Abstract
Objective
White adipose tissue (WAT) expansion regulates energy balance and overall metabolic homeostasis. The absence or loss of WAT occurring through lipodystrophy and lipoatrophy contributes to the development of hepatic steatosis and insulin resistance. We previously demonstrated that sole small ubiquitin-like modifier (SUMO) E2-conjugating enzyme Ube2i represses human adipocyte differentiation. The role of Ube2i during WAT development remains unknown.
Methods
To determine how Ube2i impacts body composition and energy balance, we generated adipocyte-specific Ube2i knockout mice (Ube2ia-KO). CRISPR/Cas9 gene editing inserted loxP sites flanking exons 3 and 4 at the Ube2i locus. Subsequent genetic crosses to Adipoq-Cre transgenic mice allowed deletion of Ube2i in white and brown adipocytes. We measured multiple metabolic endpoints that describe energy balance and carbohydrate metabolism in Ube2ia-KO and littermate controls during postnatal growth.
Results
Surprisingly, Ube2ia-KO mice developed hyperinsulinemia and hepatic steatosis. Global energy balance defects emerged from dysfunctional WAT marked by pronounced local inflammation, loss of serum adipokines, hepatomegaly, and near absence of major adipose tissue depots. We observed progressive lipoatrophy that commences in the early adolescent period.
Conclusions
Our results demonstrate that Ube2i expression in mature adipocytes allows WAT expansion during postnatal growth. Deletion of Ube2i in fat cells compromises and diminishes adipocyte function that induces WAT inflammation and ectopic lipid accumulation in the liver. Our findings reveal an indispensable role for Ube2i during white adipocyte expansion and endocrine control of energy balance.
Keywords: Ube2i, Lipodystrophy, Adipose tissue, Lipid metabolism
Highlights
-
•
A new mouse model reveals that Ube2i loss in fat cells impacts body composition.
-
•
Ube2i fat-specific knockout (Ube2ia-KO) causes fatty liver and hyperinsulinemia.
-
•
Ube2ia-KO mice develop metabolic inflexibility and cold intolerance.
-
•
Inflammation and caspase activation of cell death occur in Ube2ia-KO adipocytes.
Abbreviations
- a-KO
adipocyte-specific knockout
- BAT
brown adipose tissue
- CGL
congenital generalized lipodystrophy
- CLAMS
Comprehensive Lab Animal Monitoring System
- CRISPR
Clustered Regularly Interspace Short Palindromic Repeats
- DAPI
4′,6-diamidino-2-phenylindole
- ELISA
enzyme linked immunosorbent assay
- FGF21
fibroblast growth factor 21
- FPLD
familial partial lipodystrophy
- GFP
green fluorescent protein
- gWAT
gonadal white adipose tissue
- H/E
Hematoxylin and Eosin
- IP
intraperitoneal
- iWAT
inguinal white adipose tissue
- loxP
locus of X-over P1
- PBS
Phosphate Buffered Saline
- PPARγ
Peroxisome proliferator-activated receptor gamma
- RER
respiratory exchange ratio
- sgRNA
single guide RNA
- lssDNA
long single-stranded oligodeoxynucleotide donor
- SUMO
Small Ubiquitin-like Modifier
- SVF
stromal vascular fraction
- TG
triglyceride
- Ube2i
Ubiquitin conjugating enzyme E2 I
- WAT
White adipose tissue
1. Introduction
White adipocytes sequester lipids and protect peripheral metabolic tissues from ectopic lipid accumulation. Consequently, the failure of integral lipid metabolism responses and reduced adipose tissue expandability during chronic states of positive energy balance contribute to the development of insulin resistance, obesity, and fatty liver disease [1]. Similar metabolic abnormalities develop in patients with lipodystrophy where a lack of adipose tissue does not allow storage of surfeit energy as lipids, leading to insulin resistance, hepatic steatosis, and dyslipidemia [2]. Therefore, healthy adipose tissue development mediates key aspects of metabolic homeostasis.
Physiologic WAT expansion occurs through both increased adipocyte size (hypertrophy) and number (hyperplasia). White adipocyte differentiation requires a cascade of transcription factors that activate PPARγ, the master regulator of adipocyte differentiation [3]. Adipocyte differentiation requires PPARγ [4] to partner with distinct transcriptional co-regulators that coordinate brown and white adipocyte-specific gene expression [5,6]. Gene deletion or tissue-specific disruption of PPARγ impairs adipogenesis and results in severe lipodystrophy [4,[7], [8], [9]]. The mechanisms that enable adipose tissue development have broad basic implications for understanding energy balance disorders.
We previously showed Ube2i depletion in human subcutaneous preadipocytes accelerated fat cell differentiation [10]. In this study, we generated an adipocyte-specific Ube2i knockout (Ube2ia-KO) mouse to investigate how Ube2i regulates whole-body energy balance. Surprisingly, WAT mass failed to expand in male and female Ube2ia-KO mice. Progressive lipoatrophy and compromised adipocyte function likely provoked WAT inflammation and ectopic lipid accumulation leading to insulin resistance, impaired brown adipose tissue (BAT) function, and intolerance to cold temperatures. Taken together, our findings reveal critical roles for Ube2i in adipocyte function and expansion.
2. Methods
2.1. Animals
All procedures with animals were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine (animal protocol AN-6411). Experimental animals received humane care according to criteria in the “Guide for the Care and Use of Laboratory Animals” (8th edition, revised 2011). Experimental animals were housed (no more than four per cage) in a barrier-specific pathogen-free animal facility with 12-h dark–light cycle and free access to water and normal chow (Harlan Laboratories 2920X). All experiments were conducted using littermate-controlled male and female mice maintained on a C57BL/6J background. At the end of experiments, mice were euthanized by cervical dislocation while under isoflurane anesthesia. After euthanasia, tissues were collected, flash-frozen in liquid N2, and stored at −80 °C until use. All experiments adhered to ARRIVE Guidelines.
2.2. Generation of a conditional Ube2i allele
Ube2ifl/fl mice were generated by the Genetically Engineered Rodent Models Core at BCM using previously established methods [11]. We employed Cas9-initiated homology-driven repair (HDR) using a pair of single guide RNAs (sgRNAs) coupled with a long single-stranded oligodeoxynucleotide donor (lssDNA) template harboring short homology arms and loxP-flanked exons. The sgRNAs and the lssDNA donor were used to insert loxP sites 5′ and 3′ of exons 3 and 4 of the mouse Ube2i gene, respectively. Sequences of the sgRNAs and lssDNA donor are included in Supplemental File 1. To minimize the probability of off-target events, only sgRNAs predicted to have off-target sites with three mismatches or more were used to target Cas9 endonuclease activity to intronic sequences flanking exon 3 and 4. Two hundred C57BL/6NJ pronuclear-stage zygotes were co-injected with Cas9 mRNA, sgRNAs, and lssDNA [11]. Following micro-injection, zygotes were transferred into pseudopregnant ICR recipient females at approximately 25–32 zygotes per recipient. Sanger sequencing of cloned loxP sites and founder line genotyping from mouse genomic DNA confirmed loxP insertions and sequence fidelity. Progeny generated from putative founders were also sequence-confirmed for the fidelity of the loxP sequences and floxed exons. The targeted sequence and annotated primer locations are included in Supplemental File 2 (GenBank). Ube2ifl/fl mice were crossed with Adipoq-Cre (Jackson Laboratory #028020) to generate adipocyte-specific Ube2i knockout (Ube2ia-KO) and littermate controls (Ube2ifl/fl). Ube2ifl/fl mice were crossed with CAG-CreER™ (Jackson Laboratory #004682) to generate tamoxifen inducible Ube2i gene deletion. Ube2ifl/fl are distributed upon request.
2.3. Genotyping
DNA extracted from mouse ear clips was used in PCR reactions with primers designed to detect the 5′ (P1 - AGGTAGGGGTGGCTTAGAGG, P2 - GGTTCATTGTGCCATCAGGG) and 3′ (P3 - CAAGTCCCAGGGTAGATGCG, P4 - CAGCTCAGACCTGGCCTTAC) loxP sequences and run on agarose gels. Cre transgenic mice were genotyped according to protocols provided by the Jackson Laboratory.
2.4. Antibodies and western blotting
Tissue and whole cell lysates were prepared in Protein Extraction Reagent (Thermo Fisher) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher). Immunoblotting was performed with lysates run on 4–12% Bis-Tris NuPage gels (Life Technologies) and transferred onto Immobilon-P Transfer Membranes (Millipore) followed by antibody incubation. Immunoreactive bands were visualized by chemiluminescence. The following antibodies were used for immunoblotting: α-HSP90 (Cell Signaling #4877), α-UBE2I (Cell Signaling #4786), α-ADIPOQ (Genetex #GTX112777), α-PPARγ (Cell Signaling #2443), α-Caspase-8 (Cell Signaling #4790), and α-Cleaved Caspase-8 (Cell Signaling #8592).
2.5. Cell culture
To validate Cre-inducible deletion of Ube2i, fibroblasts were isolated from the inguinal white adipose tissue (iWAT) stromal vascular fraction (SVF) of Ube2ifl/fl mice. iWAT depots were digested in phosphate buffered saline (PBS) containing collagenase D (Roche, 1.5 U/mL) and dispase II (Sigma, 2.4 U/mL) supplemented with 10 mM CaCl2 at 37 °C for 45 min. The primary cells were filtered twice through 70 μm strainers and centrifuged to collect the SVF. For adenovirus transduction, SVF cells were incubated with adenoviral Cre recombinase or green fluorescent protein (GFP) in DMEM/F12 medium containing Glutamax (ThermoFisher) and 10% fetal bovine serum (FBS) for 24 h. After replacing the medium once, cells were cultured for 48 h before lysate preparation. Adenoviruses expressing Cre recombinase or GFP were provided by the BCM Gene Vector Core. Similarly, iWAT SVF cells were isolated from CAG-CreER;Ube2ifl/fl mice, treated with 1 μM 4-hydroxytamoxifen for 48 h followed by differentiation for eight days.
2.6. Indirect calorimetry
Ube2ia-KO and littermate controls (Ube2ifl/fl) were maintained on normal chow and housed at room temperature in Comprehensive Lab Animal Monitoring System (CLAMS) home cages (Columbus Instruments). Oxygen consumption, CO2 emission, energy expenditure, food and water intake, and activity were measured for six days (BCM Mouse Metabolic and Phenotyping Core). Mouse body weight was recorded, and body composition examined by magnetic resonance imaging (Echo Medical Systems) prior to indirect calorimetry. Statistical analysis of energy balance was performed by ANCOVA with lean body mass as a co-variate using the CalR web-based tool [12].
2.7. Glucose and insulin tolerance tests
To determine glucose tolerance, mice were fasted for 16 h and glucose was administered (1.5 g/kg body weight) by intraperitoneal (IP) injection. To determine insulin tolerance, mice were fasted 4 h prior to insulin intraperitoneal injection (1.5 U/kg body weight). Blood glucose levels were measured by a handheld glucometer. Serum was collected after fasting and during glucose tolerance tests for insulin quantification.
2.8. ELISAs and lipid assays
Fed serum levels were used to measure insulin (#EZRMI-13K; Millipore), leptin (#90030; Crystal Chem), adiponectin (#KMP0041; Thermo Fisher), and free fatty acids (#sfa-1; ZenBio), and FGF21 (#MF2100; R&D Systems). Insulin levels during glucose tolerance tests were also measured by ELISA (Millipore). Hepatic triglyceride (TG) content was quantified by Thermo Fisher Scientific Triglycerides Reagent (#TR22421) and normalized per gram of liver tissue.
2.9. Histology
Formalin-fixed paraffin-embedded adipose and liver tissue sections were stained with hematoxylin and eosin (H/E) by the BCM Human Tissue Acquisition and Pathology Core. Images were captured using a Nikon Ci-L Brightfield microscope.
2.10. Fluorescence microscopy
Following differentiation, media was aspirated and 4% formaldehyde (Electron Microscopy Sciences) in PBS was immediately added for 30 min at room temperature. Excess paraformaldehyde was quenched with 100 mM ammonium chloride. Non-specific antibody binding was blocked by pre-incubating for 30 min in 2% bovine serum albumin in PBS/0.01% saponin (which was also used as an antibody diluent) at room temperature. Anti-perilipin antibody (GP-29, Progen) was diluted at a 1:1000 concentration in antibody diluent and incubated overnight at 4 °C. Subsequently, coverslips were washed with PBS and incubated with secondary antibodies for 1 h at room temperature. AlexaFluor 647-conjugated anti-guinea pig secondary antibodies (Thermo Fisher) were used. Coverslips were then washed 3 times and incubated LipidTOX green (1:1000, Thermo Fisher) and DAPI (10 μg/mL) in PBS for 45 min at room temperature. Slides were mounted with SlowFade Gold (ThermoFisher). Imaging was performed with the DeltaVision Core Image Restoration Microscope (GE Healthcare).
2.11. qPCR
Total RNA was extracted using the Direct-zol RNA MiniPrep kit (Zymo Research). cDNA was synthesized using iScript (Bio-Rad). Relative mRNA expression was measured with SsoAdvanced Universal Probes Supermix reactions read out with a QuantStudio 3 real-time PCR system (Applied Biosystems). TATA-box binding protein (Tbp) was the invariant control. Roche Universal Probe Gene Expression Assays were used as previously described [13].
2.12. Cold tolerance test
Six-month old male Ube2ifl/fl and Ube2ia-KO mice were individually housed with water and exposed to cold temperature (4 °C) for 2.5 h. A temperature probe was placed subcutaneously on top of the intrascapular BAT two days prior to the cold tolerance test. Temperature recordings were measured in duplicate at room temperature (time = 0) and during cold exposure every 30 min.
2.13. Statistical analyses
Statistical significance was assessed by unpaired Student's t-test with a primary threshold for statistical significance set at p < 0.05. For gene expression data, statistical significance was assessed by multiple unpaired t-tests with a q-value < 0.05. Statistical analysis of energy balance was performed by ANCOVA with lean body mass as a co-variate and cumulative food intake by standard ANOVA using the CalR web-based tool [12]. All data are presented as mean ± standard error of the mean (SEM), unless otherwise stated.
3. Results
3.1. Generation of conditional Ube2i knockout mice
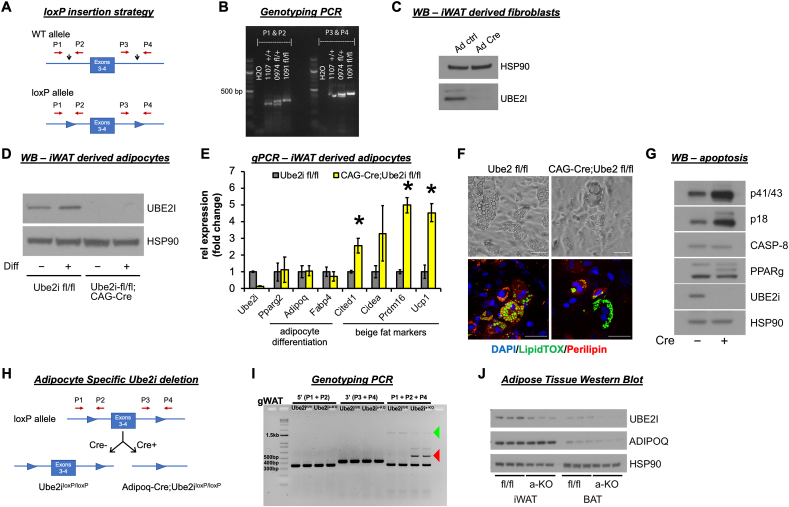
Ube2i is expressed ubiquitously across all mouse tissues [14] and knockout strategies cause lethality and sterility [[14], [15], [16], [17]]. To this end, we used CRISPR/Cas9 gene editing to generate a conditional Ube2i allele (Ube2ifl/fl) to explore tissue-specific roles for Ube2i. LoxP sites flanking exons 3 and 4 of the Ube2i locus were introduced by homology-directed repair (Figure 1A) and the in vivo presence of loxP sites in the targeted regions was confirmed by genotyping of potential founder mice (Figure 1B). We verified that the loxP sites targeted the Ube2i locus by transfecting Ube2ifl/fl iWAT-derived fibroblasts with adenovirus expressing Cre recombinase. Immunoblot analysis of whole cell lysates demonstrated near total deletion of UBE2I protein levels following Cre recombination compared to adenovirus GFP transductions (Figure 1C). To determine the cell autonomous effects of Ube2i deletion in adipocytes, CAG-CreER mice were crossed with Ube2ifl/fl mice and we isolated iWAT SVF cells for in vitro differentiation. Tamoxifen treatment knocked out UBE2I only in cells expressing CAG-Cre;Ube2ifl/fl, regardless of differentiation status (Figure 1D). Although we observed efficient UBE2I protein depletion, inducible knockout of Ube2i left representative adipocyte differentiation genes unaffected (Figure 1E). However, we observed a dramatic increase in several brown/beige adipocyte marker genes, including Ucp1, Prdm16, Cidea, and Cited1, consistent with observations from human subcutaneous adipocytes [10]. Although Western blot and qPCR showed the expression of mature adipocyte markers, we observed fewer lipid droplets in Ube2i knockout adipocytes (Figure 1F). We also observed higher cleaved Caspase-8 levels indicative of dying adipocytes (Figure 1G), which suggests an important role for UBE2I in adipocyte survival.
Figure 1.
Generation of conditional Ube2i gene deletion mice. (A)Ube2ifl/fl mice were generated using CRISPR/Cas9 gene editing. Exons 3 and 4 of the Ube2i (Ubc9) gene were targeted by sgRNAs designed complementary to intronic sequences flanking the exons, then loxP sequences were introduced by DNA donor oligonucleotides. LoxP sites were inserted before exon 3 and after exon 4 (black arrows). Primers detect the 5’ (P1, P2) and 3’ (P3, P4) loxP sequences. (B) PCR analyses of floxed alleles at the targeted loci in genomic DNA extracted from ear clips of wild-type (+/+), fl/+, and fl/fl mice. PCR products were run on agarose gels with expected band sizes for P1–P2: wild-type (+) 319 bp and loxP allele (fl) 353 bp and P3–P4: wild-type (+) 427 bp and loxP allele (fl) 461 bp. The image shows a wild-type control #1107 +/+, founder heterozygous mouse #974 fl/+, and homozygous F2 offspring #1091 fl/fl. (C) Western blot analysis of UBE2I expression in inguinal WAT (iWAT) derived fibroblasts from Ube2ifl/fl mice after transduction with adenoviral GFP or Cre. To determine cell autonomous effects of Ube2i deletion in adipocytes, tamoxifen inducible Cre-expressing mice (CAG-Cre) were crossed with Ube2ifl/fl mice. iWAT SVF cells were isolated from CAG-Cre;Ube2ifl/fl and Ube2ifl/fl control mice. All cells were treated with tamoxifen to induce Cre recombination followed by adipocyte differentiation (diff) for eight days. (D) Western blot analysis and (E) relative gene expression by qPCR were performed to assess Ube2i depletion, adipocyte maturation, and beige fat cell markers. Gray = Ube2ifl/fl, yellow = CAG-Cre;Ube2ifl/fl. Data are presented as mean +/− SEM, ∗p < 0.05. (F) Brightfield and fluorescent images of differentiated Ube2ifl/fl and CAG-Cre;Ube2ifl/fl cells stained for lipids (green, LipidTOX), perilipin (red), and nuclei (blue, DAPI). Scale bars 50 μm. (G) Whole cell lysates from differentiated cells were subjected to Western blot analysis of cleaved (p41/43, p18) and uncleaved Caspase-8. (H) Strategy for generating adipocyte-specific Ube2i gene deletion. Ube2ifl/fl mice were bred with Adipoq-Cre mice to generate adipocyte-specific Ube2i knockout (Ube2ia-KO) and Ube2ifl/fl (control) mice. (I) To validate gene deletion of Ube2i, genomic DNA was extracted from Ube2ia-KO and Ube2ifl/fl gonadal WAT (gWAT) samples and PCR products were run on an agarose gel to detect the 5’ (P1, P2 primers) and 3’ (P3, P4 primers) loxP sequences, as well as a product that spans exons 3–4 (P1+P2+P4; 1597 bp, green arrow) or the deletion product (509 bp, red arrow). (J) Western blots of whole tissue lysates from iWAT and BAT of seven-day-old mice were probed for UBE2I and adiponectin (ADIPOQ), and HSP90 served as the invariant loading control.
To specifically study the in vivo effects of Ube2i deletion in mature fat cells, we generated adipose-specific Ube2i knockout mice (Ube2ia-KO) by crossing Ube2ifl/fl animals with Adipoq-Cre transgenic mice (Figure 1H). Reproductive fitness and female nursing were unaffected by Ube2ia-KO and all pups were viable and born at the expected Mendelian ratio. PCR analysis demonstrated Cre recombination of the Ube2i locus generated a deletion product (red arrow) in the gonadal WAT (gWAT) from Ube2ia-KO mice that was absent in Ube2ifl/fl controls (Figure 1I). The full length, unrecombined product (green arrow) was also detected in Ube2ia-KO, but at lower levels than control, suggesting contributions of Cre-negative cells in the SVF to the PCR product. Similarly, UBE2I protein was reduced in iWAT and brown adipose tissue (BAT) compared to Ube2ifl/fl controls (Figure 1J).
3.2. Adipocyte-specific Ube2i deletion impairs WAT expansion during postnatal growth
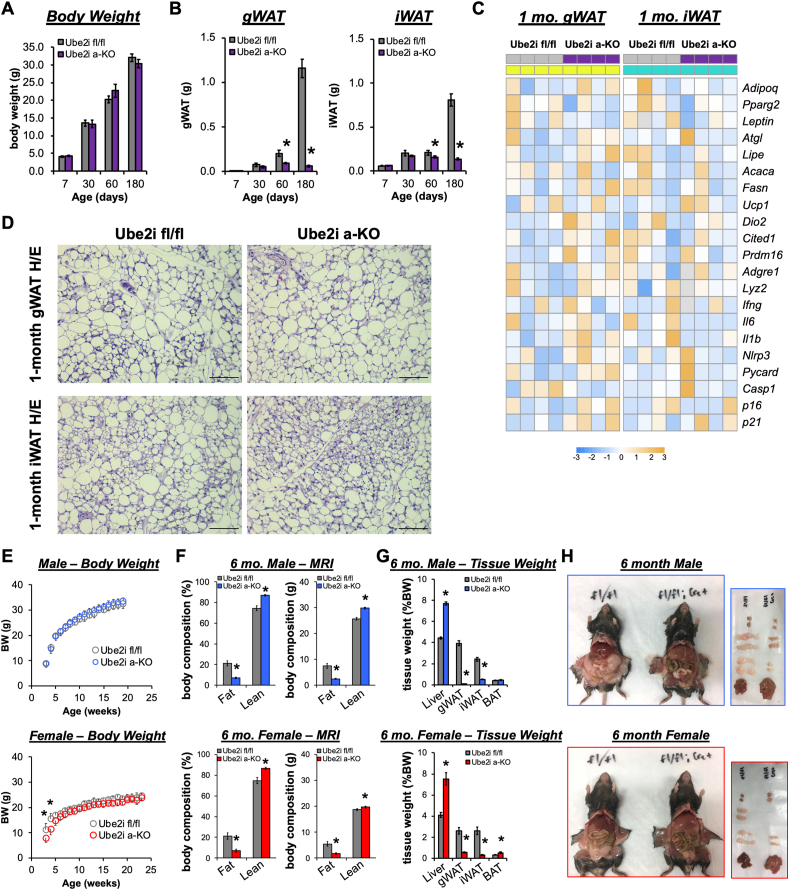
To determine the impact of adipocyte Ube2i loss in mice, we examined body weight (Figure 2A) and WAT mass (Figure 2B) at necropsy from seven days to six months of age. As expected, Ube2ifl/fl control mice exhibited progressive expansion of WAT mass with increasing age (Figure 2B). Conversely, Ube2ia-KO mice failed to expand WAT depots beginning at two months of age and by six months, WAT mass almost completely disappeared relative to Ube2ifl/fl controls. Gene expression (Figure 2C) and histological analyses (Figure 2D) of WAT at one month of age revealed similar profiles between Ube2ia-KO and Ube2ifl/fl controls, suggesting adipocyte function and morphology are normal prior to impaired WAT expansion. Despite no effects on body weight gain (Figure 2E), magnetic resonance imaging showed reduced fat mass and increased lean mass in six-month-old Ube2ia-KO male and female mice compared to controls (Figure 2F). Further examination of tissue weights at necropsy (Figure 2G) revealed grossly visible reductions (−80%) in iWAT and gWAT depots in male and female Ube2ia-KO mice (Figure 2H). Reduced fat storage in primary WAT depots resulted in significantly higher liver weights associated with gross morphological changes indicative of ectopic lipid accumulation. Similarly, lipid accumulation in BAT was apparent at necropsy, with increased BAT weight detected in female Ube2ia-KO mice. Collectively, these data demonstrate adipocyte-specific deletion of Ube2i impairs WAT expansion leading to redistribution of lipid storage.
Figure 2.
Adipocyte-specific Ube2i deletion impairs WAT expansion in male and female mice. (A) Body weight and (B) WAT weights (g) in combined male and female mice at 7 (n = 5–9), 30 (n = 4/group), 60 (n = 4/group), and 180 (n = 23–24/group) days of age for Ube2ifl/fl (gray) and Ube2ia-KO (purple) mice. Data are presented as mean +/− SEM; ∗p < 0.05. (C) Relative gene expression by qPCR in gWAT (left, yellow) and iWAT (right, cyan) from Ube2ia-KO (purple) and Ube2ifl/fl control (gray) mice for adipocyte maturation, lipid metabolism, inflammatory, and senescence genes represented as a heatmap of z-scores. Gray heatmap squares indicate outliers excluded based on Grubbs' test. (D) Representative H/E stained gWAT and iWAT sections from one-month-old Ube2ia-KO and Ube2ifl/fl control mice. Scale bar 100 μm. (E)Ube2ia-KO and Ube2ifl/fl control mice were weighed for up to 23 weeks in males (n = 12–14/group, mean +/− SD) and females (n = 6–8/group, mean +/− SD). (F) Assessment of fat and lean mass (% body weight (left) and in grams (right); male n = 11–15/group, female n = 5–7/group). (G) Tissue weights (% body weight; male n = 15-16/group, female n = 8/group) at necropsy. (H) Corresponding necropsy images from six-month-old Ube2ifl/fl and Ube2ia-KO mice. Images of excised tissues demonstrate gross morphological increases in liver size, reductions in iWAT and gWAT, and lighter coloring of liver and BAT. (E–H) Gray = Ube2ifl/fl male and female controls, blue = male Ube2ia-KO, red = female Ube2ia-KO. Data are presented as mean +/− SEM; ∗p < 0.05.
3.3. Adipocyte-specific Ube2i deletion increases WAT inflammation and apoptosis
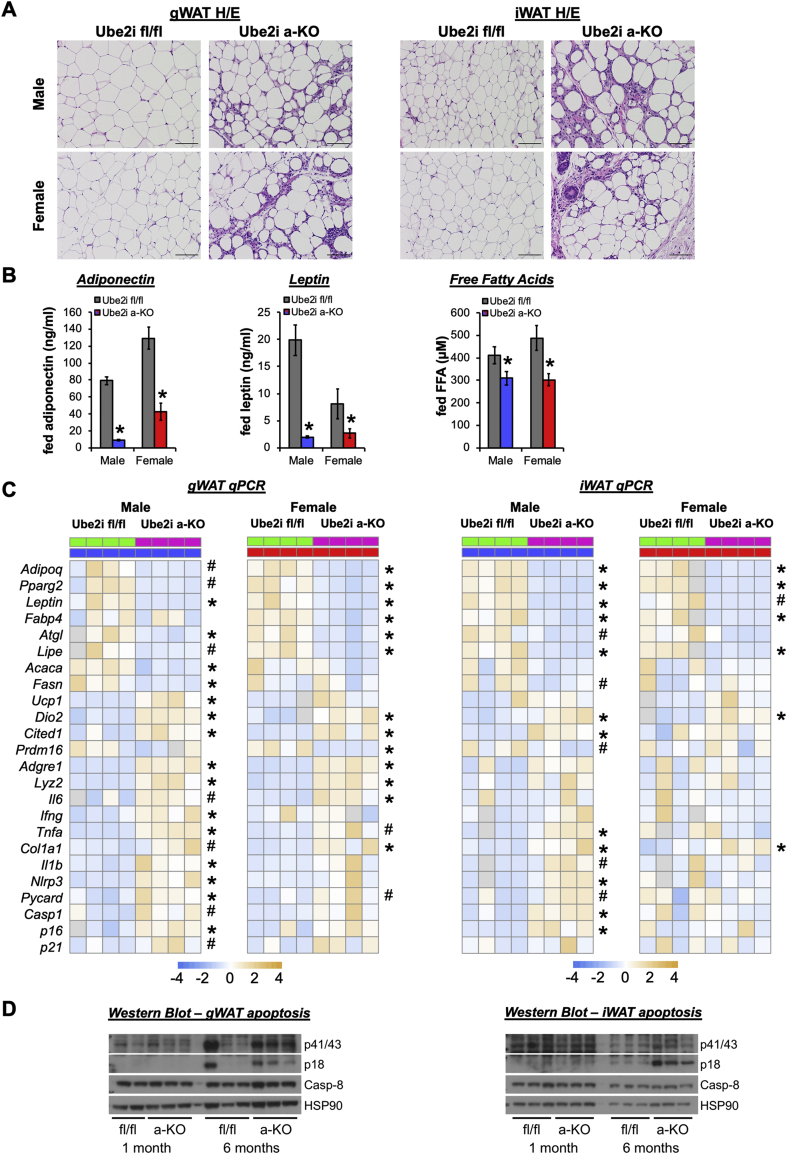
To assess the morphological changes associated with Ube2i knockout in adipose tissues, we performed H/E staining of gWAT and iWAT tissue sections in adult male and female mice. Pronounced immune cell infiltration was observed in gWAT of Ube2ia-KO mice (Figure 3A), distinct from the commonly described crown-like structures typically associated with gWAT [18]. Similarly, we observed dispersed immune cell accumulation amongst stromal cells and large adipocytes in the iWAT of Ube2ia-KO mice. Primary WAT depots from both sexes showed marked stroma invasion and few mature adipocytes. Consistent with reduced numbers of adipocytes, serum levels of adiponectin, leptin, and free fatty acids were significantly reduced in Ube2ia-KO mice compared to controls (Figure 3B). Adipocyte-specific deletion of Ube2i reduced hallmark adipocyte differentiation (Adipoq, Pparγ2, Lep, Fabp4), lipogenesis (Fasn, Acaca), and lipolysis (Atgl, Lipe) genes in WAT. Immune cell markers (F4/80, Tnfa, Ifng) and the collagen gene Col1a1 were upregulated in both the gWAT and iWAT (Figure 3C), consistent with morphological changes. However, we noted divergent levels of inflammatory and lipogenesis genes in iWAT of males and females indicative of the known influence of sex on WAT [19]. Gene expression markers of senescence (p16) and the Nlrp3 inflammasome (Il1b, Nlrp3, Pycard, and Casp1) were particularly elevated in male WAT of Ube2ia-KO, which may suggest increased innate and danger signal transduction in knockout mice. Indeed, we observed higher cleaved caspase-8 levels (Figure 3D) at six months of age, but not at one month, indicating increased cell death by apoptosis in Ube2ia-KO WAT and an important role for UBE2I in adipocyte survival of adolescent mice. Thus, impaired fat storage and WAT expansion in Ube2ia-KO mice coincides with inflammatory responses that drive adipocyte cell death.
Figure 3.
Ube2ia-KOmice display WAT dysfunction with increased inflammation and apoptosis. (A) H/E-stained sections from gWAT and iWAT of male (top row) and female (bottom row) mice show substantial immune and stromal cell infiltration in Ube2ia-KO mice. Scale bar 100 μm. (B) Fed serum adiponectin (ng/ml), leptin (ng/ml), and free fatty acids (μM) in male (blue/gray; n = 11–12/group) and female (red/gray; n = 7–9/group) Ube2ia-KO mice compared to Ube2ifl/fl controls. Data are presented as mean +/− SEM; ∗p < 0.05. (C) Relative gene expression by qPCR in gWAT (left) and iWAT (right) from male (blue) and female (red) Ube2ia-KO (pink) and Ube2ifl/fl control (green) mice for adipocyte maturation, lipid metabolism, inflammatory, and senescence genes represented as a heatmap of z-scores (∗p < 0.05, #p < 0.10). Gray heatmap squares indicate outliers excluded based on Grubbs' test. (D) Tissue lysates from gWAT (left) and iWAT (right) of Ube2ifl/fl and Ube2ia-KO mice were subjected to Western blot analysis of cleaved (p41/43, p18) and uncleaved Caspase-8. All analyses were performed in six-month-old adult mice.
3.4. Hepatosteatosis and insulin resistance in adult Ube2ia-KO mice
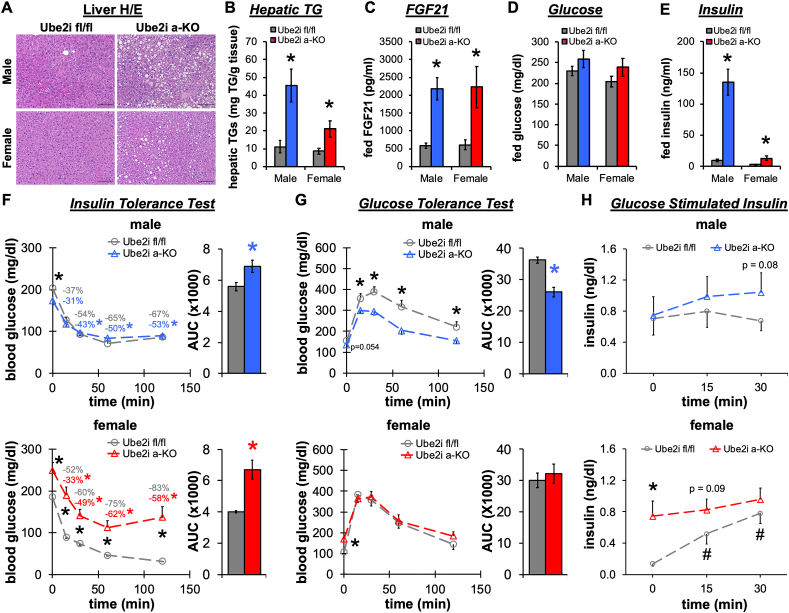
The inability of WAT depots to sequester lipids causes ectopic accumulation of energy in peripheral organs and lipodystrophic phenotypes [20]. Accordingly, six-month-old Ube2ia-KO mice developed fatty liver disease on a normal chow diet. Both male and female Ube2ia-KO mice displayed hepatic lipid droplet accumulation (Figure 4A) with significantly increased TG content (Figure 4B). FGF21 levels were increased 4-fold in Ube2ia-KO serum compared to controls (Figure 4C), indicating elevated stress responses in the liver [21]. Ad libitum fed glucose levels trended higher in Ube2ia-KO (Figure 4D), despite dramatically elevated levels of serum insulin (Figure 4E). Interestingly, we detected ∼14-fold higher serum insulin levels in male Ube2ia-KO mice while females exhibited only a 4-fold increase compared to controls. To test the consequences of adipose tissue loss on glucose homeostasis, we performed insulin (Figure 4F) and glucose tolerance tests (Figure 4G). Ube2ia-KO male and female mice were insulin resistant relative to controls. However, with a prolonged fast, male Ube2ia-KO showed improved glucose intolerance with a lower fasting glucose level. Glucose stimulated insulin secretion during glucose tolerance tests in male Ube2ia-KO mice trended higher than that in controls (Figure 4H), but importantly, demonstrated that the beta cell adapts properly during fasting compared to the fed state, as observed previously for other lipodystrophic mice [22]. Conversely, fasting glucose was elevated in female Ube2ia-KO compared to controls, with nominal effects on overall glucose tolerance due to enhanced glucose stimulated insulin secretion (Figure 4H). In summary, female Ube2ia-KO mice demonstrate fasting hyperglycemia and substantially impaired insulin sensitivity, in contrast to male Ube2ia-KO that demonstrate modest insulin resistance with improved glucose clearance during glucose tolerance tests. The sex discrepancy in glucose tolerance may be partly explained by greater skeletal muscle mass available for glucose disposal [23], as suggested by 35% more lean mass in male Ube2ia-KO than in female Ube2ia-KO mice (Figure 2F).
Figure 4.
Adipocyte-specific Ube2i knockout mice develop insulin resistance and hepatic steatosis. (A) Representative H/E stained liver sections from male (top row) and female (bottom row) Ube2ia-KO and Ube2ifl/fl control mice show lipid droplet accumulation in Ube2ia-KO mice. Scale bar 100 μm. (B) Hepatic triglycerides (TGs) per gram liver tissue (n = 7/group). (C) Fed serum FGF21 (pg/ml) (male n = 11–12/group; female n = 7–9/group). (D) Fed serum glucose (mg/dl) and (E) insulin (ng/ml) (male n = 11–12/group; female n = 7–9/group). (F) Insulin and (G) glucose tolerances tests were performed on Ube2ifl/fl and Ube2ia-KO male (n = 11–15/group) and female (n = 5–7/group) mice. Area under the curve is also shown. Values labeled on panels in (F) indicate the percentage decrease from initial fasting blood glucose. (H) Serum insulin during glucose tolerance test from Ube2ifl/fl and Ube2ia-KO male (n = 5/group) and female (n = 4–6/group) mice. Gray = Ube2ifl/fl male and female controls, blue = male Ube2ia-KO, red = female Ube2ia-KO. Data are presented as mean +/− SEM; ∗p < 0.05 between groups, #p < 0.05 versus time zero. All analyses were performed in six-month-old adult mice.
3.5. Adipocyte-specific Ube2i deletion increases energy expenditure
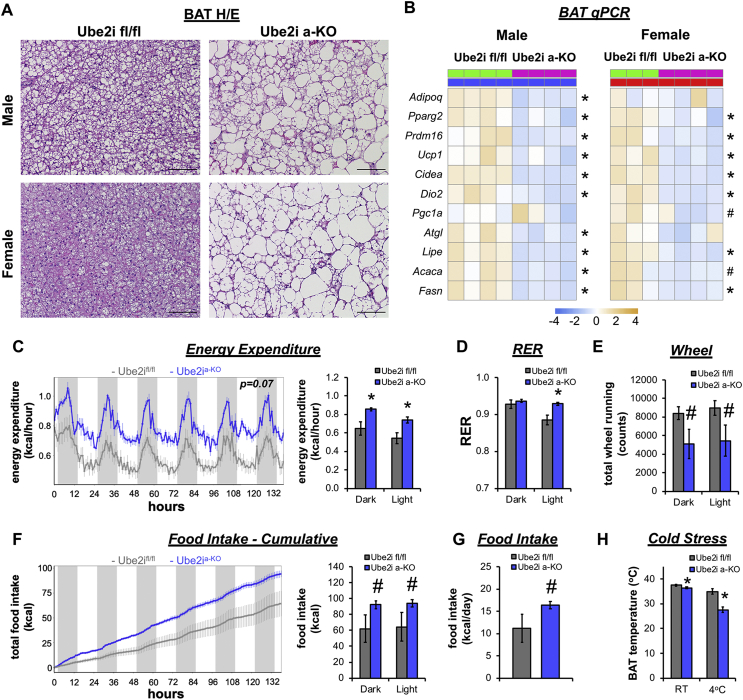
We subsequently investigated the effects of Ube2ia-KO on energy expenditure and BAT thermogenic functions. Histological assessment of H/E stained BAT sections from Ube2ia-KO male and female mice demonstrated lipid accumulation in large unilocular fat droplets, in contrast to the small multi-locular fat droplets characteristic of controls (Figure 5A). Ube2ia-KO caused dysfunctional BAT gene expression (Figure 5B) as reflected by reduced levels of hallmark brown adipocyte genes (Prdm16, Ucp1, Cidea, and Dio2) and key lipid metabolism genes (Atgl, Lipe, Acaca, and Fasn). Adult male Ube2ia-KO mice were placed in CLAMS home cages to assess heat production (energy expenditure) by indirect calorimetry. Ube2ia-KO mice exhibited increased energy expenditure (Figure 5C) and primarily used glucose as a fuel source during the light period (Figure 5D) as indicated by an elevated respiratory exchange ratio (RER). Increased energy expenditure was not associated with higher activity levels (Figure 5E), but rather poor metabolic flexibility likely stimulated greater food intake (Figure 5F,G) and consequently elevated energy expenditure in Ube2ia-KO mice. To test the functional output of BAT, we performed a cold tolerance test in the absence of food. Before cold exposure, BAT temperature was reduced in Ube2ia-KO mice compared to controls and dramatically dropped after 2.5 h at 4 °C (Figure 5H), demonstrating an inability of Ube2ia-KO mice to defend body temperature. These experiments demonstrate that Ube2ia-KO causes a hypermetabolic phenotype coupled with metabolic inflexibility and BAT thermogenic defects. Collectively, adipocyte-specific deletion of Ube2i impairs WAT expansion fostering ectopic lipid accumulation in peripheral organs, compromising insulin sensitivity and thermogenesis.
Figure 5.
Adipocyte-specific Ube2i deletion increases energy expenditure and cold intolerance. (A) H/E stained BAT sections from male and female Ube2ifl/fl and Ube2ia-KO mice show large unilocular lipid droplets in Ube2ia-KO mice. Scale bar 100 μm. (B) Relative gene expression by qPCR in BAT from male (left, blue) and female (right, red) Ube2ia-KO (pink) and Ube2ifl/fl control (green) mice for brown adipocyte and lipid metabolism genes represented as a heatmap of z-scores (∗p < 0.05, #p < 0.10). Six-month-old Ube2ifl/fl (gray) and Ube2ia-KO (blue) male mice were individually housed and monitored in CLAMS home cages for 6 days (n = 4–5/group). (C) Recorded traces of energy expenditure (kcal/hour) with mean values for dark/light periods (kcal/h). (D) Average RER during dark and light periods. (E) Activity was measured by wheel running. (F) Recorded traces of cumulative food intake (kcal) with mean values for dark/light periods (kcal) and (G) total food intake (kcal) per day. (H) Temperature probes were inserted under the skin to monitor intrascapular BAT temperature before (room temperature, RT) and after 2.5 h of cold (4 °C) exposure (n = 3–5/group). Statistical analysis of energy expenditure was performed by ANCOVA with lean body mass as a co-variate and cumulative food intake by standard ANOVA. Data are presented as mean +/− SEM; ∗p < 0.05, #p < 0.10.
4. Discussion
Our previous work identified the E2 SUMO conjugating enzyme, Ube2i, as a negative regulator of fat cell differentiation in human subcutaneous preadipocytes [10]. In the present study, we generated adipocyte-specific Ube2i knockout (Ube2ia-KO) mice to define the in vivo functions of Ube2i in mature fat cells. To our surprise, male and female Ube2ia-KO mice developed a hypermetabolic phenotype and progressive WAT lipoatrophy associated with immune cell infiltration and decreased circulating adipokines. Ube2ia-KO mice exhibited additional hallmark features of lipodystrophy, including ectopic lipid deposition in the liver, insulin resistance, and metabolic inflexibility. Ube2ia-KO WAT and BAT show broad dysfunction reflected by the expression of genes involved in energy storage and mobilization. These studies demonstrate previously unrealized functions of Ube2i in mouse WAT that are essential for mature adipocyte function and survival.
Patients with congenital general (CGL) or familial partial (FPLD) lipodystrophy exhibit profound insulin resistance, hepatic steatosis, and dyslipidemia. Female patients in particular develop more severe metabolic complications than males [24,25]. Similarly, impaired glucose and insulin tolerance in Ube2ia-KO mice were more pronounced in females, attributed in part to a greater skeletal muscle mass in male mice available for glucose disposal [23]. In terms of fat mass, CGL patients have extreme deficits at birth due to autosomal recessive mutations, while FPLD patients develop progressive fat loss beginning at puberty [2,[24], [25], [26]]. Ube2ia-KO mice resemble FPLD because WAT mass was normal in Ube2ia-KO mice after birth, but began to decrease during puberty, with as little as 2% remaining by six months. The timing of fat loss in Ube2ia-KO mice is consistent with a period of hypertrophic WAT expansion [27], impairments of which degrade adipocyte maturation and endocrine functions. Accordingly, inhibition of SUMOylation in differentiated cells [28] decreased adipocyte lipid storage and maturation genes (Pparγ, C/ebpa, Fabp4). Collectively, these observations suggest that Ube2i has a distinct role in adult adipocyte function, maturation, and survival.
A few lipodystrophic mouse models develop progressive lipoatrophy and hepatic steatosis, such as adipocyte-specific knockout of Akt1/Akt2 [29], insulin receptor [30], Raptor [31], and PPARγ [4]. Our model has many features similar to those of progressive lipoatrophy observed in PPARγ knockout models [4,8]. Lipoatrophy in fat-specific PPARγ knockout mice occurs because adipocytes lost lipids and shrank, consistent with the pivotal roles of PPARγ as a master regulator of adipose tissue formation [4,32]. PPARγ ablation in Sox2-expressing cells (PPARγΔ/Δ) skirts embryonic lethality and mice survive without WAT and BAT [8]. PPARγΔ/Δ show a hypermetabolic phenotype accompanied by higher energy expenditure and hyperphagia. Similar to Ube2ia-KO mice, PPARγΔ/Δ mice show higher RER and glucose oxidation that derives from more lean mass. Alternatively, depletion of circulating leptin in lipoatrophy would increase hyperphagia and could underlie hyperinsulinemia and increased energy expenditure. However, at least one model demonstrated that increased food intake was independent of leptin levels in lipoatrophic mice [33]. The liver dominates mass-specific metabolic rates [34] and, consequently, the hepatomegaly and hyperinsulinemia present in Ube2ia-KO likely contribute to greater glucose oxidation and metabolic inflexibility. Elevated energy expenditure and metabolic defects are similarly observed in other lipodystrophic mouse models [8,35,36]. Our observations expand on these studies and document a putative role for Ube2i and PPARγ interactions [10] in maintaining adipose tissue homeostasis during postnatal growth.
For reasons that remain unclear, Ube2i expression peaks early in differentiation of 3T3-L1 adipocytes, when adiponectin remains low, to regulate expression of the key adipogenic transcription factors PPARγ, C/EBPα, and C/EBPβ [37]. Adiponectin expression occurs late in adipogenesis [38] and Adipoq-Cre mediated ablation of Ube2i occurs in relatively mature adipocytes. While impaired adipogenesis underlies the development of lipodystrophy in some models [8,35,36,39], it is unlikely to account for lipoatrophy in Ube2ia-KO mice. The more dramatic lipodystrophy in older Ube2ia-KO mice likely results from apoptosis and inflammation, consistent with the pivotal roles of Ube2i in cell survival across many tissue types [14]. Cell survival also depends on preservation of the nuclear architecture, evident by the embryonic lethality of Ube2i deficiency [16]. Additionally, Ube2i occupies transcriptional start sites of functional genes integral for cell growth and proliferation [40]. However, loss of Ube2i induces cell growth arrest, due in part, to impaired chromosome segregation, which contributes to reduced cell viability [40]. Along these lines, the lipoatrophy caused by induction of caspase 8 in Fabp4-positive adipocytes [33] mirrors some features of Ube2ia-KO mice, including WAT inflammation and depletion of adipokines. However, recombination of alleles in other Fabp4-positive adipocytes and other tissues [41] likely contributes to some of the metabolic phenotype upon caspase-8 induction. Of note, the expression of NLRP3 inflammasome genes (Nlrp3, Il-1b, Pycard, Casp1) correlate with elevated cleaved Caspase-8 in WAT of Ube2ia-KO, suggesting a potential mechanism [42] for adipocyte cell death. New studies that use temporal deletion strategies [27] will provide insight into whether acute deletion of Ube2i in adult mice causes lipodystrophy from fat cell death or restricted adipocyte turnover.
Ube2i exerts a diverse array of cellular functions through SUMOylation and protein–protein interactions that influence gene transcription among many others roles [43]. During adipocyte differentiation, Ube2i knockdown de-represses transcription of the brown fat gene program, enabling the master transcription factor PPARγ to bind to UCP1 enhancers promoting uncoupled respiration [10]. Moreover, Ube2i interacts with the master transcription factor PPARγ to suppress DNA occupancy and ligand dependent activity in vitro [10]. In vivo, mice resistant to PPARγ SUMOylation at K107 exhibit better insulin sensitivity [44]. In some settings, SUMOylation prevents ubiquitination and protein turnover. For example, Cbx4 serves as SUMO E3 ligase that stabilizes PRDM16 to facilitate thermogenesis and glucose homeostasis [45], which may explain, at least in part, the cold intolerance and metabolic defects in Ube2ia-KO mice.
We and others demonstrated that SUMOylation of PPARγ broadly inhibits metabolic functions of white adipocytes [10,44,46]. Based on these data, we predicted greater beige fat thermogenesis and improved energy balance in Ube2ia-KO mice relative to littermate controls. The lack of exact agreement between phenotypes observed in tissue culture versus whole animals is not surprising, but perhaps expected due to the importance of Ube2i in cell survival [14,16]. Ube2i knockout in adipocytes may also cause depletion of adipocyte progenitor cells coupled with Ucp1 deregulation and induction of the fibro-inflammatory program seen in settings of accelerating ageing [47]. Regardless, the extensive roles of Ube2i suggest the mechanism underlying lipoatrophy in Ube2ia-KO is unlikely to be the result of a single SUMOylation event. It will now be important to use mass spectrometry and lineage tracing methods to identify the most critical metabolic and transcription factors downstream of Ube2i functions that explain why Ube2i allows WAT expansion during ageing.
Conditions of lipodystrophy and pathologic obesity suffer from a restricted capacity to expand white adipose depots to meet the demand for nutrient storage, which drives adipocyte dysfunction and WAT inflammation, along with ectopic lipid accumulation and insulin resistance [48]. Ultimately, identifying factors that enable healthy WAT expansion carries significant implications for treating diseases associated with fat storage and metabolism, including obesity and lipodystrophy. The striking phenotype of Ube2ia-KO mice reveals the cell-autonomous necessity of Ube2i in healthy adipose tissue maintenance and whole-body energy balance. As a rate-limiting E2 SUMO conjugating enzyme necessary for SUMOylation, this mouse model provides a valuable tool for understanding how the low-abundance post-translational modification SUMOylation [49] broadly affects mature adipocyte function and, more broadly, tissue development. In addition, Ube2ia-KO mice add to a small list of mouse models to study systemic effects of WAT loss during postnatal development and fundamental aspects of energy balance.
Funding
This work was funded by American Diabetes Association #1-18-IBS-105, and NIH R01DK 126042, R01DK114356 (to S.M.H.), R01 DK111436 (to Z.S.), R01 HD085994 and T32 HD098068 (to S.A.P). This study was also funded (in part) by an award from the Baylor College of Medicine Nutrition and Obesity Pilot and Feasibility Fund (to A.R.C.) and the Nancy Chang, PhD Award for Research Excellence at BCM (to S.M.H.). This study was also supported by the Assistant Secretary of Defense for Health Affairs endorsed by the DOD PRMRP Discovery Award (No. W81XWH-18-1-0126 to K.H.K.). Core services at BCM utilized in this project were supported with funding from NCI P30-CA125123: Genetically Engineered Rodent Models Core, Human Tissue Acquisition and Histology Core, and the Integrated Microscopy Core.
Author contributions
A.R.C. and S.M.H. conceptualized the study. P.M., N.C., A.R.C., and S.M.H. designed the experiments. A.R.C. and S.M.H. wrote the manuscript with editorial input from all authors. S.M.H. and A.R.C performed all experiments with assistance as noted: P.K.S. assisted with mouse phenotyping; A.R.C. performed qPCR analysis with assistance from J.B.F., N.C. and R.S.; D.D.M., S.M.B., and S.A.P. performed genotyping and troubleshooting; K.H.K. performed analysis of liver lipids. X.L. and Z.S. provided resources and support for body temperature experiments. All work was performed under the supervision of S.M.H. The authors thank Robb Moses (BCM) for reading drafts of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101221.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Roden M., Shulman G.I. The integrative biology of type 2 diabetes. Nature. 2019;576(7785):51–60. doi: 10.1038/s41586-019-1797-8. [DOI] [PubMed] [Google Scholar]
- 2.Garg A. Acquired and inherited lipodystrophies. New England Journal of Medicine. 2004;350(12):1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 3.Cristancho A.G., Lazar M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nature Reviews Molecular Cell Biology. 2011;12(11):722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F., Mullican S.E., DiSpirito J.R., Peed L.C., Lazar M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(46):18656–18661. doi: 10.1073/pnas.1314863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajakumari S., Wu J., Ishibashi J., Lim H.-W., Giang A.-H., Won K.-J. EBF2 determines and maintains brown adipocyte identity. Cell Metabolism. 2013;17(4):562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barak Y., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R. PPAR gamma is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 8.Gilardi F., Winkler C., Quignodon L., Diserens J.G., Toffoli B., Schiffrin M. Systemic PPARγ deletion in mice provokes lipoatrophy, organomegaly, severe type 2 diabetes and metabolic inflexibility. Metabolism. 2019;95:8–20. doi: 10.1016/j.metabol.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Molecular Cell. 1999;4(4):611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 10.Hartig S.M., Bader D.A., Abadie K.V., Motamed M., Hamilton M.P., Long W. Ubc9 impairs activation of the brown fat energy metabolism program in human white adipocytes. Molecular Endocrinology. 2015;29(9):1320–1333. doi: 10.1210/me.2015-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanza D.G., Gaspero A., Lorenzo I., Liao L., Zheng P., Wang Y. Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles. BMC Biology. 2018;16(1):69. doi: 10.1186/s12915-018-0529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mina A.I., LeClair R.A., LeClair K.B., Cohen D.E., Lantier L., Banks A.S. CalR: a web-based analysis tool for indirect calorimetry experiments. Cell Metabolism. 2018;28(4):656–666. doi: 10.1016/j.cmet.2018.06.019. e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh E.H., Chernis N., Saha P.K., Xiao L., Bader D.A., Zhu B. miR-30a remodels subcutaneous adipose tissue inflammation to improve insulin sensitivity in obesity. Diabetes. 2018;67(12):2541–2553. doi: 10.2337/db17-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demarque M.D., Nacerddine K., Neyret-Kahn H., Andrieux A., Danenberg E., Jouvion G. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology. 2011;140(1):286–296. doi: 10.1053/j.gastro.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Ding X., Wang A., Ma X., Demarque M., Jin W., Xin H. Protein SUMOylation is required for regulatory T cell expansion and function. Cell Reports. 2016;16(4):1055–1066. doi: 10.1016/j.celrep.2016.06.056. [DOI] [PubMed] [Google Scholar]
- 16.Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Developmental Cell. 2005;9(6):769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez A., Briley S.M., Patton B.K., Tripurani S.K., Rajapakshe K., Coarfa C. Loss of the E2 SUMO-conjugating enzyme Ube2i in oocytes during ovarian folliculogenesis causes infertility in mice. Development. 2019;146(23) doi: 10.1242/dev.176701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murano I., Barbatelli G., Parisani V., Latini C., Muzzonigro G., Castellucci M. Dead adipocytes, detected as crown-like structures, are prevalent in visceral fat depots of genetically obese mice. The Journal of Lipid Research. 2008;49(7):1562–1568. doi: 10.1194/jlr.M800019-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Goossens G.H., Jocken J.W.E., Blaak E.E. Sexual dimorphism in cardiometabolic health: the role of adipose tissue, muscle and liver. Nature Reviews Endocrinology. 2021;17(1):47–66. doi: 10.1038/s41574-020-00431-8. [DOI] [PubMed] [Google Scholar]
- 20.Savage D.B., Murgatroyd P.R., Chatterjee V.K., O'Rahilly S. Energy expenditure and adaptive responses to an acute hypercaloric fat load in humans with lipodystrophy. Journal of Clinical Endocrinology & Metabolism. 2005;90(3):1446–1452. doi: 10.1210/jc.2004-1494. [DOI] [PubMed] [Google Scholar]
- 21.Fisher F.M., Maratos-Flier E. Understanding the physiology of FGF21. Annual Review of Physiology. 2016;78:223–241. doi: 10.1146/annurev-physiol-021115-105339. [DOI] [PubMed] [Google Scholar]
- 22.Moitra J., Mason M.M., Olive M., Krylov D., Gavrilova O., Marcus-Samuels B. Life without white fat: a transgenic mouse. Genes & Development. 1998;12(20):3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W., Zhou H., Xuan H., Saha P., Wang G., Chen W. Novel metabolic disorders in skeletal muscle of Lipodystrophic Bscl2/Seipin deficient mice. Molecular and Cellular Endocrinology. 2019;482:1–10. doi: 10.1016/j.mce.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araújo-Vilar D., Loidi L., Domínguez F., Cabezas-Cerrato J. Phenotypic gender differences in subjects with familial partial lipodystrophy (Dunnigan variety) due to a nuclear lamin A/C R482W mutation. Hormone and Metabolic Research. 2003;35(1):29–35. doi: 10.1055/s-2003-38388. [DOI] [PubMed] [Google Scholar]
- 25.Van Maldergem L., Magré J., Khallouf T.E., Gedde-Dahl T., Delépine M., Trygstad O. Genotype-phenotype relationships in Berardinelli-Seip congenital lipodystrophy. Journal of Medical Genetics. 2002;39(10):722–733. doi: 10.1136/jmg.39.10.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunnigan M.G., Cochrane M.A., Kelly A., Scott J.W. Familial lipoatrophic diabetes with dominant transmission. A new syndrome. Quarterly Journal of Medicine. 1974;43(169):33–48. [PubMed] [Google Scholar]
- 27.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Li J., Lu D., Li J., Liu M., He Y. Ginkgolic acid, a sumoylation inhibitor, promotes adipocyte commitment but suppresses adipocyte terminal differentiation of mouse bone marrow stromal cells. Scientific Reports. 2018;8(1):2545. doi: 10.1038/s41598-018-20244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shearin A.L., Monks B.R., Seale P., Birnbaum M.J. Lack of AKT in adipocytes causes severe lipodystrophy. Molecular Metabolism. 2016;5(7):472–479. doi: 10.1016/j.molmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Softic S., Boucher J., Solheim M.H., Fujisaka S., Haering M.F., Homan E.P. Lipodystrophy due to adipose tissue-specific insulin receptor knockout results in progressive NAFLD. Diabetes. 2016;65(8):2187–2200. doi: 10.2337/db16-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee P.L., Tang Y., Li H., Guertin D.A. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Molecular Metabolism. 2016;5(6):422–432. doi: 10.1016/j.molmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He W., Barak Y., Hevener A., Olson P., Liao D., Le J. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pajvani U.B., Trujillo M.E., Combs T.P., Iyengar P., Jelicks L., Roth K.A. Fat apoptosis through targeted activation of caspase 8: a new mouse model of inducible and reversible lipoatrophy. Nature Medicine. 2005;11(7):797–803. doi: 10.1038/nm1262. [DOI] [PubMed] [Google Scholar]
- 34.Rolfe D.F., Brown G.C. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiological Reviews. 1997;77(3):731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 35.Péterfy M., Phan J., Xu P., Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nature Genetics. 2001;27(1):121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 36.Cortés V.A., Curtis D.E., Sukumaran S., Shao X., Parameswara V., Rashid S. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metabolism. 2009;9(2):165–176. doi: 10.1016/j.cmet.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cignarelli A., Melchiorre M., Peschechera A., Conserva A., Renna L.A., Miccoli S. Role of UBC9 in the regulation of the adipogenic program in 3T3-L1 adipocytes. Endocrinology. 2010;151(11):5255–5266. doi: 10.1210/en.2010-0417. [DOI] [PubMed] [Google Scholar]
- 38.Hu E., Liang P., Spiegelman B.M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. Journal of Biological Chemistry. 1996;271(18):10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 39.Chen W., Chang B., Saha P., Hartig S.M., Li L., Reddy V.T. Berardinelli-seip congenital lipodystrophy 2/seipin is a cell-autonomous regulator of lipolysis essential for adipocyte differentiation. Molecular and Cellular Biology. 2012;32(6):1099–1111. doi: 10.1128/MCB.06465-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neyret-Kahn H., Benhamed M., Ye T., Le Gras S., Cossec J.-C., Lapaquette P. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Research. 2013;23(10):1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giordano A., Murano I., Mondini E., Perugini J., Smorlesi A., Severi I. Obese adipocytes show ultrastructural features of stressed cells and die of pyroptosis. The Journal of Lipid Research. 2013;54(9):2423–2436. doi: 10.1194/jlr.M038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang H.M., Yeh E.T.H. SUMO: from bench to bedside. Physiological Reviews. 2020;100(4):1599–1619. doi: 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katafuchi T., Holland W.L., Kollipara R.K., Kittler R., Mangelsdorf D.J., Kliewer S.A. PPARγ-K107 SUMOylation regulates insulin sensitivity but not adiposity in mice. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(48):12102–12111. doi: 10.1073/pnas.1814522115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Q., Huang L., Pan D., Zhu L.J., Wang Y.X. Cbx4 sumoylates Prdm16 to regulate adipose tissue thermogenesis. Cell Reports. 2018;22(11):2860–2872. doi: 10.1016/j.celrep.2018.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dutchak P.A., Katafuchi T., Bookout A.L., Choi J.H., Yu R.T., Mangelsdorf D.J. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148(3):556–567. doi: 10.1016/j.cell.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Z., Daquinag A.C., Fussell C., Zhao Z., Dai Y., Rivera A. Age-associated telomere attrition in adipocyte progenitors predisposes to metabolic disease. Nature Metabolism. 2020;2(12):1482–1497. doi: 10.1038/s42255-020-00320-4. [DOI] [PubMed] [Google Scholar]
- 48.Kahn C.R., Wang G., Lee K.Y. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. Journal of Clinical Investigation. 2019;129(10):3990–4000. doi: 10.1172/JCI129187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamoliatte F., Caron D., Durette C., Mahrouche L., Maroui M.A., Caron-Lizotte O. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nature Communications. 2014;5:5409. doi: 10.1038/ncomms6409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.