Abstract
We developed a predictive model associated with ferroptosis to provide a more comprehensive view of esophageal squamous cell carcinoma (ESCC) immunotherapy. Gene expression data and corresponding clinical outcomes were obtained from the GEO and The Cancer Genome Atlas (TCGA) databases, and a ferroptosis-related gene set was obtained from the FerrDb database. We identified 45 ferroptosis-related genes that were differentially expressed, including enrichment in genes involved in the immune system process. We established a ferroptosis-related gene-based prognostic model based on the results of univariate Cox regression and multivariate Cox regression analyses, with an area under the curve (AUC) of 0.76 (3 years). We found that the patients with low-risk scores showed a higher proportion of CD8+ T cells, CD4+ memory activated T cells, etc. Finally, a predictive ferroptosis-related prognostic nomogram, which included the predictive values of age, gender, grade, TNM stage, and risk score, was established to predict overall survival. In sum, we developed a ferroptosis-related gene-based prognostic model that provides novel insights into the prediction of ESCC prognosis and identifies the relevance of the immune microenvironment for patient outcomes.
Keywords: esophageal squamous cell carcinoma, ferroptosis, prognostic model, immune microenvironment, TCGA
Graphical Abstract
Lu et al. developed a ferroptosis-related gene-based prognostic model that provides novel insights into the prediction of ESCC prognosis and identifies the relevance of the immune microenvironment for patient outcomes.
Introduction
Esophageal carcinoma (ESCA) is a common malignant digestive system cancer type that ranks eighth in morbidity and sixth in mortality worldwide.1 Esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC) are the two histopathological subtypes of ESCA; both subtypes differ in etiology, incidence, and clinical characteristics.2 In China, 90% of the patients have squamous cell carcinoma. The 5-year overall survival (OS) of advanced ESCC is only 18.8%.3,4 Unfortunately, most patients have advanced-stage diseases at first diagnosis, generally because of the absence of clinical symptoms.5 Recently, multidisciplinary therapy methods, including surgical resection, radiotherapy, chemotherapy, and targeted therapy, provide new options to prolong OS.6,7 Moreover, the dramatic development of immune checkpoint inhibitors, such as CTLA-4 and PD-1, suggests amazing therapeutic effects in clinical efficacy.8,9 However, the therapy conditions are not eligible for most ESCC patients, which suggests that more studies on the molecular mechanisms’ elucidation and identifying useful biomarkers for immune checkpoint inhibitors are still urgently needed for cancer immunotherapy.10,11
Ferroptosis is non-apoptotic regulated cell death (RCD) and an iron-dependent process that differs from necrosis or apoptosis and results from iron-dependent lipid peroxide accumulation.12,13 Recently, ferroptosis has been elucidated to serve multiple roles in the biological regulations and signal transduction pathways, leading to tumor initiation and progression.14, 15, 16 Many genes have been validated to promote ferroptosis in tumor cells, such as FBXW7,17 G6PD,12 and TP53,18,19 while CISD2,20 GPX4,21, 22, 23 and SLC7A1124,25 serve as suppressors to prevent ferroptosis. Wang et al.26 demonstrated that ferroptosis-specific lipid peroxidation is enhanced by CD8+ T cells and, in turn, contributes to the anti-tumor efficacy of immunotherapy. Thus, understanding the relationship between ferroptosis and immune cell infiltration may provide a more comprehensive view of cancer immunotherapy efficacy.
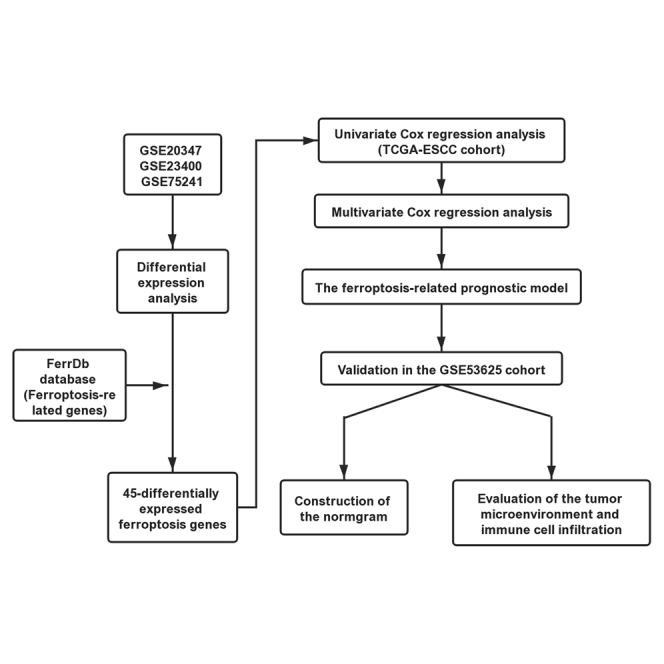
Previous studies on ferroptosis-related prognostic signatures of cancers have focused on a few relevant genes, possibly neglecting other significant factors present within the immune microenvironment.27,28 In the present study, we systematically profiled the ESCC expression data and clinical outcomes from the GEO and The Cancer Genome Atlas (TCGA) databases. Furthermore, we identified ferroptosis-related genes (drivers, suppressors, and markers) based on data from the FerrDb database.29 We analyzed the differentially expressed ferroptosis genes between ESCC and adjacent normal tissues, screened survival-associated signatures, and built a ferroptosis-genes prognostic model to predict the outcome of patients with ESCC. Our results underline the association between ferroptosis and the immune microenvironment in ESCC and add to our understanding of the interaction between ferroptosis and immune cell abundance in the tumor microenvironment (TME). Our data may suggest that targeting ferroptosis genes may improve immunotherapy efficacy.
Results
Differentially expressed ferroptosis signatures in ESCC
The information of GEO datasets used is listed in Table 1. Following the differential gene analysis, 1,367 dysregulated genes were obtained from GEO: GSE20347, with 896 genes showing upregulation and 471 showing downregulation, and 710 dysregulated genes from GEO: GSE23400, with 413 genes showing upregulation and 297 showing downregulation. Finally, we obtained 1,653 dysregulated genes from GEO: GSE75241, of which 1,125 genes were upregulated and 528 genes were downregulated (Figure 1A). Because the ferroptosis-related genes obtained from the FerrDb database were experimentally validated (Table S1), the differentially expressed genes (DEGs) obtained from the GEO datasets were intersected with the ferroptosis-gene set to obtain “differentially expressed ferroptosis genes.” The Venn diagram revealed that 45 ferroptosis-related genes intersected between four datasets (Figure 1B). Relevant details of all gene lists and overlap are available in Table S2.
Table 1.
The information of datasets from the GEO database
| Accession number | Platform | Samples | Experiment type | PMID |
|---|---|---|---|---|
| GEO: GSE20347 | GPL571 | 34 | expression profiling by array | 20955586 |
| GEO: GSE23400 | GPL96; GPL97 | 208 | expression profiling by array | 21385931 |
| GEO: GSE75241 | GPL5175 | 30 | expression profiling by array | 29682174 |
| GEO: GSE53625 | GPL18109 | 358 | expression profiling by array; non-coding RNA profiling by array | 24522499 |
Figure 1.
Overview of the differentially expressed ferroptosis signatures in ESCC
(A) Expression profiles of ferroptosis-related genes in normal and tumor samples in GEO: GSE20347, GSE23400, and GSE75241 datasets. Genes were clustered according to their expression. Red color represents high expression, and blue color represents low expression. (B) Venn diagram showing the dysregulated ferroptosis genes common to the four datasets. (C and D) Graph showing the GO and KEGG analysis based on the Metascape Online, bar plot, and network showing the distribution and relationship of the different functions. (E) PPI network and MCODE showing the hub genes in the ferroptosis gene set.
To explore the underlying mechanisms of the ferroptosis signatures in ESCC, we performed a functional analysis using Metascape Online. As shown in Figures 1C and 1D, the Gene Ontology (GO) analysis results suggest that these dysregulated ferroptosis genes are mainly enriched in response to a stimulus, immune system process, and cell proliferation. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that these dysregulated ferroptosis genes were mainly enriched in ferroptosis, interleukin-4, and interleukin-13 signaling. Thus, these data motivated us to study the relationship between the ferroptosis-gene set and tumor immune microenvironment. Furthermore, the protein-protein interaction (PPI) network and MCODE plugin based on the Metascape Online identified the significant modules in these ferroptosis genes. Module 1 included six edges and four nodes and involved SLC7A11, CD44, SLC3A2, and OTUB1. Module 2 included three edges and three nodes and involved AKR1C1, CAV1, and PGD (Figure 1E).
The establishment and verification of a ferroptosis-related prognostic model
Normalized mRNA expression data with corresponding patients’ information were obtained from TCGA-ESCC. To enhance the reliability and accuracy of the predictive model based on ferroptosis genes, GEO: GSE53625 from the GEO dataset served as a validation cohort. Univariate Cox regression analysis was first applied to detect the genes that were significantly associated with prognosis. As shown in Figures 2A–2F, six ferroptosis-related genes were identified as prognosis genes. A forest plot showing the results of the univariate Cox regression analysis is shown in Figure 2G. SCP2, MAPK1, and PRKAA1 were subsequently identified as independent prognostic signatures in a multivariate Cox regression (Table 2). Thus, a prognostic model was established based on the multivariate Cox regression. As shown in Figure 3A, a risk score for each patient was calculated as follows: 1.69 (β1) × (expression of SCP2) + (−1.05) (β2) × (expression of MAPK1) + (−0.62) (β3) × (expression of PRKAA1). Then, a high-risk group (n = 40) and a low-risk group (n = 40) were stratified based on the median of the risk score. Moreover, t-distributed stochastic neighbor embedding (t-SNE) and principal-component analysis (PCA) showed that the patients in different groups were distributed in two directions (Figures S1A and S1B). The median survival time of patients with a high risk score was significantly longer than that of the patients with a low risk score (Figure 3B). Subsequently, we created a receiver operating characteristic curve (ROC) to evaluate the prognosis prediction efficiency of the model, and we found the area under the curve (AUC) was 0.72 (1-year OS), 0.78 (3-year OS), and 0.76 (5-year OS), indicating the predictive model was well established (Figure 3C).
Figure 2.
Kaplan-Meier plots and forests plot of the prognostic ferroptosis signature
(A–F) Kaplan-Meier plots showing the ferroptosis genes with prognostic value. (G) The forest plot showing the results of the univariate Cox regression analyses.
Table 2.
Multivariate Cox regression analysis of signature in TCGA-ESCC cohort
| Variable | Coef | Exp(coef) | Se(coef) | Z | p value |
|---|---|---|---|---|---|
| SCP2 | 1.689629 | 5.417472 | 0.566988 | 2.98001 | 0.002882 |
| MAPK14 | −1.04552 | 0.351509 | 0.693474 | −1.50766 | 0.021316 |
| PRKAA1 | −0.61953 | 0.538199 | 0.373189 | −1.66009 | 0.03969 |
Figure 3.
The predictive model construction according to the multivariate Cox regression analysis
(A) The calculations for the model according to the multivariate Cox regression analyses. (B) The predictive ferroptosis-related prognostic model established from three survival-associated ferroptosis genes in TCGA-ESCC cohort. (C) The ROC curve of the three ferroptosis genes with the AUC in TCGA-ESCC cohort. (D) The predictive ferroptosis-related prognostic model was established from three survival-associated ferroptosis genes in the GEO: GSE53625 cohort. (E) The ROC curve of the three ferroptosis genes with the AUC in the GEO: GSE53625.
Furthermore, to evaluate the reliability and accuracy of the predictive model, we validated the power of the model in GEO: GSE53625. As shown in Figure 3D, the Kaplan-Meier plots revealed that the ferroptosis-related predictive model could successfully stratify patients with ESCC into a long-term and a short-term OS group. The AUC of the ROC curves of this model was 0.7 in the GEO: GSE53625 (3-year OS) (Figure 3E). Additionally, as shown in Figure S1C, we visualized the distribution of the ferroptosis genes considering the risk scores, stage, grade, gender, and age by using the R package “pheatmap.”
Assessment of the immune microenvironment in ESCC
We assigned 80 patients with gene expression profiles and clinical characteristics obtained from TCGA-ESCC to calculate the immune score, stromal score, and estimate score using the estimate algorithm. The immune scores were distributed between −1,081.35 and 2,038.01, while stromal scores ranged from −1,748.87 to 1,310.88. We then divided ESCC patients into low immune/stromal/estimate and high immune/stromal/estimate groups according to their corresponding immune/stromal/estimate scores. Then, the Kaplan-Meier plot revealed that all the groups were statistically significant, with p values of 0.033 (immune group), 0.048 (stromal group), and 0.0015 (estimate group) (Figures 4A–4C).
Figure 4.
The prognostic significance of immune score, stromal score, and estimate score in TCGA-ESCC
(A) The Kaplan-Meier plot showing the overall survival for immune score. (B) The Kaplan-Meier plot showing the overall survival for stromal score. (C) The Kaplan-Meier plot showing the overall survival for estimate score.
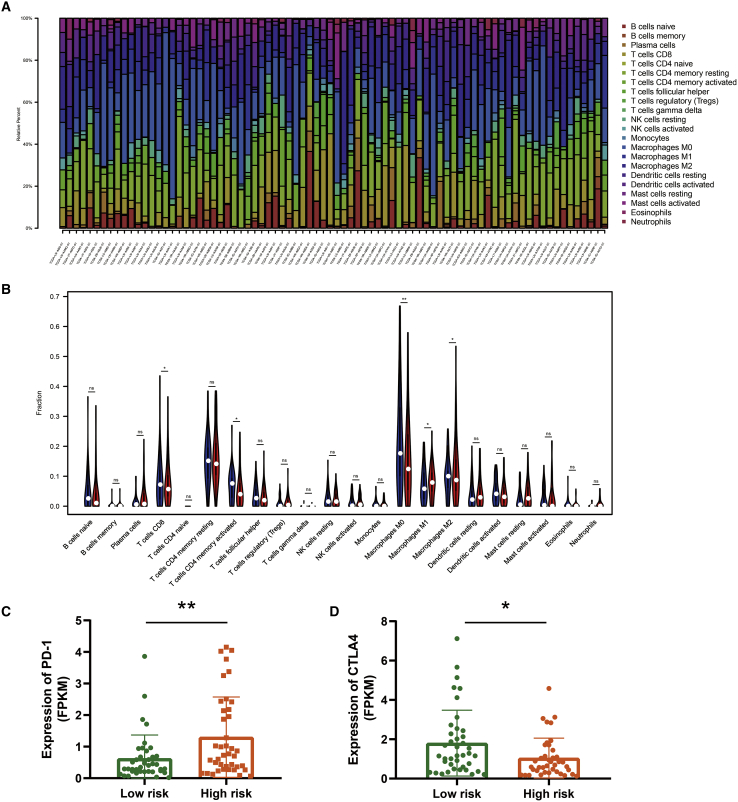
Besides calculating the relevant score of the immune microenvironment, there is a need to understand the proportion of immune landscapes in the tumor. We applied the CIBERSORT algorithm to explore the proportion of immune cells in TCGA-ESCC and GEO: GSE53625 datasets based on the gene expression data. As shown in Figure 5A, macrophages, including the M0, M1, and M2 subsets, accounted for a large proportion of infiltrating immune cells in TCGA-ESCC. Moreover, violin plots were used to visualize the immune cell subset distribution between the low- and high-risk score groups (Figure 5B). We found that the proportions of CD8+ T cells, CD4+ memory activated T cells, and M0, M1, and M2 macrophages were statistically significant. The same analysis was performed on the GEO: GSE53625 cohort. As shown in Figures S2A and S2B, macrophages are the primary immune components in the TME. Additionally, the proportions of M0, M1, and M2 macrophages were statistically significant, with a similar trend as found for TCGA-ESCC.
Figure 5.
Immune cell landscape in different ferroptosis risk score groups in TCGA-ESCC cohort
(A) The bar plot showing the proportion of infiltrated immune cells calculated by the CIBERSORT algorithm. (B) The violin plot showing the difference between 22 infiltrated immune cells in the tumor microenvironment. (C and D) Graph showing different expressions of PD-1 and CTLA4 in TCGA-ESCC cohort.
Moreover, we explored the expression of PD-1 and CTLA4 in the low- and high-risk groups and found that patients in the low-risk group exhibited a lower expression of PD-1 and a relatively higher expression of CTLA-4 in TCGA-ESCC cohort (Figures 5C and 5D). However, we noticed that the expression of CTLA4 showed no statistical significance between the two groups, and PD-1 exhibited consistency with TCGA-ESCC cohort (Figure S2C).
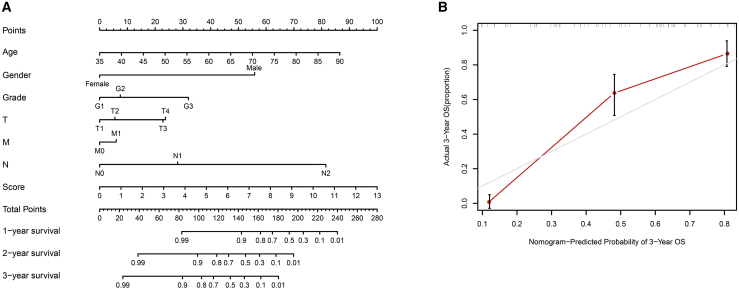
Construction of the nomogram
A predictive ferroptosis-related prognostic nomogram was established using the results of the multivariate analysis. To identify the predictive value of age, gender, grade, TNM stage, and risk score, we used nomograms to predict the 1-, 2-, and 3-year OS (Figure 6A). As shown, this nomogram was able to assess several variables to predict a patient outcome, which is based on patient characteristics, including age, gender, TNM stage, and risk score. Additionally, the predictive accuracy for OS is shown by the calibration curves. A calibration curve for the predictive probability showed an accordant agreement for the 3-year OS (Figure 6B).
Figure 6.
Proposed nomogram to predict 1-, 2-, and 3-year OS for ESCC
Locate patient’s variable on the corresponding axis. (A) To calculate survival probability, identify patient values on each axis and then for each draw a vertical line upward to the points axis. Add the points for all variables and locate this sum on the total points line. (B) Calibration curve for the probability of 3-year OS.
Discussion
Selective induction of cancer cell death is the most effective anticancer therapy.30,31 Increasing evidence has shown that ferroptosis, a recently discovered type of programmed cell death, plays a crucial role in tumorigenesis and cancer therapeutics efficacy.31 Wang et al.26 elucidated that the cell ferroptosis was regulated by CD8+ T cells and, in turn, can influence the efficacy of cancer immunotherapy. Thus, understanding the relationship between ferroptosis and the complex tumor immune microenvironment in ESCC may help identify novel biomarkers for prognosis and targeted therapy. Currently, surgical resection, chemotherapy, and radiotherapy are the three primary treatments for patients with ESCC. Although surgical resection is considered an effective treatment for improving the survival of early-stage patients, the treatment for many patients with advanced-stage disease is not satisfactory. Cancer immunotherapy provides novel therapeutic options for the comprehensive treatment of advanced resectable ESCC.
In the present study, we focused on ferroptosis-related genes and investigated their influence on prognosis. We aimed to understand the relationship between the ferroptosis-related prognostic model and the immune microenvironment to further identify potential biomarkers for prognosis detection and target therapy. First, we analyzed DEGs from the GEO: GSE20347, GSE23400, and GSE75241 datasets, assessed intersection with a validated gene set of ferroptosis obtained from the FerrDb database, and found 45 ferroptosis-related genes were received. Of note, we did not take the intersection of four datasets because few genes were obtained, but we summarized the intersection of each GEO dataset and a validated gene set of ferroptosis. Then, functional analysis was performed based on the 45 ferroptosis-related genes, and this revealed that these genes were associated with the immune system process. Univariate Cox and multivariate Cox regression analyses were used to identify the ferroptosis genes with poor prognosis and build a ferroptosis-based prognostic model. Subsequently, we studied the immune cell infiltration of the TME using the CIBERSORT and ESTIMATE algorithms. We found that the proportions of CD8+ T cells, CD4+ memory activated T cells, and M0, M1, and M2 macrophages were statistically significant.
We collected the ferroptosis-related genes from the FerrDb database. Notably, all the genes we selected had been validated. Previously, SCP2 has been shown to play significant roles in trafficking peroxidized lipids to mitochondria, and inhibition of SCP2 decreases the activity of ferroptosis.21 Poursaitidis et al.32 have demonstrated that blockade of mitogen-activated protein kinase (MAPK) signaling protects cells from ferroptosis, which in turn inhibits tumor growth. Moreover, Song et al.33 found that inhibition of PRKAA1 reduced ferroptosis through diminishing erastin-induced BECN1 phosphorylation. Thus, the ferroptosis-related genes in the prognostic model have been validated to be involved in ferroptosis in different studies.
In addition to this study, we noticed that Liang et al.28 and Liu et al.27 studied the ferroptosis-related genes in hepatocellular carcinoma and glioma, respectively. Liang et al.28 identified ferroptosis-related genes and built a predictive model for OS. Although they also studied the immune functions of the ferroptosis genes, the approaches we used were different. Additionally, Liu et al.27 studied the ferroptosis-related gene signature to predict the OS of patients with glioma and performed experiments to validate the expression and function of ferroptosis-related genes. As multiple studies have focused on ferroptosis in tumorigenesis and progression, we believe our study may help to provide novel insights into cancer immunotherapy.
In summary, our study identified a ferroptosis-related gene-based prognostic model that is independently associated with OS. Furthermore, the analysis of the ferroptosis-related prognostic model and immune profiles identifies the relevance of the immune microenvironment in affecting ESCC outcomes.
Materials and methods
Data source
The RNA expression data from accession numbers GEO: GSE20347,34 GSE23400,35 and GSE75241,36 which contained normal and tumor tissues, and the RNA expression data, as well as patients’ clinical outcomes from GEO: GSE53625,37 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo). All data were quantile normalized via log2-scale transformation to ensure standardization. The gene symbols that were detected in more than one probe were calculated using their mean expression levels. Additionally, the level three RNA sequencing (RNA-seq) data of ESCC, named TCGA-ESCC, were obtained from TCGA database (https://www.cancer.gov/tcga/). Both counts and fragments per kilobase of transcript per million mapped reads (FPKMs) and matched patients’ clinical information were downloaded for further analysis. Although the clinical information contained 96 patients, only 80 patients matched the expression information according to the sample names.
A total of 149 ferroptosis-related genes, including drivers, suppressors, and markers, obtained from the FerrDb database (http://www.zhounan.org/ferrdb/)29 were selected as candidate genes, and details are shown in Table S1. This study followed the publication guidelines of the GEO and TCGA databases.
Identification of differentially expressed ferroptosis genes and functional analysis
GEO: GSE20347, GSE23400, and GSE75241 were used to detect the DEGs between tumor and adjacent normal tissues via the R package “limma” in RStudio (version 1.2.5001), with the following cutoff for adjustment: p value < 0.05 and |log2FC| ≥ 1. The R package “pheatmap” was applied to visualize the degree range of differences in the three datasets. Then, the ferroptosis-related genes were obtained by taking the intersection of candidate genes and DEGs.
Functional analysis was performed using Metascape Online (https://metascape.org/gp/index.html#/main/step1).38 The ferroptosis-related genes were loaded into Metascape to conduct functional analysis and construct a PPI network. We performed MCODE to further reveal the densely connected regions. p <0.05 was used as a cutoff value.
Construction and validation of the ferroptosis-related gene prognostic model
TCGA-ESCC and GEO: GSE53625 datasets were used to establish a ferroptosis-related gene prognostic signature, with TCGA-ESCC being the training cohort and the GEO: GSE53625 being the validation cohort. Univariate Cox analysis of OS was first performed to identify the survival-related ferroptosis genes with a significant prognosis value. A p value <0.05 was considered statistically significant. Then, multivariate Cox regression was performed to construct the ferroptosis-related variable-based predictive model to confirm that the remaining genes were independent prognostic factors. The signature was established from the independent prognostic genes according to their corresponding coefficients.
Patients from TCGA-ESCC dataset were divided into low- and high-risk groups weighted by the risk score obtained from the multivariate Cox regression. We performed t-SNE and PCA to explore the distribution of the different groups by using the R packages “Rtsne” and “stats,” respectively. Finally, the ROC curve was created with R package “time ROC,” and the AUC was used to evaluate the efficiency of the prognostic signature.
Evaluation of the TME and infiltrated immune cells
The ESTIMATE algorithm is a method that can transform gene expression data to detect the tumor purity and activity of immune and stromal cells in the TME.39 Here, we used the R package “estimate” to calculate the immune score and used Kaplan-Meier survival curves to evaluate the relationship between immune score and patients’ survival in TCGA-ESCC dataset. The immune score could be used for the estimation of immune cells infiltrating. The two groups were selected according to the risk score of the prognostic model.
Additionally, the CIBERSORT algorithm, which is a robust method that has the capacity to characterize the composition of immune cells of complex tissues based on gene expression data, was applied to calculate the infiltrated immune cells in TCGA-ESCC tissues.40 The corresponding survival data were combined with the immune cell infiltration levels to identify the relationship between the prognostic signature and infiltrated immune cells.
Clinical relevance investigation and a nomogram construction
To study the correlation between the ferroptosis-related prognostic model and clinical characteristics, including age, gender, TNM staging system, grade, as well as PD-1 (also named PDCD1, CD279), CTLA4 expression profiles and immune cell scores were arranged for differential analysis into two groups that were determined according to the risk score of the predictive model.
Furthermore, a nomogram, which provided the survival probability of a specific outcome, was designed to integrate the risk score of the model as a prognostic factor to evaluate the predictive probability of 1-, 2-, and 3-year OS.41 A calibration curve depicting the 3-year OS was plotted to visualize the observed rates against the nomogram-predicted probabilities. The R package “rms” was used to plot the nomogram and calibration curves.
Acknowledgments
None.
Author contributions
T.L. and L.-y.Z. designed the study; T.L. and R.X. wrote the manuscript; J.-y.Z., H.Z., J.-d.G., and S.-q.Z. collected data; T.L., Q.L., H.-w.L., and J.W. analyzed data and contributed in writing the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2021.02.011.
Supplemental information
References
- 1.Goswami K.K., Ghosh T., Ghosh S., Sarkar M., Bose A., Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell. Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Soerjomataram I., Ferlay J., Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y.M., Hong P., Xu W.W., He Q.Y., Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct. Target. Ther. 2020;5:229. doi: 10.1038/s41392-020-00323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abnet C.C., Arnold M., Wei W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang F.L., Yu S.J. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J. Surg. 2018;41:210–215. doi: 10.1016/j.asjsur.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Imazeki H., Kato K. Development of chemotherapeutics for unresectable advanced esophageal cancer. Expert Rev. Anticancer Ther. 2020;20:1083–1092. doi: 10.1080/14737140.2020.1814149. [DOI] [PubMed] [Google Scholar]
- 7.Fu X., Tao L., Wu W., Zhang X. Arming HSV-Based Oncolytic Viruses with the Ability to Redirect the Host’s Innate Antiviral Immunity to Attack Tumor Cells. Mol. Ther. Oncolytics. 2020;19:33–46. doi: 10.1016/j.omto.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Liu X., Cao S., Dong X., Rao S., Cai K. Understanding Esophageal Cancer: The Challenges and Opportunities for the Next Decade. Front. Oncol. 2020;10:1727. doi: 10.3389/fonc.2020.01727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C.Y., Fan M.H., Miao C.H., Liao Y.J., Yuan R.H., Liu C.L. Engineering Chimeric Antigen Receptor T Cells against Immune Checkpoint Inhibitors PD-1/PD-L1 for Treating Pancreatic Cancer. Mol. Ther. Oncolytics. 2020;17:571–585. doi: 10.1016/j.omto.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazloom A., Ghalehsari N., Gazivoda V., Nimkar N., Paul S., Gregos P., Rateshwar J., Khan U. Role of Immune Checkpoint Inhibitors in Gastrointestinal Malignancies. J. Clin. Med. 2020;9:2533. doi: 10.3390/jcm9082533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto S., Kato K. Pembrolizumab for the treatment of esophageal cancer. Expert Opin. Biol. Ther. 2020;20:1143–1150. doi: 10.1080/14712598.2020.1792881. [DOI] [PubMed] [Google Scholar]
- 12.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Z., Song J., Yung B.C., Zhou Z., Wu A., Chen X. Emerging Strategies of Cancer Therapy Based on Ferroptosis. Adv. Mater. 2018;30:e1704007. doi: 10.1002/adma.201704007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascón S., Hatzios S.K., Kagan V.E. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stockwell B.R., Jiang X. A Physiological Function for Ferroptosis in Tumor Suppression by the Immune System. Cell Metab. 2019;30:14–15. doi: 10.1016/j.cmet.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Guo M., Li Y., Shen M., Kong D., Shao J., Ding H., Tan S., Chen A., Zhang F., Zheng S. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16:1482–1505. doi: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennis M., Kung C.P., Basu S., Budina-Kolomets A., Leu J.I., Khaku S., Scott J.P., Cai K.Q., Campbell M.R., Porter D.K. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30:918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E.H., Shin D., Lee J., Jung A.R., Roh J.L. CISD2 inhibition overcomes resistance to sulfasalazine-induced ferroptotic cell death in head and neck cancer. Cancer Lett. 2018;432:180–190. doi: 10.1016/j.canlet.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma S., Henson E.S., Chen Y., Gibson S.B. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7:e2307. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita M., Freigang S., Schneider C., Conrad M., Bornkamm G.W., Kopf M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J. Exp. Med. 2015;212:555–568. doi: 10.1084/jem.20140857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L., Hickman J.H., Wang S.J., Gu W. Dynamic roles of p53-mediated metabolic activities in ROS-induced stress responses. Cell Cycle. 2015;14:2881–2885. doi: 10.1080/15384101.2015.1068479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S.J., Li D., Ou Y., Jiang L., Chen Y., Zhao Y., Gu W. Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H.J., Hu H.M., Li G.Z., Zhang Y., Wu F., Liu X., Wang K.Y., Zhang C.B., Jiang T. Ferroptosis-Related Gene Signature Predicts Glioma Cell Death and Glioma Patient Progression. Front. Cell Dev. Biol. 2020;8:538. doi: 10.3389/fcell.2020.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J.Y., Wang D.S., Lin H.C., Chen X.X., Yang H., Zheng Y., Li Y.H. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int. J. Biol. Sci. 2020;16:2430–2441. doi: 10.7150/ijbs.45050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou N., Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford) 2020;2020:baaa021. doi: 10.1093/database/baaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan T., Nguyen V.H., A’lincourt Salazar M., Wong P., Diamond D.J., Yim J.H., Melstrom L.G. Inhibition of Autophagy Amplifies Baicalein-Induced Apoptosis in Human Colorectal Cancer. Mol. Ther. Oncolytics. 2020;19:1–7. doi: 10.1016/j.omto.2020.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang B.C., Leung P.S. Irisin Is a Positive Regulator for Ferroptosis in Pancreatic Cancer. Mol. Ther. Oncolytics. 2020;18:457–466. doi: 10.1016/j.omto.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poursaitidis I., Wang X., Crighton T., Labuschagne C., Mason D., Cramer S.L., Triplett K., Roy R., Pardo O.E., Seckl M.J. Oncogene-Selective Sensitivity to Synchronous Cell Death following Modulation of the Amino Acid Nutrient Cystine. Cell Rep. 2017;18:2547–2556. doi: 10.1016/j.celrep.2017.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song X., Zhu S., Chen P., Hou W., Wen Q., Liu J., Xie Y., Liu J., Klionsky D.J., Kroemer G. AMPK-Mediated BECN1 Phosphorylation Promotes Ferroptosis by Directly Blocking System Xc- Activity. Curr. Biol. 2018;28:2388–2399.e5. doi: 10.1016/j.cub.2018.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu N., Clifford R.J., Yang H.H., Wang C., Goldstein A.M., Ding T., Taylor P.R., Lee M.P. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H., Hu N., Yang H.H., Wang C., Takikita M., Wang Q.H., Giffen C., Clifford R., Hewitt S.M., Shou J.Z. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin. Cancer Res. 2011;17:2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolau-Neto P., Da Costa N.M., de Souza Santos P.T., Gonzaga I.M., Ferreira M.A., Guaraldi S., Moreira M.A., Seuánez H.N., Brewer L., Bergmann A. Esophageal squamous cell carcinoma transcriptome reveals the effect of FOXM1 on patient outcome through novel PIK3R3 mediated activation of PI3K signaling pathway. Oncotarget. 2018;9:16634–16647. doi: 10.18632/oncotarget.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Chen Z., Tian L., Zhou C., He M.Y., Gao Y., Wang S., Zhou F., Shi S., Feng X. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrone C.R., Kattan M.W., Tomlinson J.S., Thayer S.P., Brennan M.F., Warshaw A.L. Validation of a postresection pancreatic adenocarcinoma nomogram for disease-specific survival. J. Clin. Oncol. 2005;23:7529–7535. doi: 10.1200/JCO.2005.01.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.