Graphical abstract
Keywords: Nano-selenium, Cytotoxicity, Antioxidant, ct-DNA, Protein, DNA interaction
Highlights
-
•
Spherical SeNPs with average size 134 nm were synthesized employing Vitamin C.
-
•
The synthesized SeNPs represented great antioxidant and anticancer activity.
-
•
The interaction of SeNPs with ct-DNA, HSA, HHb and Cyt c was investigated.
-
•
Nano-selenium can bind to ct-DNA through partial intercalation binding mode.
-
•
HSA, HHb and Cyt c could keep their biological activity even in the presence of Nano-selenium.
Abstract
The aim of this study was the synthesis of selenium nanoparticles (SeNPs) employing vitamin C as a biocompatible and low toxic reducing agent. The synthesized selenium nanoparticles were characterized by using UV–vis, FT-IR, SEM-EDX, TEM, DLS, and zeta potential measurements. The results of the DPPH free radical scavenging assay demonstrate that this synthesized nano-selenium has strong potentials to scavenge the free radicals and cytotoxicity against MCF-7 and Raji Burkitt's lymphoma cancer cell lines. The interaction of calf thymus DNA (ct-DNA) with SeNPs indicated that the anticancer activity might be associated with the DNA-binding properties of nano-selenium. Finally, it was found that the synthesized nano-selenium can bind to the most important blood proteins such as human serum albumin (HSA), human hemoglobin (HHb), and Cytochrome c (Cyt c). The results showed that the secondary structure of these proteins remains unchanged, suggesting that the synthesized nano-selenium could be employed as a carrier in the drug delivery system without any cytotoxicity effect.
1. Introduction
The unique physicochemical properties of uniform and monodisperse nanometer-sized particles changed the perception of drug discovery and development by opening many hidden doors in disease pathophysiology and treatment options [1,2]. The adage, “small is the new big”, rightly fits to describe the role played by nanotechnology-based delivery systems in modern-day therapeutics. Among the different NPs, selenium nanoparticle has been an exciting area of study for many researchers that can be employed for a wide range of therapeutic applications [3]. Selenium (Se) is an essential and vital trace element of mammalian, which influences important physiological functions in the human body [4]. This kind of NPs can modify the expression of many selenoproteins such as thioredoxin reductases (TRs), iodothyroninedeiodinases (Ds), and glutathione peroxidases (GPXs) [4], which have the potential to regulate the physiological functions of the human body by acting as antioxidants, immune system function, regulating thyroid hormone metabolism, cancer prevention, and improving the production and quality of sperm, etc [5]. It has been reported that Nano-selenium demonstrates anti-tumor and anticancer activity through induction of cancer cell apoptosis with minimal side effects on normal cells [3] by different mechanisms. Moreover, Nano-selenium has a high potential to act as antiviral, antifungal, and antibacterial [6]. Antioxidant activity is one of the most fundamental features of Nano-selenium, which can remove harmful peroxides from the body through glutathione peroxidase (GSH-Px) and protect the membrane structure of organisms from damage [7]. The adequate selenium intake has been reported to be between 55 and 75 μg /day with an upper limit of ∼400 μg, which can be obtained from common foods such as mushrooms, vegetables, cereals, and food additives in a tea product that claims to possess several health benefits [8]. Nano-selenium has high biological activity, better bioavailability and low toxicity compared to organic and inorganic Se-compounds such as Se(IV) and Se(VI) [9]. One of the most widely employed methods to produce red Nano-selenium is the chemical reduction of selenium salts such as selenate, selenite, and selenium dioxide in the presence of ascorbic acid as a reducing agent [10,11]. Deoxyribonucleic acid (DNA) is the main molecular target for most of the small molecules such as nanoparticles, anticancers, antibiotics, antibacterial and antifungal drugs, which carriers hereditary information plays a crucial role in biological processes such as transcription and replication [12]. Investigation of the interaction of NPs with DNA has become an attractive research area for scientists because it has emerged as a promising target in chemotherapy. This Biomacromolecule acts as a binding template and provides a platform for metallic NPs to stick through its surface [13]. The interaction of nanoparticle can induce DNA damage and induce cell cycle arrest which in turn adversely affects other functions of DNA such as replication, transcription and finally leading to cell death [[14], [15], [16]]. Therefore, DNA-ligand interaction is a crucial issue for elucidating the mode of interaction, and provenance of some diseases to design new pharmaceutical agents that are effective in controlling gene expression [17]. A dynamic exchange between the lower and the higher affinity biomolecules over the nano surface creates a protein corona [18] with significant implication either to be positive or negative in vivo [[17], [18], [19], [20]]. This phenomenon creates a new biological identity for NPs [19] which might affect the biodistribution, immunological recognition, uptake, toxicity, and efficacy of the NPs [20]. Thereby, the interaction studies of proteins with NPs are essential to comprehend the biological implications of NPs. Human serum albumin (HSA) is one of the most important proteins in the blood plasma of humans, which processes multiple binding sites that can accommodate the adhesion of several endogenous and exogenous ligands and drugs [21]. It controls many important physiological activities like nutritionizing all over the human body [22,23]. HHb as a drug carrier can play an important role in the distribution of different compounds to the required physiological sites during therapy of many diseases [24]. Cytochrome c (Cyt c) is known as an iron-containing metalloprotein in the mitochondria of the cells with a tertiary structure that contains 104 amino acids. Covalent, electrostatic, hydrophobic, and hydrogen bonds [25] stabilize the helical structure of Cyt c, which may result in a specific tertiary arrangement. Cyt c acts as electron carrier in the mitochondria [26]. NPs can enter the mitochondria and may induce the conformational changes of Cyt c and switch on the apoptotic pathways [27]. This induced conformational change of Cyt c can be a considered as a cytotoxic effect of NPs. The main goal of this study is to investigate and elucidate the biological activity of Nano-selenium, especially their interactions with DNA and the most important blood proteins under physiological conditions (pH = 7.40). Although there are some studies on the interaction of biogenic selenium nanoparticles with ct-DNA [15] but the mechanism of the interaction of Nano-selenium with serum proteins (HSA, HHb, and cyt c) is not clear yet. Such studies need to be perused to get insights into the molecular mechanism of Nano-selenium–proteins and Nano-selenium–DNA interactions. Hence, with knowledge of the importance of the subject, the work focuses on the interaction modes of Nano-selenium with DNA and serum proteins to understand the effects of SeNPs on these important proteins and evaluate the safety of them for biomedical applications such as drug delivery, when they are injected into the blood. In this study, the synthesized SeNPs were characterized by UV–vis, FTIR, SEM-EDX, TEM, DLS, and Zeta potential measurements, and their antioxidant, and in-vitro cytotoxicity properties were assessed.
2. Materials & methods
2.1. Materials
Acridine orange (AO) (content > 98 %), Ethidium bromide (EB) (content > 95 %), Hoechst 33,258 (content > 98 %), Human serum albumin (HSA) (content > 98 %), calf thymus DNA (content > 97 %), Human hemoglobin (HHb) (content > 98 %) Cytochrome c from horse heart (Cyt c) (content > 99 %), NaH2PO4, Na2HPO4 (content > 98 %), Tris- (hydroxymethyl)-amino-methane–hydrogen chloride (content > 98 %), 2,2- diphenyl picryl hydrazyl (DPPH) (content > 99 %), 3-(4, 5- dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) (content > 98 %), l-gulutamine (content > 99 %), l-Ascorbic acid(content > 99 %), sodium selenite pentahydrate (Na2SeO3·5H2O) (content > 98 %), Dulbecco’s Modified Eagles medium (DMEM), fetal bovine serum (FBS), penicillin, and streptomycin were all purchased from Merck, Millipore and Sigma Aldrich. The Tris−HCl buffer solution was made from Tris-(hydroxymethyl)-amino-methane–hydrogen chloride (pH = 7.40), and was stored at 4 °C in the dark. The stock solution of ct-DNA was prepared by dissolving the ct-DNA powder in Tris−HCl buffer. The phosphate buffer solution was prepared from NaH2PO4, Na2HPO4, and NaOH (pH = 7.40), and was stored in the dark. The stock solution of proteins (HSA, HHb and Cyt c) was prepared in phosphate buffer. The AO, Hoechst, EB, stock solutions (1.00 × 10−3 M) were prepared in double distilled water.
2.2. Preparation of nano-selenium (SeNPs)
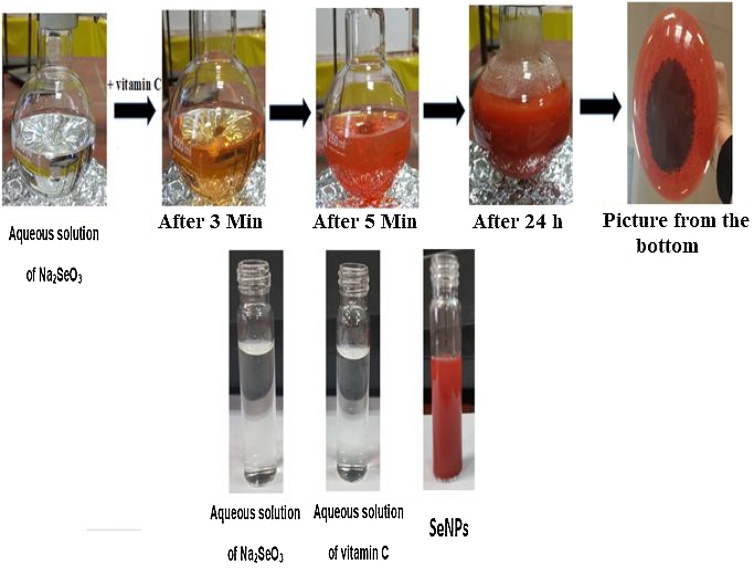
To synthesize Nano-selenium, 300 mL of vitamin C (Ascorbic acid 0.1 M) was added drop wise into the 600 mL of Na2SeO3·5H2O (5 mM). This system was gently stirred at room temperature for 24 h to prepare SeNPs. Ascorbic acid was employed as a biocompatible and low toxic reducing agent. After the addition of ascorbic acid, the color of this mixture immediately turned from colorless to yellowish-orange, and at the end of 24 h, the color changed to reddish-orange (‘brick’ red color) (Fig. 1). Then, the product was centrifuged for 15 min at 8000 rpm, the residue was removed by several times rinsing with deionized water, collected particles were overnight dried in a vacuum. Dried particles were stored in air-tight bags for subsequent experiments.
Fig. 1.
Reduction of Na2SeO3 and color change by Vitamin C.
2.3. Characterization of selenium nanoparticles
In this section, the microscopic and spectroscopic techniques were employed to characterize the synthesized SeNPs. These techniques are: Fourier transform infrared (FTIR) spectroscopy using a Bruker ALPHA FT-IR spectrometer with a wavenumber range of about 4000–400 cm−1 and a resolution of about 4–8 cm−1. The samples were prepared in FTIR discs by mixing and fine grinding of powdered SeNPs with potassium bromide (KBr) in the ratio of 1:10. The obtained peak was plotted as % of transmittance in X-axis and wavenumber (cm−1) in Y-axis. Dynamic light scattering study (DLS) and Zeta potential measurements were performed using a HORIBA and a Horiba- SZ-100-Z model, respectively. Before the measurement, 10 mg of the sample was sonicated in distilled water for 10 min. UV–vis spectra (UV–vis) were recorded, using an Agilent 8453 spectrophotometer with a 1 cm path length quartz cuvette. All the spectra were subtracted background with a baseline which was obtained from the absorption of purified water. The reduction of sodium selenite into SeNPs in the solution was monitored by the sampling of aliquots (2 mL) and measuring the UV–vis spectra of the solution at 24 h. The surface morphology of SeNPs was analyzed by scanning electron microscopy (TESCAN BRNO-Mira3 LMU) operated at an accelerating voltage of 10.0 Kv HV mode and detectors containing the second electron. The elemental composition of the SeNPs was obtained from coupled EDX spectra using a SAMx model. Thin films of the sample were prepared on a carbon-coated copper grid by putting a very small amount of the sample on the grid, which was blown with a hand drier to remove excess particles. Transmission electron microscopy (TEM) employing a Zeiss-EM10C-100 KV model. The samples were drop-casted on carbon-coated TEM copper grids and air-dried for imaging.
2.4. Cell culture conditions
The MCF-7 and Raji Burkitt's lymphoma cancer cell lines were employed and cultured in the DMEM supplemented with, 100 μ/mL penicillin, 10 mg/mL streptomycin, 10 % fetal bovine serum (FBS), and 200 mM l-glutamine at 37 °C in an incubator.
2.4.1. In vitro cytotoxicity screening
The MTT, [3-(4, 5- dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide] assay was used to distinguish the in-vitro cytotoxicity of Nano-selenium. For this purpose, the cells were cultured on 96-well plates for 24 h. After this time, the various concentrations of Nano-selenium (10, 20, 40, 80,120, and 160 μg/mL) were added and then, the cell culture plates were kept in the incubator for 24 h. Afterwards, the previous medium was removed and 75 μL of MTT solution (5 mg/mL) was added to each well. The cell culture plates were incubated for 4 h at 37 °C. Then, the medium including MTT was discarded and 100 μL DMSO was added to dissolve the dark blue crystals. After 20 min, an enzyme-linked immunosorbent assay (ELISA) reader was used at 570 nm to note the optical density of each well.
Moreover, the percentage of cell viability was computed, using the following formula:
| % Cell Viability = (OD value of treated cells)/ (OD value of untreated cells (control) ×100 | (1) |
Nano-selenium potency is expressed in terms of IC50 value (50 % inhibitory concentration) computed from the plotted dose-effect curve (through least-square regression analysis).
2.4.2. Morphological imaging (AO/EB staining)
In order to observe the cellular and nuclear morphological changes, AO/EB staining was operated. The cancer cells were seeded on 6-well plates (3 × 105 cells per well) for 24 h and different concentrations of Nano-selenium (10–160 μg/mL) were added and incubated for 12 h. After that, the cells were washed with the phosphate-buffered saline (PBS) solution. Finally, the cells were stained and labeled with an equal volume of AO (1 mg/mL) and EB (1 mg/mL) for 5 min and observed under a UV fluorescence microscope.
2.5. Biological activity
2.5.1. DNA binding experiments
2.5.1.1. Electronic absorption spectroscopy
To distinguish the interaction between ct-DNA and SeNPs, electronic absorption spectroscopy (UV–vis) was used. The absorption spectra of SeNPs (3.97 × 10−7 g/mL) with and without different concentrations of ct-DNA (2.82 × 10-5 to 2.48 × 10-4 M) were recorded in Tris-buffer solution (pH = 7.40), to evaluate the changes in absorbance of SeNPs during interaction with ct-DNA in the range of 200−350 nm.
2.5.1.2. Fluorescence studies
In order to perform the fluorescence measurements, the SeNPs concentration was fixed at 3.97 × 10−7 g/mL and the ct-DNA concentration was changed from 5.49 × 10-6 to 7.06 × 10-5 M. The Nano-selenium is excited at 280 nm and accomplished the fluorescence studies in the range of 300–500 nm at three temperatures (288.15, 298.15 and 310.15 K). The absorption of ct-DNA is around 260 nm. Hence, the fluorescence intensity for the absorption of exciting light and reabsorption of emitted light was corrected, considering the inner filter effect (IFE). The following equation was employed to eliminate the IFE.
| Fcor = Fobs × e (Aex+ Aem)/2 | (2) |
In this mentioned equation, Fobs and Fcor are observed and corrected fluorescence intensity, Aem and Aex are the absorbance values for emission and excitation wavelengths, respectively [28].
2.5.1.2.1. Dye displacement experiments
To determine the mode and nature of ct-DNA-Nano-selenium interaction, competitive displacement fluorescence assays were performed, employing two intercalator fluorescence probes (EB and AO) and a groove binder (Hoechst 33,258). The displacement experiment was carried out to elucidate the potential of Nano-selenium to displace probes from the ct-DNA structure. At first, the ct-DNA (2.73 × 10−5 M) was added to the EB, AO, and Hoechst (5.00 × 10-6 M) solutions. Then, this mixture was titrated with various concentrations of Nano-selenium (2.47 × 10-6 to 5.61 × 10-5 g/mL). The ct-DNA-probes systems were excited at 526 nm, 502 nm, and 340 nm for EB, AO, and Hoechst, respectively. Meanwhile, the scanning range of emission spectra between 530–720 nm, 505–650 nm, and 350–650 nm were set for EB, AO, and Hoechst, respectively.
2.5.1.3. Docking simulation
To decipher for the best binding sites in all conceivable cavities of ct-DNA and authenticating our experimental results, Patch Dock [29] algorithm was used. The normal form of DNA (B-DNA) was downloaded from the Protein Data Bank (PDB ID: 1BNA). To draw the Nano-selenium structure, Avogadro and Gauss View 5.0 were used. To perform docking analysis, the PDB files of Receptor (DNA) and Ligand (Nano-selenium) were uploaded to the PatchDock server, employing the cluster RMSD at a default value of 4.0. The best conformation was used with the minimum binding energy to accomplish further docking analysis. To further refining the best Patch Dock results, the Fire Dock web interface was operated. BIOVIA Discovery Studio software was used to do the visualization of docked.
2.5.2. Human serum albumin (HSA), Human Hemoglobin (HHb), Cytochrome c (Cyt c) binding experiments
2.5.2.1. Electronic absorption spectroscopy
In this section, the absorption spectra of HSA, HHb, and Cyt c (1.00 × 10−5 M) in the absence and presence of different concentrations of SeNPs (2.44 × 10−5 to 2.30 × 10-4, 9.90 × 10-6 to 9.09 × 10-5 and 2.44 × 10−5 to 2.30 × 10-4 g/mL, respectively) were recorded to evaluate the changes in absorbance of the proteins during their interactions with SeNPs in the range of 200−800 nm and phosphate buffer solution (pH = 7.40).
2.5.2.2. Fluorescence studies
In this study, a JASCO FP 6200 spectrofluorometer was used to note the emission spectra of HSA, HHb, and Cyt c (5.00 × 10−6 M) without and with various concentrations of SeNPs (2.44 × 10-5 to 2.72 × 10-4, 2.30 × 10-5 to 2.60 × 10-4, and 2.44 × 10-5 to 1.83 × 10-4 g/mL, respectively) in the range of 300−450 nm. This research was performed at 288.15, 298.15 and 310.15 K and the emission quenching signals of HSA, HHb, and Cyt c were excited at 295 nm. The synthesized SeNPs show absorption at 265 nm. So, the inner filter effect (IFE) was used.
2.5.2.3. Circular dichroism (CD) measurements
In order to evaluate the changes in the secondary structural contents of the HSA, HHb, and Cyt c during their interactions with SeNPs, the CD signals of HSA, HHb, and Cyt c solutions (5.00 × 10−6 M) were noted with and without different concentrations of SeNPs (2.49×10-6 to 1.23×10-5 g/mL), employing a JASCO spectropolarimeter J-810 in the UV region 200–260 nm.
2.5.2.4. Docking simulation
In this section, the PatchDock algorithm was used to obtain docked structures. In order to draw the Nano-selenium structure, Avogadro and GaussView 5.0 were used. Meanwhile, the crystal structures of human serum albumin (PDB ID: 1bm0), cytochrome c (PDB ID: 2b4z), and hemoglobin (PDB ID: 1fn3) were downloaded from Protein Data Bank, to perform docking analysis. The PDB files of proteins and SeNPs to the Patch Dock server were uploaded, using the cluster RMSD at a default value of 4.0. The best conformation with the minimum binding energy was used to accomplish further docking analysis. In order to further refine the best Patch Dock results, the Fire Dock web interface was employed. To do the visualization of docked the BIOVIA Discovery Studio software was used.
2.6. DPPH free radical scavenging assay
In this study, the 2,2- diphenyl picryl hydrazyl method was used to explore the antioxidant properties of SeNPs (50, 100, 200, 400, 800, and 1000 μg/mL). The mixture of 150 μL of water with 50 μL of DPPH is considered as control solution. Meanwhile, ascorbic acid and methanol are considered standard and blank, respectively. The samples were incubated in the dark for 30 min, and then the absorbance of the solutions was noted at 517 nm to accomplish the free radical scavenging assay. The increase in the percentage of antiradical activity was shown by lower absorbance in UV–vis spectroscopy. The capability of SeNPs to scavenge DPPH radical was evaluated employing the following formula:
| % Inhibition of DPPH radical = [(control OD –sample OD) / control OD] × 100 | (3) |
3. Results and discussion
3.1. Characterization studies
3.1.1. UV–vis spectra analysis
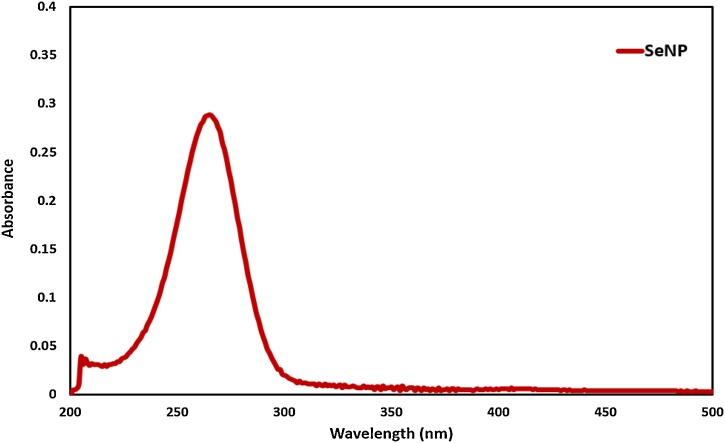
The UV–vis spectra analysis was employed to determine the surface plasmon resonance of SeNPs in the mixed solution. 2 mL aliquot of SeNPs was added in a 1 cm path-length quartz cuvette to note the UV–vis spectrum in the range of 200–800 nm. Fig. 2 exhibits the UV–vis spectrum of synthesized SeNPs with maximum absorption at 265 nm [30]. The absorbance peak of SeNPs was related to crystallizability.
Fig. 2.
UV–vis spectrum of nano-selenium.
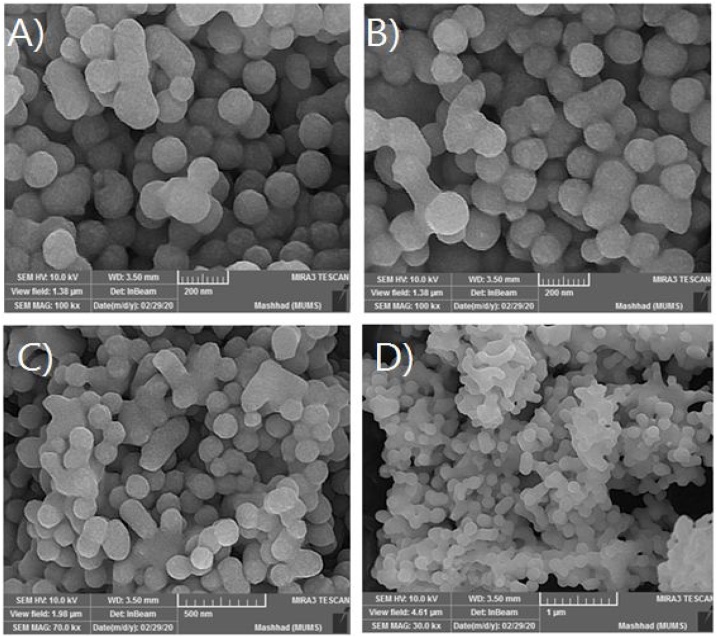
3.1.2. SEM and EDX analysis of SeNPs
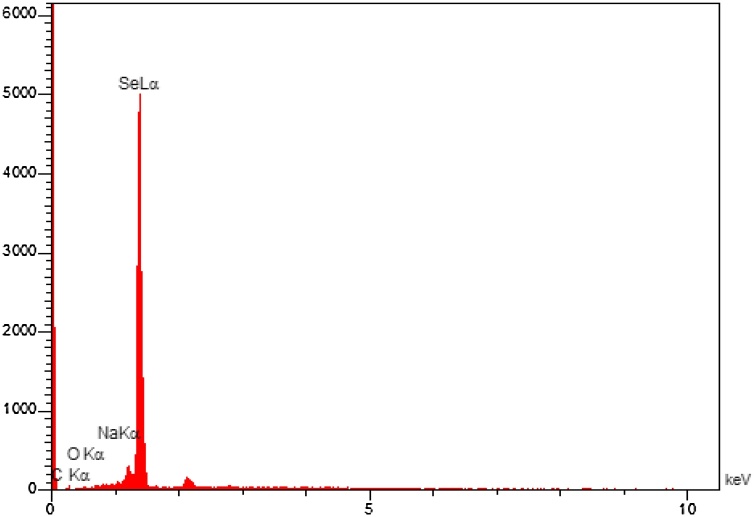
To scrutinize the morphology of the synthesized nano-selenium, SEM analysis was used. The predominant shape of nano-selenium is spherical at different magnification levels (Fig. 3A-D). Also, we used Energy-dispersive X-ray spectroscopy (EDX) to investigate the composition of SeNPs and the presence of elemental selenium. Fig. 4 represents the EDX image and the corresponding elemental energy mass ratio of the synthesized SeNPs. The elementary analysis of the surface represents a strong band of Se atoms (87.60 %), C atoms (10.11 %), O atoms (2.08 %), and N atoms (0.21 %). Carbon peaks were observed in the EDX spectrum due to the C support grid. The high amount of selenium in the spectrum authenticated the prepared Nano-selenium suspension was free of other elements.
Fig. 3.
SEM images of SeNPs with different magnification.
Fig. 4.
EDX images of SeNPs.
3.1.3. TEM analysis
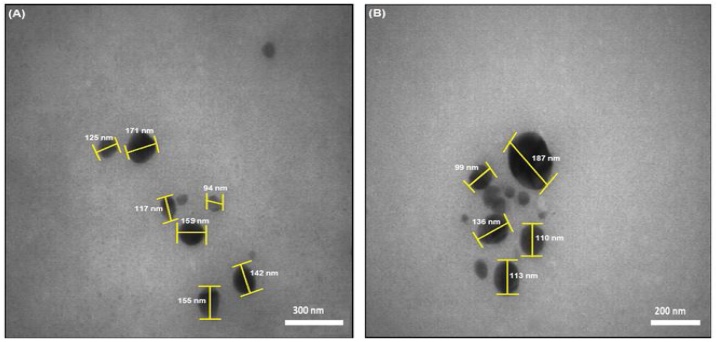
The transmission electron microscopy (TEM) was used to scrutinize the size and morphology of the synthesized nano-selenium. Fig. 5A and B demonstrate the TEM photographs of Nano-selenium at different magnification levels. It is clear from these images that the selenium nanoparticles are almost spherical. This obtained result is in line with the data obtained from SEM. The average size of selenium nanoparticles is calculated as 134 nm, using Image J software (Fig. 6).
Fig. 5.
TEM images of SeNPs with different magnification.
Fig. 6.
FTIR spectra of SeNPs.
3.1.4. FT-IR spectroscopy
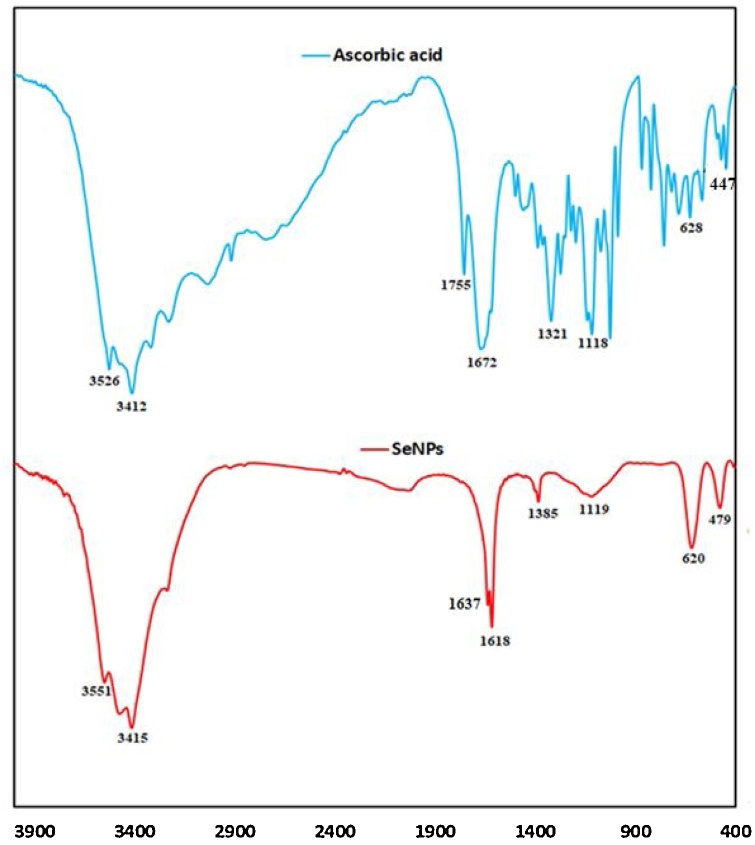
The Fourier transform infra-red (FT-IR) spectroscopy, a handy technique, was employed to further confirm the formation of Nano-selenium by ascorbic acid and identify elements involved in the fabrication of it in the range of 400–4000 cm−1. In the IR spectrum of ascorbic acid, the peaks in the range 3200−3526 cm−1 correspond to the different hydroxyl groups (—OH) or the presence of moisture in the sample [31]. The characteristic absorption peak at 1755 cm−1 in the spectrum of ascorbic acid is due to the stretching vibration of the C O of the five-membered lactone ring and this band disappeared in the IR spectrum of SeNPs. The band at 1672 cm−1 is due mainly to the C C stretching vibrations of ascorbic acid [32]. The two bands at 1637 and 1618 cm−1, as well as a weaker band at 1385 cm−1 may represent the asymmetric and symmetric stretching vibrations of carboxylates (i.e., of some carboxylic acids as products of deep AA oxidation adsorbed on SeNPs). Considerable difference in the positions of different bands of SeNPs and ascorbic acid gives direct evidence for the covalent bonding of the products of oxidation of ascorbic acid on the surface of the SeNPs [33]. The bands at 479 and 620 cm−1 representing the stretching and bending vibrations of Se-O, which may be attributed to the binding of SeNPs to the carbonyl groups from the products of oxidation of ascorbic acid, and hence, they can form a coat around SeNPs to prevent agglomeration.
3.1.5. DLS and zeta potential analysis of green synthesized SeNPs
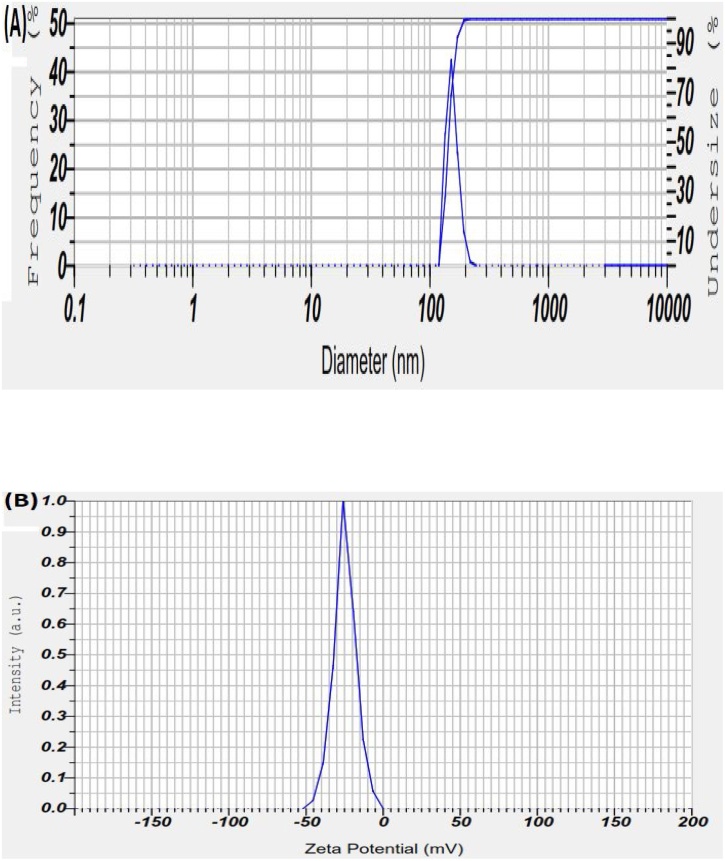
To determine the hydrodynamic size and polydispersity index (PDI) of the synthesized Nano-selenium, the dynamic light scattering (DLS) analysis in an aqueous solution was employed. Fig. 7A exhibits the graph of the size distribution of SeNPs. It was found that the average size of Nano-selenium is 146 nm with a low dispersity index (PDI) of 0.521, which authenticates the homogeneity and uniform dispersion of SeNPs. Compared with the size of particles in TEM measurements, the particles were moderately bigger in DLS measurement as in DLS; it measures the particles' hydrodynamic radius. The stability and the actual state of SeNPs in aqueous solution can be deliberated by Zeta potential measurements. Fig. 7B represents the negative zeta potential value of SeNPs (-24.8) which confirms high dispersity and stability of SeNPs with low aggregation.
Fig. 7.
DLS particle size analysis (A) and Zeta potential distribution of SeNPs (B).
3.2. Cytotoxicity (In-vitro)
3.2.1. Cytotoxicity effect of selenium nanoparticles on Raji Burkitt's lymphoma and MCF-7 cancer cell lines
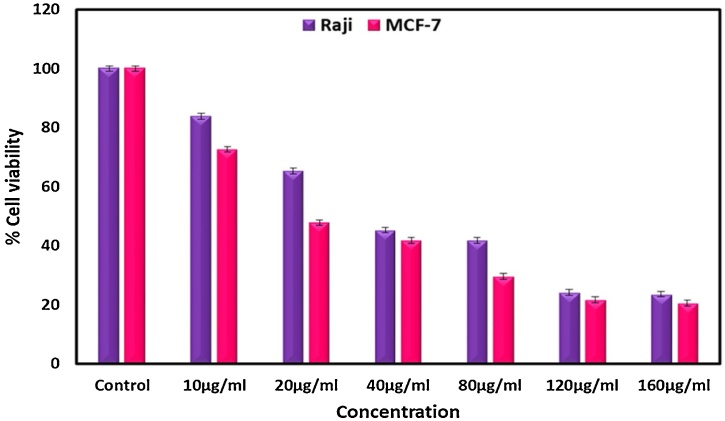
Today, one of the novel choices in the field of chemotherapy for researchers is selenium nanoparticles which have excellent anticancer activity with low toxicity [[34], [35], [36]]. In this research, the in vitro comparative anticancer properties of Nano-selenium against the MCF-7 and Raji Burkitt's lymphoma cancer cells were investigated. Fig. 8 demonstrates the dose-response curve of nano-selenium. It is evident from this curve that selenium nanoparticles show a remarkable anti-proliferative effect on both the MCF-7 and Raji Burkitt's lymphoma cancer cells. The calculated IC50 value for selenium nanoparticles in MCF-7 and Raji Burkitt's lymphoma cancer cells is 23.20 and 38.24 μg/mL, respectively. Meanwhile, it was found that 160 μg/mL of Nano-selenium was required for 79.47 % and 76.39 % growth inhibition of MCF-7 and Raji Burkitt's lymphoma cancer cells, respectively. Nano-selenium exhibits stronger effect on MCF-7 cells compared to Raji Burkitt's lymphoma cancer cells. Hassanien et al. [37] reported that the biogenic SeNPs are potent anticancer agents and inhibit the growth of the three cancer cells as indicated by the IC50 values (151, 393, and 252 μg for Caco2, HepG2, and Mcf-7, respectively). Also, Menon et al. reported the cytotoxicity effect of biosynthesized selenium nanoparticles towards the A549 lung cancer cell line. The calculated IC50 was 40 μg/mL at 48 h, and for 24 h the IC50 was 80 μg/mL. It is evident that the IC50 values of our synthesized SeNPs were significantly lower than that of the reported SeNPs. Also, the mechanism behind their cytotoxicity can be explained with the diffusion of the SeNPs to the cell membrane via the ion channels and the contact with the nitrogen bases of DNA or intracellular proteins [38].
Fig. 8.
Cytotoxic effect of SeNPs against MCF-7 and Raji at different concentration (10, 20, 40, 80, 120 and 160 μg/mL).
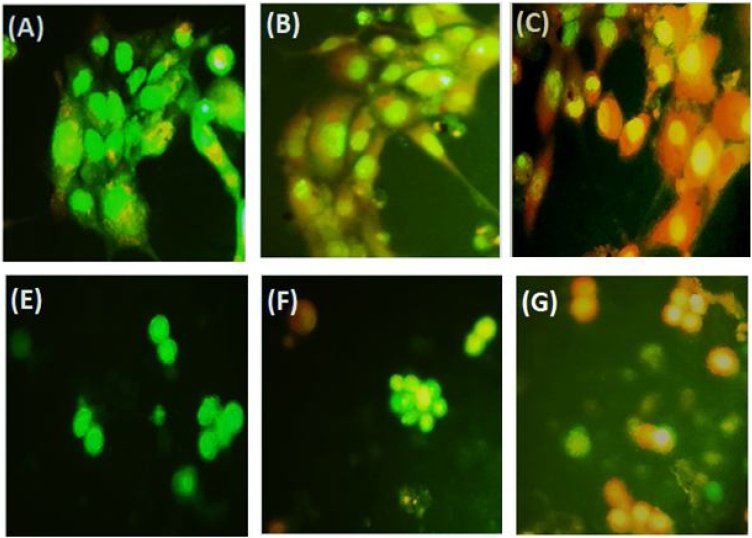
3.2.2. Morphological imaging
In this section, the DNA of the cells was stained by AO/EB to assess typical morphological apoptosis changes. Two fluorescent dyes, AO/EB, were used to determine the viable, apoptotic, and necrotic cells by their different color which can be green fluorescence, bright green fluorescence, yellow/orange fluorescence, and red fluorescence for viable, early apoptotic, late apoptotic, and non-viable cells, respectively [39]. The results of the staining assay on MCF-7 and Raji Burkitt's lymphoma cancer cells are showed in Fig. 9. The cells which did not receive Nano-selenium maintained their morphological and have normal green fluorescent nuclear architecture and an intact configuration without obvious nuclear damage. However, the MCF-7 and Raji Burkitt's lymphoma cancer cells which were treated with Nano-selenium showed extensive cell damage through cell shrinkage, condensed nuclei, and membrane blebbing in a dose-dependent manner. Also, as shown in Fig. 9, Nano-selenium exhibits stronger effect on MCF-7 cell morphology compared to Raji Burkitt's lymphoma cancer cells.
Fig. 9.
Morphological evidence of apoptosis by AO/EB dual staining MCF-7 :(A) Control (B) 10 μg/mL (C) 160 μg/mL and Raji: (A) Control (B) 10 μg/mLl (C) 160 μg/mL of SeNPs. Magnification: 200 × .
3.3. DNA binding experiments
One of the ultimate biomolecular targets for many anticancer drugs such as Nano-selenium is deoxyribonucleic acid (DNA). In this section, to investigate whether the anticancer property of Nano-selenium is related to its interaction with DNA, the interaction of Nano-selenium with ct-DNA was evaluated by multispectroscopic approach at pH = 7.40 (simulative physiological conditions) to authenticate the binding effect between them. Khurana et al. reported that SeNPs are believed to enter cancerous cells via receptor-mediated endocytosis [3]. Also, Menon et al. showed that Nano-selenium enters the receptors of cancerous cells through the nucleus, and induces DNA breakage. The breakage leads to either cell cycle arrest or apoptotic pathway [40].
3.3.1. Electronic absorption titration
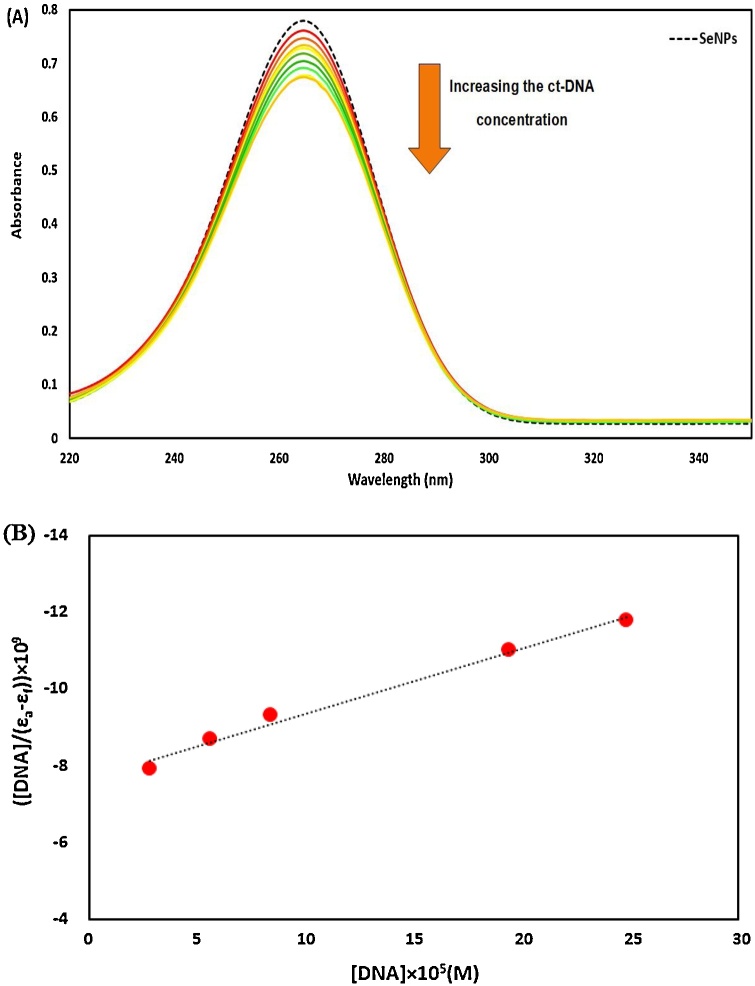
To evaluate the binding strength and the binding mode between ct-DNA with Nano-selenium during their interaction, UV–vis absorbance spectroscopy was used (Fig. 10A). The UV absorption spectra of Nano-selenium without and with successive increments of different concentrations of ct-DNA in the Tris−HCl buffer (pH = 7.40) were noted. It is evident from Fig. 10A that the UV–vis absorbance intensity of nano-selenium gradually decreases with increments of different concentrations of ct-DNA (hypochromic effect) without any shift in the absorption maxima of ct-DNA–Nano-selenium complexes. This phenomenon originated from the intercalative mode of binding of the Nano-selenium with stacked base pair of ct-DNA.
Fig. 10.
(A). Absorbance spectra of SeNPs (3.97 × 10−7 g/mL) with different concentrations of ct-DNA from 2.82 × 10-5 to 2.48 × 10-4 M. (B). Wolfe-Shimmer plot to determine the binding constant value of the DNA-SeNPs.
In order to deliberate the binding affinity of Nano-selenium to ct-DNA, the following equation (Wolfe–Shimmer) was employed:
| [DNA]/(εa-εf)= [DNA]/(εb-εf)+1/Kb(εa-εf) | (4) |
In Wolfe–Shimmer equation εa is the received extinction coefficient of absorption signal at different concentrations of ct-DNA, εb the extinction coefficient of nano-selenium when fully bound to ct-DNA, and εf is the extinction coefficient of free nano-selenium. Fig. 10B shows the plot of [DNA] / (εa–εf) against [DNA]. The (slope / intercept) of this plot was used to find the intrinsic binding constant (Kb). The obtained value of Kb for Nano-selenium-ct-DNA is 2.50 × 103 M −1, which is smaller than the corresponding value for classical intercalator ethidium bromide (EB) [41], indicating that the partial intercalation binding mode might be involved in nano-selenium.
3.3.2. Fluorescence spectroscopy of ct-DNA
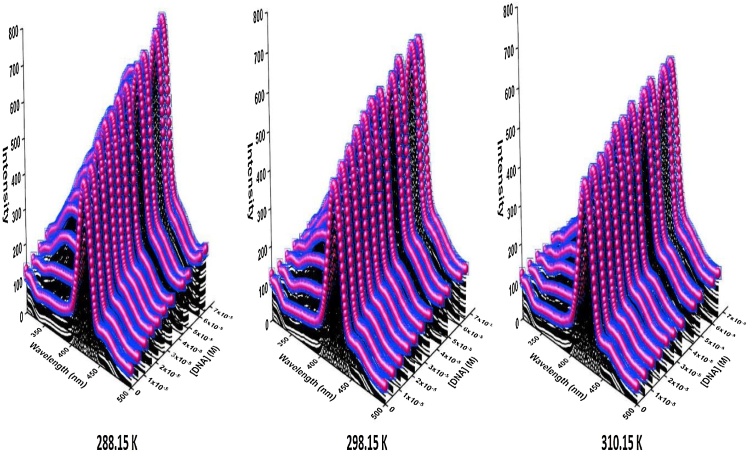
One of the fundamental techniques to deliberate the mechanism and the binding mode of the interaction between ligands with DNA is the emission experiment. Therefore, this helpful technique was used to investigate the binding mode of Nano-selenium to ct-DNA. The Nano-selenium solution in Tris-buffer emits luminescence with the maximum at around 424 nm when excited at 290 nm (approximately near to maximum absorption). Thus, we noted the emission spectra of Nano-selenium in the presence of various concentrations of ct-DNA at 288.15, 298.15, and 310.15 K (Fig. 11). The emission intensity of nano-selenium is increased steadily with increments of different concentrations of ct-DNA (hyperchromic effect) along with a spectral change (slight blue shift). This effect confirms that Nano-selenium can interact with ct-DNA base pairs and the quantum efficiency of Nano-selenium was increased [42]. Also, the efficacy of the NPs to function as a therapeutic agent is deliberate by evaluating the NPs − DNA binding on their interaction.
Fig. 11.
Fluorescence emission spectra of SeNPs in the presence of the ct-DNA at 288.15, 298.15, 310.15 K. ([SeNPs] = 3.97 × 10−7 g/mL, [ct-DNA] = 5.49 × 10-6 to 7.06 × 10-5 M).
3.3.2.1. Mechanism of binding mode
In order to compute the enhancement constant of the nano-selenium-ct-DNA system at different temperatures, Eq. (5) was used like to quenching process [43]:
| F0/F = 1 - KE [E] | (5) |
For enhancing mechanism, the Eq. (5) is reported as follows [43]:
| F0/F = 1 – KD [E] = 1- KB τo [E] | (6) |
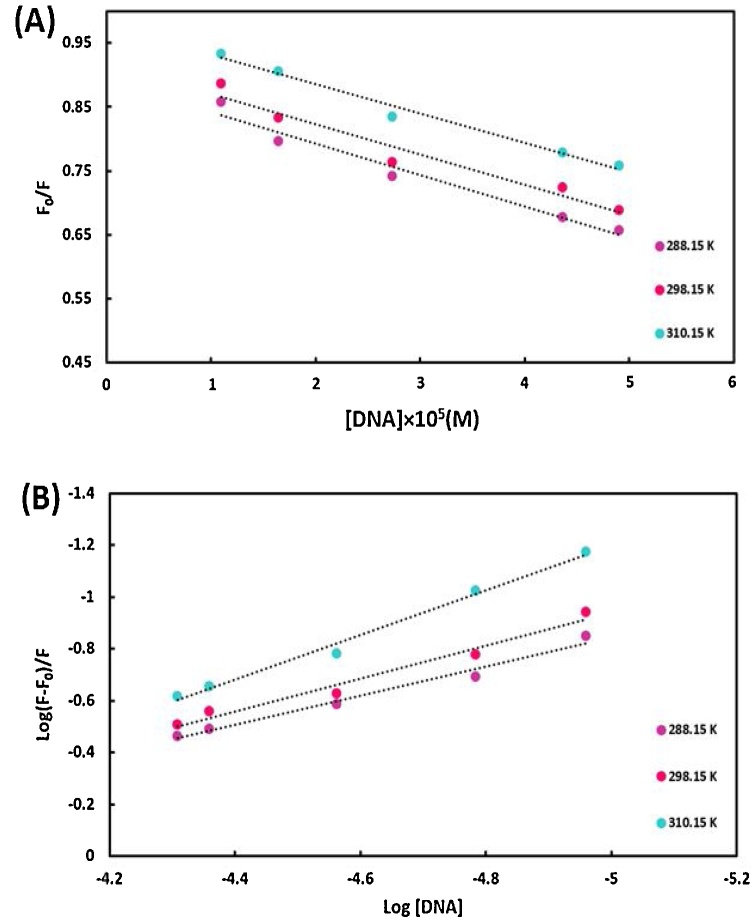
In this equation, like to dynamic quenching constant, KD is the dynamic enhancement constant, [E] is the concentration of the enhancer, [ct-DNA], τo is the lifetime of the nano-selenium without ct-DNA, like to a bimolecular quenching constant, KB is the bimolecular enhancement constant which is computed from KD = KB τo [43]. F and F0 are the intensities of Nano-selenium emission with and without ct-DNA, respectively. Fig. 12A demonstrates the plot of F0/F versus [DNA] at 288.15, 298.15, and 310.15 K, which is employed to compute the dynamic enhancement constant and the bimolecular enhancement constant. As demonstrated in Table 1, the KD and KB values decreased on enhancing the temperature, suggesting that the operative mechanism of the fluorescence enhancement is static [44]. In other words, a static process involves complex formation in the ground state.
Fig. 12.
(A) Stern–Volmer plot, (B) Scatchard plot for the binding of the SeNPs to ct-DNA at 288.15, 298.15 and 310.15 K.
Table 1.
Thermodynamic and binding parameters of the interaction of the SeNPs to DNA.
| T(K) | KD (M−1) | KB (M−1s−1) | Kf (M−1) | n | ΔG0(kJmol−1) | ΔH0(kJmol−1) | ΔS0(Jmol−1 K−1) | |
|---|---|---|---|---|---|---|---|---|
| 288.15 | 4.93 × 103 | 4.93 × 1011 | 92.40 | 0.56 | −10.28 | 91.48 | 353.14 | |
| 298.15 | 4.74 × 103 | 4.74 × 1011 | 1.68 × 102 | 0.63 | −13.81 | |||
| 310.15 | 4.58 × 103 | 4.58 × 1011 | 1.35 × 103 | 0.86 | −18.05 | |||
3.3.2.2. Binding sites and binding constants
In order to deliberate the binding stoichiometry (n) and binding constant (Kf) during complex formation between nano-selenium and ct-DNA in the ground state, the Scatchard curve (log [(F0− F)/F] vs. log[DNA]) was used, which was plotted based on Eq. (7) (Fig. 12B):
| log [(F - F0)/F] = log Kf + n log [DNA] | (7) |
Table 1 demonstrates the values of n and Kf at 288.15, 298.15, and 310.15 K, which are obtained from the slope and intercept of Eq. (7). The obtained values of n approximated to 1, indicating that a 1:1 adduct was formed between ct-DNA and nano-selenium. The values of Kf increased on elevating the temperature, suggesting that the affinity of nano-selenium to ct-DNA enhanced on increasing the temperature [45] (Fig. 13).
Fig. 13.
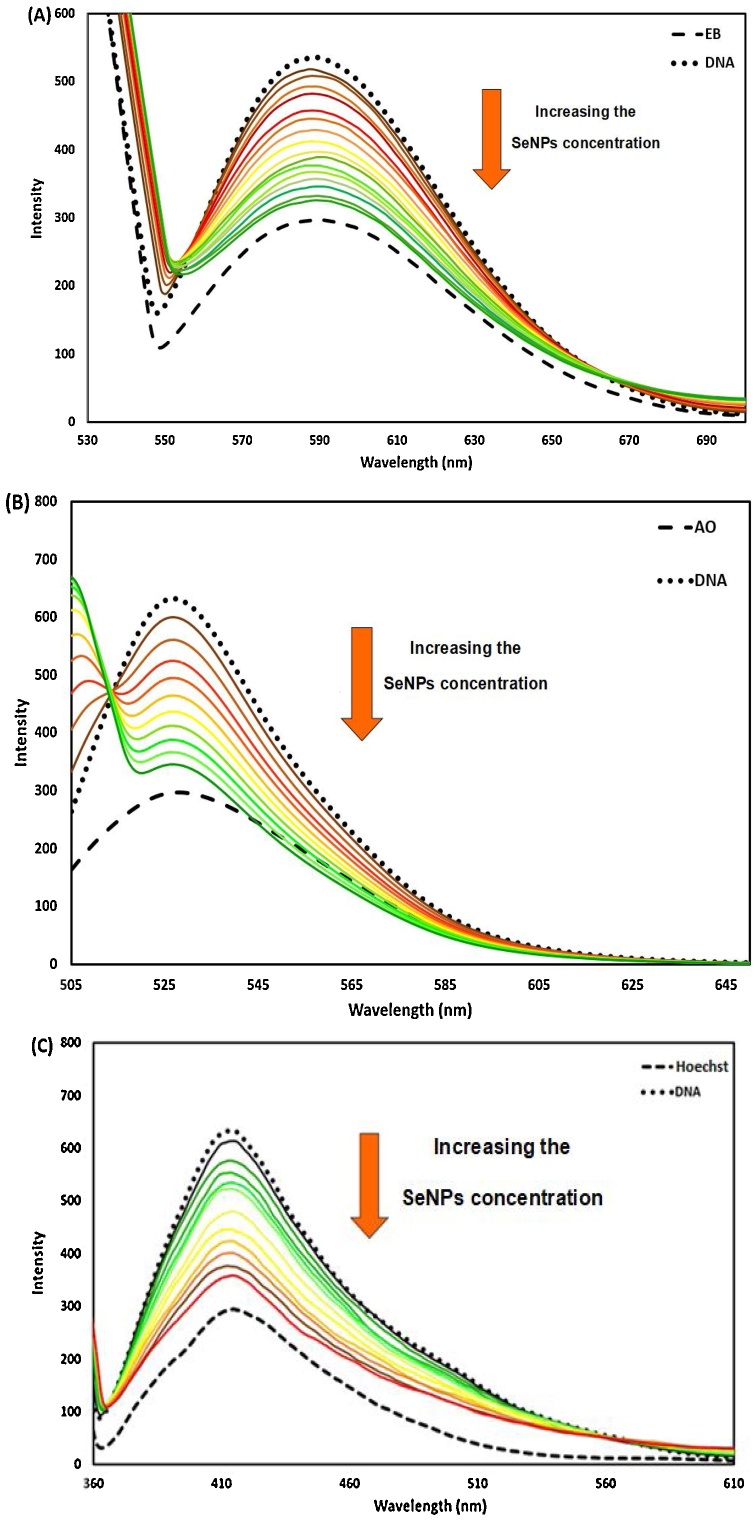
Fluorescence emission spectra of (A) EB + ct-DNA in the presence of nano-selenium CDNA= (2.73 × 10−5 M) C EB =(5.00 × 10-6M) C nano-selenium = from 2.47 × 10-6 to 5.61 × 10-5 g/mL. (B) AO + ct-DNA in the presence of nano-selenium CDNA=(2.73 × 10−5 M) CAO =(5.00 × 10-6M) C nano-selenium = from 2.47 × 10-6 to 5.61 × 10-5 g/mL. (C) Hoechst + ct-DNA in the presence of nano-selenium CDNA= (2.73 × 10−5 M) C Hoechst =(5.00 × 10-6M) C nano-selenium = from 2.47 × 10-6 to 5.61 × 10-5 g/mL.
3.3.2.3. The nature of binding forces and the thermodynamic parameters of the nano-selenium–ct-DNA interaction process
Based on Eq. (8) and Eq. (9), LnKf versus 1/T was plotted to compute the thermodynamic parameters of the nano-selenium–ct-DNA system on their interactions and deliberate the type of forces between nano-selenium and biomolecule:
| ln Kb= −ΔH/RT + ΔS/R | (8) |
| ΔG0 = ΔH0−TΔS0 | (9) |
In van’t Hoff Eq. (8) R is the gas constant. The entropy change (ΔS) and enthalpy change (ΔH) were calculated from the intercept (ΔS/R) and slope (−ΔH/R) of van’t Hoff equation. Also, the free energy change (ΔG) was obtained, employing Gibbs–Helmholtz equation Eq. (9). All of the thermodynamic parameters of the nano-selenium–ct-DNA system at 288.15, 298.15, and 310.15 K were exhibited in Table 1. We can deliberate the type of interaction forces between nano-selenium and ct-DNA based on thermodynamic parameters [46]. The positive values of ΔH° and ΔS° suggest that the hydrophobic attraction plays the main role in the binding of nano-selenium to ct-DNA [47]. Also, the values of ΔG are negative at three temperatures, indicating that the reaction between ct-DNA and nano-selenium is spontaneous.
3.3.2.4. Competitive binding studies with EB, AO, and Hoechst 33,258
Dye displacement assays were employed based on fluorescence emission. To scrutinize the nano-selenium mode of binding to double-helical ct-DNA, EB, AO, and Hoechst 33,258 as probes were used. The intensity of probes-ct-DNA complex emission was recorded with increments of different concentrations of nano-selenium. It is evident from Figs. 13A, B, and C that the intensity of probes-ct-DNA emission was changed after the addition of nano-selenium. AO and EB are fluorescence probes that intercalate to double-stranded of ct-DNA due to their planner structure [48]. A remarkable decrease in the emission intensity of the AO-ct-DNA and EB–ct-DNA system is observed on increasing the concentration of nano-selenium (Fig. 13A and B). Therefore, it can be concluded that nano-selenium was intercalated to the double-stranded helix of ct-DNA and avoided AO and EB binding and compete with AO and EB for the intercalation sites in DNA helix, therefore AO and EB were released into the solution (emission quenching). It can be suggested that the mode of nano-selenium-ct-DNA binding is intercalative mode. On the other hand, the intensity emission of the Hoechst 3325-ct-DNA was decreased on increasing the concentration of nano-selenium. Hoechst 33,258 is a well-known groove binder (external binder) of the DNA helix [48]. It can be suggested that the synthesized nano-selenium was placed on the same site of Hoechst 33,258 in the minor groove of double-stranded ct-DNA and Hoechst were released into the solution (emission quenching) indicating the presence of groove binding (external binding), too. Therefore, it can be concluded that nano-selenium interacts with ct-DNA by an unusual DNA binding mode, partial intercalation, which contains intercalation and groove properties [49].
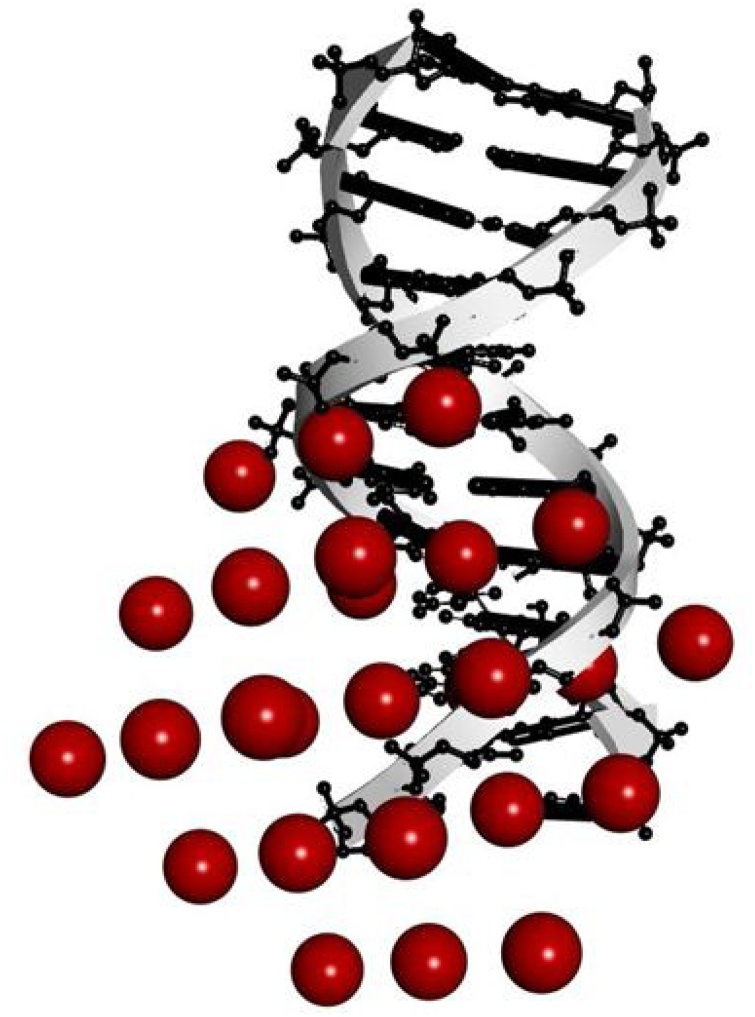
3.3.3. Analysis of molecular docking
One of the best computational tools to evaluate the NPs–DNA interactions and testify the experimental results is docking simulation. In this study, the proper bonding site of ct-DNA on the interaction with SeNPs was investigated and the energetically most favorable conformation of the docked pose structure was received employing implemented Lamarckian genetic algorithm for 20 runs. As can be seen in Fig. 14, the SeNPs were located in both intercalation and groove sites of double-helix DNA which is in line with the proposed mode of interaction. Run 6 is in agreement with the experimental results. The obtained value of Kb (7.17 × 102 M−1) and, ΔG (-16.30 kJ/mol) in 20 orientations is in line with the fluorescence experiments (1.68 × 102 M−1) binding constant.
Fig. 14.
Molecular docking perspective of SeNPs-DNA.
3.4. The interaction of SeNPs with human serum albumin, human hemoglobin, and cytochrome c
3.4.1. UV–vis absorption spectroscopy
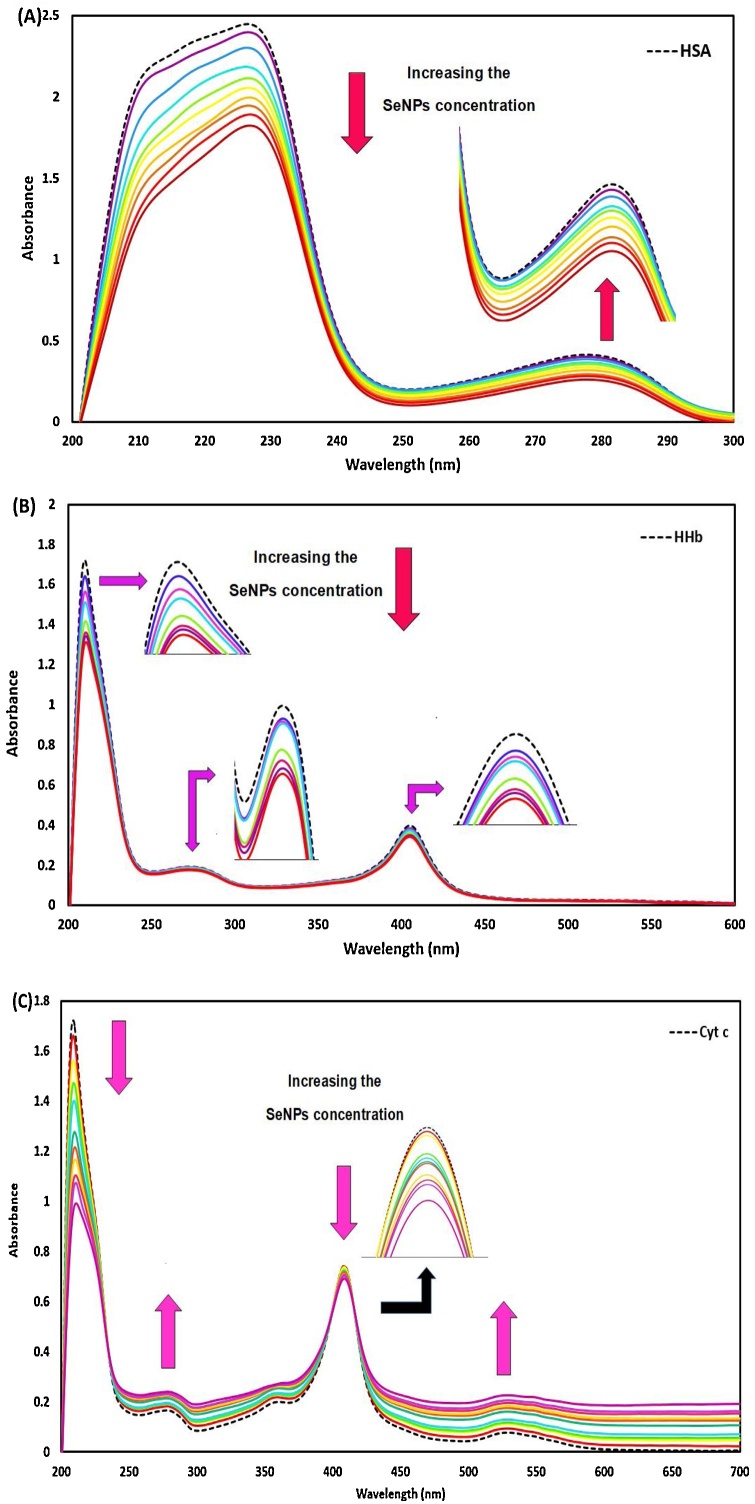
The Ultraviolet-visible absorption technique was used to distinguish the structural changes of human serum albumin, hemoglobin, and cytochrome c on their interactions with nano-selenium. Fig. 15A, B, and C demonstrate the UV–vis spectra of HSA, HHb, and Cyt c without and with various concentrations of nano-selenium. There are three characteristic absorption signals in the UV–vis spectrum of human hemoglobin at 210, 273, and 406 nm, which reflecting the protein framework conformation and concurrently to the peptide bond, the presence of the phenyl group of aromatic amino acids, like tryptophan (Trp), tyrosine (Tyr) or phenylalanine (Phe), and the porphyrin Soret band of heme located in HHb (transition of π→π* of hematoporphyrin in HHb which provides the information on the conformational integrity of the heme group region of heme protein), respectively [50]. It is evident from Fig. 15B that the intensity of HHb absorption spectra decreased (hypochromic effect) when nano-selenium is added to HHb solution without a shift in the position of characteristic peaks. The results suggest that a ground state complex was formed between nano-selenium and HHb on the interaction in an aqueous medium without any changes in the native microenvironment of the hem and amino acid residues.
Fig. 15.
(A) Absorbance spectra of HSA (1.00 × 10−5 M) with different concentrations of nano-selenium from 2.44 × 10−5 to 2.30 × 10-4 g/mL. (B) Absorbance spectra of HHb (1.00 × 10−5 M) with different concentrations of nano-selenium from 9.90 × 10-6 to 9.09 × 10-5 g/mL. (C) Absorbance spectra of Cyt c (1.00 × 10−5 M) with different concentrations of nano-selenium from 2.44 × 10−5 to 2.30 × 10-4 g/mL.
The characteristic peaks of Cyt c are located in 210, 273,408, and 530 nm, which arise from the Cyt c framework conformation and concurrently with the peptide bond, the presence of aromatic amino acids [50], the Soret band (which was caused by additive effects of the transition dipole moments in the two orbital excitations a1u-eg and a2u-eg of π→π* transitions of the porphyrin ring in Cyt c), and the Q transitions bands [51], respectively.
As can be seen in Fig. 15C, the UV–vis absorption of the peptide bond and aromatic amino acids of Cyt c undergo a severe hypochromism, and hyperchromism, respectively, which reveals the ground-state complex formation between nano-selenium and Cyt c. Meanwhile, no significant alteration was observed in the position of the signal at 210 nm, suggesting the conformation of the peptide backbone of protein didn’t change during the nano-selenium–Cyt c interaction. However, no alteration was observed in the position of the signal at 270 nm, indicating that the polarity around the amino acid residues and microenvironments of them didn’t change during the interaction.
As can be seen in Fig. 15C, the UV–vis spectra of Cyt c in the presence of different concentrations of nano-selenium are characterized by Soret and Q transitions bands of heme chromophore at 408 nm and near 530 nm. There is not any shift in the position of the peak at 408 nm. These results indicated that the nano-selenium is conjugated to Cyt c but, this conjugation does not affect the native heme microenvironment [52].
There are two characteristic signals in the absorption spectra of HSA (Fig. 15A): one of them is located at 226 nm and originates from the n→π٭ transition of the peptide bond (α-helical structure and framework conformation of HSA), and the other is located at 278 nm and refers to absorption and electron transfer (π → π*) of the aromatic amino acids (Trp, Tyr, and Phe) [53]. The UV–vis spectra of HSA in the absence and presence of different nano-selenium concentrations were noted to evaluate the nano-selenium– HSA interaction. As represented in Fig. 15A, the UV–vis absorption of the peptide bond (226 nm) and aromatic amino acids (278 nm) decreased on elevating the concentrations of nano-selenium (hypochromism), which reveals the formation of nano-selenium–HSA complex during their conjugation. As can be seen, the position of signals at 226 and 278 nm had barely changed, indicating that the conformation of peptide backbone of protein (α-helix content) and microenvironments of amino acid residues didn’t change significantly during the nano-selenium– HSA interaction which is supported by the previously reported works, Maji et al. [54,55]. In other words, the native secondary structure of HSA did not change significantly despite complex formations with nano-selenium. Such interactions may have influenced the spectroscopic properties of HSA to some extent. It has been shown by different techniques below that despite these interactions, the protein retains its structural properties. However, for some proteins, the interaction with drugs or nanoparticles can lead to a change of structure and loss of function [56]. Based on the above results, it can be concluded that denaturation of HSA upon exposure to the nano-selenium did not occur, and this protein retains its regular secondary structure and biological activity.
3.4.2. Fluorescence quenching studies
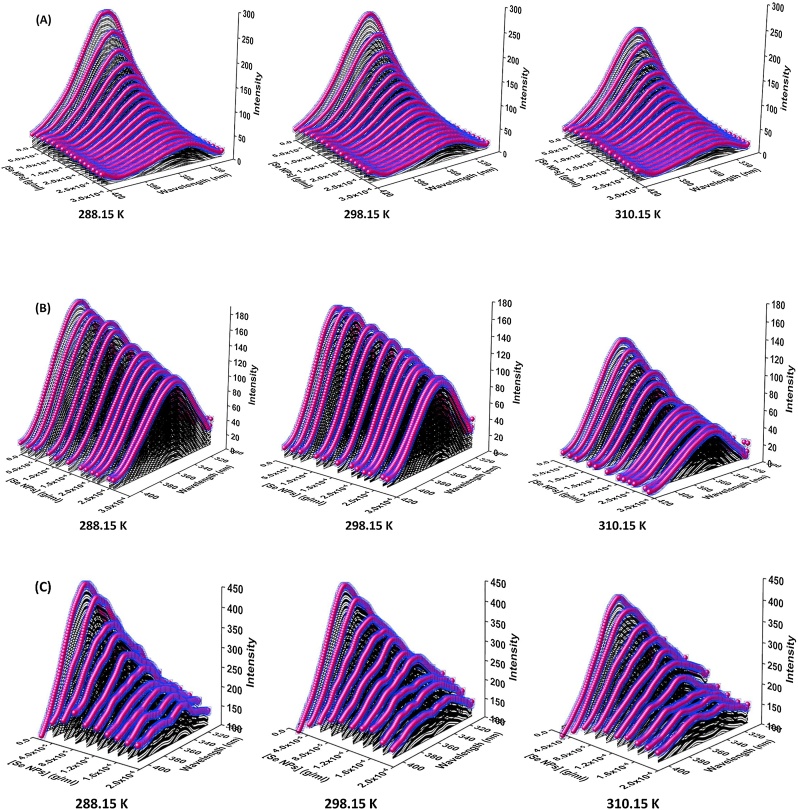
In order to investigate protein folding, assembly, dynamics and interaction, fluorescence spectral studies are a suitable method. The structure or environment changes of HSA, HHb, and Cyt c on their interactions with nano-selenium were deliberated by evaluating the inherent fluorescence of them. Most protein molecules have the residues of tryptophan, Trp-214 of HSA [57], three Trp units (α-Trp 14, ß-Trp 15 and ß-Trp 37) in each α and ß chain of HHb, and Trp-59 of Cyt c [58], which led to the fluorescence of them at approximately 346 nm [59]. The inherent fluorescence of HSA, HHb, and Cyt c in the presence of various concentrations of nano-selenium at three different temperatures under physiological conditions (pH = 7.4) was noted (Fig. 16). The maximum fluorescence emission spectra of HSA, HHb, and Cyt c quenched continuously on elevating the concentration of nano-selenium at three temperatures. This reduction in the inherent fluorescence intensity of HSA, HHb, and Cyt c can be attributed to their strong interactions with nano-selenium, which leads to conformational changes of the proteins in addition to the exposure of hydrophobic patches, as a result of ligand interaction [60]. In a hydrophobic environment, Trp and Tyr are located in the core of the protein, have a high quantum yield, and therefore high fluorescence intensity, whereas in a hydrophilic environment (exposed to solvent) their quantum yield decreases leading to low fluorescence intensity [61].
Fig. 16.
(A) The fluorescence spectra of HSA (5.00 × 10−6M) with different concentrations of nano-selenium from 2.44 × 10-5 to 2.72 × 10-4 g/mL at three temperatures. (B) The fluorescence spectra of HHb (5.00 × 10−6M) with different concentrations of nano-selenium from 2.30 × 10-5 to 2.60 × 10-4 g/mL at three temperatures. (C) The fluorescence spectra of Cyt c (5.00 × 10−6M) with different concentrations of nano-selenium from 2.44 × 10-5 to 1.83 × 10-4 g/mL at three temperatures.
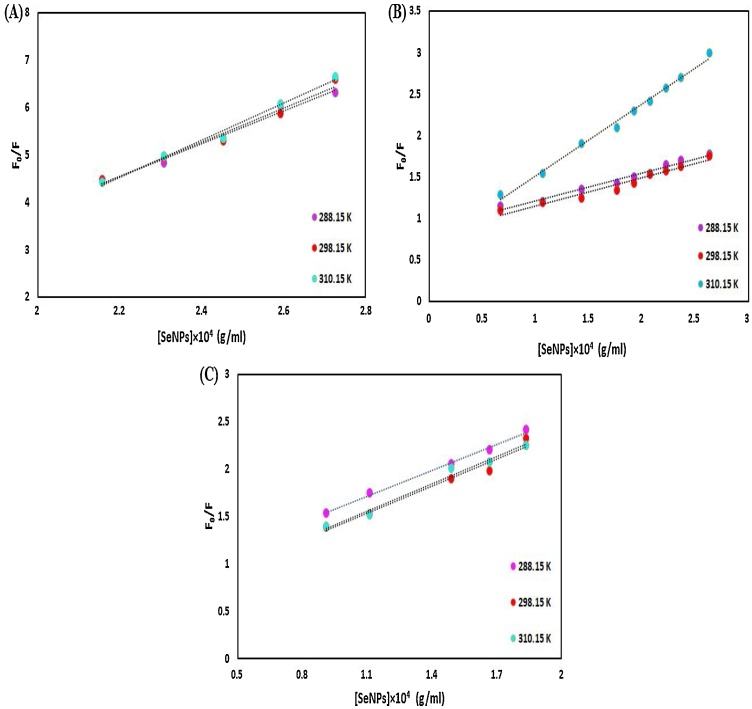
3.4.2.1. Stern–volmer analysis (binding parameters)
The nano-selenium-proteins interaction leads to quenching, which can be categorized as collision and association quenching. The collision quenching is arising from collisions between the fluorophore, and the quencher during the lifetime of the excited state and increases with elevating the temperature, which leads to an enhanced energy transfer and an increase in the quenching constant. However, association quenching refers to ground state complex formation between fluorophore and quencher and decreases with enhancing the temperature, which leads to a decrease in the stability of formed complexes, the dissociation of weakly bound complexes, and the quenching constant [62].
To clarify the mode of quenching process of the nano-selenium-proteins system on their interactions, the well-known Stern–Volmer equation was used (Eq. (10):
| F0/F = 1 + Kq τ0 [Q] = 1 + Ksv [Q] | (10) |
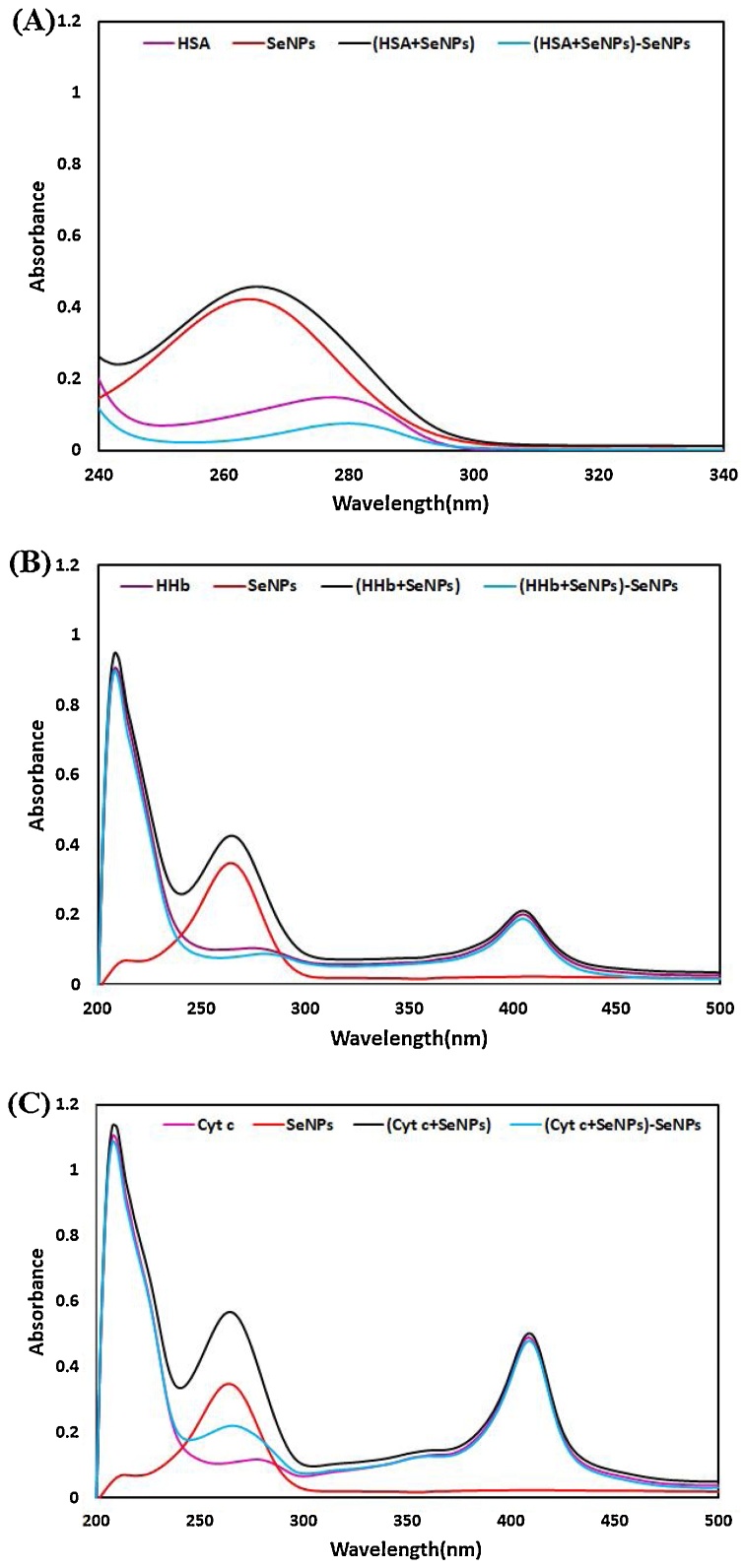
In this classical equation, [Q] is the concentration of nano-selenium, Kq is the protein bimolecular quenching rate constant, τo is the average fluorescent lifetime of the protein in the absence of nano-selenium, and Ksv is the Stern-Volmer quenching constant. The values of Ksv and Kq at 288.15, 298.15, and 310.15 K are shown in Tables 2, 3 and 4 that were calculated from the Stern-Volmer curve (F0/F against nano-selenium concentration) for HSA, HHb, and Cyt c (Fig. 17A, B and C). It is clear from these Tables that the values of Ksv and Kq increase on elevating the temperature and suggesting that the dominant quenching process in proteins-SeNPs interaction may be dynamic. One of the best techniques to scrutinize the actual nature of the quenching mechanism is careful examination of the fluorophore (proteins) UV–vis absorption spectra [63]. The dynamic quenching only affects the excited state of the quenching molecule with no function on the absorption spectrum of quenching substances (fluorophore) [64], whereas a complex of protein and nano-selenium forms in static quenching, so there will be some changes in the UV–vis spectra of proteins [65]. Fig. 18 A, B, and C represent the comparison of the absorption spectra of protein, nano-selenium, the mixture of protein and nano-selenium [protein + nano-selenium], and [protein + nano-selenium]-nano-selenium, obtained by deducting of nano-selenium spectrum from [protein + nano-selenium] spectrum. As can be seen, the intensity of the spectral band of protein (HSA, HHb, and Cyt c) was changed after the addition of nano-selenium (1:1 ratio, 1 mg/mL) and deducting. All the quoted above data oppose the involvement of dynamic quenching in the binding process observed in the nano-selenium-protein system and testify unambiguously that the overall quenching of protein fluorescence by nano-selenium can be regarded as the static quenching mechanism [[66], [67], [68]].
Table 2.
Thermodynamic and binding parameters of the interaction of the SeNPs to HSA.
| T(K) | Ksv (M−1)* | Kq (M−1s−1)* | Ka (M−1)* | n | ΔG0(kJmol−1) | ΔH0(kJmol−1) | ΔS0(Jmol−1 K−1) | |
|---|---|---|---|---|---|---|---|---|
| 288.15 | 3.43 × 104 | 3.43 × 1012 | 3.70 × 107 | 1.92 | −41.30 | 58.35 | 345.82 | |
| 298.15 | 3.57 × 104 | 3.57 × 1012 | 4.86 × 107 | 1.95 | −44.76 | |||
| 310.15 | 3.88 × 104 | 3.88 × 1012 | 2.04 × 108 | 2.12 | −48.91 | |||
M = g/mL.
Table 3.
Thermodynamic and binding parameters of the interaction of the SeNPs to HHb.
| T(K) | Ksv (M−1)* | Kq (M−1s−1)* | Ka (M−1)* | n | ΔG0(kJmol−1) | ΔH0(kJmol−1) | ΔS0(Jmol−1 K−1) | |
|---|---|---|---|---|---|---|---|---|
| 288.15 | 3.36 × 103 | 3.36 × 1011 | 2.68 × 104 | 1.27 | −25.25 | 71.89 | 337.12 | |
| 298.15 | 3.38 × 103 | 3.38 × 1011 | 1.98 × 105 | 1.52 | −28.62 | |||
| 310.15 | 8.66 × 103 | 8.66 × 1011 | 2.33 × 105 | 1.41 | −32.66 | |||
M = g/mL.
Table 4.
Thermodynamic and binding parameters of the interaction of the SeNPs to Cyt c.
| T(K) | Ksv (M−1)* | Kq (M−1s−1)* | Ka (M−1)* | n | ΔG0(KJmol−1) | ΔH0(kJmol−1) | ΔS0(Jmol−1 K−1) | |
|---|---|---|---|---|---|---|---|---|
| 288.15 | 9.02 × 103 | 9.02 × 1011 | 1.19 × 105 | 1.32 | −29.27 | 109.55 | 481.76 | |
| 298.15 | 9.54 × 103 | 9.54 × 1011 | 2.56 × 106 | 1.69 | −34.09 | |||
| 310.15 | 9.56 × 103 | 9.56 × 1011 | 3.22 × 106 | 1.71 | −39.86 | |||
M = g/mL.
Fig. 17.
Stern–Volmer plots, of (A) HSA, (B) HHb and (C) Cyt c for the binding to SeNPs.
Fig. 18.
The UV–vis spectra of (A) HSA, SeNPs, [HSA + SeNPs] and [HSA + SeNPs]-SeNPs (B) HHb, SeNPs, [HHb + SeNPs] and [HHb + SeNPs]-SeNPs (C) Cyt c, [Cyt c + SeNPs] and [Cyt c + SeNPs]-SeNPs at a 1:1 ratio (1 mg/mL).
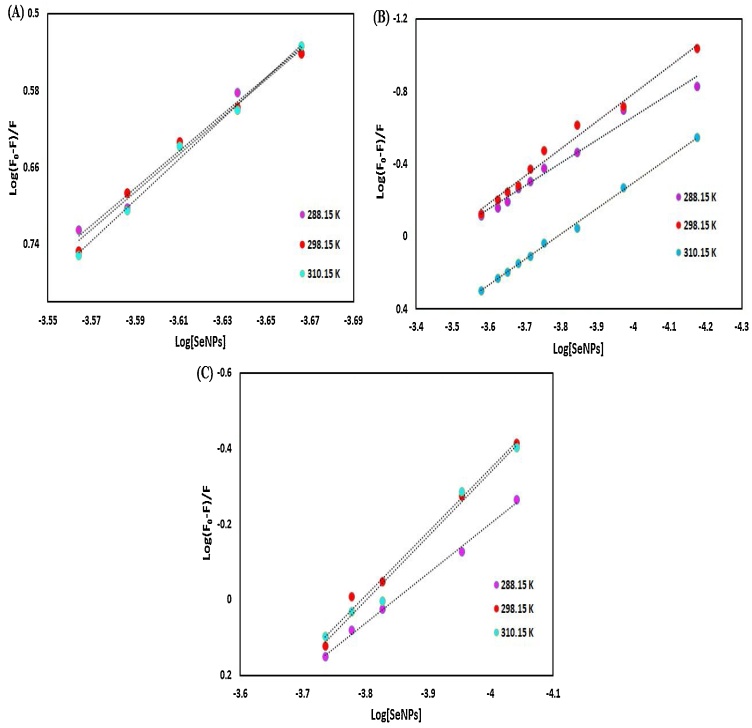
3.4.2.2. Computing the binding constants and number of binding sites
In order to calculate the binding strength (Ka) between nano-selenium and HSA, HHb, and Cyt c and the number of the binding sites per protein (n), the double logarithm regression curve were plotted (Fig. 19) according to the modified Stern-Volmer equation:
| log [(F0 - F)/F] = log Ka+ n log [Q] | (11) |
Fig. 19.
Scatchard plots, of (A) HSA, (B) HHb and (C) Cyt c for the binding to SeNPs.
Tables 2, 3 and 4 exhibit the values of n and Ka at 288.15, 298.15, and 310.15 K, which are obtained from the slope and intercept (the antilog of the y-intercept) of the Scatchard curve (log (Fo–F)/F versus log [Q]) to reveal the distribution of the SeNPs in plasma for HSA, HHb, and Cyt c (Fig. 19A, B and C). It was found that the binding constants increased by increasing temperature and the proteins- SeNPs systems could be more stable at higher temperatures [69]. In other words, the affinity of nano-selenium to proteins increased, which reduced the concentration of free nano-selenium in plasma. It was also revealed that the values of n approximately equal to 1, 1, and 2 for HHb, Cyt c, and HSA, respectively, suggesting that there was one binding site for the nano-selenium in HHb, Cyt c, and there are two binding sites for the nano-selenium in HSA.
3.4.2.3. The nature of binding forces and the thermodynamic parameters of the nano-selenium–proteins interaction process
In order to determine the acting force between nano-selenium and proteins during their interaction, the enthalpy change (ΔH0), entropy change (ΔS0), and free energy (ΔG0) were computed according to the van’t Hoff Eq. (8):
The enthalpy and entropy changes at three temperatures are represented in Tables 2, 3 and 4. These thermodynamic parameters are obtained from the slope and intercept of the linear plots of ln Ka versus 1/T, respectively. Also, Tables 2, 3 and 4 demonstrate the corresponding values of Gibbs free energy change (ΔG°) at 288.15, 298.15, and 310.15 K that computed, using Gibbs–Helmholtz equation Eq. (9):
With regards to the negative value of ΔG°, it can be suggested that the binding processes of nano-selenium-proteins interactions are spontaneous, and the formation of the selenium-proteins systems are exothermic reactions [70]. The computed values of ΔH° and ΔS° for HHb, Cyt c, and HAS are positive and indicated that the interaction of nano-selenium with proteins was governed by hydrophobic attraction [71].
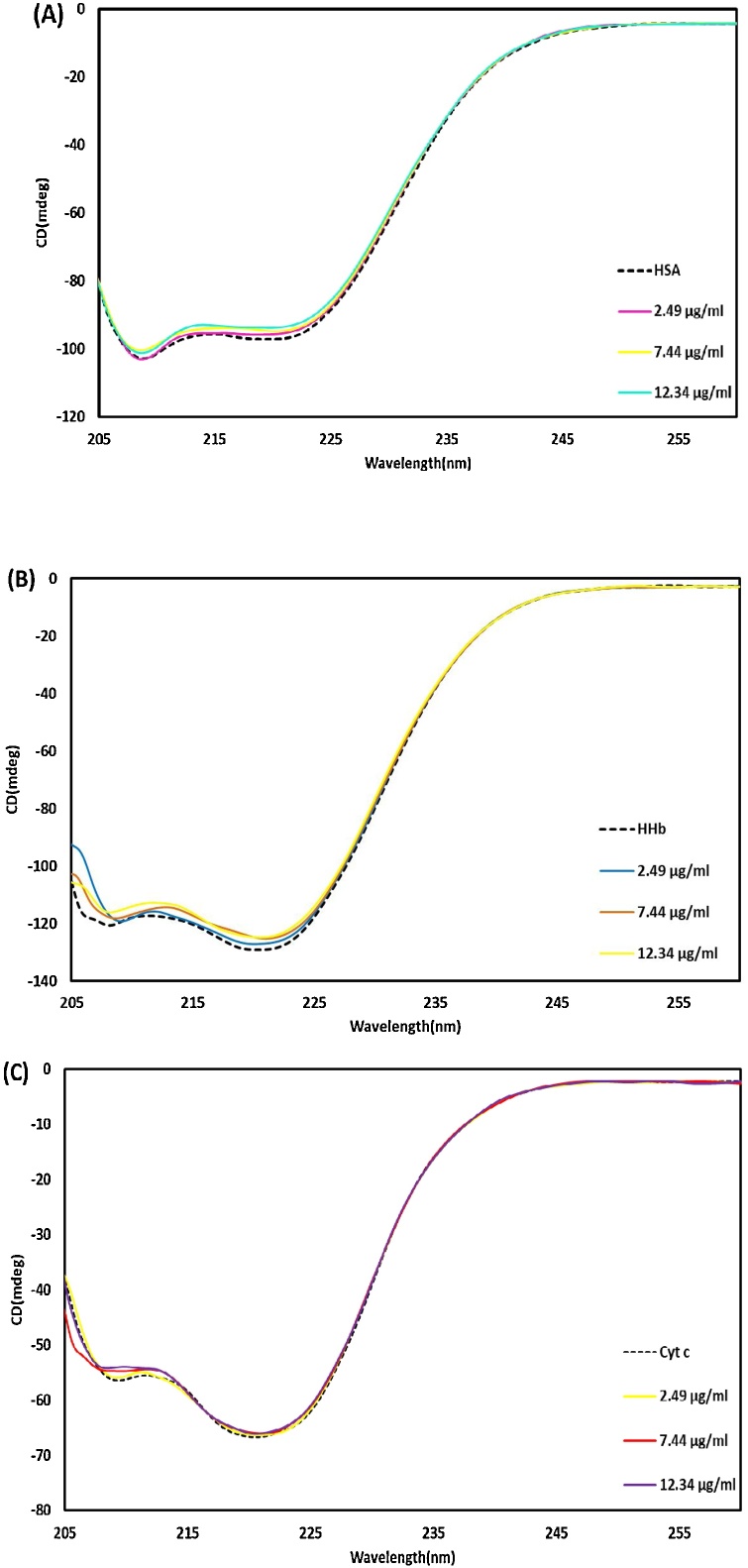
3.4.3. The secondary structural changes (CD spectra)
To investigate any changes in the secondary structures of HSA, HHb, and Cyt c, the UV CD spectrum (190−260 nm) was used, which represents two characteristic peaks at 208 nm and 222 nm. These negative signals arise from the n→π* transition for the peptide bond of α-helix structure. In this research, the UV CD spectra of proteins were noted without and with different nano-selenium concentrations to evaluate the changes in α-helix structures (Fig. 20). The native conformation of HSA, HHb, and Cyt c (secondary structure) does not change on the interaction with nano-selenium. Based on this helpful information, it can be concluded that nano-selenium is a fundamental type of NPs for medicinal applications such as in drug delivery systems without any cytotoxicity effects on biological systems.
Fig. 20.
The UV CD spectra of (A) HSA (5.00 × 10−6M) in the presence of the SeNPs from 2.49×10-6 to 1.23×10-5 g/mL. (B) HHb (5.00 × 10−6M) in the presence of the SeNPs from 2.49×10-6 to 1.23×10-5 g/mL. (C) Cyt c (5.00 × 10−6M) in the presence of the SeNPs from 2.49×10-6 to 1.23×10-5 g/mL.
3.4.4. Docking simulation
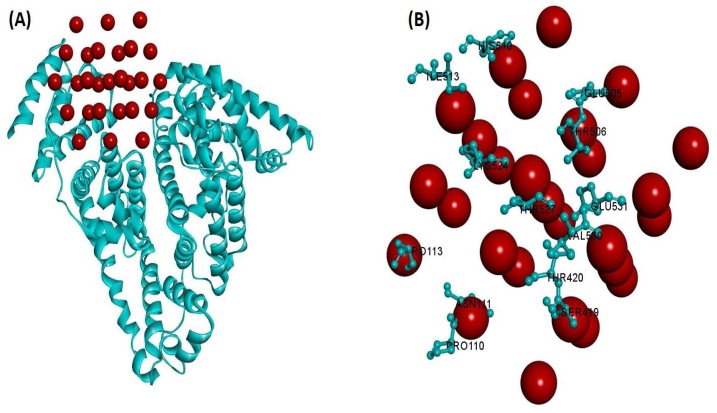
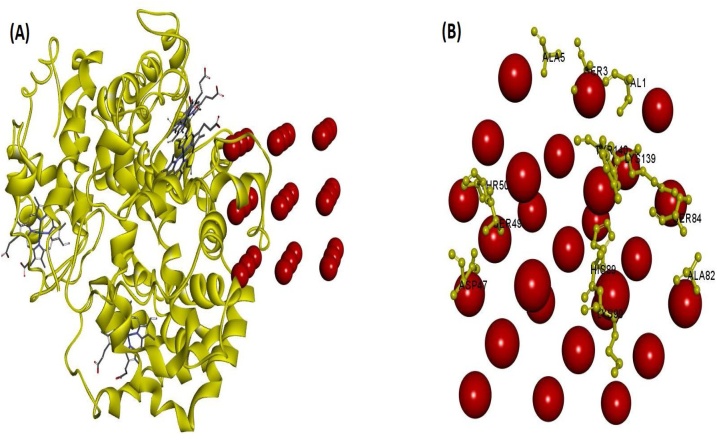
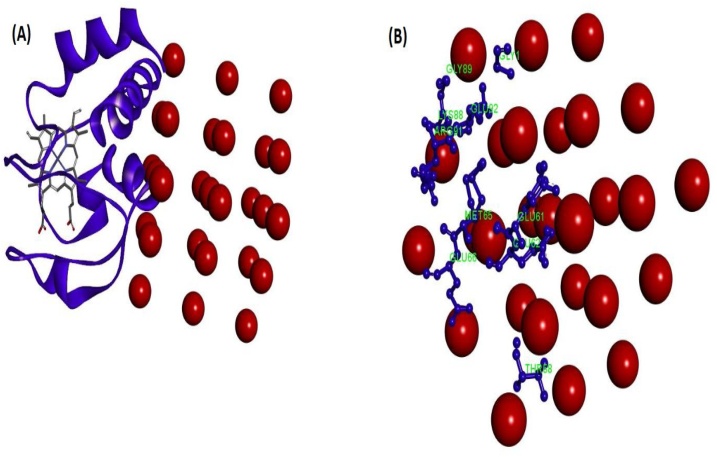
To investigate the SeNPs-proteins interaction by docking analysis, the Patch Dock Server was employed (Figs. 21, 22 and 23 ). Meanwhile, the patch Dock was used to divide the surfaces of SeNPs-proteins into patches based on the surface shape them. The shape of each system and the atomic desolvation energy [72] were further determined by the asset of scoring functions. To overcome the problem of flexibility and scoring of solutions produced by fast rigid-body docking algorithms, the Fire Dock program [73] was used. Monte Carlo minimization of the binding score function is employed to refine the side-chains rearrangement the relative position of the docking partners. Figs. 21B, 22 B, and 23 B demonstrate the amino acid residues of the three proteins on the SeNPs-proteins interaction, respectively. The results suggest that SeNPs interact to HHb through Ser3, Ala5, Val1, Lys139, Tyr140, thr50, Ser49, ser84, Asp47, Hys89, Lys90, and Ala82, to Cyt c through Gly1, Gly89, Lys88, Glu92, Arg91, met95, Glu61, Glu62, Glu66 and Thr58, and to HSA through His510, Ile513 Glu505,Thr506, Lys524, Thr527, Val530, Glu531, Pro113, Thr420, Ser419, Asn111, and Pro110. The order of protein binding was SeNPs -HSA > SeNPs -Cyt c > SeNPs −HHb and Global Energy for the binding of the SeNPs to HHb, Cyt c, and HSA were -19.98, -24.45, and -32.24 KJ/mol, respectively.
Fig. 21.
(A) Molecular docking perspective of SeNPs-HSA, (B) Amino acid residues of HSA surrounding SeNPs.
Fig. 22.
(A) Molecular docking perspective of SeNPs−HHb, (B) Amino acid residues of HHb surrounding SeNPs.
Fig. 23.
(A) Molecular docking perspective of SeNPs-Cyt c, (B) Amino acid residues of Cyt c surrounding SeNPs.
3.5. In vitro antioxidant properties
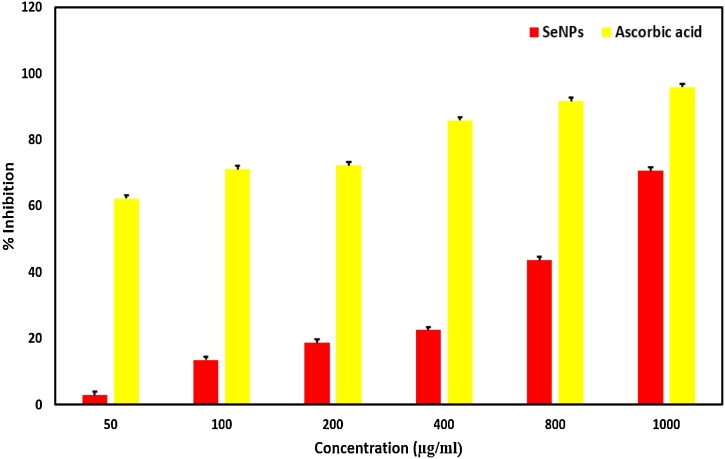
To investigate the in vitro potential of SeNPs to scavenge free radicals, the DPPH free radical scavenging assay was used. It was found that nano-selenium has strong potential (70.65 %) to scavenge the free radicals compared with the standard ascorbic acid (95.9 %) (Fig. 24). It is evident that the potential of synthesized nano-selenium increased with enhanced nano-selenium concentration through a dose-dependent manner and the deep violet color of DPPH mixtures was changed to yellow which indicates that nano-selenium can delete the DPPH free radicals in vitro. So, these SeNPs may be good choice to increase the seleno enzyme, glutathione peroxidas, which plays a leading role in preventing free radicals from damaging cells and tissue in vivo [74]. It has been reported that the nano-selenium demonstrated strong antioxidant potential without any cytotoxicity to normal cells compared to selenium and selenium dioxide [7,75].
Fig. 24.
Antioxidant activity of SeNPs.
4. Conclusion
In this study, the synthesis of SeNPs by a biocompatible reducing agent ascorbic acid was reported and its anticancer and antioxidant activity were investigated. The synthesized SeNPs are well-dispersed spherical in solution with an average diameter of approximately 134 nm. The anticancer activity of SeNPs was evaluated by MTT assay and the results demonstrate that the Nano-selenium has cytotoxicity effects on MCF-7 and Raji Burkitt's lymphoma cancer cell lines. The results of ct-DNA- SeNPs interaction showed that the SeNPs could bind to ct-DNA through partial intercalation binding mode. So the anticancer properties of SeNPs may be related to the interaction with DNA. The interaction of SeNPs with HSA, HHb, and Cyt c indicated that nano-selenium could bind to these proteins without causing alterations in the secondary structure them.
Author statement
Nahid Shahabadi; Project administration
Saba Zendehcheshm; Investigation and Methodology
Fatemeh Khademi; Data curation
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Financial support from Razi University Research Center is gratefully acknowledged.
References
- 1.Peer D., Karp J.M., Hong S., Farokhzad O.C., Margalit R., Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Davis S.S. Biomedical applications of nanotechnology—implications for drug targeting and gene therapy. Trends Biotechnol. 1997;15:217–224. doi: 10.1016/S0167-7799(97)01036-6. [DOI] [PubMed] [Google Scholar]
- 3.Khurana A., Tekula S., Saifi M.A., Venkatesh P., Godugu C. Therapeutic applications of selenium nanoparticles. Biomed. Pharmacother. 2019;111:802–812. doi: 10.1016/j.biopha.2018.12.146. [DOI] [PubMed] [Google Scholar]
- 4.Beckett G.J., Arthur J.R. Selenium and endocrine systems. Int. J. Endocrinol. 2005;184:455–465. doi: 10.1677/joe.1.05971. [DOI] [PubMed] [Google Scholar]
- 5.Fairweather-Tait S.J., Bao Y., Broadley M.R., Collings R., Ford D., Hesketh J.E., Hurst R. Selenium in human health and disease. Antioxid. Redox Signal. 2011;14:1337–1338. doi: 10.1089/ars.2010.3275. [DOI] [PubMed] [Google Scholar]
- 6.Hosnedlova B., Kepinska M., Skalickova S., Fernandez C., Ruttkay-Nedecky B., Peng Q., Baron M., Melcova M., Opatrilova R., Zidkova J., Hosnedlova G., Kepinska B., Skalickova M., Fernandez S., Ruttkay-Nedecky C., Peng B., Baron Q., Melcova M., Opatrilova M., Zidkova R., Bjørklund J. Nano-selenium and its nanomedicine applications: a critical review. Int. J. Nanomed. 2018;13:2107. doi: 10.2147/IJN.S157541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forootanfar H., Adeli-Sardou M., Nikkhoo M., Mehrabani M., Amir-Heidari B., Shahverdi A.R., Shakibaie M. Antioxidant and cytotoxic effect of biologically synthesized selenium nanoparticles in comparison to selenium dioxide. J. Trace Elem. Med. Biol. 2014;28:75–79. doi: 10.1016/j.jtemb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Martínez A., Charlet L. Selenium environmental cycling and bioavailability: a structural chemist point of view. Rev. Environ. Sci. Biotechnol. 2009;8:81–110. [Google Scholar]
- 9.Zhang S.Y., Zhang J., Wang H.Y., Chen H.Y. Synthesis of selenium nanoparticles in the presence of polysaccharides. Mater. Lett. 2004;58:2590–2594. [Google Scholar]
- 10.Lin Z.H., Wang C.C. Evidence on the size-dependent absorption spectral evolution of selenium nanoparticles. Mater. Chem. Phys. 2005;92:591–594. [Google Scholar]
- 11.Ananth A., Keerthika V., Rajan M.R. Synthesis and characterization of nano-selenium and its antibacterial response on some important human pathogens. Curr. Sci. 2019;116:285–290. [Google Scholar]
- 12.Sangeetha Gowda K.R., Mathew B.B., Sudhamani C.N., Naik H.B. Mechanism of DNA binding and cleavage. J. Biomed. Biotechnol. 2014;2:1–9. [Google Scholar]
- 13.Wahab R., Kim Y.S., Hwang I.H., Shin H.S. A non-aqueous synthesis, characterization of zinc oxide nanoparticles and their interaction with DNA. Synth. Met. 2009;159:2443–2452. [Google Scholar]
- 14.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 15.Ezhuthupurakkal P.B., Polaki L.R., Suyavaran A., Subastri A., Sujatha V., Thirunavukkarasu C. Selenium nanoparticles synthesized in aqueous extract of Allium sativum perturbs the structural integrity of Calf thymus DNA through intercalation and groove binding. Mater. Sci. Eng. C. 2017;74:597–608. doi: 10.1016/j.msec.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Cheung-Ong K., Giaever G., Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem. Biol. 2013;20:648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Sarwar T., Rehman S.U., Husain M.A., Ishqi H.M., Tabish M. Interaction of coumarin with calf thymus DNA: deciphering the mode of binding by in vitro studies. Int. J. Biol. Macromol. 2015;73:9–16. doi: 10.1016/j.ijbiomac.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Zhdanov V.P., Cho N.J. Kinetics of the formation of a protein corona around nanoparticles. Math. Biosci. 2016;282:82–90. doi: 10.1016/j.mbs.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Monopoli M.P., Åberg C., Salvati A., Dawson K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 20.Wolfram J., Yang Y., Shen J., Moten A., Chen C., Shen H., Ferrari M., Zhao Y. The nano-plasma interface: implications of the protein corona. Colloids Surf. B. 2014;124:17–24. doi: 10.1016/j.colsurfb.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moradi N., Ashrafi-Kooshk M.R., Ghobadi S., Shahlaei M., Khodarahmi R. Spectroscopic study of drug-binding characteristics of unmodified and pNPA-based acetylated human serum albumin: Does esterase activity affect microenvironment of drug binding sites on the protein? J. Lumin. 2015;160:351–361. [Google Scholar]
- 22.Zhang Y.Z., Zhou B., Liu Y.X., Zhou C.X., Ding X.L., Liu Y. Fluorescence study on the interaction of bovine serum albumin with p-aminoazobenzene. J. Fluoresc. 2008;18:109–118. doi: 10.1007/s10895-007-0247-4. [DOI] [PubMed] [Google Scholar]
- 23.Makarska-Bialokoz M. Analysis of the binding interaction in uric acid-Human hemoglobin system by spectroscopic techniques. Spectrochim. Acta A. 2017;178:47–54. doi: 10.1016/j.saa.2017.01.063. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y.Q., Zhang H.M., Zhou Q.H. Studies on the interaction of caffeine with bovine hemoglobin. Eur. J. Med. Chem. 2009;44:2100–2105. doi: 10.1016/j.ejmech.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Jafari Azad V., Kasravi S., Alizadeh Zeinabad H., Memar Bashi Aval M., Saboury A.A., Rahimi A., Falahati M. Probing the conformational changes and peroxidase-like activity of cytochrome c upon interaction with iron nanoparticles. J. Biomol. Struct. Dyn. 2017;35:2565–2577. doi: 10.1080/07391102.2016.1222972. [DOI] [PubMed] [Google Scholar]
- 26.Margoliash E., Smith E.L., Kreil G., Tuppy H. Amino-acid sequence of horse heart cytochrome c: the complete amino-acid sequence. Nature. 1961;192:1125–1127. doi: 10.1038/1921125a0. [DOI] [PubMed] [Google Scholar]
- 27.Peynshaert K., Manshian B.B., Joris F., Braeckmans K., De Smedt S.C., Demeester J., Soenen S.J. Exploiting intrinsic nanoparticle toxicity: the pros and cons of nanoparticle-induced autophagy in biomedical research. Chem. Rev. 2014;114:7581–7609. doi: 10.1021/cr400372p. [DOI] [PubMed] [Google Scholar]
- 28.Abdelhameed A.S., Alanazi A.M., Bakheit A.H., Darwish H.W., Ghabbour H.A., Darwish I.A. Fluorescence spectroscopic and molecular docking studies of the binding interaction between the new anaplastic lymphoma kinase inhibitor crizotinib and bovine serum albumin. Spectrochim. Acta A. 2017;171:174–182. doi: 10.1016/j.saa.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Schneidman‐Duhovny D., Inbar Y., Polak V., Shatsky M., Halperin I., Benyamini H., Barzilai A., Dror O., Haspel N., Nussinov R. Taking geometry to its edge: fast unbound rigid (and hinge‐bent) docking. Proteins. 2003;52:107–112. doi: 10.1002/prot.10397. [DOI] [PubMed] [Google Scholar]
- 30.Vahdati M., Moghadam T.T. Synthesis and characterization of Selenium nanoparticles-Lysozyme nanohybrid system with synergistic antibacterial properties. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-019-57333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panicker C.Y., Varghese H.T., Philip D. FT-IR, FT-Raman and SERS spectra of Vitamin C. Spectrochim. Acta A. 2006;65:802–804. doi: 10.1016/j.saa.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 32.Xiao L., Li J., Brougham D.F., Fox E.K., Feliu N., Bushmelev A., Schmidt A., Mertens N., Kiessling F., Valldor M., Fadeel B. Water-soluble superparamagnetic magnetite nanoparticles with biocompatible coating for enhanced magnetic resonance imaging. ACS Nano. 2011;5:6315–6324. doi: 10.1021/nn201348s. [DOI] [PubMed] [Google Scholar]
- 33.Sreeja V., Jayaprabha K.N., Joy P.A. Water-dispersible ascorbic-acid-coated magnetite nanoparticles for contrast enhancement in MRI. Appl. Nanosci. 2015;5:435–441. [Google Scholar]
- 34.Tan H.W., Mo H.Y., Lau A.T., Xu Y.M. Selenium species: current status and potentials in cancer prevention and therapy. Int. J. Mol. Sci. 2019;20:75. doi: 10.3390/ijms20010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collery P. Strategies for the development of selenium-based anticancer drugs. J. Trace Elem. Med. Biol. 2018;50:498–507. doi: 10.1016/j.jtemb.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 36.Li T., Xu H. Selenium-containing nanomaterials for Cancer treatment. Cell Rep. Phys. Sci. 2020 [Google Scholar]
- 37.Hassanien R., Abed‐Elmageed A.A., Husein D.Z. Eco‐friendly approach to synthesize selenium nanoparticles: photocatalytic degradation of sunset Yellow Azo Dye and anticancer activity. ChemistrySelect. 2019;4:9018–9026. [Google Scholar]
- 38.Menon S., Shanmugam V. Cytotoxicity analysis of biosynthesized selenium nanoparticles towards A549 lung Cancer cell line. J. Inorg. Organomet. Polym. Mater. 2019:1–13. [Google Scholar]
- 39.Ciniglia C., Pinto G., Sansone C., Pollio A. Acridine orange/Ethidium bromide double staining test: a simple in-vitro assay to detect apoptosis induced by phenolic compounds in plant cells. Allelopathy J. 2010;26:301–308. [Google Scholar]
- 40.Menon S., Ks S.D., Santhiya R., Rajeshkumar S., Kumar V. Selenium nanoparticles: a potent chemotherapeutic agent and an elucidation of its mechanism. Colloids Surf. B. 2018;170:280–292. doi: 10.1016/j.colsurfb.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Arjmand F., Aziz M. Synthesis and characterization of dinuclear macrocyclic cobalt (II), copper (II) and zinc (II) complexes derived from 2, 2, 2′, 2′-S, S [bis (bis-N, N-2-thiobenzimidazolyloxalato-1, 2-ethane)]: DNA binding and cleavage studies. Eur. J. Med. Chem. 2009;44:834–844. doi: 10.1016/j.ejmech.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Shahabadi N., Moeini N. Synthesis, characterization and DNA interaction studies of a new platinum (II) complex containing caffeine and histidine ligands using instrumental and computational methods. J. Coord. Chem. 2015;68:2871–2885. [Google Scholar]
- 43.Sahoo B.K., Ghosh K.S., Dasgupta S. Molecular interactions of isoxazolcurcumin with human serum albumin: spectroscopic and molecular modeling studies. Biopolymers. 2009;91:108–119. doi: 10.1002/bip.21092. [DOI] [PubMed] [Google Scholar]
- 44.Kandagal P.B., Shaikh S.M.T., Manjunatha D.H., Seetharamappa J., Nagaralli B.S. Spectroscopic studies on the binding of bioactive phenothiazine compounds to human serum albumin. J. Photochem. Photobiol. 2007;189:121–127. [Google Scholar]
- 45.Dufour C., Dangles O. Flavonoid–serum albumin complexation: determination of binding constants and binding sites by fluorescence spectroscopy. Biochim. Biophys. Acta. 2005;1721:164–173. doi: 10.1016/j.bbagen.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Izumrudov V.A., Zhiryakova M.V., Goulko A.A. Ethidium bromide as a promising probe for studying DNA interaction with cationic amphiphiles and stability of the resulting complexes. Langmuir. 2002;18:10348–10356. [Google Scholar]
- 47.Sun Y., Zhang H., Bi S., Zhou X., Wang L., Yan Y. Studies on the arctiin and its interaction with DNA by spectral methods. J. Lumin. 2011;131:2299–2306. [Google Scholar]
- 48.Westerlund F., Nordell P., Nordén B., Lincoln P. Kinetic characterization of an extremely slow DNA binding equilibrium. J. Phys. Chem. B. 2007;111:9132–9137. doi: 10.1021/jp072126p. [DOI] [PubMed] [Google Scholar]
- 49.Shahabadi N., Shiri F. Multispectroscopic studies on the interaction of a copper (ii) complex of ibuprofen drug with calf thymus DNA. Nucleosides Nucleotides Nucleic Acids. 2017;36:83–106. doi: 10.1080/15257770.2016.1223305. [DOI] [PubMed] [Google Scholar]
- 50.He W., Dou H., Li Z., Wang X., Wang L., Wang R., Chang J. Investigation of the interaction between five alkaloids and human hemoglobin by fluorescence spectroscopy and molecular modeling. Spectrochim. Acta A. 2014;123:176–186. doi: 10.1016/j.saa.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 51.Luz R.A., Crespilho F.N. Gold nanoparticle-mediated electron transfer of cytochrome c on a self-assembled surface. RSC Adv. 2016;6:62585–62593. [Google Scholar]
- 52.Šimšíková M., Antalík M., Kaňuchová M., Škvarla J. Cytochrome c conjugated to ZnO–MAA nanoparticles: the study of interaction and influence on protein structure. Int. J. Biol. Macromol. 2013;59:235–241. doi: 10.1016/j.ijbiomac.2013.04.064. [DOI] [PubMed] [Google Scholar]
- 53.Manjushree M., Revanasiddappa H.D. Interpretation of the binding interaction between bupropion hydrochloride with human serum albumin: a collective spectroscopic and computational approach. Spectrochim. Acta A. 2019;209:264–273. doi: 10.1016/j.saa.2018.10.047. [DOI] [PubMed] [Google Scholar]
- 54.Maji A., Beg M., Das S., Aktara M.N., Nayim S., Patra A., Islam M.M., Hossain M. Study on the antibacterial activity and interaction with human serum albumin of Tagetes erecta inspired biogenic silver nanoparticles. Process Biochem. 2020;97:191–200. [Google Scholar]
- 55.Maji A., Beg M., Mandal A.K., Das S., Jha P.K., Hossain M. Study of the interaction of human serum albumin with Alstonia scholaris leaf extract-mediated silver nanoparticles having bactericidal property. Process Biochem. 2017;60:59–66. [Google Scholar]
- 56.Yasrebi S.A., Takjoo R., Riazi G.H., Mague J.T. Synthesis and HSA-interaction of a new mixed ligand Cu-isothiosemicarbazonato complex with adenine nucleobase. Polyhedron. 2020;179 [Google Scholar]
- 57.Sekar G., Kandiyil S.T., Sivakumar A., Mukherjee A., Chandrasekaran N. Binding studies of hydroxylated multi-walled carbon nanotubes to hemoglobin, gamma globulin and transferrin. J. Photochem. Photobiol. B, Biol. 2015;153:222–232. doi: 10.1016/j.jphotobiol.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 58.Wu Q., Zhao H., Chen X., Cai Z. Interaction of bisphenol A 3, 4-quinone metabolite with human hemoglobin, human serum albumin and cytochrome c in vitro. Chemosphere. 2019;220:930–936. doi: 10.1016/j.chemosphere.2018.12.194. [DOI] [PubMed] [Google Scholar]
- 59.Li Z., Shang L.P., Deng H., Zhi T.X. Extraction of characteristic parameters of three-dimensional fluorescence spectra of tyrosine and tryptophan. Spectrosc. Spect. Anal. 2009;29:1925–1928. [PubMed] [Google Scholar]
- 60.Gorinstein S., Goshev I., Moncheva S., Zemser M., Weisz M., Caspi A., Libman I., Lerner H.T., Trakhtenberg S., Martín-Belloso O. Intrinsic tryptophan fluorescence of human serum proteins and related conformational changes. Int. J. Pharm. 2000;19:637–642. doi: 10.1023/a:1007192017291. [DOI] [PubMed] [Google Scholar]
- 61.Zolghadri S., Saboury A.A., Amin E., Moosavi-Movahedi A.A. A spectroscopic study on the interaction between ferric oxide nanoparticles and human hemoglobin. J. Iran. Chem. Soc. 2010;7:S145–S153. [Google Scholar]
- 62.Zhou J., Wu X., Gu X., Zhou L., Song K., Wei S., Feng Y., Shen J. Spectroscopic studies on the interaction of hypocrellin A and hemoglobin. Spectrochim. Acta A. 2009;72:151–155. doi: 10.1016/j.saa.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 63.Makarska-Bialokoz M., Borowski P. Fluorescence quenching behaviour of uric acid interacting with water-soluble cationic porphyrin. J. Lumin. 2015;160:110–118. [Google Scholar]
- 64.Grigoryan K.R., Ghazaryan A.G. Quenching mechanism of human serum albumin fluorescence by gangleron. Chem. Biol. 2013;2:6–10. [Google Scholar]
- 65.Rasoulzadeh F., Asgari D., Naseri A., Rashidi M.R. Spectroscopic studies on the interaction between erlotinib hydrochloride and bovine serum albumin. Daru. 2010;18:179. [PMC free article] [PubMed] [Google Scholar]
- 66.Shahabadi N., Zendehcheshm S. Evaluation of ct-DNA and HSA binding propensity of antibacterial drug chloroxine: multi-spectroscopic analysis, atomic force microscopy and docking simulation. Spectrochim. Acta A. 2020;230 doi: 10.1016/j.saa.2020.118042. [DOI] [PubMed] [Google Scholar]
- 67.Shahabadi N., Hadidi S. Molecular modeling and spectroscopic studies on the interaction of the chiral drug venlafaxine hydrochloride with bovine serum albumin. Spectrochim. Acta A. 2014;122:100–106. doi: 10.1016/j.saa.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 68.Sekar G., Haldar M., Kumar D.T., Doss C.G.P., Mukherjee A., Chandrasekaran N. Exploring the interaction between iron oxide nanoparticles (IONPs) and human serum albumin (HSA): spectroscopic and docking studies. J. Mol. Liq. 2017;241:793–800. [Google Scholar]
- 69.Kıpçak F., Tok T.T., Duyar H., Seferoğlu Z., Gökoğlu E. Synthesis of new morpholine containing 3-amido-9-ethylcarbazole derivative and studies on its biophysical interactions with calf thymus DNA/HSA. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1734093. [DOI] [PubMed] [Google Scholar]
- 70.Sandhya B., Hegde A.H., Kalanur S.S., Katrahalli U., Seetharamappa J. Interaction of triprolidine hydrochloride with serum albumins: thermodynamic and binding characteristics, and influence of site probes. J. Pharm. Biomed. 2011;54:1180–1186. doi: 10.1016/j.jpba.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 71.Sun Y., Zhang H., Bi S., Zhou X., Wang L., Yan Y. Studies on the arctiin and its interaction with DNA by spectral methods. J. Lumin. 2011;131:2299–2306. [Google Scholar]
- 72.Balakrishnan G., Rajendran T., Murugan K.S., Ganesan M., Sivasubramanian V.K., Rajagopal S. Synthesis, photophysics and the binding studies of rhenium (I) diimine surfactant complexes with serum albumins: a spectroscopic and docking study approach. J. Lumin. 2019;205:51–60. [Google Scholar]
- 73.Andrusier N., Nussinov R., Wolfson H.J. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 74.Zhang J., Zhang S.Y., Xu J.J., Chen H.Y. A new method for the synthesis of selenium nanoparticles and the application to construction of H2O2 biosensor. Chin. Chem. Lett. 2004;15:1345–1348. [Google Scholar]
- 75.Shakibaie M., Khorramizadeh M.R., Faramarzi M.A., Sabzevari O., Shahverdi A.R. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase‐2 expression. Biotechnol. Appl. Biochem. 2010;56:7–15. doi: 10.1042/BA20100042. [DOI] [PubMed] [Google Scholar]