Highlights
-
•
We describe a deep learning approach for thalamic measurement on clinical T2-FLAIR.
-
•
The algorithm performs comparably to FSL FIRST on high resolution T1.
-
•
Clinical relevance on disability and cognition is confirmed.
Keywords: Thalamus volume, Thalamic atrophy, Artificial intelligence, Multiple sclerosis
Abstract
Background
Thalamic volume loss is a key marker of neurodegeneration in multiple sclerosis (MS). T2-FLAIR MRI is a common denominator in clinical routine MS imaging, but current methods for thalamic volumetry are not applicable to it.
Objective
To develop and validate a robust algorithm to measure thalamic volume using clinical routine T2-FLAIR MRI.
Methods
A dual-stage deep learning approach based on 3D U-net (DeepGRAI – Deep Gray Rating via Artificial Intelligence) was created and trained/validated/tested on 4,590 MRI exams (4288 2D-FLAIR, 302 3D-FLAIR) from 59 centers (80/10/10 train/validation/test split). As training/test targets, FIRST was used to generate thalamic masks from 3D T1 images. Masks were reviewed, corrected, and aligned into T2-FLAIR space. Additional validation was performed to assess inter-scanner reliability (177 subjects at 1.5 T and 3 T within one week) and scan-rescan-reliability (5 subjects scanned, repositioned, and then re-scanned). A longitudinal dataset including assessment of disability and cognition was used to evaluate the predictive value of the approach.
Results
DeepGRAI automatically quantified thalamic volume in approximately 7 s per case, and has been made publicly available. Accuracy on T2-FLAIR relative to 3D T1 FIRST was 99.4% (r = 0.94, p < 0.001,TPR = 93.0%, FPR = 0.3%). Inter-scanner error was 3.21%. Scan-rescan error with repositioning was 0.43%. DeepGRAI-derived thalamic volume was associated with disability (r = -0.427,p < 0.001) and cognition (r = -0.537,p < 0.001), and was a significant predictor of longitudinal cognitive decline (R2 = 0.081, p = 0.024; comparatively, FIRST-derived volume was R2 = 0.080, p = 0.025).
Conclusions
DeepGRAI provides fast, reliable, and clinically relevant thalamic volume measurement on multicenter clinical-quality T2-FLAIR images. This indicates potential for real-world thalamic volumetry, as well as quantification on legacy datasets without 3D T1 imaging.
1. Introduction
Multiple sclerosis (MS) is a chronic, autoimmune disease of the central nervous system (CNS) characterized by focal and diffuse inflammation and axonal loss. (Frohman et al., 2006). Over the past two decades, imaging biomarkers of lesion burden and longitudinal lesion activity have been a vital part of MS clinical management and clinical trials, and in the last decade, measurement of brain atrophy has been recognized as an equally important biomarker.
More recently, the measurement of thalamic atrophy has emerged as a potentially sensitive and specific means to measure neurodegeneration, and has been shown to be highly related to both disability and cognition. (Houtchens et al., 2007, Batista et al., 2012, Minagar et al., 2013, Zivadinov et al., 2013b, Bergsland et al., 2016, Azevedo et al., 2018, Bisecco et al., 2019) Thalamic pathology has been shown in all MS disease types and has been suggested as an outcome in MS clinical trials because of its meaningful change, clinical relevance, early detectability, and clear delineation along its inner border. (Minagar et al., 2013, Zivadinov et al., 2016) A recent large multi-center study corroborated the clinical importance and neurodegenerative vulnerability of the thalamus in patients with MS (Eshaghi et al., 2018). Additionally, the thalamic atrophy rate is consistent throughout the entire disease duration, making it a convenient MRI biomarker to be used in clinical and research monitoring of MS patients. (Minagar et al., 2013, Azevedo et al., 2018)
However, despite its established value, quantitative assessment of thalamic atrophy is not part of standard clinical routine monitoring of MS patients, (Rovira et al., 2015, Zivadinov et al., 2016, Rocca et al., 2017), nor is it widely applied to large, real-world clinical datasets. (Zivadinov et al., 2017, Zivadinov et al., 2018) This is at least in part due to technical limitations. Although precise and reliable tools like FIRST (Patenaude et al., 2011), FreeSurfer (Fischl, 2012), and others are available to measure the thalamus, they are primarily applicable to 3D T1-weighted scans on stable scanners and protocols without hardware, software, or coil changes. Such image sets are common in research, but in many cases are not part of clinical routine, where scans often lack standardization of MRI parameters like repetition time, echo time, and acquisition matrix, and where frequent uncontrolled scanner upgrades further compound the problem. (Wattjes et al., 2015, Zivadinov et al., 2016, Zivadinov et al., 2018, Rocca et al., 2017). Therefore, although thalamic atrophy measurement remains valuable and feasible for academic centers and clinics with tight control of their MRI acquisition, it leaves behind many other clinical centers and their datasets.
This need is underscored by recent results from the Multiple Sclerosis and clinical outcome and MRI in the US (MS-MRIUS) study, a multicenter, observational, real-world investigation to assess brain MRI changes and disease progression in MS clinical that included ~ 600 relapsing-remitting MS patients across 33 participating sites. The results confirmed that that nearly every clinical routine MRI exam (99.8%) included T2-fluid attenuated inversion recovery (FLAIR), but<80% of exams had two dimensional (2D)-T1, and<40% of exams had the type of three dimensional (3D)-T1 required for standard thalamic atrophy analysis. (Zivadinov et al., 2018) Furthermore, the MS-MRIUS study found that imaging hardware or software changed in 50% of longitudinal scans pairs, remained consistent in only 30%, and was unknown in 15%. Based on this, a quantitative thalamic volume measurement tool capable of working on T2-FLAIR alone and robust to changes would greatly help the translation of thalamic volumetry to broader clinical research. Such a tool could potentially be provided by deep learning, using a domain transfer approach.
Deep convolutional neural networks (CNNs), which operate hierarchically like the human visual system and replace ad-hoc feature engineering with self-learning approaches, have enjoyed dramatic success when large amounts of training data are available, and have been transformative in the field of computer vision. (Rawat and Wang, 2017) These methods work by training a set of kernels, or filters, to respond to specific features in their input (much like the human visual system includes separate set of neurons responding to horizontal or vertical lines). Conceptually, these kernels are then scanned across input images to create a set of filter-specific response maps (one for each filter). Initial filters’ inputs are connected directly to the input image, and later (hidden layer) kernels’ inputs are connected to the response maps from prior levels, allowing for a hierarchy of semantic understanding building from simple features up to complex object recognition roughly analogous to the ventral pathway in human vision. At each transition to the next kernel set (increasing depth, similar to higher-order neurons), the spatial resolution is generally traded away for better semantic understanding. At earlier levels, the network “knows” relatively basic aspects about very specific points in space (e.g., “there is a horizontal edge at this specific voxel). At later levels, the network “knows” many more aspects (“features”) about broader areas of the input image (e.g., “there is a structure here with multiple vertical edges surrounded by a series of dark regions”). At the deepest levels, the network can recognize specific objects, with particular kernels responding to individual objects (such as thalami).
Again like the human visual system, the correct functioning of these networks depends on the proper tuning of input weights (like synaptic potentiation) to ensure that filters (neurons) only respond when they ought to. Unlike the human brain, though, these networks can be trained in a direct, relatively straightforward manner via the method of backpropagation. In this process, many examples of input images are run through the network, and the actual network outputs are compared to known correct outputs. Deviations from correct output are computed, and the sources of these deviations are propagated back through the network to adjust individual filter input weightings in the direction (up or down) that improves the output. This is a slow process, usually requiring many training examples to avoid overfitting and very small adjustment steps (learning rate) to ensure convergence. Once trained, though, such networks are capable of making predictions on previously unseen input images.
Going beyond just object recognition, CNNs with some modifications, such as U-Net, are also capable of segmentation (or delineation) of image regions/objects, which can in turn be used for quantitative volumetry. These “semantic segmentation” approaches have demonstrated high performance in many medical imaging applications, in many cases improving accuracy or precision beyond the prior state of the art, or substantially improve speed. (Henschel et al., 2020) However, it is important to note that deep learning tools can have another application: allowing domain transfer of existing tools to new datasets. By training a deep learning system to replicate the output of an existing system while using only partial or inferior data, the current state of the art can potentially be transferred to this new input domain.
Against this background, we sought to develop a semantic segmentation CNN architecture to transfer the current widely used and broadly validated FIRST thalamic segmentation on high-resolution 3D T1 scans to clinical routine T2-FLAIR MRI scans. The resulting tool, called DeepGRAI (Deep Gray Rating via Artificial Intelligence), can be readily used for real-world thalamic volume monitoring, as well as for quantification on large legacy datasets lacking research-quality MRI. In particular, we targeted the method to be suitable for widespread use on heterogeneous datasets from multiple sites, scanners, and protocols by: 1) developing a robust algorithm architecture, 2) training the classifier on a diverse dataset, 3) validating the results with independent test, clinical, scan-rescan, and inters-canner datasets, and 4) providing the classifier as an open and easily usable tool. Directly deployable docker images are available on DockerHub: https://hub.docker.com/r/buffaloneuroimaging/deepgrai.
2. Methods
2.1. Deep learning model architecture – DeepGRAI CNN
Based on preliminary experiments and established performance, we adopted a 3D U-Net semantic segmentation architecture for DeepGRAI. (Çiçek et al., 2016) The U-Net architecture is composed of two symmetric pathways – a descending contraction/encoding pathway which moves from low level voxel intensities at very high spatial resolution to abstract high-level features at coarse spatial resolution, and an ascending expansion/decoding pathway which moves from less localized high-level features to specific voxel classifications. Additionally, U-Net includes direct “skip” connections from the descending pathway into the ascending pathway, to provide the ascending pathway with low-level image feature information. In the current context, the descending pathway is essentially responsible for localizing the thalamus, and the ascending pathway is responsible for determining its precise borders.
One major issue with 3D U-Net processing is that it is computationally expensive for biomedical imaging. Brain MRI images in particular are generally on the order of 256x256x128 voxels. This leads to substantial limitations in GPU memory and potentially results in the need for training batch sizes of one. This has been addressed in many systems via a “pseudo-3D” approach, in which 3 2D classifiers are trained to separately segment axial, coronal, and sagittal slices. (Henschel et al., 2020) However, because the thalamus is a relatively small structure and because we sought to solve a single-label rather than multi-label problem, we sought to retain the benefits of a fully 3D approach. To accomplish this, we implemented a dual-stage architecture with a low-resolution stage and a high-resolution stage. The initial stage is designed to be responsible only for localizing the thalamus in order to provide a bounding box for the second stage and can therefore be trained independently to maximize resources and batch size.
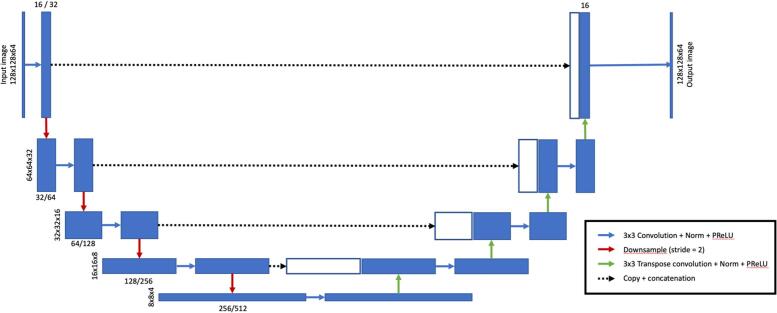
The precise architecture employed is illustrated in Fig. 1. Each of stages 1 and 2 are implemented as a 3D U-Net CNN in the PyTorch deep learning platform for tensor computation and automated gradient differentiation (https://pytorch.org), with an input size of 128x128x64. For stage 1, convolutional kernels are 3x3x3 in the spatial dimension, with the number of filters doubling at each network layer from 16 initial features to 256 features at 8x8x4 spatial resolution over 4 tiers of downsampling/upsampling operations. Stage 2 is identical except for the number of filters, which increase from 32 to 512. Non-linearities between layers are introduced via parametric rectified linear units (PReLUs), which are similar to traditional ReLUs but incorporate a small, trainable gradient in the non-active state to avoid “dying ReLUs” during training. (He et al., 2015) For the initial stage, the input image is downsampled to 2 mm isotropic voxels covering a physical volume of 256x256x128 mm, and the network is used to identify the center of mass of the resulting low-resolution thalamic probability map. This output is used to confidently isolate and resample a 0.5 mm isotropic 64x64x32mm volume of the original image centered on the thalamus, which is then fed into the final segmentation stage. This stage (stage 2) produces a probabilistic segmentation map of the thalamus that can be directly used for volumetry and/or additional analysis. The medical open network for AI (MONAI) framework was used for implementation to improve reproducibility and extendibility (https://monai.io/).
Fig. 1.
Proposed 3D U-Net DeepGRAI architecture. Our final model employed two semi-independent convolutional neural network (CNN) stages, each based on the 3D U-Net architecture shown here. Each of the two stages differs only in number of filters, with stage 2 having twice the filters of stage 1 (e.g., 16 / 32 in the figure incidates 16 filters in stage 1 and 32 filters in stage 2.
2.2. Model training
2.2.1. Training data augmentation
Because we sought to use a fully 3D architecture, each source image could only contribute one training case (in contrast to patch-based or slice-based methods, which can contribute many semi-independent training samples per subject). Therefore, to improve the robustness of the training described below, a number of data augmentation steps were employed to procedurally expand the training set. (Shorten and Khoshgoftaar, 2019) The specific transformations we applied were: random lateral flips along the left–right axis, small random translations along all three axes, small random rotations around all three axes, small random gamma and logarithmic intensity transforms, minor corruption with independent Gaussian noise. For stage two, in order to reduce the dependence on the exact field of view localized from stage one, we also included an additional intermediate augmentation randomizing the cropping location by approximately 1.5 cm in each direction.
2.2.2. Training procedure
Training was performed on an in-house AI GPU server machine with dual NVidia Titan XP GPUs (3,840 parallel CUDA cores and 16 GB on-board RAM per GPU), 128 GB RAM, and dual 10-core Intel Core i9 3.3 GHz CPUs. Each model stage was trained for 500 epochs on the training dataset (described below). Xavier initialization (Glorot and Bengio, 2010) was used to set random initial model weights based on number of inputs/outputs, in order to avoid early saturation. Weights were updated using the Adam optimizer (Kingma and Ba, 2017), which tracks the first and second moments of each individual parameter, and adapts the global learning rate to that specific parameter. An initial learning rate of 1.0e-4 was used, with exponential weight decay parameter of 1.0e-7. Additionally, to prevent stagnation, an adaptive method to reduce the learning rate by a factor of two on learning plateaus was employed (PyTorch’s ReduceLROnPlateau). For all stages, a batch size of 16 was used. As described above, each stage was trained independently. The loss function used was soft Dice. This computes the loss as the intersection of the proposed and target volumes over their union, while preserving smooth gradients to allow differentiability and backpropagation learning during the training phase. (Fidon et al., 2018). During training, the validation set (described below) was used to optimize training hyperparameters and to refine the model architecture.
2.2.3. Datasets and analyses
For this study, a number of de-identified, retrospective datasets were used. Demographic and clinical characteristics of the different datasets used in the study are reported in Table 1, while the MRI acquisition characteristics of these datasets are reported in the Table 2. The Institutional Review Board of the University of Buffalo approved the use of all multi-center de-identified datasets.
Table 1.
Demographic and clinical characteristics of four different datasets used for the various analyses.
| Demographic and clinical characteristics | Training/validation/testing MS datasets (n = 1,672) | Scan rescan dataset MS HC (n = 3) (n = 2) | Inter-scanner dataset MS HC (n = 125) (n = 52) | Clinical MS dataset (n = 49) | ||
|---|---|---|---|---|---|---|
| Number of females, n (%) | 1174 (71) | 2 (66.7) | 2 (1 0 0) | 90 (72) | 36 (69.2) | 37 (77.8) |
| Age in years, mean (SD) | 41.2 (13.8) | 32 (9.2) | 30 (1.8) | 42.9 (11.5) | 39.5 (9.4) | 30.7 (7.9) |
| Disease duration in years, mean (SD) | 12.3 (13.2) | 6.8 (9.3) | NA | 11.4 (9.8) | NA | 4.9 (5.2) |
| EDSS, median (IQR) | 2.5 (1.0–6.0) | 1.5 (0) | NA | 2.5 (2.0–4.5) | NA | 2.0 (1.5–5.0) |
| Disease subtype, mean (SD) RR SP PP |
923 (55.2) 507 (30.3) 242 (14.5) |
3 (1 0 0) |
NA |
77 (61.6) 35 (28) 13 (10.4) |
NA |
36 (73.4) 13 (26.6) 0 (0) |
| T2 lesion volume, mean (SD) | 14.1 (17.5) | 7.2 (2.1) | 0 (0) | 12.3 (12.5) | 0.2 (0) | 14.4 (17.2) |
| DeepGRAI volume, mean (SD) | 14.0 (1.6) | 15.6 (1.5) | 16.9 (1.2) | 13.2 (2.1) | 15.9 (1.5) | 14.1 (2) |
| FIRST volume, mean (SD) | 14.4 (1.8) | 15.7 (1.4) | 17.1 (1) | 13.0 (2) | 16.0 (1.4) | 14 (2.1) |
Legend: MS – multiple sclerosis; HC – healthy controls; SD – standard deviation; NA – not available; EDSS – Expanded Disability Status Scale; IQR-interquartile range; RR – relapsing-remitting; SP – secondary progressive; PP – primary progressive.
The volumes are expressed in milliliters.
Table 2.
T2-FLAIR MRI characteristics of the datasets used for analyses.
| Number of scanners | Training/validation/testing MS dataset (n = 4,590) |
Scan rescan dataset (n = 5) |
Inter-scanner dataset (n = 177) |
Clinical MS dataset (n = 49) |
||
|---|---|---|---|---|---|---|
| 2D FLAIR (n = 4288) |
3D FLAIR (n = 302) |
|||||
| 54 | 5 | 1 | 2 | 1 | ||
| TR, mean (SD) ms | 8631.5 (2336.3) | 4825.0 (1733.6) | 8500.0 (0.0) | 8002.0 (0.0) | 8500.0 (0.0) | 11000.0(0.0) |
| TE, mean (SD) ms | 124.9 (68.5) | 368.2 (93.5) | 122.0 (0.15) | 61.0 (0.12) | 122.0 (0.15) | 140.0 (0.0) |
| TI, mean (SD), ms | 2167.9 (432.6) | 1682.3 (337.8) | 2100.0 (0) | 2000.0 (0) | 2100.0 (0) | 2600.0 (0.0) |
| Slice thickness mean (SD), [min max], mm | 3.3 (1.12) [1.0 6.0] | 1.28 (0.8) [1.0 2.0] | 3.0 (0.0) [3.0 3.0] | 3.0 (0.0) [3.0 3.0] | 3.0 (0.0) [3.0 3.0] | 3.0 (0.0) [3.0 3.0] |
| Axial n (%) / Sagittal n (%) | 89 (82%) / 20 (18%) | 236 (78%) / 66 (22%) | 12 (100%) / 0 (0%) | 201 (100%) / 0 (0%) | 201 (100%) / 0 (0%) | 1725 (100%) / 0 (0%) |
Legend: MS – multiple sclerosis; SD – standard deviation; TR-repetition time; TE-echo time; TI-inversion time; mm-millimeters; ms-milliseconds.
2.2.4. Training/validation/testing with target segmentations/volumes
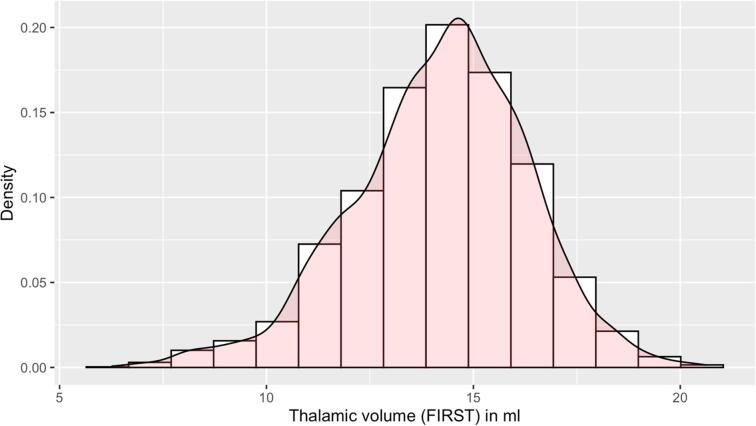
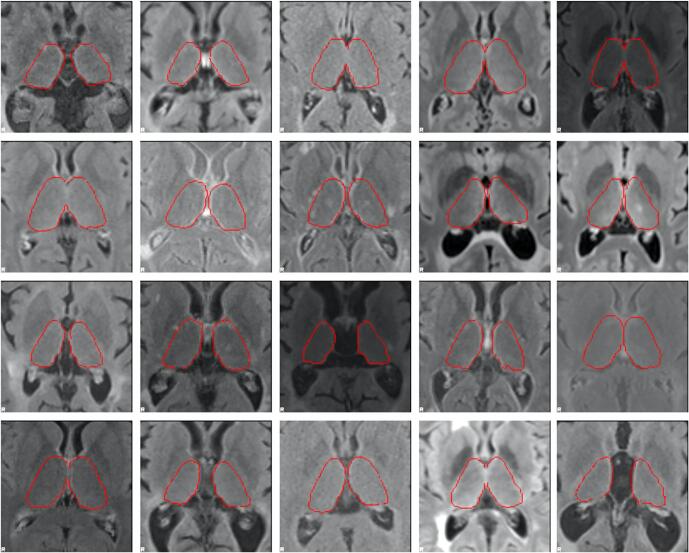
As the goal of this work was to produce a robust segmentation tool, the primary dataset we used for training, validation, and testing of the DeepGRAI model was assembled from multiple centers, scanner field strengths, and imaging protocols. This was a multi-spectral dataset consisting of 3D T1w images and 2D low-resolution T2-FLAIR images from 54 MRI scanners, 1,463 subjects, and 4,288 MRI exams. Because 3D-FLAIR is becoming more widely used (including as a recommendation of the MAGNIMS group - (Rovira et al., 2015)), we also included an additional 302 exams with 3D-FLAIR from 209 subjects across 5 scanners, bringing the total to 59 MRI scanners, 1,672 subjects, and 4,590 MRI exams. The included cases covered a broad range of acquisition types from different acquisition protocols, anatomical variants, and thalamic atrophy levels, as shown in Table 1, Table 2 and Fig. 2. Fig. 3 displays a representative sample of the diversity of image contrasts and thalamic atrophy levels. To determine ground truth for both training and testing, we began by applying FMRIB's Integrated Registration and Segmentation Tool (FIRST) deep GM segmentation tool (Patenaude et al., 2011) to create thalamic initial maps on inpainted 3D T1w images. Inpainting was performed based on co-registration of previously created T2 lesion maps, and used an in-house tool similar to that in FSL’s toolkit (Zivadinov et al., 2013a, Popescu et al., 2014). Because FIRST itself can result in segmentation errors on broad subsets of images, despite its common application in the field, we also conducted manual correction of cases by trained operators. This involved correction of common errors such as third ventricular inclusion, fornix, cavum septum pellucidum, lateral ventricles, and corpus callosum, as recently reported. (Lyman et al., 2020) After correction, we co-registered the 3D T1w images to the individual subjects’ T2-FLAIR images. This was performed using 6 degree of freedom (rigid body) alignment with FSL’s FLIRT tool. The resulting transform matrices were then used to bring the thalamic segmentation maps into the T2-FLAIR space.
Fig. 2.
Histogram of thalamic volumes (in ml) from the training and testing dataset.
Fig. 3.
Representative samples of the wide variety of underlying T2-FLAIR contrasts in real-world MRI acquisitions, as well as the broad range of thalamic atrophy levels in multiple sclerosis. Aligned FIRST segmentations derived from 3D T1-weighted images overlaid in red outlines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
After creation, this dataset was split into training (approx. 80%), validation (approx. 10%), and testing (approx. 10%) sets. Splitting was stratified at the site level, such that training, validation, and test sets did not have overlapping site data. Actual training was conducted on the training set. Model performance was regularly checked against the validation set, and the results used to inform selection of hyperparameters. The final testing dataset was held out entirely and used to assess the performance of the final model, as described below.
2.3. Statistical analyses and validation measures
2.3.1. Accuracy and agreement with manually corrected FIRST-derived volumes
Accuracy with respect to “ground truth” segmentation (FIRST with potential corrections) was assessed on the 10% of the main dataset that was fully withheld (459 exams from 13 scanners) (Table 1, Table 2). DeepGRAI was run independently on the corresponding T2-FLAIR images of this validation dataset, and the resulting automated volumes were compared to the manually corrected volumes obtained using FIRST on 3D T1w images. Association between DeepGRAI and FIRST was assessed by accuracy, true positive rate (TPR), false positive rate (FPR), coefficient of variation (CoV), pairwise correlation, Dice coefficient, Hausdorff distance (maximum shortest distance between the two segmentation borders), and symmetric surface distance (average shortest distance between the two segmentation borders). We also employed Bland-Altmann plotting to identify and correct systematic or data-specific biases. For test cases with multiple timepoints (n = 131), correlation between the two methods’ longitudinal change measures was assessed via Pearson correlation.
2.3.2. Precision via scan-rescan
To assess precision, we used a previously collected dataset of scan and rescan sessions on the same 3.0 T GE Signa Excite HD 12.0 Twin Speed 8-channel scanner. An internal dataset of MS (n = 3) and healthy control (n = 2) subjects scanned, repositioned, and then re-scanned over one week was employed. (Di Perri et al., 2009, Dwyer et al., 2017) These subjects had substantial positional changes between imaging sessions, reflecting real-world levels of consistency in positioning for clinical scans. Both 3D T1w images and 2D low-resolution T2-FLAIR images were acquired in each scanning session. For this scan-rescan dataset, DeepGRAI was run on each scan for each subject. The relationship was assessed by pairwise correlation and by coefficient of variation (CoV) analysis.
2.3.3. Interscanner stability
To assess inter-scanner stability, a previously collected dataset consisting of 125 MS patients and 52 healthy controls scanned at both 1.5 T and 3 T was used. (Di Perri et al., 2009, Dwyer et al., 2017) All subjects were examined on both scanners within one week, and the order in which subjects were scanned was randomized. The scanners used were a 1.5 T GE Signa Excite HD 12.0 8-channel scanner and a 3 T GE Signa Excite HD 12.0 Twin Speed 8-channel scanner (General Electric Milwaukee, WI). 3D T1w images and 2D low-resolution T2-FLAIR images were acquired in each scanning session. Sequences were not identical between the two scanners, but rather reflected optimizations for the specific field strengths as would be seen in clinical practice. As with the scan-rescan dataset, DeepGRAI was run on both scans for each subject, and association was again assessed by pairwise correlation and by CoV.
2.3.4. Clinical relevance
Finally, because we were interested in clinical relevance in terms of both disability and cognition, as well as predictive value of DeepGRAI, a longitudinal dataset including Expanded Disability Status Scale (EDSS) and cognitive processing speed (Symbol Digit Modalities Test, SDMT) (Smith, 1982) at baseline and 5-year follow-up was employed in 49 MS patients. (Fuchs et al., 2019) High-resolution 3D T1w images and 2D low-resolution T2-FLAIR images were acquired in each scanning session. FIRST and DeepGRAI were run on each scan (on 3D T1w and T2-FLAIR, respectively) for each subject and the correlations between clinical and MRI measures were explored with Pearson correlation. Associations with clinical and MRI measures over time were tested via age- and sex-adjusted linear regression.
2.3.5. Statistics
All statistical analyses were carried out either directly in Python/PyTorch or in R version 4.0.3. (R Development Core Team, 2019). Direct imaging measures (accuracy, TPR, FPR, Dice, Hausdorff distance, and symmetric surface distance) were assessed using the relevant Python functions provided by the scikit-learn package (https://scikit-learn.org, version 0.24) or the metrics sub-package of the MONAI toolkit (https://monai.io/, version 0.4.0). Correlations, CoVs, and linear regression models were undertaken in R. Results were considered significant at p < 0.05.
2.4. Packaging of the classifier as an easily usable tool for real-world data
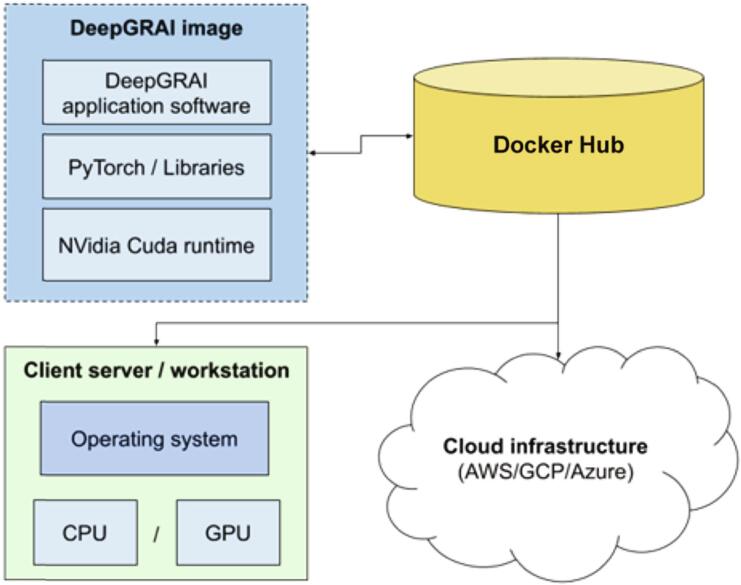
We undertook a number of steps to remove barriers to the use of the proposed DeepGRAI software. Commonly, implementation of advanced machine learning software requires a highly specialized physical environment, specific software libraries, and complex pre-processing and processing pipelines. To address this, we took advantage of modern containerization approaches to develop a deployable Docker image. Specifically, we built a container image that includes an entire python platform, CUDA runtime for both CPU and GPU, and can be easily and transparently implemented on different underlying hardware solutions (Linux/MacOS/Windows), as diagramed in Fig. 4. Instructions for use of the tool are available at https://hub.docker.com/r/buffaloneuroimaging/deepgrai.
Fig. 4.
DeepGRAI containerized architecture. The DeepGRAI application software is placed into a system-agnostic container image that includes all needed version of all libraries and supporting software. This dramatically improves deployability.
3. Results
3.1. Accuracy on independent testing dataset
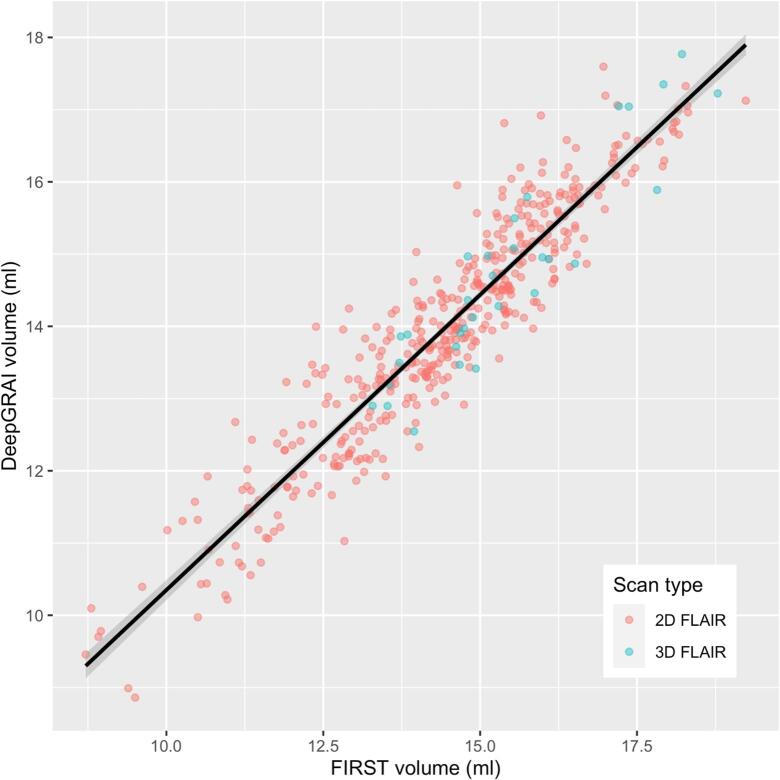
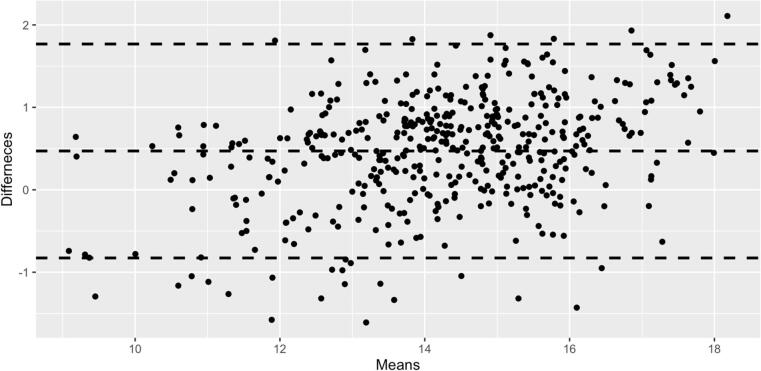
The DeepGRAI algorithm trained successfully, rapidly learning to recognize the thalamus in the first 50 epochs and then refining the delineation during later epochs. It successfully delineated the thalamus on a wide variety of cases and no visual failures were assessed (Table 1). The algorithm performed well even in cases with extreme lesion load, levels of atrophy or corruption by MRI artifacts (Fig. 6). On the combined independent testing dataset (both 2D and 3D FLAIR), no cases failed visual quality control (Table 1 and Fig. 5, Fig. 6). Agreement between corrected FIRST and DeepGRAI was very high. Accuracy relative to FIRST on 3D T1w images was 99.4% (r = 0.94, p < 0.001), as shown in Fig. 7. Mean Dice was 93.6%, TPR was 93.0%, FPR was 0.26%, Hausdorff distance was 2.4 mm, and symmetric surface distance was 0.47 mm. In the 3D-FLAIR-only subset of the testing data (n = 31), accuracy relative to FIRST on 3D T1w images was 99.4% (r = 0.92, p < 0.001). Mean Dice was 93.5%, TPR was 92.3%, FPR was 0.25%, Hausdorff distance was 2.4 mm, and symmetric surface distance was 0.49 mm. Bland-Altmann plotting did not reveal significant biases (Fig. 8). Correlation between baseline-to-followup DeepGRAI change and FIRST change was r = 0.38, p < 0.001. In the independent clinical validation dataset, longitudinal correlation between the measures was r = 0.244, p < 0.001.
Fig. 6.
Results of DeepGRAI segmentaion algorithm on cases with very high (left, thalamic volume 9.7 ml) and low (right, thalamic volume 15.9 ml) levels of atrophy. The classifier performs well in both cases despite substantial differences in brain morphology and lesion load, as well as different image contrasts.
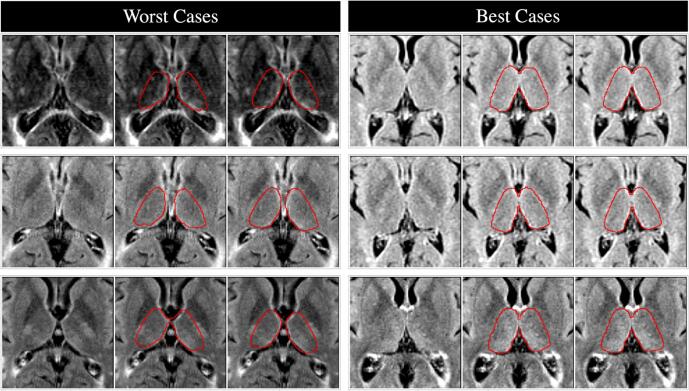
Fig. 5.
Visualization of the three worst (left) and three best (right) segmentations in the held-out test dataset (scans from untrained sites). For each figure, left panel is the raw T2-FLAIR image, middle panel shows the target FIRST segmentation in red, and right panel shows the resulting DeepGRAI segmentation. Dice scores for the worst cases were 85.9%, 86.6%, and 87.0% from top to bottom, and Dice scores for the best cases were 96.2%, 96.2%, and 96.3% from top to bottom. Average Dice across all 459 test cases was 93.6%. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Agreement / predictive value of DeepGRAI as compared to FIRST, as visualized on the independent validation dataset (n = 459 exams) comprised of heterogeneous T2-FLAIR and 3D T1w images from many individual scanners and MRI protocols.
Fig. 8.
Bland-Altman plot evaluating agreement between FIRST and DeepGRAI as a function of thalamic volume. No significant systematic biases are noted.
3.2. Scan-rescan reliability
The algorithm did not fail in any cases. Scan-rescan CoV error with repositioning was 0.43%., This is below values previously reported for FIRST scan-rescan performance (Morey et al., 2010), and very similar to the FIRST data on the same exams’ T1 images (0.40%).
3.3. Inter-scanner stability
The algorithm did not fail in any cases. Inter-scanner CoV error over one week was 3.21%. Comparatively, FIRST inter-scanner CoV on the same scans was 3.12%.
3.4. Clinical relevance
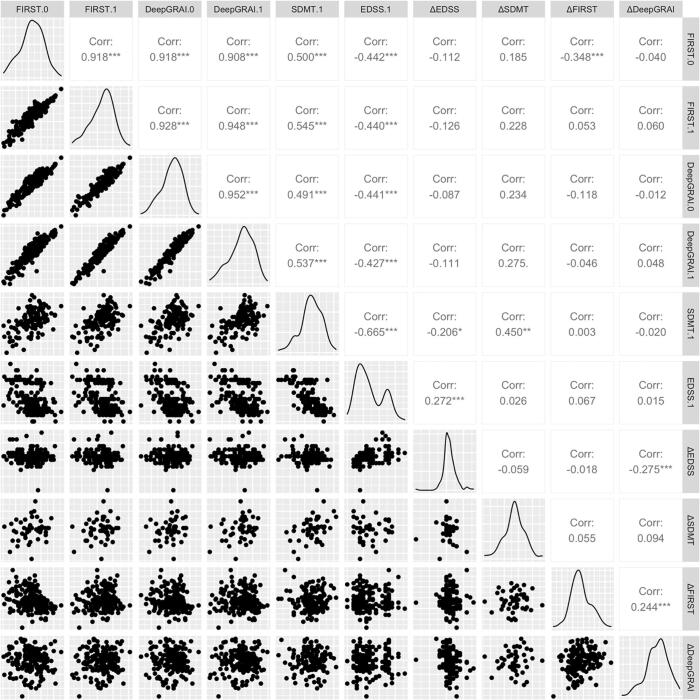
We evaluated correlations between FIRST, DeepGRAI, EDSS, and SDMT (Fig. 9). The algorithm did not fail in any cases (Table 1).
Fig. 9.
Pair-wise plot matrix for the key outcomes from the clinical validation dataset. Legend: EDSS-Expanded Disability Status Scale; SDMT-Symbol Digit Modalities Test; MS-multiple sclerosis.
We found that DeepGRAI-derived thalamic volume was significantly correlated with EDSS (r = -0.43, p < 0.01) and SDMT (r = 0.54, p < 0.01). Correlations were almost exactly in line with FIRST as assessed on 3D T1w images (r = -0.44, p < 0.01 for EDSS and r = 0.55, p < 0.01 for SDMT).
Furthermore, baseline DeepGRAI thalamic volume, controlling for age and sex, was also a significant predictor of longitudinal SDMT decline over a 5-year follow-up period (R2 = 0.081, p = 0.023; comparatively, FIRST was R2 = 0.080, p = 0.025) (Fig. 9). No significant correlations were detected between absolute EDSS changes and baseline thalamic volumetry measures for either DeepGRAI or FIRST. However, DeepGRAI change from baseline to follow-up was significantly associated with EDSS change (r = -0.275, p < 0.001).
3.5. Applicability in real-world setting
We have tested this architecture on both GPU and CPU enabled machines with different operating systems. On a standard GPU-enabled system, runtime was approximately 7 s per case. On commodity hardware with no GPU (2.8 GHz dual core Intel Core i7, 16 GB RAM), runtime was approximately 19 s.
4. Discussion
Quantitative metrics of neuroinflammation and neurodegeneration have played a vital role in our understanding of MS and have become a key component in clinical treatment trials. (Wattjes et al., 2015, Zivadinov et al., 2016, Rocca et al., 2017) GM atrophy, and thalamic atrophy in particular, have been shown to be strong predictors of disability and cognition. (Houtchens et al., 2007, Batista et al., 2012, Minagar et al., 2013, Zivadinov et al., 2013b, Azevedo et al., 2018, Eshaghi et al., 2018). However, most approaches to measurement have focused on research-quality MRI, leaving a potential gap for reliable measurement on clinical-quality scans. In particular, reliable measures of gray matter atrophy applicable to conventional T2-FLAIR images would be beneficial. In this study, we developed and validated a fully automated deep learning semantic segmentation tool to address this need.
The CNN architecture of DeepGRAI was substantially improved via a number of pre-planned and/or experimentally motivated implementation changes. We incorporated methods such as pseudo-Dice cost function, (Kleesiek et al., 2016) Xavier weight initialization, leaky ReLUs, (Szulczynski et al., 2018) bounding box restriction, and learning rate decay. We also added multi-scale iterative analysis and adaptive Adam optimization. We also included a number of data augmentations to help the trained network generalize as much as possible to new and unseen image acquisitions and scanner variations.
To assess the overall accuracy of our network’s segmentation quality, we compared it to the previously obtained “gold-standard” volumes output by FSL’s FIRST software that was run on 3D T1w images from the same exam sessions. Overall agreement with FIRST was excellent as shown in Fig. 7, Fig. 8, Fig. 9. Qualitatively, the approach was also able to handle both low and high atrophy cases, displaying robustness to a wide variety of morphological variants, as shown in Fig. 6. Taken together, these results demonstrate that the proposed architecture is capable of detecting, localizing, and quantifying the thalamus similarly to the performance of FIRST as applied to 3D T1w images, while using only low-resolution T2-FLAIR images as input. Furthermore, robustness of DeepGRAI to highly heterogeneous and multi-center data was confirmed. We also confirmed that the algorithm performs equally well on 3D-FLAIR images, which are becoming more popular due to manufacturer improvements and recent consensus recommendations (Rovira et al., 2015). It is important to note that the goal of this work was not to outperform FIRST or other current state of the art techniques, but rather to transfer the capability for reliable thalamic segmentation from the domain of 3D T1-weighted sequences to the domain of clinical quality T2-FLAIR scans. Based on the results obtained, this appears to be achievable despite the lower resolution of T2-FLAIR scans used in this study.
In addition to the primary outcome of overall segmentation accuracy, we assessed a number of areas related to real-world clinical data. These included scan-rescan and inter-scanner reliability (because many clinical scans are not consistently on the same scanner/protocol. First, a separate dataset of 177 subjects scanned at both 1.5 T and 3 T within one week was evaluated to determine inter-scanner reliability. The observed changes are very similar to those for FIRST on the same exams, and in line with previous observations for 3D T1w imaging analysis with SIENAX of approximately 3.4%. (Chu et al., 2016) Then, another dataset of subjects scanned, repositioned, and then re-scanned (n = 5) was used to evaluate scan-rescan reliability. These experiments showed that agreement of DeepGRAI (using low-resolution T2-FLAIR MRI) with a widely used and broadly validated method (FIRST, on 3D T1w MRI) is comparable to the reproducibility of that method itself on scan-rescan and inter-scanner data (2–3%).
To test external validity and to ensure that any seemingly-random small deviations from FIRST did not systematically change clinical/cognitive relationships, we also directly assessed DeepGRAI’s relationships to EDSS and SDMT. Results were very closely in line with those of FIRST on 3D T1w images. Additionally, they agree with extant data in the literature exploring thalamic volumetry. Houtchens et al. previously reported a relationship of r = -0.316 (p = 0.005) between thalamic volume and EDSS (Houtchens et al., 2007), compared to r = -0.427 (p < 0.01) here. Similarly, they reported a relationship of r = 0.658 (p < 0.001) for between thalamic volumtery and SDMT, compared to r = 0.537 (p < 0.01) here. In a later study, Batista et al. found analogous results, reporting a correlation between thalamic volume and SDMT of r = 0.543 (p < 0.001). (Batista et al., 2012) More broadly, Schoonheim et al. investigated the relationship between thalamic volume and overall cognition across domains, reporting a relationship of r = 0.551 (p < 0.001). Finally, relevance of DeepGRAI was confirmed longitudinally, as both DeepGRAI and FIRST similarly predicted cognitive decline over 5 years. Taken together, these results and broader context confirm that deep learning based thalamic volumetry on clinical routine T2-FLAIR is as relevant for cognitive and disability outcomes as traditional thalamic volumetry. This may be particularly important as thalamic atrophy has recently been shown to be a potentially modifiable outcome in clinical trials of newer disease modifying therapies. (Gaetano et al., 2018, Comi et al., 2019)
We chose to use deep learning semantic segmentation for this problem for a number of reasons. Semantic segmentation is a particular subset of deep-learning, concerned with the progression from coarse to fine inference and moving from localization / detection to complete delineation. (Long et al., 2015) In the field of medical imaging, it has already proven successful in providing fast, robust, and accurate labeling of voxels in a number of modalities and target organs. (Kamnitsas et al., 2017, Huang et al., 2018, Zhang et al., 2019). In our particular case, we implemented DeepGRAI as a fully convolutional feed-forward U-net neural network architecture. Such a network can incorporate substantial information to be more robust than traditional algorithmic approaches. This is of particular importance in a real-world setting. Real-world T2-FLAIR MRI acquisitions display a wide range of contrast characteristics, with background tissue weighting ranging from highly T2w to only marginally T2w. Furthermore, depending on the balance of inversion time with other MRI protocol parameters, the fluid attenuation (the “FLA ” in FLAIR) ranges from nearly complete to minimal, with fluid nearly isointense with tissue in some of the worst cases (Fig. 3, column 2, row 2). This dramatic variation in real-world contrast from the “ideal” FLAIR (highly T2w with perfectly suppressed cerebrospinal fluid) can result in many issues for classical, non-AI approaches.
We also chose to use the thalamus in this work for a number of reasons, both clinical and pragmatic. From a neuroanatomical perspective, the thalamus is a central relay station both to and within many brain circuits, and is therefore likely to be impacted by many elements of MS pathology. From a clinical perspective, thalamic atrophy’s roughly constant rate and very early involvement in MS have already led to its proposal as an important biomarker. (Azevedo et al., 2018) Empirically, it also strongly correlates with many key outcomes. (Minagar et al., 2013) From a pragmatic perspective, the thalamus is also an excellent target for segmentation on lower-resolution clinical-quality T2-FLAIR scans. It is a central structure, making it unlikely to be cut off or affected by artifact, as well as less susceptible to distortion due to its nearness to the magnet isocenter when scanning. It is also a topologically simple structure with a high volume-to-surface ratio, unlike the highly convoluted cortex, making it more amenable to accurate interpolation and inference in a low-resolution setting. Finally, although its lateral borders are not always fully distinct, its inner edges, bordering on ventricular CSF, are high contrast on most imaging modalities, including T2-FLAIR, reducing potential for error in measurement.
This is certainly not the first work to use deep learning for neuroimaging segmentation. Other tools have been proposed for brain extraction, tissue segmentation, and lesion identification. In many cases – and for lesions in particular – these AI methods have substantially outperformed more classical methods. In particular, the recently described FastSurfer approach even includes thalamic segmentation. (Henschel et al., 2020) However, unlike the current work, relevant approaches to structural parcellation and volumetric measurement focus on 3D T1 images as their input domain, and on improvements in accuracy or speed rather than translation to a different input domain. As such, while the current study is useful in itself, it also confirms the feasibility of a more general method for “converting” classical approaches on research-quality images to similar outcomes on clinical routine images. To accomplish this, future studies would similarly require a dataset with both research-quality and clinical-quality scans, with which they could then apply classical techniques to create masks and/or outcomes on research quality scans, and then use convolutional neural network architectures for domain transfer to clinical routine images via co-registration and independent input training.
Although this study does strongly support the feasibility of AI-based thalamic volumetry on clinical routine T2-FLAIR images, there are a number of important limitations that should be considered. Perhaps most importantly, as with many other segmentation datasets, our “ground truth” for training and evaluation was based on algorithmic outputs and visual inspection (FIRST plus manual review and corrections) rather than on histopathological ground truth. Even on high-resolution, research-quality 3D T1 imaging, the lateral borders of the thalamus are not always perfectly clear. Although sites and subjects were kept in separate training/testing/validation/splits, no other attempts were made to statistically handle the relatedness of the data for training (e.g., by de-weighting repeated subjects). We felt it better to include as many cases as possible rather than reduce to one scan per subject, given the known dependence of deep learning approaches on large datasets. Similarly, the dataset was a convenience sample including as many cases as possible, and although the data appears representative, no particular a priori balancing criteria were employed. Again, we felt that this was a reasonable approach to maximize the training set. Another potential issue is the homogeneity of our supplementary validation datasets. Although the training/testing dataset spanned across many different scanners and was likely very representative, the other validation datasets (scan/rescan, inter-scanner, and cognitive) were based on available data at our center. Ideally, such datasets would also span numerous scanners and centers. This is difficult in practice, but some initial attempts have been made (Oh et al., 2018), and such datasets could be used in future work like this to more robustly validate such secondary outcomes. A further limitation is that our data augmentation methods were based on commonly used approaches, but were certainly not exhaustive. In particular, the methods we used heavily expand the intensity and positioning domains (scanning-related factors), but with the exception of left–right flips they do not substantially expand the anatomical domain. We believe this is reasonable given the much larger variety of individual subjects included in our data as compared to scanners, but additional affine and/or warping-based augmentations might be helpful in future work. Also, it is important to note that our resampling approach cannot create data de novo that was not originally acquired. As such, the intent is not necessarily to produce a better image at 0.5 mm isotropic, but rather to allow the network to better capture partial volume and to not lose resolution from acquisitions with highly anisotropic voxels. Future work might exploit modern super-resolution techniques, though, to more completely take advantage of the inherent smoothness of the thalamus. Another limitation of the current work is the longitudinal agreement between FIRST and DeepGRAI. Although we did find highly significant associations in change measures over time in both the testing and clinical validation datasets, the magnitude of the associations was more modest than for cross-sectional data. This is likely due to insufficient follow-up time and/or relative neurodegenerative stability in the studied group of subjects (i.e., minimal actual changes to find agreement for), but it does deserve future study in more longitudinally-targeted datasets. Finally, an important limitation is that our approach suffers from a common concern with deep learning approaches – namely, the “black-box” nature of its selection process.
In conclusion, our study shows that thalamic volumetry on clinical routine images via deep learning is both possible and feasible. Our algorithm was able to successfully learn how to identify and quantify the thalamus on low-resolution T2-FLAIR images, and the resulting data was strongly related to both conventionally measured thalamic volume and to cognitive outcomes in MS. The system was packaged as a widely deployable, freely available tool via containerization technology (https://hub.docker.com/r/buffaloneuroimaging/deepgrai), and can be easily used by others for broader clinical research on real-world datasets.
CRediT authorship contribution statement
Michael Dwyer: Conceptualization, Methodology, Software. Cassondra Lyman: Writing - review & editing. Hannah Ferrari: Writing - review & editing. Niels Bergsland: Writing - review & editing, Validation. Tom A. Fuchs: Writing - review & editing. Dejan Jakimovski: Writing - review & editing. Ferdinand Schweser: Conceptualization, Writing - review & editing. Bianca Weinstock-Guttmann: Conceptualization, Writing - review & editing. Ralph H.B. Benedict: Conceptualization, Writing - review & editing. Jon Riolo: Conceptualization, Writing - review & editing. Diego Silva: Conceptualization, Writing - review & editing. Robert Zivadinov: Supervision, Writing - review & editing.
Declaration of Competing Interest
Portions of this study included research support from Bristol Myer Squibb and NIH-UL1TR001412.
Michael G. Dwyer received personal compensation from Novartis and EMD Serono, and financial support for research activities from Bristol Myers Squibb, Novartis, Mapi Pharma, Keystone Heart, Protembis, and V-WAVE Medical.
Daniel Brior, Cassondra Lyman, Hanna Ferrari, Niels Bergsland, Tom Fuchs, and Dejan Jakimovski have nothing to disclose.
Jon Riolo and Diego Silva are employees of Bristol Myers Squibb.
Bianca Weinstock- Guttman received honoraria as a speaker and as a consultant for Biogen Idec, Teva Pharmaceuticals, EMD Serono, Genzyme, Sanofi, Novartis and Acorda. Dr Weinstock-Guttman received research funds from Biogen Idec, Teva Pharmaceuticals, EMD Serono, Genzyme, Sanofi, Novartis, Acorda.
Ralph H. B. Benedict has received research support from Novartis, Genzyme, Genentech, Biogen, and Bristol Myers Squibb, and is on the speakers’ bureau for EMD Serono and Bristol Myers Squibb, and consults for Biogen, Immunic Therapeutics, Merck, Roche, Sanofi/Genzyme, Takeda, Veraci, and Novartis. Dr. Benedict also receives royalties for Psychological Assessment Resources.
Robert Zivadinov received personal compensation from Bristol Myers Squibb, EMD Serono, Sanofi, Keystone Heart, Protembis and Novartis for speaking and consultant fees. He received financial support for research activities from Sanofi, Novartis, Bristol Myers Squibb, Mapi Pharma, Keystone Heart, Protembis, Boston Scientific and V-WAVE Medical.
Acknowledgements:
We thank Daniel Brior and Dongchan Lee for their help in performing this study. Portions of the research reported in this publication were supported by Celgene Corporation (a subsidiary of Bristol Myers Squibb) and by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR001412 to the University at Buffalo. The content is solely the responsibility of the authors and does not necessarily represent the official views of Bristol Myers Squibb or the NIH.
References:
- Azevedo C.J., Cen S.Y., Khadka S., Liu S., Kornak J., Shi Y., Zheng L., Hauser S.L., Pelletier D. Thalamic Atrophy in MS: An MRI Marker of Neurodegeneration Throughout Disease. Ann. Neurol. 2018 doi: 10.1002/ana.25150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista S., Zivadinov R., Hoogs M., Bergsland N., Heininen-Brown M., Dwyer M.G., Weinstock-Guttman B., Benedict R.H. Basal ganglia, thalamus and neocortical atrophy predicting slowed cognitive processing in multiple sclerosis. J. Neurol. 2012;259:139–146. doi: 10.1007/s00415-011-6147-1. [DOI] [PubMed] [Google Scholar]
- Bergsland N., Zivadinov R., Dwyer M.G., Weinstock-Guttman B., Benedict R.H. Localized atrophy of the thalamus and slowed cognitive processing speed in MS patients. Mult Scler. 2016;22:1327–1336. doi: 10.1177/1352458515616204. [DOI] [PubMed] [Google Scholar]
- Bisecco A., Capuano R., Caiazzo G., d'Ambrosio A., Docimo R., Cirillo M., Russo A., Altieri M., Bonavita S., Rocca M.A., Filippi M., Tedeschi G., Gallo A. Regional changes in thalamic shape and volume are related to cognitive performance in multiple sclerosis. Mult Scler. 2019;1352458519892552 doi: 10.1177/1352458519892552. [DOI] [PubMed] [Google Scholar]
- Chu R., Tauhid S., Glanz B.I., Healy B.C., Kim G., Oommen V.V., Khalid F., Neema M., Bakshi R. Whole Brain Volume Measured from 1.5T versus 3T MRI in Healthy Subjects and Patients with Multiple Sclerosis. J. Neuroimaging : Official Journal of the American Society of Neuroimaging. 2016;26:62–67. doi: 10.1111/jon.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O., 2016. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Springer, Athens, Greece.
- Comi G., Kappos L., Selmaj K.W., Bar-Or A., Arnold D.L., Steinman L., Hartung H.-P., Montalban X., Kubala Havrdová E., Cree B.A.C., Sheffield J.K., Minton N., Raghupathi K., Ding N., Cohen J.A. Safety and efficacy of ozanimod versus interferon beta-1a in relapsing multiple sclerosis (SUNBEAM): a multicentre, randomised, minimum 12-month, phase 3 trial. The Lancet Neurology. 2019;18:1009–1020. doi: 10.1016/S1474-4422(19)30239-X. [DOI] [PubMed] [Google Scholar]
- Di Perri C., Dwyer M.G., Wack D.S., Cox J.L., Hashmi K., Saluste E., Hussein S., Schirda C., Stosic M., Durfee J., Poloni G.U., Nayyar N., Bergamaschi R., Zivadinov R. Signal abnormalities on 1.5 and 3 Tesla brain MRI in multiple sclerosis patients and healthy controls. A morphological and spatial quantitative comparison study. Neuroimage. 2009;47:1352–1362. doi: 10.1016/j.neuroimage.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Dwyer M.G., Silva D., Bergsland N., Horakova D., Ramasamy D., Durfee J., Vaneckova M., Havrdova E., Zivadinov R. Neurological software tool for reliable atrophy measurement (NeuroSTREAM) of the lateral ventricles on clinical-quality T2-FLAIR MRI scans in multiple sclerosis. Neuroimage Clin. 2017;15:769–779. doi: 10.1016/j.nicl.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi, A., Prados, F., Brownlee, W.J., Altmann, D.R., Tur, C., Cardoso, M.J., De Angelis, F., van de Pavert, S.H., Cawley, N., De Stefano, N., Stromillo, M.L., Battaglini, M., Ruggieri, S., Gasperini, C., Filippi, M., Rocca, M.A., Rovira, A., Sastre-Garriga, J., Vrenken, H., Leurs, C.E., Killestein, J., Pirpamer, L., Enzinger, C., Ourselin, S., Wheeler-Kingshott, C., Chard, D., Thompson, A.J., Alexander, D.C., Barkhof, F., Ciccarelli, O., group, M.s., 2018. Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann Neurol 83, 210-222. [DOI] [PMC free article] [PubMed]
- Fidon L., Li W., Garcia-Peraza-Herrera L.C., Ekanayake J., Kitchen N., Ourselin S., Vercauteren T. Springer International Publishing; Cham: 2018. Generalised Wasserstein Dice Score for Imbalanced Multi-class Segmentation Using Holistic Convolutional Networks; pp. 64–76. [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman E.M., Racke M.K., Raine C.S. Multiple sclerosis–the plaque and its pathogenesis. N. Engl. J. Med. 2006;354:942–955. doi: 10.1056/NEJMra052130. [DOI] [PubMed] [Google Scholar]
- Fuchs T.A., Benedict R.H.B., Bartnik A., Choudhery S., Li X., Mallory M., Oship D., Yasin F., Ashton K., Jakimovski D., Bergsland N., Ramasamy D.P., Weinstock-Guttman B., Zivadinov R., Dwyer M.G. Preserved network functional connectivity underlies cognitive reserve in multiple sclerosis. Hum. Brain Mapp. 2019;40:5231–5241. doi: 10.1002/hbm.24768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetano L., Häring D.A., Radue E.-W., Mueller-Lenke N., Thakur A., Tomic D., Kappos L., Sprenger T. Fingolimod effect on gray matter, thalamus, and white matter in patients with multiple sclerosis. Neurology. 2018;90 doi: 10.1212/WNL.0000000000005292. [DOI] [PubMed] [Google Scholar]
- Glorot, X., Bengio, Y., 2010. Understanding the difficulty of training deep feedforward neural networks. Proceedings of the 13th International Conference on Artificial Intelligence and Statistics (AISTATS) Chia Laguna Resort, Sardinia, Italy.
- He, K., Zhang, X., Ren, S., Sun, J., 2015. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification., Proceedings of the IEEE international conference on computer vision (ICCV) Santiago, Chile, pp. 1026-1034.
- Henschel L., Conjeti S., Estrada S., Diers K., Fischl B., Reuter M. FastSurfer - A fast and accurate deep learning based neuroimaging pipeline. Neuroimage. 2020;219 doi: 10.1016/j.neuroimage.2020.117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtchens M.K., Benedict R.H., Killiany R., Sharma J., Jaisani Z., Singh B., Weinstock-Guttman B., Guttmann C.R., Bakshi R. Thalamic atrophy and cognition in multiple sclerosis. Neurology. 2007;69:1213–1223. doi: 10.1212/01.wnl.0000276992.17011.b5. [DOI] [PubMed] [Google Scholar]
- Huang Q., Sun J., Ding H., Wang X., Wang G. Robust liver vessel extraction using 3D U-Net with variant dice loss function. Comput. Biol. Med. 2018;101:153–162. doi: 10.1016/j.compbiomed.2018.08.018. [DOI] [PubMed] [Google Scholar]
- Kamnitsas K., Ledig C., Newcombe V.F.J., Simpson J.P., Kane A.D., Menon D.K., Rueckert D., Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Kingma, D., Ba, J., 2017. Adam: A Method for Stochastic Optimization. arXiv.
- Kleesiek J., Urban G., Hubert A., Schwarz D., Maier-Hein K., Bendszus M., Biller A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. Neuroimage. 2016;129:460–469. doi: 10.1016/j.neuroimage.2016.01.024. [DOI] [PubMed] [Google Scholar]
- Long J., Shelhamer E., Darrell T. CVPR 2015 Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully Convolutional Networks for Semantic Segmentation ppt; pp. 3431–3440. [Google Scholar]
- Lyman C., Ferrari H., Fuchs T., Brior D., Bergsland N., Jakimovski D., Weinstock-Guttmann B., Zivadinov R., Dwyer M. 72nd Annual Meeting of the American Academy of Neurology. 2020. Systematic assessment of common error modes in using FIRST for MRI-based thalamic volumetry in people with multiple sclerosis. [Google Scholar]
- Minagar A., Barnett M.H., Benedict R.H., Pelletier D., Pirko I., Sahraian M.A., Frohman E., Zivadinov R. The thalamus and multiple sclerosis: Modern views on pathologic, imaging, and clinical aspects. Neurology. 2013;80:210–219. doi: 10.1212/WNL.0b013e31827b910b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey R.A., Selgrade E.S., Wagner H.R., 2nd, Huettel S.A., Wang L., McCarthy G. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum. Brain Mapp. 2010;31:1751–1762. doi: 10.1002/hbm.20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Bakshi R., Calabresi P.A., Crainiceanu C., Henry R.G., Nair G., Papinutto N., Constable R.T., Reich D.S., Pelletier D., Rooney W., Schwartz D., Tagge I., Shinohara R.T., Simon J.H., Sicotte N.L., Committee N.C.S. The NAIMS cooperative pilot project: Design, implementation and future directions. Multiple sclerosis (Houndmills, Basingstoke, England) 2018;24:1770–1772. doi: 10.1177/1352458517739990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu V., Ran N.C., Barkhof F., Chard D.T., Wheeler-Kingshott C.A., Vrenken H. Accurate GM atrophy quantification in MS using lesion-filling with co-registered 2D lesion masks. Neuroimage Clin. 2014;4:366–373. doi: 10.1016/j.nicl.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rawat W., Wang Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 2017;29:2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- Rocca M.A., Battaglini M., Benedict R.H., De Stefano N., Geurts J.J., Henry R.G., Horsfield M.A., Jenkinson M., Pagani E., Filippi M. Brain MRI atrophy quantification in MS: From methods to clinical application. Neurology. 2017;88:403–413. doi: 10.1212/WNL.0000000000003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira, A., Wattjes, M.P., Tintore, M., Tur, C., Yousry, T.A., Sormani, M.P., De Stefano, N., Filippi, M., Auger, C., Rocca, M.A., Barkhof, F., Fazekas, F., Kappos, L., Polman, C., Miller, D., Montalban, X., group, M.s., 2015. Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis-clinical implementation in the diagnostic process. Nat Rev Neurol 11, 471-482. [DOI] [PubMed]
- Shorten C., Khoshgoftaar T.M. A survey on Image Data Augmentation for Deep Learning. Journal of Big Data. 2019;6:60. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Western Psychological Services; Los Angeles, CA: 1982. Symbol Digit Modalities Test: Manual. [Google Scholar]
- Szulczynski B., Arminski K., Namiesnik J., Gebicki J. Determination of Odour Interactions in Gaseous Mixtures Using Electronic Nose Methods with Artificial Neural Networks. Sensors (Basel) 2018;18 doi: 10.3390/s18020519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattjes, M.P., Rovira, A., Miller, D., Yousry, T.A., Sormani, M.P., de Stefano, M.P., Tintore, M., Auger, C., Tur, C., Filippi, M., Rocca, M.A., Fazekas, F., Kappos, L., Polman, C., Frederik, B., Xavier, M., group, M.s., 2015. Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis--establishing disease prognosis and monitoring patients. Nat Rev Neurol 11, 597-606. [DOI] [PubMed]
- Zhang J., Saha A., Zhu Z., Mazurowski M.A. Hierarchical Convolutional Neural Networks for Segmentation of Breast Tumors in MRI With Application to Radiogenomics. IEEE Trans. Med. Imaging. 2019;38:435–447. doi: 10.1109/TMI.2018.2865671. [DOI] [PubMed] [Google Scholar]
- Zivadinov R., Bergsland N., Dolezal O., Hussein S., Seidl Z., Dwyer M.G., Vaneckova M., Krasensky J., Potts J.A., Kalincik T., Havrdova E., Horakova D. Evolution of cortical and thalamus atrophy and disability progression in early relapsing-remitting MS during 5 years. AJNR Am. J. Neuroradiol. 2013;34:1931–1939. doi: 10.3174/ajnr.A3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivadinov, R., Bergsland, N., Korn, J.R., Dwyer, M.G., Khan, N., Medin, J., Price, J.C., Weinstock-Guttman, B., Silva, D., Group, M.-M.S., 2018. Feasibility of Brain Atrophy Measurement in Clinical Routine without Prior Standardization of the MRI Protocol: Results from MS-MRIUS, a Longitudinal Observational, Multicenter Real-World Outcome Study in Patients with Relapsing-Remitting MS. AJNR Am J Neuroradiol 39, 289-295. [DOI] [PMC free article] [PubMed]
- Zivadinov R., Havrdova E., Bergsland N., Tyblova M., Hagemeier J., Seidl Z., Dwyer M.G., Vaneckova M., Krasensky J., Carl E., Kalincik T., Horakova D. Thalamic atrophy is associated with development of clinically definite multiple sclerosis. Radiology. 2013;268:831–841. doi: 10.1148/radiol.13122424. [DOI] [PubMed] [Google Scholar]
- Zivadinov R., Jakimovski D., Gandhi S., Ahmed R., Dwyer M.G., Horakova D., Weinstock-Guttman B., Benedict R.R., Vaneckova M., Barnett M., Bergsland N. Clinical relevance of brain atrophy assessment in multiple sclerosis. Implications for its use in a clinical routine. Expert Rev. Neurother. 2016;16:777–793. doi: 10.1080/14737175.2016.1181543. [DOI] [PubMed] [Google Scholar]
- Zivadinov R., Khan N., Medin J., Christoffersen P., Price J., Korn J.R., Bonzani I., Dwyer M.G., Bergsland N., Carl E., Silva D., Weinstock-Guttman B. An Observational Study to Assess Brain MRI Change and Disease Progression in Multiple Sclerosis Clinical Practice-The MS-MRIUS Study. J. Neuroimaging. 2017;27:339–347. doi: 10.1111/jon.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]