Abstract
MicroRNAs in biofluids are potential biomarkers for detecting kidney and other organ injuries. We profiled microRNAs in urine samples from patients with Russell’s viper envenoming or acute self-poisoning following paraquat, glyphosate, or oxalic acid [with and without acute kidney injury (AKI)] and on healthy controls. Discovery analysis profiled for 754 microRNAs using TaqMan OpenArray qPCR with three patients per group (12 samples in each toxic agent). From these, 53 microRNAs were selected and validated in a larger cohort of patients (Russell’s viper envenoming = 53, paraquat = 51, glyphosate = 51, oxalic acid = 40) and 27 healthy controls. Urinary microRNAs had significantly higher expression in patients poisoned/envenomed by different nephrotoxic agents in both discovery and validation cohorts. Seven microRNAs discriminated severe AKI patients from no AKI for all four nephrotoxic agents. Four microRNAs (miR-30a-3p, miR-30a-5p, miR-92a, and miR-204) had > 17 fold change (p < 0.0001) and receiver operator characteristics area-under-curve (ROC-AUC) > 0.72. Pathway analysis of target mRNAs of these differentially expressed microRNAs showed association with the regulation of different nephrotoxic signaling pathways. In conclusion, human urinary microRNAs could identify toxic AKI early after acute injury. These urinary microRNAs have potential clinical application as early non-invasive diagnostic AKI biomarkers.
Subject terms: miRNAs, Acute kidney injury, Diagnostic markers, Toxicology
Main
MicroRNAs are involved in all cellular functions, including development, differentiation, growth, metabolism and regulate numerous physiological and pathophysiological processes1,2. MicroRNAs are endogenous, non-coding RNAs, 21–25 nucleotides in size that regulate gene expression by binding to target messenger RNAs (mRNAs). Cells secrete microRNAs into the extracellular environment to function as regulators, carrying genetic information from one cell to another, or following cell injury3. These extracellular microRNAs are potential biomarkers for a variety of diseases including acute kidney injury (AKI)4 and can also be mediators of disease.
Acute kidney injury is associated with changes in the expression of specific microRNAs. While some microRNAs contribute to the pathogenesis and progression of AKI, others may serve as protective molecules5. As signalling molecules, microRNAs mediate cell–cell communications, and form a complex regulatory network2,6. They play a critical role as anti-apoptotic, pro-apoptotic and inflammatory molecules to facilitate the development of renal injury in nephrotoxic AKI7,8. Thus, microRNAs have not only emerged as novel biomarkers for AKI, but they also hold promise as potential therapeutic targets.Various microRNAs regulate different nephrotoxic pathways, for example NF-κB dependent inflammatory responses, TGF-β signalling, and oxidative stress9–11. Thus, different mechanisms of nephrotoxicity including renal tubular toxicity, inflammation, glomerular damage, crystal nephropathy, and thrombotic microangiopathy from different toxic agents would be expected to lead to variable microRNA expression.
Traditional biomarkers of AKI such as serum creatinine (SCr) or urine output are both late indicators of kidney injury12,13. Further, the rapid rise of SCr after oxalic acid, paraquat, and glyphosate surfactant herbicide (glyphosate) poisoning suggests other mechanisms occur beyond loss of renal function14–16. Protein biomarkers which reflect the underlying renal glomerular and/or tubular damage during nephrotoxicity such as β2-microglobulin, cystatin C (Cys C), albumin, neutrophil gelatinase-associated lipocalin (NGAL), kidney injury molecule-1 (KIM-1), interleukin-18 (IL-18) have highly variable diagnostic performance in these three poisonings14–16. New biomarkers that are consistent for early prediction and/or detection of diverse causes of nephrotoxic AKI are needed, whilst those specific to particular toxins may provide further understanding of the mechanisms of nephrotoxicity.
Acute kidney injury is a common clinical complication following envenoming and poisoning14,16–18. Our main aim was to explore if urinary microRNAs can be used as early renal biomarkers following nephrotoxic-AKI (snakebite envenoming, oxalic acid, paraquat, and glyphosate poisoning). We also investigated whether microRNA signatures identify some common mechanisms of nephrotoxicity using target mRNAs and pathway analysis.
Results
Global profiling of urinary microRNAs detects AKI after different causes of poisoning
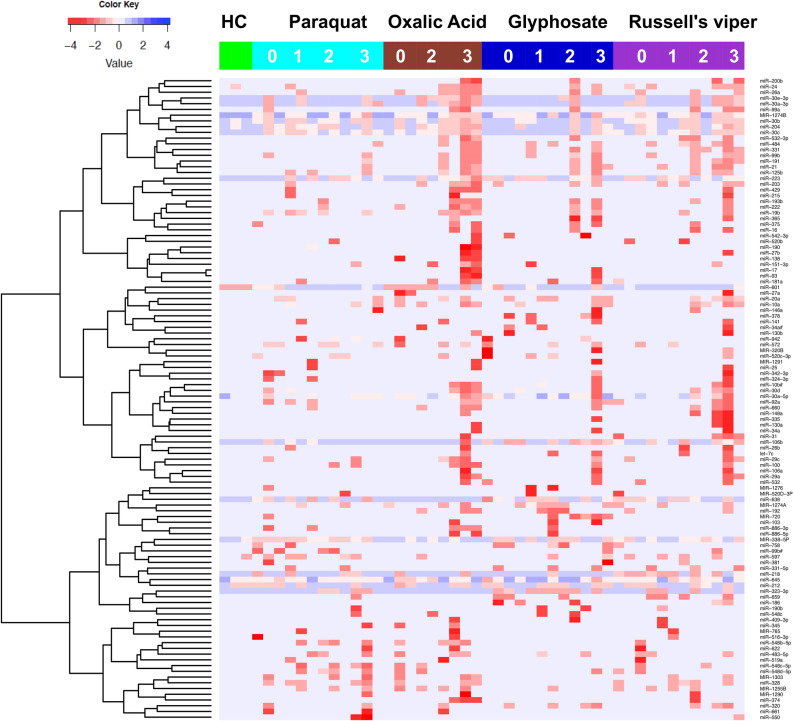
We identified 112 differentially expressed urinary microRNAs from the discovery cohort of 48 patients with Russell’s viper envenoming or acute self-poisoning following paraquat, glyphosate or oxalic acid. The cohort included those who developed no injury (NOAKI), mild injury (AKIN1), moderate injury (AKIN2) or severe injury (AKIN3) (n = 3 per group and n = 12 per each toxins) as well as healthy controls (n = 3). The expression of microRNAs was significantly upregulated in urine samples of patients who developed moderate to severe injury (AKIN2/3) compared to the NOAKI and healthy controls in the discovery cohort. Supervised hierarchical clustering heat map demonstrates the differentially expressed microRNAs between patients with moderate to severe AKI (AKIN2/3) from NOAKI and healthy controls (Fig. 1). Based on our discovery analysis or from past literature, we selected a set of 53 microRNAs (> 4 fold change and p < 0.01; Supplementary Table 1) for further validation in independent cohorts for each toxins (Russell’s viper bite = 53, paraquat = 51, glyphosate = 51, oxalic acid = 40, and 27 healthy controls) (see “Methods” section and Supplementary Table 2 for details).
Figure 1.
Urinary microRNAs detect AKI after different causes of poisoning. Supervised hierarchical clustering heat map of differentially expressed microRNAs (n=112) distinguished patients with severe AKI from NOAKI and healthy controls. Green, cyan, maroon, dark blue and purple indicates healthy controls, paraquat, Glyphosate, Oxalic acid and Russell’s viper bite respectively. AKI stages are numbered as 0, 1, 2 and 3 for NOAKI, AKIN1, AKIN2 and AKIN3 respectivly. Rows represent microRNAs (normalised Ct) and columns represent the individual samples from healthy controls and patients.
Clustering of microRNA in severe cases from different causes of AKI in the validation cohort
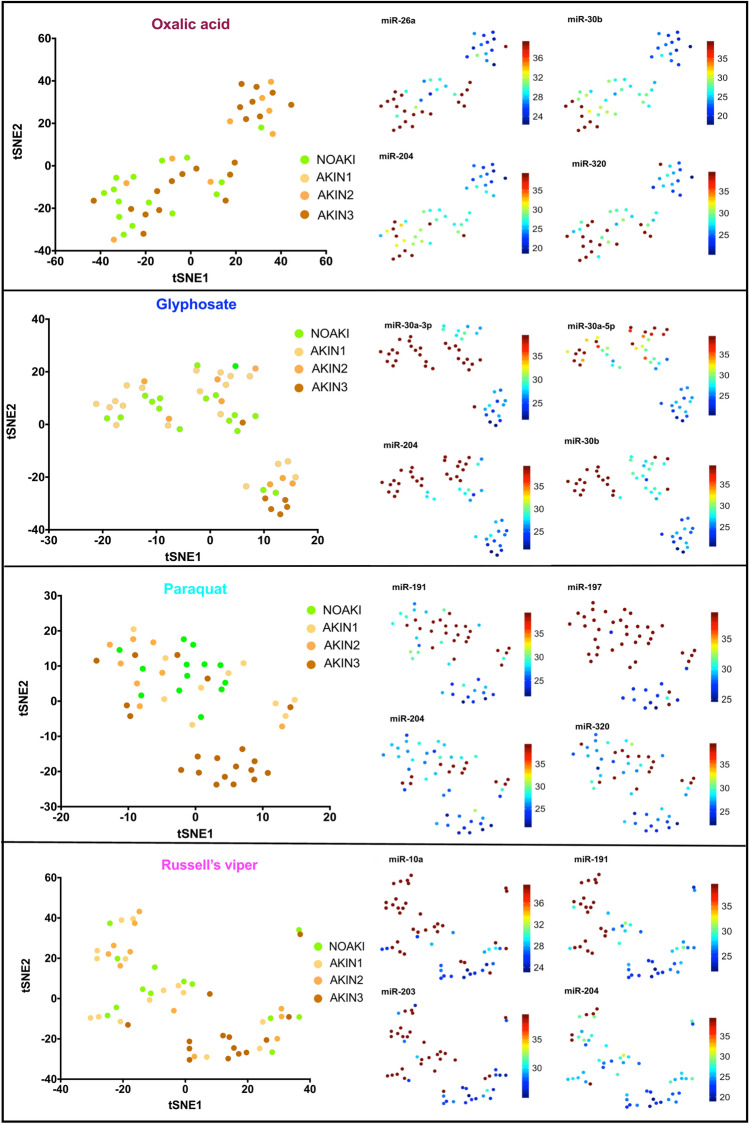
Forty four microRNAs were significantly upregulated in AKI in one or more nephrotoxic agents (oxalic acid = 25, glyphosate = 10, paraquat = 34 and Russell’s viper = 25) compared to NOAKI. Thirty two microRNAs were upregulated for two or more agents (Supplementary Table 3). Different sets of microRNAs for different toxins distinguished patients with AKI from NOAKI in the validation cohort (Fig. 2). Twelve microRNAs distinguished AKI from NOAKI for only one specific toxin [oxalic acid (n = 2), paraquat poisoning (n = 5), Russell’s viper envenoming (n = 5)] (Supplementary Table 3).
Figure 2.
tSNE plots show the clusters of individual microRNAs with NOAKI, AKIN1, AKIN2 and AKIN3 caused by different poisons. Left panels show plots of the non-linear tSNE map of samples in validation cohort using each specific set of differentially expressed microRNAs for different poisons (oxalic acid, glyphosate, paraquat) and Russell’s viper bite, (Supplementary Table 3). Plots were generated using 4-dimensional principal component space into two dimensions showing tSNE analysis. Each point represents a single sample. The right panel shows the expression of the top four most significantly altered microRNAs for each poison presented as individual tSNE-projections. Blue to red presented in the color bar indicate high to low microRNA expression (data shown in normalized Ct values, therefore a lower value represents a higher expression) respectively.
Urinary microRNAs provide signals distinguishing both kidney injury severity and toxic agents
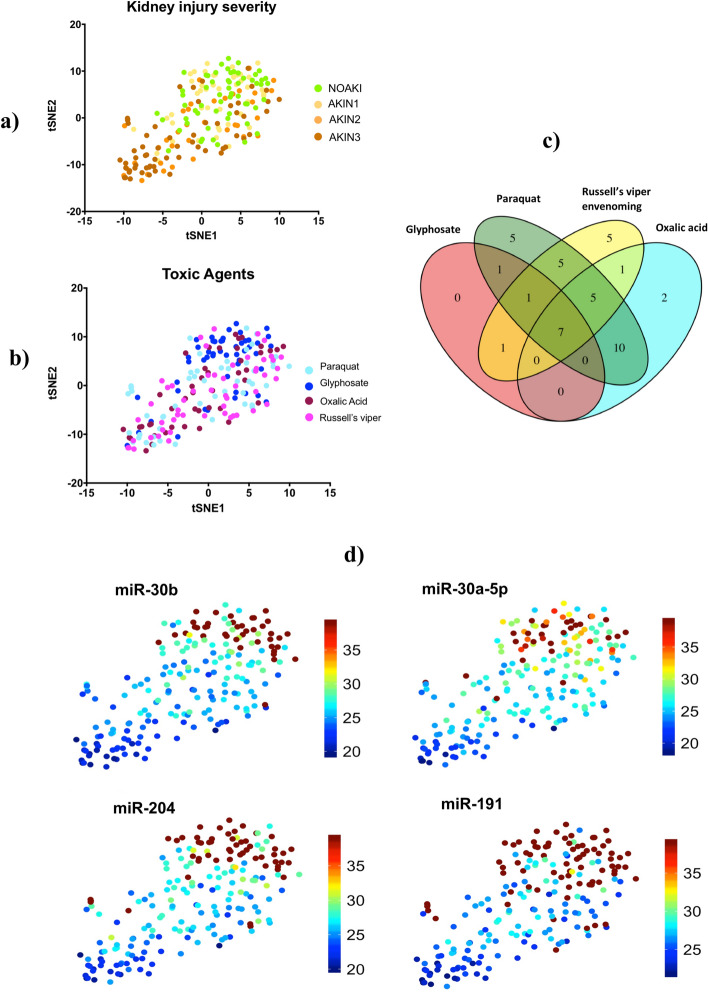
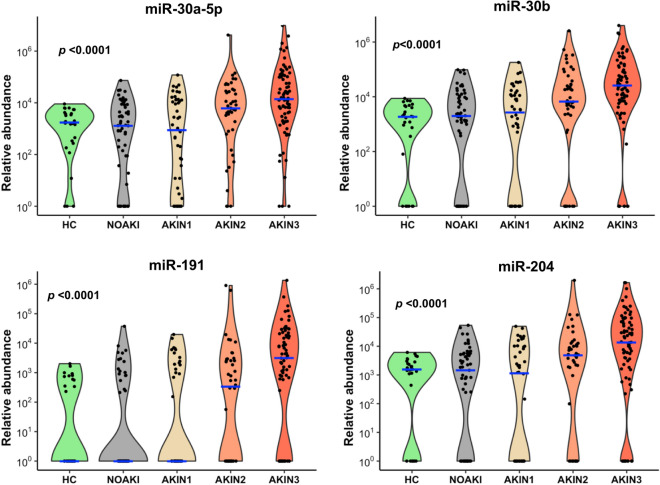
Differentially expressed microRNAs from all four nephrotoxic agents were examined together to visualize separation of patients with AKI from NOAKI using tSNE clustering analysis (Fig. 3a). Despite the selection of differentially expressed microRNAs based on comparisons across all causes of kidney injury for validation, there was still apparent glyphosate clustering in the tSNE plot (Fig. 3b). Seven common microRNAs discriminated AKIN2/3 patients from NOAKI for all four nephrotoxic agents (Fig. 3c, Supplementary Table 3). Of these seven microRNAs, four (miR-30a-5p, miR-30b, miR-191, and miR-204, Fig. 3d) had a greater than 17-fold change (p < 0.0001) and ROC-AUC = 0.72 (Table 1). Logistic regression combination of four and all seven microRNAs in NOAKI versus AKIN2/3 patients showed AUC-ROC values of 0.75 and 0.77 respectively (Supplementary Figure 1). The four microRNAs showed an association with increasing disease severity in all poisoning (Jonckheere–Terpstra nonparametric ordered test, p < 0.0001, Fig. 4). A schematic diagram shows the initial discovery, through validation and selection of four common microRNAs in all four toxic agents (Supplementary Figure 2).
Figure 3.
Plot of the non-linear tSNE map of samples from 4-dimensional principal component space into two dimensions showing tSNE analysis applied using all (n=43) differentially expressed microRNAs (Supplementary Table 3). (a) tSNE plots show the clusters of all different microRNAs in different AKI stages: NOAKI, AKIN1, AKIN2 and AKIN3 caused by all poisons. (b) tSNE plots show the clusters for different poisons: oxalic acid, glyphosate, paraquat and Russell’s viper envenoming in validation cohort. (c) Venn Diagram demonstrating overlap of 43 altered microRNAs in AKI (> 2.8 fold change, p < 0.05) amongst the nephrotoxic agents. microRNAs altered in different agents: Oxalic acid = 25, glyphosate = 10, paraquat = 34 and Russell’s viper = 25. (d) tSNE plots show best four microRNAs clusters in AKIN2/3 patients in all four nephrotoxic agents. Blue to red presented in the color bar indicates high to low microRNA expression (data shown in normalized Ct values, therefore a lower value represents a higher expression) respectively.
Table 1.
Diagnostic performance of the seven microRNAs in all causes of AKI (NOAKI vs. AKIN2/3).
| miR | Fold change (2^ΔCt) | p value | AUC (95% CI) |
|---|---|---|---|
| miR-30a-5p | 19.7 | < 0.00001 | 0.72 (0.66–0.79) |
| miR-30b | 17.1 | < 0.00001 | 0.72 (0.66–0.79) |
| miR-191 | 32.0 | < 0.00001 | 0.72 (0.66–0.78) |
| miR-204 | 17.1 | < 0.00001 | 0.72 (0.65–0.78) |
| miR-30a-3p | 22.6 | < 0.00001 | 0.70 (0.64–0.77) |
| miR-660 | 8.0 | < 0.00001 | 0.64 (0.59 0.69) |
| miR-423-5p | 5.7 | < 0.00001 | 0.60 (0.56–0.64) |
Diagnostic performance of the seven microRNAs that distinguished AKI in all poisoning was assessed using normalized Ct values. p values represent Mann–Whitney-U between NOAKI (n = 57) versus AKIN2/3 (n = 94).
Figure 4.
Top four microRNAs associated with AKI for all toxic agents. Violin plots show these microRNAs having increased abundance with increasing AKI severity in all four toxins. Blue lines indicate the medians and each black dot presents a different sample from validation cohort. The y-axis is relative abundance from the detectable limit (2(39-Ct), Ct of 39 is the threshold of detectability). MicroRNA expressions were normalised using two spike control ath-microRNAs (ath-miR-159a and ath-miR-172a). p is the alternative p value from Jonckheere–Terpstra nonparametric ordered test.
Identification of target genes and pathway analysis
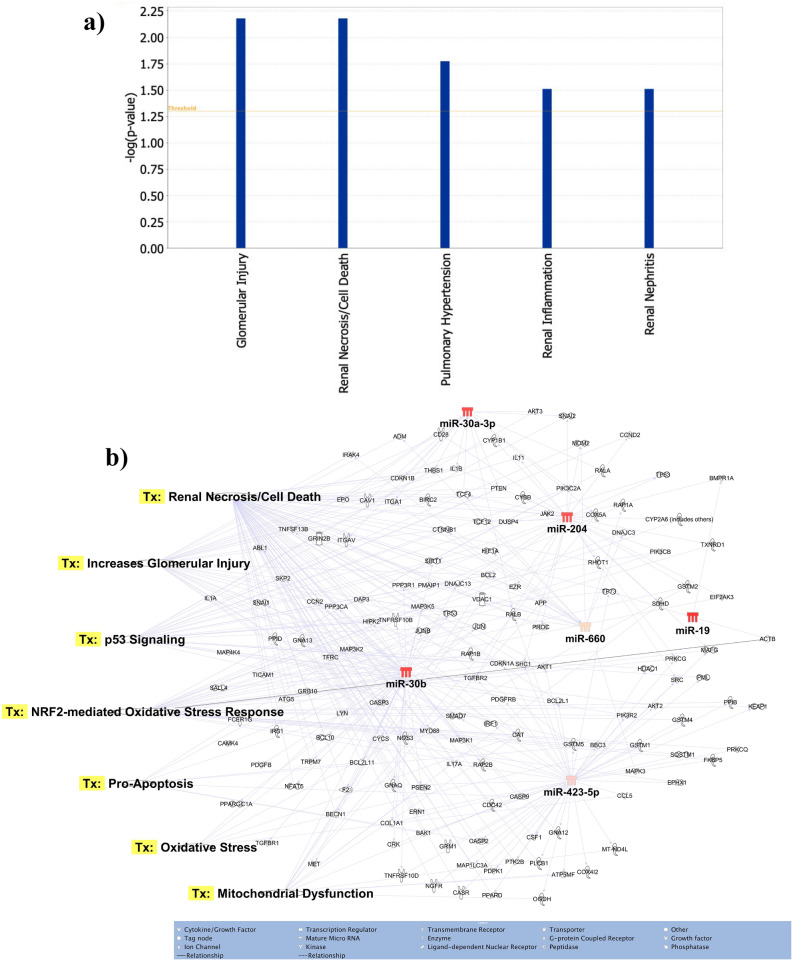
The seven most differentially expressed microRNAs target glomerular injury, renal cell death/necrosis, renal inflammation and renal nephritis as major disease/functions (Fig. 5a). For the seven differentially expressed microRNAs a total of 586 potential target mRNAs specific to kidney disease/functions were identified via Ingenuity Pathway Analysis (data not shown). These differentially expressed microRNAs targeted many genes associated with the regulation of different nephrotoxic signalling pathways (Fig. 5b). The key pathways targeted by these microRNAs were oxidative stress, renal cell death/necrosis, oxidative stress, apoptosis and mitochondrial dysfunction (Fig. 5b).
Figure 5.
Functions and nephrotoxic pathways associated with target mRNAs of seven differentially expressed microRNAs in toxic-AKI. We analyzed the expression of the seven microRNAs using co-analysis in Ingenuity Pathway Analysis (IPA), (QIAGEN, Inc., https:// targetexplorer.ingenuity.com/) and a) shows the main disease and functions of the differentially expressed microRNAs. We used IPA to identify target mRNA and microRNA-mRNA interactions. Target mRNAs were overlaid on ‘Toxic’ functions derived from select microRNAs using ‘Pathway design’ option in IPA. The top seven toxic function pathway (Tx) networks identified for microRNA-mRNA interactions are presented here.
Discussion
We report urinary microRNA signatures for nephrotoxicity in patients with four very different toxic agents (snake envenoming, pesticide and chemical poisoning) in humans. We identified seven common microRNAs that correlated with kidney injury across all four nephrotoxic agents. Another 36 microRNAs provided signals for certain nephrotoxic causes, but not for all agents. We found many of the target mRNAs of these seven common microRNAs are involved in cellular pathways that promote inflammation, apoptosis and oxidative stress.
Very few studies report on multiple agents in nephrotoxic AKI. A recent report on drug induced-AKI in rats using three very different nephrotoxic compounds, cisplatin (proximal tubule) puromycin (glomerulus) and N-phenylanthranylic acid (collecting ducts), identified microRNAs specific to each agent, but did not find any commonly expressed microRNAs across all three19. Other studies reported on single agents, for example, cisplatin-induced AKI in rats showed higher expression of urinary miR-191 and miR-30a-5p20, and urinary levels of miR-423 were significantly higher in both paracetamol overdose patients with AKI compared with healthy controls and in paracetamol overdose patients without AKI21.
We also noted some urinary microRNAs we identified are also observed in non-toxic AKI and other kidney diseases as AKI biomarkers. Urinary miR-423 expression increased in critically ill patients (intensive care unit-AKI)22. Patients with glomerular disease such as idiopathic nephrotic syndrome and focal segmental glomerulosclerosis had high levels of miR-30a-5p in urine compared to controls23–25. MiR-204 was differentially expressed in urine of patients with chronic acute cellular rejection, characterized by interstitial fibrosis and tubular atrophy, compared with patients with normal histology and functioning allografts26.
We studied four very different toxins covering a broad range of mechanisms including ischemia–reperfusion (Russell’s viper bite), uncoupling of oxidative phosphorylation (glyphosate), free radical generation (paraquat), and crystal formation (oxalic acid). However, all these poisonings or envenoming cause mitochondrial damage and inflammation triggering oxidative stress27. Oxidative stress, both directly and indirectly affects all facets of the kidney, including vascular reactivity and renal hemodynamics, glomerular filtration and tubular reabsorption and secretion in all nephron segments28. During poisoning, oxidative stress signalling alters all these processes and promotes damage pathways that lead to cellular apoptosis, necrosis, altered gene expression, progression of tissue damage, promotion of fibrosis and abnormal kidney function27,28. We identified several target mRNAs of the altered microRNAs were associated with mitochondrial dysfunction, apoptosis, oxidative stress and necrosis/cell death.
Most nephrotoxic agents induce proximal tubular injury. The increased concentration of microRNAs in urine may be due to leakage of microRNAs from proximal tubular cells with corresponding decreased levels of microRNAs in the injured tissue20, but we did not test this due to unavailability of kidney tissue. Other studies in cisplatin and gentamicin-induced proximal tubular injury showed increased miR-191 and miR-30 family in urine with corresponding decreased expression in tissue20,29,30. In human miR-30a-3p, miR-30b, miR-30c, miR-30e-3p had lower expression in acute kidney rejection biopsies compared to normal allograft biopsies31.
Interestingly, there are no studies highlighting urinary miR-204 in AKI, despite its renal specific expression32,33. MiR-204, that we found common across the four nephrotoxic agents, is involved in the regulation of epithelial-mesenchymal transition by targeting specificity protein 1 (SP1) in the tubular epithelial cells after ischemia–reperfusion injury34, and controls local inflammation by regulating interleukin-6 (IL-6) receptor expression35. MiR-204 protects interstitial tissue of renal tubules from chronic fibrotic change34.
One of our study limitations is that we had a relatively small sample size in the discovery cohort, but we validated the most differentially expressed microRNAs obtained from the discovery results in a much larger cohort. Furthermore, rather than only measuring previously studied microRNAs, we globally profiled 754 microRNAs without ‘a priori’ approach and hence successfully discovered novel microRNAs associated with nephrotoxic agents-induced AKI. We were unable to measure microRNA expression in kidney tissue as biopsies were not done on our patients limiting the study to some extent. As for most studies in patients, we did not have baseline serum creatinine measurement prior to injury and had to use the lowest level measured at any time which may underestimate the degree of injury.
The major strengths of this study are the comprehensive microRNA analysis of kidney injury with four different toxins, global profiling of microRNAs and validation in independent sets of samples using the same analysis platform. Use of multi-centre prospective study samples from five different geographic regions of Sri Lanka is also a strength of the study for generalisability of results. Another strength of the study is that most patients’ samples were taken within 8 h of the injury, a timeframe important for clinical prediction of AKI and guiding treatment plans. Therefore, microRNAs identified in our study can readily serve as potential biomarkers for AKI.
In conclusion, urinary microRNA profiling shows promise in identifying nephrotoxicity after severe AKI in humans. Signature microRNAs including miR-30a-5p, miR-30b, miR-191 and miR-204, could be promising early biomarkers for the detection of toxic AKI. Urinary microRNAs have potential clinical applications as early non-invasive biomarkers for AKI, toxic-AKI, and also for snake envenoming.
Methods
We included patients with Russell’s viper envenoming or who ingested oxalic acid, paraquat, or glyphosate from a multicenter cohort study in Sri Lanka16,17,36. These patients presented to general hospitals between October 2010 and February 2014. Participants completed a written informed consent prior to inclusion in this study. We included patients who provided admission urine samples (collected within 8 h of ingestion). Details of sample collection and storage are described16,17,36. AKI was defined by serum creatinine (SCr) and categorized based on Acute Kidney Injury Network (AKIN) criteria37. In brief, definition of AKIN stage 1 (mild): an absolute increase in SCr of more than or equal to 0.3 mg/dl or a relative increase by more than or equal to 150 to 200% (1.5 to 2 fold) from baseline, AKIN stage 2 (moderate): a relative increase in SCr of > 200 to 300% (> 2 to 3 fold) from baseline and AKIN stage 3 (severe): a relative increase in SCr of > 300% (> 3 fold) from baseline (or an absolute increase of SCr by more than or equal to 4.0 mg/dl). We used baseline SCr as the lowest level measured at any time (hospital stay or follow-up) because information on pre-exposure was not available.
This study was approved by the Human Research Ethics Committees of the University of New South Wales (Sydney), Australia and the Faculty of Medicine, University of Peradeniya, Sri Lanka. We confirm that all methods were performed in accordance with the relevant guidelines and regulations.
Sample selection for global profiling and independent validation phase
We used urine samples from patients with acute self-poisoning following paraquat, oxalic acid and glyphosate, and patients who were envenomed by Russell’s vipers. We grouped patients into those who developed no kidney injury (NOAKI), mild injury (AKIN1) moderate injury (AKIN2) or severe injury (AKIN3) (n = 3 per group, n = 12 per each toxin) as well as healthy controls (n = 3) and profiled for microRNAs using the TaqMan OpenArray quantitative real-time PCR (qPCR) platform. We selected a panel of 53 microRNAs (46 significantly different microRNAs between AKI and NOAKI in all toxins in discovery cohort with fold change > 4.0, p < 0.01; seven from literature and two were stage-specific spike-in controls and one A. thaliana (ath-miR-394a) negative microRNA control) (Supplementary Table 1). These were validated in a larger independent cohort of patients (Russell’s viper bite = 53, paraquat = 51, glyphosate = 51, oxalic acid = 40) and 27 healthy controls (Supplementary Table 2).
RNA extraction
Total RNA (including small RNA species) was extracted from 200 µl urine by first mixing with 500 µl of TRIzol (ThermoFisher Scientific, USA) and 100 µl chloroform (Sigma Aldrich, Hamburg, Germany), followed by using the RNeasy-HT Kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions on a QiaCube-HT robotic RNA isolation system. Samples were also spiked in with 2.5 µl of 50 nM synthetic control microRNA ath-miR-172a (Sigma Aldrich, Hamburg, Germany) during RNA extraction procedure. The quantity of total RNA was measured using the NanoDrop spectrophotometer.
OpenArray panels—quantitative real-time PCR
Complementary (c)DNA was synthesised from total RNA using Megaplex RT Primers, Human Pool A and Pool B (ThermoFisher Scientific, USA) and reagents from the TaqMan microRNA Reverse Transcription Kit (ThermoFisher Scientific, USA). Each reverse transcription (RT) reaction contained a final volume of 7.5 μl (100 ng total RNA input). Pre-amplification was performed using Megaplex PreAmp Primers, Human Pool A and B (ThermoFisher Scientific, USA) and TaqMan PreAmp Master mix (ThermoFisher Scientific, USA). Pre-amplified product was diluted 1:40 in 0.1X TE buffer (pH 8.0). Diluted product was then combined with TaqMan OpenArray PCR master mix at a 1:1 ratio and loaded onto TaqMan OpenArray Human microRNA Panel (ThermoFisher Scientific, USA) using the AccuFill system. RT-qPCR was completed using the QuantStudio 12 K Flex System (Life Technologies, Foster City, CA, USA)38. Validation PCR was carried out using the Custom TaqMan OpenArray (ThermoFisher Scientific, USA) according to the manufacturer’s ‘low-sample-input’ protocol with custom microRNA RT primer pools, RT spike-in microRNA (ath-miR-159a), custom PreAmp Primer Pools and PCR master mix (supplied with the custom OpenArray panel).
Data analysis
High throughput data generated from TaqMan OpenArray RT-qPCR data were uploaded into ThermoFisher Connect for global normalisation and to set a threshold point. MicroRNAs that were undetectable or had an Amp Score < 1.24 and Cq confidence < 0.6 were eliminated, as per the pre-defined QC criteria for this study. A cut-off threshold cycle (Ct) value of 39 was defined. Validation panel data were directly imported into Excel and normalised using the two spike controls (ath-miR-159a and ath-miR-172a).
Statistical analysis
The relative cycle change (ΔCt = CtHealthy Control − CtPatient Sample) between each group was calculated using normalised Ct-values and the statistical significance (p value) was calculated by Student t-tests in Excel. We used semi-supervised (samples-supervised and microRNA expression-unsupervised) cluster heatmaps to show the microRNA expression in different toxic agents by grouping the samples as toxic agents with the severity of AKI stages (supervised). That allows clustering of microRNAs according to expression in each sample. MicroRNAs showing statistically significant changes over ± 4 fold at p < 0.01 were selected and measured on the custom OpenArray panel. R statistical program was used to plot heatmaps and t-distributed Stochastic Neighbour Embedding (tSNE) plots to visualise differentially expressed microRNAs, through using R package Heatplus and Rtsne respectively, along with other supportive packages for graphics and resampling. Area under the receiver operator characteristics curve (AUC-ROC) analysis was calculated using GraphPad Prism 8 to evaluate the diagnostic value of candidate microRNAs. Logistic AUC-ROC was calculated to evaluate the diagnostic performance of the seven microRNAs using R statistical program (Proc package). Relative abundance was calculated from the detectable limit (2^(39-Ct)). The Jonckheere-Terpstra non-parametric ordered test (with ordering of the populations as follows: Healthy controls, NOAKI, AKIN1, AKIN2, AKIN3) was performed to identify statistically significant trend between independent groups.
Pathway analysis
We analyzed the expression of the seven microRNAs using co-analysis in Ingenuity Pathway Analysis (IPA), (QIAGEN, Inc., https://targetexplorer.ingenuity.com/) to find the main disease association and functions of these differentially expressed microRNAs. Target mRNAs of these differentially expressed microRNAs were examined using microRNA target filter option in IPA (filter criteria: Renal and urological disease, all molecular types, Pathways: transport, apoptosis, cellular growth, development and proliferation, cellular stress and injury, ingenuity toxicity list). The target mRNAs were then overlaid on ‘Disease and function’ and examined for significant functions and involvement in signalling pathways using Tox function in pathway analysis. Finally, the top pathway and the targets were overlayed with the significantly altered microRNAs to plot the network diagram.
Supplementary information
Acknowledgements
This project was funded by the National Health and Medical Research Council (NHMRC) (Grant ID1055176 and ID1011772). We thank Hardikar Laboratory (Islet Biology and Diabetes Group) at NHMRC Clinical Trials Centre, University of Sydney for the infrastructure support to conduct laboratory experiments. FS received a University of Sydney International Research Scholarship. AAH, MVJ and WKMW were supported through JDRF Australia Clinical Research Network CDA, JDRF International Advanced Post-doctoral fellowships and JDRF Australia/Helmsley Charitable Trust funding respectively.
Author contributions
Conceived the study: N.B., D.S., G.I., A.A.H. and F.S. Patient recruitment and sample collections: I.G. and F.M. Performed experimental work: F.S. and M.J. microRNA data collection and analysis: F.S., M.J., A.A.H., and W.W. Drafted the first manuscript: F.S. Critically revised and approved manuscript: all authors.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Anandwardhan A. Hardikar, Devanshi Seth and Nicholas A. Buckley.
Contributor Information
Fathima Shihana, Email: fhan0023@sydney.edu.au.
Nicholas A. Buckley, Email: nicholas.buckley@sydney.edu.au
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87918-0.
References:
- 1.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1:31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzen JM, Thum T. Circulating and urinary microRNAs in kidney disease. Clin. J. Am. Soc. Nephrol. 2012;7:1528–1533. doi: 10.2215/CJN.01170212. [DOI] [PubMed] [Google Scholar]
- 4.Shihana F, Joglekar MV, Raubenheimer J, Hardikar AA, Buckley NA, et al. Circulating human microRNA biomarkers of oxalic acid-induced acute kidney injury. Arch. Toxicol. 2020;94:1725–1737. doi: 10.1007/s00204-020-02679-5. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z, Wang S, Mi QS, Dong Z. MicroRNAs in pathogenesis of acute kidney injury. Nephron. 2016;134:149–153. doi: 10.1159/000446551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang W. MicroRNAs: biomarkers, diagnostics, and therapeutics. Methods Mol. Biol. 2017;1617:57–67. doi: 10.1007/978-1-4939-7046-9_4. [DOI] [PubMed] [Google Scholar]
- 7.Wei Q, Mi QS, Dong Z. The regulation and function of microRNAs in kidney diseases. IUBMB Life. 2013;65:602–614. doi: 10.1002/iub.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohamed DI, Khairy E, Saad SST, Habib EK, Hamouda MA. Potential protective effects of Dapagliflozin in gentamicin induced nephrotoxicity rat model via modulation of apoptosis associated miRNAs. Gene. 2019;707:198–204. doi: 10.1016/j.gene.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Liao W, Fu Z, Zou Y, Wen D, Ma H, et al. MicroRNA-140-5p attenuated oxidative stress in Cisplatin induced acute kidney injury by activating Nrf2/ARE pathway through a Keap1-independent mechanism. Exp. Cell Res. 2017;360:292–302. doi: 10.1016/j.yexcr.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Bhatt K, Zhou L, Mi QS, Huang S, She JX, et al. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol. Med. 2010;16:409–416. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu HY, Liu MY, Hong Q, Zhang D, Geng WJ, et al. Role of microRNA-181a in the apoptosis of tubular epithelial cell induced by cisplatin. Chin. Med. J. (Engl.) 2012;125:523–526. [PubMed] [Google Scholar]
- 12.Devarajan P. Biomarkers for the early detection of acute kidney injury. Curr. Opin. Pediatr. 2011;23:194–200. doi: 10.1097/MOP.0b013e328343f4dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coca SG, Parikh CR. Urinary biomarkers for acute kidney injury: perspectives on translation. Clin. J. Am. Soc. Nephrol. 2008;3:481–490. doi: 10.2215/CJN.03520807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohamed F, Buckley NA, Jayamanne S, Pickering JW, Peake P, et al. Kidney damage biomarkers detect acute kidney injury but only functional markers predict mortality after paraquat ingestion. Toxicol. Lett. 2015;237:140–150. doi: 10.1016/j.toxlet.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Wijerathna TM, Gawarammana IB, Dissanayaka DM, Palanagasinghe C, Shihana F, et al. Serum creatinine and cystatin C provide conflicting evidence of acute kidney injury following acute ingestion of potassium permanganate and oxalic acid. Clin. Toxicol. (Phila) 2017;55:970–976. doi: 10.1080/15563650.2017.1326607. [DOI] [PubMed] [Google Scholar]
- 16.Mohamed F, Endre ZH, Pickering JW, Jayamanne S, Palangasinghe C, et al. Mechanism-specific injury biomarkers predict nephrotoxicity early following glyphosate surfactant herbicide (GPSH) poisoning. Toxicol. Lett. 2016;258:1–10. doi: 10.1016/j.toxlet.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Ratnayake I, Mohamed F, Buckley NA, Gawarammana IB, Dissanayake DM, et al. Early identification of acute kidney injury in Russell's viper (Daboia russelii) envenoming using renal biomarkers. PLoS Negl. Trop. Dis. 2019;13:e0007486. doi: 10.1371/journal.pntd.0007486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wijerathna TM, Mohamed F, Dissanayaka D, Gawarammana I, Palangasinghe C, et al. Albuminuria and other renal damage biomarkers detect acute kidney injury soon after acute ingestion of oxalic acid and potassium permanganate. Toxicol. Lett. 2018;299:182–190. doi: 10.1016/j.toxlet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Glineur SF, Hanon E, Dremier S, Snelling S, Berteau C, et al. Assessment of a urinary kidney microRNA panel as potential nephron segment-specific biomarkers of subacute renal toxicity in preclinical rat models. Toxicol. Sci. Off. J. Soc. Toxicol. 2018;166:409–419. doi: 10.1093/toxsci/kfy213. [DOI] [PubMed] [Google Scholar]
- 20.Kanki M, Moriguchi A, Sasaki D, Mitori H, Yamada A, et al. Identification of urinary miRNA biomarkers for detecting cisplatin-induced proximal tubular injury in rats. Toxicology. 2014;324:158–168. doi: 10.1016/j.tox.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Pavkovic M, Robinson-Cohen C, Chua AS, Nicoara O, Cardenas-Gonzalez M, et al. Detection of drug-induced acute kidney injury in humans using urinary KIM-1, miR-21, -200c, and -423. Toxicol. Sci. 2016;152:205–213. doi: 10.1093/toxsci/kfw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, et al. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin. Chem. 2013;59:1742–1752. doi: 10.1373/clinchem.2013.210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Zhang C, Chen H, Li L, Tu Y, et al. Evaluation of microRNAs miR-196a, miR-30a-5P, and miR-490 as biomarkers of disease activity among patients with FSGS. Clin. J. Am. Soc. Nephrol. 2014;9:1545–1552. doi: 10.2215/CJN.11561113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, Wang C, Chen X, Zhong T, Cai X, et al. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin. Chem. 2013;59:658–666. doi: 10.1373/clinchem.2012.195297. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzen JM, Thum T. MicroRNAs in idiopathic childhood nephrotic syndrome. Clin. Chem. 2013;59:595–597. doi: 10.1373/clinchem.2013.202671. [DOI] [PubMed] [Google Scholar]
- 26.Scian MJ, Maluf DG, David KG, Archer KJ, Suh JL, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am. J. Transplant. 2011;11:2110–2122. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamed F, Endre ZH, Buckley NA. Role of biomarkers of nephrotoxic acute kidney injury in deliberate poisoning and envenomation in less developed countries. Br. J. Clin. Pharmacol. 2015;80:3–19. doi: 10.1111/bcp.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratliff BB, Abdulmahdi W, Pawar R, Wolin MS. Oxidant mechanisms in renal injury and disease. Antioxid. Redox Signal. 2016;25:119–146. doi: 10.1089/ars.2016.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassirpour R, Homer BL, Mathur S, Li Y, Li Z, et al. Identification of promising urinary microRNA biomarkers in two rat models of glomerular injury. Toxicol. Sci. 2015;148:35–47. doi: 10.1093/toxsci/kfv167. [DOI] [PubMed] [Google Scholar]
- 30.Nassirpour R, Mathur S, Gosink MM, Li Y, Shoieb AM, et al. Identification of tubular injury microRNA biomarkers in urine: comparison of next-generation sequencing and qPCR-based profiling platforms. BMC Genomics. 2014;15:485. doi: 10.1186/1471-2164-15-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Koo S, White N, Peralta E, Esau C, et al. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81:617–627. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 34.Chen SJ, Wu P, Sun LJ, Zhou B, Niu W, et al. miR-204 regulates epithelial-mesenchymal transition by targeting SP1 in the tubular epithelial cells after acute kidney injury induced by ischemia-reperfusion. Oncol. Rep. 2017;37:1148–1158. doi: 10.3892/or.2016.5294. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Qiu J, Chen B, Lin Y, Chen Y, et al. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-kappaB pathway. Int. Immunopharmacol. 2018;59:252–260. doi: 10.1016/j.intimp.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Mohamed F, Endre Z, Jayamanne S, Pianta T, Peake P, et al. Mechanisms underlying early rapid increases in creatinine in paraquat poisoning. PLoS ONE. 2015;10:e0122357. doi: 10.1371/journal.pone.0122357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta RL, Kellum JA, Shah SV, Molitoris BA, Ronco C, et al. Acute kidney injury network: report of an initiative to improve outcomes in acute kidney injury. Crit. Care. 2007;11:R31. doi: 10.1186/cc5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong W, Farr R, Joglekar M, Januszewski A, Hardikar A. Probe-based real-time PCR approaches for quantitative measurement of microRNAs. J. Vis. Exp. 2015;98:e52586. doi: 10.3791/52586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.