Abstract
Lymphocyte cytosolic protein 2 (LCP2) is one of the SLP-76 family of adapters, which are critical intermediates in signal cascades downstream of several receptors. LCP2 regulates immunoreceptor signaling (such as T-cell receptors) and is also required for integrin signaling in neutrophils and platelets. However, the role of LCP2 in the tumor microenvironment is still unknown. In this study, we found a significant increase of mRNA and protein expression of LCP2 in metastatic skin cutaneous melanoma compared to normal skin. The upregulation of LCP2 was associated with good overall survival of patients with metastatic skin cutaneous melanoma, who received pharmacotherapy and radiation. GSEA signaling pathways analysis showed that LCP2 was involved in multiple pathways of immune response and correlation analysis revealed LCP2 was positively correlated with molecules in TCR signaling and 11 immune checkpoints, while LCP2 negatively correlated with 2 immune checkpoints in the metastatic skin cutaneous melanoma. According to the different expressions of LCP2, high LCP2 expression was positively correlated with more tumor-infiltrating CD8+ T cells. Furthermore, Kaplan–Meier plot indicated that LCP2 acted as a prognostic biomarker for progression-free survival of patients with metastatic skin cutaneous melanoma receiving anti-PD1 immunotherapy. In conclusion, our results integrated both the expression and function of LCP2 in melanoma using multiple tools, shedding light on the potential role of LCP2 in melanoma, and suggesting LCP2 serves as a prognostic biomarker and therapeutic target in anti-tumor immunity.
Subject terms: Biomarkers, Medical research
Introduction
Studies of immune checkpoints, such as PD-1 and CTLA4, provide novel immunomodulatory targets to treat malignant tumors1. Immune checkpoint inhibitors have revolutionized the treatment for metastatic melanoma and substantially improved patients’ survival rate. Tumor-infiltrating immune cells play essential roles in affecting disease prognosis and are indicators of anti-tumor responses in patients with metastatic melanoma receiving immune checkpoint inhibitors2. Although some prognostic biomarkers3–5 for overall survival (OS) and response have been mentioned in melanoma, most of them are not sensitive and/or specific enough to be used for evaluating the infiltrating-immune cells based anti-tumor response. Novel prognostic biomarkers that are associated with tumor-infiltrating immune cells are required.
Our previous study indicated that lymphocyte cytosolic protein 2 (LCP2) protein tightly interacted with UBASH3B protein, which was identified as a novel prognostic biomarker and correlated with immune cell infiltration in the tumor microenvironment6. LCP2 is an adapter protein-encoding gene that is involved in T cell antigen receptor (TCR)-activated protein tyrosine kinase pathway7 and is essential for normal T-cell development and activation by serine phosphorylation8. LCP2 is involved in mediating the second signal from CD28/B7 families to thus regulate TCR signaling pathways9. In addition to T cells, there are several studies detailing the function of LCP2 in other immune cells. LCP2 plays an important role in NK-cell mediated recognition of missing-self targets10 and positively regulates antigen-induced mast cell activation by recruiting BCR11. Quantitative reductions of LCP2 trigger immune dysregulation with the excessive production of proinflammatory cytokines and autoantibodies12. These studies indicate that LCP2 deeply participates in immune responses through the regulation of immune cells. The function of LCP2 might vary from diseases, and further studies are required according to the gene profile alteration of immune cells in the tumor microenvironment. With this regard, deciphering the role of LCP2 in melanoma and the interaction between LCP2 and immune cells in melanoma will be of great interest.
In our study, we used bioinformatic methods to present a schematic diagram of LCP2 in skin cutaneous melanoma (SKCM). The highly expressed LCP2 in metastatic skin cutaneous melanoma (SKCM-Metastasis) suggested it was associated with good clinical outcomes. Differential LCP2 expression levels were able to affect the infiltration levels of immune cells. Furthermore, molecules of TCR signaling downstream and B7-CD28 family members were correlated with LCP2 expression in metastatic skin cutaneous melanoma. Based on our findings, we identified LCP2 as a prognostic biomarker for progressive-free survival (PFS) of patients with metastatic melanoma receiving anti-PD-1 immunotherapy. This study provides new insights into further exploration of the potential function and the mechanism of LCP2 in anti-tumor immunity. Understanding the complex interplay of LCP2 and the immune cells in melanoma could predict the therapeutic response of immune checkpoint-related therapy.
Results
LCP2 has different expression level in Pan-cancer
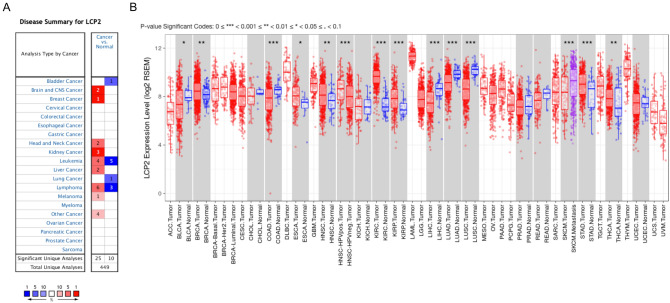
To explore the expression of LCP2 in malignant tumors, we first investigated the Oncomine database, which contains abundant microarray data of GEO and TCGA database. In comparison with normal tissue, there were 25 studies showing that LCP2 was upregulated in cancer, whereas 10 studies reporting downregulated LCP2 in cancer (Fig. 1A). LCP2 expression in the TIMER database including 29 types of tumors was sequentially evaluated. In comparison with corresponding normal tissues, LCP2 was upregulated in 7 types of cancers, while downregulated in 5 types of cancers (Fig. 1B). Furthermore, HPV-positive head and neck cancer (HNSC-HPV positive) had higher LCP2 expression than HPV-negative head and neck cancer. LCP2 expression in metastatic skin cutaneous melanoma (SKCM-Metastasis) was higher than primary skin cutaneous melanoma (SKCM-Primary) (Fig. 1B).
Figure 1.
The gene expression of LCP2 in different types of cancers. (A) Oncomine database showed LCP2 was expressed in 11 types of cancers. (B) TIMER database displayed LCP2 was expressed in multiple cancers based on TCGA database. TCGA The Cancer Genome Atlas, TIMER Tumor IMmune Estimation Resource, BRCA breast cancer, ESCA esophageal cancer, KIRC kidney renal clear cell cancer, KIRP kidney renal papillary cell cancer, STAD stomach adenocarcinoma, THCA thyroid cancer, BLCA bladder cancer, COAD colon adenocarcinoma, LIHC liver hepatocellular cancer, LUAD lung adenocarcinoma, LUSC lung squamous cell cancer, HNSC head and neck squamous cell cancer, SKCM skin cutaneous melanoma.
High expression of LCP2 in metastatic skin cutaneous melanoma indicates a good prognosis
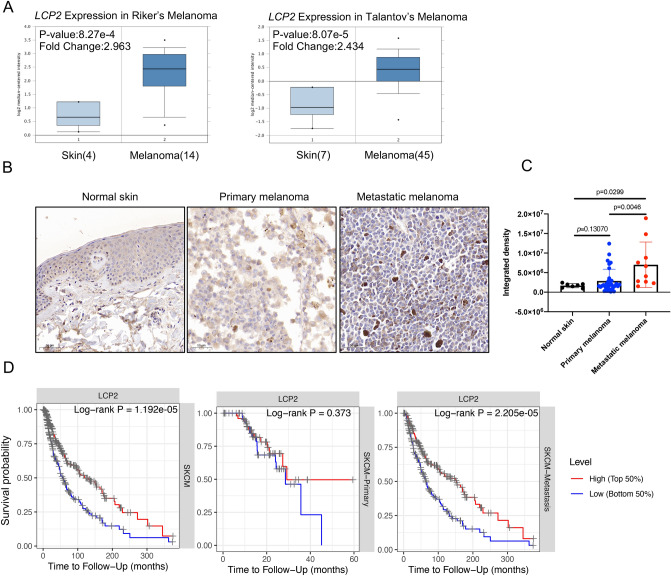
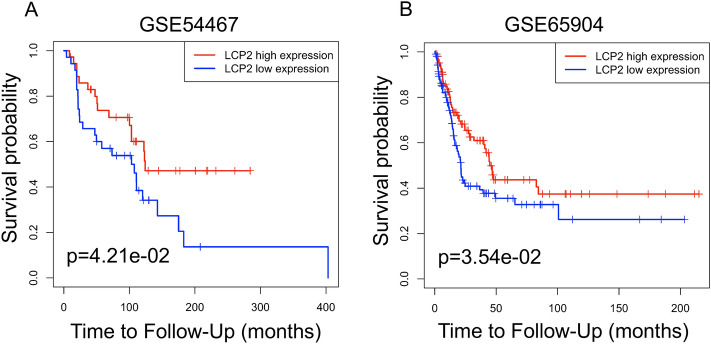
To explore the most obvious correlation between LCP2 and overall survival, survival analysis was performed in TCGA Pan-cancer. We found that LCP2 expression was significantly correlated with the overall survival in 9 cohorts (SKCM, SKCM-Metastasis, UVM, LGG, UCEC, THYM, SARC, GBM, HNSC-HPV positive). Among these cohorts, SKCM cohort exhibited the strongest correlation between LCP2 expression and overall survival (Supplementary Table S1). Therefore, we focused on investigating the role of LCP2 in the SKCM cohort, especially in the SKCM-Metastasis metastatic cohort. In comparison with normal skin, LCP2 was upregulated in skin cutaneous melanoma in Riker’s study13 (p value: 8.27 × 10−4; Fold Change: 2.963) and in Talantov’s study14 (p value: 8.07 × 10−5; Fold Change: 2.434) (Fig. 2A). Quantitative analysis of immunohistochemistry indicated that LCP2 protein was also highly expressed in metastatic melanoma tissue compared to normal skin and primary skin cutaneous melanoma (Fig. 2B,C). To investigate the prognostic value of LCP2 in the cohort, in which patients were received pharmacotherapy and radiation, TCGA-SKCM, GSE54467 and GSE65904 were included in this study. Kaplan–Meier curves showed that high LCP2 expression was significantly associated with a good overall survival in the SKCM and the SKCM-Metastasis cohorts, but not significant in the SKCM-Primary cohort (Fig. 2D). Based on two metastatic melanoma datasets (GSE5446715 and GSE6590416), the prognostic value of LCP2 expression was consistent with the result from the SKCM-Metastasis cohort (Fig. 3A,B).
Figure 2.
The expression and prognostic value of LCP2 in melanoma. (A) LCP2 expression level in human normal skin tissue and skin cutaneous melanoma. Based on the Oncomine database, box-plot diagrams were displayed to compare the LCP2 level in human normal skin tissue with that in skin cutaneous melanoma from studies reported by Riker et al. (p value: 8.27e−4, Fold Change: 2.963) and Talantov al. (p value: 8.07e−5, Fold Change: 2.434). (B) LCP2 protein expression in normal skin, primary melanoma and metastatic melanoma tissue. (C) Quantitative analysis of immunohistochemistry indicated that LCP2 protein was highly expressed in metastatic melanoma tissue compared with normal skin and primary melanoma. (D) Based on differential LCP2 expression level, patients with high-LCP2 expression had a better prognosis as compared with those with low-LCP2 expression in skin cutaneous melanoma and in cutaneous melanoma with metastasis, but not in primary sites of skin cutaneous melanoma.
Figure 3.
Prognostic value of LCP2 in melanoma. Kaplan–Meier curves display survival fraction for different LCP2 expression groups of patients based on GSE54467 (A) and GSE65904 (B) datasets.
Based on UALCAN analysis, we investigated the relationship between the clinical characteristics and LCP2 expression. There were significant differences of LCP2 expression in the different SKCM cohorts (primary vs. metastasis, p = 1.94 × 10−8) and individual cancer stages (stage 1 vs. stage 2, p = 2.42 × 10−3; stage 2 vs. stage 3, p = 1.97 × 10−5) (Supplementary Figure S1).
LCP2 and LCP2-correlated genes are involved in a variety of processes of immune response
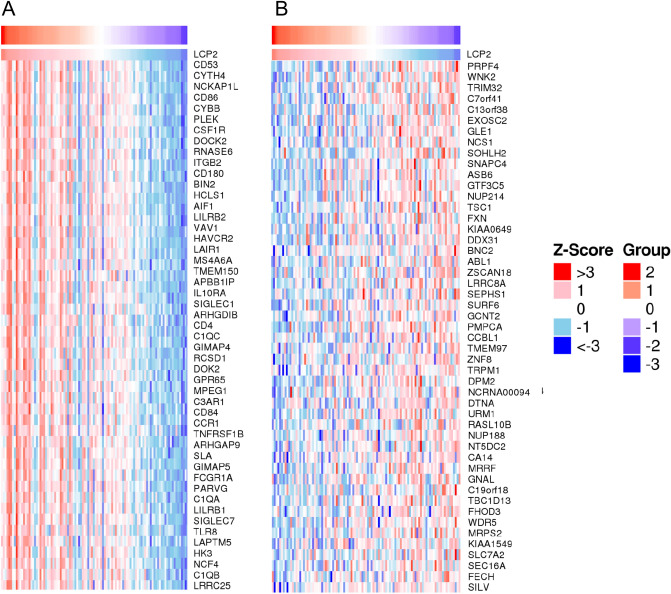
To further explore the genes interacting with LCP2, LinkedOmics analysis was performed. Top 10 genes that were positively correlated with LCP2 were CD53, CYTH4, NCKAP1L, CD86, CYBB, PLEK, CSF1R, DOCK2, RNASE6 and ITGB2. Top 10 genes that were negatively correlated with LCP2 were PRPF4, WNK2, TRIM32, C7orf41, C13orf38, EXOSC2, GLE1, NCS1, SOHLH2 and SNAPC4. Heatmaps of top 50 LCP2-positively correlated genes and LCP2-negatively correlated genes are displayed in Fig. 4A,B, respectively. These LCP2-correlated genes, such as CD86, HAVCR2 and SIGLEC1 were involved in multiple immune responses.
Figure 4.
Heatmaps of LCP2-correlated genes. (A) Heatmap of top 50 positively correlated genes. (B) Heatmap of top 50 negatively correlated genes.
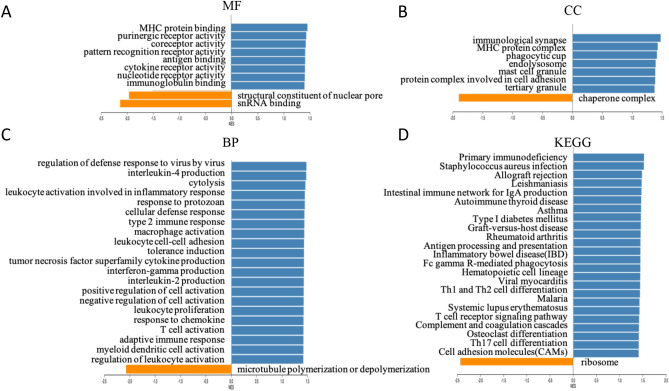
To further understand the functional enrichment of LCP2-correlated genes, GSEA method was used to analyze GO and KEGG signaling pathways. The major molecular function was located in “MHC protein binding” (Fig. 5A). In the cellular component, the highest normalized enrichment score was “immunological synapse” (Fig. 5B). In the biological process, “regulation of defense response to virus by virus” had the largest normalized enrichment score, with the maximum enrichment (Fig. 5C). For KEGG signaling pathways, several main pathways were enriched, including “antigen processing and presentation”, “Fc gamma R-mediated phagocytosis”, “Th1 and Th2 cell differentiation”, “T cell receptor signaling pathway” and “Th17 cell differentiation” (Fig. 5D). Collectively these results suggest LCP2 plays a critical role in signal transmission of immune responses.
Figure 5.
LCP2-correlated genes were involved in the GO and KEGG pathways. Based on the expression of LCP2 in melanoma, GSEA analysis was performed and the enrichments of MF (A) CC (B), BP (C) and KEGG signaling pathways (D) were shown. MF molecular function, CC cellular component, BP biological process.
LCP2 is positively correlated with TCR signaling activation and B7/CD28 immune checkpoint families in metastatic melanoma
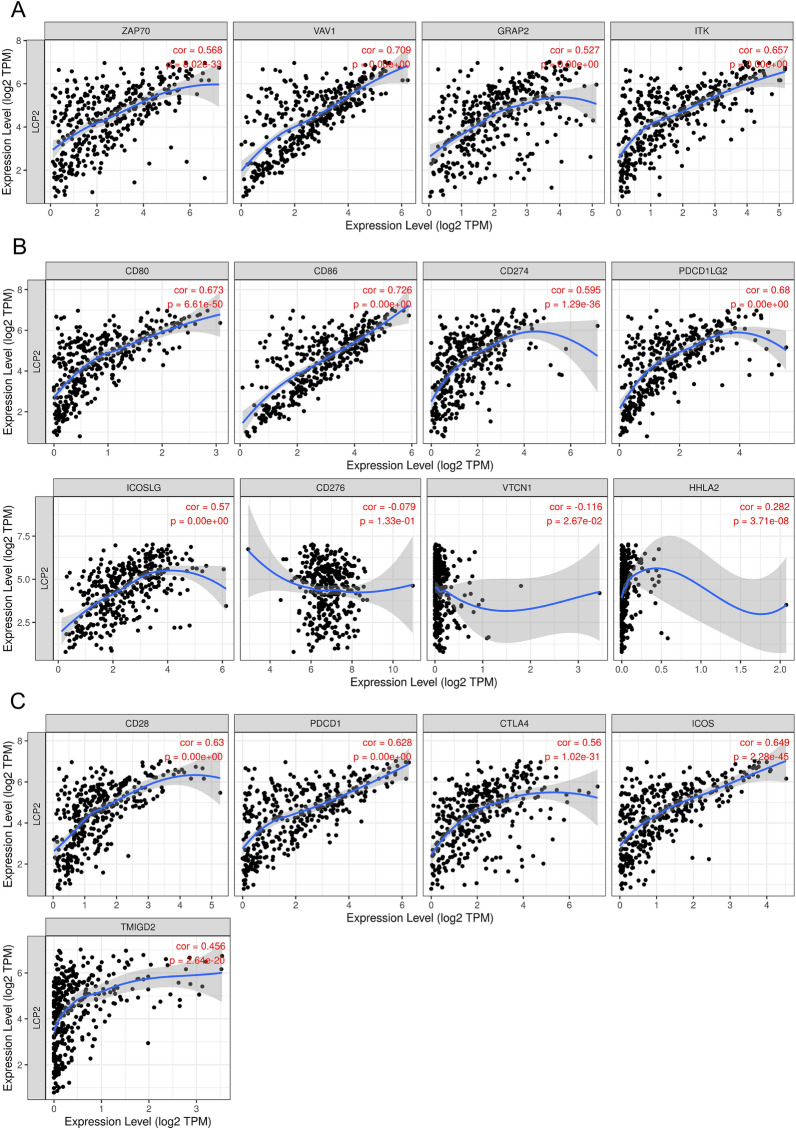
LCP2 is an important gene which is directly involved in the activation of TCR signaling and indirectly affects TCR signaling through CD28 and B7 families9. We investigated the correlation between LCP2 and recruited TCR signaling genes (ZAP70, VAV1, GRAP2, ITK). The results showed positive correlations between LCP2 and ZAP70 (cor = 0.568, p = < 0.001), LCP2 and VAV1 (cor = 0.709, < 0.001), LCP2 and GRAP2 (cor = 0.527, < 0.001), LCP2 and ITK (cor = 0.657, p < 0.001) (Fig. 6A). Based on T cell co-signaling pathway (ligand–receptor interactions in PathCards, https://pathcards.genecards.org), 35 out of 58 immune checkpoint genes were revealed in the LCP2-correlated gene sets (Supplementary Table S2). These results suggested that LCP2 had a potentially correlated expression pattern with immune checkpoints. When studying B7 and CD28 immune checkpoint families, LCP2 expression was positively correlated with CD80 (cor = 0.673, p < 0.001), CD86 (cor = 0.726, p < 0.001), CD274 (cor = 0.595, p < 0.001), PDCD1LG2 (cor = 0.68, p < 0.001), ICOSLG (cor = 0.57, p < 0.001), HHLA2 (cor = 0.282, p < 0.001), CD28 (cor = 0.63, p < 0.001), CTLA4 (cor = 0.56, p < 0.001), PDCD1 (cor = 0.628, p < 0.001), ICOS (cor = 0.649, p < 0.001), and TMIGD2 (cor = 0.456, p < 0.001) in SKCM-Metastasis cohorts. Whereas LCP2 was negatively correlated with VTCN1 (cor = -0.116, p = 0.0267) in SKCM-Metastasis cohorts but was not significantly correlated with CD276 (cor = -0.079, p = 0.133) (Fig. 6B,C). Further analysis revealed that LCP2 was positively correlated with other immune exhausting genes and immunosuppressive genes (TIM3 also called HAVCR2, LAG3, TOX, TIGIT) (Supplementary Figure S2).
Figure 6.
The correlation between LCP2 and signal molecules of TCR/CD28-B7 checkpoint families. (A) LCP2 expression level was correlated with ZAP70, VAV1, GRAP2, ITK in SKCM-Metastasis. (B) LCP2 expression level was correlated with CD80, CD86, CD274, PDCD1LG2, ICOSLG, CD276, VTCN1, HHLA2 in the SKCM-Metastasis cohort. (C) LCP2 expression level was correlated with CD28, CTLA4, PDCD1, ICOS and TMIGD2 in the SKCM-Metastasis cohort. SKCM skin cutaneous melanoma.
LCP2 is mainly correlated with tumor-infiltrating CD8+ T cells in metastatic melanoma
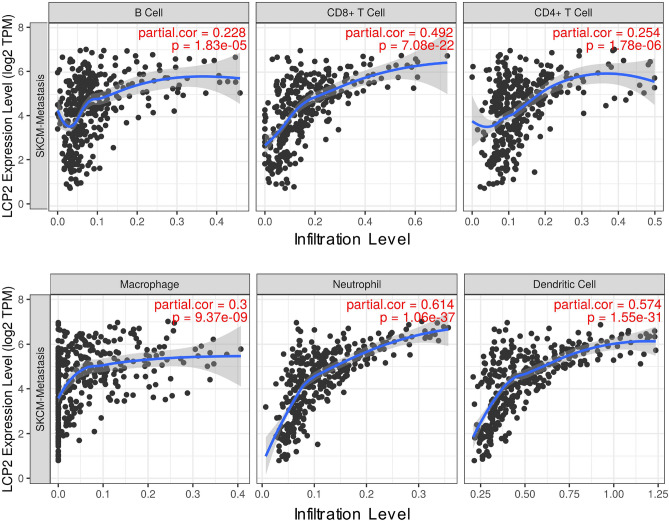
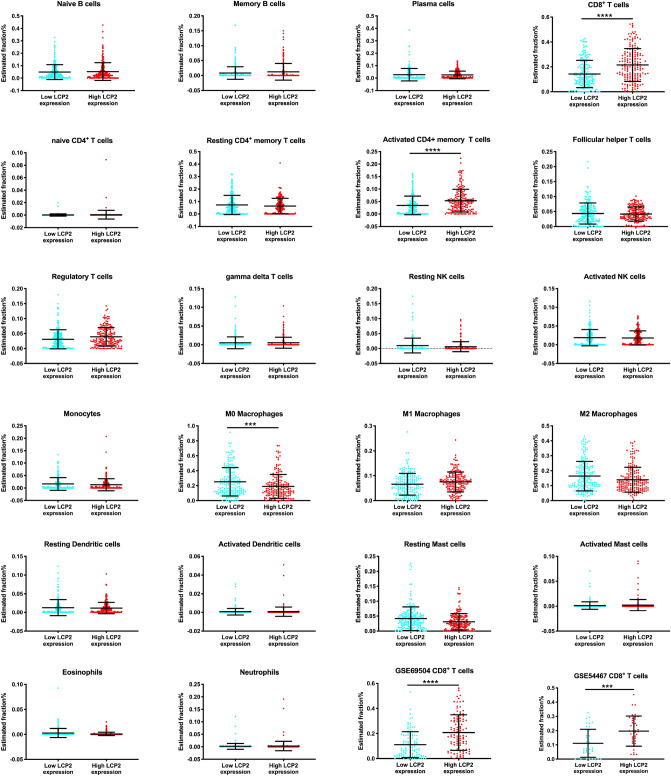
The correlations between LCP2 expression and 6 types of melanoma-infiltrating-immune cells were evaluated via TIMER. LCP2 expression was correlated with tumor-infiltrating B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil and dendritic cell in SKCM-Metastasis cohorts (Fig. 7). To further investigate more immune cell subtypes, gene signature file LM22 which defined 22 immune cell subtypes was applied and CIBERSORT was performed. Based on SKCM-Metastasis, there were significantly higher estimated fractions of CD8+ T cells and activated CD4+ T cells in the high-LCP2 expression group, whereas lower estimated fractions of M0 macrophages in the high-LCP2 expression group (Fig. 8). In the validation cohort of GSE65904 dataset, more CD8+ T cells were infiltrated in the high-LCP2 expression group (Fig. 8), a similar infiltration pattern of CD8+ T cells was observed in the second validation cohort of GSE54467 dataset (Fig. 8). Although the infiltration of more CD8+ T cells in the high-LCP2 expression group was not significantly observed in the TCGA-SKCM primary melanoma cohort, the trend observed was similar with metastatic melanoma (Supplementary Figure S3).
Figure 7.
The correlation between LCP2 and tumor-infiltrating immune cells. LCP2 expression was correlated with tumor-infiltrating B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil and dendritic cell in the SKCM-Metastasis cohort.
Figure 8.
CIBERSORT analysis of 22 immune cells based on LCP2 expression level. Significantly higher estimated fractions of CD8+ T cells and activated memory CD4+ T cells occurred in the high-LCP2 expression group, whereas lower estimated fractions of M0 macrophages existed in the high-LCP2 expression group. Validation cohorts (GSE65904 and GSE54467) indicated that significantly higher estimated fractions of CD8+ T cells occurred in the high-LCP2 expression group.
LCP2 acts as a novel prognostic biomarker of progression-free survival for patients who received anti-PD1 immunotherapy
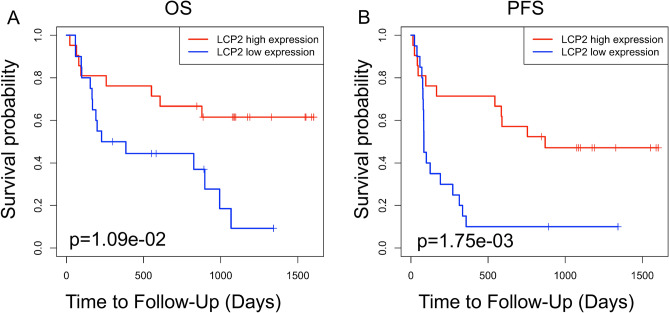
To further explore the prognostic value of LCP2 in patients who received immune checkpoint inhibitors, the Gide2019_PD1 and Gide2019_PD1+CTLA4 cohorts were included in this study17. Kaplan–Meier curves of overall survival and progression-free survival were plotted in these two cohorts. In the Gide2019_PD1 cohort of patients receiving anti-PD-1 immunotherapy, we found that high-LCP2 expression was associated with a good overall survival (Fig. 9A, p = 1.09 × 10−2) and progression-free survival (PFS) (Fig. 9B, p = 1.75 × 10−3). To strengthen results obtained in Kaplan–Meier analysis, survival (PFS and OS) data were also evaluated by univariate and multivariate Cox regression analyses, preferably using LCP2 expression as continuous variable to avoid possible bias due to cutoff point selection. Tumor-infiltrating immune cells were analyzed with LCP2 expression as variables. The results indicated that LCP2 was a prognostic factor of progression-free survival (p = 0.01) (Supplementary Table 3), but not overall survival (p = 0.072) (Supplementary Table 4). In addition, LCP2 expression was unable to predict the overall survival (p = 0.511) and progression-free survival (p = 0.150) in the Gide2019_PD1 + CTLA4 cohort of patients receiving anti-PD-1 and anti-CTLA4 combination immunotherapy17 (Supplementary Figure S4).
Figure 9.
Prognostic value of LCP2 in melanoma patients receiving immune checkpoint blockade. Kaplan–Meier curves of OS (A) and PFS (B) for patients with low and high expression of LCP2 in the Gide2019_PD1 cohort who received anti-PD-1 immunotherapy. OS overall survival, PFS progression-free survival.
Discussion
In the present study, we analyzed RNA-seq data by using multiple tools and performed immunohistochemistry of tissue microarray to investigate the expression of LCP2 in melanoma. We found mRNA and protein of LCP2 was highly expressed in SKCM-Metastasis. The high expression of LCP2 was associated with good survival of patients in SKCM-Metastasis cohort, in which patients received pharmacotherapy and radiation. Furthermore, LCP2 was involved in some immune-responses-related signaling pathways and high expression of LCP2 increased the infiltration of anti-tumor immune cells and thus helped to predict the progression-free survival of patients with metastatic skin cutaneous melanoma receiving anti-PD-1 immunotherapy.
LCP2 protein plays an important role in the TCR-mediated signal transduction pathway and tumor malignancy. High LCP2 protein expression is correlated with aggressive behavior in chronic lymphocytic leukemia cells18. LCP2 is involved in colon cancer metastasis as a differentially expressed gene19 and plays a critical role in inflammation of colorectal cancer20. The high LCP2 expression pattern also occurred in metastatic skin cutaneous melanoma and was correlated with good overall survival. In addition, LCP2 protein tightly interacted with UBASH3B protein, which was identified as a novel prognostic biomarker and correlated with immune cell infiltration in the tumor microenvironment6. The function of LCP2 protein in normal T cell differentiation and activation had been reported that serine phosphorylation of LCP2 protein was essential to T cell differentiation by modulating Th1 and Th2 cells function8. GSEA indicated LCP2 was correlated with “T cell receptor signaling pathway”, “Th1 and Th2 cell differentiation” and “Th17 cell differentiation”. These results were consistent with previous studies8. Although mislocalization of LCP2 protein caused CD4+ T cells to skew towards the inflammatory Th1 and Th17 lineages21, the role of LCP2 in Th17 cell differentiation is still unclear and is worth investigating.
Activation of the TCR signaling pathway is important for anti-tumor immune response. In this study, LCP2 was found to be associated with TCR signal molecules. These results are in accordance with recent studies indicating that LCP2 was involved in TCR-mediated signal transduction22. Additionally, the receptor-ligand interactions of immune checkpoints regulated TCR signal transduction and affected anti-tumor immune response23. Interestingly, we found a few immune checkpoint genes that were correlated with LCP2. The suppressive immune checkpoints and “exhausting genes” regulated functions of immune cells in the tumor microenvironment, resulting in CD8+ T cells dysfunction24. In addition, a large number of studies have reported the different roles of various tumor-infiltrating immune cells in melanoma2,24–26. In the SKCM metastasis cohort, the increase in the amount of CD8+ T cells and activated CD4+ memory T cells may have more potential to kill cancer cells after undergoing anti-PD-1 immunotherapy. High-LCP2 expression patients in our study had more infiltrations of CD8+ T cells, which was associated with a good prognosis27. Kaplan–Meier Curve indicated that LCP2 was associated with a good overall survival of patients with skin cutaneous metastatic melanoma, who did not receive immunotherapy. It is suggested that LCP2 may serve as a favorable prognostic biomarker and played an important role in skin cutaneous metastatic melanoma through the main function of CD8+ T cells. Excitingly, LCP2 was a good prognostic biomarker of progression-free survival for patients who received anti-PD1 immunotherapy.
There were some limitations in this study. First, the potential mechanisms of LCP2 involving anti-PD1 immunotherapy and combination immunotherapy are still unclear. Second, the interactions between LCP2 and immune checkpoints are required by further analysis in melanoma.
In summary, our results offer new insights into the role of LCP2 associated immune responses in metastatic skin cutaneous melanoma. The integrated analysis of LCP2 expression datasets and the utilization of multiple methods help us translate bioinformatics data into biomarkers identification for clinical application in the prognostic prediction of melanoma. Altogether our results established a novel role of LCP2 as an immune regulator in melanoma and identified LCP2 as a rational prognostic biomarker in patients with metastatic skin cutaneous melanoma receiving anti-PD-1 immunotherapy.
Methods
Expression datasets and clinical information
RNA transcriptome data of all melanoma samples (n = 472) (SKCM), primary skin cutaneous melanoma (SKCM-Primary) (n = 104) and metastatic SKCM (SKCM-Metastasis) (n = 368) were downloaded from TCGA (https://portal.gdc.cancer.gov/).
Transcriptome and clinical survival data of validation cohorts (GSE54467 (n = 71) and GSE65904 (n = 210)) were included in this study. Raw data were normalized by log(X + 1) for further analysis.
2 cohorts of metastatic melanoma with immunotherapy (Gide2019_PD1 (n = 41), Gide2019_PD1 + CTLA4 (n = 32)) were included to evaluate the prediction ability of LCP2.
Expression analysis
The mRNA expression levels of LCP2 in different types of cancer were analyzed in the Oncomine database (https://www.oncomine.org) and mRNA expression level of LCP2 in skin cutaneous melanoma and normal skin tissue was investigated in a serial of melanoma studies, including Riker and Talantov’s studies.
This study was approved by the Clinical Research Ethics Committee of the Second Xiangya Hospital of Central South University. Immunohistochemistry (IHC) was used to evaluate the expression of LCP2 protein. The protocol of immunohistochemistry was detailed in the previous study6. The anti-LCP2 antibody (#DF7020, Biosciences, China) was used as the primary antibody. A human melanoma tissue microarray (K063Me01, Bioaitech, China) which contained 63 samples was purchased from commercial company and used in the study. Our inclusive criteria were normal skin and skin cutaneous melanoma. 7 normal skin, 32 primary melanoma and 10 metastatic melanoma samples were included as three groups and an IHC Toolbox plug-in was used to quantitative the brown positive particles. The integrated density of the brown positive particles was used to compare the expression of LCP2 protein in different groups.
UALCAN analysis
UALCAN database (http://ualcan.path.uab.edu/index.html)28 is allowed to analyze the relationship between relative expression of LCP2 and different types of skin tissue (normal skin tissue, primary skin cutaneous melanoma and metastatic skin cutaneous melanoma), as well as clinical characteristics of melanoma patients, such as age, gender, stages, weight and promoter methylation level.
LinkedOmics analysis
LinkedOmics (http://www.linkedomics.org)29 is a publicly available portal that includes multi-omics data from all 32 TCGA Cancer types. We used RNA-seq data of SKCM and chose the Pearson test to calculate the correlation coefficient for LCP2 correlated significant genes. In the LinkInterpreter module, Gene set enrichment analysis (GSEA) was applied to generate Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways30. “Rank Criteria (from LinkFinder Result)” was FDR. “Minimum Number of Genes (Size)” was 3 and “Simulations” was 10,000.
TIMER
The different mRNA expression of LCP2 was analyzed in all types of cancers in Tumor IMmune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer)31 and three melanoma cohorts: SKCM, SKCM-Primary, SKCM-Metastasis were used to evaluate the association between LCP2 mRNA expression level and overall survival. In the SKCM cohorts, patients with primary and metastatic skin cutaneous melanoma were included. The correlations between LCP2 and immune cells (B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil and dendritic cell) were evaluated. Furthermore, the correlations between LCP2 and downstream signaling molecules of TCR and B7-CD28 immune checkpoint family members were studied.
CIBERSORT
CIBERSORT32 is an online website for analyzing 22 immune cell types infiltrated in different kinds of cancers. Fragments per kilobase million (FPKM) data of SKCM-Metastasis were downloaded from the TCGA database and the data were extracted and merged by Perl software as a matrix file. FPKM value was converted to TPM value by R Studio Version 1.1.463. In addition, matrixes of GSE54467 and GSE65904 were downloaded and annotated by R studio. The matrix files were uploaded into the CIBERSORT network (https://cibersort.stanford.edu) as a Mixture file. LM22 was selected as “Signature gene file” and “Permutations” run 1000. Disable quantile normalization was only recommended for RNA-seq of SKCM-Metastasis. Deconvolution results were expressed as relative fractions and the analysis was performed in the SKCM-Primary, SKCM-Metastasis, GSE54467 and GSE65904 datasets, respectively. Based on the median expression of LCP2 as a dynamical cutoff, melanoma patients were divided into two groups, high-LCP2-expression group and low-LCP2-expression group. Data were imported into GraphPad Prism 7 and the relative value of 22 types of immune cells was evaluated in both high- and low-LCP2 expression groups.
Kaplan–Meier curve analysis
The “survival” R package was used to plot the Kaplan–Meier curve. The median value of LCP2 expression was settled as a cut-off. The patients were divided into two groups: high-LCP2-expression group and low-LCP2-expression group. Overall survival and progression-free survival were used to evaluate the prognostic value of LCP2 expression in melanoma. GSE54467, GSE65904, Gide2019_PD1, Gide2019_PD1 + CTLA4 cohorts were used to evaluate the prognostic value of LCP2.
Statistics analysis
The normally distributed quantitative data are shown as mean ± standard deviation (SD). The Student’s t-test is used to compare immune cell fractions in high- and low- LCP2-expression groups for statistical analysis. KM plot was used to evaluate the correlation between LCP2 and OS/PFS through the two-sided Wald test in a Cox proportional hazards regression analysis. A Bonferroni test was displayed as a correction for multiple comparisons. When p < 0.05, statistical differences are considered.
Supplementary information
Acknowledgements
We thank Phillip M. Galbo Jr from Albert Einstein College of Medicine as native English speaker for language editing. We thank Kanehisa laboratory for formal permission of the KEGG pathway database.
Abbreviations
- LCP2
Lymphocyte cytosolic protein 2
- TCR
T cell antigen receptor
- KIRC
Kidney renal clear cell cancer
- KIRP
Kidney renal papillary cell cancer
- SKCM-Metastasis
Skin cutaneous melanoma with metastasis
- STAD
Stomach adenocarcinoma
- THCA
Thyroid cancer
- BLCA
Bladder cancer
- COAD
Colon adenocarcinoma
- LIHC
Liver hepatocellular cancer
- HNSC
Head and neck squamous cell cancer
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell cancer
Author contributions
Z.W. and M.P. wrote the main manuscript text. Z.W. performed the IHC experiment and analyzed the data. M.P. designed and supervised the project. All authors reviewed the manuscript.
Funding
This study was supported by Hunan Provincial Natural Science Foundation of China (2018JJ3762), the Fundamental Research Funds for the Central Universities of Central South University (No. 2018zzts262) and China Scholarship Council (Nos. 201806370101, 201806375066).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88676-9.
References
- 1.Perez-Ruiz E, et al. Prophylactic TNF blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature. 2019 doi: 10.1038/s41586-019-1162-y. [DOI] [PubMed] [Google Scholar]
- 2.Balatoni T, et al. Tumor-infiltrating immune cells as potential biomarkers predicting response to treatment and survival in patients with metastatic melanoma receiving ipilimumab therapy. Cancer Immunol. Immunother. 2018;67:141–151. doi: 10.1007/s00262-017-2072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner NB, et al. Tumor microenvironment-derived S100A8/A9 is a novel prognostic biomarker for advanced melanoma patients and during immunotherapy with anti-PD-1 antibodies. J. Immunother. Cancer. 2019;7:343. doi: 10.1186/s40425-019-0828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng L, Chen Z, Chen Y, Wang X, Tang N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8:7161–7173. doi: 10.1002/cam4.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner NB, Forschner A, Leiter U, Garbe C, Eigentler TK. S100B and LDH as early prognostic markers for response and overall survival in melanoma patients treated with anti-PD-1 or combined anti-PD-1 plus anti-CTLA-4 antibodies. Br. J. Cancer. 2018;119:339–346. doi: 10.1038/s41416-018-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Wang Y, Peng M, Yi L. UBASH3B is a novel prognostic biomarker and correlated with immune infiltrates in prostate cancer. Front. Oncol. 2019;9:1517. doi: 10.3389/fonc.2019.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr VA, et al. Development of nanoscale structure in LAT-based signaling complexes. J. Cell Sci. 2016;129:4548–4562. doi: 10.1242/jcs.194886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navas VH, Cuche C, Alcover A, Di Bartolo V. Serine phosphorylation of SLP76 is dispensable for T cell development but modulates helper T cell function. PLoS ONE. 2017;12:e0170396. doi: 10.1371/journal.pone.0170396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel F, Attal-Bonnefoy G, Mangino G, Mise-Omata S, Acuto O. CD28 as a molecular amplifier extending TCR ligation and signaling capabilities. Immunity. 2001;15:935–945. doi: 10.1016/s1074-7613(01)00244-8. [DOI] [PubMed] [Google Scholar]
- 10.Lampe K, et al. Slp-76 is a critical determinant of NK-cell mediated recognition of missing-self targets. Eur. J. Immunol. 2015;45:2072–2083. doi: 10.1002/eji.201445352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bounab Y, et al. Proteomic analysis of the SH2 domain-containing leukocyte protein of 76 kDa (SLP76) interactome in resting and activated primary mast cells [corrected] Mol. Cell Proteomics. 2013;12:2874–2889. doi: 10.1074/mcp.M112.025908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siggs OM, et al. Quantitative reduction of the TCR adapter protein SLP-76 unbalances immunity and immune regulation. J. Immunol. 2015;194:2587–2595. doi: 10.4049/jimmunol.1400326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riker AI, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talantov D, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 15.Cabrita R, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 16.Jayawardana K, et al. Determination of prognosis in metastatic melanoma through integration of clinico-pathologic, mutation, mRNA, microRNA, and protein information. Int. J. Cancer. 2015;136:863–874. doi: 10.1002/ijc.29047. [DOI] [PubMed] [Google Scholar]
- 17.Gide TN, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell. 2019;35:238–255.e236. doi: 10.1016/j.ccell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Dezorella N, et al. SLP76 integrates into the B-cell receptor signaling cascade in chronic lymphocytic leukemia cells and is associated with an aggressive disease course. Haematologica. 2016;101:1553–1562. doi: 10.3324/haematol.2015.139154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu S, Wang H, Yu M. A putative molecular network associated with colon cancer metastasis constructed from microarray data. World J. Surg. Oncol. 2017;15:115. doi: 10.1186/s12957-017-1181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, et al. Inflammatory genes are novel prognostic biomarkers for colorectal cancer. Int. J. Mol. Med. 2018;42:368–380. doi: 10.3892/ijmm.2018.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnenberg GF, et al. Mislocalization of SLP-76 leads to aberrant inflammatory cytokine and autoantibody production. Blood. 2010;115:2186–2195. doi: 10.1182/blood-2009-08-237438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo WL, et al. Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat. Immunol. 2018;19:733–741. doi: 10.1038/s41590-018-0131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sankin A, et al. The expanding repertoire of targets for immune checkpoint inhibition in bladder cancer: what lies beneath the tip of the iceberg, PD-L1. Urol. Oncol. 2018;36:459–468. doi: 10.1016/j.urolonc.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D. CAR-T "the living drugs", immune checkpoint inhibitors, and precision medicine: a new era of cancer therapy. J. Hematol. Oncol. 2019;12:113. doi: 10.1186/s13045-019-0819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dabrosin N, et al. Innate immune cell infiltration in melanoma metastases affects survival and is associated with BRAFV600E mutation status. Melanoma Res. 2019;29:30–37. doi: 10.1097/CMR.0000000000000515. [DOI] [PubMed] [Google Scholar]
- 26.Antonio N, et al. The wound inflammatory response exacerbates growth of pre-neoplastic cells and progression to cancer. EMBO J. 2015;34:2219–2236. doi: 10.15252/embj.201490147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards J, et al. CD103(+) Tumor-resident CD8(+) T cells are associated with improved survival in immunotherapy-naive melanoma patients and expand significantly during anti-PD-1 treatment. Clin. Cancer Res. 2018;24:3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- 28.Chandrashekar DS, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Can. Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.