Abstract
A cardinal feature of COVID-19 is lung inflammation and respiratory failure. In a prospective multi-country cohort of COVID-19 patients, we found that increased Notch4 expression on circulating regulatory T (Treg) cells was associated with disease severity, predicted mortality, and declined upon recovery. Deletion of Notch4 in Treg cells or therapy with anti-Notch4 antibodies in conventional and humanized mice normalized the dysregulated innate immunity and rescued disease morbidity and mortality induced by a synthetic analog of viral RNA or by influenza H1N1 virus. Mechanistically, Notch4 suppressed the induction by interleukin-18 of amphiregulin, a cytokine necessary for tissue repair. Protection by Notch4 inhibition was recapitulated by therapy with Amphiregulin and, reciprocally, abrogated by its antagonism. Amphiregulin declined in COVID-19 subjects as a function of disease severity and Notch4 expression. Thus, Notch4 expression on Treg cells dynamically restrains amphiregulin-dependent tissue repair to promote severe lung inflammation, with therapeutic implications for COVID-19 and related infections.
Keywords: COVID-19, SARS-CoV-2, influenza, Notch4, regulatory T cells, IL-6, IL-18, amphiregulin
Graphical abstract
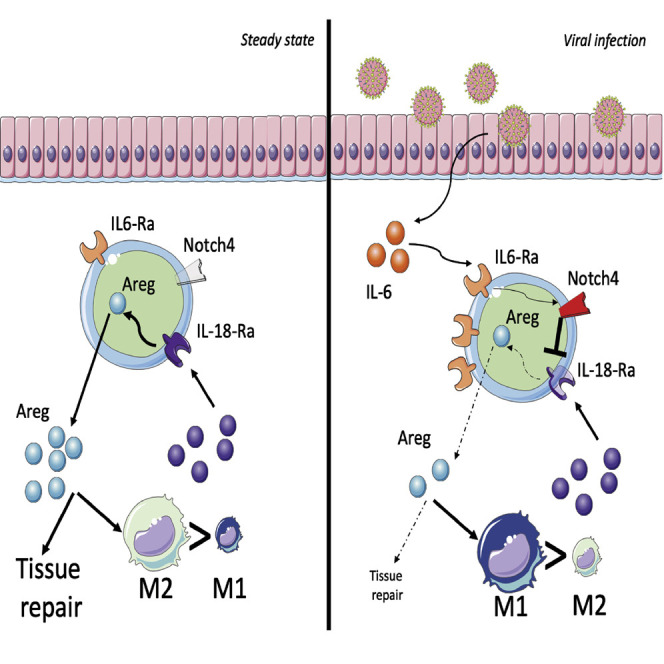
Harb, Benamar, et al. find that interleukin-6 increases Notch4 expression on lung regulatory T cells, which, in turn, restrains production of the tissue repair cytokine amphiregulin and promotes severe lung inflammation. Their findings have implications for treatment of COVID-19 and other respiratory viral infections.
Introduction
The current pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has resulted in massive morbidity and mortality in the United States and globally (Cucinotta and Vanelli, 2020). Although many infected subjects are asymptomatic or have a mild form of the disease, a subset suffers from more severe disease with pneumonia and marked hypoxia, leading to acute respiratory distress syndrome (Berlin et al., 2020; Richardson et al., 2020; Zhou et al., 2020a). In many of those individuals, the disease proves fatal despite intensive respiratory support. Uncontrolled activation of the immune response, leading to a cytokine storm, is a key risk factor for mortality (Henderson et al., 2020; Lucas et al., 2020; Vabret et al., 2020). The molecular mechanisms governing lung disease severity are not yet well understood. Notably, innate immune cell hyperactivation plays a critical role in the pathogenesis of severe coronavirus disease 2019 (COVID-19) (Del Valle et al., 2020; Vabret et al., 2020; Zhou et al., 2020b), a finding also recapitulated in emerging mouse models (Winkler et al., 2020). Among the innate immune cytokines, interleukin-6 (IL-6) has been proposed to play a pathogenic role because of its increased serum concentrations in subjects with severe COVID-19 and reports of favorable clinical responses to anti-IL-6 receptor (IL-6R) monoclonal antibody (mAb) therapy (Copaescu et al., 2020; The REMAP-CAP Investigators, 2021; Salama et al., 2021; Toniati et al., 2020; Xu et al., 2020). Other studies, however, have failed to confirm such a benefit (Hermine et al., 2021; Rosas et al., 2021; Salvarani et al., 2021; Veiga et al., 2021).
An IL-6-dependent pathway subverts lung regulatory T (Treg) cells to promote tissue inflammation in severe asthma by increasing Treg cell expression of the receptor Notch4 (Harb et al., 2020; Xia et al., 2015, 2018). In this setting, Notch4 acts via downstream pathways, including Hippo and Wnt, to disrupt Treg cell regulation of the T helper type 2 (Th2) and Th17 adaptive immune responses. In case studies, treatment of individuals with severe asthma with the anti-IL-6R mAb tocilizumab decreased Notch4 expression on circulating Treg cells and improved disease outcomes (Esty et al., 2019).
The NOTCH4 locus is associated with critical illness in COVID-19 (Pairo-Castineira et al., 2021). Given this and the evidence supporting a pathogenic role for high levels of IL-6 in COVID-19, we examined the effect of inducible Notch4 expression on Treg cells in lung viral infection, including COVID-19. We found increased Notch4 expression in subjects with COVID-19 as a function of disease severity. In mouse models of respiratory viral infections, Notch4 enabled virus-induced tissue inflammation by mechanisms distinct from those involved in its regulation of adaptive immunity in allergic airway inflammation. Rather, Notch4 expression inhibited Treg-cell-mediated regulation of innate immune responses and promotion of tissue repair. The protective function of Notch4 inhibition involved increased production by Treg cells of the epidermal growth factor-like cytokine amphiregulin, which plays a critical role in in mediating tissue repair by Treg cells in lung viral infections (Arpaia et al., 2015). These findings identify Notch4 as an effector of COVID-19 disease severity and point to interventions along the Notch4-amphiregulin axis as a viable therapeutic strategy to restore immune regulation in severe viral respiratory infections, including COVID-19.
Results
Treg cell Notch4 expression is independently predictive of mortality from COVID-19
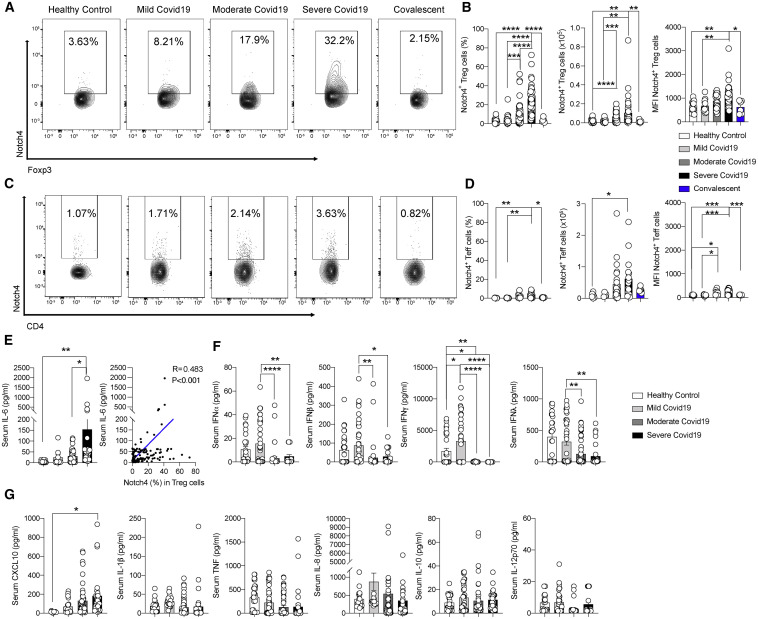
To determine the association between Notch4 and disease severity in individuals with COVID-19, we recruited three cohorts of people with COVID-19 as well as healthy control individuals from Boston, Massachusetts (n = 38 and n = 10, respectively); Genoa, Italy (n = 44 and n = 10); and Istanbul, Turkey (n = 36 and n = 20) (Table S1). The subjects were segregated into three disease severity groups (mild, moderate, and severe) based on the need for hospitalization and advanced respiratory support as well as a convalescent group, following criteria detailed in the STAR Methods. Older male individuals with a history of malignancy, cardiac disease, or endocrine disease had more severe disease (Table S1). Frequencies, absolute numbers, and mean fluorescence intensity (MFI) of Notch4 expression on peripheral blood CD4+CD25+Foxp3+ Treg cells of COVID-19 subjects in the different cohorts increased progressively as a function of disease severity and declined precipitously in the convalescent subjects (Figures 1A and 1B; Table S1). In contrast, there was a modest increase in Notch4 expression on CD4+CD25–Foxp3– T effector (Teff) cells of subjects with moderate and severe disease and otherwise minimal expression in the other subject groups (Figures 1C and 1D). Also, there was no difference in expression of the other Notch receptors, including Notch1, Notch2, and Notch3, on circulating Treg cells among subjects in the different study groups (Figure S1). In multivariate logistic regression adjusting for age, gender, comorbidities, glucocorticoid treatment, and serum IL-6 levels, each 1% increase in Notch4 expression in Treg cells was independently associated with 1.046 higher odds of mortality (univariate odds ratio 1.052 [95% confidence interval (CI) 1.020–1.088], p = 0.002; multivariate odds ratio 1.046 [95% CI 1.008–1.090], p = 0.023) (Table S1). For example, an increase in Treg cells expressing Notch4 by 10% in a 65-year-old male individual with a history of malignancy treated with steroids would lead to a rise in predicted mortality from 28.4% to 38.3%. Overall, serum IL-6 concentrations were correlated positively with Treg cell Notch4 expression (Figure 1E). Moreover, the serum concentrations of interferon alpha (IFNα), IFNβ, and IFNγ were decreased in individuals with moderate and severe disease compared with those with mild disease (Figure 1F; Blanco-Melo et al., 2020; Hadjadj et al., 2020). In contrast, the serum concentrations of other cytokines, including IL-1β, tumor necrosis factor (TNF), IL-8, IL-10, and IL-12p70, were unchanged, whereas those of CXCL10 were slightly increased in subjects with severe disease (Figure 1G).
Figure 1.
Increased expression of Notch4 on circulating Treg cells of subjects with COVID-19
(A–D) Flow cytometry analysis, cell frequencies, absolute numbers, and MFI of Notch4 expression in Treg cells (A and B) and Teff cells (C and D) of control subjects and subjects with mild, moderate, severe, or resolved COVID-19 (healthy control subjects, n = 37; mild disease, n = 20; moderate disease, n = 54; severe disease, n = 36; convalescent subjects, n = 6).
(E) Serum concentrations of IL-6 in the different subject groups (healthy control subjects, n = 37; mild disease, n = 18; moderate disease, n = 45; severe disease, n = 21).
(F) Correlation analysis of Notch4 expression on Treg cells of affected and control subjects as a function of serum IL-6 concentrations (n = 121).
(G) Serum concentrations of IFNα, IFNβ, IFNγ IFNλ, CXCL10, IL-1β, IL-8, IL-10, IL-12, and TNF in control and affected subjects (healthy control subjects, n = 37; mild disease, n = 18; moderate disease, n = 45; severe disease, n = 21).
Each symbol represents one subject. Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by one-way ANOVA with Dunnett’s post hoc analysis (A–E and G) and Pearson correlation analysis (D). Data represent a pool of two or three independent experiments.
Treg cell Notch4 promotes lung tissue inflammation induced by polyinosinic:polycytidylic acid (poly(I:C))
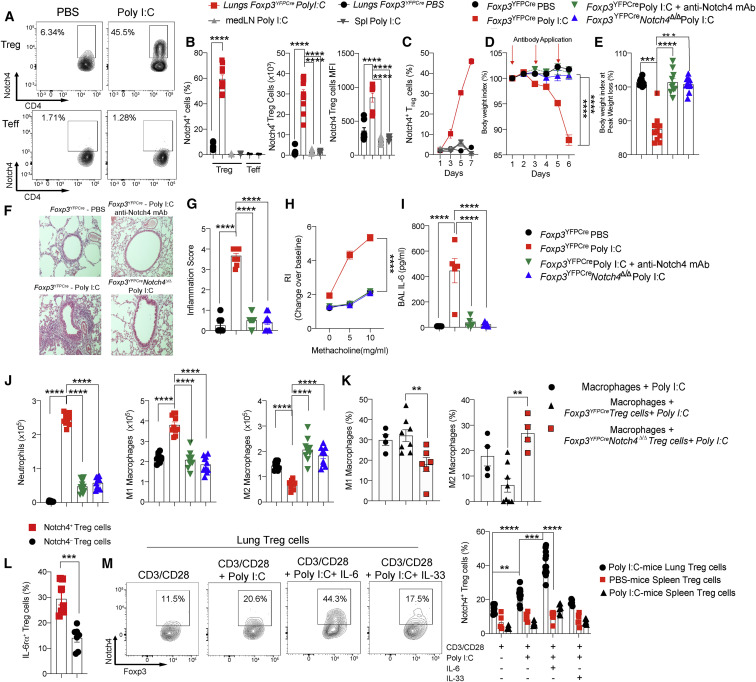
To investigate the mechanism by which Notch4 licenses lung tissue inflammation in viral respiratory infections, we first examined its role in poly(I:C)-induced airway inflammation. Poly(I:C) stimulates Toll-like receptor 3 and the downstream viral RNA sensors cytoplasmic retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), providing a proxy model of RNA viral infection (Broggi et al., 2020; Iwasaki and Pillai, 2014; Kato et al., 2006). Daily intratracheal instillation of poly(I:C) in mice for 6 consecutive days resulted in sharp progressive increases in the frequencies and absolute numbers of Notch4-expressing lung tissue Treg cells as well as in the MFI of Notch4 on those cells (Figures 2 A–2C). In contrast, minimal expression of Notch4 was noted on CD4+ Teff cells. Treg cell-specific deletion of Notch4 using a Foxp3-driven Cre recombinase (Foxp3 YFPCre) and floxed Notch4 allele (Foxp3 YFPCre Notch4 Δ/Δ) or treatment with an anti-Notch4 neutralizing mAb, but not an isotype control mAb, protected mice from weight loss induced by poly(I:C) treatment (Figures 2D and 2E). Histopathological analysis revealed intense inflammation in the lungs of poly(I:C)-treated mice that was suppressed by Notch4 antagonism (Figures 2F and 2G). Similarly, airway hyperresponsiveness (AHR), which was increased in poly(I:C)-treated mice, was also normalized (Figure 2H). Analysis of bronchoalveolar lavage (BAL) fluid revealed increased IL-6 concentrations in poly(I:C)-treated mice that were suppressed upon Notch4 inhibition, consistent with decreased inflammation (Figure 2I). A similar increase was also noted for a number of other cytokines, including IFNγ, IL-1α, IL-1β, TNF-α, IFNβ, MCP1, GM-CSF, IL-17, oncostatin M, leukemia inhibitory factor, and CXCL5, most of which were also suppressed by Notch4 antagonism (Figure S3A). Further analysis revealed that Notch4 inhibition suppressed lung tissue neutrophil infiltration induced by poly(I:C) treatment and reversed the increase in lung M1 macrophages while increasing M2 macrophages (Figure 2J). In vitro co-cultures of poly(I:C)-treated macrophages with lung Treg cells derived from poly(I:C)-treated mice revealed that Foxp3 YFPCre Notch4 Δ/Δ but not Foxp3 YFPCre Treg cells reversed the M1 skewing and promoted M2 macrophage differentiation (Figure 2K). These results confirmed a critical role of Treg cell Notch4 expression in the evolution of lung inflammation induced by poly(I:C).
Figure 2.
Protective effect of Notch4 inhibition in poly(I:C)-induced lung injury
(A and B) Flow cytometry analysis (A) and cell frequencies, absolute numbers, and MFI (B) of Notch4 expression in lung, mediastinal lymph node (medLN), and spleen Treg and Teff cells of Foxp3YFPCre mice treated with PBS or poly(I:C) once daily for 6 days.
(C) Notch4 expression on lung, medLN, and spleen Treg cells of Foxp3YFPCre mice.
(D and E) Weight index (D) and peak weight loss (E) of Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice treated with PBS or poly(I:C) together with an isotype control or anti-Notch4 mAb.
(F) Hematoxylin and eosin-stained sections of lung tissues (200× magnification).
(G) Inflammation scores.
(H) AHR in response to methacholine. RI, responsiveness index (a measure of airway resistance).
(I and J) BAL fluid IL-6 concentrations (I) and graphical representation of lung tissue neutrophils and M1 and M2 macrophages.
(K) M1 and M2 macrophage frequencies in cultures of poly(I:C)-treated lung macrophages incubated with Treg cells from the indicated poly(I:C)-treated mice.
(L) Flow cytometry analysis of IL-6Rα expression in lung Notch4+ or Nottch4– Treg cells of Foxp3YFPCre mice treated with poly(I:C).
(M) In vitro induction of Notch4 expression in Treg cells from the lungs or spleens of poly(I:C) or PBS-treated Foxp3YFPCre mice. Each symbol represents one mouse (n = 5–15 per group).
Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: Student’s t test (B), one-way ANOVA with Dunnett’s post hoc analysis (E, G, and I–K), and two-way ANOVA with Sidak’s post hoc analysis (C, D, H, and L). ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Data were pooled from two or three independent experiments.
Different Notch receptors control Treg cell function (Charbonnier et al., 2015; Ostroukhova et al., 2006; Perumalsamy et al., 2012). However, the role of Notch4 in licensing lung tissue inflammation by poly(I:C) was highly specific in that Treg cell-specific deletion of the other Notch genes, including Notch1, Notch2, and Notch3, failed to protect against tissue inflammation. In contrast, Treg cell-specific deletion of Rbpj, encoding the Notch canonical pathway transcription factor RBPJ, largely reproduced the protective effect of Notch4 deletion (Figures S2A–S2F). Downstream of Notch4, our previous studies have implicated the Hippo and Wnt pathways in Treg cell regulation of Th17 and Th2 responses in allergic airway inflammation, respectively (Harb et al., 2020). Poly(I:C) treatment potently induced expression of the Hippo pathway effector Yap in lung tissue Treg cells, whereas expression of the Wnt pathway effector β-catenin was unchanged (Figures S2G and S2H). Treg cell-specific deletion of Yap1 and Wwtr1, encoding the Hippo pathway effectors Yap and Taz; Ctnnb1, encoding the Wnt pathway effector β-catenin; or all three genes failed to protect against poly(I:C)-induced inflammation. These findings indicate that Notch4 regulation of the acute inflammatory responses to viral infection proceeds by distinct mechanisms (Figures S2I–S2N).
We have previously identified a key role of IL-6 in upregulating Notch4 expression on Treg cells in asthmatic inflammation (Harb et al., 2020). Consistent with these results, Notch4+ lung Treg cells from poly(I:C)-treated mice expressed higher levels of IL-6 receptor alpha chain (IL-6Rα) compared with their Notch4– counterparts (Figure 2L). To further elucidate the mechanisms of Notch4 induction in Treg cells, we conducted in vitro studies to examine the capacity of poly(I:C) and cytokine treatment to upregulate Notch4 expression in Treg cells isolated from PBS or poly(I:C)-treated mice. The results showed that poly(I:C) treatment of lung but not splenic Treg cells increased Notch4 expression in synergy with IL-6 but not IL-33 (Figure 2M). In vivo, Treg cell-specific deletion of Il6ra attenuated poly(I:C)-induced Notch4 expression on lung Treg cells and the attendant tissue inflammatory response, including the decline in body weight, increase in AHR, influx of neutrophils, and alteration in macrophage populations (Figures S3B–S3G). These results indicate that IL-6 and additional signals contribute to Notch4 upregulation on lung Treg cells in poly(I:C)-treated mice.
Notch4 promotes lung inflammation induced by H1N1 influenza A virus
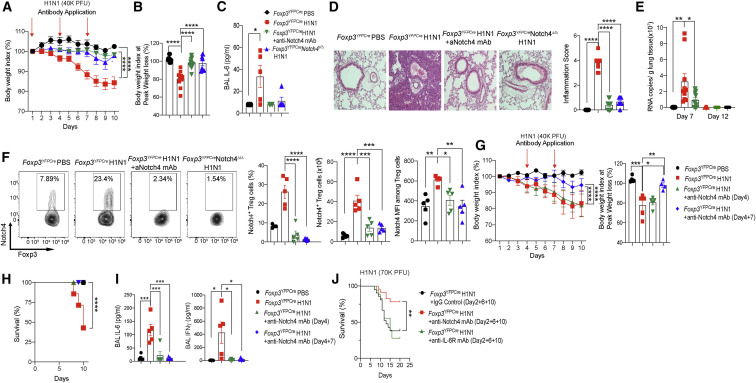
To extend these findings to an RNA virus model, we analyzed the role of Notch4 expression on Treg cells in licensing lung tissue inflammation in a mouse model of H1N1 influenza A virus infection (Woodham et al., 2020). In studies where mice were infected with H1N1 virus at 4 × 104 plaque-forming units (PFUs)/mouse, the weight loss induced by the infection was abrogated by prior treatment with a neutralizing anti-Notch4 mAb or upon Treg cell-specific deletion of Notch4 (Figures 3A and 3B). Notch4 antagonism suppressed the virus-induced increase in BAL fluid IL-6 concentration (Figure 3C). It also suppressed induction by viral infection of a number of other inflammatory cytokines, similar to what was noted for with poly(I:C) treatment (Figure S4A). It also reversed the lung tissue inflammatory response in infected mice (Figure 3D). Antagonism of Notch4 expression did not result in increased viral loads, as measured on days 7 and 12 after infection (Figure 3E). Analysis of Notch4 expression on lung Treg cells in the respective mouse groups revealed that H1N1 infection increased Notch4 expression, an effect abrogated by treatment with an anti-Notch4 mAb or by Treg cell-specific Notch4 deletion (Figure 3F). Both interventions greatly attenuated lung tissue neutrophil influx, restored the alveolar macrophage population, and reversed skewing of tissue macrophages away from the pro-inflammatory M1 phenotype toward an anti-inflammatory M2 phenotype (Figures S4B–S4D). Notch4 inhibition did not affect the frequencies of IFNγ+CD4+ or IFNγ+CD8+ T cells or IFNγ+ macrophages infiltrating the lungs of infected mice, whereas those of IFNγ+ innate lymphoid cells 1 (ILC1s) were decreased. Nevertheless, the absolute numbers of all four populations were decreased sharply, consistent with the markedly attenuated inflammatory response (Figures S5A–S5F). Notch4 inhibition also decreased the frequencies and absolute numbers of lung tissue IL-17+ CD4 and CD8 T cells (Figures S5G and S5H).
Figure 3.
Protective effect of Notch4 inhibition in influenza A H1N1 virus infection
(A and B) Weight index (A) and peak weight loss (B) of Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice that were sham treated or infected with a 40,000-PFU dose of H1N1 virus alone or together with an anti-Notch4 mAb, as indicated. Arrows indicate the time of antibody treatment.
(C) BAL fluid IL-6 concentrations in the indicated groups.
(D) Hematoxylin and eosin-stained sections and inflammation score of lung tissue isolated from the indicated mouse groups (200× magnification).
(E) Viral load (viral copies per gram of tissue) in the respective mouse groups (n = 5 per group).
(F) Flow cytometry analysis and graphical representation of Notch4 expression in lung tissue Treg cells of the respective Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mouse groups.
(G and H) Weight index and peak weight loss (G) and survival (H) of Foxp3YFPCre mice that were sham treated or infected with a 40,000-PFU dose of H1N1 virus alone or together with an anti-Notch4 mAb, as indicated. Arrows indicate the time of antibody treatment.
(I) BAL fluid IL-6 and IFNγ concentrations.
(J) Survival curve of Foxp3YFPCre mice that were sham treated or infected with a lethal dose of H1N1 virus alone or together with an anti-Notch4 or anti-IL-6Rα mAb, as indicated.
Each symbol represents one mouse (n = 5–23). Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: one-way ANOVA with Dunnett’s post hoc analysis (B, C, F, I, and J) and two-way ANOVA with Sidak’s post hoc analysis (A, D, E, G, and K). ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Data were pooled from two or three independent experiments.
To determine the capacity of Notch4 antagonism to rescue active disease, mice were infected with the same dose of H1N1 virus and were treated with an isotype control mAb or with an anti-Notch4 mAb following onset of infection. The results revealed that treatment of mice with the anti-Notch4 mAb on day 4 or on days 4 and 7 after infection rescued mice from death, whereas only day 4 and 7 treatment suppressed the weight loss and innate immune activation associated with infection (Figures 3G and 3H). Both anti-Notch4 mAb treatment regimens suppressed the increase in BAL fluid IL-6 and IFNγ concentrations induced by the viral infection (Figure 3I). We further compared different anti-Notch4 mAb treatment regimens for their capacity to rescue mice from a lethal dose 75% (LD75) of H1N1 virus (at 7 × 104 PFU/mouse). Treatment with the anti-Notch4 mAb on days 2, 6, and 10 was associated with decreased mortality (22%). In contrast, treatment with an isotype control mAb or with an anti-IL-6Rα mAb failed to protect the mice from exacerbated mortality (62% and 72% mortality, respectively) (Figure 3J). These results indicate that therapy with an anti-Notch4 mAb protects mice from active influenza virus infection.
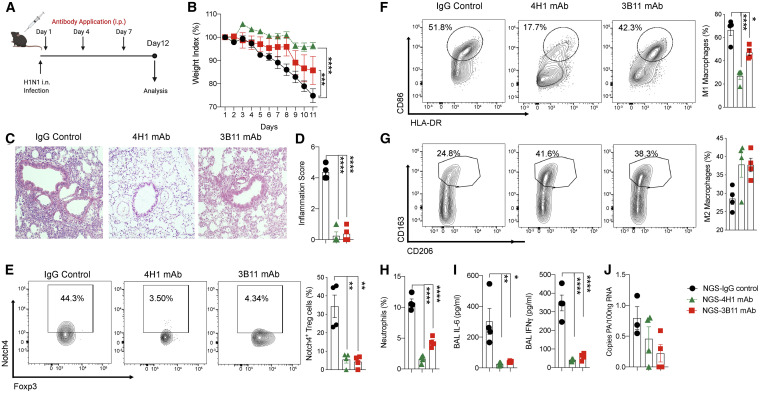
To extend the above results to the setting of a humanized immune system mouse model, we analyzed the capacity of neutralizing anti-human Notch4 mAbs to rescue disease in non-obese diabetic (NOD)-Prkdc scid Il2rg tmiwjl/Sz (NSG) mice reconstituted with human peripheral blood mononuclear cells (PBMCs) and infected with the H1N1 virus (Verma et al., 2017). Neutralizing mAbs specific for Notch4, but not other Notch proteins, were derived by immunizing mice with a recombinant N-terminal human Notch4 protein fragment (amino acids 1–637) (Figures S6A and S6B). Treatment of mice with one neutralizing anti-human Notch4 mAb (clone 4H1), but not an isotype control mAb, protected against H1N1 virus infection-induced weight loss and tissue inflammation (Figures 4 A–4D). Another neutralizing antibody (clone 3B11) was partially protective. Both antibodies inhibited Notch4 expression on lung Treg cells (Figure 4E) and inhibited macrophage skewing, neutrophil infiltration, and production of the cytokines IL-6 and IFNγ (Figures 4F–4I). The antibody treatment did not affect virus clearance, as measured by virus copy numbers in lung tissue (Figure 4J). These results confirm that Notch4 regulates the magnitude of the human inflammatory response to H1N1 infection and that it can be targeted successfully for therapeutic intervention.
Figure 4.
Notch4 licenses viral lung inflammation in humanized mice
(A) Schematic of humanized mouse infection with H1N1 influenza virus and treatment with neutralizing anti-human Notch4 mAbs.
(B) Weight index of mice that were infected with a sublethal dose of H1N1 virus and treated with an isotype control mAb or anti-Notch4 mAb 3B11 or 4H1, as indicated.
(C and D) Hematoxylin and eosin-stained sections and inflammation score of lung tissue isolated from the indicated mouse groups (200× magnification).
(E) Flow cytometry analysis and frequencies of Notch4 expression in lung Treg cells of the respective groups.
(F and G) Flow cytometry analysis and graphical representation of lung tissue M1 (F) and M2 macrophages (G).
(H) Neutrophil infiltration in the lungs of the respective mouse groups.
(I) BAL fluid IL-6 and IFNγ concentrations.
(J) Viral load in the respective mouse groups, measured as viral copies per 100 ng lung tissue RNA.
Each symbol represents one mouse (n = 4–5 per group). Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: two-way ANOVA with Sidak’s post hoc analysis (B) and one-way ANOVA with Dunnett’s post hoc analysis (D–J). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
Protection by Notch4 antagonism is amphiregulin dependent
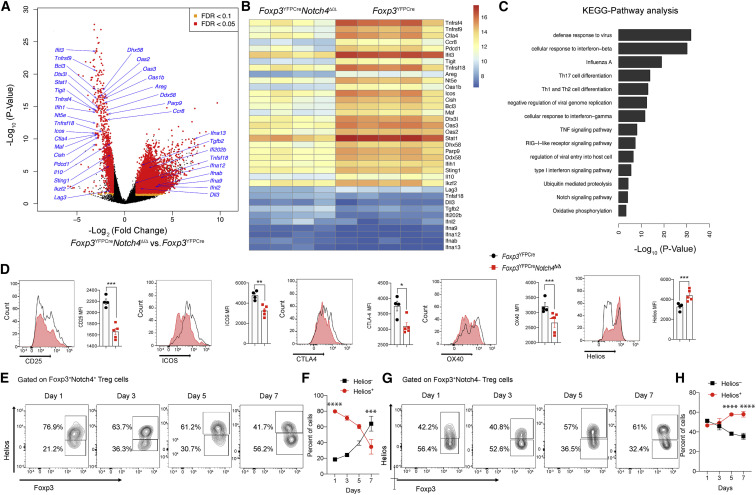
To better understand the mechanisms by which Notch4 deficiency protected against lung inflammation, we analyzed the transcriptomes of lung Treg cells of Foxp3 YFPCre and Foxp3 YFPCre Notch4 Δ/Δ mice treated with poly(I:C) for 6 days. The results revealed that lung Treg cells of poly(I:C)treated Foxp3 YFPCre mice exhibited an activated effector Treg cell signature, with upregulation of many canonical Treg cell transcripts, including Ctla4, Ikzf2, Il10, and Tigit and the TNF receptor superfamily members Tnfrsf4, Tnfrsf9, and Tnfrsf18 (Arpaia et al., 2015; Figures 5A and 5B). Poly(I:C) treatment also imparted an exhausted T cell-like signature with increased Pdcd1, Icos, Lag3, and other related transcripts that may impair their suppressive function (Lowther et al., 2016). In contrast, Treg cells of poly(I:C)-treated Foxp3 YFPCre Notch4 Δ/Δ mice increased several type I IFN genes. More broadly, key pathways enriched in Foxp3 YFPCre versus Foxp3 YFPCre Notch4 Δ/Δ lung Treg cells included those involved in innate virus sensing and response, type I IFN signaling, TH cell differentiation, and ubiquitin-mediated proteolysis (Figure 5C). Although the RNA and protein expression of several key canonical markers was increased concordantly in Foxp3 YFPCre lung Treg cells, exceptions included the transcription factor Helios, encoded by Ikzf2, whose expression was decreased despite increased transcript levels (Figure 5D). Further analysis localized the loss of Helios expression to the Notch4+ fraction of Foxp3 YFPCre lung Treg cells, suggesting an expansion of Helioslow Notch4+-induced Treg cells, similar to our observation in the asthma model (Harb et al., 2020), or loss of Helios expression leading to an increased potential for destabilization (Thornton et al., 2019; Figure 5E). The latter results suggest that Notch4 initiated a program of post-translational regulation by pathways such as ubiquitination and proteolysis, both of which were increased in Treg cells of poly(I:C)-treated Foxp3 YFPCre mice.
Figure 5.
Notch4 deficiency reprograms the lung Treg cell transcriptome of poly(I:C)-treated mice
(A–C) Volcano plot (A), heatmap (B), and pathway analysis (C) of gene transcripts of lung Treg cells isolated from Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice treated with poly(I:C) (n = 4 and n = 5, respectively).
(D) Flow cytometry histograms and graphical representation of lung tissue Treg cell expression of CD25, Helios, CTLA4, ICOS, and OX40 in Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice sampled on day 7 after poly(I:C) treatment (n = 5 for each time point).
(E–H) Flow cytometry analysis (E and G) and graphical representation (F and H) of Helios expression in Foxp3+Notch4+ and Foxp3+Notch4– lung tissue Treg cells in Foxp3YFPCre mice sampled at the indicated dates after poly(I:C) treatment (n = 5 for each time point).
Each symbol represents one mouse. Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: pairwise comparisons of differential gene expression were computed using DESeq2 (A–C); Student’s unpaired two-tailed t test (D) and two-way ANOVA with Sidak’s post hoc analysis (F and H). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
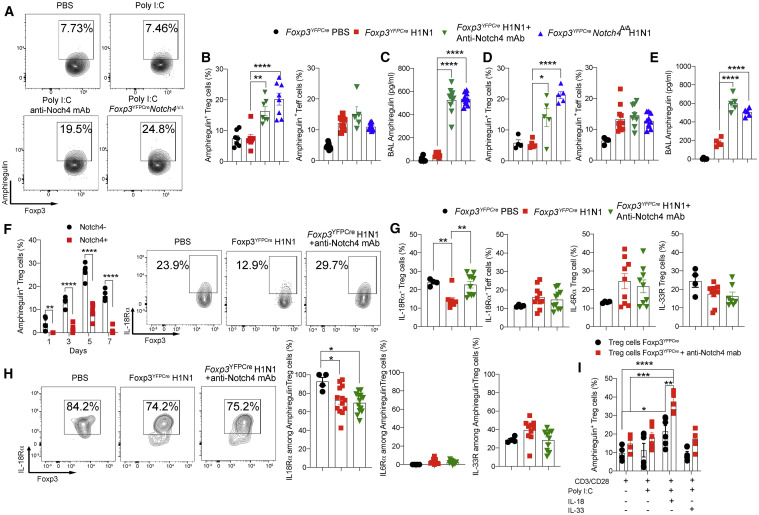
The function of Treg cells in lung tissue repair in H1N1 virus infection involves production of the epidermal growth factor-like cytokine amphiregulin (Arpaia et al., 2015). Treg cells are the main source of amphiregulin in the lungs, whereas Treg cell-specific deletion of Areg, the gene encoding amphiregulin, worsens disease outcome (Arpaia et al., 2015). Analysis of lung Treg cells revealed that, despite markedly increased Areg transcripts in Foxp3 YFPCre compared with Foxp3 YFPCre Notch4 Δ/Δ Treg cells following poly(I:C) treatment (Figure 6 A), amphiregulin protein expression failed to increase in Treg cells and BAL fluid of Foxp3 YFPCre mice. However, it was increased markedly in lung Treg cells and BAL fluid of Foxp3 YFPCre Notch4 Δ/Δ mice and in Foxp3 YFPCre mice treated with an anti-Notch4 mAb (Figures 6A–6C). In contrast, amphiregulin expression in lung tissue Teff cells was unaffected by Notch4 antagonism (Figure 6B). Similar results were also found in the H1N1 infection model (Figures 6D and 6E). Consistent with these results, amphiregulin expression was found to be differentially enriched in Notch4– versus Notch4+ lung Treg cell in poly(I:C)-treated mice (Figure 6F). Similar results were also found in lung Treg cells of influenza H1N1 virus-infected mice (data not shown).
Figure 6.
Notch4 inhibition promotes an amphiregulin-dependent immunoregulatory program
(A) Flow cytometry analysis of amphiregulin expression in Treg cells from the lungs of Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice sham treated or treated with poly(I:C) alone or together with an anti-Notch4 mAb, as indicated.
(B) Graphical representation of amphiregulin expression in lung tissue Treg and Teff cells.
(C) BAL fluid amphiregulin concentrations.
(D) Amphiregulin expression in lung tissue Treg and Teff cells of Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice sham treated or infected with H1N1 influenza virus alone or together with an anti-Notch4 mAb, as indicated.
(E) BAL fluid amphiregulin concentrations.
(F) Flow cytometric analysis and graphical representation of amphiregulin expression in Notch4+ or Notch4– Treg cells from the lungs of Foxp3YFPCre mice treated with PBS or poly(I:C) at the indicated time points.
(G and H) Flow cytometry analysis and graphical representation of IL18Rα, IL6Rα, and ST2 expression in total (G) or amphiregulin+ (H) Treg cells from the lungs of PBS- or poly(I:C)-treated Foxp3YFPCre mice with an isotype control or an anti-Notch4 mAb, as indicated.
(I) In vitro induction of amphiregulin expression in Treg cell from the lungs or spleens of poly(I:C)- or PBS-treated Foxp3YFPCre mice.
Each symbol represents one mouse (n = 5–8 per group). Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: one-way ANOVA with Dunnett’s post hoc analysis (B–D, F, and G) and two-way ANOVA with Sidak’s post hoc analysis (E and H). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Induction of amphiregulin expression in lung Treg cells is mediated primarily by the action of IL-18 with an ancillary effect by IL-33 (Arpaia et al., 2015). We examined the expression of IL-18Rα, IL-6Rα, and IL-33R on lung Treg cells in sham-infected versus H1N1-infected mice, the latter without or with anti-Notch4 mAb therapy. The results showed that expression of IL-18Rα on Treg cells declined upon H1N1 infection but recovered upon anti-Notch4 mAb treatment. In contrast, IL-18Rα expression on lung tissue Teff cells was unaffected by anti-Notch4 mAb treatment (Figure 6G). Expression of IL-33R on lung Treg cells was unchanged by anti-Notch4 mAb treatment, whereas that of IL-6Rα was increased by poly(I:C) treatment (Figure 6G). Further analysis revealed that most of the amphiregulin-expressing Treg cells strongly expressed IL-18Rα and, to a lesser extent, IL-33R but virtually no IL-6Rα (Figure 6H). The role of Notch4 in modulating IL-18-induced amphiregulin was analyzed in in vitro experiments employing lung Treg cells isolated from poly(I:C)-treated mice. Although amphiregulin expression in these Treg cells was induced minimally by IL-18 treatment, it was increased upon co-treatment with the anti-Notch4 mAb (Figure 6I). These results suggest that Notch4 inhibits amphiregulin expression in Treg cells at least in part by antagonizing its upregulation by IL-18.
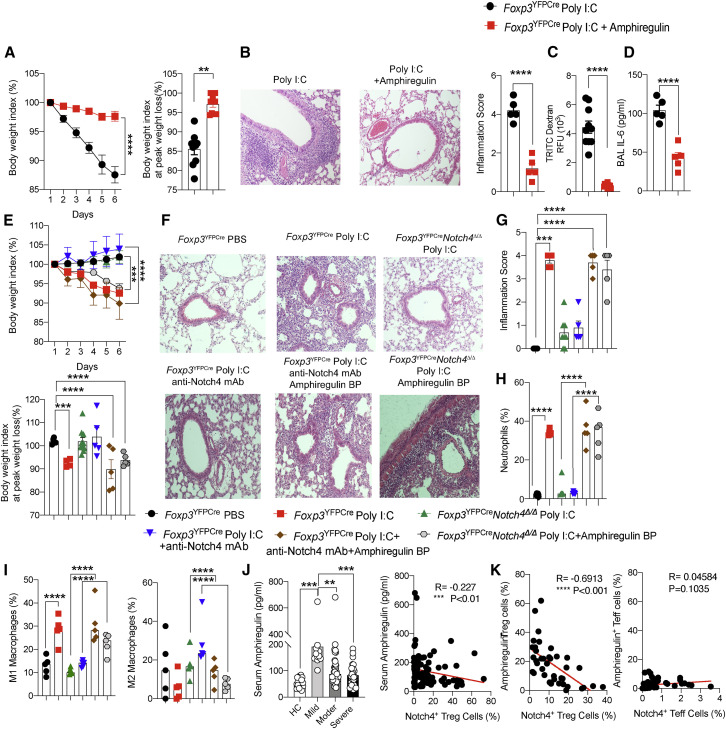
To establish the role of amphiregulin in protection against lung inflammation induced by Notch4 antagonism, and in light of previous studies demonstrating a protective effect of amphiregulin treatment in respiratory infections (Jamieson et al., 2013), we examined the capacity of recombinant amphiregulin therapy to rescue poly(I:C)-induced lung inflammation. Treatment of Foxp3 YFPCre mice with recombinant amphiregulin largely abrogated the weight loss and the inflammatory responses induced by poly(I:C) treatment in association with restoration of intact epithelial barrier function (Figures 7 A–7D). Recombinant amphiregulin also reversed skewing of lung macrophages toward an M1 phenotype upon their in vitro treatment with poly(I:C) (Figure S6H), mirroring the same effect of Foxp3 YFPCre Notch4 Δ/Δ Treg cells (Figure 2J).
Figure 7.
Lung protection by Notch4 antagonism is amphiregulin dependent
(A) Weight index and peak weight loss of Foxp3YFPCre mice sham treated or treated with poly(I:C) alone or together with recombinant amphiregulin.
(B) Hematoxylin and eosin-stained sections and inflammation score of lung tissue isolated from the indicated mouse groups (200× magnification.
(C) Serum TRITC dextran in poly(I:C) + recombinant amphiregulin-treated Foxp3YFPCre mice 1 h after intratracheal instillation.
(D) BAL fluid concentration of IL-6.
(E) Weight index and peak weight loss of Foxp3YFPCre and Foxp3YFPCreNotch4Δ/Δ mice sham treated or treated with poly(I:C), anti-Notch4 mAb, and/or amphiregulin-blocking peptide (bp), as indicated.
(F) Hematoxylin and eosin-stained lung sections (200× magnification).
(G) Inflammation scores (n = 5 per group).
(H and I) Lung neutrophils (H) and M1 and M2 macrophages (I).
(J) Left panel: serum amphiregulin concentrations in healthy control subjects and COVID-19 subject groups (control subjects, n = 17; mild disease, n = 20; moderate disease, n = 49; severe disease, n = 32). Right panel: Pearson correlation of Treg cell Notch4 expression and serum amphiregulin concentration in affected and control subjects (n = 99).
(K) Pearson correlation of Notch4 and amphiregulin expression in Treg cells (right panel) and Teff cells (left panel) of individuals with COVID-19 (n = 39).
Each symbol represents one mouse (n = 5–10 per group). Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: one-way ANOVA with Dunnett’s post hoc analysis (B–E and G–J) and two-way ANOVA with Sidak’s post hoc analysis (A and E). ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Reciprocally, we analyzed the effect of co-treatment with an amphiregulin peptide spanning the amino-terminal heparin binding domain, which is necessary for amphiregulin activation of the epidermal growth factor receptor (EGFR) (Johnson and Wong, 1994), on the protective effects of Notch4 antagonism. The peptide was validated to competitively inhibit amphiregulin activation of EGFR, as measured by the latter’s tyrosine autophosphorylation (Figure S6C). The results showed that the amphiregulin-blocking peptide abrogated the protective effects of anti-Notch4 mAb treatment or Treg cell-specific Notch4 deletion in poly(I:C)-induced lung injury, resulting in exacerbated weight loss, tissue inflammation, neutrophil infiltration, and M1 macrophage skewing (Figures 7E–7I). These results were reproduced in experiments utilizing a similarly validated neutralizing anti-amphiregulin mAb (Figure S6D), which also abrogated the protection imparted by Treg cell-specific Notch4 deficiency against poly(I:C)-induced weight loss, lung tissue neutrophil infiltration, and increased M1 versus M2 macrophage skewing (Figure S6E). The anti-amphiregulin mAb co-treatment also reversed restoration by Notch4 deficiency of airway epithelial barrier integrity, as measured by increased leakage of high-molecular-weight dextran instilled in the airways into the blood stream (Figure S6F).
Consistent with the above results, analysis of the sera of subjects with COVID-19 revealed increased amphiregulin concentrations in subjects with mild disease compared with those of healthy controls, which declined in the sera of individuals with moderate and severe disease in a manner inversely proportional to Notch4 expression (Figure 7J). This inverse correlation between amphiregulin and Notch4 was particularly pronounced when comparing Notch4 and amphiregulin expression specifically in circulating Treg cells (Figure 7K). In contrast, there was no correlation between Notch4 and amphiregulin expression on circulating Teff cells (Figure 7K). Overall, these results are consistent with suppression of amphiregulin production by Treg cells as a key mechanism of Notch4 inhibitory function in viral infections.
Discussion
In this study, we addressed the role of a tissue-specific immunoregulatory mechanism involving inducible Notch4 expression on Treg cells in mediating severe lung tissue inflammation in COVID-19 and related viral infections. We found that Notch4 was increased on circulating Treg cells of individuals with COVID-19 as a function of disease severity and associated with heightened mortality, indicating that this mechanism is operative in SARS-CoV-2 infection and may be critically involved in the pathogenesis of acute respiratory failure in this disorder. Furthermore, by employing conventional and humanized immune system mouse models of respiratory viral infections, we found that the evolution of severe viral lung inflammation was contingent on induction of Notch4 on lung Treg cells, which acted as a molecular switch to alter their regulatory and tissue repair activities. Thus, Notch4 emerges as a critical regulator of disease severity in different respiratory viral infections.
Notch4 was increased on tissue Treg cells very early in lung inflammation and acted to promote the innate immune response at a time when adaptive immunity had not yet been mobilized effectively. In particular, the downstream mechanisms by which Notch4 promoted innate immune cell activation in viral infection were different from those involved in promoting the TH2 and TH17 cell-adaptive immune responses in asthma: the Wnt and Hippo pathways, respectively (Geng et al., 2017; Harb et al., 2020). Thus, Treg cell-specific deletion of Yap1 and Wwtr1, encoding the Hippo pathway mediators Yap and Taz, or Ctnnb1, encoding the Wnt pathway effector protein β-catenin, showed little effect on disease induced by poly(I:C) treatment. This bifurcation in the mechanisms by which Notch4 promotes innate versus adaptive immunity points to distinct functions served by this pathway in the respective immune responses. In the context of an acute virus-mediated injury, Notch4 allows evolution of an innate immune response instigated by lung epithelium-derived danger signals that is unencumbered by tonic Treg cell suppression (Whitsett and Alenghat, 2015), consistent with the previously noted role of Treg cells in coordinating early protective immunity to viral infection (Lund et al., 2008). This physiological function of Notch4 becomes detrimental in the context of severe respiratory viral infections, resulting in uncontrolled pathological activation of innate immunity, leading to damaging lung tissue injury.
Analysis of Notch4 expression on Treg cells in the context of respiratory viral infection revealed its restriction to lung tissue Treg cells, suggesting a lung tissue origin of circulating Notch4+ Treg cells in individuals with COVID-19. Induction of Notch4 on lung Treg cells was in part IL-6 dependent, as evidenced by in vitro studies showing that co-treatment of lung but not splenic Treg cells with IL-6 and poly(I:C) increased Notch4 expression. These results are consistent with partial suppression of poly(I:C)-induced Notch4 expression and tissue inflammation upon Treg cell-specific deletion of Il6ra. They are also consistent with the positive correlation of increased Notch4 expression in individuals with COVID-19 with serum IL-6 concentrations. Notwithstanding these findings, Notch4 but not IL-6 predicted mortality in the affected population. Direct comparison of genetic and functional interventions that target Notch4 versus IL-6R pathways revealed the superiority of the former approach in mediating protection against poly(I:C) and influenza H1N1 virus-induced lung injury. This difference may reflect previously noted additional positive functions of IL-6 in protecting against severe respiratory viral infections, separate from its role in Notch4 induction (Dienz et al., 2012; Pyle et al., 2017; Yang et al., 2017).
A key mechanism by which Notch4 impairs lung tissue Treg cell function involved amphiregulin, whose expression is enriched in tissue-resident Treg cells, especially under conditions of active tissue repair (Arpaia et al., 2015; Burzyn et al., 2013; Schiering et al., 2014; Zaiss et al., 2013, 2015). Amphiregulin protein expression was increased markedly in poly(I:C)-treated and H1N1 influenza virus-infected mice upon Treg cell-specific Notch4 deletion or treatment with an anti-Notch4 mAb. The protective function of Notch4 antagonism in viral lung injury was reproduced by treatment with recombinant amphiregulin and abrogated by co-treatment with an amphiregulin-blocking peptide or neutralizing antibody. Amphiregulin may thus serve as a key effector mechanism by which lung tissue Treg cells suppress danger signals emanating from the epithelium and other innate immune cells, including macrophages. A previous study by Arpaia et al. (2015) has shown that Treg cells are the predominant source of amphiregulin in the course of H1N1 infection in the lungs. Furthermore, amphiregulin expression specifically in Treg cells is critical for lung tissue repair in the context of H1N1 infection, highlighting the privileged role of Treg cell-derived amphiregulin in this process (Arpaia et al., 2015). Amphiregulin expression in Treg cells is induced primarily by IL-18 and secondarily by IL-33 (Arpaia et al., 2015). Our study demonstrated that Notch4 inhibits the capacity of IL-18 to upregulate amphiregulin expression in lung Treg cells, providing a mechanism for suppression of amphiregulin protein expression by Notch4. Amphiregulin mobilization upon Notch4 antagonism would then allow recovery from virus-induced lung injury.
Our results identify the Notch4-amphiregulin nexus as a putative target of therapy in viral respiratory infections, including SAR-CoV-2 and influenza. The remarkable capacity of anti-Notch4 mAb therapy to abrogate poly(I:C) and H1N1 influenza virus-induced disease morbidity and mortality in conventional and humanized mice makes Notch4 an attractive candidate for intervention in severe respiratory viral infections. That Notch4 also plays an essential role in promoting tissue inflammation in asthma further denotes it as a hub common to different inflammatory responses in the lungs that may serve as a target of therapy. Furthermore, the essential role of amphiregulin in mediating the therapeutic outcome of Notch4 inhibition in viral lung injury points to the amphiregulin-EGFR pathway as an additional locus of intervention. The full spectrum of Notch4-licensed diseases and the role of different downstream pathways in such disorders will require further intensive investigation.
Limitations of study
There are limitations to this study that should be noted. The individuals with COVID-19 were part of an observational cohort of adults who were predominantly of European and Turkic ethnicities. The study was not specifically designed or powered to examine differences in Notch4 expression among affected individuals of different racial and ethnic backgrounds, nor did it extend to pediatric subjects with COVID-19 or people with other lower respiratory viral infections. The mouse studies employed proxies of SARS-CoV2 infection, including poly(I:C) treatment and influenza virus infection. They were relatively short in duration and thus do not explore additional immune-regulatory mechanisms that may be operative in long-term sequalae of COVID-19. These limitations highlight the urgent task of elucidating the full range of tissue-inflammatory licensing mechanisms operative in COVID-19 and related viral infections as a requisite step for designing precision therapies to restore immune tolerance and effective tissue repair in these disorders.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| eBioscience Fixable Viability Dye eFluor 506 and eFluor 780 | Thermo Fischer | Cat# 65-0866-18 Cat# 65-0865-14 |
| APC-Cy7 and FITC anti-mouse CD3 Antibody | Biolegend | Cat# 100222; RRID AB_2242784 Cat# 100204; RRID AB_312661 |
| Brilliant Violet 605 anti-mouse CD4 Antibody | Biolegend | Cat# 100451; RRID AB_2564591 |
| Brilliant Violet 605 anti-mouse CD86 Antibody | Biolegend | Cat# 105036; RRID AB_2686973 |
| PE anti-mouse IL-4 Antibody | Biolegend | Cat# 504104; RRID AB_315318 |
| FOXP3 Monoclonal Antibody (FJK-16 s), eFluor 450, eBioscience | Thermo Fischer | Cat# 48-5773-82; RRID AB_1518812 |
| PE/Cyanine7 anti-mouse IFN-γ Antibody | Biolegend | Cat# 505825; RRID AB_1595591 |
| IL-13 Monoclonal Antibody (eBio13A), APC-eFluor 780, eBioscience | Thermo Fischer | Cat# 47-7133-82; RRID AB_2716964 |
| PE/Cyanine7 anti-mouse IL-17A Antibody | Biolegend | Cat# 506922l AB_2125010 |
| APC anti-mouse IFN-γ Antibody | Biolegend | Cat# 505810; RRID AB_315404 |
| PE Anti-Mouse TNF | BD Biosciences | Cat# 554419; RRID AB_395380 |
| HELIOS Monoclonal Antibody (22F6), APC-eFluor 780, eBioscience | Thermo Fischer | Cat# 47-9883-42; AB_2573998 |
| PE/Cyanine7 anti-mouse CD11c Antibody | Biolegend | Cat# 117317; RRID AB_493569 |
| FITC anti-mouse I-Ad Antibody | Biolegend | Cat# 115006; RRID AB_313621 |
| Brilliant Violet 605 anti-mouse CD45 Antibody | Biolegend | Cat# 103140; RRID AB_2562342 |
| PE anti-mouse CD152 (CTLA-4) Antibody | Biolegend | Cat# 106306; RRID AB_313255 |
| Alexa Fluor® 647 anti-mouse CD279 (PD-1) Antibody | Biolegend | Cat# 109118; RRID AB_2566550 |
| PerCP/Cyanine5.5 anti-mouse/human CD11b Antibody | Biolegend | Cat# 101227; RRID AB_893233 |
| Brilliant Violet 605 anti-mouse CD4 Antibody | Biolegend | Cat# 100451; RRID AB_2564591 |
| PE/Cyanine7 anti-mouse CD206 (MMR) Antibody | Biolegend | Cat# 141719; RRID AB_2562247 |
| APC anti-mouse CD163 Antibody | Biolegend | Cat# 155305; RRID AB_2814059 |
| APC/Cyanine7 anti-mouse CD68 Antibody | Biolegend | Cat# 137023; RRID AB_2616812 |
| eFluor 450 anti-mouse CD44 Antibody | Thermo Fischer | Cat# 48-0441-82; RRID AB_1272246 |
| APC anti-mouse CD62L Antibody | Thermo Fischer | Cat# 17-0621-82; RRID AB_469410 |
| PE/Cyanine5 anti-mouse CD80 Antibody | Biolegend | Cat# 104711; RRID AB_313132 |
| F4/80 Monoclonal Antibody (BM8), eFluor 450, eBioscience | Thermo Fischer | Cat# 48-4801-82; RRID AB_1548747 |
| PerCP/Cyanine5.5 anti-mouse IL-33Rα (IL1RL1, ST2) Antibody | Biolegend | Cat# 145311; RRID AB_2565635 |
| FITC anti-mouse Lineage Cocktail | Biolegend | Cat# 133301 |
| PE/Cyanine7 anti-mouse CD25 Antibody | Thermo Fischer | Cat# 25-0251-82; RRID AB_469608 |
| APC-eFluor 780 anti-mouse HELIOS Antibody | Thermo Fischer | Cat# 47-9883-42; RRID AB_2573998 |
| PE/Cyanine7 anti-mouse IL-17A Antibody | Biolegend | Cat# 506921; RRID AB_2125011 |
| PE anti-mouse Ki-67 Antibody | Thermo Fischer | Cat# 12-5698-82; RRID AB_11150954 |
| FITC anti-mouse Ly-6G/Ly-6C (Gr-1) Antibody | Biolegend | Cat# 108405; RRID AB_313370 |
| Brilliant Violet 605 anti-mouse CD45 Antibody | Biolegend | Cat# 103139; RRID AB_2562341 |
| PE anti-mouse Notch 4 Antibody | Biolegend | Cat# 128407; RRID AB_1133997 |
| CD134 (OX40) Monoclonal Antibody (OX-86), APC, eBioscience | Thermo Fischer | Cat# 17-1341-82; RRID AB_10717260 |
| APC anti-mouse CD152 Antibody | Biolegend | Cat# 106309; RRID AB_2230158 |
| CD223 (LAG-3) Monoclonal Antibody (eBioC9B7W (C9B7W)), PE, eBioscience | Thermo Fischer | Cat# 12-2231-83; RRID AB_494214 |
| CD279 (PD-1) Monoclonal Antibody (J43), PE, eBioscience | Thermo Fischer | Cat# 12-9985-82; RRID AB_466295 |
| InVivomAb anti-mouse IL-6R | BioxCell | Cat# BE0047; RRID AB_1107588 |
| PE anti-mouse CD339 (Jagged 1) Antibody | Biolegend | Cat# 130907; RRID AB_2561302 |
| AREG antibody | BiorByt | orb676838 |
| PE/Cyanine7 anti-mouse CD126 (IL-6Rα chain) Antibody | Biolegend | Cat# 115813; RRID AB_2565574 |
| Phospho-EGF Receptor (Tyr1068) (D7A5) XP® Rabbit mAb #3777 | CST | Cat# 3777T |
| TruStain FcX (anti-mouse CD16/32) Antibody | Biolegend | Cat# 101319; RRID AB_1574973 |
| PerCP/Cyanine5.5 anti-human CD4 Antibody | Biolegend | Cat# 300529; RRID AB_893328 |
| FOXP3 Monoclonal Antibody (PCH101), eFluor 450, eBioscience | Thermo Fischer | Cat# 48-4776-42; RRID AB_1834364 Cat# 56-4716-41 |
| HELIOS Monoclonal Antibody (22F6), APC-eFluor 780, eBioscience | Thermo Fischer | Cat# 47-9883-42; RRID AB_2573998 |
| BB515 Mouse Anti-Human Notch1 | BD Biosciences | Cat# 566023 |
| BV605 Mouse Anti-Human Notch2 | BD Biosciences | Cat# 742291 |
| BV421 Mouse Anti-Human Notch3 | BD Biosciences | Cat# 744828 |
| PE Mouse Anti-Human Notch4 | BD Biosciences | Cat# 563269 |
| YAP (D8H1X) XP® Rabbit mAb (Alexa Fluor® 488 Conjugate) | CST | Cat# 14729S |
| Human/Mouse/Rat beta-Catenin Alexa Fluor® 405-conjugated Antibody | R&D | Cat# IC13292V |
| PE/Cyanine7 anti-human CD127 (IL-7Rα) Antibody | Biolegend | Cat# 351320; RRID AB_10897098 |
| CD25 Monoclonal Antibody (BC96), Alexa-Fluor 647, eBioscience | Thermo Fischer | Cat# 51025941 |
| APC/Cyanine7 anti-human CD3 Antibody | Biolegend | Cat# 300317; RRID AB_314053 |
| Brilliant Violet 605 anti-human IL-4 Antibody | Biolegend | Cat# 500827; RRID AB_2562311 |
| PerCP-Cyanine5.5 anti-human CD4 Antibody | Thermo Fischer | Cat# 45-0049-42; RRID AB_1518744 |
| PerCP/Cyanine5.5 anti-human IL-13 Antibody | Biolegend | Cat# 501912; RRID AB_2124283 |
| Brilliant Violet 605 anti-human CD194 (CCR4) Antibody | Biolegend | Cat# 359417; RRID AB_2562482 |
| APC anti-human CD183 (CXCR3) Antibody | Biolegend | Cat# 353707; RRID AB_10962949 |
| FITC anti-human CD294 (CRTH2) Antibody | Biolegend | Cat# 350107; RRID AB_11204250 |
| PE-Cy7 Mouse Anti-Human IFN-γ | BD Biosciences | Cat# 560741 |
| Brilliant Violet 510 anti-human CD196 (CCR6) Antibody | Biolegend | Cat# 353423; RRID AB_2561936 |
| InVivoMAb anti-mouse Notch4 | BioxCell | Cat# BE0129; RRID AB_10948996 |
| Critical commercial assays | ||
| recombinant Amphiregulin | R&D | Cat# 989-AR-100/CF |
| Dynabeads Mouse T-Activator CD3/CD28 | Thermo Fischer | Cat# 11452D |
| Poly(I:C) HMW | Invivogen | Cat# tlrl-pic-5 |
| recombinant IL-6 | Peprotech | Cat# 500-P56 |
| recombinant IL-33 | Peprotech | Cat# 210-33 |
| recombinant IL-18 | R&D | Cat# 9139-IL-010/CF |
| Notch4-His protein | Sino-Biologicals | Cat# 11869-H08B |
| complete Freund’s adjuvant | Sigma Chemical | Cat# F5881-10ML |
| Mouse IL-6 Quantikine ELISA Kit | R&D | Cat# M6000B |
| Human IL-6 Quantikine ELISA Kit | R&D | Cat# D6050 |
| Human Amphiregulin Quantikine ELISA Kit | R&D | Cat# DAR00 |
| Mouse Amphiregulin Quantikine ELISA Kit | R&D | Cat# DY989 |
| LEGENDplex Human Anti-Virus Response Panel | Biolegend | Cat# 740349 |
| LEGENDplex Mouse Inflammation Panel | Biolegend | Cat# 740150 |
| Mouse Oncostatin M (OSM) Quantikine ELISA Kit | R&D | Cat# MSM00 |
| Mouse LIF Quantikine ELISA Kit | R&D | Cat# MLF00 |
| Mouse LIX Quantikine ELISA Kit | R&D | Cat# MX000 |
| Mouse Amphiregulin Antibody | R&D | Cat# AF989 |
| Normal Goat IgG Control | R&D | Cat# AB-108-C |
| AREG blocking peptide | Mybiosource | Cat# MBS3224924 |
| RNAeasy mini kit | QIAGEN | Cat# 74104 |
| SuperScript III First-Strand Synthesis System | Thermo Fischer | Cat# 18080051 |
| TRITC-Dextran | Sigma Chemical | Cat# 42874-1G |
| Experimental models: Organisms/strains | ||
| Il6rfl/fl (B6;SJL-Il6ratm1.1Drew/J) | Jax Laboratory | Stock No: 012944 |
| Notch1fl/fl (B6.129X1-Notch1tm2Rko/Grid/J) | Jax Laboratory | Stock No: 007181 |
| Notch2fl/fl (B6.129S-Notch2tm3Grid/J) | Jax Laboratory | Stock No: 010525 |
| Notch3fl/fl | In this paper | N/A |
| Wwtr1fl/fl (B6.129(Cg)-Wwtr1tm1Hmc/J) | Jax Laboratory | Stock No: 032669 |
| Yap1fl/fl (B6.129P2(Cg)-Yap1tm1.1Dupa/J) | Jax Laboratory | Stock No: 032192 |
| NOD-PrkdcscidIl2rgtmiwjl/Sz (NSG) | Jax Laboratory | Stock No: 005557 |
| (Ctnnb1fl/fl) (B6(Cg)-Ctnnb1tm1Knw/J) | Jax Laboratory | Stock No: 022775 |
| Notch4tm1c(NCOM)Mfgc | Canadian mutant mouse repository | |
| Rbpj1fl/fl (B6.129P2-Rbpjtm1Hon/HonRbrc) | Pamela Stanley and Tasuku Honjo, | |
| Foxp3YFPcre | Jax | Stock No: 016959 |
| Influenza A/PR/8/34 (H1N1) | Charles River | |
| Deposited data | ||
| GEO | GSE159584: A Regulatory T Cell Notch4 Switch Governs Lung Inflammation in Viral Infections | |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad Software | N/A |
| FlowJo 10.4.2 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Talal A. Chatila (talal.chatila@childrens.harvard.edu)
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The data presented in the manuscript, including de-identified patient results, will be made available to investigators following request to the corresponding author. Any data and materials to be shared will be released via a material transfer agreement. RNA sequencing datasets have been deposited in the Gene Expression Omnibus, GSE159584: A Regulatory T Cell Notch4 Switch Governs Lung Inflammation in Viral Infections.
Experimental model and subject details
Human subjects
We recruited three prospective cohorts of patients with COVID-19 from the following institutions: (1) Massachusetts General Hospital in Boston, Massachusetts, USA; (2) San Martino Hospital in Genoa, Italy; (3) Istanbul Medical Faculty Hospitals in Istanbul, Turkey. The patient demographics are shown in Table S1. Inclusion criterion included a clinical syndrome consistent with COVID-19 disease and a positive SARS-CoV-2 test from a nasal swab based on RT-PCR. Severity of illness was defined as follows: (1) Mild for patients who did not require inpatient hospitalization; (2) Moderate for patients requiring inpatient hospitalization and who did not require therapies for acute respiratory failure such as high flow oxygen (defined as a flow rate of more than 15 l per minute), non-invasive positive pressure ventilation, mechanical ventilation, and who did not require therapies for other types of organ failure such as renal replacement therapy or shock; (3) Severe for patients with organ failure requiring supportive therapies typically administered in the intensive care unit such as high flow oxygen, non-invasive positive pressure ventilation, mechanical ventilation, vasopressors, renal replacement therapy; (4) Convalescent for patients who have recovered from their acute illness and discharged from the hospital. A cohort of country-matched healthy controls were also recruited (Table S1).
Clinical data including patient characteristics, therapies received, and clinical outcomes was abstracted from the medical record into a password-protected REDCap (Research Electronic Data Capture) database. A peripheral blood draw was obtained at the time of enrollment and weekly thereafter (if available) for flow cytometry and cytokine analysis.
Study approval
Recruitment of Boston study participants was approved by the Institutional Review Board at Massachusetts General Hospital and Boston Children’s Hospital. Recruitment of Genoa study participants was approved by the Institutional Review Board at Genoa University. Recruitment of Istanbul study participants was approved by the Institutional Review Board at Istanbul University. All animal studies were reviewed and approved by the Boston Children’s Hospital office of Animal Care Resources and Massachusetts General Hospital office of Animal Care Resources.
Mice
The following mouse strains were obtained from the JAX Laboratories: Foxp3 YFPCre (B6.129(Cg)-Foxp3 tm4(YFP/icre)Ayr/J) (Rubtsov et al., 2008), Il6r fl/fl (B6;SJL-Il6ra tm1.1Drew/J) (McFarland-Mancini et al., 2010), Notch1 fl/fl (B6.129X1-Notch1 tm2Rko/Grid/J) (Yang et al., 2004), Notch2 fl/fl (B6.129S-Notch2 tm3Grid/J) (McCright et al., 2006), Notch3 fl/fl Wwtr1 fl/fl (B6.129(Cg)-Wwtr1 tm1Hmc/J) and Yap1 fl/fl (B6.129P2(Cg)-Yap1 tm1.1Dupa/J) (Reginensi et al., 2013; Zhang et al., 2010), NOD-Prkdc scid Il2rg tmiwjl/Sz (NSG) (Verma et al., 2017). and (Ctnnb1 fl/fl) (B6(Cg)-Ctnnb1 tm1Knw/J) (Messerschmidt et al., 2016) mice. Notch4fl/fl (Notch4 tm1c(NCOM)Mfgc) were obtained from the Canadian mutant mouse repository. Rbpj1 fl/fl (B6.129P2-Rbpj tm1Hon/HonRbrc) were kind gifts of Pamela Stanley and Tasuku Honjo, respectively (Han et al., 2002; Shi and Stanley, 2003). Notch3 floxed allele mice (Notch3 fl/fl) were derived by Crispr/Cas 9 targeting with LoxP sequences inserted in introns 4 and 9 to target exons 5-9 by Cre-mediated excision.
Method details
Regulatory T cell co-cultures with macrophages
Macrophages cells were isolated from lungs of Poly I:C-treated mice. Macrophages were isolated based on CD45+ F4/80+ MHCII+. Macrophages were then co-cultured (at 1:1 concentration) with lungs Treg cells from Foxp3 YFPCre or Foxp3 YFPCre Notch4 Δ/Δ mice Poly I:C-treated mice. Cells were treated with Poly I:C at a concentration of 10ug/ml for three days. After 72h, M1 (F4/80+ MHCII+ CD68+ CD80+CD86+) and M2 (F4/80+ MHCII+ CD163+ CD206+) polarization was measured by flow cytometric analysis.
In vitro Macrophages–Amphiregulin co-cultures
Macrophages cells were isolated from the lungs of Poly I:C treated Foxp3YFPCre mice. These cells were incubated with recombinant Amphiregulin (R&D) (10 μg/ml). 72 hours later, the expression of different M1 and M2 markers were measured by flow cytometry.
In vitro Notch4 induction
Lung and spleen Treg cells from Poly I:C or PBS-treated Foxp3 YFPCre mice were isolated by cell sorting (Sony Sorter, MA900). Treg cells were seeded at 1 × 104 cells in 96-well plates then stimulated with CD3/CD28 Dynabeads (ThermoFisher) in presence of Poly I:C (10 μg/ml) alone or in combination with recombinant IL-6 and IL-33 (10 μg /ml; Peprotech) for 72h. Notch4 expression on Foxp3+ Treg cells was then assessed by Flow cytometry.
In vitro amphiregulin induction
Lung Treg cells from Poly I:C or PBS-treated Foxp3 YFPCre mice were seeded at 1 × 104 cells then stimulated with CD3/CD28 Dynabeads with Poly I:C (10 μg/ml) either alone or in combination with recombinant IL-18 and IL-33 (R&D systems, Peprotech, respectively, 10 μg/ml) for 72h. Amphiregulin expression was then assessed by Flow cytometry.
Mouse anti-human Notch4 monoclonal antibody generation
Notch4-deficient mice were immunized with 50 μg of recombinant Notch4-His protein (Sino-Biologicals, Wayne, PA) suspended in Dulbecco’s phosphate buffered saline (PBS; GIBCO, Grand Island, NY) and emulsified with an equal volume of complete Freund’s adjuvant (Sigma Chemical Co., St. Louis, MO). Fourteen days after the initial immunization, the mice were given a booster immunization i.p. with 25 μg of recombinant Notch4-His protein suspended in PBS and emulsified with an equal volume of incomplete Freund’s adjuvant. A second booster of 25 μg of recombinant Notch-His protein in PBS was given after another 14 days. The mice were re-boosted 2X with 25 μg of recombinant Notch4-His protein to raise their antibody titers, then the mouse with the best titer was rested for 3 weeks from the last immunization, then boosted by intraperitoneal and subcutaneous injection of 25 ug recombinant Notch4-His protein in PBS. Three days later the mouse was euthanized, and the spleen and lymph nodes were collected and made into a cell suspension, then washed with DMEM. The spleen/lymph node cells were counted and mixed with SP 2/0 myeloma cells (ATCC No. CRL8-006, Rockville, MD) that are incapable of secreting either heavy or light chain immunoglobulin chains (Kearney et al., 1979) using a spleen:myeloma ratio of 2:1. Cells were fused with polyethylene glycol 1450 (ATCC) in eight 96-well tissue culture plates in HAT selection medium according to standard procedures (Köhler and Milstein, 1975). Between 10 and 21 days after the fusions, hybridoma colonies became visible and culture supernatants were harvested then screened by ELISA for Notch4. Potential positive wells were expanded and re-screened against Notch1-His, Notch2-His, Notch3-His/Gst, Notch4-His and an irrelevant protein by ELISA. Hybridomas showing specificity for Notch4 alone were identified and subcloned twice by limiting dilution to obtain stable, clonal hybridomas.
Isolation of Human peripheral blood mononuclear cells (PBMCs)
Human PBMCs were isolated from whole blood from either healthy control, mild COVID-19, moderate COVID-19 or severe COVID-19 probands via density gradient using Ficoll (GE Healthcare). PBMCs were then stored frozen in Fetal Calf Serum (FCS) (Sigma Aldrich) and 15% Dimethyl sulfoxide (DMSO) (Sigma Aldrich). The cells were later thawed for analysis of their Notch, Yap1, β-Catenin or different chemokine and cytokine expression by flow cytometry.
IL-6 and Amphiregulin ELISA
Genoa patients IL-6 concentrations were measured using ELISA (R&D, USA) per manufacturer’s protocol. As for Amphiregulin, Amphiregulin concentrations for the whole cohort were measured using Amphiregulin ELISA kit (R&D, USA).
Cytokine measurements
IL-1β, IL-6 (For Boston and Istanbul cohorts), IL-8, IFNα, IFNβ, IFNγ, IFNλ, CXCL10 and TNF were measured using Legendplex (Biolegend) per manufacturer’s protocol. Mouse cytokine were assessed using Legendplex (Biolegend) per manufacturer’s protocol on BAL samples from Poly I:C treated mice (Day 7) or on BAL samples from H1N1-infected mice (Day 12). OSM, LIF and CXCL5 were also measured on the same samples using Quantikine Elisa kits (R&D systems).
Polyinosinic-polycytidylic acid (Poly I:C) mouse model
Mice were treated intratracheally with 2.5 mg/kg of Poly I:C HMW (InvivoGen) daily for six consecutive days. The weight of the mice was recorded daily upon application of the Poly I:C. The mice were subjected to airway hyperresponsiveness at day 7, then euthanized and analyzed. For blockading amphiregulin, mice were treated with a peptide spanning amino acids 91-140 of the middle region of the human amphiregulin preproprotein (amphiregulin91-140 peptide; Mybiosource). The peptide was given intratracheally at 10 μg/ml in PBS in a final volume of 100 μl. For the amphiregulin neutralizing antibody tests, the mice were given intraperitoneally 20μg of goat anti-mouse amphiregulin mAb (clone AF989; R&D systems) or isotype control mAb (clone AB-108-C; R&D systems) daily for the duration of the experiment.
H1N1 influenza A virus preparation
Mouse-adapted H1N1 Influenza A virus (PR/8/34) was obtained from Charles River (Catalogue no. 10100374). Viral stocks were calculated to contain 40,000 infectious units (IU) per mouse and were diluted to a volume of 20 μl/mouse in PBS. For lethal dose (LD75), the viral stocks were calculated to contain 60,000 IU per mouse and diluted to a volume of 20 μl/mouse in PBS as well.
H1N1 Influenza A viral infection model
Mice were treated intranasally on day 0 of the experiments with either a 40,000 pfu dose of the H1N1 virus or 70,000pfu dose, equivalent to a lethal dose 75 (LD75), as indicated. The mice were monitored on a daily basis to see signs of infection. The weights of the mice were recorded and once a mouse weight loss exceeded 20%–25%, the mouse was euthanized. The endpoint of the experiments was set at day 12, to capture the peak of inflammation. For antibody treatment experiments, mice received anti-Notch4 mAb (10 μg/ml) (clone HMN-4-14; Bio-X-Cell), anti-IL-6Rα mAb (10 μg/ml) (clone 15A7, Bio-X-Cell) or their respective isotype control antibodies, as indicated.
Humanized mice H1N1 viral infection
NOD-Prkdc scid Il2rg tmiwjl/Sz (NSG) humanized mice were reconstituted with PBMCs from healthy control. Then, mice were treated with a sublethal dose of the virus as indicated intranasally on day 0. The mice were monitored on a daily basis to see signs of infection. The weights of the mice were recorded and once a mouse weight loss exceeded 20%–25%, the mouse was euthanized. For sublethal experiments, the endpoint of the experiment was set at day 12, to capture the peak of inflammation.
Viral Load
Viral titers were determined by quantification of viral transcripts using reverse transcription (RT)-quantitative (q) PCR as previously described (Cho et al., 2012; Glenn et al., 2017; Moltedo et al., 2011). The collected lung tissue was weighed, and total RNA was isolated using RNAeasy mini kit (QIAGEN). Purified RNA was converted into cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen) following the manufacturer’s recommended procedures. Quantification of viral load was performed in 96-well plates and run on a QuantStudio 3 Real-Time PCR instrument (Applied Biosystems). The amplification was carried out using H1N1 nucleoprotein region specific primers: forward primer, 5′-GGGGGAAGCTCTCCACTAGA-3′ and reverse primer, 5′-GTTGTGTGCTGATTTGGCCC-3′ in the presence of the Power SYBR Green Master Mix (Applied Biosystems). The PCR amplification cycle was set as 95°C for 5 min for initial denaturation, followed by 50 cycles of 95°C for 15 s, 60°C for 1min. The standard curve was generated using 10-fold serial dilutions of constructed plasmid pcDNA3-NP (Addgene) by plotting each cycle threshold (CT) value against the log quantity of each standard plasmid copy number. Viral RNA copies were extrapolated from the standard curve using the sample CT value and represented as viral RNA copies per gram of tissue.
For determination of the viral load in the humanized mouse model, viral transcripts were quantitated using reverse transcription (RT)-quantitative (q) PCR as previously described (Cho et al., 2012; Glenn et al., 2017; Moltedo et al., 2011). Briefly, RNA was isolated from right upper lung lobe homogenates prepared in TRIzol (Sigma-Aldrich) and 1 μg RNA as determined by a Nanodrop 2000 spectrophotometer (ThermoFisher) was converted to cDNA by reverse transcription using the protocol provided by the manufacturer (TaqMan Reverse Transcription Reagents, ThermoFisher). qPCR reactions in the presence of SYBR Green (FastStart Essential DNA Green Master Mix, Roche) were performed on a Light-Cycler 96 Instrument (Roche). Primers: Polymerase gene (PA), 5′-CGGTCCAAATTCCTGCTGA-3′ (forward) and 5′-CATTGGGTTCCTTCCATCCA-3′(reverse).
Measurement of airway functional responses
Poly I:C-induced airway hyperreactivity (AHR) was measured, as previously described (Xia et al., 2018). Anesthetized mice were exposed to doubling concentrations of aerosolized acetyl-β-methacholine (Sigma-Aldrich) by using a Buxco small-animal ventilator (Data Sciences International). The relative peak airway resistance for each methacholine dose, normalized to the saline baseline, was calculated.
Lung histopathology staining
Paraffin-embedded lung sections were stained with hematoxylin and eosin (H&E) or Paraffin-acid-Schiff staining (PAS). The lung pathology was scored by blinded operators. Inflammation was scored separately for cellular infiltration around blood vessels and airways: 0, no infiltrates; 1, few inflammatory cells; 2, a ring of inflammatory cells 1 cell layer deep; 3, a ring of inflammatory cells 2–4 cells deep; 4, a ring of inflammatory cells of > 4 cells deep (Tachdjian et al., 2009). A composite score was determined by the adding the inflammatory scores for both vessels and airways.
Dextran leakage assay
25 μg of TRITC-Dextran (MW 40,000; Sigma) in 80 μl saline was instilled intratracheally in each mouse. After one hour of dextran instillation, blood from the mice were collected retro-orbitally in sodium citrate solution, and the plasma was separated by centrifugation (2000 rpm, 15 minutes at 4C). Leakage of dextran in the bloodstream is calculated as TRITC fluorescence in the plasma compared to plasma from mock-treated mice.
Flow cytometric analysis of mouse and human cells
Antibodies against the following murine antigens were used for flow cytometric analyses: IL-4 (clone 11B11, catalog no: 504104 1:300 dilution, Biolegend), Foxp3 (FJK-16S, catalog no: 48-5773-82 1:300, Thermofisher), IFNγ (XMG1.2, catalog no: 505825 1:300, Biolegend), IL-13 (eBio13a, catalog no.: 47-7133-82 1:300, Thermofisher), TNF (MP6-XT22, Catalogue no: 554419, 1:200, BD Biosciences), Helios (22F6, catalog no: 47-9883-42 1:200, Thermofisher, CD11c (N418, catalog no: 117318 1:500, Biolegend), CD11b (M1/70, catalog no: 101228 1:500, Biolegend), CD4 (GK1.5, catalog no: 100451, 1:500, Biolegend), CD206 (C068C2, Catalogue no: 141719, 1:300, Biolegend), CD163 (S15049l, Catalogue no: 155305 1:200, Biolegend), CD68 (FA-11, Catalogue no: 137023 1:200, Biolegend), CD80 (16-10A1, Catalogue no: 104711 1:200, Biolegend), F4/80 (BM8, Catalogue no: 48-4801-82 1:300, Thermofisher), MHCII (39-10-8, Catalogue no: 115006, 1:300, Biolegend), IL-33Ralpha (DIH9, Catalogue no: 145312 1:300, Biolegend), Lineage (anti-CD3/GR-1/CD11B/CD45-B220/TER 119) (Biolegend, 1:300 catalog no: 78022), CD3 (17A2, catalog no: 100203, 1:500, Biolegend), IL-17 (TC11-18H10.1, catalog no: 506922, 1:200, Biolegend), GR-1 (RB6-8C5, catalog no: 108406, 1:500, Biolegend), CD45 (30-F11, catalog no: 103140, 1:300, Biolegend), Notch4 (HMN4-14, catalog no: 128407 1:400, Biolegend), CD134 (DX-86, catalog no:17-1341-82, 1:400 Thermofisher), CTLA-4 (CD152) (UC10-4B9, catalog no: 106310, 1:400 Biolegend), CD223 (Ebio-C9B7W, catalog no:12-2231-83, 1:400 Therrmofisher), CD279 (J43, catalog no:12-9985-82, 1:400 Thermofisher), IL-6Rα (15A7, catalog no: BE0047, 1:200 Bio-X-Cell), Jagged1 (CD339) (HMJ1-29, catalog no:130908, 1:400 Biolegend), Amphiregulin polyclonal antibody (orb7378, 1:200, Biorbyt), IL-6Rα (D7715A7, catalog 115813, 1:400 Biolegend), IL-18Rα (BG/IL18RA, catalog 132903, 1:400 Biolegend), P-EGFR(D7A5, catalog no:D7A5, 1:200 Cell Signaling Technology), Anti-CD16/CD32 (93, catalog no: 101319, 1:1000 Biolegend). Antibodies against the following human antigens were used: CD3 (HIT3a, catalog no: 300318, 1:200,Biolegend), CD4 (RPA-T4, catalog no: 300530, 1:200,Biolegend), Foxp3 (PCH-101,catalog no: 48-4776-42,56-4716-41, 1:100 Thermofisher), Helios (22F6, catalog no: 47-9883-42 1:100, Thermofisher), Notch1 (HMN1-519, catalog no: 566023, 1:300, BD PharMingen), Notch2 (HMN2-25, catalog no: 742291, 1:300, BD PharMingen), Notch3 (HMN3-21, catalog no: 744828, 1:300, BD PharMingen), Notch4 (HMN4-2, Catalogue no: 563269, 1:300, BD PharMingen), Yap1 (D8H1X, Catalogue no: 14729S, 1:100, Cell Signaling Technology), beta-Catenin (196624, Catalogue no: IC13292V, 1:200, R&D system), CD25 (BC96, Catalogue no: 51-025-941 1:300, Thermofisher), IL-4 (MP4-25D2, Catalogue no: 500828 1:200, Biolegend), IL-13 (JES10-5A2, Catalogue no: 501912, 1:200, Biolegend), CCR4 (L291H4, Catalogue no: 359417 1:300, Biolegend), CXCR3 (G025H7, Catalogue no: 353708, 1:300 Biolegend), CRTH2 (BM16, Catalogue no: 350108 1:300, Biolegend), CD127 (A019D5, Catalogue no: 351320 1:300, Biolegend), IFNγ (4S.B3, Catalogue no: 560741 1:200, BD Biosciences) and Amphiregulin polyclonal antibody (orb7378 1:200, Biorbyt), CD196 (G034E3, Catalogue no: 353423, 1:300 Biolegend). The specificity and optimal dilution of each antibody was validated by testing on appropriate negative and positive controls or otherwise provided on the manufacturer’s website. Intracellular cytokine staining was performed as previously described (Charbonnier et al., 2015). Dead cells were routinely excluded from the analysis based on the staining of eFluor 780 Fixable Viability Dye (1:1000 dilution) (Thermofisher). Stained cells were analyzed on a BD LSR Fortessa cell analyzer (BD Biosciences) and data were processed using Flowjo (Tree Star Inc.).
M1 and M2 gating strategy
M1 was defined as follows: CD45+CD4– F4/80+ MHCII+ CD68+ CD80+CD86+ while M2 macrophages were defined as follows: CD45+CD4–F4/80+ MHCII+CD163+CD206+.
Transcriptome profiling
Treg cells were isolated from either Foxp3 YFPcre or Foxp3YFPCre Notch4 Δ/Δ mice that were treated with Poly I:C as shown above. mRNA was isolated using QIAGEN RNeasy mini kit (QIAGEN). RNA was then converted into double-stranded DNA (dsDNA), using SMART-Seq v4 Ultra Low Input RNA kit (Clontech). dsDNA was then fragmented to 200-300 bp size, using M220 Focused-ultrasonicator (Covaris), and utilized for construction of libraries for Illumina sequencing using KAPA Hyper Prep Kit (Kapa Biosystems). Libraries were then quantified using Qubit dsDNA HS (High Sensitivity) Assay Kit on Agilent High Sensitivity DNA Bioanalyzer.
Gene-level read counts were quantified using feature Counts and the latest Ensambl mouse annotation (GRCm38.R101). Raw data were trimmed using Trimmomatic (version 0.39, default parameters), tool for Illumina NGS data. To identify differentially expressed genes, we used 3 algorithms: DESeq2 (version 1.26.0), edgeR (version 3.28.1) and Lima (3.42.2) Bioconductor packages with default parameters. Count tables were normalized to TPM (Transcripts per Million) for visualizations and QC. Sample clustering, path analyses and integration of the results were performed using a custom-made pipeline available upon request (Variant Explorer RNaseq). Transcripts were called as differentially expressed when the adjusted p values were below 0.05, fold-changes over ± 1.5 and false discovery rate (FDR) were below 0.05. For our path analyses, we tested 10,715 biological pathways from KEGG and GO annotations. We filtered the results using an adjusted p value below 0.01.
Quantification and statistical analysis
Student’s two-tailed t test, one- and two-way ANOVA and repeat measures two-way ANOVA with post-test analysis and log-rank test of groups were used to compare test groups, as indicated. Linear Regression was used for correlation analysis. For analysis of the human data, summary statistics were calculated using number (percentage) for binary and categorical data and mean (standard deviation) or median (interquartile range) for continuous data depending on the normality of the distribution. Logistic regression was performed to determine the association between Notch4 expression and mortality. Covariates including age, gender, and study site were selected based on a priori knowledge. In the case of highly correlated covariates, we chose the predictor that had the strongest univariate association between the potential predictor and outcome. In the final model, the outcome was death at any time after study enrollment, the predictors of interest were Notch4 expression and serum interleukin-6 levels, and covariates included age, gender, history of malignancy, and corticosteroid treatment. Analyses were performed in R version 3.6.1. Two-sided p values of < 0.05 were considered statistically significant.
Acknowledgments
We thank Jacob Vieira, Hui-Yu Sui, and Naci Senkal for data collection and entry. This work was supported by National Institutes of Health grants R01 AI115699, R01 AI065617, and R01 AI06561720S1 (to T.A.C.) and 1R01AI121066, 1R01DK115217, and NIAID-DAIT-NIHAI201700100 (to I.Z.); German Research Society grant HA 8465/1-1 (to H.H.); and Istanbul University BAP Project TSG2020-36834 (to G.D.). I.Z. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. T.A.C. is the recipient of a grant from Digitalis Ventures.
Author contributions
H.H., M.B., P.S.L., R.D.P., and T.A.C. conceived the project and designed experiments. H.H., M.B., P.S.L., P.C., J.W.G., Q.C., J.F., G.D.Z., N.P., S.K., A.B., S.G., M.Y.G., F.B.O., E.A.C., Z.W., Y.C., E.S.-V., and L.-M.C. performed experiments. I.Z., H.P., G.D., R.D.P., and T.A.C. supervised the experimental studies. P.S.L., G.D., and R.D.P. supervised subject recruitment and sample collection at the respective centers. X.G.Y., J.Z.L., and L.B. provided additional samples. P.S.L., L.M., G.F., A.K., and M.K. collected and analyzed clinical data. K.S.-A. analyzed the RNA-seq data. P.S.L., R.D.P., G.D., E.C., X.G.Y., and J.Z.L. obtained institutional review board approval at the respective centers. H.H., M.B., and T.A.C. wrote the manuscript.
Declaration of interests
T.A.C., H.H., M.B., P.S.L., P.C., and R.D.P. are inventors on provisional patent application US 63/038,186 titled “Methods and Compositions for treating coronavirus infectious disease.” H.H. and T.A.C. are co-founders of and hold equity in Alcea Therapeutics.
Published: April 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.04.002.
Supplemental information
References
- Arpaia N., Green J.A., Moltedo B., Arvey A., Hemmers S., Yuan S., Treuting P.M., Rudensky A.Y. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier L.M., Wang S., Georgiev P., Sefik E., Chatila T.A. Control of peripheral tolerance by regulatory T cell-intrinsic Notch signaling. Nat. Immunol. 2015;16:1162–1173. doi: 10.1038/ni.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.L., Roche M.I., Sandall B., Brass A.L., Seed B., Xavier R.J., Medoff B.D. Enhanced Tim3 activity improves survival after influenza infection. J. Immunol. 2012;189:2879–2889. doi: 10.4049/jimmunol.1102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copaescu A., Smibert O., Gibson A., Phillips E.J., Trubiano J.A. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J. Allergy Clin. Immunol. 2020;146:518–534.e1. doi: 10.1016/j.jaci.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienz O., Rud J.G., Eaton S.M., Lanthier P.A., Burg E., Drew A., Bunn J., Suratt B.T., Haynes L., Rincon M. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 2012;5:258–266. doi: 10.1038/mi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esty B., Harb H., Bartnikas L.M., Charbonnier L.M., Massoud A.H., Leon-Astudillo C., Visner G., Subramaniam M., Phipatanakul W., Chatila T.A. Treatment of severe persistent asthma with IL-6 receptor blockade. J. Allergy Clin. Immunol. Pract. 2019;7:1639–1642.e4. doi: 10.1016/j.jaip.2019.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Yu S., Zhao H., Sun X., Li X., Wang P., Xiong X., Hong L., Xie C., Gao J., et al. The transcriptional coactivator TAZ regulates reciprocal differentiation of TH17 cells and Treg cells. Nat. Immunol. 2017;18:800–812. doi: 10.1038/ni.3748. [DOI] [PubMed] [Google Scholar]
- Glenn J.D., Smith M.D., Xue P., Chan-Li Y., Collins S., Calabresi P.A., Horton M.R., Whartenby K.A. CNS-targeted autoimmunity leads to increased influenza mortality in mice. J. Exp. Med. 2017;214:297–307. doi: 10.1084/jem.20160517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Tanigaki K., Yamamoto N., Kuroda K., Yoshimoto M., Nakahata T., Ikuta K., Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Harb H., Stephen-Victor E., Crestani E., Benamar M., Massoud A., Cui Y., Charbonnier L.M., Arbag S., Baris S., Cunnigham A., et al. A regulatory T cell Notch4-GDF15 axis licenses tissue inflammation in asthma. Nat. Immunol. 2020;21:1359–1370. doi: 10.1038/s41590-020-0777-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L.A., Canna S.W., Schulert G.S., Volpi S., Lee P.Y., Kernan K.F., Caricchio R., Mahmud S., Hazen M.M., Halyabar O., et al. On the Alert for Cytokine Storm: Immunopathology in COVID-19. Arthritis Rheumatol. 2020;72:1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermine O., Mariette X., Tharaux P.L., Resche-Rigon M., Porcher R., Ravaud P., CORIMUNO-19 Collaborative Group Effect of Tocilizumab vs Usual Care in Adults Hospitalized With COVID-19 and Moderate or Severe Pneumonia: A Randomized Clinical Trial. JAMA Intern. Med. 2021;181:32–40. doi: 10.1001/jamainternmed.2020.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson A.M., Pasman L., Yu S., Gamradt P., Homer R.J., Decker T., Medzhitov R. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science. 2013;340:1230–1234. doi: 10.1126/science.1233632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G.R., Wong L. Heparan sulfate is essential to amphiregulin-induced mitogenic signaling by the epidermal growth factor receptor. J. Biol. Chem. 1994;269:27149–27154. [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kearney J.F., Radbruch A., Liesegang B., Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J. Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Lowther D.E., Goods B.A., Lucca L.E., Lerner B.A., Raddassi K., van Dijk D., Hernandez A.L., Duan X., Gunel M., Coric V., et al. PD-1 marks dysfunctional regulatory T cells in malignant gliomas. JCI Insight. 2016;1:e85935. doi: 10.1172/jci.insight.85935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J.M., Hsing L., Pham T.T., Rudensky A.Y. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCright B., Lozier J., Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44:29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- McFarland-Mancini M.M., Funk H.M., Paluch A.M., Zhou M., Giridhar P.V., Mercer C.A., Kozma S.C., Drew A.F. Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J. Immunol. 2010;184:7219–7228. doi: 10.4049/jimmunol.0901929. [DOI] [PubMed] [Google Scholar]
- Messerschmidt D., de Vries W.N., Lorthongpanich C., Balu S., Solter D., Knowles B.B. β-catenin-mediated adhesion is required for successful preimplantation mouse embryo development. Development. 2016;143:1993–1999. doi: 10.1242/dev.133439. [DOI] [PubMed] [Google Scholar]
- Moltedo B., Li W., Yount J.S., Moran T.M. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog. 2011;7:e1002345. doi: 10.1371/journal.ppat.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroukhova M., Qi Z., Oriss T.B., Dixon-McCarthy B., Ray P., Ray A. Treg-mediated immunosuppression involves activation of the Notch-HES1 axis by membrane-bound TGF-beta. J. Clin. Invest. 2006;116:996–1004. doi: 10.1172/JCI26490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. GenOMICC Investigators. ISARIC4C Investigators. COVID-19 Human Genetics Initiative. 23andMe Investigators. BRACOVID Investigators. Gen-COVID Investigators Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Perumalsamy L.R., Marcel N., Kulkarni S., Radtke F., Sarin A. Distinct spatial and molecular features of notch pathway assembly in regulatory T cells. Sci. Signal. 2012;5:ra53. doi: 10.1126/scisignal.2002859. [DOI] [PubMed] [Google Scholar]
- Pyle C.J., Uwadiae F.I., Swieboda D.P., Harker J.A. Early IL-6 signalling promotes IL-27 dependent maturation of regulatory T cells in the lungs and resolution of viral immunopathology. PLoS Pathog. 2017;13:e1006640. doi: 10.1371/journal.ppat.1006640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi A., Scott R.P., Gregorieff A., Bagherie-Lachidan M., Chung C., Lim D.S., Pawson T., Wrana J., McNeill H. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 2013;9:e1003380. doi: 10.1371/journal.pgen.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., et al. the Northwell COVID-19 Research Consortium Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas I.O., Brau N., Waters M., Go R.C., Hunter B.D., Bhagani S., Skiest D., Aziz M.S., Cooper N., Douglas I.S., et al. Tocilizumab in Hospitalized Patients with Severe Covid-19 Pneumonia. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2028700. Published online February 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R., Jr., et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Salama C., Han J., Yau L., Reiss W.G., Kramer B., Neidhart J.D., Criner G.J., Kaplan-Lewis E., Baden R., Pandit L., et al. Tocilizumab in Patients Hospitalized with Covid-19 Pneumonia. N. Engl. J. Med. 2021;384:20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvarani C., Dolci G., Massari M., Merlo D.F., Cavuto S., Savoldi L., Bruzzi P., Boni F., Braglia L., Turrà C., et al. RCT-TCZ-COVID-19 Study Group Effect of Tocilizumab vs Standard Care on Clinical Worsening in Patients Hospitalized With COVID-19 Pneumonia: A Randomized Clinical Trial. JAMA Intern. Med. 2021;181:24–31. doi: 10.1001/jamainternmed.2020.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering C., Krausgruber T., Chomka A., Fröhlich A., Adelmann K., Wohlfert E.A., Pott J., Griseri T., Bollrath J., Hegazy A.N., et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–568. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachdjian R., Mathias C., Al Khatib S., Bryce P.J., Kim H.S., Blaeser F., O’Connor B.D., Rzymkiewicz D., Chen A., Holtzman M.J., et al. Pathogenicity of a disease-associated human IL-4 receptor allele in experimental asthma. J. Exp. Med. 2009;206:2191–2204. doi: 10.1084/jem.20091480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The REMAP-CAP Investigators Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2100433. Published online February 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton A.M., Lu J., Korty P.E., Kim Y.C., Martens C., Sun P.D., Shevach E.M. Helios+ and Helios- Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires. Eur. J. Immunol. 2019;49:398–412. doi: 10.1002/eji.201847935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toniati P., Piva S., Cattalini M., Garrafa E., Regola F., Castelli F., Franceschini F., Airò P., Bazzani C., Beindorf E.A., et al. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: A single center study of 100 patients in Brescia, Italy. Autoimmun. Rev. 2020;19:102568. doi: 10.1016/j.autrev.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga V.C., Prats J.A.G.G., Farias D.L.C., Rosa R.G., Dourado L.K., Zampieri F.G., Machado F.R., Lopes R.D., Berwanger O., Azevedo L.C.P., et al. Coalition covid-19 Brazil VI Investigators Effect of tocilizumab on clinical outcomes at 15 days in patients with severe or critical coronavirus disease 2019: randomised controlled trial. BMJ. 2021;372:n84. doi: 10.1136/bmj.n84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M.K., Clemens J., Burzenski L., Sampson S.B., Brehm M.A., Greiner D.L., Shultz L.D. A novel hemolytic complement-sufficient NSG mouse model supports studies of complement-mediated antitumor activity in vivo. J. Immunol. Methods. 2017;446:47–53. doi: 10.1016/j.jim.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett J.A., Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 2015;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodham A.W., Zeigler S.H., Zeyang E.L., Kolifrath S.C., Cheloha R.W., Rashidian M., Chaparro R.J., Seidel R.D., Garforth S.J., Dearling J.L., et al. In vivo detection of antigen-specific CD8(+) T cells by immuno-positron emission tomography. Nat. Methods. 2020;17:1025–1032. doi: 10.1038/s41592-020-0934-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Viera-Hutchins L., Garcia-Lloret M., Noval Rivas M., Wise P., McGhee S.A., Chatila Z.K., Daher N., Sioutas C., Chatila T.A. Vehicular exhaust particles promote allergic airway inflammation through an aryl hydrocarbon receptor-notch signaling cascade. J. Allergy Clin. Immunol. 2015;136:441–453. doi: 10.1016/j.jaci.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Harb H., Saffari A., Sioutas C., Chatila T.A. A Jagged 1-Notch 4 molecular switch mediates airway inflammation induced by ultrafine particles. J. Allergy Clin. Immunol. 2018;142:1243–1256.e17. doi: 10.1016/j.jaci.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Klein R., Tian X., Cheng H.T., Kopan R., Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Yang M.L., Wang C.T., Yang S.J., Leu C.H., Chen S.H., Wu C.L., Shiau A.L. IL-6 ameliorates acute lung injury in influenza virus infection. Sci. Rep. 2017;7:43829. doi: 10.1038/srep43829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss D.M., van Loosdregt J., Gorlani A., Bekker C.P., Gröne A., Sibilia M., van Bergen en Henegouwen P.M., Roovers R.C., Coffer P.J., Sijts A.J. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity. 2013;38:275–284. doi: 10.1016/j.immuni.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss D.M.W., Gause W.C., Osborne L.C., Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42:216–226. doi: 10.1016/j.immuni.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bai H., David K.K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R.A., Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the manuscript, including de-identified patient results, will be made available to investigators following request to the corresponding author. Any data and materials to be shared will be released via a material transfer agreement. RNA sequencing datasets have been deposited in the Gene Expression Omnibus, GSE159584: A Regulatory T Cell Notch4 Switch Governs Lung Inflammation in Viral Infections.