Abstract
The Src-homology 2 (SH2) domain-containing phosphatase 2 (SHP2), encoded by the PTPN11 proto-oncogene, is a key mediator of receptor tyrosine kinase (RTK)-driven cell signaling, promoting cell survival and proliferation. In addition, SHP2 is recruited by immune check point receptors to inhibit B and T cell activation. Aberrant SHP2 function has been implicated in the development, progression, and metastasis of many cancers. Indeed, small molecule SHP2 inhibitors have recently entered clinical trials for the treatment of solid tumors with Ras/Raf/ERK pathway activation, including tumors with some oncogenic Ras mutations. However, the current class of SHP2 inhibitors is not effective against the SHP2 oncogenic variants that occur frequently in leukemias, and the development of specific small molecules that target oncogenic SHP2 is the subject of current research. A common problem with most drug discovery campaigns, with those involving cytosolic proteins like SHP2, is that the primary assays that drive chemical discovery are often in vitro assays that do not report on the cellular target engagement of candidate compounds. To provide a platform for measuring cellular target engagement, we developed both wild-type and mutant SHP2 cellular thermal shift assays. These assays reliably detect target engagement of SHP2 inhibitors in cells. Here, we provide a comprehensive protocol of this assay, which provides a valuable tool for the assessment and characterization of novel SHP2 inhibitors.
Keywords: cellular target engagement assay, cellular thermal shift, protein tyrosine phosphatase, Src-homology 2 domain-containing phosphatase, SHP2, PTPN11, allosteric inhibitor, small molecule, anticancer drug, drug discovery, protein drug interaction
SUMMARY:
The ability to assess target engagement by candidate inhibitors in intact cells is crucial for drug discovery. This protocol describes a 384 well format cellular thermal shift assay that reliably detects cellular target engagement of inhibitors targeting either wild-type SHP2 or its oncogenic variants.
INTRODUCTION:
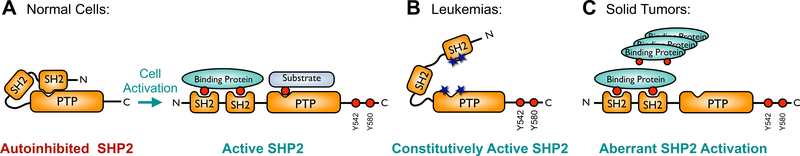
Tyrosine phosphorylation plays an important role in signal transduction in cells1,2. This post-translational modification is catalyzed by protein tyrosine kinases (PTKs) and reversed by protein tyrosine phosphatases (PTPs). Therefore, aberrant PTK or PTP function leads to many inherited or acquired human diseases3–6. The Src-homology 2 (SH2) domain-containing phosphatase 2 (SHP2) is a widely expressed non-receptor type PTP encoded by the proto-oncogene PTPN117 and is a key regulator of numerous physiological processes that involve signal transduction by activation of the Ras/Raf/ERK, PI3K/Akt, or JAK/STAT signaling pathways8. Normally, SHP2 activity is tightly regulated in order to prevent aberrant signaling. Under basal conditions SHP2 is autoinhibited by its N-terminal SH2 domain, which blocks access to the active site within the catalytic phosphatase domain (Figure 1A)9,10. Upon cell activation, tyrosine phosphorylated binding proteins recruit SHP2, causing it to adopt its active conformation, in which the active site is now accessible to its substrates. In many cancers SHP2 activity is elevated. Somatic gain-of-function (GOF) mutations in PTPN11 have been identified mainly in leukemias and prevent binding of the N-SH2 domain to the phosphatase domain, resulting in constitutively active SHP2 (Figure 1B)11. Germline GOF mutations in PTPN11 are responsible for ~50% of cases of Noonan syndrome, a developmental disorder with an increased risk of malignancy12. In solid tumors, where PTPN11 mutations are rare, greater levels of phosphorylated binding proteins lead to enhanced SHP2 activity (Figure 1C). SHP2 is also important for immune checkpoint signaling, as checkpoint receptors such as BTLA or PD-1 recruit SHP2 to dephosphorylate key signaling molecules, preventing immune cell activation13–15.
Figure 1. Regulation of SHP2 activity.
(A) SHP2 activity is tightly regulated in normal cells. Under basal conditions, its N-terminal SH2 domain occludes the active site within the phosphatase (PTP) domain, resulting in autoinhibition. RTK growth factor and cytokine stimulation leads to tyrosine phosphorylation of adapter proteins, which then bind to the SH2 domains, causing a conformational change whereby the SHP2 active site becomes accessible to its substrates. (B) Somatic SHP2 mutations, frequently found in leukemias, are located at the interface between the N-SH2 and phosphatase domains and prevent SHP2 from adopting the closed, autoinhibited conformation, resulting in a constitutively active phosphatase. (C) In solid tumors, amplification or overexpression of growth factors, RTKs, or scaffolding adapters result in aberrant activation of SHP2.
Targeting PTPs with small molecules has been a challenge, because the active site of PTPs is highly conserved and highly charged; inhibitors that target the active site are often potent, but exhibit poor selectivity and oral bioavailability16–22. Indeed, many reported SHP2 inhibitors suffer from poor selectivity and lack of efficacy in cells23. Recently, allosteric inhibitors of SHP2 with good potency and excellent selectivity have been reported (e.g., SHP09924 and RMC-455025) and have sparked renewed interest in SHP2 inhibitors. Compounds based on SHP099 and RMC-4550 are currently in phase I clinical trials to treat solid tumors with receptor tyrosine kinase (RTK) pathway activation26,27. While groundbreaking, these compounds are ineffective against many of the SHP2 oncogenic mutants that drive leukemogenesis in a significant number of blood cancer patients28–30. This lack of potency of SHP099-like compounds toward the SHP2 oncogenic variants stems from their unique allosteric mechanism, as they inhibit SHP2 activity by binding and stabilizing the inactive, closed conformation, which is disrupted in SHP2 mutants. Further, based on a recent report31, adaptive resistance mechanisms in patients treated with SHP099-like inhibitors are quite conceivable. Consequently, the development of next generation SHP2 inhibitors that target its active, open state is a subject of intense research.
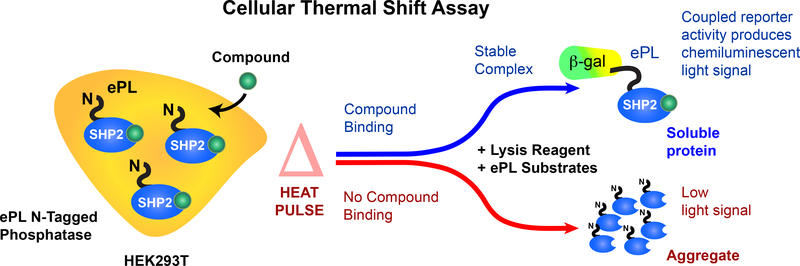
The characterization of novel SHP2 inhibitors in cells is an essential aspect of the lead optimization process. Critically, proven target engagement of the inhibitor under physiological conditions provides an additional level of confidence that resources for medicinal chemistry are efficiently deployed on compounds with promising cellular efficacy. In the past, several methods to assess the binding of small molecule inhibitors to their targets have been developed, primarily for protein kinases32. To develop a SHP2 cellular target engagement assay, we utilized a cellular thermal shift assay33. This assay, similar to the in vitro thermal shift (PTS) assay for proteins34, monitors the target protein thermal stability, which is typically altered by the binding of small molecules. The original assay is a low throughput assay that utilizes antibodies to quantify target protein levels. Alternatively, we chose a recently reported variant of the thermal shift assay that utilizes a β-galactosidase enzyme fragment complementation (EFC) assay (Figure 2)35. For these experiments, the protein of interest is expressed in cells as an N- or C-terminal fusion protein carrying an enhanced ProLabel tag (ePL, a 42 amino acid fragment of β-galactosidase). Cells are then transferred to 384 well PCR compatible plates and incubated with compounds of interest. A thermocycler is utilized to apply a temperature gradient to the cells, whose proteins will denature and aggregate as the temperature increases based upon their thermal stability. The ability of a candidate compound to bind and stabilize the protein of interest will result in an increased thermal stability of that protein. Therefore, following the lysis of the cells, those tagged proteins that have been stabilized by a candidate compound will remain in solution at higher temperatures than tagged proteins of cells incubated with vehicle control. Reporter enzyme acceptor (EA) is able to complement the soluble ePL-tagged proteins, resulting in detectable β-galactosidase activity using a luminescence substrate.
Figure 2. SHP2 cellular thermal shift principles.
Our cellular target engagement assay is based on a β-galactosidase (β-gal) enzyme fragment complementation (EFC) assay. Cells (HEK293T), expressing full-length SHP2 with an ePL fusion tag (42 amino acid fragment of β-gal), are treated with a probe compound and are subjected to a short heat pulse that causes SHP2 to denature and form inaccessible insoluble aggregates, rendering the ePL tag inaccessible. Specific binding of the probe compound to SHP2 can enhance its thermal stability. Thermal stabilization provides more soluble target protein with the ePL tag for complementation in the reporter enzyme chemiluminescence system. Aggregates result in reduced complementation and luminescence. (This figure has been modified from Ref. 36)
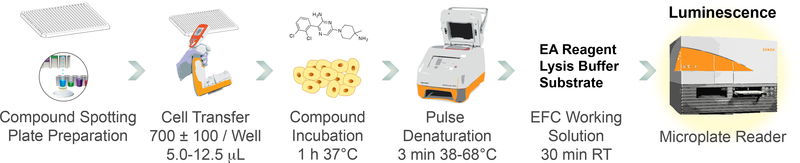
We recently developed a robust cellular thermal shift assay for wild-type SHP2 (SHP2-WT) and a frequent SHP2 oncogenic variant (SHP2-E76K) in a miniaturized 384-well format36. Here, we report a detailed protocol of this assay, which reliably detects target engagement by SHP2 inhibitors in cells and demonstrates a high degree of correlation between inhibitor potency and cellular thermal shift data. The general assay workflow is illustrated in Figure 3. Our platform uses N-terminally tagged full-length ePL-SHP2 fusion proteins. For the generation of the corresponding pICP-ePL-N-SHP2-WT and pICP-ePL-N-SHP2-E76K expression plasmids, please refer to our recent publication36. This assay can be performed using a thermal gradient to establish SHP2 thermal profiles and determine SHP2 melting temperatures in the presence or absence of inhibitor. Once thermal profiles have been established, it can also be performed under isothermal conditions, allowing inhibitor dose-response assessment. Both types of experiments are described below.
Figure 3. Miniaturized SHP2 equipment workflow.
Routine performance of the assay makes use of the equipment indicated. Assay plates are prepared using an acoustic liquid handler that delivers nanoliter volumes of compounds. Cell transfer can be performed manually, using a multichannel pipette, or by using an automatic liquid dispenser. Heat pulse denaturation uses a thermocycler that can deliver thermal gradients to a 384-well plate. A microplate reader is used to detect the β-gal EFC assay chemiluminescence.
PROTOCOL:
1. Preparation of cell culture and reagents
1.1. Formulate a 500 mL bottle of growth media with 10% fetal bovine serum, 1x antibiotic/antimicotic, 20 mM HEPES, and 1 mM sodium pyruvate. Store at 4 °C.
1.2. Thaw cellular thermal shift reagents (EA reagent, lysis buffer, and substrate) from frozen original stock bottles.
1.3. Dispense reagents and buffer as 2 mL aliquots and store at −20 °C.
NOTE: Avoid freeze/thaw for reproducibility and use only that volume of reagent required for the assay procedure.
2. Growth and maintenance of HEK293T cells
2.1. Obtain low passage adherent HEK293T cells from cryo storage.
2.2. Maintain HEK293T cells in growth media at 37 °C, 5% CO2.
2.3. Split cells every 4 days in a 1:14 ratio.
NOTE: For best performance, HEK293T cells are not allowed to passage greater than 25 times.
3. HEK293T cell preparation for transient transfection
3.1. Detach HEK293T cells from 16 mL plates using 3 mL of the cell detachment reagent.
3.2. Dilute with 12 mL of growth media. Collect cells by centrifugation at 1,400 x g for 4 min.
3.3. Resuspend the pellet in 10 mL growth media. Measure the concentration and viability of cells using trypan blue and a cell counter.
3.4. Plate 7.0 × 105 exponentially growing HEK293T cells per well in a 6 well cell culture plate approximately 24 h before transfection.
NOTE: Reserve one well for each plasmid to be transfected.
3.5. Incubate for 24 h at 37 °C, 5% CO2.
4. Transfection of HEK293T cells
4.1. From a purified plasmid stock of pICP-ePL-N-SHP2-WT or pICP-ePL-N-SHP2-E76K (~200 ng/μL) dilute 2 μg of plasmid DNA into 200 μL of transfection buffer.
4.2. Vortex for 10 s and centrifuge at 1,400 x g for 4 min.
4.3. Add 4 μL of transfection reagent to the diluted DNA. Vortex for 10 s and centrifuge at 1,400 x g for 4 min.
4.4. Incubate at 23 °C for 10 min.
4.5. Remove the 6 well plate containing growing HEK293T cells from incubator. Add the transfection mix to the attached cells in the 6-well plate. Incubate for 24 h at 37 °C, 5% CO2.
5. Preparation of assay plates
5.1. Prepare 10 mM stock solutions in DMSO of compounds to be tested.
5.2. Dispense inhibitor solutions into a 384 well low dead volume source plate for immediate use.
5.3. Spot the desired volume of inhibitors or vehicle (DMSO) using a liquid handler into 384 well real-time PCR plates at a target final volume of less than 0.5% DMSO (v/v). Seal the plate using a plate sealer with inert gas purging.
NOTE: For inhibitor dose-response assays, make sure to backfill DMSO accordingly so that equal amounts of DMSO are used for each inhibitor concentration. For best results, store plates at 23 °C and use plates within 24 h.
6. Transfected cell detachment and preparation
6.1. Preincubate growth media and cell detachment reagent in a 37 °C water bath. Remove transfected cells from the incubator. Gently aspirate media from the wells of the plate.
NOTE: Use a new aspirator for every well that contains a different transfected plasmid.
6.2. Add 0.3 mL of the cell detachment reagent. Gently rock the plate back and forth to thoroughly cover the surface of the plate bottom. Incubate at 23 °C for 2 min.
6.3. Add 1 mL of growth media to each well. Gently pipette media and cells in the well and transfer to a 15 mL Falcon centrifuge tube. Collect cells by centrifugation at 1,400 x g for 4 min.
6.4. Gently but thoroughly aspirate off the media.
NOTE: Residual phenol red in cell detachment reagent can interfere with the assay.
6.5. Carefully resuspend cell pellet in 2 mL of growth media. Measure the concentration and viability of cells using trypan blue and a cell counter.
NOTE: Cell viability should be > 90% for best results.
6.6. Dilute cells to a concentration of 125 cells/μL. For example, for a 5 μL assay this is ~625 cells/μL. For optimal viability keep cells in suspension for no more than 2 h.
7. Incubation of cells with SHP2 inhibitors
7.1. Dispense cells into a sterile single channel solution trough.
7.2. Centrifuge 384 well real-time PCR plate that has been pre-prepared by SHP2 inhibitor deposition at 2,500 x g for 5 min at 23 °C.
7.3. Remove seal from the compound plate. Using a 125 μL multichannel pipette, add 5 μL of the diluted cells to desired wells.
7.4. Centrifuge the plate at 42 x g for 30 s without a lid on the plate. Attach a lid seal to the plate after the plate is spun down and incubate the assay plate at 37 °C, 5% CO2 for 1 h.
8. Preparation of chemiluminescent reagent master mix
8.1. Remove the necessary quantity of detection reagents (EA reagent, lysis buffer, and substrate) from −20 °C storage after plating cells. Thaw reagents at 23 °C.
NOTE: For convenience in transfer, prepare a volume that is 1.5 X the total volume of cells that have been deposited to the plate. Do not reuse reagents after use.
8.2. Prepare a master mix of the reagents using condition EA-10, which consists of component and volume fraction as follows: EA reagent (0.17), lysis buffer (0.17), substrate (0.67).
NOTE: Reagent ratios are based on optimization experiments performed as specified in the supplier’s manual.
9: Isothermal or thermal profile gradient heat pulse
9.1. Program the gradient capable thermocycler for the delivery of heat pulse.
9.1.1. For thermal profile gradient experiments, preprogram the thermocycler with the following example specifications:
9.1.2. Heat pulse: 3 min desired melting temperature (vertical or horizontal gradient +/− 15 °C; example 38–68 °C spread across 24 wells yields temperature increments of 1.25 °C).
Equilibration recovery step: 3 min 20 °C with ramp speed = 1 °C/s
9.2.1. For isothermal experiments set up the protocol set up as follows:
9.2.2. Heat pulse: 3 min 55 °C. Equilibration recovery step: 3 min 20 °C with ramp speed = 1°C/s
NOTE: For increased reproducibility, as it can be difficult to place the plate precisely at the time the thermocycler reaches its desired temperature, set up the program to count down 15 s before beginning the 3 min pulse. For the thermocycler used in this experiment see Table of Materials, the lid is kept up during the heat pulse.
10. Lysis detection and measurement
10.1. Supplement assay plates with lysis detection master mix by addition of equal volumes (5 μL) to each well to be analyzed using a multichannel pipette.
10.2. Centrifuge the plate 42 x g for 30 s. Store at 23 °C in darkness for 30–60 min.
10.3. Measure chemiluminescence using a microplate reader capable of detecting luminescence in 384-well format. Perform luminescence measurement with the plate type and integration time optimized (integration time 1000 ms, settle time 0 s). Measure values as counts/s and output for further analysis.
11. Data analysis
11.1. Analyze the luminescence data using a scientific 2D graphing and statistics software.
11.2. Generate thermal profiles by analyzing the normalized luminescence data (normalized to the vehicle control) using nonlinear regression and a Boltzmann sigmoidal model.
NOTE: In thermal profile experiments, the calculated V50, the temperature that is the halfway point between bottom and top of the curve, defines the melting temperature of SHP2. In isothermal experiments, EC50 defines the concentration of the inhibitor that gives half-maximal response.
REPRESENTATIVE RESULTS:
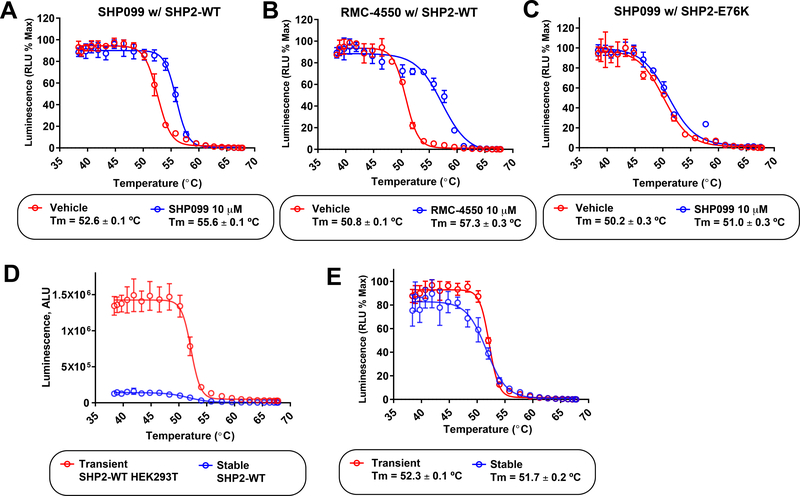
The thermal gradient experiment for SHP2-WT resulted in a sigmoidal cellular thermal profile with a narrow melting transition that is typical and consistent for a folded protein (Figure 4A). SHP2 consists of three independent domains: two SH2 domains and the catalytic domain (Figure 1). In the autoinhibited closed conformation these domains self-associate; the melting transition that was observed in the thermal profile experiment presumably reflected this state in the cell. Incubation of SHP2-WT with the allosteric inhibitor SHP099 at 10 μM stabilized the SHP2-WT thermal profile to a significant and measurable degree (Figure 4A). This is reflective of the known binding mode of SHP099-like inhibitors which act as “molecular glue”37 between the regulatory binding SH2 domains and the catalytic subunit. Importantly, our cellular thermal shift assay confirmed that SHP099 can penetrate the cell and bind to the labeled SHP2-WT. The stabilization of SHP2 also tracked with the potency of SHP099-like allosteric compounds. At 10 μM, the more potent RMC-4550 (reported IC50 = 0.6 nM) produced a greater degree of stabilization than SHP099 (reported IC50 = 70 nM) for SHP2-WT (Figure 4B).
Figure 4. Cellular target engagement of allosteric SHP2 inhibitors with SHP2-WT and SHP2-E76K proteins.
(A) The SHP2-WT protein in the presence of vehicle (DMSO, red) produced a well-behaved cellular thermal profile with a narrow melting transition consistent with a folded protein in the autoinhibited state. Incubation of SHP2-WT expressing cells with 10 μM SHP099 stabilized the protein in the bound state (blue). (B) Cellular target engagement thermal profiles of SHP2-WT in the absence (red) or presence of the SHP099-like allosteric inhibitor RMC-4550 (10 μM, blue). RMC-4550 produced an increased cellular stabilization of SHP2-WT than that observed for SHP099. This increased target engagement tracks with its greater biochemical potency. (C) The SHP2-E76K mutant had a less pronounced melting curve consistent with a state that is largely in the “open” active form (red). SHP099 at 10 μM only marginally increased the SHP2-E76K melting temperature (blue), which is in agreement with biochemical inhibition and cellular efficacy data. (D) The observed chemiluminescence signal corresponds to the expression level of ePL-SHP2. Compared to transiently transfected HEK293T cells, HEK293 cells in which the ePL tagged SHP2-WT was stably integrated into the genome, had a reduced level of ePL-SHP2 expression. Consequently, the raw unnormalized chemiluminesence thermal profiles showed a greatly reduced luminescence signal for the stably expressing HEK293 cells (blue) compared to the transiently transfected HEK293 cells (red). E) Normalization of the raw chemiluminescence data for stable (HEK293) and transiently transfected (HEK293T) produced comparable thermal profiles, showing the sensitivity of the measurements. Data points and error bars (±SD) represent duplicate measurements.
We observe a different behavior in the thermal profile of the target engagement assay with the oncogenic mutant SHP2-E76K (Figure 4C). The thermal profile for SHP2-E76K has a melting transition that is less narrow, reflecting the destabilization imparted on the tertiary structure of SHP2 by the E76K mutation, which is known to disrupt interactions between the SH2 domains and the catalytic subunit. Therefore, the observed melting temperature was reduced compared to the WT protein. Importantly, when incubated with the allosteric inhibitor SHP099 at 10 μM, only marginal thermal stabilization was observed, consistent with the known properties of SHP099 and similar inhibitors28–30.
The expression level of the ePL-tagged protein to be probed can dramatically influence the signal intensity. Shown (Figure 4D) are thermal profiles for transient and stably integrated SHP2-WT cells under identical assay conditions. Normalization of these disparate curves affords comparable thermal profiles (Figure 4E), indicating the wide range of the EFC detection system. It is important to note that we have employed both transient transfection and stable integration SHP2 expressing cells with comparable results. For the generation of stable cell lines it should be noted that HEK293 cells rather than HEK293T cells should be used, as HEK293T cells already have a neomycin resistance marker that would prevent selection of cells with integrated pICP-ePL-N-SHP2 plasmid, which contains a neomycin/kanamycin resistance gene for selection in mammalian cells.
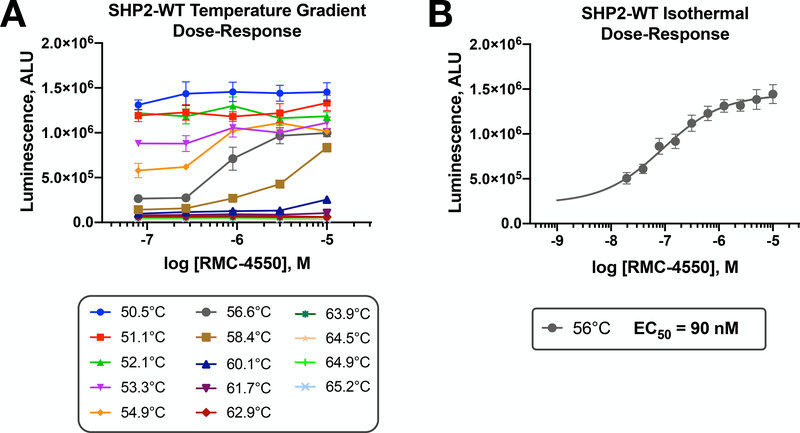
To obtain inhibitor dose-response measurements, one can perform compound titrations on cells under isothermal conditions. To do so, we suggest to first determine the temperature optimum for these measurements. Taking advantage of the thermocycler ability to produce thermal gradients on the “short” axis of a 384-well plate, a series of isothermal titrations can be performed on a single plate. Figure 5A shows representative results of this optimization for SHP2-WT using a limited five-point dose-response of RMC-4550. Temperatures that were too low to denature the protein produced a high signal that was independent of the drug. Similarly, temperatures that were too high afforded little capacity for the compound to stabilize the protein. However, at an optimal temperature, as determined by this experiment, an isothermal titration using a full 10-point dose-response yielded a useful EC50 for RMC-4550 (Figure 5B). In addition to the representative data shown here, we have determined that experiments using optimized isothermal conditions can be employed to efficiently screen novel SHP2 inhibitors for their ability to cross cell membranes and engage their target in cells36.
Figure 5. Cellular target engagement isothermal dose-response assay for SHP2-WT.
A) In order to establish optimal isothermal conditions to evaluate the dose-dependent target engagement of SHP2 inhibitors, a thermal gradient across the short axis of a 384-well plate is first performed. This allows efficient assessment of five-point dose-responses of inhibitors (RMC-4550; 0.08–10 μM) to identify the optimal temperature. B) A full cellular isothermal dose-response (10-point) for RMC-4550 at an optimal temperature of 56°C. The EC50 at this temperature is indicated. Data points and error bars (±SD) represent quadruplicate measurements.
DISCUSSION:
We have presented a target engagement assay that can confirm direct binding of small molecules to the SHP2 phosphatase in cells. The assay can discriminate between low and high affinity inhibitors and, importantly, confirm a lack of potency by the allosteric inhibitors of the SHP099-type for the GOF oncogenic SHP2-E76K mutant. A strength of this miniaturized assay is its ability to be integrated into a SHP2 inhibitor screening campaign. The ability of the assay to confirm intracellular binding to SHP2 by unknown chemical matter is an important capacity to assure that screening resources can be efficiently deployed. Overall, the miniaturized protocol is robust and reliable.
A critical aspect of the assay is timing; best results are obtained 24 h after transfection of the HEK293T cells. The SHP2-WT and SHP2-E76K proteins do not appear to be toxic; however, long-term high expression could reduce the sensitivity of the experiment and would require re-optimization. Careful handling of cells to detach and prepare them for the assay is an additional critical consideration. However, standard cell culture methods are sufficient to produce high-quality data. We have incorporated a number of centrifugation steps into the workflow because of the small volumes utilized in this assay. These steps have the purpose to assure quantitative mixing, so adherence to the acceleration and time of centrifugation is recommended. Of note, the assay reagents can suffer after poor handling. Therefore, we recommend that they be aliquoted and stored frozen until use.
The most commonly encountered problem in the performance of this assay is an erratic (noisy) or low luminescence signal. To determine the source of this problem, it is routine to perform Western blot analysis on transfected HEK293T cells using anti-ePL antibodies to assure a reliable expression level of SHP2-WT and SHP2-E76K. We have obtained best results using the reagent protocol outlined in step 4. Once transfection conditions are established, and a single stock of SHP2 expression plasmid is obtained, we have found the assay to be highly reliable.
Assay development requires optimization of the chemiluminescent signal window to assure reliable results are obtained. To optimize the assay window, the most significant parameter is the formulation of the detection reagent master mix as described in the supplier’s manual. This optimization is performed by systematically changing the EA reagent to substrate ratio in the master mix, while the lysis buffer component is held constant (four different conditions are usually sufficient for this measurement). A condition with zero EA reagent serves as the background. The assay is performed as described above, and the ratio of the observed signal to background (S/B) is calculated. Reliable results are obtained when the S/B ratio is >50. Master mix conditions that result in extraordinarily high signal are to be avoided, as they will result in depletion of the chemiluminescence substrate.
A potential limitation of the assay is its sensitivity. In our experience, inhibitors with at least a low micromolar potency in SHP2 phosphatase inhibition assays are required for producing measurable effects in the cellular thermal shift assay. As a general guideline, we suggest to first assess candidate compounds in in vitro PTS assays using recombinant SHP236. Based on results from our ongoing SHP2 inhibitor discovery program, a compounds ability to shift the SHP2 melting temperature in vitro tracks well with its capacity to stabilize (or sometimes destabilize) the SHP2 protein in cells. Interestingly, the relative stabilization of SHP2-WT by SHP099-like allosteric inhibitors appears to be greater than that caused by equally potent SHP2 active site-directed inhibitors, likely reflecting the ability of the allosteric inhibitors to effectively stabilize a physiologically relevant conformation of the SHP2-WT protein.
In addition to the SHP2 phosphatase, we have gained experience in developing cellular target engagement assays for other phosphatases and kinases. A primary consideration with a different protein target is to utilize a combination of positive and negative control experiments to establish the validity of the assay, before fully committing resources to the interpretation of ligand binding data. Further, there are other technologies available for assessing cellular target engagement. We have explored a number of alternatives for the SHP2 phosphatase but have found this assay to be highly useful in our efforts. Finally, this assay is independent of SHP2 enzymatic activity and therefore allows for the evaluation and optimization of small molecules without regard to their binding mode.
Supplementary Material
ACKNOWLEDGMENTS:
This work was supported by National Institutes of Health Grant 1R21CA195422 (to L. T.), Epstein Family Foundation Award (to N. D. P. C.), and NCI Cancer Center Support Grant P30CA030199. Additionally, this project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Chemical Biology Consortium Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DISCLOSURES:
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES:
- 1.Hunter T Tyrosine phosphorylation: thirty years and counting. Current Opinion in Cell Biology. 21 (2), 140–146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A et al. Protein tyrosine phosphatases in the human genome. Cell. 117 (6), 699–711 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Cohen P Protein kinases—the major drug targets of the twenty-first century? Nature Reviews Drug Discovery. 1 (4), 309 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Ferguson FM, Gray NS Kinase inhibitors: the road ahead. Nature Reviews: Drug Discovery. 17 (5), 353–377 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Stanford SM, Bottini N Targeting Tyrosine Phosphatases: Time to End the Stigma. Trends in Pharmacological Sciences. 38 (6), 524–540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonks N Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS Journal. 280 (2), 346–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan R, Feng G PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 109 (3), 862–867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan G, Kalaitzidis D, Neel B The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27 (2), 179–192 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hof P, Pluskey S, Dhe-Paganon S, Eck M, Shoelson S Crystal structure of the tyrosine phosphatase SHP-2. Cell. 92 (4), 441–450 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Barford D, Neel B Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 6 (3), 249–254 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Neel BG, Chan G, Dhanji S in Handbook of Cell Signaling (Second Edition) Vol. 2 Pages 771–809 Amsterdam: Academic Press; (2010). [Google Scholar]
- 12.Tartaglia M et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nature Genetics. 29 (4), 465–468 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Yokosuka T et al. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. Journal of Experimental Medicine. 209 (6), 1201–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proceedings of the National Academy of Sciences U. S. A. 98 (24), 13866–13871 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe N et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nature Immunology. 4 (7), 670–679 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Bialy L, Waldmann H Inhibitors of protein tyrosine phosphatases: next-generation drugs? Angewandte Chemie International Edition England. 44 (25), 3814–3839 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, Liang F, Lawrence D, Zhang Z Small molecule approach to studying protein tyrosine phosphatase. Methods. 35 (1), 9–21 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Tautz L, Pellecchia M, Mustelin T Targeting the PTPome in human disease. Expert Opinion in Thereapeutics Targets. 10 (1), 157–177 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Tautz L, Mustelin T Strategies for developing protein tyrosine phosphatase inhibitors. Methods. 42 (3), 250–260 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Vintonyak V, Antonchick A, Rauh D Waldmann H The therapeutic potential of phosphatase inhibitors. Current Opinion in Chemical Biology. 13 (3), 272–283 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Barr A Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Future Medicinal Chemistry. 2 (10), 1563–1576 (2010). [DOI] [PubMed] [Google Scholar]
- 22.He R, Zeng L, He Y, Zhang S, Zhang Z Small molecule tools for functional interrogation of protein tyrosine phosphatases. FEBS Journal. 280, 731–750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsutsumi R, Ran H, Neel BG Off-target inhibition by active site-targeting SHP2 inhibitors. FEBS Open Biology. 8 (9), 1405–1411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YN et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 535 (7610), 148–152 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Nichols RJ et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nature Cell Biology. 20 (9), 1064–1073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical Trial NCT03114319, <https://clinicaltrials.gov/ct2/show/NCT03114319?term=NCT03114319&rank=1> (2019).
- 27.Clinical Trial NCT03634982, <https://clinicaltrials.gov/ct2/show/NCT03634982?term=NCT03634982&rank=1> (2019).
- 28.Sun X et al. Selective inhibition of leukemia-associated SHP2(E69K) mutant by the allosteric SHP2 inhibitor SHP099. Leukemia. 32 (5), 1246–1249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRochelle JR et al. Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition. Nature Communication. 9 (1), 4508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padua RAP et al. Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2. Nature Communication. 9 (1), 4507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu H et al. Resistance to allosteric SHP2 inhibition in FGFR-driven cancers through rapid feedback activation of FGFR. Oncotarget. 11 (3), 265–281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth LA, Collins I Measuring and interpreting the selectivity of protein kinase inhibitors. Journal of Chemical Biology. 2 (3), 131–151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez Molina D et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 341 (6141), 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Ericsson U, Hallberg B, Detitta G, Dekker N, Nordlund P Thermofluor-based high-throughput stability optimization of proteins for structural studies. Analytical Biochemistry. 357 (2), 289–298 (2006). [DOI] [PubMed] [Google Scholar]
- 35.McNulty DE et al. A High-Throughput Dose-Response Cellular Thermal Shift Assay for Rapid Screening of Drug Target Engagement in Living Cells, Exemplified Using SMYD3 and IDO1. SLAS Discovery. 23 (1), 34–46 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Romero C et al. A cellular target engagement assay for the characterization of SHP2 (PTPN11) phosphatase inhibitors. Journal of Biological Chemistry. 295 (9), 2601–2613, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ran H, Tsutsumi R, Araki T & Neel BG Sticking It to Cancer with Molecular Glue for SHP2. Cancer Cell. 30 (2), 194–196, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.