Abstract
Eukaryotic sliding clamp proliferating cell nuclear antigen (PCNA) plays a critical role as a processivity factor for DNA polymerases and as a binding and acting platform for many proteins. The ring-shaped PCNA homotrimer and the DNA damage checkpoint clamp 9-1-1 are loaded onto DNA by clamp loaders. PCNA can be loaded by the pentameric replication factor C (RFC) complex and the CTF18-RFC-like complex (RLC) in vitro. In cells, each complex loads PCNA for different purposes; RFC-loaded PCNA is essential for DNA replication, while CTF18-RLC-loaded PCNA participates in cohesion establishment and checkpoint activation. After completing its tasks, PCNA is unloaded by ATAD5 (Elg1 in yeast)-RLC. The 9-1-1 clamp is loaded at DNA damage sites by RAD17 (Rad24 in yeast)-RLC. All five RFC complex components, but none of the three large subunits of RLC, CTF18, ATAD5, or RAD17, are essential for cell survival; however, deficiency of the three RLC proteins leads to genomic instability. In this review, we describe recent findings that contribute to the understanding of the basic roles of the RFC complex and RLCs and how genomic instability due to deficiency of the three RLCs is linked to the molecular and cellular activity of RLC, particularly focusing on ATAD5 (Elg1).
Subject terms: DNA synthesis, Checkpoints
Genetic stability: DNA clamp proteins
The attachment and removal of clamp proteins that encircle DNA as it is copied and assist its replication and maintenance is mediated by DNA clamp loader and unloader proteins; defects in loading and unloading can increase the rate of damaging mutations. Kyoo-young Lee and Su Hyung Park at the Institute for Basic Science in Ulsan, South Korea, review current understanding of the activity of clamp loading and unloading proteins. They examine research on the proteins in eukaryotic cells, those containing a cell nucleus, making their discussion relevant to understanding the stability of the human genome. They focus particular attention on a protein called ATAD5, which is involved in unloading the clamp proteins. Deficiencies in ATAD5 function have been implicated in genetic instability that might lead to several different types of cancer.
Introduction
Eukaryotic chromosomal DNA is duplicated by replicative DNA polymerases (Pols), Pol δ and Pol ε, which are tethered to a sliding clamp, proliferating cell nuclear antigen (PCNA). The DNA-encircling PCNA homotrimer increases the processivity of replicative DNA polymerases1. Repair DNA synthesis is the final step in excision repair and homologous recombination (HR) and is also carried out by DNA polymerases bound to PCNA. In addition to DNA polymerases, PCNA functions as a platform for recruiting many other proteins involved in different DNA transactions, such as lagging strand maturation, sister chromatid cohesion establishment, nucleosome reassembly, and DNA damage checkpoint activation. In addition, PCNA, when modified by ubiquitination and SUMOylation under specific cellular conditions, recruits proteins specialized in DNA damage tolerance pathways or anti-recombination activity2.
The closed ring-shaped PCNA homotrimer is abundant in the nucleus in its nucleoplasmic free form and DNA-encircling form; of these two forms, the latter participates in cellular activities regulating DNA metabolism. The two forms undergo transition through two processes: PCNA loading and unloading3. A closed PCNA homotrimer in the nucleoplasm is opened and loaded onto DNA by a clamp loader complex at a single-strand DNA/double-strand DNA junction (ssDNA/dsDNA) with a 3′-OH end. After PCNA completes this task, it is unloaded from the DNA by a clamp-unloading complex. To meet the high demand for PCNA during DNA replication, efficient cycling between nucleoplasmic PCNA and DNA-loaded PCNA through the loading/unloading process is critical.
In eukaryotes, another ring-shaped clamp, RAD9–RAD1–HUS1 (the 9-1-1 complex, Ddc1–Rad17–Mec3 in budding yeast), plays a role in ataxia telangiectasia-mutated (ATM)- and rad3-related (ATR)-mediated checkpoint activation after being loaded at damage sites4,5. Similar to the PCNA-loading process, the 9-1-1-loading process requires ring opening, followed by recruitment to DNA, which is performed by a specialized checkpoint clamp loader complex6.
PCNA is loaded onto DNA by the replication factor C (RFC) complex, which is a pentameric AAA+ ATPase complex composed of a large subunit, RFC1, and four small RFC proteins, RFC2, 3, 4, and 57–9. In eukaryotes, three RFC-like complexes (RLCs) exist with overlapping, as well as distinct, cellular tasks (Table 1). Each RLC complex is composed of CTF18, ATAD5 (Elg1 in yeast), or RAD17 (Rad24 in budding yeast) as the large subunit and four small subunits, RFC2–5. CTF18 is stably associated with DCC1 and CTF8 and forms a heptameric complex with RFC2–5. The CTF18–DCC1–CTF8–RLC complex (hereafter referred to as CTF18-RLC) participates as a single entity in various DNA transactions, such as PCNA loading10,11, sister chromatid cohesion12,13, and DNA replication checkpoint activation14,15, although DCC1 and CTF8 are not essential for the PCNA loading/unloading activity of CTF18-RLC in vitro. ATAD5 (Elg1)-RLC is a primary PCNA unloader10,16,17. In contrast to the RFC complex and two RLCs, RAD17 (Rad24)-RLC functions as a 9-1-1 damage clamp loader6.
Table 1.
A summary of the primary activities of the eukaryotic RFC complex and the three RLCs.
| Biochemical and cellular activity | RFC | CTF18-RLC | RAD17 (Rad24)-RLC | ATAD5 (Elg1)-RLC | ||||
|---|---|---|---|---|---|---|---|---|
| Human | Yeasta | Human | Yeast | Human | Yeast | Human | Yeast | |
| PCNA loading | O | O | O | O | X | X | X | X |
| PCNA unloading | O (weak)b | O (weak)b | X | Xb/Oc | X | X | O | O |
| Cohesion establishment | Unknown | X | O | O | X | X | X | X |
| 9-1-1 loading | X | X | X | X | O | O | Xb | Xb |
| Damage checkpoint | Unknownd | Unknownd | Unknown | X | O | O | Xe | Of |
| Replication checkpoint | Unknownd | Unknownd | Unknown | O | O | X | Unknown | Xf |
Considering the multiple essential roles of PCNA in cellular processes, PCNA loading/unloading needs to be accurately regulated. The importance of PCNA loading/unloading regulation is strongly supported by the unviability of cells with deficient PCNA-loading processes and by the severe genomic instability in cells defective with PCNA unloading (in species from yeast to humans). In this review, we first describe the primary roles of eukaryotic clamp loaders/unloaders while focusing on recent findings that reinforce the original concept and help establish a composite understanding of the interrelationships between these basic roles. Next, building on recent findings, we outline the genomic instability observed in yeast and mammals with deficient clamp loaders/unloaders and describe in detail how genomic instability due to deficiency of the three RLCs is linked to their cellular activity, particularly focusing on ATAD5 (Elg1). We do not discuss PCNA modifications, such as SUMOylation and phosphorylation, or checkpoint regulation by Elg1-RLC in this review, since they are well described in other reviews, and their relevance to mammals is still being investigated18,19.
Primary roles of the RFC complex and RLCs
PCNA loading
The RFC complex is a primary PCNA loader. The ATP-bound RFC complex binds to and opens a PCNA homotrimer ring; the ring-opened RFC complex–PCNA intermediates bind to gapped or nicked DNA, and the binding is augmented by ATP binding. DNA binding of the intermediates leads to ATP hydrolysis, which triggers the release of PCNA from the RFC complex, followed by closure of the PCNA ring on the DNA. In vitro mutational studies have shown that the ordered hydrolysis of ATP in the RFC complex subunits is required for PCNA loading7.
PCNA is loaded primarily at ssDNA/dsDNA junctions with a 3′-OH end in different ways during DNA replication and repair. For lagging strand synthesis, the primer–template junction is repeatedly generated by DNA Pol α/primase. PCNA is loaded onto the primer–template junction, after which Pol δ synthesizes the discontinuous lagging strand. For leading strand synthesis, the first Okazaki fragment (passing over the origin of replication) is synthesized by Pol δ, followed by continuous leading strand synthesis performed by Pol ε20,21. In DNA repair pathways, the PCNA-loadable structures are provided by the 3′ ends in gaps formed during the incision/excision process or by the 3′ overhang that invades the sister chromatid during HR.
At the replication fork, it has been widely accepted that the leading and lagging strands are mainly replicated by Pol ε and Pol δ, respectively, based on strand-specific ribonucleotide incorporation mapping in yeast22,23. However, one report has suggested that Pol δ synthesizes both strands at the replication fork24.
Both yeast and human CTF18-RLC can load PCNA on gapped DNA with lower efficiency than the RFC complex but not on nicked DNA10,11. In the same in vitro system, RAD24-RLC and ATAD5-RLC cannot load PCNA. In both yeast and human cells, CTF18-RLC interacts with Pol ε via DCC125,26. When complexed with nonsynthesizing Pol ε in this way, human CTF18–DCC1–CTF18 RLC can load PCNA more efficiently than pentameric CTF18-RLC25,27. A recent genome-wide analysis of PCNA occupation on the leading and lagging strands consistently showed that Ctf18-RLC preferentially loads PCNA on the leading strand, and that the RFC complex preferentially loads PCNA on the lagging strand28. This study, along with other studies, showed that interaction with Pol ε contributes to the recruitment of Ctf18-RLC to replication forks28,29. However, Ctf18 is not essential for bulk DNA replication and cannot substitute for Rfc1 deletion. Instead, PCNA loading by Ctf18 is known to be important for cohesion establishment and DNA replication checkpoint activation, which are discussed below.
PCNA unloading
After completion of DNA synthesis during DNA replication and repair, PCNA is unloaded from DNA by the eukaryotic clamp unloader ATAD5-RLC (Elg1-RLC in yeast)10,16,17,30. The RFC complex can unload PCNA in vitro9, even though its activity is significantly lower than that of ATAD5-RLC10. It has been reported that yeast Ctf18-RLC can unload PCNA in vitro when the single-strand region of the DNA substrate is coated by heterotrimeric replication protein A (RPA)11. However, another recent study has shown that Ctf18-RLC catalyzes PCNA loading instead of PCNA unloading10. In addition, the physiological relevance of PCNA unloading by the RFC complex and CTF18-RLC has not yet been addressed. It has been shown that small-interfering RNA-mediated CTF18 depletion does not increase the amount of chromatin-bound PCNA in human cells16. The observed PCNA unloading by the RFC complex or Ctf18-RLC might represent the activity of subcomplexes of the small RFC subunits because PCNA unloading by RFC2, 3, 4, and 5 and RFC2, 5 subcomplexes has been reported in vitro31. However, ATAD5 (Elg1) is not an essential protein for cell viability32,33, and the amount of chromatin-bound PCNA is reduced after ATAD5-depleted cells enter the M phase30. Therefore, it is possible that the RFC complex or CTF18-RLC functions as a back-up PCNA unloader.
Similar to PCNA loading by the RFC complex, PCNA unloading by ATAD5-RLC requires a functional ATPase domain and interaction with small RFC subunits10,16. ATAD5 with mutations in the ATPase domain or RFC2–5-binding motif fails to reduce chromatin-bound PCNA accumulation in ATAD5-depleted cells. Single-molecule experiments have identified mechanical similarities and differences between PCNA loading and unloading processes10. In both processes, the binding of ATP to the RFC complex or ATAD5-RLC opens the PCNA ring. The RFC complex–PCNA intermediate binds to ssDNA/dsDNA junctions to initiate the PCNA loading process. However, ATAD5-RLC–PCNA intermediates are released from the DNA, and this step is not dependent on ATP hydrolysis. This suggests that ATP hydrolysis triggers the dissociation of PCNA from the bound RFC or ATAD5-RLC, followed by ring closure to complete the loading or unloading process, respectively. The following questions remain to be addressed in relation to the PCNA unloading process: (1) how and when do the ATAD5-RLC-PCNA intermediates dissociate from DNA and (2) how is the ATP hydrolysis of ATAD5-RLC triggered to separate PCNA from ATAD5-RLC.
During DNA replication, PCNA increases the processivity of tethered replicative DNA polymerases. In addition, PCNA recruits FEN1 and DNA ligase I for Okazaki fragment maturation and CAF-1 for nucleosome assembly21,34,35. Chromatin duplication is completed before PCNA unloading. In yeast, the depletion of the DNA ligase Cdc9 prevents the ligation of Okazaki fragments and results in PCNA accumulation on chromatin. The depletion of the histone chaperone CAF-1 or ASF1 in human cells delays PCNA unloading17,36,37. These results suggest that the PCNA-unloading process is tightly coordinated with DNA replication and nascent chromatin assembly. Pol δ inhibits RFC complex-mediated PCNA unloading in vitro38. FEN1, DNA ligase I, and Pol δ also inhibit ATAD5-RLC-mediated PCNA unloading to different degrees10. Therefore, it is possible that the PCNA-interacting replisome proteins compete with a PCNA unloader during DNA synthesis to prevent premature unloading of PCNA. Recently, another regulatory mechanism for PCNA unloading has been suggested. Bromodomain and extra-terminal domain (BET) family proteins are preferentially associated with chromatin enriched in histone H4 acetylated at lysine residues 5 and 12 and facilitate RNA Pol II-mediated transcription39,40. Kang et al. showed that after DNA replication, BET family proteins bind to nascent chromatin through interactions with newly synthesized histones that are acetylated at H4 lysine residues 5 and 1241. Nascent chromatin-bound BRD4 interacts with ATAD5 through its ET domain and inhibits the PCNA unloading activity of ATAD5-RLC to prevent premature PCNA unloading41,42.
S-phase sister chromatid cohesion
In eukaryotes, replicated DNA is paired and held together upon synthesis until pair separation in mitosis. This sister chromatid cohesion is essential for the faithful transmission of replicated chromosomes during cell division43,44. Sister chromatid cohesion is achieved by the establishment of a large ring-shaped cohesin complex (which contains two coiled-coil subunits Smc1 and Smc3) concomitant with DNA replication. Cohesin loading onto chromosomes is assisted by the Scc2–Scc4 cohesin loader complex (NIPBL-MAU2 in humans) during the G1 phase. Cohesin release is facilitated by the Wapl-Pds5 dimer45. Cohesin is destabilized by Wapl and thus does not stay on DNA for a long time before chromosome duplication. During or after cohesin encircles the newly replicated sister chromatid, Smc3 acetylation by Eco1 acetyltransferase (ESCO1 and ESCO2 in humans) establishes stable cohesion by inhibiting Wapl-mediated cohesin destabilization46–48.
Several nonessential replisome proteins, such as Ctf4 and Chl1, contribute to cohesion establishment43. In addition, Ctf18-RLC is required for sister chromatid cohesion in yeast12. It has also been reported that human CTF18-RLC is important for sister chromatid cohesion and processive fork movement based on studies using DCC1-knockout retinal pigment epithelial cells, in which a marked reduction in CTF18 has also been reported49. In this report, SMC3 acetylation-mediated dissociation of WAPL-PDS5 was shown to be required for processive fork movement. The binding of EcoI acetyltransferase to PCNA is crucial for cohesion establishment during the S phase and for cell viability50. It has been suggested that during DNA replication, the PCNA mainly associated with Eco1 binding and subsequent Smc3 acetylation is loaded on the leading strand by Ctf18-RLC; the role of this PCNA is distinct from the primary role of the PCNA that is loaded by the RFC complex for DNA synthesis28.
The involvement of Elg1 in cohesion is not yet clearly defined. Smc3 acetylation is not reduced but actually slightly increased in elg1∆ mutants28. This finding suggests that Elg1 does not have a role in cohesion establishment. In the same report, reduced Smc3 acetylation in ctf18∆ mutants was rescued upon elg1 loss. This finding suggests that the PCNA accumulated on the lagging strand upon elg1 loss can support cohesion establishment in the absence of Ctf18-RLC-dependent PCNA loading on the leading strand. This result is consistent with the observation that elg1 loss partially suppresses the cohesion defect and temperature sensitivity of eco1-1 mutants51. Elg1∆ mutants exhibit varying degrees of cohesion defect in several reports; however, in each report, the defect was milder than that observed in ctf18∆ mutants28,51,52. It is likely that Elg1 has a role in cohesion establishment that is different from the well-established Smc3 acetylation-mediated mechanism.
During undisturbed DNA replication, PCNA is evenly distributed at both the leading and lagging strands28. Pol ε-mediated continuous leading strand synthesis is highly processive. Therefore, it remains to be elucidated how PCNA is frequently loaded onto leading strands. Because the ssDNA/dsDNA junction is required for PCNA loading by the RFC complex and CTF18-RLC, it is likely that at least the catalytic domain of Pol ε is temporally displaced from the 3′ end of the leading strand for CTF18-RLC-mediated PCNA loading. It has been proposed that Pol ε utilizes both the CDC45–MCM2–7-GINS (CMG) replicative helicase complex and PCNA as processivity factors to facilitate normal replication rates21,53. Tethering by the CMG complex might allow Pol ε to dissociate from the 3′ end of the leading strand but stays at the replication fork until leading strand synthesis restarts. The following remaining questions include: (i) if Pol ε is frequently halted, how is the new PCNA loaded after temporal Pol ε halting? (ii) How do extra PCNA molecules away from the replication fork function during cohesion establishment? (iii) How is PCNA unloading coordinated with cohesion establishment on leading strands?
Checkpoint activation
When DNA damage sensors recognize damage, checkpoint pathways are activated to arrest the cell cycle and promote the DNA damage response and DNA repair. In human cells, ATM (Tel1 in yeast) and ATR (Mec1 in budding yeast and Rad3 in fission yeast) kinases are master regulators of major checkpoint pathways5,54. ATM is recruited to the broken DNA ends and then activated. ATR is activated when ssDNA levels increase due to various types of DNA damage and replication stress. ATR is recruited to RPA-coated ssDNA at the sites of DNA damage through its regulatory partner ATRIP (Ddc2 in budding yeast and Rad26 in fission yeast)55,56. Full activation of ATR activity at the damage sites requires TopBP1 (Dpb11 in budding yeast and Rad4 in fission yeast) and/or Ewing’s tumor-associated antigen 1 (ETAA1). TopBP1 is recruited to the damage site through its interaction with the 9-1-1 complex, and ATR is autophosphorylated55,57. RAD17-RLC (Rad24-RLC in yeast) loads the 9-1-1 complex at the dsDNA/ssDNA junctions of DNA damage sites in an RPA-dependent manner. The loaded 9-1-1 complex and RAD17-RLC bind with TopBP1, which is critical for TopBP1-mediated ATR activation. ETAA1 is recruited to RPA-coated ssDNA by directly binding to RPA and then activates ATR independently of TopBP1 but in parallel with TopBP1 recruitment58–60.
During DNA replication in budding yeast, stalled forks and DNA lesions, such as postreplication gaps and fork breaks, activate the Mec1–Rad53 (ATR-CHK2 in humans) checkpoint pathway but require different signal mediators, Mrc1 (CLASPIN in humans) and Rad9 (53BP1 in humans), respectively, for full Rad53 activation61. Ctf18Δ mutant cells are highly sensitive to replication stress induced by treatment with hydroxyurea, which depletes the nucleotide pool in cells62. Ctf18-RLC, but neither Rad24-RLC nor Elg1-RLC, is required for Mrc1-mediated DNA replication checkpoint activation upon hydroxyurea treatment14,15. Consistently, in ctf18Δ mutants, checkpoint activation in response to hydroxyurea-induced fork stalling is delayed and depends on the Rad9-mediated DNA damage checkpoint14.
Ctf18-RLC interacts with Pol ε via Dcc126, and this interaction contributes to the recruitment of Ctf18-RLC to replication forks28,29. Ctf18 mutants defective with Pol ε-binding were originally reported to be sensitive to hydroxyurea and defective in the replication checkpoint, as shown by reduced Rad53 phosphorylation63. On the other hand, two recent reports show that hydroxyurea sensitivity and reduced Rad53 phosphorylation are not observed in either Pol ε-binding-defective dcc1 mutants or Dcc1-binding-defective Pol ε mutants28,29. However, the same paper showed that Rad52 foci, markers for recombination, and Psf1 phosphorylation, a marker for late origin firing, both of which are normally suppressed by checkpoint activation, are increased in Pol ε-binding-defective ctf18 mutants treated with hydroxyurea. This suggests the importance of Ctf18-RFC recruitment to the leading strand for activation of the replication checkpoint29. Human CTF18-RLC interacts with Pol ε via DCC126, but it is unknown whether this interaction is also important for checkpoint activation in human cells.
The RFC complex and RLCs in cell viability and genomic stability
In eukaryotes, all five subunits of the RFC complex are essential for cell survival33,64. The other large subunits of RLCs, Rad24 (Rad17 in fission yeast and humans), Ctf18, and Elg1 (ATAD5 in humans), are not essential for cell viability in yeast32,65 or mammals33. Although these RLC genes are not essential for normal cell proliferation, a portion of human cancer cells depends on each gene for survival, according to genome-scale clustered–regularly interspaced–short-palindromic-repeat (CRISPR)–Cas9 screen data33.
Rad24Δ mutants display sensitivity to ultraviolet radiation and methyl methanesulfonate (MMS)66. RAD17-knockout HCT116 human colon cancer cells display defects in damage-induced CHK1 phosphorylation by ATR. Consequently, these cells are defective in mitotic and S-phase damage checkpoint functions, as seen in the case of yeast rad24 deletion, and show chromosomal aberration and endoreduplication67. Rad17-knockout mice are embryonically lethal68. However, embryonic stem cells can be recovered from knockout mice; these knockout embryonic stem cells show DNA damage-dependent aberrant recombination and hypersensitivity to various DNA‐damaging agents.
Ctf18 was first identified as a suppressor of chromosome missegregation in budding yeast9,19. Ctf18Δ mutants display chromosome loss, elevated recombination frequency, sensitivity to MMS, and telomere mislocalization. Later, its roles in sister chromatid cohesion, checkpoint activation, and PCNA regulation were revealed in both yeast and mammals. Chtf18 (mouse CTF18)-knockout mice are viable but display defects in meiotic recombination, leading to chromosome missegregation and ultimately an abnormal chromosome number69.
Elg1 was first identified as a suppressor of gross chromosomal rearrangements in budding yeast19. Elg1Δ mutants display gross chromosomal rearrangements, chromosome loss, an elevated recombination rate, telomere lengthening, a high mutation rate, and sensitivity to MMS19. In human ATAD5-depleted cells, there is an increase in spontaneous HR but a reduction in double-strand DNA break (DSB)-induced HR70. A recent paper showed that the centrosome is overduplicated upon ATAD5 depletion71. These findings suggest that ATAD5 (Elg1) is critical for maintaining genome stability. Atad5 homozygous mutant mice are embryonically lethal72. Atad5 heterozygote mutant mice exhibit genome instability and develop tumors72. Somatic ATAD5 mutations are found in human patients with endometrial cancer and intraocular melanoma10,72. In addition, a genome-wide analysis identified the ATAD5 gene as a susceptibility locus for breast and ovarian cancers73,74. These observations suggest that ATAD5 functions as a tumor suppressor.
Mechanisms by which ATAD5 (Elg1)-RLC maintains genomic stability
Most of the genomic instability observed in cells depleted of CTF18 or RAD17 (Rad24 in budding yeast) can be explained or inferred by their primary activity. In the case of ATAD5 (Elg1), considering that PCNA unloading is the final step after DNA synthesis and, in case of DNA replication, the final step after postreplication processes is completed, a defect in PCNA unloading cannot be easily linked to the severe genomic instability observed in ATAD5 (Elg1)-depleted cells. However, accumulating data suggest that accumulated PCNA or PCNA that remains on the DNA for a long time due to depletion of ATAD5 (Elg1)16,17 can be the primary cause of genome instability. Johnson et al. showed that alleviating PCNA accumulation by either disassembly-prone PCNA mutants or G2/M-specific Elg1 expression rescues genome instability, such as hyperrecombination, telomere lengthening, and MMS sensitivity, in elg1∆ mutants75. Using site-specific mutations in Elg1 with different PCNA unloading activities, Shemesh et al. showed that DNA damage sensitivity and recombination rates correlate with the level of PCNA accumulation on DNA76. The effect of long-term PCNA residence on DNA on other processes is discussed below.
The C-terminal region of ATAD5, which contains the ATPase domain and the small RFC interaction domain, is sufficient for PCNA unloading in vitro and in cells10. The N-terminal region of ATAD5 interacts with many other proteins and these interactions are important for different activities regulating DNA metabolism. For example, ATAD5 interacts with the ubiquitin-specific protease 1 (USP1)-associated factor (UAF1) complex through the N-terminal domain and contributes to the PCNA deubiquitination process that terminates error-prone translesion synthesis (TLS)77. The biological importance of these protein–protein interactions is discussed below based on recent reports.
Taken together, timely PCNA unloading and proper protein binding or recruitment to replication forks by ATAD5-RLC contribute to many cellular activities regulating DNA metabolism, either alone or in concert, to maintain genomic stability.
S-phase progression
S-phase progression is delayed in ATAD5 (Elg1)-depleted cells16,78,79. In human cells, it has been shown that this results from a slow DNA replication rate (measured by 5-ethynyl-2′-deoxyuridine incorporation into nascent DNA) but not checkpoint activation16. A ATAD5 mutant with defective PCNA unloading fails to restore the reduced DNA replication rate upon ATAD5 depletion10, which suggests that PCNA remaining on DNA might be the reason for the slow replication rate. However, DNA fiber experiments have revealed that there is no change in the replication fork speed or interorigin distance upon ATAD5 depletion16. A slow replication rate can result from defects in the formation of new replication factories upon capture of related proteins by accumulated PCNA due to the role of PCNA as an intrinsic scaffold. Many replication proteins are enriched on chromatin in ATAD5-depleted cells, and this accumulation is attenuated when PCNA is depleted16.
Two recent reports have suggested that PCNA retention on DNA can lead to defects in replication-coupled nucleosome assembly in elg1∆ mutants. Gali et al. showed that postreplication nucleosome assembly, which is measured by the Okazaki fragment lengths and micrococcal nuclease sensitivity of newly replicated DNA, is defective in elg1∆ mutants80. Janke et al. showed that transcriptional silencing at specific loci maintained by histone chaperones is defective in elg1∆ mutants, and transcription is rescued upon the overexpression of the histone chaperone CAF-181. In both reports, defects caused by elg1 loss were not observed in the disassembly-prone PCNA background. This outcome suggests that the PCNA remaining on DNA physically inhibits nucleosome assembly or that histone chaperones required for nucleosome assembly are trapped by PCNA accumulated on DNA. Depletion of CAF-1 or ASF1 has been reported to reduce DNA replication rate36. Therefore, defects in postreplication nucleosome assembly can be another cause of slow S-phase progression in ATAD5 (Elg1)-depleted cells.
Mismatch repair
The DNA mismatch repair (MMR) pathway recognizes and corrects base mispairs, insertions, and deletions generated during replication and that escape proofreading by replicative DNA polymerases82,83. PCNA participates in several steps during the DNA MMR process, including mismatch recognition by MSH2–MSH6 or MSH2–MSH3, strand discrimination, strand excision, and repair DNA synthesis. Other negative effects of PCNA accumulation on DNA metabolism have recently been reported in the MMR pathway84. The mutation rate is increased in elg1∆ mutants, but this is not observed in a disassembly-prone PCNA or msh2∆/msh6∆-mutant background. In contrast, pcna mutants in which PCNA is overretained on DNA up to the G2/M phase display an increase in the mutation rate, regardless of Elg1 level. In addition, Msh6 accumulates on chromatin in a PCNA-interaction-dependent manner in elg1∆ mutants. In human cells, chromatin-bound MSH2 levels are increased upon ATAD5 depletion and reduced upon simultaneous PCNA depletion16. This finding suggests that MMR defects also occur in ATAD5-depleted cells via a similar mechanism. Interestingly, the same report showed that the mutation rate also increases through MMR-independent but accumulated PCNA-dependent processes in elg1∆ mutants84. Based on the results in human cells, one such possible candidate process has been identified as monoubiquitinated PCNA-directed error-prone translesion DNA synthesis77.
PCNA is monoubiquitinated by the E2–E3 RAD6–RAD18 complex or by the CRL4Cdt2 ubiquitin ligase complex when a replication fork is stalled by DNA lesions or intrinsic replication blocks85,86. The resulting monoubiquitinated PCNA recruits error-prone TLS polymerases to bypass the DNA lesion (Fig. 1a). This potentially mutagenic TLS activity needs to be minimized in unperturbed cells and terminated immediately after lesion bypass by TLS polymerases. TLS termination is carried out by ubiquitin-specific proteases USP1 and/or USP1087,88. UAF1 is required for the optimal activity and protein stability of USP189. ATAD5 interacts with UAF1, and this interaction is important for USP1-mediated PCNA deubiquitination (Fig. 1a)77. Consequently, in ATAD5-depleted human cells, the level of monoubiquitinated PCNA increases without exogenous DNA damage, and the mutation frequency is increased, as indicated by SupF assay77. Collectively, dysregulated MMR and translesion DNA synthesis can contribute to increased mutations in elg1∆ mutants and ATAD5-depleted cells.
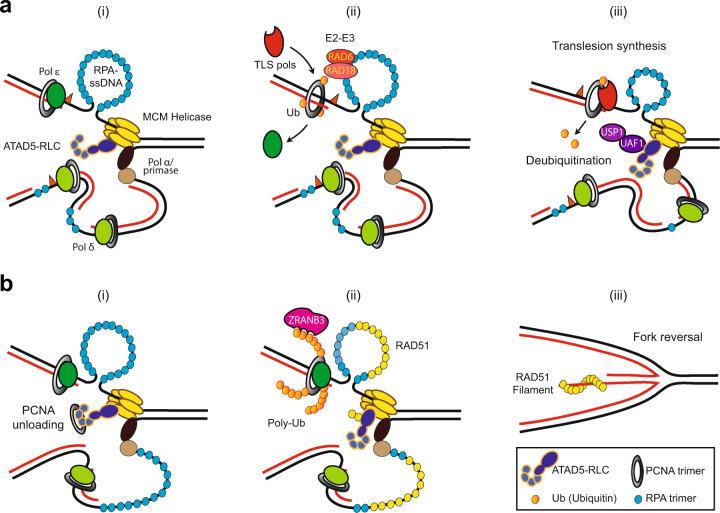
Fig. 1. Graphical model of cellular mechanisms for preserving stalled fork integrity and the roles of ATAD5-RLC in this process.
DNA lesions (orange triangle) and replication stress stall PCNA-tethered replicative polymerases physically (a, i) or chemically (e.g., nucleotide depletion) (b, i). This stalling causes the uncoupling of helicase and replicative polymerases, leading to the generation of a long single-strand DNA that is immediately coated by the trimeric RPA complex. a, ii, iii PCNA, monoubiquitinated by the E2–E3 RAD6–RAD18 complex, recruits translesion polymerases (TLS pols), and if the lesions are manageable by any of the TLS pols, the lesions are effectively bypassed. a, iii The USP1/UAF1 deubiquitinase complex removes ubiquitin from monoubiquitinated PCNA in an ATAD5 interaction-dependent manner, which reduces error-prone TLS-induced mutations. b, ii, iii Many DNA damage-inducing drugs lead to PCNA polyubiquitination by E2–E3 MMS2/UBC13–RAD5 protein. RAD51 recombinase and the translocase activity of HLTF and ZRANB3, which bind to the polyubiquitin chain of PCNA, cooperatively inducing fork reversal upon replication stress. ATAD5-RLC unloads PCNA in a timely manner and subsequently facilitates the recruitment of RAD51 to stalled forks, a process mediated by a replication stress-enhanced interaction between the two proteins. The RAD51 filament protects the reversed fork from nucleolytic attacks (iii).
Fork reversal
When DNA damage slows or stalls the progression of the replication fork, template DNA and nascent DNA reannealing can generate a four-way junction structure in a process referred to as fork reversal (Fig. 1b, iii), which can stabilize the replication fork90. RAD51 recombinase is important for both fork reversal and the stability of reversed forks91–93. In addition, three translocases, ZRANB3, SMARCAL1, and HLTF, have been shown to have fork reversal activity in vitro; of these proteins, ZRANB3 and HLTF have been shown to display this activity in cells94–96.
Damage-induced lysine 63 (K63)-linked PCNA polyubiquitination mediates the error-free DNA damage tolerance pathway through template switching. Production of a polyubiquitin chain for en bloc chain transfer to PCNA at K164 or sequential addition of ubiquitin to monoubiquitinated PCNA at K164 is mediated by the cooperative activity of the MMS2/UBC13 ubiquitin-conjugating dimer and either of the two yeast Rad5 ubiquitin ligase homologs, SHPRH and HLTF, or a currently unidentified E3 ligase85,97–100. The requirement of K63-linked PCNA polyubiquitination and polyubiquitinated PCNA interaction with ZRANB3 for damage-induced fork reversal suggests that fork reversal is closely linked to template switching95,101,102.
ATAD5 directly interacts with RAD51 through the N-terminal domain of ATAD5103. ATAD5 facilitates RAD51 recruitment to stalled forks by replication stress-enhanced protein–protein interactions (Fig. 1b)103. ATAD5 also removes PCNA from stalled forks in a timely manner for RAD51 recruitment. Consistently, ATAD5 depletion inhibits the deceleration of fork progression and reduces the native 5-bromo-2ʹ-deoxyuridine signal upon replication stress, which suggests inhibited fork reversal. This effect eventually leads to increased genomic instability, both in cells and mice undergoing replication stress. Since ATAD5 depletion increases the abundance of monoubiquitinated PCNA on DNA77, polyubiquitinated PCNA can also be increased in ATAD5-depleted cells. Therefore, conceptually, facilitated fork reversal by increased PCNA polyubiquitination through ZRANB3 recruitment might be expected; however, the actual observations are different in ATAD5-depleted cells103. This finding suggests that the RAD51 recombinase and the ZRANB3 translocase can cooperate to drive fork reversal, but depending on the type of genotoxicity, one of these proteins may be dominant. ZRANB3-depleted cells display higher sensitivity to camptothecin compared to other drugs, which suggests a high demand for ZRANB3 for processing camptothecin-induced DNA damage102.
R-loop regulation
R-loops are reversible nucleic acid structures that feature a DNA/RNA hybrid and the resulting nonhybridized ssDNA104,105. R-loops are formed temporarily to regulate many cellular processes. However, persistent R-loops make the genome vulnerable to DNA damage due to exposure of ssDNA regions. R-loops can be formed during DNA replication when replicative forks collide with transcriptional machinery. Consistently, replication stress due to nucleotide depletion or DNA polymerase inhibition increases R-loop formation106. Transcription–replication conflicts interfere with replication fork progression, resulting in potential threats to genome stability104.
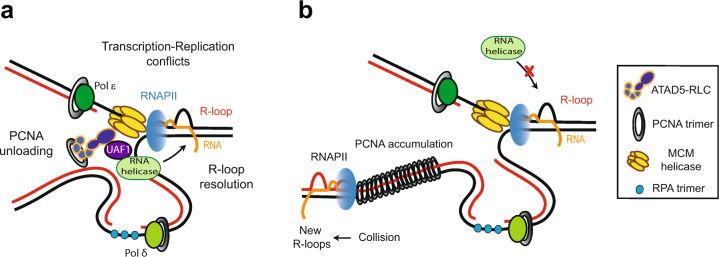
Recently, dual roles have been suggested for ATAD5-RLC in R-loop regulation during DNA replication (Fig. 2)107. ATAD5-RLC prevents the generation of new R-loops behind replication forks by unloading PCNA. Without ATAD5-RLC, PCNA accumulates and persists on the lagging strand DNA for a long time, causing a collision with transcription machinery, which can lead to R-loop formation. In addition, ATAD5 recruits DEAD/DExH-box DNA/RNA helicases at the replication forks through UAF1-mediated protein–protein interactions. Under replication stress, the same DNA/RNA helicases are additionally recruited to the replication forks in a process that is also dependent on helicase interaction with ATAD5/UAF1. These recruited DNA/RNA helicases then resolve R-loops under normal and replication stress conditions and facilitate replication fork progression.
Fig. 2. Graphical model for R-loop regulation by ATAD5-RLC and ATAD5/UAF1-interacting DNA/RNA helicases.
a During normal replication, ATAD5-RLC and ATAD5/UAF1-interacting DNA/RNA helicases migrate with a replication fork. ATAD5/UAF1-interacting DNA/RNA helicases resolve R-loops ahead of the replication fork and facilitate replication fork progression. Upon replication stress, which increases transcription–replication conflicts and unscheduled R-loop formation, additional DNA/RNA helicases are recruited to the replication fork, which resolves R-loops to ensure faithful replication fork progression. Under both normal and replication stress conditions, the recruitment of helicases at the replication fork is dependent on the ATAD5/UAF1 interaction. b In ATAD5-depleted cells, reduced R-loop resolution by ATAD5/UAF1-interacting DNA/RNA helicases leads to defects in replication fork progression. In addition, PCNA and its interacting proteins accumulated on lagging strand DNA behind the forks collide with transcription machinery, which consequently increases R-loop formation at the collision site.
DNA double-strand break repair
DSB is one of the most dangerous types of DNA lesions. Nonhomologous end joining (NHEJ) and HR are the two primary double-strand DNA break repair (DSBR) pathways, while microhomology-mediated end joining and single-strand annealing are back-up mechanisms. A primary determinant for the pathway choice between NHEJ and HR is end resection, which predominantly occurs during the S/G2 phase to generate a long tract of 3′-OH ssDNA. End resection is required for forming the ssDNA-coated RAD51 filament that invades the homologous sister chromatid during HR.
The first evidence for the role of Elg1 (ATAD5) in DSBR was found because of the sensitivity of elg1∆ mutants to phleomycin, a compound that generates DSBs, and reduced damage-induced recombination repair in elg1∆ mutants108. The same report also showed that Elg1 is recruited to DSB sites independent of Rad52 (a key factor for HR in yeast) and that repair DNA synthesis but not PCNA recruitment is slightly defective upon elg1 loss. In human cells, DSB-induced HR frequency is also reduced upon ATAD5 depletion70. A recent report showed that ATAD5-depleted cells are sensitive to MMS, bleomycin, and camptothecin, all of which can generate DSBs109. Consistent with the requirement of poly(ADP-ribose) polymerase (PARP)-mediated repair for the survival of HR-deficient cells110,111, ATAD5-depleted cells are highly sensitive to PARP inhibitors. Collectively, the positive role of ATAD5 (Elg1) in HR is evident, but the molecular mechanism remains to be studied. The Rad52-independent recruitment of Elg1 to DSB sites in yeast and the rapid localization of RFC complex proteins and PCNA to DSB sites in human cells suggest the involvement of ATAD5 (Elg1) in the early steps of HR108,112.
In addition to S-phase sister chromatid cohesion formed during DNA replication, genome-wide cohesion is generated by DSBs in both yeast and human cells113,114. In both human and yeast cell lines, damage checkpoint kinase-mediated phosphorylation and subsequent Eco1-mediated acetylation are required; the targets for both these protein modifications are Scc1 in yeast and SMC3 in human cells115. It is not clear whether the CTF18-RLC and PCNA are also involved in DSB-induced cohesion as they are in S-phase sister chromatid cohesion. However, a few reports support this possibility. Ogiwara et al. showed that Ctf18 is recruited to DSB sites; ctf18∆ mutants are sensitive to DSB-inducing drugs, and damage-induced recombination between sister chromatids and between homologous chromosomes is defective in the same mutants116. In addition, it has been reported that PCNA moves rapidly to DSB sites even when RFC1 is depleted in human cells112, which also suggests that Ctf18-RLC serves as a PCNA loader at DSB sites.
Concluding remarks
To date, elaborate efforts have elucidated many aspects of eukaryotic clamp loaders and unloaders, such as their basic biochemical and cellular activities and the molecular mechanism of genomic instability when their activities are diminished. As described in this review, recent findings have increased our composite understanding of how RFC/RLCs cooperatively ensure genomic integrity by spatiotemporally regulating PCNA, in addition to properly recruiting the necessary factors. However, it is still unclear how the knockout phenotypes in mice, especially embryonic lethality and tumor incidence in ATAD5-knockout mice, are related to the molecular and cellular activities of these proteins. In addition, several phenotypes, such as lengthened telomeres, a high recombination rate, and sister chromatid exchange in ATAD5 (Elg1)-depleted cells, require mechanistic explanations to understand how PCNA accumulation on DNA leads to these defects.
Acknowledgements
We thank Kyungjae Myung and Sukhyun Kang for their helpful discussions and comments on the paper. This work was supported by the Institute for Basic Science (IBS-R022-D1).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 4.Navadgi-Patil VM, Burgers PM. Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase. Biochem. Soc. Trans. 2011;39:600–605. doi: 10.1042/BST0390600. [DOI] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermudez VP, et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao NY, O’Donnell M. The RFC clamp loader: structure and function. Subcell. Biochem. 2012;62:259–279. doi: 10.1007/978-94-007-4572-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 9.Shiomi, Y. & Nishitani, H. Control of genome integrity by RFC complexes; conductors of PCNA loading onto and unloading from chromatin during DNA replication. Genes8, 10.3390/genes8020052 (2017). [DOI] [PMC free article] [PubMed]
- 10.Kang MS, et al. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat. Commun. 2019;10:2420. doi: 10.1038/s41467-019-10376-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol. Cell Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 13.Bermudez VP, et al. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl Acad. Sci. USA. 2003;100:10237–10242. doi: 10.1073/pnas.1434308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabbe L, et al. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat. Struct. Mol. Biol. 2010;17:1391–1397. doi: 10.1038/nsmb.1932. [DOI] [PubMed] [Google Scholar]
- 15.Kubota T, Hiraga S, Yamada K, Lamond AI, Donaldson AD. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol. Cell Proteom. 2011;10:M110 005561. doi: 10.1074/mcp.M110.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KY, Fu H, Aladjem MI, Myung K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J. Cell Biol. 2013;200:31–44. doi: 10.1083/jcb.201206084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Sau S, Kupiec M. A role for the yeast PCNA unloader Elg1 in eliciting the DNA damage checkpoint. Curr. Genet. 2020;66:79–84. doi: 10.1007/s00294-019-01020-7. [DOI] [PubMed] [Google Scholar]
- 19.Gazy I, Liefshitz B, Parnas O, Kupiec M. Elg1, a central player in genome stability. Mutat. Res. Rev. Mutat. Res. 2015;763:267–279. doi: 10.1016/j.mrrev.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Garbacz MA, et al. Evidence that DNA polymerase delta contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat. Commun. 2018;9:858. doi: 10.1038/s41467-018-03270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeeles JTP, Janska A, Early A, Diffley JFX. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell. 2017;65:105–116. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016;26:640–654. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase delta in replication of both the leading and lagging DNA strands. Mol. Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabarczyk DB, Silkenat S, Kisker C. Structural basis for the recruitment of Ctf18-RFC to the replisome. Structure. 2018;26:137–144. e133. doi: 10.1016/j.str.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Murakami T, et al. Stable interaction between the human proliferating cell nuclear antigen loader complex Ctf18-replication factor C (RFC) and DNA polymerase {epsilon} is mediated by the cohesion-specific subunits, Ctf18, Dcc1, and Ctf8. J. Biol. Chem. 2010;285:34608–34615. doi: 10.1074/jbc.M110.166710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujisawa R, Ohashi E, Hirota K, Tsurimoto T. Human CTF18-RFC clamp-loader complexed with non-synthesising DNA polymerase epsilon efficiently loads the PCNA sliding clamp. Nucleic Acids Res. 2017;45:4550–4563. doi: 10.1093/nar/gkx096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu HW, et al. Division of labor between PCNA loaders in DNA replication and sister chromatid cohesion establishment. Mol. Cell. 2020;78:725–738. e724. doi: 10.1016/j.molcel.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stokes K, et al. form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication. Nucleic Acids Res. 2020;48:8128–8145. doi: 10.1093/nar/gkaa541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiomi Y, Nishitani H. Alternative replication factor C protein, Elg1, maintains chromosome stability by regulating PCNA levels on chromatin. Genes Cells. 2013;18:946–959. doi: 10.1111/gtc.12087. [DOI] [PubMed] [Google Scholar]
- 31.Yao NY, Johnson A, Bowman GD, Kuriyan J, O’Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J. Biol. Chem. 2006;281:17528–17539. doi: 10.1074/jbc.M601273200. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, et al. Contrasting effects of Elg1-RFC and Ctf18-RFC inactivation in the absence of fully functional RFC in fission yeast. Nucleic Acids Res. 2005;33:4078–4089. doi: 10.1093/nar/gki728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behan FM, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 34.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 35.Boehm EM, Gildenberg MS, Washington MT. The many roles of PCNA in eukaryotic DNA replication. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mejlvang J, et al. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota T, Katou Y, Nakato R, Shirahige K, Donaldson AD. Replication-coupled PCNA unloading by the Elg1 complex occurs genome-wide and requires Okazaki Fragment ligation. Cell Rep. 2015;12:774–787. doi: 10.1016/j.celrep.2015.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedglin M, Perumal SK, Hu Z, Benkovic S. Stepwise assembly of the human replicative polymerase holoenzyme. eLife. 2013;2:e00278. doi: 10.7554/eLife.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu SY, et al. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang MS, et al. PCNA unloading is negatively regulated by BET proteins. Cell Rep. 2019;29:4632–4645 e4635. doi: 10.1016/j.celrep.2019.11.114. [DOI] [PubMed] [Google Scholar]
- 42.Wessel SR, Mohni KN, Luzwick JW, Dungrawala H, Cortez D. Functional analysis of the replication fork proteome identifies BET proteins as PCNA regulators. Cell Rep. 2019;28:3497–3509 e3494. doi: 10.1016/j.celrep.2019.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhlmann F. SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 44.Peters, J. M. & Nishiyama, T. Sister chromatid cohesion. Cold Spring Harb. Perspect. Biol.4, 10.1101/cshperspect.a011130 (2012). [DOI] [PMC free article] [PubMed]
- 45.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Unal E, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 48.Rolef Ben-Shahar T, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 49.Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Maradeo ME, Skibbens RV. The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS ONE. 2009;4:e4707. doi: 10.1371/journal.pone.0004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parnas O, et al. The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS ONE. 2009;4:e5497. doi: 10.1371/journal.pone.0005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langston LD, et al. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl Acad. Sci. USA. 2014;111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marechal, A. & Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol.5, 10.1101/cshperspect.a012716 (2013). [DOI] [PMC free article] [PubMed]
- 55.Yazinski SA, Zou L. Functions, regulation, and therapeutic implications of the ATR checkpoint pathway. Annu. Rev. Genet. 2016;50:155–173. doi: 10.1146/annurev-genet-121415-121658. [DOI] [PubMed] [Google Scholar]
- 56.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017;18:622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma M, Rodriguez A, Sugimoto K. Activation of ATR-related protein kinase upon DNA damage recognition. Curr. Genet. 2020;66:327–333. doi: 10.1007/s00294-019-01039-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee YC, Zhou Q, Chen J, Yuan J. RPA-binding protein ETAA1 is an ATR activator involved in DNA replication stress response. Curr. Biol. 2016;26:3257–3268. doi: 10.1016/j.cub.2016.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haahr P, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016;18:1196–1207. doi: 10.1038/ncb3422. [DOI] [PubMed] [Google Scholar]
- 60.Bass TE, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016;18:1185–1195. doi: 10.1038/ncb3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 62.Naiki T, Kondo T, Nakada D, Matsumoto K, Sugimoto K. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell Biol. 2001;21:5838–5845. doi: 10.1128/MCB.21.17.5838-5845.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Rodriguez LJ, et al. A conserved Pol binding module in Ctf18-RFC is required for S-phase checkpoint activation downstream of Mec1. Nucleic Acids Res. 2015;43:8830–8838. doi: 10.1093/nar/gkv799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, et al. Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev. 2003;17:965–970. doi: 10.1101/gad.1065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Budzowska M, et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berkowitz KM, et al. Disruption of CHTF18 causes defective meiotic recombination in male mice. PLoS Genet. 2012;8:e1002996. doi: 10.1371/journal.pgen.1002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sikdar N, et al. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle. 2009;8:3199–3207. doi: 10.4161/cc.8.19.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim SJ, et al. ATAD5 suppresses centrosome over-duplication by regulating UAF1 and ID1. Cell Cycle. 2020;19:1952–1968. doi: 10.1080/15384101.2020.1785724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bell DW, et al. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet. 2011;7:e1002245. doi: 10.1371/journal.pgen.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maleva Kostovska I, et al. Rare ATAD5 missense variants in breast and ovarian cancer patients. Cancer Lett. 2016;376:173–177. doi: 10.1016/j.canlet.2016.03.048. [DOI] [PubMed] [Google Scholar]
- 74.Kuchenbaecker KB, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015;47:164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson C, Gali VK, Takahashi TS, Kubota T. PCNA retention on DNA into G2/M phase causes genome instability in cells lacking Elg1. Cell Rep. 2016;16:684–695. doi: 10.1016/j.celrep.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shemesh K, et al. A structure-function analysis of the yeast Elg1 protein reveals the importance of PCNA unloading in genome stability maintenance. Nucleic Acids Res. 2017;45:3189–3203. doi: 10.1093/nar/gkw1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee KY, et al. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 79.Bellaoui M, et al. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 2003;22:4304–4313. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gali VK, et al. Identification of Elg1 interaction partners and effects on post-replication chromatin re-formation. PLoS Genet. 2018;14:e1007783. doi: 10.1371/journal.pgen.1007783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Janke R, King GA, Kupiec M, Rine J. Pivotal roles of PCNA loading and unloading in heterochromatin function. Proc. Natl Acad. Sci. USA. 2018;115:E2030–E2039. doi: 10.1073/pnas.1721573115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paul Solomon Devakumar LJ, Gaubitz C, Lundblad V, Kelch BA, Kubota T. Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex. Nucleic Acids Res. 2019;47:6826–6841. doi: 10.1093/nar/gkz441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoege C, et al. repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 86.Terai K, Abbas T, Jazaeri AA, Dutta A. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell. 2010;37:143–149. doi: 10.1016/j.molcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Park JM, et al. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol. Cell. 2014;54:626–638. doi: 10.1016/j.molcel.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 88.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 89.Cohn MA, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 90.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 91.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zellweger R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhat KP, Cortez D. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018;25:446–453. doi: 10.1038/s41594-018-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poole LA, Cortez D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem Mol. Biol. 2017;52:696–714. doi: 10.1080/10409238.2017.1380597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vujanovic M, et al. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell. 2017;67:882–890 e885. doi: 10.1016/j.molcel.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bai GS, et al. HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol. Cell. 2020;78:1237-+. doi: 10.1016/j.molcel.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl Acad. Sci. USA. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krijger PH, et al. HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: existence of an alternative E3 ligase. DNA Repair. 2011;10:438–444. doi: 10.1016/j.dnarep.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Unk I, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl Acad. Sci. USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masuda Y, et al. Regulation of HLTF-mediated PCNA polyubiquitination by RFC and PCNA monoubiquitination levels determines choice of damage tolerance pathway. Nucleic Acids Res. 2018;46:11340–11356. doi: 10.1093/nar/gky943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weston R, Peeters H, Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 2012;26:1558–1572. doi: 10.1101/gad.193516.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ciccia A, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park SH, et al. ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress. Nat. Commun. 2019;10:5718. doi: 10.1038/s41467-019-13667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gomez-Gonzalez, B. & Aguilera, A. Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev.10.1101/gad.324517.119 (2019). [DOI] [PMC free article] [PubMed]
- 105.Crossley MP, Bocek M, Cimprich KA. R-Loops as cellular regulators and genomic threats. Mol. Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-replication conflict orientation modulates R-Loop levels and activates distinct DNA damage responses. Cell. 2017;170:774–786 e719. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim, S. et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res.10.1093/nar/gkaa501 (2020). [DOI] [PMC free article] [PubMed]
- 108.Ogiwara H, Ui A, Enomoto T, Seki M. Role of Elg1 protein in double strand break repair. Nucleic Acids Res. 2007;35:353–362. doi: 10.1093/nar/gkl1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Giovannini S, et al. ATAD5 deficiency alters DNA damage metabolism and sensitizes cells to PARP inhibition. Nucleic Acids Res. 2020;48:4928–4939. doi: 10.1093/nar/gkaa255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Farmer H, et al. Targeting the NA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 111.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 112.Hashiguchi K, Matsumoto Y, Yasui A. Recruitment of DNA repair synthesis machinery to sites of DNA damage/repair in living human cells. Nucleic Acids Res. 2007;35:2913–2923. doi: 10.1093/nar/gkm115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Strom L, et al. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 114.Kim BJ, et al. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J. Biol. Chem. 2010;285:22784–22792. doi: 10.1074/jbc.M110.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ogiwara H, et al. Ctf18 is required for homologous recombination-mediated double-strand break repair. Nucleic Acids Res. 2007;35:4989–5000. doi: 10.1093/nar/gkm523. [DOI] [PMC free article] [PubMed] [Google Scholar]