muscle invasive bladder cancer (MIBC, pT2-pT4a stage) tends to metastasize and usually requires the combination of surgical resection and chemoradiotherapy. Chemotherapy is associated with significant toxicity and complications especially in frail patients. Understanding the mechanism of chemoresistance and searching for chemosensitizing agent remains an unmet clinical need.
The hallmark of high-grade bladder cancer is genome instability and defective DNA repair [2]. DNA repair mechanisms appear to play out both in the development and chemoresistance of bladder cancer. In this project, we focus on human apurinic/apyrimidinic (AP) endonuclease 1 (APE1). As reflected by its acronym, APE1 is an essential protein in DNA base excision repair (BER) pathway. The BER is the main pathway responsible for the repair of DNA damages caused by oxidation, irradiation and alkylating agents [3]. Upon removal of damaged bases by DNA glycosylase and thus the formation of apurinic/apyrimidinic (AP) site, APE1 hydrolyzes the phosphodiester backbone 5’ adjacent to the AP site [4, 5]. This incision generates a normal 30-hydroxyl group and an abasic deoxyribose-5-phosphate, which is processed by subsequent enzymes of the BER pathway, β-polymerase and DNA Ligase III/XRCC1. In addition, APE1 also possesses redox ability to restore multiple transcription factors controlling gene expression and cell survival. It has been shown to activate various transcription factors and facilitate their DNA binding via the reduction of a cysteine residue [6].
Multiple reports have shown that overexpression of APE1 is observed in various types of cancer (reviewed in [7]). Previously we have showed that APE1 is acetylated by p300 upon binding to chromatin at damage sites and acetylation increased its DNA repair activity [8]. Recently, we have shown that FACT (facilitates chromatin transcription) complex, a histone chaperone comprising of SSRP1 and SPT16, interacts with APE1 and facilitates its access to chromatin. SSRP1 and SPT16 are multi-domain proteins, and both of them contain dimerization domains (DD), middle domains (MD), and acidic C-terminal domains (CTD) [9]. The heterodimer helps to preserve chromatin structure which, in turn, prevents transcription initiation from cryptic promoters. It also interacts with DNA-binding surfaces of H2A/H2B dimers, facilitating uncoiling of DNA from the histone octamer and promoting nucleosome survival during transcription [10]. Previously we have demonstrated that targeting the FACT complex with small molecule curaxins/CBL0137 significantly improves the efficacy of 5-fluorouracil colon cancer in vitro and in vivo [11]. In this study, we sought to investigate the expression profiles of APE1 and FACT complex in low- and high-grade bladder cancer, and to investigate the correlation of APE1 and FACT expression with prognosis. The goal of this study is to provide fundamental evidence of the application of FACT complex inhibitor in the treatment of advanced bladder cancer in the following studies.
2. Materials and methods
Bladder cancer tissues used in this study were obtained (with IRB approval #012-17-EP) by urologic surgeons in the Division of Urology at University of Nebraska Medical Center. Archive tissues from 2008-2012 were retrieved from Department of Pathology and Microbiology. We chose to examine the paraffin-embedded blocks of bladder cancer from the Paraffin Tissue Bank who had undergone cystectomy or transurethral resection of bladder tumor (TURBT) and in whom we had sufficient follow-up to determine treatment outcome. A total of 69 samples were available for this project.
Tissue slides were immunostained with anti-APE1 (1:800) and SSRP1 (1:100) antibody [11, 12] following standard IHC protocol and analyzed using a blinded coding system by pathologists such that staining procedures and microscopic assessments were performed without knowledge of the histopathological diagnosis. Any appreciable brown staining will be considered positive and graded as 1+ if barely detectable, 2+ if easily seen fine granules were present diffusely throughout the nucleus or cytoplasm, and 3+ when dark course granules are observed. Also, the percentage of cells exhibiting positive staining was quantitated. H-score was calculated as the product of the actual percentage of positive stained cells and intensity score. The nuclear and cytoplasmic staining will be recorded separately. The H&E slides will then be reviewed to determine diagnosis and to map the location of the various histological patterns, such as carcinoma, atrophy, inflammation, and normal, to correlate with the staining patterns observed in the IHC preparations.
Medical records of bladder cancer patients were reviewed to gather the following data, including age, family history, diagnosis, tumor size, grade, TNM stage, and overall survival.
2.1. R2 genomics analysis and visualization platform
The R2 Genomics Platform (https://r2.amc.nl) is a free, publicly accessible web-based genomics analysis and visualization platform allowing biomedical researchers, without bioinformatics training, to integrate, analyze and visualize clinical and genomics data. There are two bladder cancer datasets available for survival analysis. One of them is TCGA – Tumor Bladder Urothelial Carcinoma – 408, which has an independent portal [13] developed by the National Cancer Institute. It contains expression data for a total of 408 patients, including T1 (n = 3), T2 (n = 119), T3 (n = 194), T4 (n = 58), and others (n = 34). The other dataset is Tumor Bladder – Hoglund, which includes 308 patients. It contains Ta (n = 116), T1 (n = 97), T2 (n = 85), T3 (n = 7), T4 (n = 1), and Tx (n = 2). TNM staging system which was maintained by AJCC (American Joint Committee on Cancer) and UICC (Union for International Cancer Control) was used. The T score is a rating of the extent of the primary tumor. In bladder cancer, the T staging is as follows (AJCC Cancer Staging Manual, 8th edition):
T0:No evidence of primary tumor
Ta:Noninvasive papillary carcinoma
Tis:Carcinoma in situ: “flat tumor”
T1:Tumor invades lamina propria (subepithelial connective tissue)
T2:Tumor invades muscularis propria
T3:Tumor invades perivesical tissue
T4:Tumor invades any of the following: prostatic stroma, seminal vesicles, uterus, vagina, pelvic wall, abdominal wall
We used the integrated plotting function to generate Kaplan-Meier curve by gene expression with default setting. The genes being analyzed were APEX1 and SSRP1.
2.2. Statistical analysis
Normality of variables was tested with the Kolmogorov-Smirnov test. Nonparametric data were presented with the median value and the interquartile range (IQR). For comparison of multiple categorical variables, the Chi-square test was used. Survival probabilities were analyzed using Kaplan-Meier curves. In the Kaplan-Meier survival analysis, patients were divided into two sets: H-score ≥50 percentile vs H-score <50 percentile. Correlation was analyzed in Graphpad Prism 7.0. Cox proportional-hazards model was used to investigate the impact of APE1 and SSRP1 expression when adjusting for covariates. Statistical analysis was performed by using SPSS 18.0 for Windows (SPSS Inc., Chicago, IL). For all statistical tests, p < 0.05 is considered to indicate a statistically significant result.
3. Results
3.1. Subcellular localization of APE1 is altered in high grade tumor
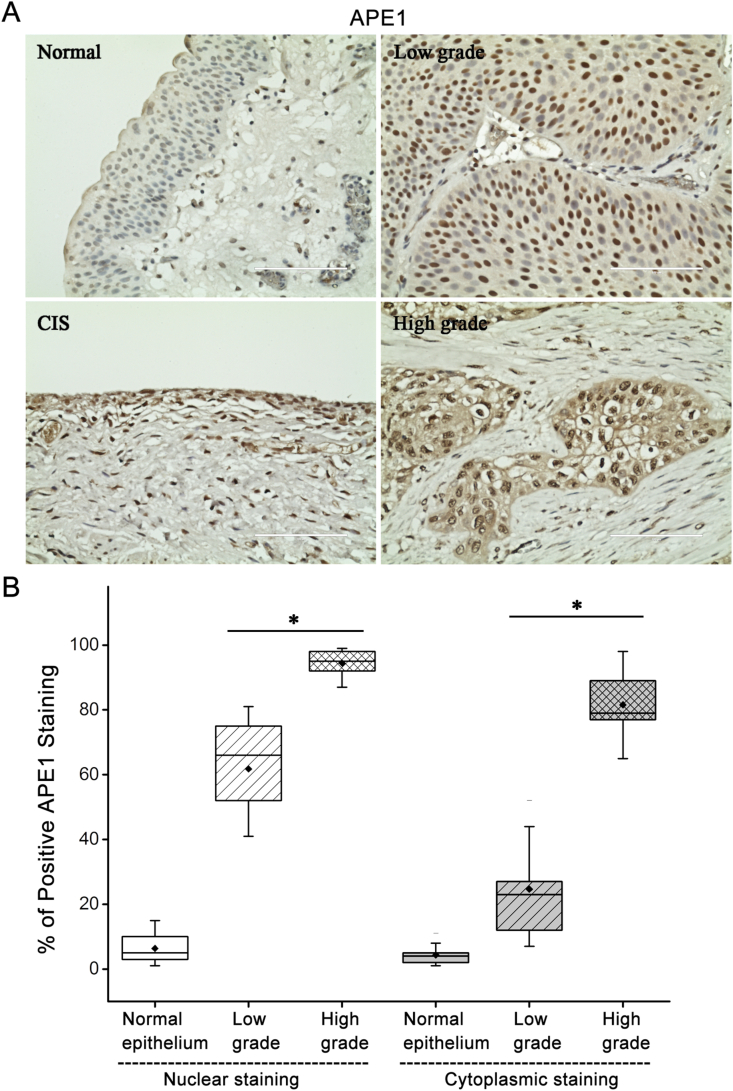
To investigate the expression and localization of APE1 in bladder cancer, we started with staining the surgical specimens with APE1 antibody. Scattered positive staining was observed only at the nuclear level of normal urothelial epithelium and inflammatory cells. In low grade papillary tumor, the expression of APE1 increased as compared to normal epithelium, but it remained at the nuclear level. In contrast, APE1 was detected not only in the nucleus but not also in the cytoplasm in high grade tumors including carcinoma in situ (CIS) and MIBC (Figure 1A). While normal bladder exhibited only 4.2% positive staining nuclei, low- and high-grade tumors had 66% and 98% positive staining at nuclear level (all p < 0.01). Compared to low-grade tumors, high grade tumors had a significantly higher percentage of positive staining nuclei (98% vs 66%, p < 0.05). Regarding to cytoplasmic staining, high grade tumors had a much higher percentage of positive brown staining (81%) while nearly none or minimal staining was observed in low grade tumors (23%) and normal bladder (2.7%) which was statistically significant (all p < 0.01) (Figure 1B).
Figure 1.
Alteration of APE1 subcellular localization in high grade bladder cancer. A) Immunohistochemical staining of APE1 in primary bladder cancer tissues (original magnification x400). All samples were obtained from tissue bank in UNMC after IRB approval. Representative images of normal urothelium, low-grade tumor, carcinoma in situ and high-grade tumors were shown. B) Box chart of the APE1 nuclear and cytoplasmic immunohistochemical reactivity in normal bladder epithelium, low- and high-grade bladder cancers. Data report the median, 25th and 75th percentiles of APE1 at the nuclear and cytoplasmic levels. ∗p < 0.05.
3.2. APE1 and SSRP1 expression is elevated concurrently in invasive bladder cancer
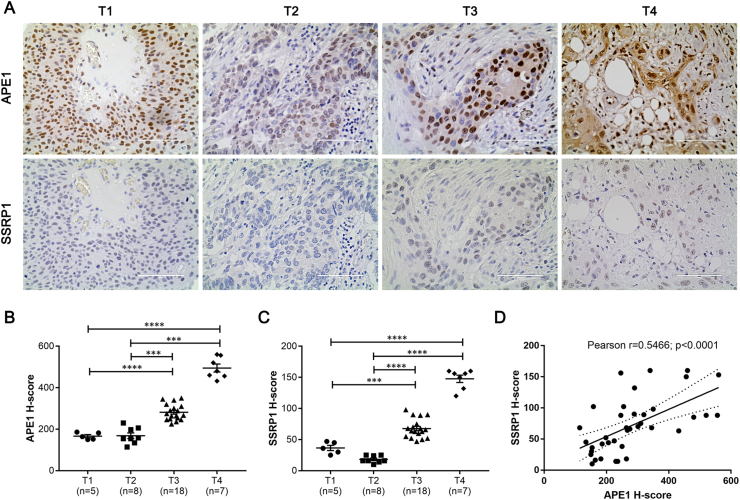
As chemotherapy is applied clinically in invasive bladder cancer, we turned our attention to the invasive cancer patients in our cohort which included 38 patients. Based on our previous study, FACT complex facilitates the access and acetylation of APE1 in chromatin, and their expression appears to be correlated in clinical specimens in colon cancer [11]. To examine the expression of FACT complex and its correlation with APE1 in invasive bladder cancer, we stained SSRP1 using consecutive slides from tumor specimen. The expression of SSRP1 was undetectable in normal tissue (data not shown), consistent with prior study [14]. On the other hand, the SSRP1 level increased as tumor invaded deep into tissue. We compared the expression of APE1 and SSRP1 in different T stages and found that the expression of both proteins increased as tumor progressed (Figure 2A). H-score was calculated to semi-quantify the expression level of APE1 and SSRP1 in tumors of various T stages. We found that there was no significant difference between T1 and T2, while T3 and T4 exhibited a significant increase of both proteins (Figure 2B and 2C). Given our biochemical evidence of the interaction of APE1 and SSRP1, we then use H-score to examine the correlation of APE1 and SSRP1 in these tumor tissues. There was a significant correlation between APE1 and SSRP1 in invasive tumors, with Spearman r of 0.5466 that was statistically significant (Figure 2D).
Figure 2.
APE1 correlates with SSRP1 in high grade tumors. A) Tumors at various T stages were stained with APE1 and SSRP1. Representative images were shown. B) & C) H-score of staining intensity was calculated based on percentage of positive staining in nucleus and cytoplasm. ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. D) H-score of APE1 and SSRP1 of each sample were entered in pairs in Graphpad Prism to perform correlation analysis. Spearman r = 0.5466, p < 0.0001.
3.3. Elevated APE1 and SSRP1 is associated with poor overall survival
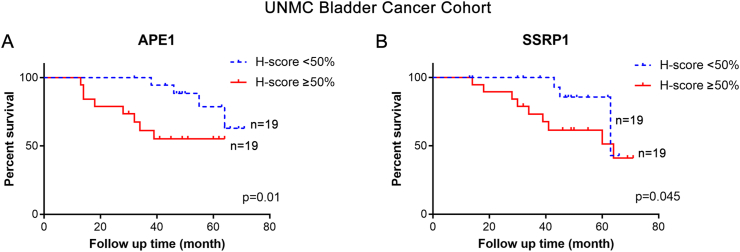
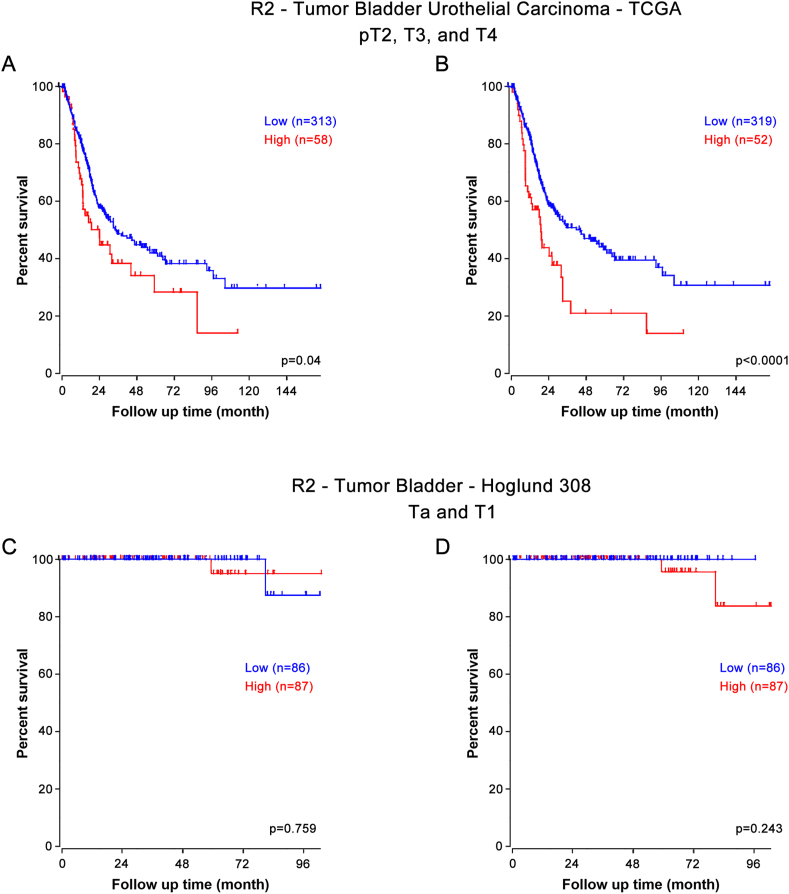
To examine whether expression of APE1 and SSRP1 in tumor tissue correlates with patient survival, we calculated the H-score based on cytoplasmic and nuclear staining in patients with invasive bladder cancer. As shown in Figure 3A, we noted a significant difference in patient survival based on expression level of APE1 and SSRP1. Elevated expression of APE1 was associated with poor overall survival. Similar pattern was observed with SSRP1 expression (Figure 3B). In the multivariate Cox analysis, high APE1 and SSRP1 expression remained a significant risk factor for poor survival when adjusting for covariates including age, gender, cardiovascular diseases, history of stroke, tumor stage, and tumor size, with hazard ratio (HR) of 1.14 (95% CI 1.08–1.27) and 1.39 (95% CI 1.17–1.52). Given the fact that our patient cohort has limited number of patients, we sought out to validate our findings in larger dataset R2. Tumor Bladder Urothelial Carcinoma – TCGA is an independent genome database that is included in R2. It has 408 patients with 371 patients are categorized as T2 or above. Using the default setting in R2, we noted a significant association of high APEX1 and SSRP1 with poor overall survival (Figure 4A and 4B). On the other hand, Tumor Bladder – Hoglund dataset has 308 patients, with the majority of which are non-muscle invasive tumors (116 Ta and 97 T1 patients). There was no statistical difference in the Kaplan-Meier survival curve when the cohort was divided based on low vs high APEX1 (Figure 4C). Similarly, SSRP1 expression did not appear to be associated with overall survival in T1 or T1 patients (Figure 4D). Together these data suggest that upregulation of APEX1 and SSRP1 were associated with poor survival in invasive bladder cancers but not in T1 or Ta tumors.
Figure 3.
Elevated APE1 and SSRP1 expression is associated with poor survival. A) & B) Patients with high grade tumors were divided into 2 groups based on H-score percentile: ≥50 vs < 50 percentile. Kaplan-Meier curves of APE1 and SSRP1 expression were plotted using Graphpad Prism 7.0.
Figure 4.
Upregulation of APEX1 and SSRP1 was associated with poor survival in MIBC in R2 genomic database. A) & B) In R2 – TCGA database, survival analysis was performed using pT2, pT3 and pT4 patients regarding APEX1 and SSRP1. This captures only muscle invasive bladder cancer and has 371 (91%) patients in this cohort that has survival data for analysis. C) & D) In R2 – Hoglund database, survival analysis was performed using Ta and T1 patients regarding APEX1 and SSRP1. This captures only non-muscle invasive bladder cancer and has 213 (69%) patients in this cohort. Final analysis contains 173 (56%) patients with survival data available for analysis.
4. Discussion
In this study, we report the expression profile of APE1 and SSRP1 in bladder cancer and examine their prognostic significance. APE1 is overexpressed in bladder cancer compared to normal tissues. Additionally, the pattern of APE1 expression shifts to more cytoplasmic staining in invasive cancer, while there is no cytoplasmic staining detected on low grade, non-invasive tumors. The expression of SSRP1 is only observed in bladder cancer tissues. There is a correlation of APE1 and SSRP1 in invasive bladder cancers. Furthermore, the overexpression of APE1 and SSRP1 is associated with poor overall survival in our cohort. This finding is validated in a publicly available database, R2 Genomics Analysis and Visualization Platform.
APE1 is a multifunctional protein that plays a critical role in BER pathway. It has been shown to be overexpressed in various tumors and associated with chemoresistance. Knock down of APE1 by siRNA in tumor cells demonstrates enhanced cytotoxicity and induction of apoptosis when combined with alkylating agents, hydrogen peroxide and other DNA damaging agents [15, 16, 17, 18]. Shin et al. found that serum levels of APE1 in bladder cancer patients were significantly elevated and its levels were associated with the tumor stage, grade, muscle invasion, and recurrence [19]. Choi et al. measured urinary APE1 level using ELISA and found that urine APE1 was elevated in bladder cancer patients, and its elevation was correlated with advanced tumor stage and recurrence [20]. On the contrary, Sak et al. showed that APE1 was elevated in MIBC and for those who were treated with radiotherapy the increased expression was associated with better cancer-specific survival [21]. Chantre-Justino et al. demonstrated that reduced levels of APE1 transcripts were significantly associated with cancer-specific mortality [22]. The authors proposed that a reduced expression of APE1 reflects the poorly differentiated nature of tumor cells in more aggressive tumors. The cells from aggressive tumors with extensive genomic instability may harbor chromosomal aberrations that result in failure of gene transcription, including DNA repair genes (in this case, APE1), resulting in lower protein expression of the gene products. The discrepancy of correlation between APE1 and prognosis in bladder cancer has led us to investigate the expression of APE1 in bladder cancer in this study. We found that the overall expression of APE1 is increased in bladder cancer compared to normal adjacent tissues. Moreover, the staining pattern changed between low- and high-grade tumors. We noticed an apparent cytoplasmic staining only in high grade and invasive cancer.
Alterations in subcellular distribution of APE1 in tumor tissues compared to normal tissues have been demonstrated in several tumors. Kakolyris et al. first described the differential expression pattern in a wide spectrum of cells [23]. The classic example is colon cancer. In normal colorectal mucosa, the predominant staining is nuclear in the less differentiated cells of the lower part of the crypt, but is cytoplasmic in the more differentiated and superficial colonic epithelium. During tumorigenesis this distribution is completely disrupted and the nuclear restricted pattern is lost in both adenoma and carcinoma, which display nuclear and cytoplasmic localization with a predomination of the latter, in front of a prominent nuclear localization in the normal tissue [24]. Similar pattern has been observed in breast cancer, HCC, thyroid carcinomas, epithelial ovarian cancers and NSCLC (reviewed in [25]). The dysregulation in nuclear versus cytoplasmic ratio toward a more cytoplasmic staining correlates well with aggressiveness and prognosis of the tumor: nuclear localization was always associated with better prognostic features. Our findings, along with others’ previous report, suggest that the APE1 in different location, reflected by its multifunctional nature, may play a different role in cell proliferation and survival. It is proposed that the endoribonuclease activity, role in mtBER, and its redox activity on newly synthesized transcription factors are major functions of cytoplasmic APE1, and it may provide an explanation why cytoplasmic localization is associated with poor survival: cells are more sustainable in oxidative stress and coactivation of transcription factors may contribute to tumor proliferation and progression.
APE1 and its endonuclease activity has been well studied in naked DNA in vitro. However, how APE1 accesses to DNA damages sites in the context of chromatin remains largely unknown. Previously we have shown that FACT complex interacts with APE1 and facilitates its access to chromatin and subsequent acetylation, which as a result promotes the BER function [8]. This has led us to investigate the feasibility of using FACT as an indirect target to interfere BER function. In fact, we found that using siRNA of FACT complex significantly decreased APE1 acetylation, and cells retain DNA damage despite allowing to recover after withdrawal of insult. A group of small molecules called curaxins that target FACT complex and cause chromatin trapping [26] is currently in multiple phase I and phase II clinical trials for metastatic or unresectable cancers (NCT01905228, NCT02931110 and NCT03727789). We have shown that curaxins interfere BER function and increase sensitivity significantly of colon cancer cells to chemotherapy. FACT complex is an appealing target for the following reasons. First, FACT complex is involved in multiple DNA repair pathways [27], and share a common role in nucleosome modulation. Second, reports have shown that FACT complex is undetectable in normal cells of adult mammalian tissues, except for undifferentiated and stem-like cells. It is upregulated during in vitro transformation and promotes survival and growth of established tumor cells [14]. Such differential expression among normal and tumoral tissues is advantageous in lowering side effects when it comes to treatment. Third, there is no available molecule that targets APE1 except for APX3330, which targets APE1's redox function but not DNA repair activity [28]. Here we demonstrated that APE1 and FACT complex are overexpressed in MIBC, and their expression is correlated with Spearman r of 0.5466. Their overexpression has been shown to be associated with poor overall survival in our patient cohort and in TCGA bladder cancer database. This provides fundamental evidence of the usage of curaxins to sensitize bladder cancer to chemotherapy, while further study would be warranted.
In conclusion, APE1 is overexpressed in low- and high-grade bladder tumors. The expression pattern shifts in high grade tumors where a cytoplasmic staining is observed. The critical interacting partner of APE1, SSRP1, is noted to be upregulated in MIBC, and is correlated with APE1 expression. The overexpression of APE1 and SSRP1 is associated with poor overall survival in MIBC patients. This suggests the usage of FACT inhibitor curaxins in MIBC to target FACT complex and APE1 to improve chemosensitization after further validation.
Declarations
Author contribution statement
Jiping Zeng: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Heyu Song: Performed the experiments.
Subodh Lele: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Chad A. LaGrange: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Kishor Kumar Bhakat: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Heyu Song is supported by UNMC graduate assistant fellowship. Jiping Zeng is supported in part by UNMC Department of Surgery Student Research Program of Excellence and Alpha Omega Alpha Carolyn L. Kuckein Student Research Fellowship.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Antoni S., Ferlay J., Soerjomataram I., Znaor A., Jemal A., Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur. Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Knowles M.A., Hurst C.D. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat. Rev. Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 3.Tell G., Damante G., Caldwell D., Kelley M.R. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxidants Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 4.Abbotts R., Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Fishel M.L., Kelley M.R. The DNA base excision repair protein Ape 1/Ref-1 as a therapeutic and chemopreventive target. Mol. Aspect. Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Thakur S., Dhiman M., Tell G., Mantha A.K. A review on protein-protein interaction network of APE1/Ref-1 and its associated biological functions. Cell Biochem. Funct. 2015;33:101–112. doi: 10.1002/cbf.3100. [DOI] [PubMed] [Google Scholar]
- 7.Abbotts R., Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Canc. Treat Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Roychoudhury S., Nath S., Song H., Hegde M.L., Bellot L.J., Mantha A.K., Sengupta S., Ray S., Natarajan A., Bhakat K.K. Human apurinic/apyrimidinic endonuclease (APE1) is acetylated at DNA damage sites in chromatin, and acetylation modulates its DNA repair activity. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00401-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valieva M.E., Gerasimova N.S., Kudryashova K.S., Kozlova A.L., Kirpichnikov M.P., Hu Q., Botuyan M.V., Mer G., Feofanov A.V., Studitsky V.M. Stabilization of nucleosomes by histone tails and by FACT revealed by spFRET microscopy. Cancers. 2017;9 doi: 10.3390/cancers9010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh F.K., Kulaeva O.I., Patel S.S., Dyer P.N., Luger K., Reinberg D., Studitsky V.M. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7654–7659. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song H., Zeng J., Roychoudhury S., Biswas P., Mohapatra B., Ray S., Dowlatshahi K., Wang J., Band V., Talmon G., Bhakat K.K. Molecular Cancer Therapeutics; 2019. Targeting Histone Chaperone FACT Complex Overcomes 5-Fluorouracil Resistance in Colon Cancer. molcanther.0600.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhakat K.K., Sengupta S., Adeniyi V.F., Roychoudhury S., Nath S., Bellot L.J., Feng D., Mantha A.K., Sinha M., Qiu S., Luxon B.A. Regulation of limited N-terminal proteolysis of APE1 in tumor via acetylation and its role in cell proliferation. Oncotarget. 2016;7:22590–22604. doi: 10.18632/oncotarget.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia H., Miecznikowski J.C., Safina A., Commane M., Ruusulehto A., Kilpinen S., Leach R.W., Attwood K., Li Y., Degan S., Omilian A.R., Guryanova O., Papantonopoulou O., Wang J., Buck M., Liu S., Morrison C., Gurova K.V. Facilitates chromatin transcription complex is an "accelerator" of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013;4:159–173. doi: 10.1016/j.celrep.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Luo M., Kelley M.R. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Therapeut. 2004;3:679–686. [PubMed] [Google Scholar]
- 16.Walker L.J., Craig R.B., Harris A.L., Hickson I.D. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D.S., Olkowski Z.L. Biological responses of human apurinic endonuclease to radiation-induced DNA damage. Ann. N. Y. Acad. Sci. 1994;726:306–308. doi: 10.1111/j.1749-6632.1994.tb52834.x. [DOI] [PubMed] [Google Scholar]
- 18.Silber J.R., Bobola M.S., Blank A., Schoeler K.D., Haroldson P.D., Huynh M.B., Kolstoe D.D. The apurinic/apyrimidinic endonuclease activity of Ape 1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin. Cancer Res. 2002;8:3008–3018. [PubMed] [Google Scholar]
- 19.Shin J.H., Choi S., Lee Y.R., Park M.S., Na Y.G., Irani K., Lee S.D., Park J.B., Kim J.M., Lim J.S., Jeon B.H. APE1/Ref-1 as a serological biomarker for the detection of bladder cancer. Cancer Res. Treat. 2015;47:823–833. doi: 10.4143/crt.2014.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi S., Shin J.H., Lee Y.R., Joo H.K., Song K.H., Na Y.G., Chang S.J., Lim J.S., Jeon B.H. Urinary APE1/ref-1: a potential bladder cancer biomarker. Dis. Markers. 2016;2016:7276502. doi: 10.1155/2016/7276502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sak S.C., Harnden P., Johnston C.F., Paul A.B., Kiltie A.E. APE1 and XRCC1 protein expression levels predict cancer-specific survival following radical radiotherapy in bladder cancer. Clin. Cancer Res. 2005;11:6205–6211. doi: 10.1158/1078-0432.CCR-05-0045. [DOI] [PubMed] [Google Scholar]
- 22.Chantre-Justino M., Alves G., Britto C., Cardoso A., Scherrer L., Moreira Ados S., Quirino R., Ornellas A., Leitao A., Lage C. Impact of reduced levels of APE1 transcripts on the survival of patients with urothelial carcinoma of the bladder. Oncol. Rep. 2015;34:1667–1674. doi: 10.3892/or.2015.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakolyris S., Kaklamanis L., Giatromanolaki A., Koukourakis M., Hickson I.D., Barzilay G., Turley H., Leek R.D., Kanavaros P., Georgoulias V., Gatter K.C., Harris A.L. Expression and subcellular localization of human AP endonuclease 1 (HAP1/Ref-1) protein: a basis for its role in human disease. Histopathology. 1998;33:561–569. doi: 10.1046/j.1365-2559.1998.00541.x. [DOI] [PubMed] [Google Scholar]
- 24.Kakolyris S., Kaklamanis L., Engels K., Turley H., Hickson I.D., Gatter K.C., Harris A.L. Human apurinic endonuclease 1 expression in a colorectal adenoma-carcinoma sequence. Cancer Res. 1997;57:1794–1797. [PubMed] [Google Scholar]
- 25.Shah F., Logsdon D., Messmann R.A., Fehrenbacher J.C., Fishel M.L., Kelley M.R. Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: from bench to clinic. NPJ Precis Oncol. 2017;1 doi: 10.1038/s41698-017-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang H.-W., Valieva M.E., Safina A., Chereji R.V., Wang J., Kulaeva O.I., Morozov A.V., Kirpichnikov M.P., Feofanov A.V., Gurova K.V., Studitsky V.M. Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci. Adv. 2018;4:eaav2131. doi: 10.1126/sciadv.aav2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler D.D., Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley M.R., Luo M., Reed A., Su D., Delaplane S., Borch R.F., Nyland R.L., 2nd, Gross M.L., Georgiadis M.M. Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxidants Redox Signal. 2011;14:1387–1401. doi: 10.1089/ars.2010.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.