Abstract
OBJECTIVE.
The virtual imaging trial is a unique framework that can greatly facilitate the assessment and optimization of imaging methods by emulating the imaging experiment using representative computational models of patients and validated imaging simulators. The purpose of this study was to show how virtual imaging trials can be adapted for imaging studies of coronavirus disease (COVID-19), enabling effective assessment and optimization of CT and radiography acquisitions and analysis tools for reliable imaging and management of COVID-19.
MATERIALS AND METHODS.
We developed the first computational models of patients with COVID-19 and as a proof of principle showed how they can be combined with imaging simulators for COVID-19 imaging studies. For the body habitus of the models, we used the 4D extended cardiac-torso (XCAT) model that was developed at Duke University. The morphologic features of COVID-19 abnormalities were segmented from 20 CT images of patients who had been confirmed to have COVID-19 and incorporated into XCAT models. Within a given disease area, the texture and material of the lung parenchyma in the XCAT were modified to match the properties observed in the clinical images. To show the utility, three developed COVID-19 computational phantoms were virtually imaged using a scanner-specific CT and radiography simulator.
RESULTS.
Subjectively, the simulated abnormalities were realistic in terms of shape and texture. Results showed that the contrast-to-noise ratios in the abnormal regions were 1.6, 3.0, and 3.6 for 5-, 25-, and 50-mAs images, respectively.
CONCLUSION.
The developed toolsets in this study provide the foundation for use of virtual imaging trials in effective assessment and optimization of CT and radiography acquisitions and analysis tools to help manage the COVID-19 pandemic.
Keywords: coronavirus disease, COVID-19, CT, radiography, virtual imaging trials
Coronavirus disease (COVID-19) is an infectious disease that emerged in late 2019. In March 2020, the World Health Organization proclaimed the COVID-19 outbreak to be a pandemic [1]. By April 12, more than 1.8 million patients were reported to have tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) worldwide, with a rapidly rising trend [2, 3]. The pandemic has led to dramatic changes in societies and economies and has placed a tremendous load on health care systems worldwide.
To effectively manage a pandemic, it is essential to have reliable diagnostic methods to detect the disease and to follow up with patients. The reverse transcription–polymerase chain reaction (RT-PCR) test is considered the reference standard diagnostic tool for the identification of patients with SARS-CoV-2. However, RT-PCR tests tend to be slow, are not available in all affected regions, are not highly sensitive, and cannot be used to manage follow-up care or to assess the extent of the disease in the lungs.
Recent studies have shown that CT is highly sensitive for detecting the pulmonary abnormalities related to COVID-19 [4]. However, interpretations, analysis, and imaging acquisitions have not yet been optimized to differentiate COVID-19 from other lung abnormalities, particularly viral or bacterial pneumonia [4–13]. In addition, the throughput, radiation dose, and clinical practicability of CT still require careful consideration. Compared with CT, chest radiography offers the advantages of having widespread access, high throughput, a low radiation dose, and practicality (upright examination, a lower room-disinfection burden, and portable application), but there is currently a lack of data on the validity of chest radiography for the assessment of patients with COVID-19. Therefore, there is a need to investigate how either of the imaging modalities can detect or differentiate COVID-19 from other diseases and to do so through optimized protocols that enhance the task performance and minimize the radiation dose to patients.
Although essential, such investigations cannot be easily performed in vivo when the health care providers’ efforts are focused on curbing the spread of a pandemic disease. Furthermore, such studies are not feasible with real patients because of health concerns, especially with modalities such as CT and radiography that involve ionizing radiation. Instead, the assessment and optimization of imaging methods can be facilitated through virtual imaging trials. In virtual imaging trials (also known as virtual clinical trials or in silico trials), the imaging experiment is emulated using computational models of patients combined with validated imaging simulators. Virtual imaging trials are highly advantageous in that the same subjects can be imaged with different scanners and settings, and the exact truth of the anatomic and physiologic features of the subjects are a priori known.
Virtual imaging trials have been developed over the past decades and used in various medical imaging applications but have not been specifically targeted for patients with COVID-19 [14]. The purpose of this article is to report how virtual imaging trials can be adapted and used for imaging studies of COVID-19, which will enable effective assessment and optimization of CT and radiography acquisitions and analysis tools for reliable imaging and management of COVID-19.
Materials and Methods
Virtual imaging trials have two main components: representative models of targeted subjects and realistic models of imaging scanners. In this study, we developed computational models of patients with COVID-19 and showed, as a proof of principle, how they can be combined with imaging simulators for COVID-19 imaging studies. We focused on CT and chest radiography because they are currently considered the most promising imaging modalities for the diagnosis and management of the disease.
Patient Models
There are three major parts in modeling a COVID-19 computational phantom: modeling the body habitus, modeling the morphologic features of the abnormality, and modeling the texture and material of the abnormality.
Body habitus—
Existing computational phantoms that model the human anatomy can be used to provide the body habitus for the patient models. Many different phantoms have been developed over the years by various groups [14–16]. For this work, we use the 4D extended cardiac-torso (XCAT) model developed at Duke University [17–19]. The XCAT series includes highly detailed human models representing both sexes at varying age, height, and weight combinations. The XCAT phantoms are constructed using actual patient data and include thousands of anatomic structures as well as models for cardiac and respiratory motions and statistically defined textures to model tissue material heterogeneity [20–23]. Figure 1 shows an example of a female XCAT model highlighting the stroma and parenchyma anatomy within the lungs. The detailed models of organs and intraorgan structures make the XCAT a suitable phantom population for use in COVID-19 studies.
Fig. 1—
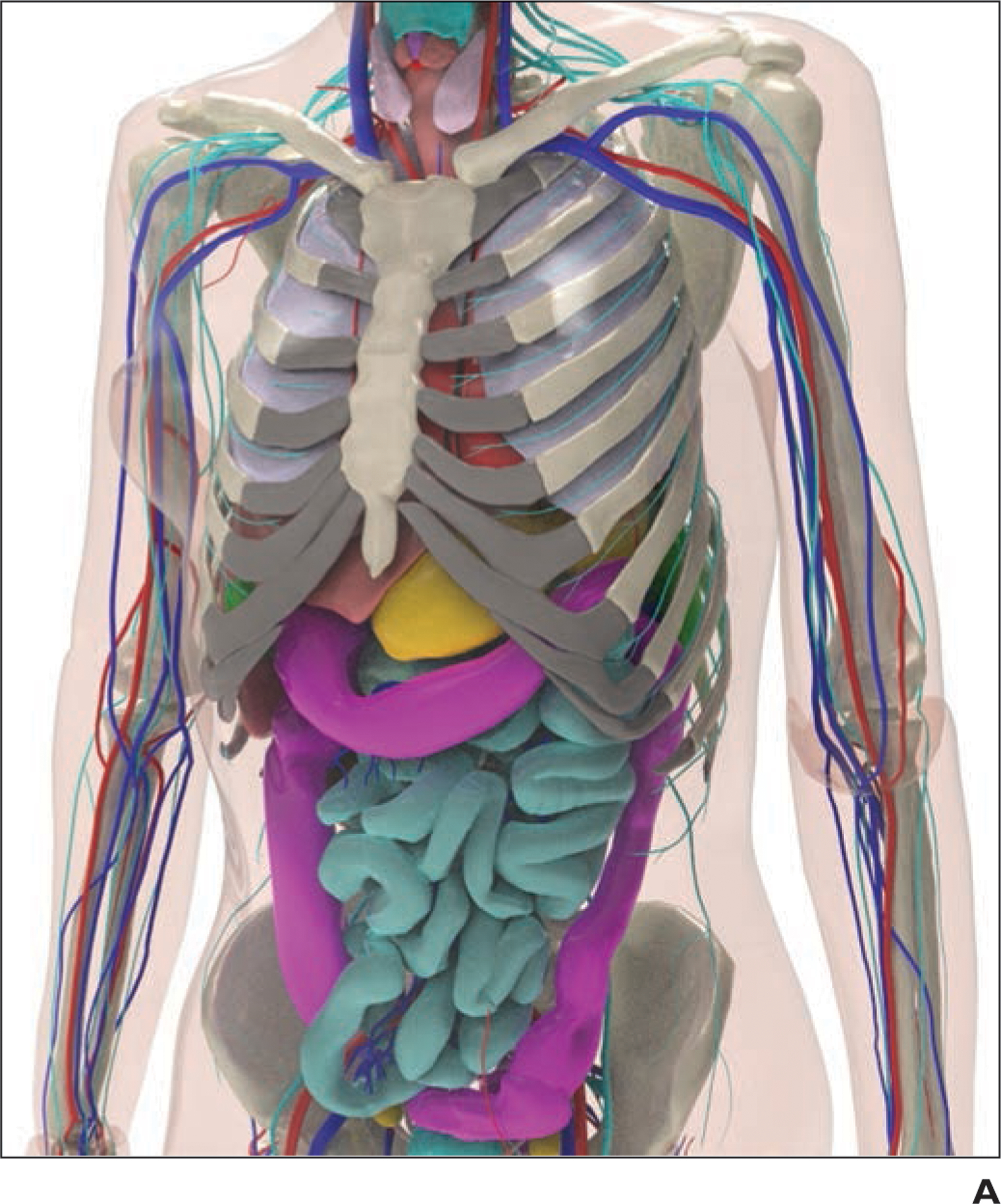
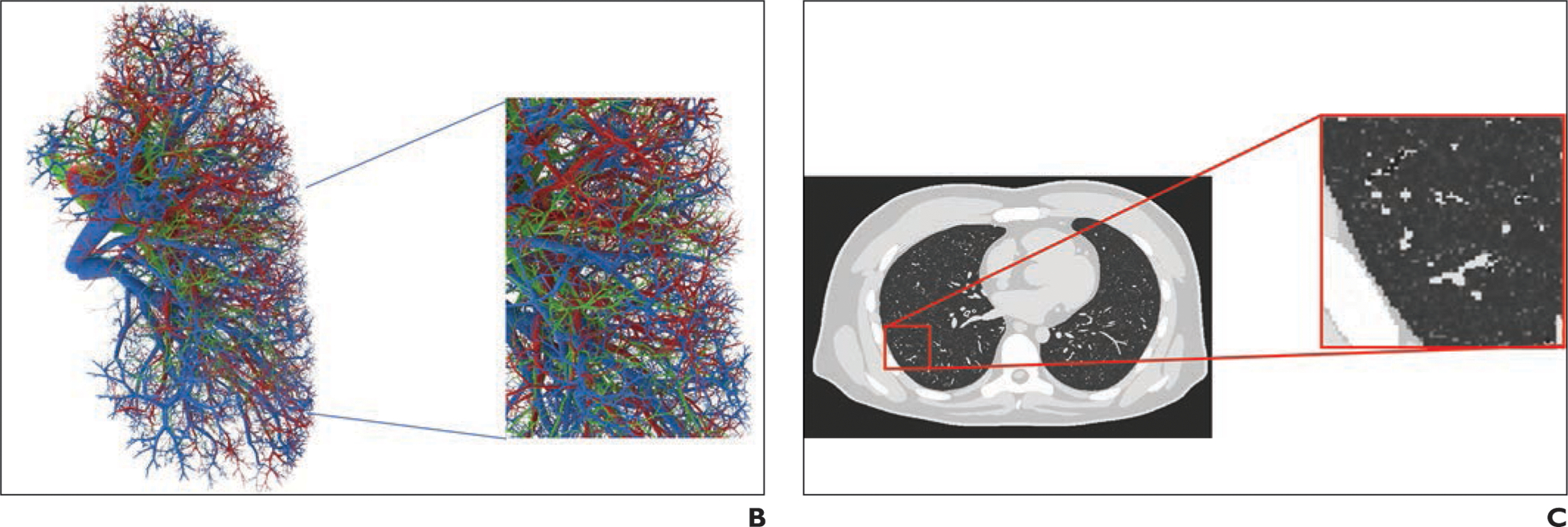
Example of 4D extended cardiac-torso (XCAT) phantom developed at Duke University.
A, Representative computational model shows female XCAT phantom with anthropomorphic organs and structures.
B, Representative computational model shows lung stroma intraorgan structure of XCAT phantom that was developed using anatomically informed mathematic model. Inset shows enlarged view for better visibility of details and small structures.
C, Voxelized rendition (ground truth) of XCAT phantom highlights detailed model of lung parenchyma. Inset shows enlarged view for better visibility of details and small structures.
Morphologic features of the abnormality—
The most common CT manifestations of COVID-19 are ground-glass opacities (GGOs), consolidations within the lower lobe, and peripheral distribution [9]. Findings are typically multilobar and bilateral.
With institutional review board approval, we collected and analyzed 20 clinical cases with multidiagnostic confirmation of SARS-CoV-2 infection. To define models of COVID-19, CT images of multiple confirmed cases were manually segmented to create a series of surfaces representing the size and shape of the manifestations. These surfaces were then placed within a selected XCAT phantom to define the disease areas. To find the closest XCAT for each patient, they were matched according to body diameter, sex, and age.
Texture and material—
Within a given disease area, the texture and material of the underlying lung parenchyma within the phantom were modified to match the observed properties. For GGOs, the predefined texture in each secondary pulmonary lobule was combined with fluid to match the mean linear attenuation coefficient that was measured in each segmented abnormality. The material was modified for consolidation in a similar manner as with GGO modeling; however, the texture was changed to be uniform because those regions are filled with fluid. For the less likely manifestation of crazy paving pattern, the secondary pulmonary lobules were modified to be 2–3 times thicker than normal anatomic values [24].
Using the above methods, we created three COVID-19 models on the basis of CT images of patients who had tested positive for SARS-CoV-2 infection and incorporated them into an adult male XCAT phantom.
CT and Radiography Simulation
The developed COVID-19 computational phantoms can be virtually imaged using accurate simulation models of imaging devices. Such simulators are described in detail in our earlier work [14].
In this study, we used a validated radiographic simulator, DukeSim [25, 26], to create CT and chest radiographic images of three COVID-19 computational models. DukeSim is a hybrid simulator that generates radiographic images using ray tracing (for primary signal) and Monte Carlo simulation (for scatter signal and radiation dose estimations). DukeSim can generate scanner-specific CT or chest radiographic images for predefined acquisitions and protocols incorporating the physics (for source and detector) of image formation for specific imaging devices.
The CT simulations in this study were performed by modeling a commercial CT scanner (Definition Flash, Siemens Healthineers) with a tube voltage of 120 kV and pitch of 1. The images were reconstructed at pixel size of 0.4 mm and slice thickness of 0.6 mm. The radiographic images were simulated with a tube voltage of 120 kV and source-to-detector distance of 180 cm, for a typical indirect-capture flat-panel system with 0.2-mm pixel size.
Results
Figure 2 shows images from three patients with GGO and consolidation findings and the segmented masks that were used to model COVID-19 abnormalities within the XCAT model. Figure 3 shows an adult male XCAT phantom with COVID-19 abnormalities with different shapes that model GGOs and consolidation. These examples show the detailed pulmonary structures that are modeled within the lungs of each XCAT phantom. It also highlights how the underlying normal parenchyma texture was modified to realistically model the disease (Fig. 1).
Fig. 2—
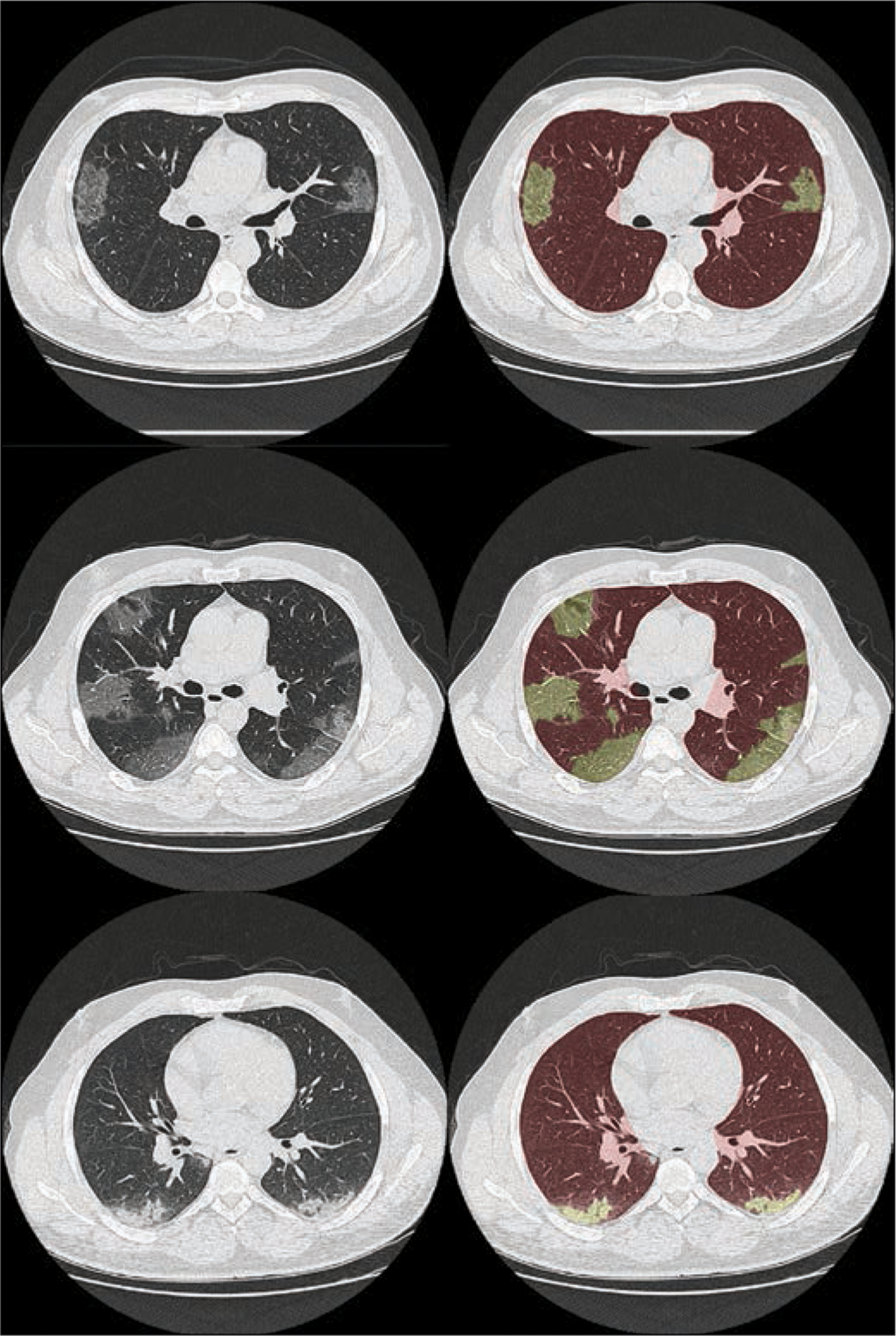
CT images (left) of patients who had tested positive for coronavirus disease (COVID-19) with ground-glass opacity (top and middle) and consolidation (bottom) abnormalities. Right images show overlay of lung and abnormality segmentation on corresponding CT images.
Fig. 3—
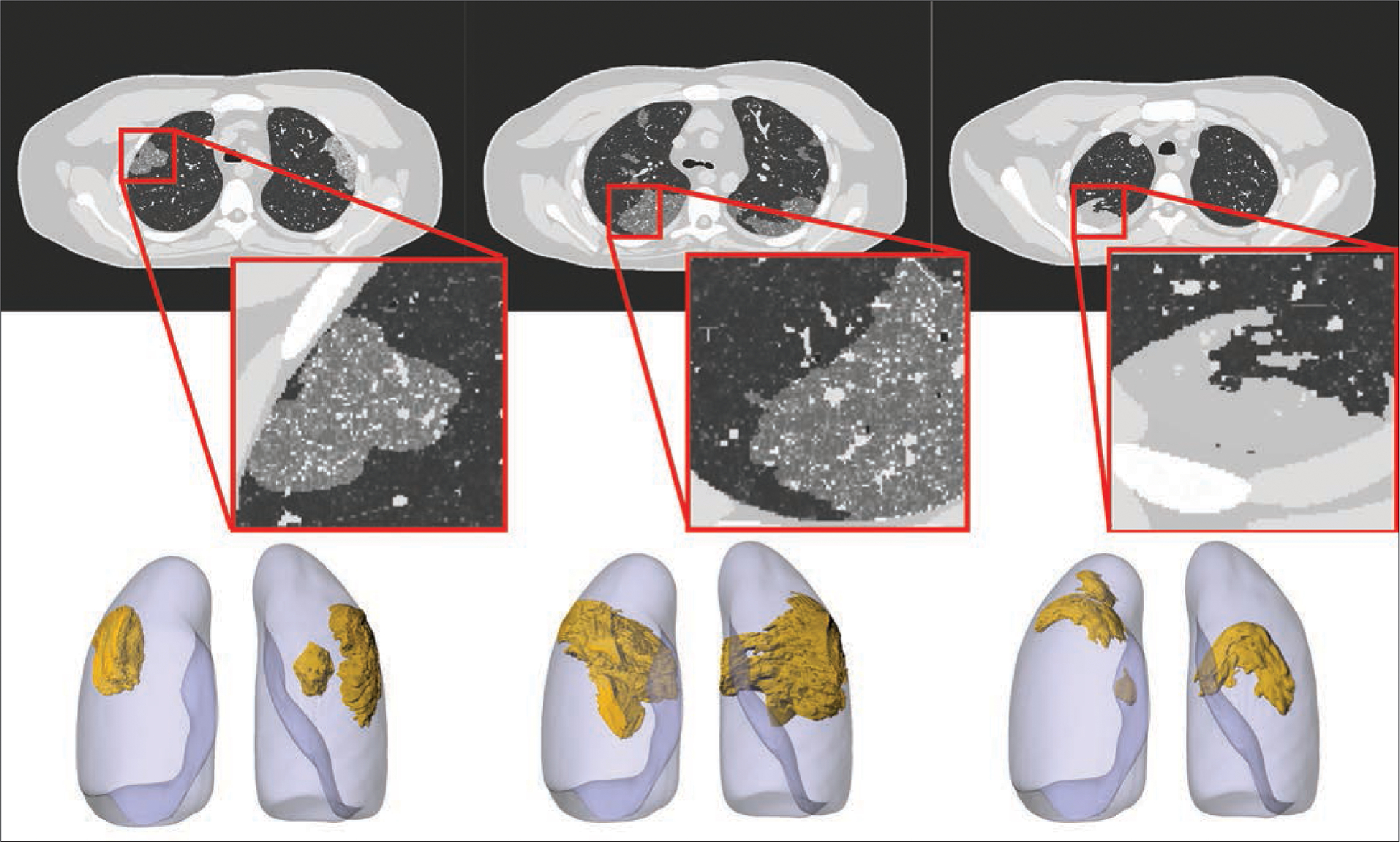
Four-dimensional extended cardiac-torso (XCAT) phantom developed at Duke University shows three coronavirus disease (COVID-19) abnormalities with different shapes. Left and center phantoms show ground-glass opacities and right phantom shows consolidation. Top images show voxelized renditions (ground truth) of phantoms, and insets show enlarged view for better visibility of details and small structures. Bottom images show surface-based rendition of same COVID-19 abnormalities incorporated into adult male XCAT phantom.
Figure 4 shows the ground-truth phantom images of three XCAT phantoms with COVID-19 abnormalities, compared against their simulated CT and chest radiographic images, which include noise and artifacts from the imaging processes. The ground-truth images enable quantitative comparison of how imaging techniques affect the representation of the abnormalities.
Fig. 4—
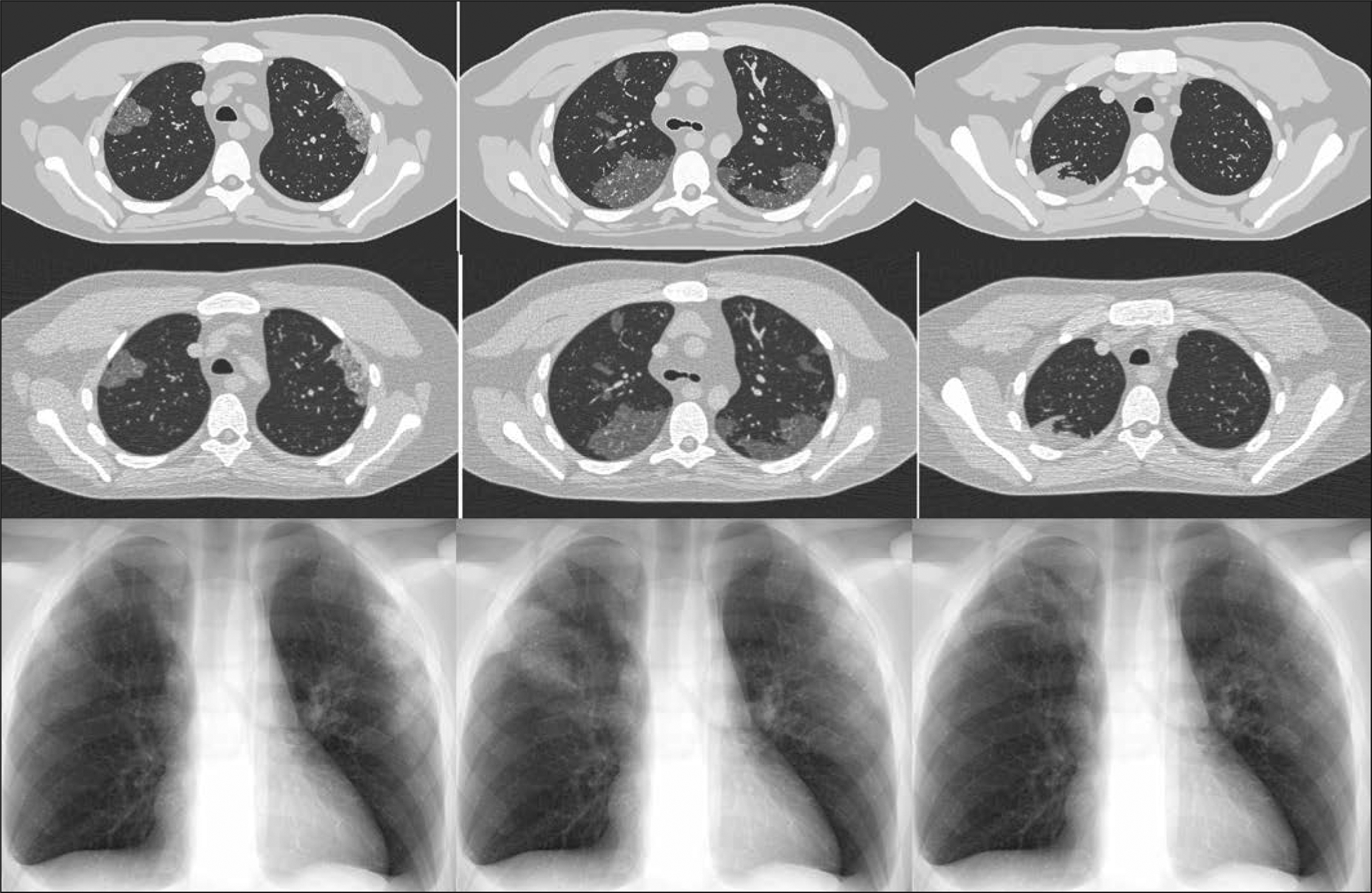
Ground-truth phantom images (top) of 4D extended cardiac-torso phantom developed at Duke University show different coronavirus disease (COVID-19) abnormalities. Simulated CT (middle) and radiographic (bottom) images were generated using DukeSim, also developed at Duke University. Ground-glass opacity (left and middle) and consolidation (right) abnormalities are visible on both CT and radiographic images.
Subjectively, the simulated abnormalities look realistic in terms of their shape and texture. The abnormalities simulated in Figure 4 (GGOs and consolidation) are visible in both simulated CT and radiographic images. The findings in the chest radiographs are more subtle. Figure 4 shows the promising potential of virtual imaging trials to scan a population of COVID-19 models with either chest CT or chest radiography and compare the ability of the two modalities to reliably detect the disease and differentiate it from other abnormalities.
Figure 5 shows simulated images of a COVID-19 XCAT model at three different dose levels (representing 5-, 25-, and 50-mAs settings), as well as a chest radiographic image of the same phantom showing that our developed platform can be used to image COVID-19 phantoms at different dose values to determine the optimal required radiation dose for CT and radiography. Our preliminary results show that the contrast-to-noise ratios in the abnormal regions were 1.6, 3.0, and 3.6 for 5-, 25-, and 50-mAs images, respectively.
Fig. 5—
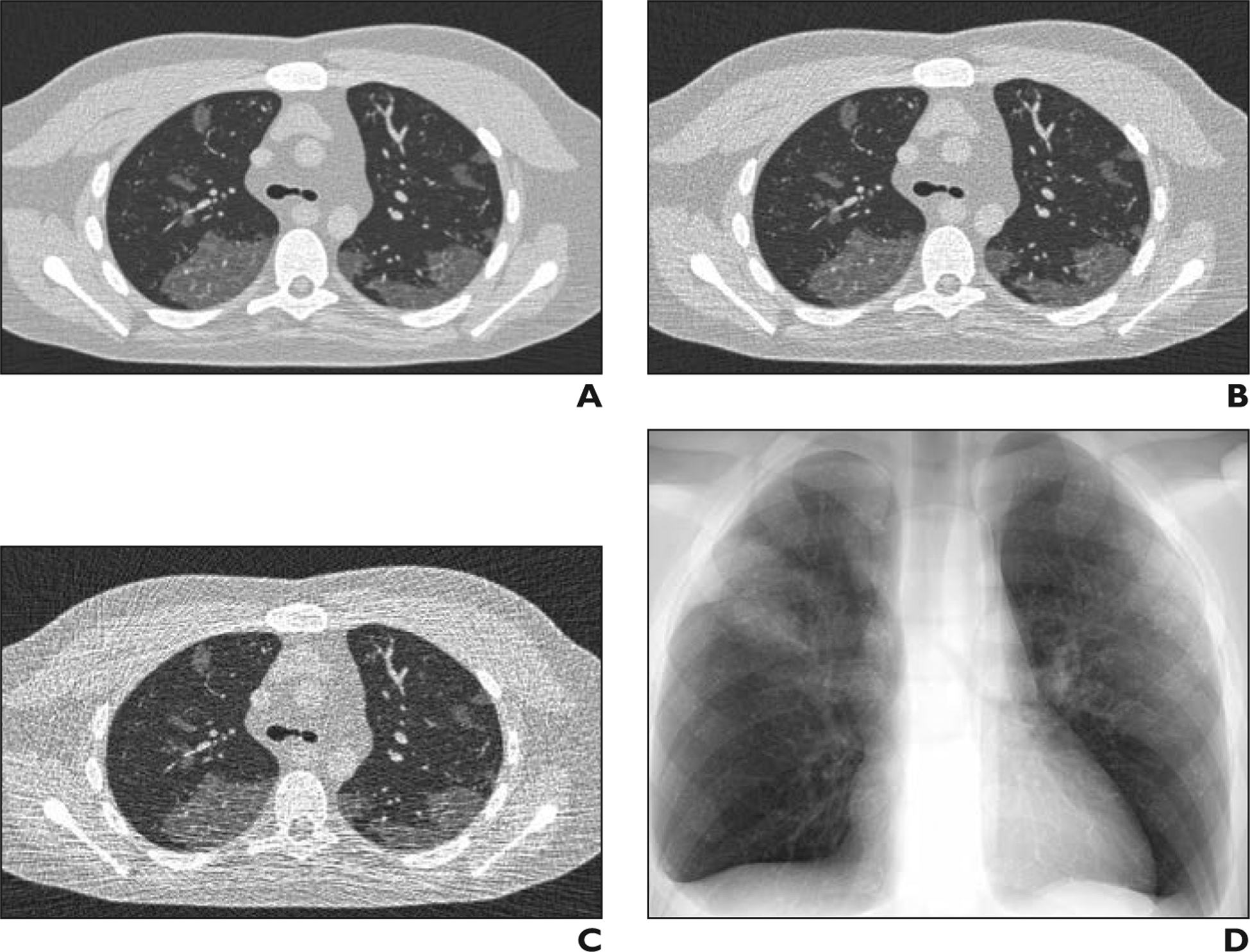
Simulated CT images of coronavirus disease (COVID-19) on 4D extended cardiac-torso phantom developed at Duke University.
A–D, Images of same phantom show simulated CT at 50 (A), 25 (B), and 5 (C) mAs as well as simulated chest radiograph (D).
Discussion
Chest CT and radiography are promising modalities for screening COVID-19. A virtual imaging trial has the potential to investigate and optimize the role of these imaging modalities in diagnosis of COVID-19 and follow-up. In a virtual imaging trial, quality and performance of imaging methods can be comprehensively assessed and compared. Further, radiation dose and image quality can be quantitatively optimized for each modality [27, 28]. Because virtual imaging trials have predefined models of the disease, they can provide a valuable tool to train and evaluate either principle-informed or machine-learning algorithms designed for disease classification and quantification.
In this study we showed the potential usefulness of virtual imaging trials in the development and assessment of imaging techniques for the COVID-19 pandemic. As a proof of principle, we developed computational models of patients with COVID-19 and virtually imaged them using CT and chest radiography simulations. Building on this framework, ongoing work will focus on modeling a larger population of COVID-19 XCAT phantoms using more patient CT examinations. The phantoms are intended to represent various stages and manifestations of the disease. These models will be informed by CT images acquired at different stages and clinical information of patients with COVID-19 and by longitudinal studies [29]. It is crucial to have these models at different stages of the disease because they are envisioned to be used for various imaging studies from screening to treatment evaluations and follow-up care. Further ongoing work will include systematic simulation of imaging protocols and task-based characterizations of the resulting images. With these tools, we will perform virtual imaging trials to compare radiography and CT methods and protocols with the goal of defining COVID-19 protocols for optimized screening and follow-up of patients, with the confidence of knowing the expected performance. One potential application is to compare the performance of chest radiography and CT for screening the patients. Another application is to determine the effects of radiation dose on detection accuracy and further optimize this parameter in imaging patients with COVID-19. Our virtual toolsets can also be used to optimize the imaging and analysis tools in terms of sensitivity and specificity for the detection and classification of the disease.
Conclusion
We have developed strategies to adapt and use virtual imaging trials for imaging studies of COVID-19, which will provide the foundation for the effective assessment and optimization of CT and radiography acquisitions and analysis tools to manage the COVID-19 pandemic.
Acknowledgments
We thank our collaborators Francesco Ria, Hamid Soltanian-Zadeh, Juan Carlos Ramirez, Rafael Fricks, and Pegah Khoshpouri for their assistance in our ongoing projects on COVID-19.
Supported in part by grant R01EB001838 from the National Institutes of Health.
Footnotes
The authors declare that they have no disclosures relevant to the subject matter of this article.
References
- 1.World Health Organization website. WHO director-general’s opening remarks at the media briefing on COVID-19: 11 March 2020. www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020. Accessed April 10, 2020
- 2.Worldometer website. COVID-19 coronavirus pandemic. www.worldometers.info/coronavirus/. Accessed April 9, 2020
- 3.Johns Hopkins University Center for Systems Science and Engineering website. Coronavirus COVID-19 global cases. gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6. Accessed April 9, 2020
- 4.Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020. February 26 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Qin L, Xu Z, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020. March 19 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology 2020. February 20 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 2020. February 19 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer WJ. ACR releases CT and chest X-ray guidance amid COVID-19 pandemic. Diagnostic Imaging; website. www.diagnosticimaging.com/coronavirus/acr-releases-ct-and-chest-x-ray-guidance-amid-covid-19-pandemic. Accessed March 20, 2020 [Google Scholar]
- 9.Chung M, Bernheim A, Mei X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020; 295:202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei J, Li J, Li X, Qi X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 2020; 295:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai HX, Hsieh B, Xiong Z, et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology 2020. March 10 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mossa-Basha M, Meltzer CC, Kim DC, Tuite MJ, Kolli KP, Tan BS. Radiology department preparedness for COVID-19: Radiology scientific expert panel. Radiology 2020. March 16 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inui S, Fujikawa A, Jitsu M, et al. Chest CT findings in cases from the cruise ship “Diamond Princess” with coronavirus disease 2019 (COVID-19). Radiol Cardiothorac Imaging 2020; 2:e200110 [Erratum in Radiol Cardiothorac Imaging 2020; 2:e204002] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abadi E, Segars WP, Tsui BMW, et al. Virtual clinical trials in medical imaging: a review. J Med Imaging (Bellingham) 2020. April 11 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kainz W, Neufeld E, Bolch WE, et al. Advances in computational human phantoms and their applications in biomedical engineering: a topical review. IEEE Trans Radiat Plasma Med Sci 2019; 3:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu XG. An exponential growth of computational phantom research in radiation protection, imaging, and radiotherapy: a review of the fifty-year history. Phys Med Biol 2014; 59:R233–R302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW. 4D XCAT phantom for multimodality imaging research. Med Phys 2010; 37:4902–4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segars WP, Norris H, Sturgeon GM, et al. The development of a population of 4D pediatric XCAT phantoms for imaging research and optimization. Med Phys 2015; 42:4719–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segars WP, Bond J, Frush J, et al. Population of anatomically variable 4D XCAT adult phantoms for imaging research and optimization. Med Phys 2013; 40:043701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abadi E, Segars WP, Sturgeon GM, Roos JE, Ravin CE, Samei E. Modeling lung architecture in the XCAT series of phantoms: physiologically based airways, arteries and veins. IEEE Trans Med Imaging 2018; 37:693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abadi E, Segars WP, Sturgeon GM, Harrawood B, Kapadia A, Samei E. Modeling textured bones in virtual human phantoms. IEEE Trans Radiat Plasma Med Sci 2019; 3:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abadi E, Sturgeon G, Agasthya G, et al. , Airways, vasculature, and interstitial tissue: anatomically-informed computational modeling of human lungs for virtual clinical trials. In: Flohr TG, Lo JY, Schmidt TG, eds. Medical Imaging 2017: Physics of Medical Imaging—Proceedings of SPIE, vol. 10132. Orlando, FL: SPIE, 2017:101321Q [Google Scholar]
- 23.Chang Y, Lafata K, Segars WP, Yin FF, Ren L. Development of realistic multi-contrast textured XCAT (MT-XCAT) phantoms using a dual-discriminator conditional-generative adversarial network (D-CGAN). Phys Med Biol 2020. March 19 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb WR. Thin-section CT of the secondary pulmonary lobule: anatomy and the image—the 2004 Fleischner lecture. Radiology 2006; 239:322–338 [DOI] [PubMed] [Google Scholar]
- 25.Abadi E, Harrawood B, Sharma S, Kapadia A, Segars WP, Samei E. DukeSim: a realistic, rapid, and scanner-specific simulation framework in computed tomography. IEEE Trans Med Imaging 2019; 38:1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abadi E, Harrawood B, Rajagopal JR, et al. Development of a scanner-specific simulation framework for photon-counting computed tomography. Biomed Phys Eng Express 2019; 5:055008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ria F, Davis JT, Solomon JB, et al. Expanding the concept of diagnostic reference levels to noise and dose reference levels in CT. AJR 2019; 213:889–894 [DOI] [PubMed] [Google Scholar]
- 28.Ria F, Solomon JB, Wilson JM, Samei E. Technical note: validation of TG 233 phantom methodology to characterize noise and dose in patient CT data. Med Phys 2020; 47:1633–1639 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Dong C, Hu Y, et al. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology 2020. March 19 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


