Abstract
Despite numerous viral outbreaks in the last decade, including a devastating global pandemic, diagnostic and therapeutic technologies remain severely lacking. CRISPR-Cas systems have the potential to address these critical needs in the response against infectious disease. Initially discovered as the bacterial adaptive immune system, these systems provide a unique opportunity to create programmable, sequence-specific technologies for detection of viral nucleic acids and inhibition of viral replication. This review summarizes how CRISPR-Cas systems—in particular the recently discovered DNA-targeting Cas12 and RNA-targeting Cas13, both possessing a unique trans-cleavage activity—are being harnessed for viral diagnostics and therapies. We further highlight the numerous technologies whose development has accelerated in response to the COVID-19 pandemic.
Keywords: CRISPR, class 2 Cas proteins, viruses, CRISPR-based antivirals, CRISPR-based detection, viral diagnostics
In this review, Freije et al. describe the ways CRISPR-Cas systems have been harnessed for the detection and inhibition of mammalian viruses. They focus on Cas12- and Cas13-based detection technologies, including those developed in response to the COVID-19 pandemic and studies of Cas9 and Cas13’s antiviral activity.
Introduction
In 2020, we were directly faced with the question of whether we were prepared for a global pandemic—and we were not. The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed that comprehensive diagnostic testing at a global scale and rapid discovery of potent therapeutics were not achievable with current approaches. For diagnostics, the limits lie in cost, turn-around time, and laboratory equipment required for commonly used technologies; diagnostic reagents and testing infrastructure can also become strained when demand is high (Jayamohan et al., 2021). Additionally, the United States Food and Drug Administration (FDA) has approved antivirals and vaccines for only a small number (<25) of known viruses (FDA, 2020a, 2020b). Without the means to detect emerging and re-emerging viruses in the early stages of an outbreak, and without a toolkit of antivirals, viruses will continue to threaten human health.
The expanse and diversity of viral species with pandemic potential makes it challenging to prepare for a pandemic. Genomic characterization of our microbial world has identified thousands of viruses circulating in animal or insect species; some of these microbes are known to cause human disease while others could potentially infect humans (Morse et al., 2012; Nieto-Rabiela et al., 2019; Olival et al., 2017; Woolhouse and Adair, 2013). Within the thousands of viral species, there is further extensive genomic sequence diversity. For example, there are three types of influenza (A, B, and C), and within influenza A virus (FLUAV) there are 18 hemagglutinin subtypes and 11 neuraminidase subtypes (Bouvier and Palese, 2008; Tong et al., 2013). Moreover, viruses evolve at a heightened rate compared with their hosts. These mutations can lead to tolerance of new hosts, drop out of diagnostic targets as has been observed for the SARS-CoV-2 B1.1.7 variant initially discovered in the United Kingdom, or resistance to current therapies (Behind The Bench Staff, 2020; Holmes, 2009; Sanjuán et al., 2010). As a result, there is great uncertainty in what viral species, variant, or virus exposure will lead to the appearance of a more pathogenic variant of an existing virus or a zoonotic spillover event. Given this uncertainty, diagnostics and therapeutics would ideally not require an in-depth understanding of viral biology, and these technologies should be robust to viral mutations or be easy to re-design when necessary.
Traditional diagnostic assays suffer from a trade-off between programmability and field deployability. Polymerase chain reaction (PCR) represents a gold standard for molecular diagnosis, and design of PCR primers and probes relies on sequence alone. However, the required laboratory infrastructure limits PCR’s use outside well-equipped diagnostic laboratories. Antigen-based tests represent a promising alternative for point-of-care testing, but their sensitivity is often lower and assay design can be much slower (Jayamohan et al., 2021).
Current approved antivirals require knowledge of the optimal viral or host protein targets and are largely bespoke. The effectiveness of small-molecule inhibitors has led to many being approved for human use, yet both the development and repurposing of previously tested inhibitors is a slow and tedious process (De Clercq and Li, 2016). Recently, large-scale proteomic discovery studies have been used to identify potential SARS-CoV-2 therapeutics. These approaches can help refine the list of drug candidates that could be repurposed and were used when our understanding of SARS-CoV-2 biology was still limited (Gordon et al., 2020a, 2020b). Nucleic-acid-based antivirals such as small-interfering RNAs (RNAi) have been tested in cell culture and animal models, but no RNAi-based therapeutics approved to date are antivirals (Kole et al., 2012; Warren et al., 2010).
Therefore, additional technologies that only require knowledge of the genomic sequence of a virus could be critical for successfully responding to future outbreaks. Clustered regularly interspaced short palindromic repeat (CRISPR)-associated (CRISPR-Cas) systems represent one such biological tool, as they primarily rely on nucleic acid sequence information for their activity. These systems have revolutionized our understanding of numerous biological systems and have already demonstrated extensive therapeutic potential for treating previously untreatable genetic diseases. Furthermore, continued assessment of microbial diversity has led to the discovery of additional CRISPR-Cas systems with different nucleic acid targets and enzymatic properties. These systems could be key for the development of both point-of-care diagnostics and programmable, sequence-based antiviral therapeutics.
This review describes how CRISPR offers an alternative approach for the detection of viral nucleic acids and the inhibition of viral replication, with a focus on the recent, accelerated development of these technologies due to the COVID-19 pandemic.
CRISPR-Cas systems
To protect against pathogens and foreign plasmids, prokaryotes have evolved systems to degrade foreign nucleic acids, to record these invasions as a form of immune memory, and to detect and destroy these invaders when they return. Most prokaryotic genomes contain CRISPR-Cas systems, which are characteristically composed of repetitive sequences (CRISPR arrays) and nearby CRISPR-associated (Cas) proteins (Barrangou et al., 2007; Marraffini and Sontheimer, 2008). A CRISPR array is defined as a series of palindromic direct repeats (DRs) separated by short, unique sequences called spacers. CRISPR arrays are bordered by Cas proteins that provide the enzymatic activity necessary for the three stages of CRISPR immunity: (1) adaptation, (2) CRISPR RNA (crRNA) biogenesis and expression, and (3) interference. During the adaptation stage, spacers derived from newly encountered foreign phages or plasmids are inserted into the CRISPR array. During crRNA biogenesis, CRISPR arrays are processed into individual, mature crRNAs (i.e., a single spacer and DR) that surveil the cell for foreign nucleic acids. Finally, during the interference stage, Cas proteins complexed with mature crRNAs cleave foreign nucleic acid molecules with sequence complementarity to the crRNA’s spacer sequence (see Hille et al., 2018 for a comprehensive review).
A number of CRISPR-Cas systems have been identified in nature; these have been organized into many categories defined by 2 classes, 6 types, and 33 subtypes. To date, all CRISPR-based technologies for viral nucleic acid detection and therapeutics use class 2 Cas effector proteins. Class 2 systems are simple to implement in mammalian systems because a single, multidomain effector protein is responsible for nucleic acid cleavage. This class is further divided into type II, V, or VI based on the Cas effector protein that mediates interference, and these types represent Cas9, Cas12, and Cas13, respectively (Makarova et al., 2020). Each Cas effector protein has distinct modes of action, described in detail below, that factor into their relative applications (Figure 1 ).
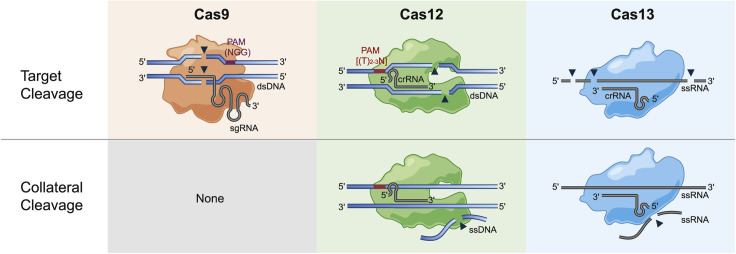
Figure 1.
Cleavage activity of class 2 CRISPR-Cas effector proteins
Purple box, Cas9 PAM; red box, Cas12 PAM; gray lines and curves, RNA; blue lines and curves, DNA; black triangles, cleavage; shaded object, Cas effector protein. Cas9 in complex with its single guide RNA (sgRNA) cleaves double stranded DNA (dsDNA) to create a double-stranded break at a precise location. Cas12 in complex with a crRNA creates a staggered break in dsDNA and the presence of a dsDNA target results in collateral cleavage of single-stranded DNA (ssDNA). The complex of Cas13 and its crRNA cleaves single-stranded RNA (ssRNA) often in multiple locations and in the presence of the ssRNA target Cas13 collaterally cleaves ssRNA. The orientation of Cas13’s crRNA is representative of subtypes VI-A/C/D. Cas9 does not possess collateral cleavage activity. See text for more detail.
crRNA-directed activity—target cleavage
Cas9 remains the most well known, characterized, and used class 2 Cas effector protein. The ability of Cas9 to edit DNA with exquisite specificity has transformed molecular biology, with applications including epigenome editing, mutagenesis screens, nucleic acid tracking, among others (see Pickar-Oliver and Gersbach, 2019 for a detailed review). Cas9 possesses a RuvC-like endonuclease and HNH nuclease domain that together generate a blunt double-stranded DNA (dsDNA) break in target sites with a 3′-NGG protospacer adjacent motif (PAM) (Jinek et al., 2012; Mojica et al., 2009). Breaks are precisely three nucleotides upstream of the PAM and repaired by host DNA repair pathways. Repair of these breaks results in insertions or deletions (indel) at the cleaved location. Cas9 activity also requires a trans-activating CRISPR RNA (tracrRNA) in addition to the crRNA; together, the crRNA and tracrRNA can be produced as a single RNA molecule termed the single guide RNA (sgRNA) (Jinek et al., 2012). As a result, Cas9-sgRNA complexes represent a simple, programmable tool for specific editing and disruption of dsDNA targets.
Cas12 is a more recently discovered RNA-guided, DNA-targeting Cas protein in which cleavage of either dsDNA or single-stranded DNA (ssDNA) is mediated by a single RuvC endonuclease domain. Cas12-based cleavage of dsDNA requires a 5′-(T)TTN PAM (Zetsche et al., 2015), yet Cas12 protein engineering has been used to change or increase the flexibility of its PAM sequence (Gao et al., 2017; Kleinstiver et al., 2019). Cas12 cleavage results in a staggered dsDNA break, distant from the PAM, with a 4–5 nucleotide overhang. Host DNA repair pathways ultimately create an indel at the cut site (Zetsche et al., 2015). Cas9 and Cas12 subtypes almost completely represent the known DNA-targeting class 2 Cas effector proteins, and their discovery has expanded the DNA sequences that can be cleaved and edited due to their differing PAM requirements.
While DNA-targeting Cas effector proteins represent the majority of currently described class 2 systems, the recently discovered, type VI effector Cas13 specifically cleaves single-stranded RNA (ssRNA). Cas13 was initially predicted to be an RNA-targeting protein because of the presence of two higher eukaryotes and prokaryotes nucleotide (HEPN)-binding domains. These domains have been associated with both antiviral and abortive infection mechanisms and are present in class 1 Cas effectors with RNA-targeting activity (Anantharaman et al., 2013; Shmakov et al., 2015). Cas13’s single-stranded RNA (ssRNA)-targeting activity was demonstrated via its ability to protect bacteria against infection by the RNA bacteriophage MS2 and its ability to cleave synthetic ssRNA targets (Abudayyeh et al., 2016).
Unlike Cas9 and Cas12, Cas13 targets RNA, and cleavage sites are not crRNA-location specific. Cas13 RNA cleavage does not require a PAM, yet initial studies showed conflicting evidence for a required sequence motif adjacent to the target site, named the protospacer flanking site (PFS) (Abudayyeh et al., 2016; East-Seletsky et al., 2016). Subsequent characterization of Cas13 illustrated that six nucleotides of perfect complementarity between the target RNA and the direct repeat sequence adjacent to the spacer completely inhibited Cas13 cleavage (Meeske and Marraffini, 2018). Given this sequence length and need for complete complementarity, the locations of Cas13 cleavage sites are not heavily restricted. In addition, Cas13 cleaves RNA at multiple locations. Identified cut sites are not at a precise location relative to the spacer but instead are dependent on RNA structure and nucleotide identity with specific preferences varying by Cas13 homolog (Abudayyeh et al., 2016; East-Seletsky et al., 2017). The discovery and characterization of Cas13’s RNA-targeting activity was essential for expanding CRISPR-Cas’s applications to programmable manipulation of RNA. As a result, the diversity of both nucleic acid targets and cleavage behavior discovered within class 2 CRISPR-Cas systems has enabled tractable applications for both viral dsDNA and ssRNA targeting.
Target-binding-mediated activity—collateral cleavage
Further characterization of type V and type VI Cas proteins, Cas12 and Cas13, identified a unique, target-activated, non-specific cleavage activity termed collateral cleavage. Collateral cleavage activity was first discovered for Cas13. Following RNA target recognition, Cas13 cleaved ssRNA in trans, and this trans-cleaved RNA did not need to include a sequence complementary to the provided Cas13 crRNA (Abudayyeh et al., 2016; East-Seletsky et al., 2016). Since this initial discovery, collateral cleavage activity was identified for multiple subtypes of Cas12. In the presence of a DNA target, Cas12 was shown to cleave ssDNA in trans. This cleaved ssDNA did not require a sequence complementary to the Cas12 crRNA (Chen et al., 2018; Harrington et al., 2018). For both Cas13 and Cas12, collateral cleavage activity was not observed when a complementary nucleic acid target was absent.
The discovery of collateral cleavage activity led to the development of numerous CRISPR-based diagnostics and viral detection technologies. Collateral cleavage has been harnessed for these technologies by introducing short synthetic ssRNA or ssDNA reporter molecules to the Cas protein-containing reactions. Fluorescent detection is achieved using a synthetic reporter flanked by a fluorescent molecule and quencher, and fluorescence is measured as the quencher becomes physically separated from the fluorescent protein in the presence of collateral cleavage (East-Seletsky et al., 2016). Alternatively, synthetic reporters flanked by fluorescein amidites (FAMs) and biotin have been paired with commercially available lateral flow test strips. Here, accumulation of FAM-bound gold nanoparticles (AuNPs) enables visualization of colored bands at one or two locations on the test strip: (1) the streptavidin location and (2) the FAM-binding antibody location. Separation of the biotin and FAM by collateral cleavage results in AuNP accumulation at the both locations instead of being entirely sequestered by streptavidin bound to the biotin (Gootenberg et al., 2018). For both formats, the signal generated by collaterally cleaved reporters is then used as a readout for the presence of a target sequence of interest.
CRISPR-based detection of viruses
Today, nucleic-acid-based diagnostics involve amplification and detection of a viral sequence by either PCR or isothermal amplification. Quantitative or qualitative PCR (qPCR) or reverse-transcriptase qPCR (RT-qPCR) is considered a gold-standard nucleic-acid-based method. The design of qPCR assays is straightforward and only requires knowledge of the viral sequence. However, the cost, sample-to-answer time, and personnel and equipment requirements limit widespread deployability. To eliminate the need for expensive thermal cycling equipment, numerous isothermal amplification methods have been developed and applied for virus detection. These approaches include loop-mediated amplification (LAMP) (Notomi et al., 2000), nucleic-acid-sequence-based amplification (NASBA) (Compton, 1991), recombinase polymerase amplification (RPA) (Piepenburg et al., 2006), and nicking enzyme amplification reaction (NEAR) (Abbott, 2020). Each approach has trade-offs in performance characteristics related to multiplexibility, available readouts, sensitivity, specificity, and testing throughput (Kurosaki et al., 2017; Notomi et al., 2000; Patel et al., 2016).
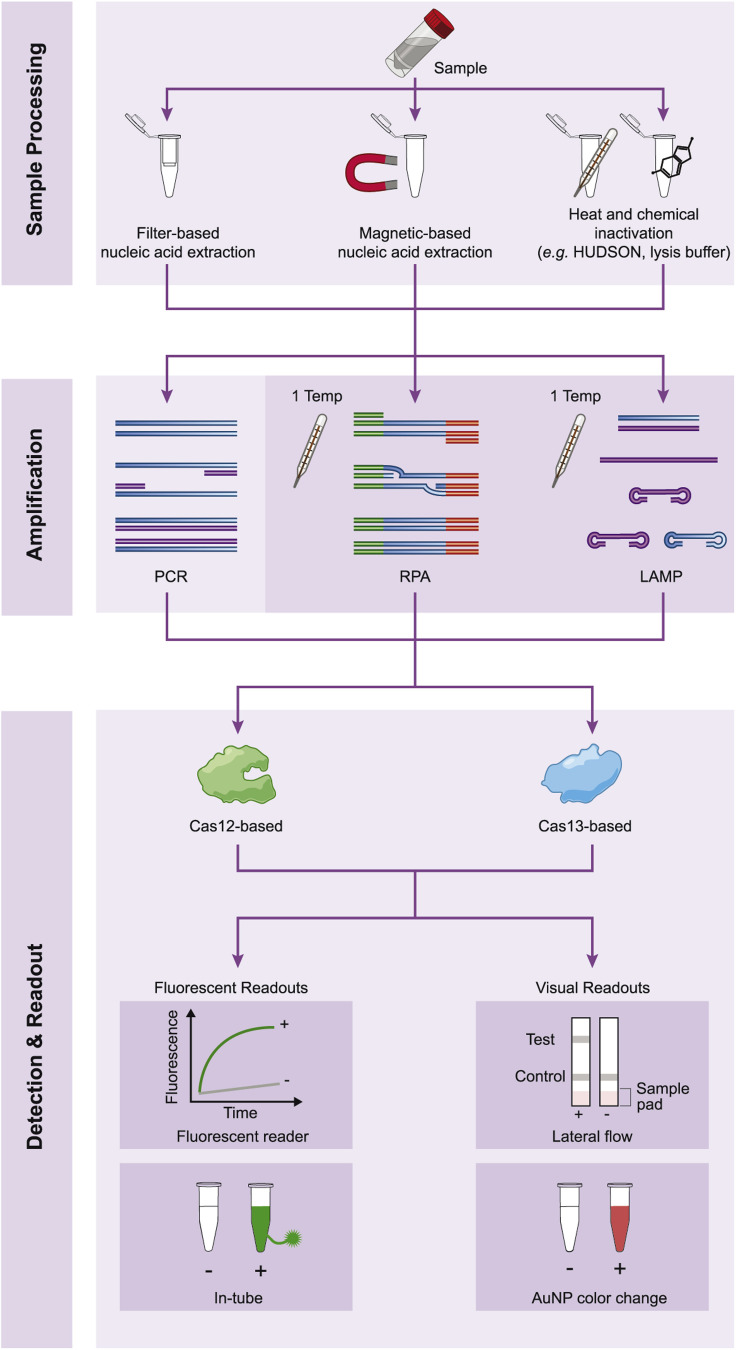
CRISPR-based diagnostic technologies can supplement current approaches because they are highly specific and sensitive but do not require expensive laboratory equipment. The sequence-specific nature of Cas-crRNA complexes inherently allows CRISPR-based technologies to have similar specificity to that of PCR. Cas13 and Cas12’s collateral cleavage does not require temperature cycling (Chen et al., 2018; East-Seletsky et al., 2016). Furthermore, collateral cleavage can be observed using reporters compatible with visual readouts, eliminating the need for expensive equipment like thermocyclers and fluorescent readers (Gootenberg et al., 2018). CRISPR-based detection technologies developed to date utilize different sample processing and amplification methods, Cas effector proteins, and readouts, and have been designed for a wide array of both DNA and RNA viruses, underscoring the versatility of these systems for detecting viral nucleic acids (Figure 2 ).
Figure 2.
Different experimental methods for CRISPR-based detection of viral nucleic acids
Most CRISPR-based detection methods require sample processing, amplification, and Cas12- or Cas13-mediated detection with a particular readout. Sample processing can be performed by filter-based or magnetic-based extraction or various chemical and/or heat inactivation approaches. Amplification is performed using PCR or isothermal methods such as RPA and LAMP. Detection by Cas12 and Cas13 can have either a fluorescent or visual readout.
Cas9-guided detection of viruses
Prior to the discovery of Cas effector proteins with collateral cleavage activity, inventive methods were developed that use Cas9’s specificity for identification of viral genes or differentiation between viral strains. In order to identify specific viral targets, Cas9 cleavage was combined with PCR in a method called CARP (Cas9/sgRNAs-associated reverse PCR), also known as ctPCR (CRISPR-typing PCR). This method involved amplifying a particular target sequence containing two physically distanced Cas9 PAM sites with PCR. The initial PCR resulted in sufficient levels of dsDNA that could then be targeted with two Cas9 sgRNAs. Iterative improvements of ctPCR (versions 1.0, 2.0, and 3.0) varied the reactions required after Cas9 cleavage to determine the presence or absence of the target. Target identification was achieved by PCR amplification of ligated adapters (Wang et al., 2018a), PCR amplification using reverse primers that only amplified a cleaved and ligated sequence (Zhang et al., 2018a), or qPCR amplification where relative efficiency between reactions with or without Cas9 were compared (Zhang et al., 2018b). In addition to ctPCR, NASBA-CRISPR cleavage (NASBACC) was developed, which combines NASBA amplification, toe-hold sensors, and Cas9 cleavage. Implementation of NASBACC required at least one divergent site between the sequences to be differentiated that disrupts Cas9’s PAM. If Cas9’s PAM was present in the target, Cas9 cleaved the toe-hold sensor binding site allowing for differential signals between the two targets (Pardee et al., 2016).
These Cas9 detection systems have been applied to a few viruses and viral strains. One key example is ctPCR’s use to detect human papillomavirus (HPV) genes in multiple subtypes of HPV16 and HPV18. Additionally, NASBACC was used to differentiate between Zika virus (ZIKV) American and Asian strains. Although these studies highlight the versatility of Cas9 for detecting and differentiating viruses, these methods require multiple reactions—either for amplification or for manipulation of the amplified products—which limits their deployability.
Cas13- and Cas12-based detection technologies
The characterization of Cas13 and Cas12’s collateral cleavage activity has led to the development of a number of user-friendly, CRISPR-based detection methods with potential for field deployability as well as massive scalability.
Following the characterization of Cas13’s collateral activity, the technology SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) was developed (Gootenberg et al., 2017; Kellner et al., 2019). SHERLOCKv1 harnessed the programmable, RNA-guided activity of Leptotrichia wadei (LwaCas13a), which had the optimal target-activated collateral cleavage activity known at the time. LwaCas13a alone could not detect the wide range of potential viral titers in patient samples, so the isothermal amplification method RPA (Piepenburg et al., 2006) was used to increase assay sensitivity. Because RPA results in a dsDNA product, SHERLOCK required T7-mediated in vitro transcription of the amplified product to generate RNA that can be detected by Cas13. Detection of the amplified target was achieved through the introduction of a synthetic ssRNA molecule flanked by a quencher and an attached fluorescent dye, and fluorescent signal generated by collateral cleavage was measured by a fluorescent reader.
Soon after, Cas12 was used for nucleic acid detection in other technologies such as DETECTR (DNA endonuclease targeted CRISPR trans reporter) (Chen et al., 2018) and HOLMES (1-h low-cost multipurpose highly efficient system) (Li et al., 2018). These methods eliminated the need for in vitro transcription after amplification because Cas12 can detect the direct product of amplification reactions (i.e., dsDNA). When Cas12’s collateral cleavage activity was triggered by the sensed dsDNA target, an introduced ssDNA quenched fluorescent reporter molecule was cleaved in trans. These two Cas12-based methods used different amplification approaches; in their initial publications, DETECTR employed RPA while HOLMES used PCR. These CRISPR-based detection methods can be applied across numerous contexts ranging from cancer to human genotyping, described in more detail in Wang et al. (2020a).
Cas13- and Cas12-based detection of a single virus
Cas13- and Cas12-based diagnostics have been shown to sensitively detect a wide range of viral targets with singleplex assays. SHERLOCK’s utility for detecting viral nucleic acids was first demonstrated amidst the Zika epidemic through its ability to detect low quantities of ZIKV in contrived lentivirus samples at known concentrations and in patient samples with a range of viral titers (Gootenberg et al., 2017). DETECTR was first used to detect HPV16 and HPV18 DNA in a set of 25 patient samples. When this technology was compared with gold-standard qPCR, DETECTR produced concordant results in all but two samples (Chen et al., 2018). A growing number of CRISPR-based assays have since been developed and validated for human viruses: Japanese encephalitis virus (JEV), Epstein-Barr virus (EBV) (Wu et al., 2019), Powassan virus (Normandin et al., 2020), H7N9 influenza virus (Liu et al., 2019), hantavirus (Curti et al., 2020), Ebola virus (EBOV), and Lassa virus (LASV; causative agent of Lassa fever) (Barnes et al., 2020). Due to the flexibility of these platforms, CRISPR-based assays could be developed for any viral pathogen with sufficient genomic information.
The emergence of SARS-CoV-2 in late 2019 highlighted how rapidly CRISPR-based detection methods can be tested and validated for a new virus once its genomic sequence is known. Almost immediately after the first SARS-CoV-2 genomes were published, new assays were being developed and posted on social media and preprint servers (Broughton et al., 2020a; Lucia et al., 2020; Metsky et al., 2020; Zhang et al., 2020), followed shortly by peer-reviewed publications. A SARS-CoV-2 assay was developed using the DETECTR method and validated on >70 patient samples, highlighting the speed at which these assays can be developed (Broughton et al., 2020b). Similarly, a SHERLOCK assay was validated on >150 patient samples in Thailand having strong concordance with RT-qPCR (Patchsung et al., 2020). Many more publications soon followed (Ackerman et al., 2020; Ali et al., 2020; Arizti-Sanz et al., 2020; Chen et al., 2020; Fozouni et al., 2021; Guo et al., 2020; Hou et al., 2020; Huang et al., 2020; Joung et al., 2020; Ning et al., 2020; Ramachandran et al., 2020; Wang et al., 2020b) with methods summarized in Table 1 .
Table 1.
SARS-CoV-2 RNA Detection Technologies
| Technology | Sample Processing | Amplification Method | Cas Effector Protein | Assay Readout | Companion Smartphone App | SARS-CoV-2 Target Gene | Patient Sample Validation |
Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of + Samples | No. of – Samples | Sensitivity (%) | Specificity (%) | ||||||||
| DETECTR | Nucleic acid extraction | RT-LAMP | Cas12 | Lateral flow | E | 36 | 42 | 95 | 100 | Broughton et al., 2020b | |
| N | 36 | 42 | 95 | 100 | |||||||
| CARMEN | Nucleic acid extraction | PCR | Cas13 | Fluorescent | Orf1ab | n.d. | n.d. | n.a. | n.a. | Ackerman et al. 2020 | |
| CASdetec | Chemical treatment | RT-RPA | Cas12 | Fluorescent | Orf1ab | n.d. | n.d. | n.a. | n.a. | Guo et al. 2020 | |
| SHERLOCK | Nucleic acid extraction | RT-RPA | Cas13 | Fluorescent | S | 81 | 453 | 100 | 100 | Patchsung et al., 2020 | |
| Lateral flow | S | 81 | 453 | 97 | 100 | ||||||
| CRISPR-COVID | Nucleic acid extraction | RT-RPA | Cas13 | Fluorescent | Orf1ab | 52 | 36 | 100 | 100 | Hou et al. 2020 | |
| CRISPR-FDS-v1.0 | Nucleic acid extraction | RT-RPA or RT-PCR | Cas12 | Fluorescent | N or Orf1ab | 19 | 10 | 100 | 71.4 | Huang et al. 2020 | |
| CRISPR/Cas12a-NER | Nucleic acid extraction | RT-RPA | Cas12 | Fluorescent | E | 16 | 15 | 100 | 100 | Wang et al. 2020 | |
| STOPCovidv1 | Chemical and heat treatment | RT-LAMP | Cas12 | Lateral flow | N | 62 | 22 | 84 | 100 | Joung et al. 2020 | |
| STOPCovidv2 | Chemical, heat, and magnetic treatment | Fluorescent | N | 200 | 200 | 93.1 | 98.5 | ||||
| iSCAN | Nucleic acid extraction | RT-LAMP | Cas12 | Fluorescent | E | 26 | 5 | 50 | 100 | Ali et al. 2020 | |
| Fluorescent | N | 26 | 5 | 88 | 100 | ||||||
| Lateral flow | N | 21 | 3 | 81 | 100 | ||||||
| Electric field-mediated SARS-CoV-2 detection | Chemical and heat treatment followed by isotachophoresis extraction | RT-LAMP | Cas12 | Fluorescent | N | 32 | 32 | 90.6 | 100 | Ramachandran et al. 2020 | |
| E | 32 | 32 | 90.6 | 100 | |||||||
| SHINE | Chemical and heat treatment (HUDSON) | RT-RPA | Cas13 | Fluorescent | X | Orf1ab | 42 | 40 | 93.3 | 100 | Arizti-Sanz et al. 2020 |
| Lateral flow | Orf1ab | 4 | 3 | 100 | 100 | ||||||
| Contamination-free, visual Cas12 assay | Nucleic acid extraction | RT-LAMP | Cas12 | Fluorescent | X | Orf1ab | 7 | 3 | 100 | 100 | Chen et al. 2020 |
| E | n.d. | n.d. | n.a. | n.a. | |||||||
| N | n.d. | n.d. | n.a. | n.a. | |||||||
| Direct, Cas13a detection | Nucleic acid extraction | n.a. | Cas13 | Fluorescent | X | E & N combined | 5 | 1 | 100 | 100 | Fozouni et al. 2020 |
| CRISPR-FDS-v2.0 | Chemical and heat treatment of saliva | RT-RPA | Cas12 | Fluorescent | Orf1ab | 44 | 59 | 100 | 100 | Ning et al. 2020 | |
| Fluorescent | X | Orf1ab | 44 | 59 | 97.7 | 100 | |||||
| Chemical and heat treatment of nasal swab | RT-RPA | Cas12 | Fluorescent | Orf1ab | 27 | 76 | 100 | 97.7 | |||
| Fluorescent | X | Orf1ab | 27 | 76 | 100 | 98.7 | |||||
The COVID-19 pandemic also created a path for emergency use authorization (EUA) by the FDA for CRISPR-based SARS-CoV-2 diagnostics. Soon after these CRISPR-based detection technologies were published, SHERLOCK Biosciences received an EUA for a CRISPR-based diagnostic for SARS-CoV-2, and Mammoth Biosciences soon followed (FDA, 2020c, 2020d)—the first FDA authorizations of a CRISPR-based diagnostic—underscoring the future potential of CRISPR-based diagnostics for becoming part of the standard selection of molecular assays for viral diagnosis.
Cas13- and Cas12-based detection of viral mutations
The sensitivity of Cas13 and Cas12 cleavage activity to mismatches between the target and crRNA enabled development of CRISPR-based assays that can identify single-nucleotide polymorphisms (SNPs). It was experimentally determined that the presence of a single mismatch between Cas13’s crRNA and the target RNA did not eliminate cleavage, but if a second mismatch was present, target cleavage and consequently collateral cleavage was disrupted. CRISPR-based SNP assays were then developed by designing crRNAs with a synthetic mismatch near the SNP, so the SNP would then act as the second mutation between the crRNA and the target sequence. In this way, there would be differential collateral cleavage signal depending on the presence or absence of the SNP in the target (Gootenberg et al., 2017).
CRISPR-based SNP assays have been developed for a variety of viruses and viral mutations. Early studies showed that Cas13 crRNAs could differentiate ZIKV strains, two dengue virus (DENV) serotypes, functional viral mutations, and drug resistance mutations (Gootenberg et al., 2017; Myhrvold et al., 2018). In an additional study, Cas13-based assays were developed for 27 common HIV drug resistance mutations: six reverse-transcriptase inhibitor mutations and 21 integrase inhibitor mutations (Ackerman et al., 2020). Similarly, Cas12 crRNAs can be designed for detection of specific mutations, and Cas12-based assays were developed to identify a SNP that differs between HPV16 and HPV18 (Teng et al., 2019). The specificity of Cas13 and Cas12 has therefore unlocked a new diagnostic application that was not possible for most simple molecular diagnostics and has significant implications for monitoring viral strains and drug resistance.
High-throughput and multiplexed CRISPR-based viral diagnostics
Diagnostic methods that can test many samples in parallel or surveil many viral targets have substantial advantages for outbreak testing and viral surveillance. Initial multiplex assays had amplification primers designed against conserved viral regions and Cas13 crRNAs that would specifically bind a specific viral species or subtype. A single amplification reaction could be used for all targets, and separate detection reactions were run in parallel. This approach was used to create a flavivirus panel as well as an assay to differentiate among the four serotypes of DENV (Myhrvold et al., 2018). However, the requirement of separate detection reactions limited this multiplexed method for high-throughput testing.
Detection reactions could be performed in a single tube per sample by harnessing different cleavage preferences of multiple Cas proteins or orthologs. Upon characterization of the nucleotide-specific cleavage preferences of Cas proteins, SHERLOCK reactions were developed that can detect up to four targets in a single reaction (Gootenberg et al., 2018). These assays applied the discovery that LwaCas13a cleaves at Us while different Cas13b orthologs cleave at As or UA, and Cas12 collaterally cleaves ssDNA. Two-target assays have been used to detect ZIKV and/or DENV in the same detection reaction (Gootenberg et al., 2018) or to identify the F260L mutation that differentiates the chronic or acute strains of the mammalian arenavirus, lymphocytic choriomeningitis virus (LCMV) (Freije et al., 2019). It is evident that varied cleavage preferences have important implications for the development of multiplexed CRISPR-based assays.
The integration of microfluidic approaches and CRISPR-based detection methods enabled both automated approaches and versatile methods to test hundreds of samples or targets in parallel. To eliminate reaction steps and increase the ease of testing, a portable microfluidic device was developed to detect EBOV RNA in up to 24 samples in parallel without amplification (Qin et al., 2019). In addition, microwell arrays and Cas13-based detection were combined in a highly multiplexed detection method called combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN)-Cas13 (Ackerman et al., 2020). Color-coded, nanoliter-sized droplets of pre-amplified samples and Cas13 detection reactions are generated, and all potential pairs of samples and detection reagents are arranged at random in a customized microfluidic chip. Paired droplets are imaged to map target and crRNA locations, and droplet pairs are then merged to initiate the detection reactions. In its initial instantiation, CARMEN-Cas13 was able to run nearly 5,000 tests at a time. It was also designed to be modular, so that thousands of samples could be simultaneously tested against a few viral targets or a few samples could be tested against all viral targets.
The scale of CARMEN-Cas13’s multiplexing was highlighted in the design of a pan-viral assay that allows for testing samples for all viruses known or suspected to infect humans with sufficient viral sequence data. Beyond this pan-viral assay, CARMEN-Cas13 was used for discriminating among all hemagglutinin (H1–H16) and neuraminidase (N1–N9) subtypes of FLUAV. The power of these microfluidic and other miniaturization approaches will enable more high-throughput approaches to diagnostic testing that could be useful both for active outbreak tracking and continuous viral surveillance.
Expanding the utility of CRISPR-based diagnostics beyond the laboratory
The potential for CRISPR-based diagnostics to be used at the point of care is a driving force in the enhancement of these technologies. To achieve this goal, limited (or even no) laboratory equipment or trained personnel should be necessary from sample collection to the result. Several advances are critical to bring point-of-care testing to fruition.
Simple sample inactivation methods are key to reducing hands-on and sample-to-answer time and achieving truly deployable diagnostics. New sample inactivation technologies include commercial lysis buffers and heat and chemical treatments, which eliminate the need for time consuming, filter- or robotic-based extraction methods. HUDSON (heating unextracted diagnostic samples to obliterate nucleases) uses chemical and heat treatment to inactivate nucleases in patient samples and lyse viral particles once these nucleases were inactivated (Myhrvold et al., 2018). Sample types tested included urine, saliva, and several blood products. HUDSON-treated samples could be directly added to SHERLOCK reactions without impairing sensitivity or specificity. HUDSON was initially applied to detect ZIKV and DENV from unextracted patient samples, but the method was later shown to be appropriate for other viruses and sample types. A modified HUDSON protocol was shown to sufficiently inactivate LASV with viral inactivation tested by plaque assay (Barnes et al., 2020). In addition, a 10-min HUDSON method could be performed in saliva and nasal swab collection media (Arizti-Sanz et al., 2020). Proteinase k and heat treatment has also been used for simplified sample processing, and this approach was sufficient to isolate HPV DNA from patient samples and used for DETECTR assays (Chen et al., 2018). Unlike many filter-based extraction methods, these early methods do not concentrate nucleic acids in a sample. However, recent SARS-CoV-2 detection methods have combined room-temperature lysis buffers and magnetic beads to concentrate nucleic acids with a simple and fast method (Joung et al., 2020).
Reducing the number of user manipulations following sample processing is essential for further simplification of these methods and reduction of the risk of target contamination. There are two approaches to combining amplification and detection reactions in a single tube: (1) physical separation of the reaction components within the same tube that can be mixed later or (2) identification of reaction conditions that eliminate the need for amplification or are compatible for both amplification and CRISPR-based cleavage. The former approach still requires the user to manipulate the tube, albeit without opening it, and has been tested using mineral oil (Chen et al., 2020) or placing reagents on the side of the tube (Ali et al., 2020) or in the tube’s cap (Guo et al., 2020), followed by shaking the tube after the initial incubation. Thermophilic Cas12 orthologs were used in HOLMESv2 and STOPCovid so that RT-LAMP and Cas12 detection could be performed in a single tube. These one-step methods were used for detection of JEV (Li et al., 2019) and SARS-CoV-2 (Joung et al., 2020). A single-step SHERLOCK method, as part of SHINE (streamlined highlighting of infections to navigate epidemics), combined RT-RPA and Cas13 cleavage into a single reaction to detect SARS-CoV-2 RNA. The ability to use a single reaction was achieved through optimization of the buffer conditions, the reporter, and enzymatic components (Arizti-Sanz et al., 2020). Finally, combining crRNAs and using a more active Cas13a ortholog, Leptotrichia buccalis Cas13a, has been shown to eliminate the need for amplification (Fozouni et al., 2021). These methods reduced hands-on time, and amplified products remained contained in a single tube, reducing the risk of contamination without reducing assay sensitivity.
The use of inexpensive, portable fluorometers and colorimetric readouts in the form of a lateral flow paper strip or color change are another key to field deployability. Green fluorescent signal can be measured using a portable, blue-light-emitting device and has been used to detect SARS-CoV-2 using both Cas12- and Cas13-based approaches (Arizti-Sanz et al., 2020; Fozouni et al., 2021; Guo et al., 2020; Ning et al., 2020; Wang et al., 2020b). Fluorescent readouts with portable readers combined with simple sample processing would allow these technologies to be implemented in pop-up labs.
Colorimetric readouts require no equipment for readout and are thus even more tractable for at-home testing. In the development of SHERLOCKv2, lateral flow readouts were paired with Cas13 detection of ZIKV, DENV, and viral SNPs by replacing the fluorescent reporter with a FAM-biotin ssRNA reporter. Results were then visualized when AuNPs accumulated at the control and/or test line of commercially available lateral flow strips (Gootenberg et al., 2018; Myhrvold et al., 2018). This same strategy has been applied for lateral flow-based detection of African swine fever virus (ASFV) using Cas12-based detection of amplified samples (Lu et al., 2020). Lateral flow readouts have also been used with Cas9-based detection methods. CASLFA (CRISPR-Cas9-mediated lateral flow nucleic acid assay) used lateral flow strips to detect ASFV. Samples were amplified by PCR or RPA using biotin-containing primers and an AuNP-coupled probe was introduced that could bind to an extended hairpin of Cas9’s sgRNA. When Cas9 and the sgRNA bound the biotin-containing amplified product, a visual band appeared on a lateral flow test strip (Wang et al., 2020c).
Colorimetric readouts that do not require individual paper test strips per sample would enhance the scalability of these equipment-free approaches. The isothermal, nucleic-acid-based amplification method LAMP can be performed with an in-tube, colorimetric readout. Samples, processed by either nucleic acid extraction or lysis, are heated at a single temperature, and amplification of the target nucleic acid causes a drop in pH visualized using a pH indicator (Poole et al., 2017). A pH-driven colorimetric readout has yet to be shown for CRISPR-based detection. However, in-tube colorimetric readouts for CRISPR-based assays can be accomplished by visualizing collateral cleavage activity using nucleic acid-coupled AuNP because the visual appearance of AuNPs is dictated by their proximity—aggregated AuNPs are clear while disaggregated AuNPs are red. Therefore, the color of a sample can be representative of the presence or absence of collateral cleavage activity. AuNP-based colorimetric assays have harnessed Cas12 to detect ASFV. Although Cas13-based assays developed in this study were not developed to detect ASFV, it is likely Cas13-based methods can extend to viral detection as well (Yuan et al., 2020). Fluorescent and AuNP readouts are more scalable, less prone to contamination, and thus could have increased utility in pop-up or clinical laboratories since a test strip does not need to be inserted into each reaction; yet for the at-home setting lateral-flow-based tests may be sufficient because sample numbers are small.
Software tools for CRISPR-based detection
Automated analysis of detection results using smartphone applications removes the risk of user bias in results interpretation. Both Cas13- and Cas12-based diagnostics have methods that capture fluorescent collateral cleavage via a portable commercial or a 3D-printed, blue-light-emitting device. In order to streamline analysis, smartphone applications have been developed and used to interpret and report results (Arizti-Sanz et al., 2020; Chen et al., 2020; Fozouni et al., 2021; Ning et al., 2020). Furthermore, for low-titer samples, the signal of the lateral flow test band is often difficult to distinguish from the background signal. Therefore, similar smartphone applications have been developed that can determine, in real time, the lateral flow results, and this application was validated for EBOV and LASV assays (Barnes et al., 2020). Most software tools developed to date report qualitative results per sample (i.e., positive or negative) and are used with single-target assays. As a result, companion smartphone applications for CRISPR-based detection assays are not yet amenable to multiplex assays, assays with in-sample internal controls, and often do not relay quantitative information such as virus levels in a given sample all of which can be achieved with laboratory-based tests like qPCR. Software tools are essential for standardization of these methods especially for assays that are used outside of laboratory settings.
The ability to deploy sensitive and specific diagnostics paired with companion smartphone applications would have immense benefits for viral surveillance and outbreak mitigation. The use of smartphone applications for infectious disease diagnostics allows for the combining of results with symptom reporting, geospatial mapping, and could ultimately enhance testing and tracing capabilities through real-time reporting of exposures and sharing of information with public health agencies. Altogether, this could give an integrated view of what and how viruses are circulating in a community and consequently inform public health responses. Achieving this goal, however, relies on tools that ensure patient data are secure and on broader access to mobile technologies and cellular networks (see Wood et al., [2019] for a comprehensive review).
CRISPR-based inhibition of viruses
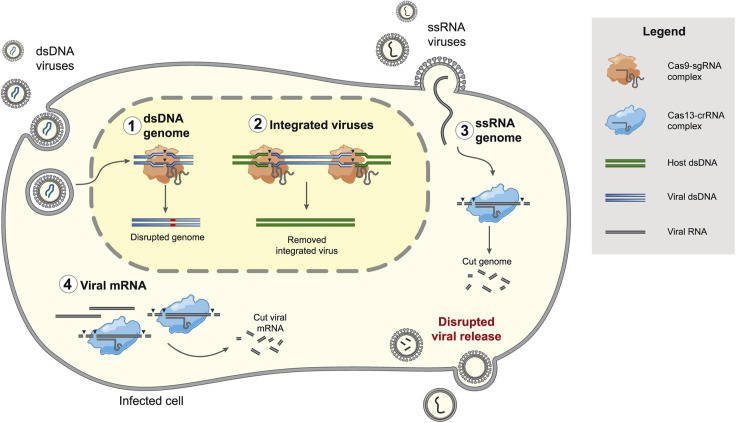
The antiviral activity of CRISPR-Cas systems, evolved in nature to protect prokaryotes against foreign nucleic acids, can be repurposed to inhibit mammalian viruses. Traditionally, the identification of antivirals requires screening many molecules, discovering antibodies, or an advanced understanding of the underlying viral biology. For CRISPR-Cas systems to target viruses, only knowledge of the viral sequence is required for crRNA design. Depending on the targeting properties of the Cas effector protein, CRISPR-Cas systems can be used to cleave the genome and/or intermediate nucleic acid species produced as part of the viral life cycle (Figure 3 ). As a result, pathogenesis of numerous DNA and RNA viruses has been disrupted by either Cas9 or Cas13.
Figure 3.
The stages of a viral life cycle inhibited by CRISPR-based targeting
Cas9, orange-shaded object; Cas13, blue-shaded object; black triangles, cleavage; blue and green lines, DNA; gray lines and curves, RNA; red line, indel. Cas9 and Cas13 inhibit viral replication through the specific targeting of either the viral genome or replication intermediates. Cas9 directly targets (1) dsDNA genomes or (2) integrated viruses. Cas13 targets (3) ssRNA viral genomes or any ssRNA intermediate created during the life cycle of either RNA or DNA viruses, such as (4) viral mRNAs. All targeting activity occurs within the cell once the nucleic acid is accessible to Cas9-sgRNA or Cas13-crRNA complexes and can result in disrupted viral release.
Targeting viruses with DNA genomes and intermediates using Cas9
Numerous DNA viruses represent potential targets for Cas9-mediated targeting because Cas9 can directly disrupt essential genes through the introduction of insertions or deletions. Multiple herpesviruses (EBV, herpes simplex virus 1 [HSV-1], human cytomegalovirus [HCMV]) have been inhibited in cell culture by Cas9-based cleavage of viral genomic DNA. This approach was more consistently effective when essential viral genes were targeted (van Diemen et al., 2016). In some cases, indels that disrupted Cas9 sgRNA binding still were translated into effective proteins (i.e., escape mutations). Therefore, delivery of multiple sgRNAs proved more effective. Cas9’s ability to target HSV-1 was tested in a latent infection model, but Cas9 could not restrict newly produced viral genomes in this model system. In another study, Cas9 disruption of the HSV-1 E3-ubiquitin ligase protein ICP0 strongly inhibited HSV-1 because ICP0 was no longer able to disrupt promyelocytic leukemia bodies, an essential component of HSV-1’s life cycle. Disruption of ICP0 by Cas9 was achieved through both plasmid transfection and lentivirus transduction (Roehm et al., 2016). These studies highlight the efficiency of Cas9-based disruption of DNA virus infection in cell culture and suggest that this approach could be applied to numerous DNA viruses.
Latent hepatitis B virus (HBV) infection cannot be treated with conventional antivirals, and Cas9 targeting offers an alternative approach. Cas9 targeting studies have focused on developing sgRNAs that target the four predominant HBV genotypes, A–D. Cas9 with sgRNAs designed against conserved targets across these genotypes reduced expression of multiple HBV proteins and the intermediate covalently closed circular DNA (Wang et al., 2015). Further, this study showed that delivery of multiple sgRNAs had an additive effect on the reduction of HBV infection. Cas9-mediated inhibition of HBV infection has also been demonstrated in multiple mouse models including a de novo infection model (Li et al., 2016; Liu et al., 2015; Ramanan et al., 2015; Zhen et al., 2015). Of note, another hepatitis virus, hepatitis C virus, which instead has an RNA genome, has been targeted with an RNA-targeting ortholog of Cas9, Francisella novicida Cas9 (Price et al., 2015). It is evident that hepatitis viruses can be effectively inhibited by Cas9 in both cell culture and mouse models of infection, and there are multiple target sites that are effective in reducing protein levels and consequently infection.
Although highly active antiretroviral therapy (HAART) has proven very effective at managing HIV-1 levels and disease, Cas9-based approaches represent a potential avenue to eliminate integrated HIV provirus. Initial studies of Cas9-targeting of HIV-1 used long terminal repeat (LTR)-targeting sgRNAs and demonstrated that HIV-1 proviral DNA could be disrupted. This disruption resulted in less HIV-1 RNA following reactivation in Jurkat cells (Ebina et al., 2013). Cas9 targeting of HIV-1 proviral DNA was also effective in latently infected microglia, monocytes, and T cells (Hu et al., 2014). It was later demonstrated that even when sites of HIV-1 integration were transcriptionally silent, Cas9 could still target the LTRs and reduce levels of reactivated virus (Zhu et al., 2015). These initial successes prompted the study of Cas9’s effectiveness against reducing or eliminating HIV-1 levels in mouse models of infection (Bella et al., 2018; Dash et al., 2019; Yin et al., 2017).
Cas9 delivery is a key area of work for assessing the effectiveness of Cas9-based antivirals. Early studies required the use of smaller orthologs of Cas9, such as Staphylococcus aureus (saCas9), in order to package Cas9 into adeno-associated viruses (AAVs). Delivery of saCas9 with four sgRNAs successfully excised proviral HIV-1 DNA from multiple tissues and organ types using multiple mouse models of HIV-1 infection including humanized bone-marrow-liver-thymus (BLT) mice (Yin et al., 2017). Combination therapeutic approaches have been the most effective at removing proviral HIV-1 in vivo. When sequential long-acting slow-effective release antiviral therapy (LASER ART) and AAV-delivered Cas9 were used to treat HIV-1-infected humanized mice, proviral RNA was eliminated, and HIV-1 was not detected in blood or tissue. Adoptive transfer of cells into non-infected mice did not result in any viral progeny. However, complete elimination was only achieved for half of the dual-treated animals, and Cas9 treatment alone was not sufficient to eliminate HIV-1 completely (Dash et al., 2019). Lentiviral delivery systems have also been tested since they do not have the same size limitations as AAVs. HIV-1 provirus was successfully excised by the larger, more commonly used Streptococcus pyogenes Cas9 (SpCas9) from human PBMCs engrafted into NON-SCID mice. SpCas9 targeting resulted in lower levels of full-length proviral DNA and reduced viral outgrowth measured in the blood and spleen (Bella et al., 2018). In addition, lentivirus transduction of tat- and rev-targeting Cas9 sgRNAs reduced HIV-1 expression significantly and had strong activity in latent CD4+ T cells (Ophinni et al., 2018). Therefore, Cas9 represents a promising strategy for removing integrated HIV-1, a target difficult to disrupt using standard antiviral approaches.
Cas9 can also be used to target host factors as an alternative strategy for curing disease. The possibility of targeting host factors to cure HIV-1 gained much attention with the finding that complete elimination of HIV-1 from individuals could be achieved using transplantation of HIV co-receptor-null, specifically CCR5-null, hematopoietic stem cells (Gupta et al., 2019; Hütter et al., 2009). HIV-1 entry into cells requires engagement with CD4 and either CCR5 or CXCR4. Cas9 was successfully used to knockout CCR5 in hematopoietic stem cells, and adoptive transfer into mouse models of HIV-1 infection resulted in resistance to infection (Xu et al., 2017). Further, Cas9 can be used to generate CCR5/CXCR4 double knockout primary T cells, and these cells were then resistant to both CCR5- and CXCR4-tropic HIV infection (Yu et al., 2018). Approved use of Cas9 for treatment of disease in humans is occurring sparingly as the safety of Cas9 gene editing is still being evaluated and can have ethical dangers of misuse (Cyranoski and Ledford, 2018). Yet a recent study investigated the effects of transplanting CCR5-disrupted hematopoietic stem and progenitor cells (HSPCs), edited ex vivo, into an HIV-1-infected individual with acute lymphoblastic leukemia. CCR5-disrupted cells successfully engrafted and differentiated, and no adverse effects of the Cas9-edited cells were observed within the first 19 months. However, transplantation was not sufficient to cure the patient of infection (Xu et al., 2019).
Disrupting viral proteins using Cas9 can also reveal unknown functional components of viral infection. Functional investigation of EBV in multiple cell lines was tested by Cas9-based removal of the promoter region upstream of latent-transcript BART (BamHI A rightward transcript). Removal of this region by Cas9 consequently disrupted BART expression and was used to identify BART’s promoter sequence (Yuen et al., 2015). In another study, potential resistance mutations in HIV-1’s primer-binding sites were identified by introducing indels at this site using Cas9. These experiments identified multiple escape mutations and variable escape mechanisms which informed both Cas9 sgRNA design and HIV-1 biology (Wang et al., 2018b).
Whether Cas9-based targeting of DNA virus genomes or of essential host factors will be practical clinically involves further investigation of its effectiveness and off-target effects. Directly targeting viruses in infected cells requires efficient expression and minimal off-target activity, especially because any off-target effects will create a permanent change in the treated cells. Cas9-based antiviral activity will need to be enhanced as both mouse and human studies have shown that Cas9 alone is not sufficient to eliminate disease, although combined with other antivirals could result in a cure. Cas9 immunogenicity is also a concern for clinical use. As a result, most advanced Cas9 human clinical studies to date involve Cas9’s use ex vivo followed by transplantation of edited cells (Mehta and Merkel, 2020; Pickar-Oliver and Gersbach, 2019).
Targeting viral mRNA or RNA genomes with Cas13
A majority of pathogenic viruses have ssRNA genomes, making Cas13 a powerful tool for disrupting RNA virus genomes and consequently inhibiting viral replication. Earlier studies illustrated that Cas13a (Abudayyeh et al., 2017) and Cas13b (Cox et al., 2017) could be used to specifically reduce expression of host mRNA in mammalian cells. Building upon this work, Cas13 was used to reduce levels of viral RNA for three distinct RNA viruses: LCMV, FLUAV, and vesicular stomatitis virus. This work demonstrated Cas13’s ability to efficiently reduce RNA levels even when RNA levels were quickly increasing (Freije et al., 2019). This study also reported the results of a tiled screen against both strands of LCMV, illuminating targeting hotspots and principles that could inform crRNA design such as sequence conservation and enrichment of cleavage nucleotides. An additional systematic evaluation of Cas13-based targeting of green fluorescent protein mRNA and multiple host transcripts determined the following factors for effective targeting in cells: (1) limited secondary structure of the crRNA, (2) enrichment of cytosines nearby the target, and (3) enrichment of the preferred cleavage nucleotide, in this study Us, upstream of the target (Wessels et al., 2020).
Subsequent studies have expanded our understanding of Cas13 targeting and demonstrated Cas13’s ability to inhibit additional RNA viruses. Cas13a has since been shown to reduce levels of HIV-1 RNA, viral particles, integrated viral DNA, and the RNA from reactivated provirus (Yin et al., 2020). DENV2 viral RNA levels and infectivity were reduced using Cas13a and NS3-targeting crRNAs (Li et al., 2020). In addition, Cas13b expression in multiple mosquito cell lines reduced luciferase expression in a Chikungunya virus-split replication system, using individually expressed crRNAs and the crRNA expressed from multiple positions within a CRISPR array (Tng et al., 2020).
Because Cas13’s antiviral activity can be tested as soon as viral sequence information is known, Cas13’s ability to reduce SARS-CoV-2 infection was quickly investigated both in vitro and in vivo. A few months after the start of the COVID-19 pandemic, PACMAN (prophylactic antiviral CRISPR in human cells) was developed (Abbott et al., 2020). This method harnessed the potent activity of Cas13d (i.e., CasRx), an ortholog that is small enough to be packaged into AAVs (Konermann et al., 2018). PACMAN was used to reduce FLUAV levels in human lung epithelial cells. In addition, crRNAs were designed against conserved regions of human-infecting coronaviruses, including SARS-CoV-2, and Cas13 activity was assessed using a SARS-CoV-2 reporter system (Abbott et al., 2020). Subsequent work demonstrated that mRNA-encoding Cas13a and virus-targeting crRNAs, formulated with a PBAE-based polymer, could be delivered to mice or hamsters using a nebulizer. The effectiveness of this mRNA-based delivery was tested in mouse models of FLUAV infection and hamster models of SARS-CoV-2 infection. For both in vivo models, Cas13a and crRNA delivery followed by viral infection lead to reduced weight loss and reduced viral loads in harvested lung tissue albeit with modest effect sizes (Blanchard et al., 2021).
The implications of Cas13’s collateral cleavage activity in the context of cellular mRNA or viral RNA targeting is still unfolding. It has been shown for several Cas13 orthologs that targeting host mRNA (Abudayyeh et al., 2017; Cox et al., 2017; Konermann et al., 2018) or viral RNA (Blanchard et al., 2021) has limited to no off-target transcriptomic effects, and cell viability is not affected when a non-cytotoxic virus is targeted by Cas13 (Freije et al., 2019). However, collateral cleavage has been observed in glioma cells where Cas13a was overexpressed using lentiviral transduction and when Cas13a targeted an overexpressed RNA (Wang et al., 2019). Despite the collateral cleavage observed in glioma cell culture, use of these Cas13-expressing cells in mouse models of intracranial glioblastoma was still effective in inhibiting tumor growth. Given the methodological differences between these studies related to the Cas13 ortholog used, Cas13 expression, and target RNA expression, further work is needed to evaluate which contexts lead to collateral cleavage in mammalian cells and whether this has implications for the safety of Cas13 as a therapeutic.
Cas13-based antivirals could be a promising platform for counteracting risks of antiviral resistance, but their effectiveness and safety require further investigation before realizing their clinical impact. Designing Cas13 crRNAs to target conserved regions is one such way to select target sites where resistance is less likely to emerge. Furthermore, Cas13’s ability to process its own CRISPR array (Shmakov et al., 2015) makes multiplexing simple, increases viral RNA reduction, and could reduce the likelihood of mutations eliminating Cas13’s antiviral activity. The propensity for Cas13 resistance to emerge has been moderately studied in cell culture. Viral RNA in the supernatant of Cas13-treated cells infected with LCMV (Freije et al., 2019) or DENV2 (Li et al., 2020) was sequenced. Mutations were not observed within the crRNA-binding site; however, in supernatant from DENV2 infected cells insertions and deletions flanking the target site were found. Similar studies should be performed in vivo and could inform crRNA design. Ultimately, Cas13’s application as an antiviral will be dependent on efficient delivery to infected cells and limited to no off-target effects of Cas13 expression and cleavage in the host. Also, studies to date have primarily shown Cas13’s effect when expressed prior to infection and in vivo results had quite modest effect sizes. Therefore, more studies assessing whether Cas13’s activity is sufficient to overcome replication at various stages of infection are needed. Lastly, for viruses with approved antivirals, Cas13’s effectiveness would need to equal or surpass that of current approaches.
Outlook and future directions
The ongoing discovery and characterization of CRISPR-Cas systems is continuing to expand our toolkit for the study and benefit of mammalian systems, including our efforts to combat viral infectious disease. The importance of alternative technologies for diagnostics and therapeutics have been underscored in the face of COVID-19, as diagnostic testing fails to be readily available and antiviral candidates identified thus far improve outcomes but do not cure disease.
Substantial efforts have been made to expand the utility of CRISPR-based diagnostics with the ultimate goal of testing at the point of care, anywhere in the world, or for comprehensive surveillance. In order to fully achieve this goal, integrating simple sample processing methods, single-step assays that do not require heat, and a visual readout that can be interpreted by a small device, are essential. For diagnostic technologies, it is important to consider sample-to-answer time, the limit of detection, cold-chain requirements, and assay and equipment cost, as each plays a role in the ultimate global adoption of these technologies for clinical diagnosis. The sensitivity, specificity, and adaptability of CRISPR-based tests alongside advances in miniaturization also pave the way for massively multiplexed testing. Further development and extensive validation on patient samples is still needed for these nascent technologies.
In order for CRISPR-based antivirals to be used clinically, many future studies are needed to determine the ideal approach for delivery and expression in relevant tissues. Viruses infect numerous cell types and tissues, and the specifics differ by viral pathogen. They replicate in different compartments of the cell, and in some cases, antivirals may need to be readministered. These characteristics represent a challenge for delivery and will ultimately require testing various modalities and dosages (see Xu et al. [2021] for a review on in vivo delivery of CRISPR-Cas systems). Although viral vectors remain a common strategy for delivery, there remain questions in regard to their safety in humans. For example, recent delivery of high doses of AAVs have been fatal in a small number of individuals (Nature Biotech News, 2020), and AAVs are not practical if transient expression of the Cas effector protein is preferred. RNA-based delivery is a promising approach for transient Cas protein and crRNA or sgRNA expression and can be delivered using lipid nanoparticles (LNPs) for which high-throughput methods to identify tissue-specific LNPs have been developed (Sago et al., 2018). Two challenges of particular interest—delivery of ribonucleoprotein complexes (RNPs) and delivery to epithelial tissues—were recently overcome using engineered cell-penetrating peptides. These peptides delivered Cas9-sgRNA and Cas12-crRNA RNPs to airway epithelial cells (Krishnamurthy et al., 2019). Continued study and the development of new delivery modalities is essential for the adoption and use of in vivo CRISPR-Cas therapeutics.
Even if optimal delivery approaches are identified, the safety of Cas expression and efficacy as an antiviral in vivo need further investigation. As mentioned above, transplantation of ex vivo Cas9-edited cells into an HIV-infected patient was not sufficient to cure disease (Xu et al., 2019), and in vivo demonstrations of Cas13 antiviral activity only modestly reduced virus levels in mice and hamsters (Blanchard et al., 2021). Continued evaluation and improvement of the efficacy of these approaches and how viruses evolve in response to CRISPR-based targeting are essential next steps. Larger effect sizes and reduced off-target cleavage effects could be mitigated by additional understanding of sgRNA and crRNA design principles and Cas protein engineering, both already being investigated for Cas9 (Kleinstiver et al., 2016; Slaymaker et al., 2016). Because direct targeting of viral genomes relies on in vivo expression, obstacles related to protein immunogenicity will need to be overcome. In addition, CRISPR-based antiviral technologies will need to be compared with the standard of care for those viruses with approved therapeutics or vaccines.
In summary, it is evident that in face of current and future viral outbreaks CRISPR-based technologies have significant potential to add to our toolbox of diagnostics and antivirals, and these sequence-based countermeasures could be a key to lasting and effective pandemic preparedness.
Acknowledgments
We would like to thank C. Boehm, C. Myhrvold, J. Arizti-Sanz, and N. Welch for reading the manuscript and offering comments and suggestions. This work was supported by Merck KGaA’s Future Insight Prize, Defense Advanced Research Projects Agency (D18AC00006), Flu Lab, and a cohort of generous donors through TED’s Audacious Project, including the ELMA Foundation, MacKenzie Scott, the Skoll Foundation, and Open Philanthropy. The views, opinions and/or findings expressed are those of the author(s) and should not be interpreted as representing the official views or policies of the Department of Defense or the US Government.
Declaration of interests
C.A.F. and P.C.S. are inventors on patent filings related to SHERLOCK, SHINE, and Cas13-based antiviral technologies. P.C.S. is a co-founder of, shareholder in, and advisor to SHERLOCK Biosciences, as well as a Board member of and shareholder in Danaher Corporation.
References
- Abbott Id NOW™ COVID-19. 2020. https://www.globalpointofcare.abbott/en/product-details/id-now-covid-19.html
- Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., Ye S.H., Boehm C.K., Kosoko-Thoroddsen T.-S.F., Kehe J., et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M., et al. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Makarova K.S., Burroughs A.M., Koonin E.V., Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.-S.F., Kemball M.E., et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K.G., Lachenauer A.E., Nitido A., Siddiqui S., Gross R., Beitzel B., Siddle K.J., Freije C.A., Dighero-Kemp B., Mehta S.B., et al. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat. Commun. 2020;11:4131. doi: 10.1038/s41467-020-17994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bella R., Kaminski R., Mancuso P., Young W.B., Chen C., Sariyer R., Fischer T., Amini S., Ferrante P., Jacobson J.M., et al. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol. Ther. Nucleic Acids. 2018;12:275–282. doi: 10.1016/j.omtn.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard E.L., Vanover D., Bawage S.S., Tiwari P.M., Rotolo L., Beyersdorf J., Peck H.E., Bruno N.C., Hincapie R., Michel F., et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00822-w. [DOI] [PubMed] [Google Scholar]
- Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Singh J., Streithorst J., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., et al. medRxiv; 2020. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay.https://www.medrxiv.org/content/10.1101/2020.03.06.20032334v2 [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shi Y., Chen Y., Yang Z., Wu H., Zhou Z., Li J., Ping J., He L., Shen H., et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: a promising method in the point-of-care detection. Biosens. Bioelectron. 2020;169:112642. doi: 10.1016/j.bios.2020.112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curti L.A., Pereyra-Bonnet F., Repizo G.D., Fay J.V., Salvatierra K., Blariza M.J., Ibañez-Alegre D., Rinflerch A.R., Miretti M., Gimenez C.A. CRISPR-based platform for carbapenemases and emerging viruses detection using Cas12a (Cpf1) effector nuclease. Emerg. Microbes Infect. 2020;9:1140–1148. doi: 10.1080/22221751.2020.1763857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D., Ledford H. Genome-edited baby claim provokes international outcry. Nature. 2018;563:607–608. doi: 10.1038/d41586-018-07545-0. [DOI] [PubMed] [Google Scholar]
- Dash P.K., Kaminski R., Bella R., Su H., Mathews S., Ahooyi T.M., Chen C., Mancuso P., Sariyer R., Ferrante P., et al. Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice. Nat. Commun. 2019;10:2753. doi: 10.1038/s41467-019-10366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A., O’Connell M.R., Burstein D., Knott G.J., Doudna J.A. RNA targeting by functionally orthogonal Type VI-A CRISPR-Cas enzymes. Mol. Cell. 2017;66:373–383.e3. doi: 10.1016/j.molcel.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H.D., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial High-dose AAV gene therapy deaths. Nat. Biotechnol. 2020;38:910. doi: 10.1038/s41587-020-0642-9. [DOI] [PubMed] [Google Scholar]
- FDA New drugs at FDA. 2020. https://www.fda.gov/drugs/development-approval-process-drugs/new-drugs-fda-cders-new-molecular-entities-and-new-therapeutic-biological-products
- FDA Vaccines licensed for use in the United States. 2020. https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states
- FDA Sherlock crispr SARS-CoV-2 kit. 2020. https://www.fda.gov/media/137746/download
- FDA SARS-CoV-2 RNA DETECTR assay. 2020. https://www.fda.gov/media/139937/download
- Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D’Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333.e9. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije C.A., Myhrvold C., Boehm C.K., Lin A.E., Welch N.L., Carter A., Metsky H.C., Luo C.Y., Abudayyeh O.O., Gootenberg J.S., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837.e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Cox D.B.T., Yan W.X., Manteiga J.C., Schneider M.W., Yamano T., Nishimasu H., Nureki O., Crosetto N., Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370 doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K., Abdul-Jawad S., McCoy L.E., Mok H.P., Peppa D., Salgado M., Martinez-Picado J., Nijhuis M., Wensing A.M.J., Lee H., et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;568:244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F., Richter H., Wong S.P., Bratovič M., Ressel S., Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- Holmes E.C. The evolutionary genetics of emerging viruses. Annu. Rev. Ecol. Evol. Syst. 2009;40:353–372. [Google Scholar]
- Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J., et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O., et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H., Beeman M., Mahmood T., Nze U., Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021;413:49–71. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.G., Woolley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., et al. Detection of SARS-CoV-2 with Sherlock one-pot testing. N. Engl. J. Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Sherlock: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I., et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S., Wohlford-Lenane C., Kandimalla S., Sartre G., Meyerholz D.K., Théberge V., Hallée S., Duperré A.M., Del’Guidice T., Lepetit-Stoffaes J.-P., et al. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 2019;10:4906. doi: 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Y., Martins D.B.G., Kimura M., Catena A.D.S., Borba M.A.C.S.M., Mattos S.D.S., Abe H., Yoshikawa R., de Lima Filho J.L., Yasuda J. Development and evaluation of a rapid molecular diagnostic test for Zika virus infection by reverse transcription loop-mediated isothermal amplification. Sci. Rep. 2017;7:13503. doi: 10.1038/s41598-017-13836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Sheng C., Liu H., Liu G., Du X., Du J., Zhan L., Li P., Yang C., Qi L., et al. An effective molecular target site in hepatitis B virus S gene for Cas9 cleavage and mutational inactivation. Int. J. Biol. Sci. 2016;12:1104–1113. doi: 10.7150/ijbs.16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang S., Dong X., Li Q., Li M., Li J., Guo Y., Jin X., Zhou Y., Song H., Kou Z. CRISPR-Cas13a cleavage of dengue virus NS3 gene efficiently inhibits viral replication. Mol. Ther. Nucleic Acids. 2020;19:1460–1469. doi: 10.1016/j.omtn.2020.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-Cas12b-Assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hao R., Chen S., Guo D., Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J. Gen. Virol. 2015;96:2252–2261. doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xu H., Liu C., Peng L., Khan H., Cui L., Huang R., Wu C., Shen S., Wang S., et al. CRISPR-Cas13a nanomachine based simple technology for avian influenza A (H7N9) virus on-site detection. J. Biomed. Nanotechnol. 2019;15:790–798. doi: 10.1166/jbn.2019.2742. [DOI] [PubMed] [Google Scholar]
- Lu S., Li F., Chen Q., Wu J., Duan J., Lei X., Zhang Y., Zhao D., Bu Z., Yin H. Rapid detection of African swine fever virus using Cas12a-based portable paper diagnostics. Cell Discov. 2020;6:18. doi: 10.1038/s41421-020-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucia C., Federico P.-B., Alejandra G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.02.29.971127v1 [Google Scholar]
- Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske A.J., Marraffini L.A. RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell. 2018;71:791–801.e3. doi: 10.1016/j.molcel.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Merkel O.M. Immunogenicity of Cas9 protein. J. Pharm. Sci. 2020;109:62–67. doi: 10.1016/j.xphs.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsky H.C., Freije C.A., Kosoko-Thoroddsen T.-S.F., Sabeti P.C., Myhrvold C. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.02.26.967026v2 [Google Scholar]
- Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading) 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Morse S.S., Mazet J.A.K., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Rabiela F., Wiratsudakul A., Suzán G., Rico-Chávez O. Viral networks and detection of potential zoonotic viruses in bats and rodents: A worldwide analysis. Zoonoses Public Health. 2019;66:655–666. doi: 10.1111/zph.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B., Yu T., Zhang S., Huang Z., Tian D., Lin Z., Niu A., Golden N., Hensley K., Threeton B., et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2020;7 doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normandin E., Solomon I.H., Zamirpour S., Lemieux J., Freije C.A., Mukerji S.S., Tomkins-Tinch C., Park D., Sabeti P.C., Piantadosi A. Powassan virus neuropathology and genomic diversity in patients with fatal encephalitis. Open Forum Infect. Dis. 2020;7:ofaa392. doi: 10.1093/ofid/ofaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophinni Y., Inoue M., Kotaki T., Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci. Rep. 2018;8:7784. doi: 10.1038/s41598-018-26190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Patel P., Abd El Wahed A., Faye O., Prüger P., Kaiser M., Thaloengsok S., Ubol S., Sakuntabhai A., Leparc-Goffart I., Hufert F.T., et al. A field-deployable reverse transcription recombinase polymerase amplification assay for rapid detection of the Chikungunya virus. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole C.B., Li Z., Alhassan A., Guelig D., Diesburg S., Tanner N.A., Zhang Y., Evans T.C., Jr., LaBarre P., Wanji S., et al. Colorimetric tests for diagnosis of filarial infection and vector surveillance using non-instrumented nucleic acid loop-mediated isothermal amplification (NINA-LAMP) PLoS One. 2017;12 doi: 10.1371/journal.pone.0169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.A., Sampson T.R., Ratner H.K., Grakoui A., Weiss D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA. 2015;112:6164–6169. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Park M., Alfson K.J., Tamhankar M., Carrion R., Patterson J.L., Griffiths A., He Q., Yildiz A., Mathies R., Du K. Rapid and fully microfluidic Ebola virus detection with CRISPR-Cas13a. ACS Sens. 2019;4:1048–1054. doi: 10.1021/acssensors.9b00239. [DOI] [PubMed] [Google Scholar]
- Ramachandran A., Huyke D.A., Sharma E., Sahoo M.K., Huang C., Banaei N., Pinsky B.A., Santiago J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V., Shlomai A., Cox D.B.T., Schwartz R.E., Michailidis E., Bhatta A., Scott D.A., Zhang F., Rice C.M., Bhatia S.N. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehm P.C., Shekarabi M., Wollebo H.S., Bellizzi A., He L., Salkind J., Khalili K. Inhibition of HSV-1 replication by gene editing strategy. Sci. Rep. 2016;6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sago C.D., Lokugamage M.P., Paunovska K., Vanover D.A., Monaco C.M., Shah N.N., Gamboa Castro M., Anderson S.E., Rudoltz T.G., Lando G.N., et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA. 2018;115:E9944–E9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behind the bench staff Solutions for surveillance of the S gene mutation in the B.1.1.7 SARS-CoV-2 variant. Thermo Fisher Scientific, 31 December, 2020. 2020. https://www.thermofisher.com/blog/behindthebench/solutions-for-surveillance-of-the-s-gene-mutation-in-the-b117-501yv1-sars-cov-2-strain-lineage/#:∼:text=Next%2Dgeneration%20sequencing%20(NGS),of%20SARS%2DCoV%2D2
- Teng F., Guo L., Cui T., Wang X.G., Xu K., Gao Q., Zhou Q., Li W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20:132. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tng P.Y.L., Carabajal Paladino L., Verkuijl S.A.N., Purcell J., Merits A., Leftwich P.T., Fragkoudis R., Noad R., Alphey L. Cas13b-dependent and Cas13b-independent RNA knockdown of viral sequences in mosquito cells following guide RNA expression. Commun. Biol. 2020;3:413. doi: 10.1038/s42003-020-01142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., Yang H., Chen X., Recuenco S., Gomez J., et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diemen F.R., Kruse E.M., Hooykaas M.J.G., Bruggeling C.E., Schürch A.C., van Ham P.M., Imhof S.M., Nijhuis M., Wiertz E.J.H.J., Lebbink R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xu Z.W., Liu S., Zhang R.Y., Ding S.L., Xie X.M., Long L., Chen X.M., Zhuang H., Lu F.M. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J. Gastroenterol. 2015;21:9554–9565. doi: 10.3748/wjg.v21.i32.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang R., Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 2020;165:112430. doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu X., Zhou J., Yang C., Wang G., Tan Y., Wu Y., Zhang S., Yi K., Kang C. The CRISPR-Cas13a gene-editing system induces collateral cleavage of RNA in glioma cells. Adv. Sci. (Weinh) 2019;6:1901299. doi: 10.1002/advs.201901299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang B., Xu X., Long F., Wang J. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci. Rep. 2018;8:14126. doi: 10.1038/s41598-018-32329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H., Zhang G., Sun J., Zhou X. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14:2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhong M., Liu Y., Ma P., Dang L., Meng Q., Wan W., Ma X., Liu J., Yang G., et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout. CRISPR/Cas12a-NER. Sci. Bull. (Beijing) 2020;65:1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang W., Cui Y.C., Pan Q., Zhu W., Gendron P., Guo F., Cen S., Witcher M., Liang C. HIV-1 employs multiple mechanisms to resist Cas9/single guide RNA targeting the viral primer binding Site. J. Virol. 2018;92 doi: 10.1128/JVI.01135-18. e01135–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Warfield K.L., Wells J., Swenson D.L., Donner K.S., Van Tongeren S.A., Garza N.L., Dong L., Mourich D.V., Crumley S., et al. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat. Med. 2010;16:991–994. doi: 10.1038/nm.2202. [DOI] [PubMed] [Google Scholar]
- Wessels H.H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020;38:722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C.S., Thomas M.R., Budd J., Mashamba-Thompson T.P., Herbst K., Pillay D., Peeling R.W., Johnson A.M., McKendry R.A., Stevens M.M. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature. 2019;566:467–474. doi: 10.1038/s41586-019-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E.J., Adair K. The diversity of human RNA viruses. Future Virol. 2013;8:159–171. doi: 10.2217/fvl.12.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liu S.X., Wang F., Zeng M.S. Room temperature detection of plasma Epstein–Barr virus DNA with CRISPR–Cas13. Clin. Chem. 2019;65:591–592. doi: 10.1373/clinchem.2018.299347. [DOI] [PubMed] [Google Scholar]
- Xu C.F., Chen G.J., Luo Y.L., Zhang Y., Zhao G., Lu Z.D., Czarna A., Gu Z., Wang J. Rational designs of in vivo CRISPR-Cas delivery systems. Adv. Drug Deliv. Rev. 2021;168:3–29. doi: 10.1016/j.addr.2019.11.005. [DOI] [PubMed] [Google Scholar]
- Xu L., Wang J., Liu Y., Xie L., Su B., Mou D., Wang L., Liu T., Wang X., Zhang B., et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- Xu L., Yang H., Gao Y., Chen Z., Xie L., Liu Y., Liu Y., Wang X., Li H., Lai W., et al. CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol. Ther. 2017;25:1782–1789. doi: 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Zhang T., Qu X., Zhang Y., Putatunda R., Xiao X., Li F., Xiao W., Zhao H., Dai S., et al. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Zhao F., Sun H., Wang Z., Huang Y., Zhu W., Xu F., Mei S., Liu X., Zhang D., et al. CRISPR-Cas13a inhibits HIV-1 infection. Mol. Ther. Nucleic Acids. 2020;21:147–155. doi: 10.1016/j.omtn.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Yao Y., Xiao H., Li J., Liu Q., Yang Y., Adah D., Lu J., Zhao S., Qin L., Chen X. Simultaneous knockout of CXCR4 and CCR5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both X4- and R5-tropic human immunodeficiency virus type 1 infection. Hum. Gene Ther. 2018;29:51–67. doi: 10.1089/hum.2017.032. [DOI] [PubMed] [Google Scholar]
- Yuan C., Tian T., Sun J., Hu M., Wang X., Xiong E., Cheng M., Bao Y., Lin W., Jiang J., et al. Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Anal. Chem. 2020;92:4029–4037. doi: 10.1021/acs.analchem.9b05597. [DOI] [PubMed] [Google Scholar]
- Yuen K.S., Chan C.P., Wong N.-H.M., Ho C.H., Ho T.H., Lei T., Deng W., Tsao S.W., Chen H., Kok K.-H., Jin D.-Y. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wang Q., Xu X., Xia Q., Long F., Li W., Shui Y., Xia X., Wang J. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal. Bioanal. Chem. 2018;410:2889–2900. doi: 10.1007/s00216-018-0873-5. [DOI] [PubMed] [Google Scholar]
- Zhang B., Xia Q., Wang Q., Xia X., Wang J. Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique. Anal. Biochem. 2018;561–562:37–46. doi: 10.1016/j.ab.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Zhang F., Abudayyeh O.O., Gootenberg J.S. A protocol for detection of COVID-19 using CRISPR diagnostics. 2020. https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf
- Zhen S., Hua L., Liu Y.H., Gao L.C., Fu J., Wan D.Y., Dong L.H., Song H.F., Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- Zhu W., Lei R., Le Duff Y., Li J., Guo F., Wainberg M.A., Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]