Summary
Improved stem cell-derived pancreatic islet (SC-islet) differentiation protocols robustly generate insulin-secreting β cells from patient induced pluripotent stem cells (iPSCs). These advances are enabling in vitro disease modeling studies and the development of an autologous diabetes cell replacement therapy. SC-islet technology elucidates key features of human pancreas development and diabetes disease progression through the generation of pancreatic progenitors, endocrine progenitors, and β cells derived from diabetic and nondiabetic iPSCs. Combining disease modeling with gene editing and next-generation sequencing reveals the impact of diabetes-causing mutations and diabetic phenotypes on multiple islet cell types. In addition, the supply of SC-islets, containing β and other islet cell types, is unlimited, presenting an opportunity for personalized medicine and overcoming several disadvantages posed by donor islets. This review highlights relevant studies involving iPSC-β cells and progenitors, encompassing new conclusions involving cells from patients with diabetes and the therapeutic potential of iPSC-β cells.
Keywords: diabetes, stem cells, differentiation, disease modeling, cell therapy, CRISPR, islets, beta cells, iPS cells, pluripotency
Graphical abstract
Highlights
Improved differentiation protocols generate pancreatic islet from patient stem cells
Diabetic stem cell-derived islet studies identified key markers for cell function
Gene editing aims to address unmet needs for stem cell therapy field
Stem cell-derived islets are a promising source for diabetes stem cell therapy
Maxwell and Millman review the current literature involving induced pluripotent stem cell-derived pancreatic islets from patients with diabetes. Advances in differentiation protocols have enabled disease modeling, exhibited the benefits of gene editing, and conveyed the potential to leverage stem cells for personalized diabetes therapy.
Introduction
Diabetes is a disease diagnosed in >451 million patients worldwide.1 This epidemic is caused by the death and dysfunction of pancreatic, insulin-secreting β cells. The current treatments for patients with type 1 diabetes, constituting ∼10% of diabetes mellitus diagnoses, include exogenous insulin injections and cadaveric donor islet transplantation.2,3 Long-term exogenous insulin is commonly accompanied by hyperglycemia and hypoglycemia events, which can cause cardiovascular and kidney complications.4 A cell therapy pipeline developed from cadaveric islet transplantation emphasizes the potential power of cell replacement technology to enhance the quality of life for patients with diabetes. However, there is a limited supply of cadaveric donors and a need for immunosuppression.5,6 An alternate promising source for diabetes cell therapy is self-renewable human pluripotent stem cells (hPSCs).
Stem cell-derived definitive endoderm (DE), pancreatic progenitors (PPs), and endocrine progenitors (EPs) were created starting in 2006.7, 8, 9 These initial differentiation stages were critical to create stem cell-derived β (SC-β) cells and continue to be the initial stages in several differentiation protocols. SC-β cells were first generated in 2014 from embryonic stem cells (ESCs), with several key β cell characteristics capable of reversing diabetes in mouse models several weeks after transplantation.10,11 Recent advances in differentiation protocols have enabled the robust generation of SC-β cells from patient induced pluripotent stem cells (iPSCs).12, 13, 14 The new, refined protocols generated cell populations commonly called SC-islets due to the composition consisting of almost all endocrine (chromogranin A+) cells, including glucagon+ alpha (SC-α) and somatostatin+ delta (SC-δ) cells. Patient-derived diabetic iPSC-β cells allow for disease progression studies of pancreatic and EPs during stages 4 and 5 of differentiation, benefiting many groups studying diabetes from patient stem cells due to the unknown presence and progression of diabetes at early development stages. In addition, studying patient SC-β cells with a known diabetic genotype at the final cell fate in stage 6 is effective for the diabetes field, drug screening, and future therapeutics. Furthermore, disease modeling with patient cells allows for a better understanding of multiple forms of diabetes, such as type 1 (T1D), type 2 (T2D), cystic fibrosis-related diabetes (CFRD), and monogenetic diabetes (MD).3 Genetic engineering tools have been applied to patient iPSC-islets to generate nondiabetic (ND) phenotype and transgenic cell lines. Specifically, our group and colleagues used gene editing to correct MD variants found in neonatal diabetes (NeoD), maturity-onset diabetes of the young (MODY), and Wolfram syndrome (WS).15, 16, 17 Results from these studies demonstrated the potential for an autologous diabetes cell therapy. Microfluidic devices also present an opportunity for improved culture systems and drug screening platforms to study diabetes in vitro.18 Stem cell technology holds immense potential as a cell therapy to combat diabetes. This review summarizes the present literature on iPSC-islets from patients with diabetes by analyzing diabetes modeling and cell therapy development to highlight the potential of personalized therapy.
Generation efficiency of iPSC-islets from multiple differentiation protocols
Differentiation protocols for SC-islets generally target the same developmental stages, beginning with DE and primitive gut tube, then narrowing cell fate to pancreatic and EPs, and ultimately targeting the final differentiated β cell.7,8,10, 11, 12, 13, 14 Small molecules and growth factors are typically used to navigate the pathways needed for the differentiation stages, aiming to mimic embryonic development. β cell and progenitor stem cell differentiation protocols have been meticulously developed over the past decade, beginning with DE and leading to functional SC-β cells, which have been reviewed previously.19, 20, 21, 22, 23 Alternatively, Sui et al.24 used nuclear transfer embryonic stem (NT-ES) cells to generate NT-ES-β cells. Favorable off targets were commonly produced during the protocol, including α and δ cells. The presence of all main islet endocrine cell types led the field to rename SC-β cell clusters to SC-islets.17,25,26 Enterochromaffin (SC-EC) cells were recently identified as an off target generated in SC-islet differentiations that persist in vivo, validated by multiple groups.25,26 SC-EC cells are an endocrine (CHGA+) cell type, and their effect on SC-islet function and maturation is unknown.25 In addition, adverse off target cell types, such as neural progenitor cells (NPCs), pancreatic exocrine (mesenchyme, acinar), and polyhormonal (PH) cells, can contaminate the final cell cluster. Differentiation efficiency, genetic background, and disease state of the hPSCs influenced the composition proportions of these off targets.15,17,25
Before the recent β cell differentiation protocols, iPSCs were unable to differentiate into SC-β cells as efficiently as ESCs. Protocols were typically optimized for a single ESC line, producing iPSC-β cells at lower efficiencies.10,11,14 SC-β or SC-β-like cells ranged from 17%27 to 73%14 C-Peptide+, a proxy marker for insulin, at the final stage of differentiation. Of note, C-Peptide+ cells can include both functional (NKX6.1+C-Peptide+) cells and PH (GCG+C-Peptide+ and SST+C-Peptide+) cells, which are nonfunctional and immature endocrine cells. Protocols with smaller cell yields used GFP reporter cell lines to create higher-purity β cell clusters based on β cell markers such as insulin, NKX6.1, or CD177.13,28,29 Purifying methods enabled the study of diabetes-relevant β cells for disease modeling in vitro and transplanting β cells in vivo.
Innovative refinements to remove off target cells during the EP stage used latrunculin A, an actin cytoskeleton modulator, to enrich the endocrine population.12,17 This protocol provided an opportunity to perform the SC-islet differentiation in planar in 6 stages, reducing technical expertise and costly equipment for three-dimensional (3D) culture. In addition, the removal of function-inhibiting small molecules and reaggregation in the final differentiation stage enhanced the function of SC-islets and showcased the rapid reversal of diabetes in rodent models with patient SC-β cells.13,14 The improved protocols required minimal optimization of multiple patient iPSC lines for successful differentiations. The SC-islets contained almost pure endocrine cell populations.12 We and our colleagues can now readily investigate SC-islets from patients with diabetes.
Disease modeling with iPSCs from patients with diabetes
SC-islets from patients with diabetes are critical to gain a better understanding of the disease and its progression. Thus, iPSCs have been reprogrammed from patients with T1D, T2D, MODY, CFRD, WS, and NeoD (Figure 1; Tables 1 and 2). Typically, the Yamanaka factors (OCT4, KLF4, SOX2, c-MYC) were used to reprogram the somatic cells through either retrovirus, Sendai virus, or episomal reprogramming.30,31 Alternatively, Sui et al.24 used somatic cell NT to generate T1D NT-ESCs from skin fibroblasts. Studies differentiated patient iPSCs into SC-β cells and progenitors, and investigated cell maturation, differentiation efficiencies, and insulin secretion function. When differences were found, they typically occurred after pancreatic cell fate was designated. The cells were commonly compared to either nondiabetic or gene-edited isogenic controls. Genetic engineering is used to reverse these effects and correct the known diabetes, causing mutations.
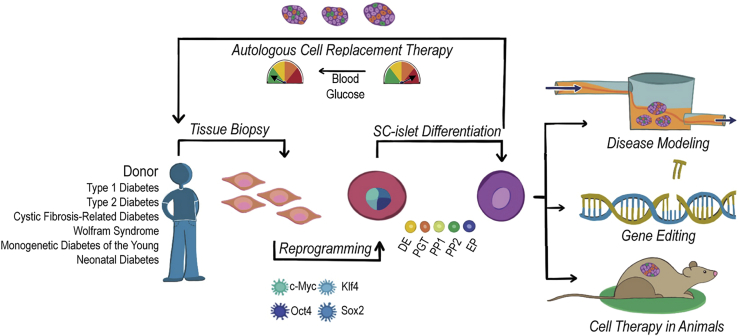
Figure 1.
Applications of SC-islets from patients with diabetes
Patient tissue biopsies can be reprogrammed into induced pluripotent stem cells (iPSCs) and differentiated into stem cell-derived islets (SC-islets) that contain β (SC-β) cells. These SC-islets have applications in disease modeling, gene editing, and cell therapy in animal models. They also provide a source for autologous cell replacement therapy for patients with diabetes. DE, definitive endoderm; EP, endocrine progenitor; PGT, primitive gut tube; PP, pancreatic progenitor.
Table 1.
Summary of T1D and T2D patient iPSC cell line studies for diabetes-in-a-dish
| Study reference | Diabetic state | Cell type generated | In vivo transplantation | Additional notes |
|---|---|---|---|---|
| Leite et al. (2020)32 | T1D | SC-β and SC-α cells | no | isogenic co-culture with immune cells |
| Wang et al. (2019)33 | T2D | β-like cells | no | microarray, RNA-seq, and ChIP-seq data from PPs |
| Dwivedi et al. (2019)34 | T2D | β-like cells | – | RNA-seq |
| Sui et al. (2018)24 | T1D | NT-iPSC-β cells | yes, prevent diabetes onset | NT patient SCs |
| Cosentino et al. (2018)27 | T2D | β-like cells | no | – |
| Amin et al. (2018)35 | T1D, T2D | SC-β cells | yes, study in vivo cells | chemical screen, RNA-seq of sorted β sorted cells |
| Millman et al. (2016)36 | T1D | SC-β cells | yes, prevent diabetes onset | microarray data |
| Thatava et al. (2013)37 | T1D | Pancreatic islet-like cells | – | microarray data of iPSCs |
Table 2.
Summary of studies investigating known diabetes variants for T1D, T2D, and MD
| Study reference | Diabetic state | Variant of interest | Cell type generated | Genetic engineering | In vivo transplantation | Additional notes |
|---|---|---|---|---|---|---|
| Lithovius et al. (2020)38 | CHI/MODY | SUR1 | β-like cells | yes, CRISPR/Cas9 to correct mutation | yes, study in vivo cells | – |
| Maxwell et al. (2020)17 | WS | WFS1 | SC-β cells | yes, CRISPR/Cas9 to correct variant | yes, reverse preexisting diabetes | Single-cell RNA-seq |
| Cardenas-Diaz et al. (2019)39 | MODY3 | HNF1A | β-like cells | yes, CRISPR/Cas9 to introduce HET and homozygous KO mutations | no | microarray of INS-GFP+NKX6.1+ cells |
| Wang et al. (2019)33 | T2D | PDX1 | β-like cells | yes, CRISPR/Cas9 to introduce HET and homozygous KO mutations in iPSC line | no | microarray, RNA-seq, and ChIP-seq data from PPs |
| Dwivedi et al. (2019)34 | T2D | ZnT8 | β-like cells | yes, CRISPR/Cas9 to introduce protective alleles into SLC30A8 locus | yes, study in vivo cells | RNA-seq |
| Balboa et al. (2018)15 | NeoD | INS | β-like cells | yes, CRISPR/Cas9 to correct variant | yes, study in vivo cells | single-cell RNA-seq |
| Kishore et al. (2020)40 | adult-onset diabetes | GATA6 | β-like cells | yes, CRISPR/Cas9 to correct variant | no | ChIP-qPCR |
| Amin et al. (2018)35 | T1D, T2D, NeoD | GLIS3 | SC-β cells | yes, CRISPR/Cas9 to introduce GLIS3 mutations | yes, study in vivo cells | chemical screen, RNA-seq of sorted β sorted cells |
| Ma et al. (2018)16 | NeoD | INS | β-like cells | yes, CRISPR/Cas9 correction of variant | yes, prevent diabetes onset | – |
| Stepniewski et al. (2015)41 | MODY3 | HNF1A | β-like cells | – | yes, tested iPSC tumor formation | – |
| Shang et al. (2014)42 | WS | WFS1 | insulin-producing cells | WT WFS1 lentivirus to correct variant | yes, prevent diabetes onset | tested ER chaperone drugs |
| Teo et al. (2013)43 | MODY1 | HNF4A | – | – | – | – |
| MODY1 | HNF4A | – | – | – | – | |
| MODY2 | GCK | – | – | – | – | |
| MODY3 | HNF1A | – | – | – | – | |
| MODY5 | HNF1B | – | – | – | – | |
| MODY5 | HNF1B | – | – | – | – | |
| MODY8 | CEL | – | – | – | – |
T1D and T2D are the most common forms of diabetes mellitus, constituting ∼10% and 90% of patients with diabetes, respectively.3 Understanding the pathophysiology of these diseases is complex due to risk factors, including epigenetics, environment, and lifestyle. Many rodent models of T1D and T2D exist; however, there are several physiologic differences between rodents and humans, specifically in the islets.44,45 Therefore, we and others used stem cell technology to create iPS cells from human patients to model T1D and T2D and gain a better understanding of disease pathogenesis (Table 1). Early studies used PDX1+NKX6-1+ PPs, which give rise to all pancreatic cell fates. T1D iPSC-β cells functioned and had differentiation efficiencies similar to ND iPSC-β cells,36 which is consistent with previous reports comparing T1D and ND SC-PPs.37 Under further investigation, in vitro modeling systems revealed that T1D patient-derived endocrine cells respond to stress36 and react to autologous immune cells in a cell-type-specific manner, providing a better understanding of T1D pathogenesis.32 There is no literature investigating SC-islets from patients with T2D; however, T2D iPSCs are available.30 There are multiple studies using patient iPSCs with T2D-risk variants including SLC30A8,34 TRMT10A,27 and PDX1.33 To further study T2D, next-generation sequencing (NGS) identified gene expression differences in T2D primary human islets compared to healthy controls.46,47 Xin et al.48 reported differentially expressed genes found in the main endocrine islet cell types. Specifically, in β cells, PAXBP-1, GLS2, and FXYD23 are upregulated and G6PC2, GLRA1, and IGFBPL1 are downregulated.
CFRD and MD are rarer forms of diabetes that limit the availability of primary samples. Thus, patient iPSCs are critical for uncovering disease pathogenesis and therapeutic development. Pancreatic ductal epithelial cells were derived from iPSCs with a CF genetic background, creating a drug screening platform in vitro.49 MD is caused by a single gene mutation that occurs in roughly 2% of patients who are diagnosed with diabetes mellitus (reviewed by Harris et al.50). Patient iPSCs from MODY, NeoD, and WS backgrounds have allowed for human MD modeling. Recent publications investigated mutations in PDX1, GLIS3, HNF1A, HNF1B, GCK, INS, GATA6, SUR1, and WFS1, which are each related to a form of MD (Table 2). Genetic engineering tools were commonly applied to correct the known mutation in an isogenic cell line, which often led to a healthy phenotype (Table 2).15, 16, 17,33,34,35,38,39,42 Diabetic SC derivatives were typically less efficient in generating SC-islets, SC-β cells, and SC-PPs. This occurred when evaluating mutations in GATA6,40 WFS1,17,42 and PDX133 (Table 2). However, the SUR1 mutation caused the inactivation of the KATP channel, resulting in congenital hyperinsulinism (CHI) and eventually MODY, producing an increased number of SC-β-like cells with greater function.38 Transplantation was commonly performed to observe cell maturation in vivo and examine whether cells could either reverse preexisting diabetes or prevent diabetes induction in mice (Table 2). In summary, human iPSC differentiation protocols enable the study of diabetic β cells and progenitors in vitro.
Insights in stem cell-derived islets from NGS of iPSCs
Several iPSC studies used NGS strategies15,17,33 and microarrays36,37,40,51 to study differentially expressed genes in specific islet cell types, such as α and β cells. Balboa et al.15 and Maxwell et al.17 used single-cell NGS, and both studies observed increases in β cell genes for gene-edited SC-β-like and SC-β cells. Wang et al.33 identified downregulated PDX1-bound genes that reduce differentiation efficiency, including MNX1, CES1, and MEG3. Millman et al.36 identified TAP1 as the largest differentially expressed gene in T1D SC-β cells compared to healthy patients. Recently, Augsornworawat et al.26 evaluated the transcriptome of transplanted SC-islets with single-cell RNA sequencing from both ESCs and iPSCs to show the strong resemblance of in vivo SC-islets and primary human islets. A maturation list for β and α cells was curated from the sequencing data, providing a resource of targets for β cell maturation in vitro.26 NGS of primary human islets and SC-islets was previously reviewed and highlighted key findings for the differences between diabetic and nondiabetic human islets, as well as important islet cell type-defining gene signatures.52 Of note, Muraro et al.53 and Segerstolpe et al.46 characterized human islet samples from ND and T2D patients. Krentz et al.54 and Hrvatin et al.51 compared progenitor cell types to human islets. Lastly, Veres et al.25 profiled the various stages of differentiation to yield a potential cell sorting mechanism with CD49a and identified SC-EC cells. Xin et al.48 detailed the unfolded protein response in primary β cells from multiple donors. These studies used innovative biotechnology to perturb the transcriptome of primary and stem cell-derived human islets.
Insights on diabetes through disease-in-a-dish studies
-
(1)
SC-islets and progenitor derivatives are used to model diabetes-in-a-dish. Endoplasmic reticulum (ER) stress experiments are commonly applied to ND and diabetic SC-islets to represent diabetes in vitro.55 In all forms of diabetes, ER stress is found in β cells, causing dysfunction and eventual unfolded protein response (UPR)-mediated cell death.56 Balboa et al.,15 Shang et al.,42 and Maxwell et al.17 studied SC-β cells and progenitors from patients with mutations linked to diabetes and known to cause ER stress. The results revealed a combination of increased protein and gene expression of ER stress markers, dysfunctional insulin processing, and reduced insulin secretion in response to glucose. Shang et al.42 and Maxwell et al.17 also showed dilated ER with electron microscopy, indicating ER stress. Balboa et al.15 and Maxwell et al.17 used NGS to identify increased ER stress (ATF6, BiP/HSPA5), apoptosis (CHOP, CASP3), and oxidative stress transcripts (TXNiP). SC-β cells with greater maturation compared to progenitors influenced ER stress and study results. As portrayed when comparing WS studies, SC-β cells17 were more mature than insulin-producing cells,42 indicated by the attainment of β cell markers and functional insulin secretion in response to glucose. Specifically, for WS studies, C-Peptide+ insulin-producing cell yields were not reduced with a WFS1 mutation; however, SC-β cells had lower yields when comparing the unedited and corrected iPSC differentiations. Greater insulin production supported higher functional capacity, and in return, increased ER stress due to increased insulin production and protein folding. The inability to handle elevated protein folding activated the unfolding protein response at a basal level and likely influenced differentiation from EPs to SC-β cells. There are few articles available examining ER stress in human islets48,57; therefore, human SC-islets are a critical resource to understand diabetes-in-a-dish.
-
(2)
Transgenic cell lines are generated through editing technologies that insert and express foreign DNA. For diabetes research, transgenic stem cells were created using biotechnology to either study diabetes through the insertion of a known diabetes-causing variant or monitor differentiation with a reporter cell line. This technology advancement was critical to study known variants associated with T1D and T2D. Amin et al.35 used a GLIS3 mutation associated with T1D, T2D, and NeoD to generate SC-PP2 and SC-PP2-β cells. With the mutation, they observed increased cell death and impaired differentiation compared to isogenic controls, developing a drug screening platform. Shang et al.42 transfected WT WFS1 lentivirus in their patient iPS-derived insulin-producing cells to model WS in vitro. Dwivedi et al.34 inserted a ZnT8 loss-of-function (LoF) allele, expected to protect against T2D, using CRISPR/Cas9 in patient iPSCs and found increased glucose responsiveness, making ZnT8 a promising target for increased insulin secretion as a T2D treatment. Transgenic stem cell lines were also used to study neurologic, epidermal, and cardiomyocyte disorders.
-
(3)
Microfluidic devices are used to mimic the in vivo pancreas and islet microenvironment. Engineered devices perturbed islet function by enhancing insulin sensing, monitoring islet hormone secretion in response to glucose, and quantifying pulsatile insulin release. Misun et al.58 generated a device to monitor dynamic insulin secretion of a single human islet. Lenguito et al.59 combined convective fluidic devices and computational modeling of glucose-stimulated insulin secretion (GSIS) to develop a competitive alternative to commercially available perifusion systems with the opportunity to image cells while in the machine. Jun et al.60 and Sankar et al.61 developed a dynamic culture system as an alternative to typical static culture by integrating interstitial flow to better mimic in vivo islet conditions in vitro. Glieberman et al.62 designed a microfluidic device incorporating the native islet microenvironment by delivering pulsatile insulin through arteriole and capillary channels on a chip. This system also continuously monitored insulin release, quantified with fluorescence anisotropy. Future protocols may benefit from differentiating PSCs in these engineered culture devices, such as the integrated platform developed by Ishahak et al.63 that incorporated continuous perfusion culture with the ability to monitor functional maturation. These newly developed devices are competitive platforms for drug screening and studying dynamic insulin secretion in vitro.
-
(4)
Sorting SC-β cells and progenitors are helpful in studying specific cell types of interest and removing off target cell types from differentiation protocols. Several studies used NKX6.1GFP/w and INSGFP/w stem cell lines to enhance SC-β cell yields.16,24,28,64 Mahaddalkar et al.65 identified anterior DE markers CD177 and CD275 to specify pancreatic and liver fates, respectively Thus, they sorted for CD177+CD275− after DE was defined in the first stage of SC-islet differentiation, improving the percentage of INS+NKX6.1+ β-like cells at stage 7. To enrich functional cells, Li et al.66 sorted human β-like cells for CD9, a cell-surface marker found in immature human β cells, to negatively enrich glucose-responsive cells. In addition, Veres et al.25 recently identified CD49a/ITGA1 as a surface marker for β cells in SC-islet clusters, thereby increasing the insulin secretion of individual islets. Sorting strategies are beneficial to study β cells for disease-in-a-dish modeling of diabetes.
Insights in disease mechanism through gene editing
Editing of genes important for β cell differentiation is commonly performed in the SC-β cell field to determine the effect on β cell insulin secretion. The impact of PDX1, MAFA, and NEUROG3 have been extensively studied in SC-β cells (review by Zhu et al.67). Gene editing has also been proposed for cell therapy efforts in both correction of diabetes-causing mutations and resistance to immune rejection. We and other colleagues used CRISPR/Cas9 and lentiviral transduction to correct known diabetes-causing mutations in patient iPSCs. Maxwell et al.17 and Ma et al.16 used gene correction to reverse and prevent diabetes in mice, respectively. These results are very promising for the potential of an autologous cell therapy. Lithovius et al.38 corrected the SUR1 mutant KATP activator and identified a role for the KATP channel in development by revealing increased insulin secretion and proliferation of the mutant compared to the isogenic gene-corrected SC-β-like cells. Enhanced function was expected as patients with CHI, and eventually MODY, are unable to initiate membrane depolarization and continually secrete insulin.38 Eichstadt et al.68 used gene-corrected autologous cell therapy in a Phase I/IIa clinical trial for recessive dystrophic epidermolyis bullosa and observed no adverse effects. Río et al.69 published results from a Phase I/II clinical trial of gene-corrected hematopoietic gene therapy for patients with Fanconi anemia and also detected no immune rejection effects. However, there remains concern that gene editing of autologous cells may cause immune rejection due to the autoimmune reactive nature of diabetes. Therefore, the current efforts investigating immune rejection resistance in allogeneic cell therapy may also be needed to a lesser extent for autologous cell therapy. Genome engineering to address immune intervention was previously reviewed.70 For SC-β cell therapy, key targets for the genome engineering of diabetes cell therapy included human leukocyte antigen (HLA) modification to expand SC-islet utility and cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) and programmed death-1/programmed death ligand-1 (PD-1/PD-L1) adjustment to target T cell immune rejection upon transplantation.71,72 Alternate iPSC-derived engineered cell types (endothelial, smooth muscle, cardiomyocytes) described by Deuse et al.73 used major histocompatibility complex (MHC)-mismatched allogeneic sources to inactivate the MHC class I and II genes and overexpress CD47 with CRISPR/Cas9 to successfully evade immunosuppression in recipients. These advances in SC-β cell and alternate cell genetic engineering aim to address an unmet need in the autologous and allogeneic stem cell therapy field.
Insights on in vivo cell therapy with animal models
To develop an adequate cell therapy, in vivo testing is needed in animal models for therapeutic validation. Our group and colleagues have established several studies confirming the use of SC-islets, SC-β cells, and PPs for cell therapy in rodent models. Validation methods included prevention of diabetes16,24,42,74,75 and diabetes reversal10,11,13,14,17 by transplanting cells in the kidney capsule before or after diabetes induction, respectively. Streptozotocin and alloxan induced diabetes in mouse and rat models. Nephrectomies were also commonly performed to confirm that the transplanted cells were the source of euglycemia in kidney capsule transplants.9,12,17,24,75,76 Several groups confirmed the maturation of SC-islets, SC-β cells, and PPs post-transplantation with immunostaining. Transplantation sites ranged from portal vein, intramuscular, and subcutaneous to kidney capsule.12,16,24,75 The impact on pregnancy and sex between mouse recipients revealed no differences between male and female mice.75,77 In vivo models were used to test encapsulation devices with stem cell-derived β-like cells and progenitors.75,78, 79, 80, 81 Transplantation was typically assessed in immunocompromised mice due to immune rejection of SC-islets; however, Yoshihara et al.72 successfully transplanted and maintained glucose homeostasis in diabetic immunocompetent mice for 50 days, with their human islet-like organoids genetically engineered to overexpress PD-L1. These combined efforts for in vivo testing are encouraging that a diabetes cell therapy will soon be produced.
Personalized cell replacement therapy
The standard care of treatment for patients with T1D is exogenous insulin. Risks and complications involved with this treatment include hypoglycemia, swelling and infection at injection site, and weight gain. In addition, long-term exposure may increase cardiovascular complications, stroke risk, and kidney damage, due to uncontrolled hyperglycemic and hypoglycemic events.82 Experimental treatment methods currently in practice include pancreas and cadaveric islet transplantation.83, 84, 85 These methods were only used when a patient was unresponsive to standard treatment or kidney damage was present and the patient concurrently underwent kidney transplantation surgery. Cadaveric islet transplantations have limited donor availability and require multiple donors, resulting in donor-to-donor variation. Clinical trials are also under way byViaCyte using allogeneic stem cell-derived PPs and an encapsulation device (NCT02239354 and NCT03163511) and planned by Vertex Pharmaceuticals using allogeneic SC-islets (NCT04786262). The transplantation of allogeneic cells and organs requires the recipient to be taking immunosuppressive drugs, which can be problematic for patients with T1D. SC-islets overcome several of these limitations, providing an unlimited cell source with robust function for use as a cell therapy. Autologous diabetes cell therapy is made possible with patient iPSC-islets, which contain their missing insulin-secreting β cells, and has the potential to remove the need for immunosuppressive drugs post-transplant. However, there is a financial concern regarding autologous cell therapies for a wide range of diseases. Once cell therapy is established as a safe and viable treatment for disease, measures to reduce costs will be used, allowing for mass cell product manufacturing. Protocol updates will enable the generation of large cell batches, enabling the cryopreservation of functional SC-β cells.75 An additional concern is de novo mutations in mitochondrial DNA occurring in autologous iPSCs and leading to the production of neoantigens, dependent on MHC genotype.86 With advances in stem cell technology, diabetes cell therapy can be used in a wider patient population, including insulin-independent patients with T1D and T2D and those undergoing a pancreatectomy.
Future outlook
In general, there are technical difficulties and financial concerns related to iPSC therapy. To reduce the financial burden, biobanks of genome-stable iPSCs capable of matching the majority of HLA types87 and rarer blood types88 are being explored. This format would create more personalized medicine than allogeneic while also reduce the immunogenicity through HLA matching.87 Biobanks would also ease the technical and costly difficulties of personalized medicine by creating banks of HLA-matched differentiated cell types using mass production. Genomic instability is an additional obstacle in iPSC therapy clinical translation. When reprogramming somatic cells to iPSCs, chromosomal aberrations or mutations often occur.89 Furthermore, tumor formation remains a concern. While diabetes stem cell replacement therapy has several established protocols that have little to no detectable teratoma-forming cells,12 many organs and differentiated cell types necessary to address other diseases remain preliminary and will require several additional iterations before clinical translation can occur. However, genetic engineering of suicide genes to destroy transplanted cells upon tumor formation is being investigated.90 After these obstacles are overcome and stem cell therapy is available in the clinical setting, the quality of life will be greatly increased for patients worldwide.
SC-islets are a promising renewable cell source for diabetes cell therapy that can be produced with recent differentiation protocols.12,13,25 While advances in SC-islet differentiation protocols are encouraging, a universal differentiation protocol adaptable to all iPSC lines would better facilitate autologous cell therapy. This would eliminate the costly and time-consuming optimization and difficulties that are often encountered with new cell lines. Genetic engineering tools could aid in advancing autologous diabetes cell therapy research. These tools could identify iPSC or early-stage markers that define suitability for an efficient SC-islet differentiation65 or genes to overexpress in iPSCs to initiate or improve differentiation.91 In addition, reporters to enable robust readouts of function and maturation after cell fate specification would help both cell therapy development and drug screening applications.92, 93, 94 Ideally, SC-islets would be indistinguishable from primary islets for transplantation, specifically in terms of glucose-stimulated insulin secretion, but further protocol improvements are needed to achieve this.
Acknowledgments
J.R.M. was supported by the NIH (R01DK114233 and R01DK127497), a JDRF Career Development Award (5-CDA-2017-391-A-N), and startup funds from the Washington University School of Medicine Department of Medicine. We thank Sarah Gale and Michelle Kim for editorial assistance and Cassandra Pinhas for graphic design.
Author contributions
K.G.M. and J.R.M. wrote, edited, and reviewed the manuscript.
Declaration of interests
K.G.M. and J.R.M. are inventors on licensed patents and patent applications related to the SC-β cell technology described in this manuscript. J.R.M. is a consultant for Sana Biotechnology. K.G.M. is the chief operations officer and co-founder of Salentra Biosciences. J.R.M. is the chief scientific officer and co-founder of Salentra Biosciences.
References
- 1.Cho N.H., Shaw J.E., Karuranga S., Huang Y., da Rocha Fernandes J.D., Ohlrogge A.W., Malanda B. IDF Diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 2.Meloche R.M. Transplantation for the treatment of type 1 diabetes. World J. Gastroenterol. 2007;13:6347–6355. doi: 10.3748/wjg.v13.i47.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharroubi A.T., Darwish H.M. Diabetes mellitus: the epidemic of the century. World J. Diabetes. 2015;6:850–867. doi: 10.4239/wjd.v6.i6.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S.S., Epstein S., Corkey B.E., Grant S.F.A., Gavin J.R., III, Aguilar R.B., Herman M.E. A unified pathophysiological construct of diabetes and its complications. Trends Endocrinol. Metab. 2017;28:645–655. doi: 10.1016/j.tem.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Hering B.J., Clarke W.R., Bridges N.D., Eggerman T.L., Alejandro R., Bellin M.D., Chaloner K., Czarniecki C.W., Goldstein J.S., Hunsicker L.G., Clinical Islet Transplantation Consortium Phase 3 trial of transplantation of human islets in type 1 diabetes complicated by severe hypoglycemia. Diabetes Care. 2016;39:1230–1240. doi: 10.2337/dc15-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan E.A., Paty B.W., Senior P.A., Bigam D., Alfadhli E., Kneteman N.M., Lakey J.R., Shapiro A.M. Five-year follow-up after clinical islet transplantation. Diabetes. 2005;54:2060–2069. doi: 10.2337/diabetes.54.7.2060. [DOI] [PubMed] [Google Scholar]
- 7.D’Amour K.A., Bang A.D., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 8.D’Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 9.Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 10.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O’Dwyer S., Quiskamp N., Mojibian M., Albrecht T. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 12.Hogrebe N.J., Augsornworawat P., Maxwell K.G., Velazco-Cruz L., Millman J.R. Targeting the cytoskeleton to direct pancreatic differentiation of human pluripotent stem cells. Nat. Biotechnol. 2020;38:460–470. doi: 10.1038/s41587-020-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair G.G., Liu J.S., Russ H.A., Tran S., Saxton M.S., Chen R., Juang C., Li M.L., Nguyen V.Q., Giacometti S. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat. Cell Biol. 2019;21:263–274. doi: 10.1038/s41556-018-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velazco-Cruz L., Song J., Maxwell K.G., Goedegebuure M.M., Augsornworawat P., Hogrebe N.J., Millman J.R. Acquisition of Dynamic Function in Human Stem Cell-Derived β Cells. Stem Cell Reports. 2019;12:351–365. doi: 10.1016/j.stemcr.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balboa D., Saarimäki-Vire J., Borshagovski D., Survila M., Lindholm P., Galli E., Eurola S., Ustinov J., Grym H., Huopio H. Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. eLife. 2018;7:e38519. doi: 10.7554/eLife.38519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma S., Viola R., Sui L., Cherubini V., Barbetti F., Egli D. β cell replacement after gene editing of a neonatal diabetes-causing mutation at the insulin locus. Stem Cell Reports. 2018;11:1407–1415. doi: 10.1016/j.stemcr.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxwell K.G., Augsornworawat P., Velazco-Cruz L., Kim M.H., Asada R., Hogrebe N.J., Morikawa S., Urano F., Millman J.R. Gene-edited human stem cell-derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med. 2020;12:eaax9106. doi: 10.1126/scitranslmed.aax9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran R., Moraes C., Hoesli C.A. Developmentally-Inspired Biomimetic Culture Models to Produce Functional Islet-Like Cells From Pluripotent Precursors. Front. Bioeng. Biotechnol. 2020;8:583970. doi: 10.3389/fbioe.2020.583970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nostro M.C., Keller G. Generation of beta cells from human pluripotent stem cells: potential for regenerative medicine. Semin. Cell Dev. Biol. 2012;23:701–710. doi: 10.1016/j.semcdb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velazco-Cruz L., Goedegebuure M.M., Millman J.R. Advances Toward Engineering Functionally Mature Human Pluripotent Stem Cell-Derived β Cells. Front. Bioeng. Biotechnol. 2020;8:786. doi: 10.3389/fbioe.2020.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theis F.J., Lickert H. A map of β-cell differentiation pathways supports cell therapies for diabetes. Nature. 2019;569:342–343. doi: 10.1038/d41586-019-01211-9. [DOI] [PubMed] [Google Scholar]
- 22.Aguayo-Mazzucato C., Bonner-Weir S. Stem cell therapy for type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2010;6:139–148. doi: 10.1038/nrendo.2009.274. [DOI] [PubMed] [Google Scholar]
- 23.Tremmel D.M., Mitchell S.A., Sackett S.D., Odorico J.S. Mimicking nature-made beta cells: recent advances towards stem cell-derived islets. Curr. Opin. Organ Transplant. 2019;24:574–581. doi: 10.1097/MOT.0000000000000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sui L., Danzl N., Campbell S.R., Viola R., Williams D., Xing Y., Wang Y., Phillips N., Poffenberger G., Johannesson B. β-Cell Replacement in Mice Using Human Type 1 Diabetes Nuclear Transfer Embryonic Stem Cells. Diabetes. 2018;67:26–35. doi: 10.2337/db17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H., Harb G., Poh Y.C., Sintov E., Gürtler M., Pagliuca F.W. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Augsornworawat P., Maxwell K.G., Velazco-Cruz L., Millman J.R. Single-Cell Transcriptome Profiling Reveals β Cell Maturation in Stem Cell-Derived Islets after Transplantation. Cell Rep. 2020;32:108067. doi: 10.1016/j.celrep.2020.108067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosentino C., Toivonen S., Diaz Villamil E., Atta M., Ravanat J.L., Demine S., Schiavo A.A., Pachera N., Deglasse J.P., Jonas J.C. Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018;46:10302–10318. doi: 10.1093/nar/gky839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nostro M.C., Sarangi F., Yang C., Holland A., Elefanty A.G., Stanley E.G., Greiner D.L., Keller G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports. 2015;4:591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russ H.A., Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M., Guo T., Puri S., Haataja L., Cirulli V. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudva Y.C., Ohmine S., Greder L.V., Dutton J.R., Armstrong A., De Lamo J.G., Khan Y.K., Thatava T., Hasegawa M., Fusaki N. Transgene-free disease-specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem Cells Transl. Med. 2012;1:451–461. doi: 10.5966/sctm.2011-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Leite N.C., Sintov E., Meissner T.B., Brehm M.A., Greiner D.L., Harlan D.M., Melton D.A. Modeling Type 1 Diabetes In Vitro Using Human Pluripotent Stem Cells. Cell Rep. 2020;32:107894. doi: 10.1016/j.celrep.2020.107894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Sterr M., Ansarullah, Burtscher I., Böttcher A., Beckenbauer J., Siehler J., Meitinger T., Häring H.U., Staiger H. Point mutations in the PDX1 transactivation domain impair human β-cell development and function. Mol. Metab. 2019;24:80–97. doi: 10.1016/j.molmet.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dwivedi O.P., Lehtovirta M., Hastoy B., Chandra V., Krentz N.A.J., Kleiner S., Jain D., Richard A.M., Abaitua F., Beer N.L. Loss of ZnT8 function protects against diabetes by enhanced insulin secretion. Nat. Genet. 2019;51:1596–1606. doi: 10.1038/s41588-019-0513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amin S., Cook B., Zhou T., Ghazizadeh Z., Lis R., Zhang T., Khalaj M., Crespo M., Perera M., Xiang J.Z. Discovery of a drug candidate for GLIS3-associated diabetes. Nat. Commun. 2018;9:2681. doi: 10.1038/s41467-018-04918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millman J.R., Xie C., Van Dervort A., Gürtler M., Pagliuca F.W., Melton D.A. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat. Commun. 2016;7:11463. doi: 10.1038/ncomms11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thatava T., Kudva Y.C., Edukulla R., Squillace K., Genebriera De Lamo J., Krotova Khan Y., Sakuma T., Ohmine S., Terzic A., Ikeda Y. Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Mol. Ther. 2013;21:228–239. doi: 10.1038/mt.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lithovius V., Saarimäki-Vire J., Balboa D., Ibrahim H., Montaser H., Barsby T., Otonkoski T. SUR1-mutant iPS cell-derived islets recapitulate the pathophysiology of congenital hyperinsulinism. Diabetologia. 2021;64:630–640. doi: 10.1007/s00125-020-05346-7. [DOI] [PubMed] [Google Scholar]
- 39.Cardenas-Diaz F.L., Osorio-Quintero C., Diaz-Miranda M.A., Kishore S., Leavens K., Jobaliya C., Stanescu D., Ortiz-Gonzalez X., Yoon C., Chen C.S. Modeling Monogenic Diabetes using Human ESCs Reveals Developmental and Metabolic Deficiencies Caused by Mutations in HNF1A. Cell Stem Cell. 2019;25:273–289.e5. doi: 10.1016/j.stem.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishore S., De Franco E., Cardenas-Diaz F.L., Letourneau-Freiberg L.R., Sanyoura M., Osorio-Quintero C., French D.L., Greeley S.A.W., Hattersley A.T., Gadue P. A Non-Coding Disease Modifier of Pancreatic Agenesis Identified by Genetic Correction in a Patient-Derived iPSC Line. Cell Stem Cell. 2020;27:137–146.e6. doi: 10.1016/j.stem.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stepniewski J., Kachamakova-Trojanowska N., Ogrocki D., Szopa M., Matlok M., Beilharz M. Induced pluripotent stem cells as a model for diabetes investigation. Sci Rep. 2015;5:8597. doi: 10.1038/srep08597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang L., Hua H., Foo K., Martinez H., Watanabe K., Zimmer M., Kahler D.J., Freeby M., Chung W., LeDuc C. β-cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes. 2014;63:923–933. doi: 10.2337/db13-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teo A.K., Windmueller R., Johansson B.B., Dirice E., Njolstad P.R., Tjora E. Derivation of Human Induced Pluripotent Stem Cells from Patients with Maturity Onset Diabetes of the Young. J Biol Chem. 2013;288:5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steiner D.J., Kim A., Miller K., Hara M. Pancreatic islet plasticity: interspecies comparison of islet architecture and composition. Islets. 2010;2:135–145. doi: 10.4161/isl.2.3.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eizirik D.L., Pipeleers D.G., Ling Z., Welsh N., Hellerström C., Andersson A. Major species differences between humans and rodents in the susceptibility to pancreatic beta-cell injury. Proc. Natl. Acad. Sci. USA. 1994;91:9253–9256. doi: 10.1073/pnas.91.20.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.M., Andréasson A.C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M.K. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 48.Xin Y., Dominguez Gutierrez G., Okamoto H., Kim J., Lee A.H., Adler C., Ni M., Yancopoulos G.D., Murphy A.J., Gromada J. Pseudotime ordering of single human β-cells reveals states of insulin production and unfolded protein response. Diabetes. 2018;67:1783–1794. doi: 10.2337/db18-0365. [DOI] [PubMed] [Google Scholar]
- 49.Simsek S., Zhou T., Robinson C.L., Tsai S.Y., Crespo M., Amin S., Lin X., Hon J., Evans T., Chen S. Modeling Cystic Fibrosis Using Pluripotent Stem Cell-Derived Human Pancreatic Ductal Epithelial Cells. Stem Cells Transl. Med. 2016;5:572–579. doi: 10.5966/sctm.2015-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris A.G., Letourneau L.R., Greeley S.A.W. Monogenic diabetes: the impact of making the right diagnosis. Curr. Opin. Pediatr. 2018;30:558–567. doi: 10.1097/MOP.0000000000000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hrvatin S., O’Donnell C.W., Deng F., Millman J.R., Pagliuca F.W., DiIorio P., Rezania A., Gifford D.K., Melton D.A. Differentiated human stem cells resemble fetal, not adult, β cells. Proc. Natl. Acad. Sci. USA. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Augsornworawat P., Millman J.R. Single-cell RNA sequencing for engineering and studying human islets. Curr. Opin. Biomed. Eng. 2020;16:27–33. doi: 10.1016/j.cobme.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muraro M.J., Dharmadhikari G., Grün D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M.A., Carlotti F., de Koning E.J.P., van Oudenaarden A. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krentz N.A.J., Lee M.Y.Y., Xu E.E., Sproul S.L.J., Maslova A., Sasaki S., Lynn F.C. Single-Cell Transcriptome Profiling of Mouse and hESC-Derived Pancreatic Progenitors. Stem Cell Reports. 2018;11:1551–1564. doi: 10.1016/j.stemcr.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demine S., Schiavo A.A., Marín-Cañas S., Marchetti P., Cnop M., Eizirik D.L. Pro-inflammatory cytokines induce cell death, inflammatory responses, and endoplasmic reticulum stress in human iPSC-derived beta cells. Stem Cell Res. Ther. 2020;11:7. doi: 10.1186/s13287-019-1523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonseca S.G., Gromada J., Urano F. Endoplasmic reticulum stress and pancreatic β-cell death. Trends Endocrinol. Metab. 2011;22:266–274. doi: 10.1016/j.tem.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hakonen E., Chandra V., Fogarty C.L., Yu N.Y., Ustinov J., Katayama S., Galli E., Danilova T., Lindholm P., Vartiainen A. MANF protects human pancreatic beta cells against stress-induced cell death. Diabetologia. 2018;61:2202–2214. doi: 10.1007/s00125-018-4687-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misun P.M., Yesildag B., Forschler F., Neelakandhan A., Rousset N., Biernath A., Hierlemann A., Frey O. In Vitro Platform for Studying Human Insulin Release Dynamics of Single Pancreatic Islet Microtissues at High Resolution. Adv. Biosyst. 2020;4:e1900291. doi: 10.1002/adbi.201900291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenguito G., Chaimov D., Weitz J.R., Rodriguez-Diaz R., Rawal S.A., Tamayo-Garcia A., Caicedo A., Stabler C.L., Buchwald P., Agarwal A. Resealable, optically accessible, PDMS-free fluidic platform for ex vivo interrogation of pancreatic islets. Lab Chip. 2017;17:772–781. doi: 10.1039/c6lc01504b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jun Y., Lee J., Choi S., Yang J.H., Sander M., Chung S., Lee S.H. In vivo-mimicking microfluidic perfusion culture of pancreatic islet spheroids. Sci. Adv. 2019;5:eaax4520. doi: 10.1126/sciadv.aax4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sankar K.S., Green B.J., Crocker A.R., Verity J.E., Altamentova S.M., Rocheleau J.V. Culturing pancreatic islets in microfluidic flow enhances morphology of the associated endothelial cells. PLoS ONE. 2011;6:e24904. doi: 10.1371/journal.pone.0024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glieberman A.L., Pope B.D., Zimmerman J.F., Liu Q., Ferrier J.P., Kenty J.H.R., Schrell A.M., Mukhitov N., Shores K.L., Tepole A.B. Synchronized stimulation and continuous insulin sensing in a microfluidic human islet on a chip designed for scalable manufacturing. Lab Chip. 2019;19:2993–3010. doi: 10.1039/c9lc00253g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishahak M., Birman L., Carbonero D. Integrated platform for operating and interrogating organs-on-chips. Anal. Methods. 2019;11:5645–5651. [Google Scholar]
- 64.Wesolowska-Andersen A., Jensen R.R., Alcántara M.P., Beer N.L., Duff C., Nylander V., Gosden M., Witty L., Bowden R., McCarthy M.I. Analysis of Differentiation Protocols Defines a Common Pancreatic Progenitor Molecular Signature and Guides Refinement of Endocrine Differentiation. Stem Cell Reports. 2020;14:138–153. doi: 10.1016/j.stemcr.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahaddalkar P.U., Scheibner K., Pfluger S., Ansarullah, Sterr M., Beckenbauer J., Irmler M., Beckers J., Knöbel S., Lickert H. Generation of pancreatic β cells from CD177+ anterior definitive endoderm. Nat. Biotechnol. 2020;38:1061–1072. doi: 10.1038/s41587-020-0492-5. [DOI] [PubMed] [Google Scholar]
- 66.Li X., Yang K.Y., Chan V.W., Leung K.T., Zhang X.B., Wong A.S., Chong C.C.N., Wang C.C., Ku M., Lui K.O. Single-Cell RNA-Seq Reveals that CD9 Is a Negative Marker of Glucose-Responsive Pancreatic β-like Cells Derived from Human Pluripotent Stem Cells. Stem Cell Reports. 2020;15:1111–1126. doi: 10.1016/j.stemcr.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y., Liu Q., Zhou Z., Ikeda Y. PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res. Ther. 2017;8:240. doi: 10.1186/s13287-017-0694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eichstadt S., Barriga M., Ponakala A., Teng C., Nguyen N.T., Siprashvili Z., Nazaroff J., Gorell E.S., Chiou A.S., Taylor L. Phase 1/2a clinical trial of gene-corrected autologous cell therapy for recessive dystrophic epidermolysis bullosa. JCI Insight. 2019;4:130554. doi: 10.1172/jci.insight.130554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Río P., Navarro S., Wang W., Sánchez-Domínguez R., Pujol R.M., Segovia J.C., Bogliolo M., Merino E., Wu N., Salgado R. Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat. Med. 2019;25:1396–1401. doi: 10.1038/s41591-019-0550-z. [DOI] [PubMed] [Google Scholar]
- 70.Lee J., Bayarsaikhan D., Bayarsaikhan G., Kim J.S., Schwarzbach E., Lee B. Recent advances in genome editing of stem cells for drug discovery and therapeutic application. Pharmacol. Ther. 2020;209:107501. doi: 10.1016/j.pharmthera.2020.107501. [DOI] [PubMed] [Google Scholar]
- 71.Sackett S.D., Rodriguez A., Odorico J.S. The nexus of stem cell-derived beta-cells and genome engineering. Rev. Diabet. Stud. 2017;14:39–50. doi: 10.1900/RDS.2017.14.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshihara E., O’Connor C., Gasser E., Wei Z., Oh T.G., Tseng T.W., Wang D., Cayabyab F., Dai Y., Yu R.T. Immune-evasive human islet-like organoids ameliorate diabetes. Nature. 2020;586:606–611. doi: 10.1038/s41586-020-2631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deuse T., Hu X., Gravina A., Wang D., Tediashvili G., De C., Thayer W.O., Wahl A., Garcia J.V., Reichenspurner H. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 2019;37:252–258. doi: 10.1038/s41587-019-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Millman J.R., Pagliuca F.W. Autologous Pluripotent Stem Cell-Derived β-Like Cells for Diabetes Cellular Therapy. Diabetes. 2017;66:1111–1120. doi: 10.2337/db16-1406. [DOI] [PubMed] [Google Scholar]
- 75.Stock A.A., Manzoli V., De Toni T., Abreu M.M., Poh Y.C., Ye L., Roose A., Pagliuca F.W., Thanos C., Ricordi C., Tomei A.A. Conformal Coating of Stem Cell-Derived Islets for β Cell Replacement in Type 1 Diabetes. Stem Cell Reports. 2020;14:91–104. doi: 10.1016/j.stemcr.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu S., Russ H.A., Wang X., Zhang M., Ma T., Xu T., Tang S., Hebrok M., Ding S. Human pancreatic beta-like cells converted from fibroblasts. Nat. Commun. 2016;7:10080. doi: 10.1038/ncomms10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saber N., Bruin J.E., O’Dwyer S., Schuster H., Rezania A., Kieffer T.J. Sex Differences in Maturation of Human Embryonic Stem Cell-Derived β Cells in Mice. Endocrinology. 2018;159:1827–1841. doi: 10.1210/en.2018-00048. [DOI] [PubMed] [Google Scholar]
- 78.Bruin J.E., Rezania A., Xu J., Narayan K., Fox J.K., O’Neil J.J., Kieffer T.J. Maturation and function of human embryonic stem cell-derived pancreatic progenitors in macroencapsulation devices following transplant into mice. Diabetologia. 2013;56:1987–1998. doi: 10.1007/s00125-013-2955-4. [DOI] [PubMed] [Google Scholar]
- 79.Robert T., De Mesmaeker I., Stangé G.M., Suenens K.G., Ling Z., Kroon E.J., Pipeleers D.G. Functional beta cell mass from device-encapsulated hESC-derived pancreatic endoderm achieving metabolic control. Stem Cell Reports. 2018;10:739–750. doi: 10.1016/j.stemcr.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang R., Faleo G., Russ H.A., Parent A.V., Elledge S.K., Bernards D.A., Allen J.L., Villanueva K., Hebrok M., Tang Q., Desai T.A. Nanoporous immunoprotective device for stem-cell-derived β-cell replacement therapy. ACS Nano. 2017;11:7747–7757. doi: 10.1021/acsnano.7b01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Desai T., Shea L.D. Advances in islet encapsulation technologies. Nat. Rev. Drug Discov. 2017;16:338–350. doi: 10.1038/nrd.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nathan D.M. Long-term complications of diabetes mellitus. N. Engl. J. Med. 1993;328:1676–1685. doi: 10.1056/NEJM199306103282306. [DOI] [PubMed] [Google Scholar]
- 83.Scharp D.W., Lacy P.E., Santiago J.V., McCullough C.S., Weide L.G., Falqui L., Marchetti P., Gingerich R.L., Jaffe A.S., Cryer P.E. Insulin independence after islet transplantation into type I diabetic patient. Diabetes. 1990;39:515–518. doi: 10.2337/diab.39.4.515. [DOI] [PubMed] [Google Scholar]
- 84.Shapiro A.M., Ricordi C., Hering B.J., Auchincloss H., Lindblad R., Robertson R.P., Secchi A., Brendel M.D., Berney T., Brennan D.C. International trial of the Edmonton protocol for islet transplantation. N. Engl. J. Med. 2006;355:1318–1330. doi: 10.1056/NEJMoa061267. [DOI] [PubMed] [Google Scholar]
- 85.Shapiro A.M.J., Lakey J.R.T., Ryan E.A., Korbutt G.S., Toth E., Warnock G.L., Kneteman N.M., Rajotte R.V. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 2000;343:230–238. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- 86.Deuse T., Hu X., Agbor-Enoh S., Koch M., Spitzer M.H., Gravina A., Alawi M., Marishta A., Peters B., Kosaloglu-Yalcin Z. De novo mutations in mitochondrial DNA of iPSCs produce immunogenic neoepitopes in mice and humans. Nat. Biotechnol. 2019;37:1137–1144. doi: 10.1038/s41587-019-0227-7. [DOI] [PubMed] [Google Scholar]
- 87.de Rham C., Villard J. Potential and limitation of HLA-based banking of human pluripotent stem cells for cell therapy. J. Immunol. Res. 2014;2014:518135. doi: 10.1155/2014/518135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park Y.J., Jeon S.H., Kim H.K., Suh E.J., Choi S.J., Kim S., Kim H.O. Human induced pluripotent stem cell line banking for the production of rare blood type erythrocytes. J. Transl. Med. 2020;18:236. doi: 10.1186/s12967-020-02403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turinetto V., Orlando L., Giachino C. Induced Pluripotent Stem Cells: Advances in the Quest for Genetic Stability during Reprogramming Process. Int. J. Mol. Sci. 2017;18:E1952. doi: 10.3390/ijms18091952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Penney J., Ralvenius W.T., Tsai L.H. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry. 2020;25:148–167. doi: 10.1038/s41380-019-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ng A.H.M., Khoshakhlagh P., Rojo Arias J.E., Pasquini G., Wang K., Swiersy A., Shipman S.L., Appleton E., Kiaee K., Kohman R.E. A comprehensive library of human transcription factors for cell fate engineering. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cardenas-Diaz F.L., Leavens K.F., Kishore S., Osorio-Quintero C., Chen Y.J., Stanger B.Z., Wang P., French D., Gadue P. A Dual Reporter EndoC-βH1 Human β-Cell Line for Efficient Quantification of Calcium Flux and Insulin Secretion. Endocrinology. 2020;161:bqaa005. doi: 10.1210/endocr/bqaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kracht M.J.L., de Koning E.J.P., Hoeben R.C., Roep B.O., Zaldumbide A. Bioluminescent reporter assay for monitoring ER stress in human beta cells. Sci. Rep. 2018;8:17738. doi: 10.1038/s41598-018-36142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burns S.M., Vetere A., Walpita D., Dančík V., Khodier C., Perez J., Clemons P.A., Wagner B.K., Altshuler D. High-throughput luminescent reporter of insulin secretion for discovering regulators of pancreatic Beta-cell function. Cell Metab. 2015;21:126–137. doi: 10.1016/j.cmet.2014.12.010. [DOI] [PubMed] [Google Scholar]