Highlights
-
•
Identify novel autophagy-related signature for OS and DSS in HNSCC patients.
-
•
The role of autophagy in tumor immune microenvironment.
-
•
Provide promising targets that can enhance the efficacy of cancer immunotherapy.
Keywords: TCGA, Autophagy-related genes, Overall survival, Disease-specific survival, Prognostic risk signature, TIME
Abstract
Background
Many studies have demonstrated that autophagy plays a significant role in regulating tumor growth and progression. However, the effect of autophagy-related genes (ARGs) on the prognosis have rarely been analyzed in head and neck squamous cell carcinoma (HNSCC).
Methods
We obtained differentially expressed ARGs from HNSCC mRNA data in The Cancer Genome Atlas (TCGA) database. And then we performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to explore the autophagy-related biological functions. The overall survival (OS)-related and disease specific survival (DSS)-related ARGs were identified by univariate Cox regression analyses. With these genes, we established OS-related and DSS-related risk signature by LASSO regression method, respectively. We validated the reliability of the risk signature with receiver operating characteristic (ROC) analysis, Kaplan-Meier survival curves, clinical correlation analysis, and nomogram. Then we analyzed relationships between risk signature and immune cell infiltration.
Results
We established the prognostic signatures based on 14 ARGs for OS and 12 ARGs for DSS. The ROC curves, survival analysis, and nomogram validated the predictive accuracy of the models. Clinic correlation analysis showed that the risk group was closely related to Stage, pathological T stage, pathological N stage and human papilloma virus (HPV) subtype. Cox regression demonstrated that the risk score was an independent predictor for the prognosis of HNSCC patients. Furthermore, patients in low-risk score group exhibited higher immunescore and distinct immune cell infiltration than high-risk score group. And we further analysis revealed that the copy number alterations (CNAs) of ARGs-based signature affected the abundance of tumor-infiltrating immune cells.
Conclusion
In this study, we identified novel autophagy-related signature for the prediction of OS and DSS in patients with HNSCC. Meanwhile, our study provides a novel sight to understand the role of autophagy and elucidate the important role of autophagy in tumor immune microenvironment (TIME) of HNSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC) refer to a heterogeneous group of cancers originating from the mouth, pharynx and the larynx. As one of the most common malignant tumors, HNSCC ranks 6th in the global cancer burdens, with ~600 000 new cases are diagnosed and ~350 000 cancer-related deaths annually worldwide [1]. Poor prognosis and lack of accurate and reliable prognostic indicators for monitoring HNSCC development results in the high mortality rate of patients.
Despite the rapid improvement of medical techniques for early diagnosis of HNSCC, nearly 50% of patients are diagnosed at an advanced stage. Although the comprehensive treatment of HNSCC (including surgical resection, radiotherapy, and chemotherapy) continues to develop in the past few decades, but the 5-year survival rate of HNSCC still remains dismal, especially for the advanced patients [2]. Thus, the identification of disease-related prognostic biomarkers in HNSCC is necessary to help optimize the level of early diagnosis and develop novel therapeutic approach to enhance the survival rate of patients.
In recent years, screening for useful genetic and epigenetic biomarkers by bioinformatic technologies has been an important means for diagnosis and prognosis of cancer [3], in which the impact of autophagy-related genes (ARGs) in the pathogenesis of cancer has received increasing attention.
Autophagy is a vital catabolic pathway that regulate homeostasis of the intracellular environment by reducing and recycling components of aged or damaging organelles, misfolded proteins [4, 5]. The upregulation and deregulation of autophagic level have been implicated in the pathogenesis of several diseases, including immune-related diseases, neurodegenerative diseases, and tumors. For example, ATG16L2 is an autophagy-related gene proposed as a systemic lupus erythematosus risk locus [6], and is associated with multiple sclerosis [7] and Crohn's disease [8]. Upregulation of autophagy may protect against neurodegeneration [9]. Interestingly, autophagy plays a contradictory role in all stages of tumor. During early stages of tumorigenesis, it can delay or prevent the formation of tumor. However, once tumor has formed, elevated autophagy promotes growth of cancer cell, and protect cancer cells from the damage of chemotherapy drugs [10], [11], [12]. Therefore, the levels of autophagy proteins play a significant role in regulating tumor cell growth and progression [13].
Several previous studies have conclusively illustrated the role of autophagy in HNSCC. Zhou et al. reported that radiation-induced autophagy enhanced the survival of cancer cell, by autophagy inhibition inducing cell death [14]. An experiment included 195 oral cancer tissues revealed that increased level of LC3-II in patients was associated with poor outcome, supporting an autophagy paradox when autophagy dysregulation in early stages cancers impedes tumorigenesis [15, 16]. Furthermore, autophagy inhibition may be beneficial for head and neck cancer patients [17, 18]. However, these studies simply clarified the correlation between a single autophagy gene and the prognosis of HNSCC or constructed a model with ARGs to predict the prognosis outcomes, few studies investigated the function of ARGs in tumor immune microenvironment (TIME).
This study aimed to systematically assess the correlations of ARGs with prognosis and TIME in HNSCC. The risk models of ARGs were established to facilitate treatment decision-making for HNSCC patient's outcome, which may represent a potential prognostic indicator. Subsequently, the relationships between risk models, immunoscores, and immune cell infiltration were thoroughly analyzed based on the autophagy-related signatures to further explore the effect of ARGs on TIME. This study also contributes to elucidate the regulatory mechanisms associated with TIME.
Materials and methods
Data collection
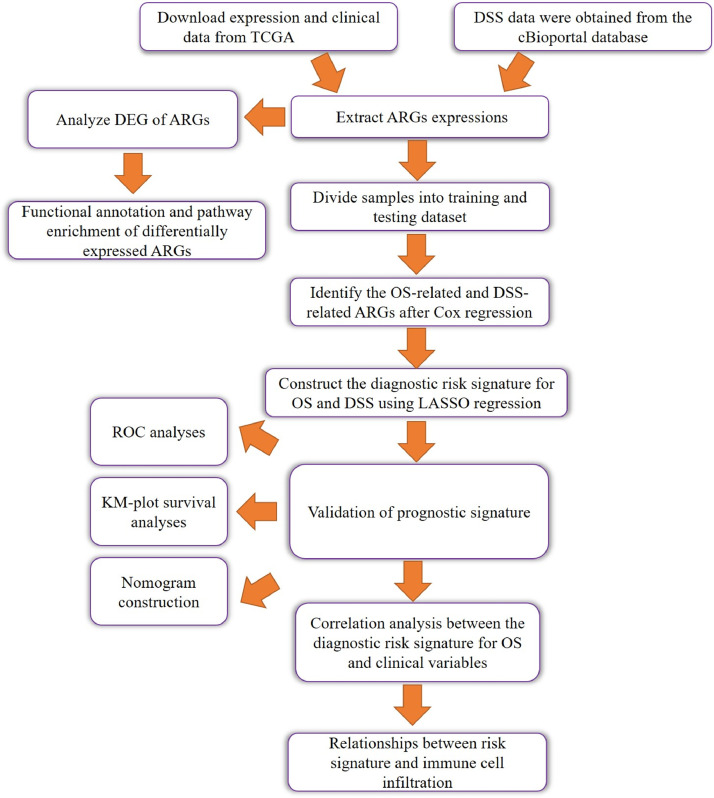
The detailed workflow of our study is shown as a chart (Fig. 1). The transcriptome profiles (HTSeq-Counts) of 546 HNSCC patients were downloaded from the TCGA database (https://portal.gdc.cancer.gov/), and the clinical data, including OS and DSS, was identified from the cBioPortal database (https://www.cbioportal.org/).Then, patients with follow-up times of less than 30 days were excluded. Finally, we obtained complete expression data and OS data from 487 patients and DSS data from 470 patients. A list of 233 ARGs were obtained from the Human Autophagy Database (HADb, http://www.autophagy.lu/) [19].
Fig. 1.
The workflow of study process.
Differentially expressed autophagy-related genes and enrichment analysis
The analysis of differentially expressed ARGs between HNSCC and corresponding normal samples was identified by edgeR package in R software. The cut-off criterion for differentially expressed ARGs was set as | log2FC |> 1 and false discovery rate (FDR) < 0.05. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed to better understand the key roles of differentially expressed ARGs using the “clusterProfiler” package [20] and “GOplot” package [21].
Construction and validation of ARGs-related risk model
All patients in TCGA were randomly divided into the training and testing cohort t a 3:2 ratio using the “caret” package. Firstly, univariate Cox regression was conducted in the TCGA training cohort to identify ARGs associated with OS and DSS. Then we performed LASSO, the least absolute shrinkage and selection operator, based on univariate Cox result to establish prognostic predictive module in the training cohort. The following formula is used to predict the risk score of each HNSCC patient: risk score =, where the exp was the expression value of gene, and coef was the coefficient of gene in the LASSO analysis. The risk score was calculated for each patient in the training, validation and whole cohorts, respectively. The median risk score was set as the cut-off value. Kaplan-Meier survival analysis assessed the differences between groups. We performed univariate and multivariate Cox regression to confirm whether the two signatures could be an independent prognostic factor for patients. Finally, the value of prediction models was evaluated by the receiver-operator characteristic (ROC) and corresponding areas under the curve (AUC).
Construction and assessment of the nomogram
To assist clinical decision making, the risk score and other clinicopathological factors were integrated to construct a nomogram, a quantitative tool to evaluate clinical outcome. Calibration curves were used to study the prognostic performance.
Relationships between risk signature and immune cell infiltration
The immunoscores of each sample were calculated with R package ESTIMATE [22]. To evaluate proportions of 22 immune cells, the cell type identification by estimating relative subset of known RNA transcripts (CIBERSORT) method was applied to TCGA cohort [23]. The analysis was run with 1000 permutations, and batch adjusted data. Samples whose P value of CIBERSORT results were ≥ 0.05 were eliminated in the subsequent analysis. Then, we further analyzed the effect of CNAs of the risk related genes in signature on immune cell infiltration levels via the TIMER database(https://cistrome.shinyapps.io/timer/) [24].
Statistical analysis
Statistical analysis was performed with R software (version 4.0.2), GraphPad Prism 8 (San Diego, CA) [25]. The volcano plot and heatmaps were draw using the “ggplot2” package in R. Chi-square test was used to evaluate the differences of clinicopathological parameters between the high- and low-risk groups. Survival curves were generated by the Kaplan-Meier method [26]. Univariate and LASSO regression analyses were performed to explore prognostic risk model. Time-dependent ROC analysis was applied to compare the predictive accuracy of the models.
Results
Detection of differentially expressed ARGs in HNSCC and normal human tissues
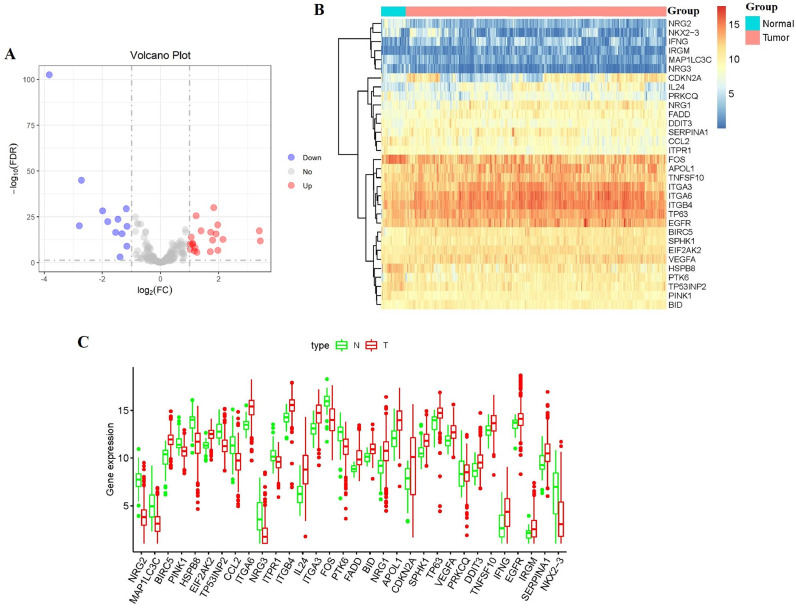
The study procedure is presented in Fig. 1. The RNA expression and clinical data of 546 HNSCC patients were downloaded from the TCGA database. Then we extract the expression values of 233 ARGs. On the basis of the screening criteria of DEGs, we obtained 32 differentially expressed ARGs (included 12 downregulated ARGs and 20 upregulated ARGs) (Fig. 2A and 2B). Furthermore, the box plot was generated to display the expression pattern of differentially expressed ARGs between cancers and normal tissues (Fig. 2C).
Fig. 2.
Differentially expressed autophagy-related genes (ARGs) between HNSCC and normal head and neck tissues. (A) The volcano plot for the 233 ARGs in HNSCC. Red indicates high expression and blue indicates low expression. Gray shows those genes showed no difference. (B) Heatmap of the 32 differentially expressed ARGs in head and neck cancer. (C) Boxplot showing the expression pattern of differentially expressed ARGs in HNSCC.
Functional enrichment analysis of the differentially expressed ARGs
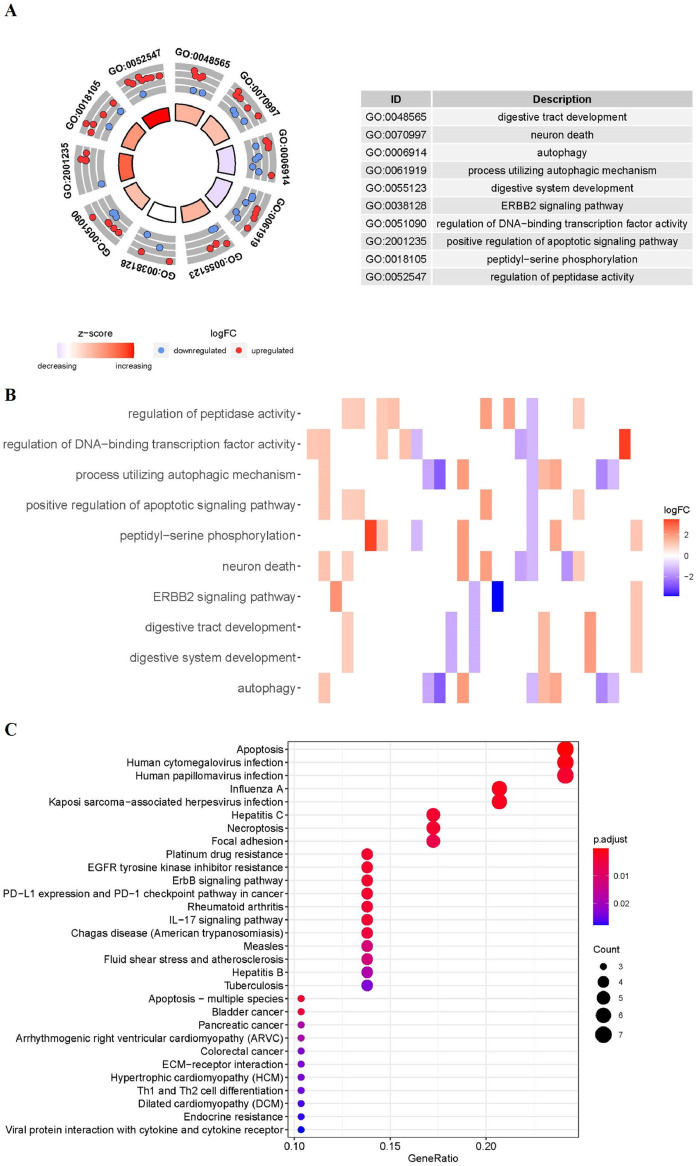
Functional enrichment analysis was performed according to the 32 differentially expressed ARGs. The GO terms and KEGG pathway enrichment results of these genes were summarized in Fig. 3. GO results show that these ARGs were mainly enriched in digestive tract development, neuron death, autophagy (Fig. 3A). The relation between ARGs and GO enrichment results was also displayed in the heatmap (Fig. 3B). KEGG enrichment results revealed that the 32 ARGs were mainly involved in apoptosis, human cytomegalovirus infection, and human papillomavirus infection (Fig. 3C).
Fig. 3.
Functional enrichment of the ARGs. (A) Circos plot of the GO enrichment results. (B) Heatmap of the GO enrichment results. The depth of the bar depends on the logFC values. (C) KEGG shows the potential pathway involved in differential ARGs. The size of the circle indicates the counts of enriched genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FC, fold change.
Construction and validation of the risk signature based on the prognosis-related ARGs for OS
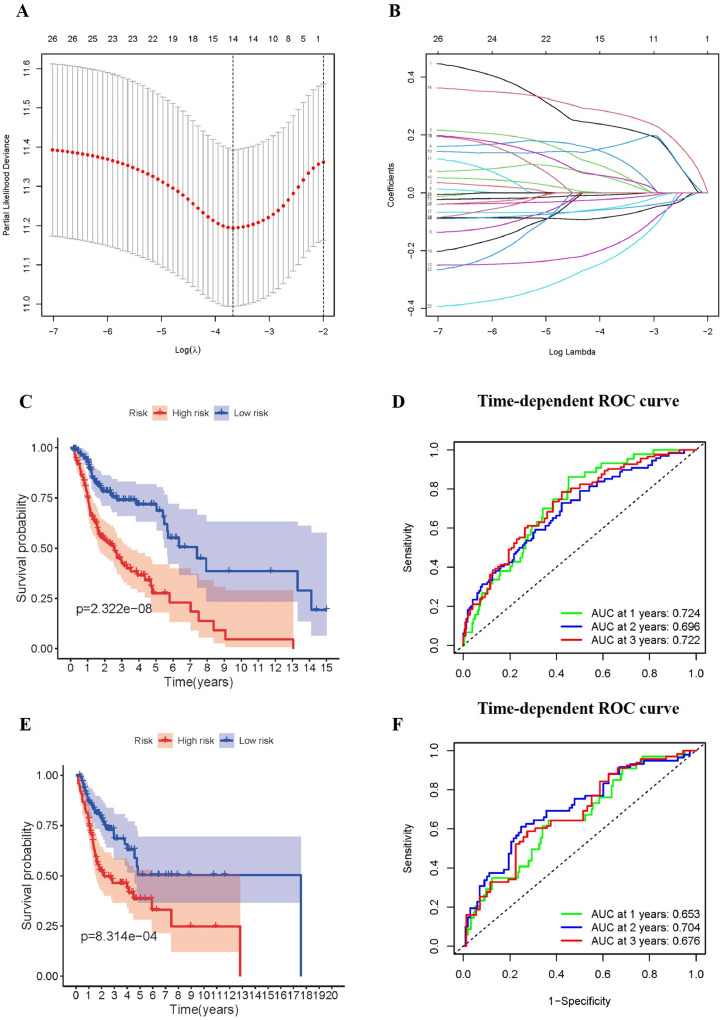
Next, we explored the prognostic role of ARGs. The 487 patients were randomly divided into training cohort and testing cohort according to the ratio of 3 to 2. To screen out the genes that significantly associated with OS, we performed univariate Cox regression analysis using the data from TCGA training cohort. 27 genes were significantly associated with OS based on univariate Cox regression (p < 0.05, full data are presented in Supplementary Table S1). Then the LASSO regression used above 27 genes to identify prognosis associated genes, namely GABARAPL2, GOPC, MAPK9, TP73, HSP90AB1, ST13, IKBKB, IFNG, GAPDH, NKX2–3, CCR2, VAMP7, CAPN10, CDKN2A, and establish the signature of OS for patients. (Fig. 4A and Fig. 4B). The risk scores of the TCGA training, testing and whole cohorts were calculated. Afterward, patients were classified into high-/low-risk groups based on the cut-off of the risk scores, respectively. Survival curve was used to compare the OS outcomes in patient groups. The patients in the low-risk group had poorer OS than those in high-risk group in training and testing cohorts (p < 0.001, Fig. 4C and Fig. 4E). To assess the efficiency of the model, we conducted 1-, 2- and 3-year ROC curve analyses by comparing the respective AUC values. In the training cohort, the1-, 2- and 3-year AUC values for the risk signatures were 0.724, 0.696 and 0.722, respectively (Fig. 4D). In the testing cohort, the 1-, 2- and 3-year AUC values for the risk signatures were 0.653, 0.704 and 0.676, respectively (Fig. 4F). Results show that the prognostic signature based on ARGs had certain prediction abilities in predicting survival outcome.
Fig. 4.
Construction and validation of the prognostic risk signature for OS. (A-B) The Lasso regression analyses of ARGs using the OS model. The Lasso regression was performed using prognosis-significant ARGs in the training cohort of HNSCC. Survival curve of OS in the training cohort (C), and testing cohort (E). ROC curves in the training cohort (D), and the testing cohort (F).
The correlation between the OS-related prediction model and prognosis of HNSCC patients
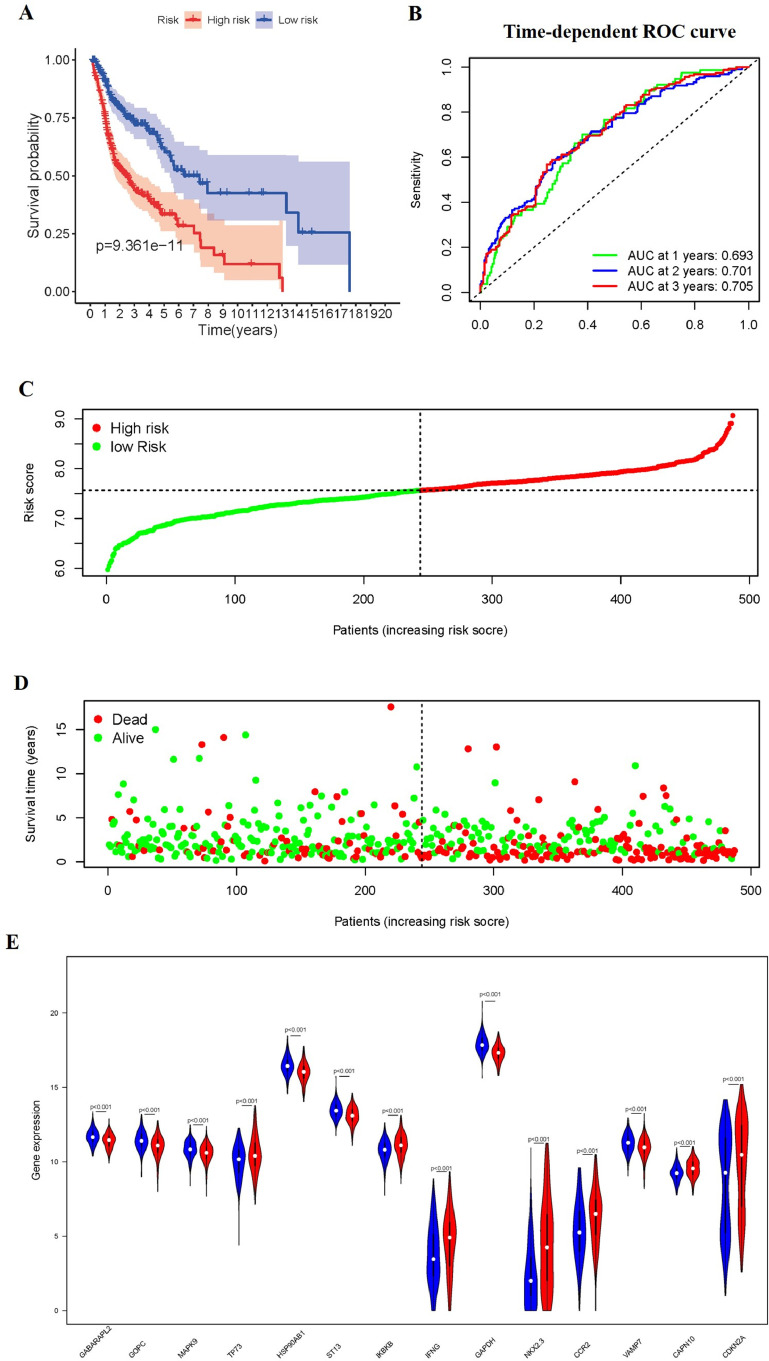
According to the same formula mentioned above, we calculate the risk score for patients, and divided them into high-risk group and low-risk group in the whole set. The Kaplan-Meier survival plot, the distribution of the risk scores, vital status, and corresponding 14 ARGs expression level of patients in TCGA whole cohort is displayed in Fig. 5.
Fig. 5.
The correlation between the ARGs prognostic signature and the prognosis. (A) Kaplan-Meier curves of the high-risk and low-risk groups. (B) The ROC of OS for the ARGs-based prognostic signature. (C) Distribution of the risk scores. (D) Survival status of patients in different groups. (E) Expression of 14 autophagy-related genes in the high-risk and low-risk groups.
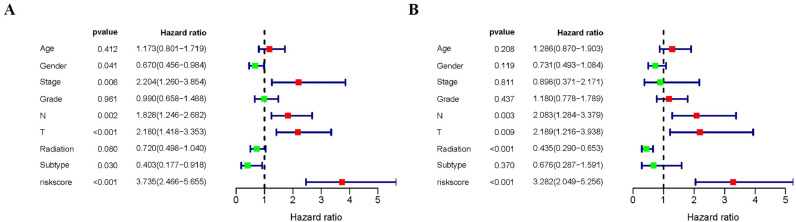
Furthermore, we evaluated the clinical significance of the signature by analyzing the correlation among clinical parameters. Univariate Cox regression revealed that the Stage, Gender, pathological T stage, pathological N stage, HPV subtype and risk score were correlated with OS in HNSCC patients (P < 0.05) (Fig. 6A). Multivariate Cox analysis showed that pathological T stage, pathological N stage, Radiation therapy and risk score were correlated with OS in HNSCC patients (P < 0.05) (Fig. 6B).
Fig. 6.
Univariate and multivariate Cox regression analyses of OS. Forest plots showing the univariate (A) and multivariate cox regression results (B) of OS in HNSCC.
And then the survival analyses were performed based on the OS-related genes and the results were exhibited in Fig. S1. The low expression level of GABARAPL2, GOPC, MAPK9, HSP90AB1, ST13, GAPDH in HNSCC were significantly associated with better OS rate. However, this association did not hold true among these genes-VAMP7, CAPN10, IKBKB, CDKN2A (Supplementary Fig.S1).
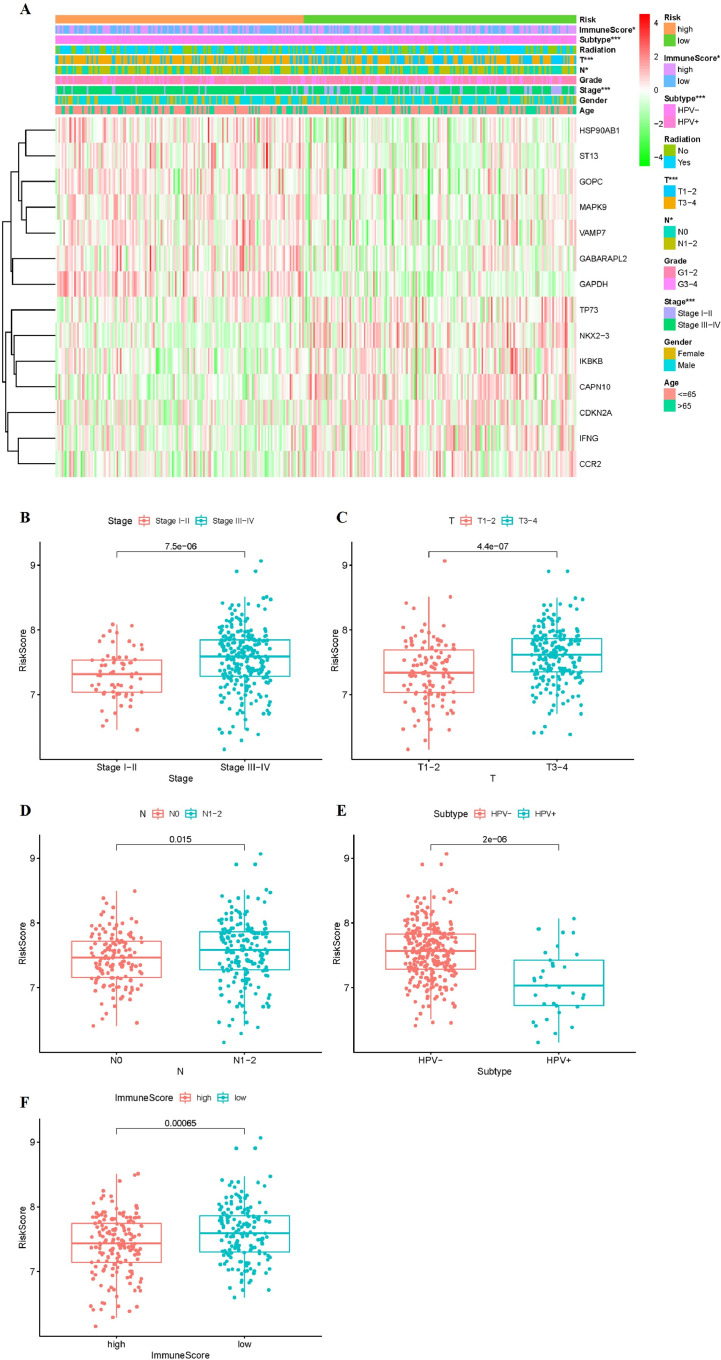
The OS-related risk scores correlated with clinicopathological characteristics, immunoscore
Further evaluation of the correlation between risk scores and clinicopathological characteristics was conducted. The expression of 14 ARGs in the low- and high-risk groups in the TCGA whole cohort was also demonstrated in the heatmap (Fig. 7A). The expression of HSP90AB1, ST13, GOPC, MAPK9, VAMP7, GABARAPL2, GAPDH in the low-risk group were lower than those of the high-risk group. The expression of TP73, NKX2–3, IKBKB, CAPN10, CDKN2A, IFNG, CCR2 were high in the low-risk group. We further found high risk score is closely related to HPV subtype (p < 0.001), stage (p < 0.001), pathological stage T (p < 0.001), pathological stage N (p < 0.05), immunescore (p < 0.05), but it had no correlation with gender, age, grade and radiation therapy (Fig. 7A). We also investigated a possible relationship between risk score, and HPV subtype, stage, pathological T stage, pathological N stage, and immunescore.
Fig. 7.
Risk Scores Correlated with Clinicopathological Characteristics and Immunoscore. (A) Clinicopathologic characteristics of low- and high-risk groups were generated in the heatmap (B-F) Distribution of risk scores stratified by stage (B), pathological stage T (C), pathological stage N (A), HPV subtype (E) and immunescore (F). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
The risk score of the stage Stage III-IV was higher than that of the Stage I-II (p < 0.001, Fig. 7B). The risk score was positively correlated to the pathological T stage (p < 0.001) and pathological N stage (p < 0.05) (Fig. 7C and Fig. 7D). The patients whose HPV are negative had higher risk than the patients whose HPV are positive (p < 0.001, Fig. 7E). There were significant differences between the high-risk and low-risk patients in terms of immunescore (p < 0.001, Fig. 7F). These findings indicated that the risk score was positively associated with higher stage, pathological T stage, pathological N stage and immunescore level.
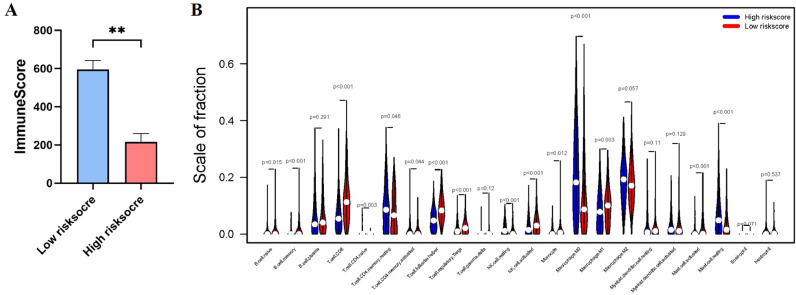
The OS-related signature associated with immune cell infiltration
In addition, the different risk socres between low-immunescore and high-immunescore groups prompt us to investigate which type of immune cell has association with t high-risk and low-risk patients (Fig. 8 and Supplementary Fig. S2). The two groups revealed a significant difference in immunescore (Fig. 8A). Subsequently, we analyzed the fraction of 22 immune cells in the low/high risk groups. High-risk group showed higher infiltration levels of T cell CD4 naïve, T cell CD4 memory resting, NK cell resting, Macrophage M0, Mast cell resting (Fig. 8B and Supplementary Fig. S2), whereas low-risk group was more correlated with B cell naïve, B cell memory, T cell CD8, T cell CD4 memory activated, T cell follicular helper, T cell regulatory Tregs, NK cell activated, Monocyte, Macrophage M1, Mast cell activated (Fig. 8B and Supplementary Fig. S2).
Fig. 8.
Immune Cell Infiltration in TCGA Cohort. (A) Immunescore in the low/high risk groups. (B) The infiltrating levels of 22 immune cells. ∗p < 0.05 and ∗∗p < 0.01.
Genetic alterations of the OS-related signature on immune cell
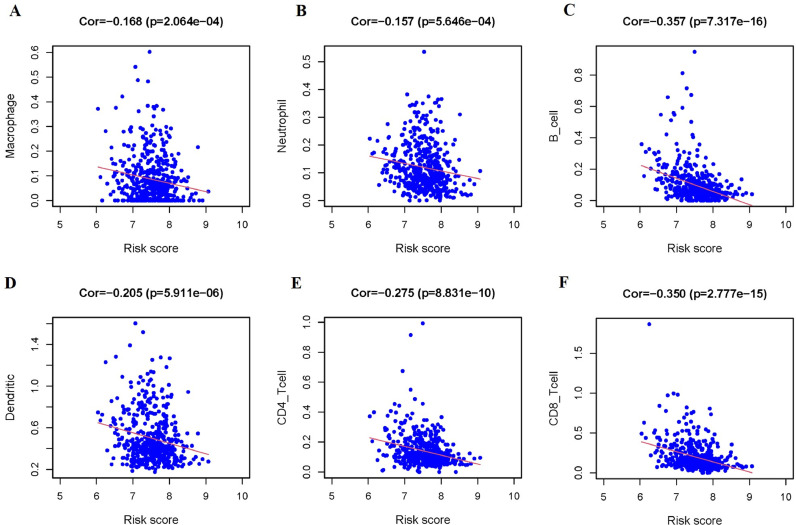
Furthermore, the correlation between risk score and infiltration abundances of six types of immune cells was calculated to estimate the effect of the OS-related signature on the TIME. Correlation analysis revealed negative correlation between the risk score and infiltration of the Macrophage (p < 0.001), Neutrophil (p < 0.001), B Cell (p < 0.001), Dendritic Cell (p < 0.001), CD4+ T Cell (p < 0.001), and CD8+ T Cell (p < 0.001, Fig. 9).
Fig. 9.
The Relationships between the Risk Score and Tumor-Infiltrating Immune Cells. (A-F) Macrophage (A), Neutrophil (B), B Cell (C), Dendritic Cell (D), CD4+ T Cell (E), and CD8+ T Cell (F).
To explore the mechanisms underlying of ARGs on immune cell infiltration, we detect somatic copy-number alterations (CNAs) in TIMER. The infiltration levels of immune cells in risk-related genes were significantly affected by the CNAs mainly because of arm-level deletion/gain (Supplementary Fig. S3). These results demonstrated that ARGs may be pivotal regulators in the tumor immune microenvironment for HNSCC patients.
Construction and validation of prognostic signature for DSS
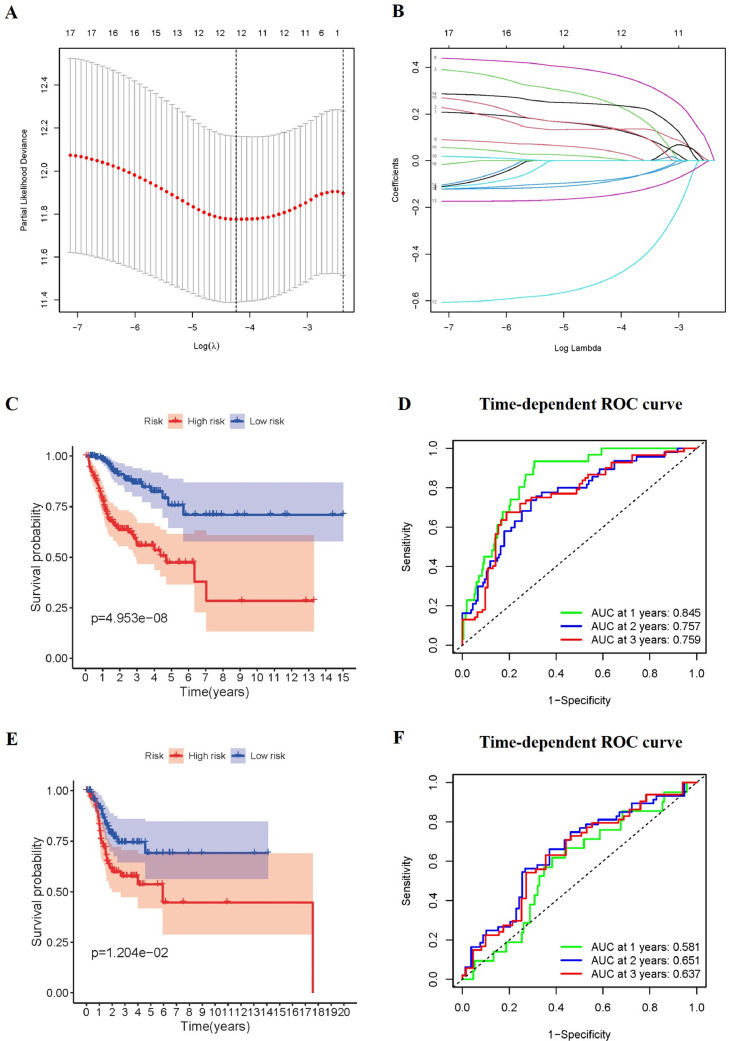
Further evaluations of the prognostic signature based on ARGs for disease-specific survival (DSS) were analyzed. 470 patients were randomly divided into a training set (n = 282) and a testing set (n = 188). Among 233 ARGs, a total of 19 DSS-related ARGs were found in the univariate Cox regression analysis using the training set (p < 0.05, details are given in Supplementary Table S2). Subsequently, these DSS-related genes were subjected to a LASSO regression analysis using the training set. Finally, 12 prognostic ARGs were obtained and the prognostic signature composed of these genes was established (Fig. 10A and Fig. 10B). Based on the median value of risk score, the patients were stratified into high-risk and low-risk groups. According to the Kaplan-Meier curve, patients in the high-risk groups had significantly poorer DSS outcomes than those in the low-risk group (p < 0.05, Fig. 10C and Fig. 10E). In the training cohort, the AUC for predicting DSS were 0.845, 0.757, and 0.759, respectively (Fig. 10D). In the testing cohort, the AUC for predicting DSS were 0.581, 0.651, and 0.637, respectively (Fig. 10F).
Fig. 10.
Construction and validation of the prognostic signature for DSS. (A-B) Lasso regression analyses of ARGs using the DSS model. The Lasso regression was performed using prognosis-significant ARGs in the training cohort of HNSCC. Kaplan-Meier survival curve for DSS in the training cohort (C), and testing cohort (E). ROC curves in the training cohort (D), and the testing cohort (F).
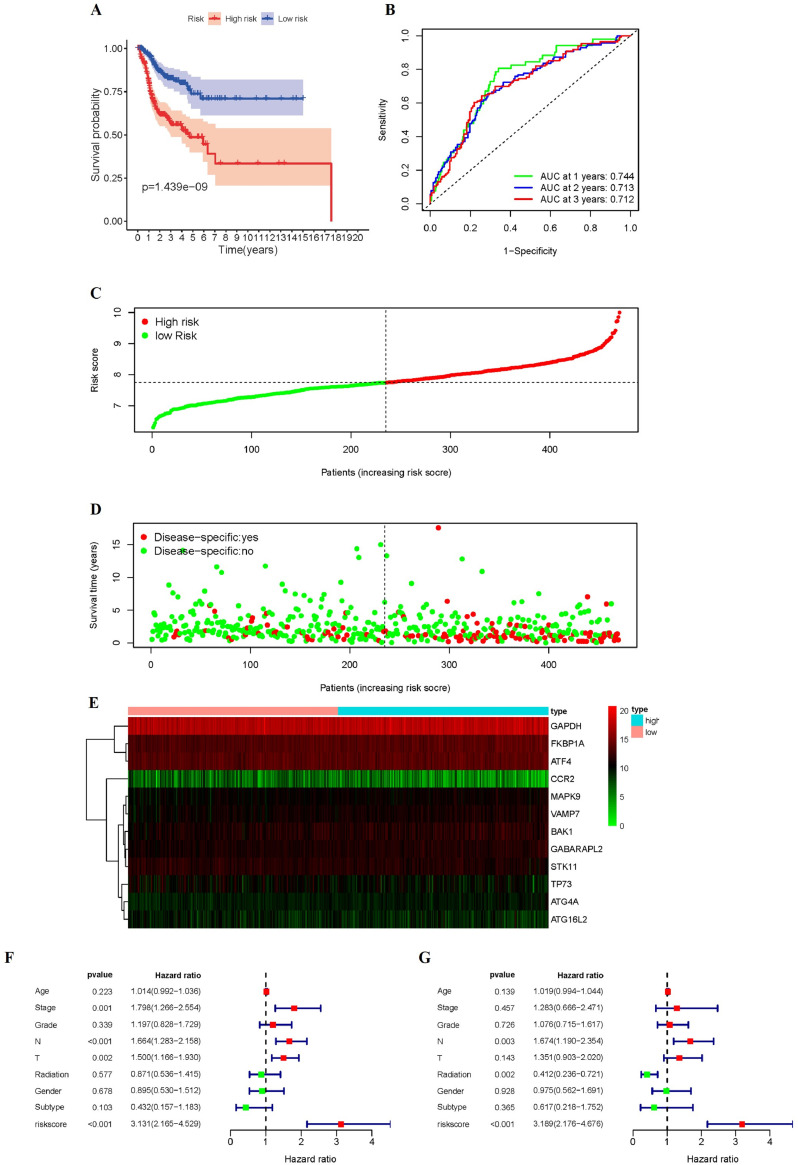
Consistent with the results obtained from the training and testing cohorts, patients in the high-risk group in the whole dataset (p < 0.01, Fig. 11A) had shorter DSS times than the low-risk groups. In the whole cohort, the AUC for predicting DSS were 0.744, 0.713, and 0.712, respectively (Fig. 11B). Fig. 11C-D showed the DSS-related distribution of risk scores and the relation between risk scores and DSS outcomes. The expression of 12 genes in high risk or low risk group was shown in the heatmap (Fig. 11E).
Fig. 11.
Characteristics of DSS-related signatures. (A) Kaplan-Meier curves for the high-/low-risk groups. (B) The ROC of DSS for the ARGs-based prognostic signature (C-D) Distribution of risk score and patient survival time, and status of HNSCC. (E) Heatmap of DSS-related ARGs expressions in the HNSCC. Forest plots showing the univariate (F) and multivariate cox regression results (G) of DSS in HNSCC.
By combining the results of univariate (HR = 3.131, 95% CI = 2.165–4.529, P < 0.001) (Fig. 11F) and multivariate Cox regression (HR = 3.189, 95% CI = 2.176–4.676, P < 0.001) (Fig. 11G) analysis, we proved that the risk signature of DSS can be an independent prognostic factor.
In the DSS-related prediction model, high expression of GAPDH, BAK1, FKBP1A, GABARAPL2, and MAPK9 genes were associated with worse DSS rate (Supplementary Fig. S4). In addition, high expression of TP73 and CCR2 was associated with better DSS rate (Supplementary Fig. S4). Furthermore, no associations between the expression level of VAMP7, ATGA4A, ATGA16L2, ATF4 and DSS rate were found (Supplementary Fig. S4).
In order to further clarify what type of regulation of autophagy (upregulation or downregulation) could have a positive impact on HNSCC patient's survival, we comprehensively analyzed the expression level of DSS-related and OS-related ARGs. Based on the screening criteria of | log2FC |> 1 and false discovery rate (FDR) <0.05, we found that the expression level of HSP90AB1 and IFNG genes were highly expressed in tumor tissues compared to adjacent normal tissues, while the expression level of NKX2–3 in tumor tissues was lower than that in adjacent normal tissues. The expression of other genes had no significant difference (Fig. 2C). In the results of survival analysis, the high expression level of HSP90AB1 in HNSCC were significantly associated with poorer survival rate. The high expression level of IFNG and NKX2–3 in HNSCC were significantly associated with better survival rate (Supplementary Fig.S1 and S4). These results indicated that the upregulation of some ARGs could have a positive impact on HNSCC patient's survival, and the downregulation of other ARGs might have the similar positive impact on HNSCC patient's survival.
Nomogram
By combining risk signature and other clinicopathological parameters, we performed nomogram to provide a risk prediction of 1-year, 2-years, 3-years OS and 1-year, 2-years, 3-years DSS for an individual patient (Supplementary Fig.S5). The calibration curves showed that the nomogram we established showed good performance for predicting the 1-year OS, 2-year OS, 3-year OS (Supplementary Fig. S5) and the 1-year DSS, 2-year DSS, 3-year DSS (Supplementary Fig. S5) of HNSCC.
Discussion
HNSCC carries a poor prognosis and has a high invasive tendency, with a low 5-year survival rate in advanced stages. Although the specific causes of HNSCC are unclear, various factors and clinical features, including smoking, alcohol as well as human papillomavirus status, are known to be associated with the disease [27], [28], [29]. Despite significant advances in the understanding of cancer, the prognosis of patients with HNSCC has not improved much vary from person-to-person because of their genetic conditions. Therefore, finding novel molecular biomarkers can improve the effectiveness of diagnosis and treatment strategies in the early stages of HNSCC. Thus, it is urgent to investigate effective diagnostic and therapeutic biomarkers for HNSCC patients. An increasing number of studies have shown that defects in autophagy is significantly related to the occurrence and progression of HNSCC, but most researchers focused on the role of single gene related to autophagy and its potential effects. And few explored the role of ARGs in tumor immune microenvironment. Feng et al. [30] constructed a model of ARGs and showed the interface between risk signature and OS. However, a comprehensive exploration of ARGs in the TIME, the value of HPV status in risk signature, the analysis of DSS, is lacking. Based on the transcriptome expression profiles of patients, we screened autophagy-related genes and identified 32 differentially expressed ARGs in HNSCC patient tumor samples. Considering the potential molecular mechanisms and biological pathways of these genes may contribute to the occurrence and development of HNSCC, we further carried out biological enrichment analyses, including GO and KEGG pathway analyses, to outline the potential functions of ARGs. The GO and KEGG analyses revealed that the majority of these differentially expressed ARGs are involved in autophagy, process utilizing autophagic mechanism, apoptosis and HPV infection. Autophagy is deeply associated with proliferation of HNSCC and poor prognosis, so it is worthwhile to research useful prognostic signature to assist clinician. We used the TCGA training cohort to constructed OS-related model and DSS-related model, respectively. Firstly, we identified some OS-related ARGs in univariate Cox regression analysis. Then, we constructed a prognostic model using LASSO regression. Our OS-related risk signature included 14 genes (GABARAPL2, GOPC, MAPK9, TP73, HSP90AB1, ST13, IKBKB, IFNG, GAPDH, NKX2–3, CCR2, VAMP7, CAPN10, CDKN2A). Our DSS-related risk signature included 12 genes (BAK1, GABARAPL2, MAPK9, TP73, FKBP1A, ATG4A, GAPDH, STK11, CCR2, VAMP7, ATF4, and ATG16L2).
The TP53 family member gene TP73 is a crucial tumor suppressor inducing cell cycle arrest and apoptosis [31]. Ge et al. [32] found that a chemical small molecule, 3-benzyl-5-((2-nitrophenoxy) methyl)-dihydrofuran-2(3H)-one, that activated mTOR by targeting FKBP1A.In colorectal cancer, ST13 suppressed cell growth and migration in vitro and tumorigenic ability in vivo [33]. Wang et al. observed that the tumor-associated macrophages infiltration and CCR2 expression was associated with the tumor metastasis in human lung cancer [34]. VAMP7 is a core functional gene in the vesicular fusion machinery of primary. And VAMP7 knock-down sharply reduced the killing effects of T cells, without suppressing T-cell mediated immunity [35]. MAPK9 is upregulated in non-small-cell lung carcinoma, which is closely linked to T stage in NSCLC patients. Furthermore, circRNF20 stimulates NSCLC proliferation by activating MAPK9 [36].
Univariate Cox regression analysis was performed to find genes significantly associated with OS and DSS of HNSCC. And then, the LASSO regression was used to build up risk signatures. According to the risk formula, we calculated risk score of each patient and divided patients into high/low-risk groups according to the median risk score. The result of KM survival analysis revealed that the OS for patients in the high-risk group was significantly poorer than that of low-risk group, and ROC analysis reflected the predictive signature performed well. The next finding implied that the ARGs-related signature for OS could be a satisfactory independent predictor of poor prognosis in HNSCC. The internal testing set was used to assess the predictive performance of the signature, revealing that the autophagy-related signature for OS we constructed had good prospect for application. The clinic correlation analysis confirmed that the risk score was closely related to Stage, pathological T stage, pathological N stage and HPV subtype. Notably, the prognostic evaluation model for DSS we also established and underwent validation. The ARGs-related signature for DSS can predict 1-year, 2-year, and 3-year survival rate accurately for HNSCC patients. Furthermore, we built nomogram model by incorporating clinical risk factors and risk score. The nomogram developed in our study accurately predicts the OS and DSS for HNSCC and calibration graphs added strong confirmation. Meanwhile, we illustrated the association between the OS-related risk score and immune cell infiltration level via CIBERSORT method and TIMMER database, which showed that the tumor microenvironment may affect the growth and metastasis in tumor development. Analysis of our study also demonstrated that the CNAs of risk-related genes had an impact on the immune cell infiltration. However, undeniably, our study still have some limitations. First, the two signatures we established were only obtained and processed from TCGA data. Although we have performed internal verification, the reliability and robustness of the model still needs to be further assessed using external datasets. Secondly, the specific and detailed of the genes underlying the prognostic signature in HNSCC are still unclear. Additionally, further investigations regarding the regulatory mechanism of risk-related genes in TIME is warranted.
In conclusion, our study established the ARGs-related signature for OS and DSS that were all independent prognostic indicators for HNSCC patients. And then, we firstly present a deep understanding of ARGs in the TIME, but further large scale studies and experiments are needed to verify our hypothesis. Meanwhile, our study may provide promising targets that can enhance the efficacy of cancer immunotherapy.
Ethics approval and consent to participate
In this study, all gene expression data were downloaded from TCGA database (https://portal.gdc.cancer.gov/) and all clinical data were identified from the cBioPortal database (https://www.cbioportal.org/). There are no restrictions on the use of TCGA and cBioPortal data for research and data analysis purposes. All datasets can be downloaded and used freely, and not require an ethics statement.
Funding
This research was supported with fund from the Joint Funds of Health Commission of Hubei Province (WJ2019H017), and the Independent Research projects of Wuhan University (2042019kf0128).
Availability of data and materials
The datasets generated during the current study were TCGA (https://portal.gdc.cancer.gov/).
CRediT authorship contribution statement
Jiayu Fang: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. Zhiqiang Yang: Conceptualization, Methodology, Writing – review & editing. Jing Xie: Formal analysis, Software. Ziang Li: Methodology. Chang Hu: Formal analysis. Minlan Yang: Resources, Supervision. Xuhong Zhou: Conceptualization, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101094.
Contributor Information
Minlan Yang, Email: yangml@whu.edu.cn.
Xuhong Zhou, Email: zhouxuhong66@126.com.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Adelstein D., Gillison M.L., Pfister D.G., Spencer S., Adkins D., Brizel D.M. NCCN guidelines insights: head and neck cancers, version 2.2017. J. Natl. Compr. Canc. Netw. 2017;15:761–770. doi: 10.6004/jnccn.2017.0101. [DOI] [PubMed] [Google Scholar]
- 3.Lakhani S.R., Ashworth A. Microarray and histopathological analysis of tumours: the future and the past? Nat. Rev. Cancer. 2001;1:151–157. doi: 10.1038/35101087. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018;20:521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 5.Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 6.Lessard C.J., Sajuthi S., Zhao J., Kim K., Ice J.A., Li H. Identification of a systemic lupus erythematosus risk locus spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol. 2016;68:1197–1209. doi: 10.1002/art.39548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igci M., Baysan M., Yigiter R., Ulasli M., Geyik S., Bayraktar R. Gene expression profiles of autophagy-related genes in multiple sclerosis. Gene. 2016;588:38–46. doi: 10.1016/j.gene.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 8.Yang S.-.K., Hong M., Zhao W., Jung Y., Baek J., Tayebi N. Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63:80–87. doi: 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]
- 9.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janku F., McConkey D.J., Hong D.S., Kurzrock R. Autophagy as a target for anticancer therapy. Nat. Rev. Clin. Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 12.Parikh N.D., Singal A.G., Hutton D.W. Cost effectiveness of regorafenib as second-line therapy for patients with advanced hepatocellular carcinoma. Cancer. 2017;123:3725–3731. doi: 10.1002/cncr.30863. [DOI] [PubMed] [Google Scholar]
- 13.Yun C.W., Lee S.H. The roles of autophagy in cancer. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z.-.R., Zhu X.-.D., Zhao W., Qu S., Su F., Huang S.-.T. Poly(ADP-ribose) polymerase-1 regulates the mechanism of irradiation-induced CNE-2 human nasopharyngeal carcinoma cell autophagy and inhibition of autophagy contributes to the radiation sensitization of CNE-2 cells. Oncol. Rep. 2013;29:2498–2506. doi: 10.3892/or.2013.2382. [DOI] [PubMed] [Google Scholar]
- 15.Liu J.L., Chen F.F., Lung J., Lo C.H., Lee F.H., Lu Y.C. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br. J. Cancer. 2014;111:944–954. doi: 10.1038/bjc.2014.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White E., Karp C., Strohecker A.M., Guo Y., Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr. Opin. Cell Biol. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn M.Y., Yoon H.-.E., Kwon S.-.M., Lee J., Min S.-.K., Kim Y.-.C. Synthesized Pheophorbide a-mediated photodynamic therapy induced apoptosis and autophagy in human oral squamous carcinoma cells. J. Oral Pathol. Med. 2013;42:17–25. doi: 10.1111/j.1600-0714.2012.01187.x. [DOI] [PubMed] [Google Scholar]
- 18.Zang Y., Thomas S.M., Chan E.T., Kirk C.J., Freilino M.L., DeLancey H.M. Carfilzomib and ONX 0912 inhibit cell survival and tumor growth of head and neck cancer and their activities are enhanced by suppression of Mcl-1 or autophagy. Clin. Cancer Res. 2012;18:5639–5649. doi: 10.1158/1078-0432.CCR-12-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Y., Zhu L., Cai H., Wang G., Liu B. Autophagic compound database: a resource connecting autophagy-modulating compounds, their potential targets and relevant diseases. Cell Prolif. 2018;51:e12403. doi: 10.1111/cpr.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G., Wang L.-.G, Han Y., He Q.-.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter W., Sánchez-Cabo F., Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–ee10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkman S.J., Roscoe E.M., Bourret J.C. Comparing self-directed methods for training staff to create graphs using Graphpad Prism. J. Appl. Behav. Anal. 2019;52:188–204. doi: 10.1002/jaba.522. [DOI] [PubMed] [Google Scholar]
- 26.Saluja R., Cheng S., Delos Santos K.A., Chan K.K.W. Estimating hazard ratios from published Kaplan-Meier survival curves: a methods validation study. Res. Synth. Methods. 2019;10:465–475. doi: 10.1002/jrsm.1362. [DOI] [PubMed] [Google Scholar]
- 27.Jethwa A.R., Khariwala S.S. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017;36:411–423. doi: 10.1007/s10555-017-9689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawakita D., Matsuo K. Alcohol and head and neck cancer. Cancer Metastasis Rev. 2017;36:425–434. doi: 10.1007/s10555-017-9690-0. [DOI] [PubMed] [Google Scholar]
- 29.Chow L.Q.M. Head and neck cancer. N. Engl. J. Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 30.Feng H., Zhong L., Yang X., Wan Q., Pei X., Wang J. Development and validation of prognostic index based on autophagy-related genes in patient with head and neck squamous cell carcinoma. Cell. Death Discov. 2020;6:59. doi: 10.1038/s41420-020-00294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scian M.J., Carchman E.H., Mohanraj L., Stagliano K.E.R., Anderson M.A.E., Deb D. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27:2583–2593. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- 32.Ge D., Han L., Huang S., Peng N., Wang P., Jiang Z. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014;10:957–971. doi: 10.4161/auto.28363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu D.B., Zhong S.Y., Yang M., Wang Y.G., Qian Q.J., Zheng S. Potent antitumor activity of double-regulated oncolytic adenovirus-mediated ST13 for colorectal cancer. Cancer Sci. 2009;100:678–683. doi: 10.1111/j.1349-7006.2009.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmall A., Al-Tamari H.M., Herold S., Kampschulte M., Weigert A., Wietelmann A. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am. J. Respir. Crit. Care Med. 2015;191:437–447. doi: 10.1164/rccm.201406-1137OC. [DOI] [PubMed] [Google Scholar]
- 35.Chitirala P., Ravichandran K., Galgano D., Sleiman M., Krause E., Bryceson Y.T. Cytotoxic granule exocytosis from human cytotoxic T lymphocytes is mediated by VAMP7. Front. Immunol. 2019;10:1855. doi: 10.3389/fimmu.2019.01855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z.X., Zhao Y., Wang Y.B., Zhang Q., Zou Q.X., Liang F.H. CircRNF20 aggravates the progression of non-small-cell lung carcinoma by activating MAPK9. Eur. Rev. Med. Pharmacol. Sci. 2020;24:9981–9989. doi: 10.26355/eurrev_202010_23211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study were TCGA (https://portal.gdc.cancer.gov/).