Abstract
Here we report the development of our Next Generation Automated Multiplexed Oligonucleotide Synthesizer (NG-1536-AMOS), capable of producing 1536 samples in a single run using a multi-well filtered titer plate. With the potential to synthesize up to 3456 samples per plate, we converted the BioRAPTR Flying Reagent Dispenser into an open-well system where spent reagents are drained to waste under vacuum. During synthesis, reagents are delivered on-the-fly to each micro-titer well at volumes ≤5 μl with plate speeds up to 150 mm/s. Using gas-phase cleavage and deprotection, a full plate of 1536 60mers may be processed with same-day turnaround with an average yield per well at 3.5 nmol. Final product at only $0.00277/base is eluted into a low-volume collection plate for immediate use in downstream application (e.g. Biomek FX for versatile sample handling). Also, crude oligonucleotide quality is comparable to that of commercial synthesis instrumentation, with an error rate on the NG-1536-AMOS platform of 1.53/717 bases. Furthermore, mass spectral analysis on strands synthesized up to 80 bases showed high purity with an average coupling efficiency of 99.5%.
Keywords: Oligonucleotide, Synthesis automation, Chemical DNA synthesis, Synthetic biology
1. Introduction
The Stanford Genome Technology Center (SGTC) has been at the forefront of oligonucleotide DNA synthesis automation since the early 1990s with its development of the first 96-well Automated Multiplexed Oligonucleotide Synthesizer (96-AMOS) (Lashkari et al., 1995). Capable of generating 6000 25-base primers per month on a single machine, it made large-scale projects such as sequencing the Saccharomyces cerevisiae and human genomes timely and economically feasible (Dietrich et al., 1997; Lander et al., 2001).
While industry costs at the time for low-throughput column synthesis were $2.00–3.00 per base (Hager et al., 1999; Goforth, 2002), 96-AMOS was synthesizing DNA 10–15-fold cheaper within the first year of production. Currently at $0.06 per base, this platform remains competitive with other column and titer plate-based synthesizers that have since been introduced (Cheng et al., 2002; Livesay et al., 2002; Rayner et al., 1998; Lebl et al., 2007; Gan, 1990; Jensen et al., 2012).
However, with today’s multiplexed, small volume assays (Hardenbol et al., 2003; Dahl et al., 2007), there is a greater demand for even higher throughput and lower cost oligonucleotide synthesis. As such high density synthesis arrays and micro-fluidic devices (Gao et al., 2004; Livesay et al., 2002; Lee et al., 2010; Kong et al., 2007) offer mass production at a fraction of the cost. And though array densities can be as high as 4 million features, low yield and heterogeneous oligonucleotide pooling may present a challenge for specific downstream applications, in particular synthetic biology (Kim et al., 2012; Borovkov et al., 2010; Ma et al., 2012a,b; Quan et al., 2011; Mueller et al., 2009).
In contrast to the microarray which may be limited to 106 molecules per feature, multi-well/column platforms generally give very high yields (>25 nmol); but this is often a wasteful over-production of sample. As a consequence, excess product is either disposed of or kept in deep-well collection plates (volumes up to 1.2 ml/well) for long term storage thus taking up valuable freezer space.
Focusing on the strengths of both the array and multi-well/column platforms, we describe the development of our Next Generation AMOS capable of producing 1536 samples on a single titer plate (NG-1536-AMOS). Based on the original 96-AMOS, we combined its technology with that of the commercial BioRAPTR Flying Reagent (non-contact) Dispenser (BioRAPTR FRD, Beckman Coulter), intended for ultra high-throughput delivery (up to 3456 wells) of aqueous buffers, diluents and dye solutions (Yasgar et al., 2012; Auld et al., 2008).
Using on-the-fly dispense, NG-1536-AMOS delivers reagents ≤5 μl per well at plate speeds up to 150 mm/s. Following each reaction step, vacuum is applied to draw spent reagents to waste two rows (96 wells) at a time. This feature allows for either full or partial plate synthesis as well as synthesis of varying length strands simultaneously, something not possible with top pressure drain systems (e.g. 96-AMOS).
NG-1536-AMOS has several other advantages which include (i) post-synthesis amplification is not required (on average, yield per well is 3.5 nmol), (ii) product is eluted into shallow-well collection plates for maximum space-saving storage, and (iii) collection plates are fully compatible with existing robotic devices such as the Biomek FX for versatile sample handling (e.g. cherry picking and for creating combinatory oligonucleotide libraries). Also, fragments used in synthetic biology to construct genes de novo can be swapped without having to re-synthesize the entire plate/array.
In addition to mass spectral confirmation of 80-base strands synthesized on our NG-1536-AMOS, we sequence-verified 40-base fragments used to assemble the green fluorescent protein (GFP) gene. With error rates comparable to commercial synthesis methodologies, to date we have processed several 1536-well titer plates in our general synthesis production lab. Moreover, with implementation of a modified gas-phase cleavage and deprotection protocol (based on previous work (Jensen et al., 2010a)), it is possible to process an entire plate (60 bases/well) with same-day turnaround for immediate use in downstream application (Hardenbol et al., 2003; Dahl et al., 2007; Cobb et al., 2013; Weber and Fussenegger, 2012).
2. Materials and methods
2.1. Instrument design and setup
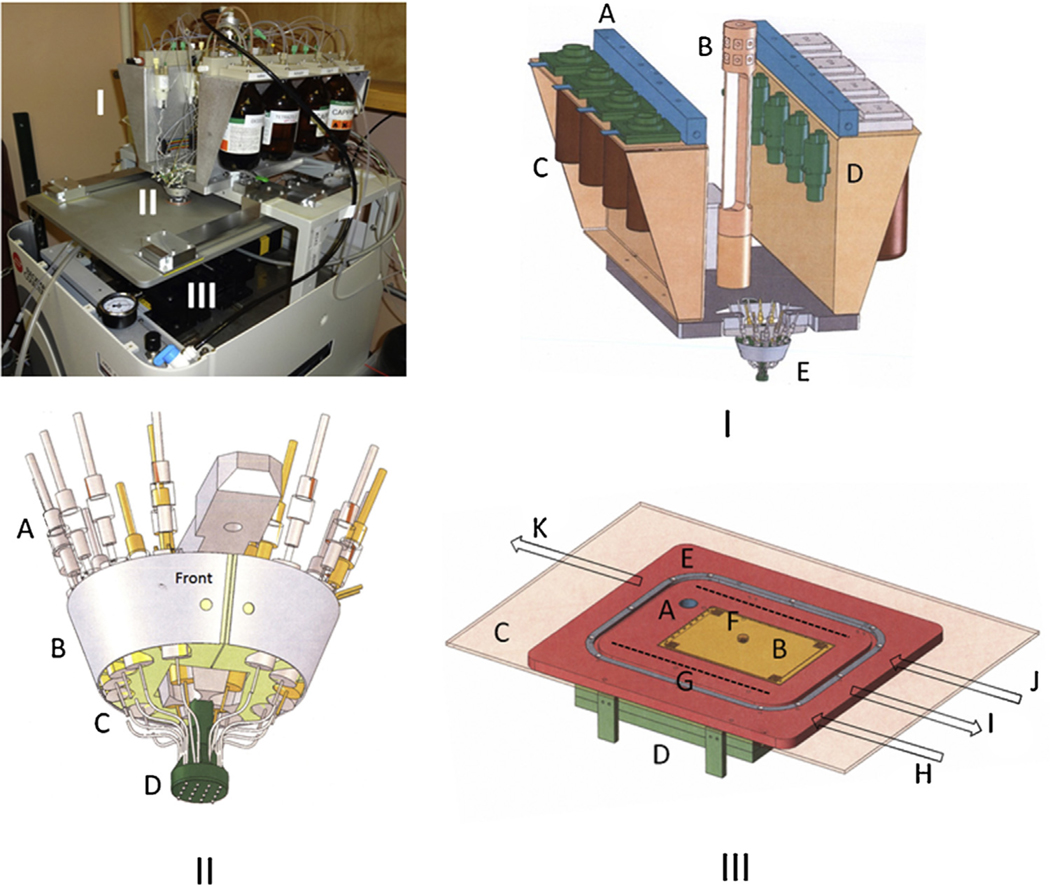
The basic principles of the original 96-AMOS platform were applied to the BioRAPTR FRD to create our custom next generation oligonucleotide synthesizer. The BioRAPTR FRD was converted into an open reaction well system (all modifications to the original platform were designed in-house (SGTC) and fabricated by Nevarez Machining and SGTC)). The 16-nozzle dispense head (Fig. 1I and II) is stationary while the titer plate (Fig. 1III) moves below in the X and Y directions during reagent delivery. Argon is used to (i) pressurize the bottles (9 psi), (ii) provide for an inert gas layer over the reaction wells and (iii) allow for vacuum-assisted low pressure reagent drain-to-waste.
Fig. 1.
NG-1536-AMOS (top left): (I) reagent assembly, (II) 16-tip valve head, and (III) titer plate holder with drain block underneath. The drive system below the plate holder (III D) consists of X, Y and Z axes (motor-driven tracks); (I) reagent and gas supply assembly (tubing and wires omitted for clarity): (A) wash manifolds for purging reagent lines after a completed run, (B) central tubing manifold for argon gas supply to (C) the reagent bottles, (D) 3-way valves for switching from wash (acetonitrile) to reagent and vice versa, and (E) valve assembly and dispense head. Each phosphoramidite position is designed to hold Applied Biosystems 2 g bottles, and oxidizer, activator, and capping reagents are in 200 ml bottles (Applied Biosystems); (II) valve assembly and dispense head: (A) tubing from reagent bottles to dedicated valves, (B) valve block outer shell, (C) valve-nozzle junction, and (D) nozzle head (4 × 4 configuration); (III) top view of titer plate assembly: (A) reagent purge and prime manifold for dispensing reagents, (B) 1536-well titer plate (C) top plate that sits above the titer plate assembly for sealing in argon gas (a strip of silicone rubber and Teflon are bracketed in around the titer plate for maximum seal between the top and bottom plates during synthesis to minimize gas leakage), (D) waste basin for collecting spent reagents during drain steps of each synthesis cycle, (E) bottom plate, (F) center hole where the 16-nozzle head is positioned above the titer plate during synthesis, (G) argon gas jet inlets, (H and J) gas line-in, (I) reagent-to-waste out (vacuum), and (K) prime station purge-to-waste. Note: arrows indicate direction of gas/vacuum/waste flow through the system.
2.2. Drain block design
To manage reagent flow-through to waste during synthesis, the titer plate (NG-1536-AMOS) is seated over a 1536-well drain block (Supplementary Figure 1). Sealed with a 0.5 mm silicone gasket (or comparable), the bottom drip directors of the titer plate extend inside individual drain holes (Supplementary Figure 2) where each two rows (96 wells) feed into a single channel. From here, valves control the vacuum on each two-row block allowing spent reagents to drain to waste over a given time (e.g. 1 s) according to a specified protocol step (a synthesis protocol file is a list of reagent dispenses, reaction hold times and drain steps repeated for each cycle/base addition).
2.3. Instrument specifications
The key features of this platform include (i) plate speeds 80–150 mm/s (accuracy within 45 μm at 80 mm/s) with on-the-fly dispense and repeatability (%CV) of ±5 μm, (ii) 0.1 μl and ≥0.8 μl accuracy of ±15% and ±10%, respectively, (iii) dispense range from 0.1 μl to 60 μl, and (iv) pre-defined variable plate configurations (96, 384, 1536 and 3456-well).
2.4. Titer plate and solid support
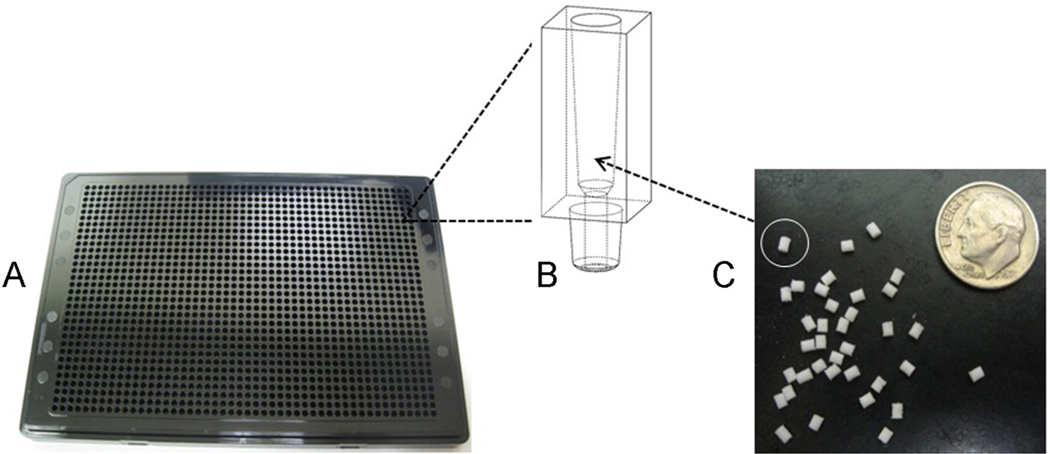
Custom titer plates with 1536-well configuration were designed in-house (SGTC) and molded (Zeonor 1420 R 4%) by Plastic Design Corporation. Custom 1.2 mm × 2.0 mm synthesis plugs (1000 Å controlled pore glass (CPG) with UnyLinker) were manufactured by Chemgenes Corporation, and pressure fitted into the wells (Fig. 2).
Fig. 2.
(A) Shows the custom 1536-well titer plate, (B) exploded view of single reaction well with external drip director (0.6 mm exit hole), and (C) individual synthesis plugs (1.5 mm × 2.0 mm, cylindrical) that are pressure-fitted inside each titer plate well.
2.5. NG-1536-AMOS user interface (UI)
Based on the original 96-AMOS UI (Jova Solutions), software for our next generation platform is a multifaceted design giving the end-user complete control over how the instrument functions (Supplementary Figure 3). Here, synthesis is initiated by uploading the protocol and sequence files (*.PRO, and *.SEQ), then selecting the START button (below “protocol file”, Supplementary Figure 3B). The protocol file is displayed in real-time, highlighting the current step. Indicator bars show the current step, cycle and total synthesis progress. A 1536-well grid also displays which bases are currently being synthesized for all active well positions.
2.6. Oligonucleotide synthesis
All samples were generated in-house (SGTC) with our NG-1536-AMOS platform. General synthesis steps (e.g. similar to Applied Biosystems AB3900 synthesizer protocol) were followed using trichloroacetic acid (TCA) (3% in dichloromethane) (American International Chemicals (AiC)), acetonitrile, also referred to as the “Wash” reagent (AiC), 0.02 M oxidizing solution (Sigma–Aldrich), capping reagents A/B (Glen Research), 0.10 M solutions of dA, dG, dC and dT phosphoramidites (Thermo Scientific), 0.25 M 5-Ethylthio-1H-tetrazole (Glen Research), used as an activator during nucleoside phosphoramidite coupling steps, and argon gas (Prax-air).
2.7. Post-synthesis
Inside a small aluminum box (150 mm × 115 mm × 75 mm, Supplementary Figure 4A), the titer plate (with fully synthesized oligonucleotides) is placed top down over a trough (Supplementary Figure 4B) filled with 50 ml ammonium hydroxide (28–30% NH4OH, Fischer Scientific). A cover then seals the chamber, and under elevated pressure (12.75 psi) and temperature (70 °) for 4 h, gaseous ammonia cleaves and deprotects oligonucleotides embedded within the solid support matrix. This procedure is adapted from of our prior work studying universal linker-bound oligonucleotide cleavage and dephosphorylation in the gas-phase (Jensen et al., 2010a). The reaction chamber is then removed from the incubator and allowed to cool to room temperature. A release valve at the bottom of the apparatus is opened to vent residual ammonia under the fume hood. The titer plate is then seated over a low volume collection plate (Greiner Bio-one, 18 μl capacity per well) with drip directors positioned inside the collection wells. Five μl of deionized water are added to the wells, then fully processed samples are eluted from the synthesis plugs through centrifugation (e.g. 1 min at 1000 RPM). This process can be repeated to maximize yield.
2.8. Oligonucleotide quality analysis
To validate the mass (Daltons) of 80-base strands (Supplementary Table 1) generated on our NG-1536-AMOS platform, Electrospray Ionization Mass Spectrometry (ESI MS) was carried out by Integrated DNA Technologies (Fig. 3). We also synthesized and assembled the green fluorescent protein (GFP) gene de novo for sequence validation of a set of 40mers (Supplementary Table 2). We used the ligase chain reaction (LCR) method to assemble the GFP gene (Fuhrmann et al., 1999). For all steps regarding assembly, PCR amplification and gel analysis, we referenced our previous work (Jensen et al., 2010b). Left and right primers, 5′-TCAGCATGAGTAAAGGAGAA and 5′-CGCTCTATTTGTATAGTTCA, were used to amplify the GFP construct. Target bands (717 bp) were then extracted (QIAquick Gel Extraction Kit, Qiagen), 5′adenylated (PCR extension 72 °C for 10 min with regular Amplitaq polymerase), ligated and cloned (TOPO TA Cloning kit, Invitrogen), then plasmid purified (R.E.A.L. Prep 96, Qiagen). Samples were sent to GenScript for sequencing, while trace analysis to determine error rate (Supplementary Table 3) was done in-house (SGTC).
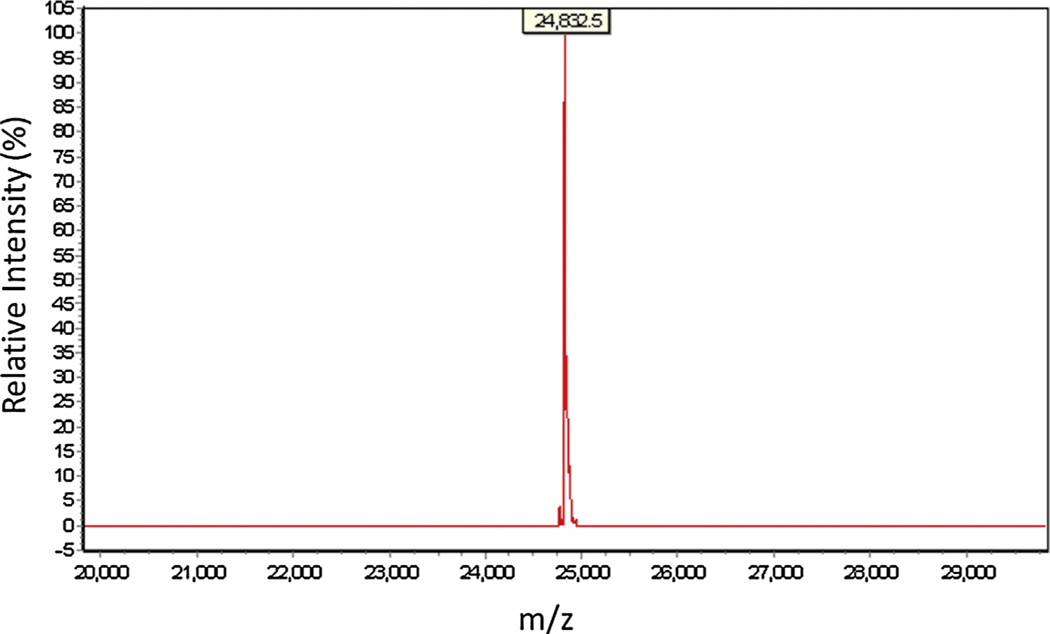
Fig. 3.
Electrospray Ionization Mass Spectrometry (ESI MS) characterization of GFP sequence R640 (80mer, Supplementary Table 1). Several oligonucleotide sequences (Supplementary Tables 1 and 2) were analyzed with ESI MS, all of which showed an average coupling efficiency of 99.5%. In this particular example, R640 is measured at 24,832.5 g/mol while its calculated mass is 24,830.1 g/mol (Note: Y axis = relative intensity (%); X axis = charge-to-mass ratio (m/z))
3. Results and discussion
3.1. Synthesis cost, yield and cycle time
Compared to the synthesis cost of $0.06 per base on 96-AMOS, cost is decreased to $0.00277 on our NG-1536-AMOS platform (Table 1). Calculated average yield per sample/well for our 96-AMOS platform is 28 nmol compared to 3.5 nmol on NG-1536-AMOS; final product yield depends on the maximum allowable volume of the sample collection plate and the number of product elution steps.
Table 1.
NG-1536-AMOS reagent consumption, yield and cost per base.
| Reagent | μl/step | Steps | μl/cycle | μl/20 cycles | ml/platea of 1536 samples | Fold-lower than 96-AMOS |
|---|---|---|---|---|---|---|
| TCA | 3.5 | 3 | 10.5 | 210 | 322.56 | 27 |
| Base | 1 | 1 | 1 | 20 | 30.72 | 20 |
| Activator | 1.5 | 1 | 1.5 | 30 | 46.08 | 18 |
| Cap | 2 | 1 | 2 | 40 | 61.44 | 13 |
| Oxidizer | 3 | 1 | 3 | 60 | 92.16 | 7 |
| Wash | 4 | 5 | 20 | 400 | 614.40 | 58 |
| Argon | 4b | 113 | ||||
| Yieldc | 3.5 | |||||
| Cost/based | 0.00277 |
Reagent consumption on NG-1536-AMOS platform compared with 96-AMOS. For example, 3.5 μl of TCA are delivered per well (10.5 μl over 3 steps). A total of 210 μl of TCA are delivered over 20 cycles (to generate a 20-base strand); 322.56 ml total TCA are delivered to 1536 wells over 20 cycles. Reagent consumption of TCA on NG-1536-AMOS compared to 96-AMOS is 27-fold lower.
20-Base strands per well.
Pounds of argon per 20 cycles.
Average yield (nanomoles) over 50 samples.
Cost/base including labor; the washing reagent “Wash” used for synthesis is acetonitrile.
Each capping step is composed of Cap A and B at 1 μl each (total 2 μl).
Current cycle time for NG-1536-AMOS is 11.85 min. This is largely determined by the dispense volume and number of passes each reagent makes across the plate during a given step (Table 2). With the 16-valve capacity, high volume reagents such as acetonitrile (wash) and TCA (deblock) can have multiple nozzle assignment for maximum plate coverage (Supplementary Figure 5B and D); current setup has acetonitrile and TCA delivered three rows at a time. Because dA, dG, dC and dT are delivered in series one row at a time with activator trailing in the previous row, nucleoside phosphoramidite coupling is the rate limiting step of the protocol.
Table 2.
Calculated synthesis cycle time on NG-1536-AMOS.
| Reagent | Steps | Well coverage | Total travel (min) | Total Reaction hold (min) | Total drain (min) | Total cycle (min)/step |
|---|---|---|---|---|---|---|
| TCA | 3 | 144 | 2.67 | 0.00 | 0.05 | 2.72 |
| Activatora | 1 | 96 | 1.07 | 0.67 | 0.02 | 1.76 |
| Cap | 1 | 96 | 1.07 | 0.33 | 0.02 | 1.42 |
| Oxidizer | 1 | 96 | 1.07 | 0.33 | 0.02 | 1.42 |
| Wash | 5 | 144 | 4.45 | 0.00 | 0.08 | 4.53 |
| Total cycle time (min) | 11.85 |
Here it is shown how the total synthesis cycle time on the NG-1536-AMOS platform is calculated. For example, during the deblock step, TCA is delivered to three rows simultaneously covering 144 wells. The total time it takes the titer plate to move continuously over all 32 rows while delivering TCA (e.g. 3.5 μl/well) is 0.89 min. After a reaction hold period of 0.00 s, TCA is then drained from the wells (vacuum-to-waste valve is open for 1 s). If all these steps are repeated two more times, it will take 2.72 min to complete the deblock portion of the synthesis cycle. Coupling, capping and oxidation steps are also factored in the same way to derive at 11.85 min total cycle time.
This step includes phosphoramidite addition (activator + base per coupling step). The washing reagent “Wash” used for synthesis is acetonitrile.
To increase overall speed of oligonucleotide production, additional steps can be taken which include (i) further protocol optimization, (ii) test of various activators (e.g. activator-42, DCI and benzoyl 1-H tetrazole) to lower coupling reaction times, and (iii) introduction of fast-deprotecting groups (PAC (phenoxyacetyl)-dA, dmf-dG and acetyl-dC) into the gas-phase cleavage and deprotection step; this could decrease the incubation period by 10-fold (Reddy et al., 1994).
3.2. Support loading
While loading base-specific support for large-scale commercial synthesizers such as the Applied Biosystems AB3900 (48-column) or 96-AMOS (plate synthesizer) is straightforward, there is minimal risk of manual error compared with support loading in denser well-configurations (≥384); therefore, to mainstream the titer plate setup for a 1536-well array, a universal linker substrate is required. We chose the UnyLinker (Ravikumar et al., 2008) (Chemgenes), which is highly stable and user-friendly with standard post-synthesis conditions (liquid and gas-phase processes). On our NG-1536-AMOS, 1000 Å CPG support is ideal for generating product in the target range of 25–90 bases. For longer strands (up to 200 bases), synthesis plugs can be loaded with either 1400 Å or 2000 Å support. With the larger pore-size, however, synthesis plugs will require extra support loading to compensate for the decreased surface area (actual yields for the various loading capacities have yet to be determined). The universal linker also allows for synthesis diversity on strands with 3′ modifications (e.g. amino and biotin).
3.3. NG-1536-AMOS synthesis protocol
The protocol file (Supplementary Figure 3C) dictates (i) the reagent to be delivered, (ii) its dispense volume, (iii) the reaction hold time, and (iv) reagent purge-to-waste duration. There are several additional parameters that may be useful in tailoring a protocol file to a particular run. For example, synthesis of ultra long oligonucleotides may benefit from multiple, integrated cycles where the coupling hold time is increased after the first forty base additions, then further increased after eighty, etc.
Because the dispense nozzles/valves on our system have an inner diameter of only 0.25 mm (0.81 mm for respective tubing), reagents such as the activator (tetrazole) and dG (Jensen et al., 2010b) are more susceptible to crystallization. Therefore, we integrated 3-way valves (Fig. 1I D) where the end-user can switch to acetonitrile to wash/purge the reagent lines. This is necessary following synthesis, especially if the instrument will be left idle for two or more days.
3.4. Features of UI
With a 1536-well plate density, it is essential to maintain accuracy with simultaneous plate movement and reagent delivery; for a well pitch of 2.25 mm, on-center nozzle placement may be negatively affected if the plate is misaligned by just a few microns. Similarly, for a small-scale synthesis (2–5 nmol), dispensing volumes between 0.5 and 5.0 μl is routine, and therefore, necessitates stringent valve calibration. The BioRAPTR FRD has an automated calibration method built into the system. Here reagents from each valve are dispensed onto a sponge and weighed with an analytical balance. Because liquids used on the BioRAPTR FRD are water-based, little to no evaporation occurs during the weighing process. However, our modified platform uses volatile chemicals, and at micro-/sub-microliter volumes (especially for TCA), measurements by weight cannot be taken with great accuracy due to rapid evaporation. Instead each valve is manually calibrated by dispensing directly into a 0.5 ml eppendorf tube, weighing it, then plotting its value on a graph to generate a standard curve. In addition, the NG-1536-AMOS UI allows the end-user to adjust each valve separately by increasing or decreasing dispense volumes by a certain percentage (factor) of the actual volume. This is of particular importance when optimizing the coupling step. In this case, the end-user may program one valve (e.g. dA) to dispense more or less reagent than another valve (dG, dC or dT) in order to maximize synthesis efficiency.
3.5. Advantages of 16-channel waste basin
The objective of the 16-channel waste basin design (Supplementary Figures 1 and 2) is to (i) provide for a uniform drain per two-row block (96 wells) (vacuum and/or top pressure applied to 1536 wells simultaneously causes uneven plate drainage, thus sacrificing overall sample quality), (ii) permit synthesis of varying length strands, (iii) have the option of partial plate synthesis, and (iv) allow for better control over reaction conditions. By segregating the drain-to-waste steps (two rows at a time), the end-user can optimize how each block behaves during synthesis. For example, while TCA is delivered to rows 5–8, vacuum to channels 1 and 2 (rows 1–4) will open after the specified reaction hold step. This will prevent overexposure to acid while the plate travels all the way to row 32 before finally draining (each titer plate is arranged by 32 rows and 48 columns in a 1536-well array).
3.6. Gas-phase post-synthesis production
Manually processing all 1536 samples in the liquid-phase (NH4OH) using a multi-channel pipette at very small volumes (5 μl) would permit significant ammonia loss through evaporation in an open system. Based on our previous work studying optimal conditions for cleavage and dephosphorylation of the universal linker in gas-phase (Jensen et al., 2010a), we developed a modified protocol using a compact reaction chamber (closed system, Supplementary Figure 4) to house a single titer plate. Under elevated pressure and temperature, samples (80mers with standard protecting groups) are fully processed in 4 h.
3.7. Oligonucleotide quality analysis
We have demonstrated that oligonucleotide sample purity on our NG-1536-AMOS platform is comparable to that generated on either the AB3900 (Applied Biosystems) or 96-AMOS instruments (SGTC). Based on sample percent abundance determined by ESI MS, we calculated an average coupling efficiency of 99.5% for both sets of 40 and 80mers (Supplementary Tables 1 and 2, Fig. 3); no additional means of sample purification other than standard desalting (Bio-Rad P-6 columns) were applied prior to analysis.
We also performed de novo synthesis and assembly of the GFP gene for sequence validation using 40-base overlapping fragments assembled with LCR (Jensen et al., 2010b; Wiedmann et al., 1994) (Supplementary Table 2). In sequencing 70 clones, we found on average 1.53 errors out of 717 bases (Supplementary Table 3).
3.8. Potential scale-down to a 3456-well synthesis platform
Because the original BioRAPTR FRD instrument is set up to run the pre-defined plate configurations, 96, 384, 1536 and 3456-well, it is possible to generate as many as 3456 unique oligonucleotide samples on one titer plate. However, further discussion of its development is beyond the scope of this article.
4. Conclusion
Based on the commercial BioRAPTR FRD platform, we developed a custom, next generation oligonucleotide synthesizer capable of producing 1536 80mers in a single run, with a potential scale-down to 3456 samples/titer plate. At $0.00277/base with an average yield/well of 3.5 nmol, there is greater flexibility for gene assembly, creating combinatorial libraries, and for mass sample handling using compatible fluidic workstations (e.g. Biomek FX). In addition, sequencing analysis confirms high sample quality on our NG-1536-AMOS platform at only 1.53 errors per 717 bases (on average) using 40mers to construct the GFP gene. Furthermore, mass spectral analysis of 80mers demonstrates an average coupling efficiency of 99.5%. Also, with implementation of a modified gas-phase cleavage and deprotection method, a plate of 60mers could have same-day turnaround; and with low-volume sample collec-tion, oligonucleotides are ready for immediate use in downstream application.
Supplementary Material
Acknowledgements
We would like to thank Lauren Jauregui, Christiana Santiago, Michael Proctor, Anuj Mohan, Maureen Hillenmeyer and Gergana Vandova for their generous contributions to this project.
Funding
This work was supported by the National Institutes of Health (P01-HG000205).
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jbiotec.2013.11.027.
References
- Auld DS, Thorne N, Nguyen DT, Inglese J, 2008. A specific mechanism for non-specific activation in reporter-gene assays. ACS Chem. Biol. 3, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovkov AY, Loskutov AV, Robida MD, Day KM, Cano JA, Le Olson T, Patel H, Brown K, Hunter PD, Sykes KF, 2010. High-quality gene assembly directly from unpurified mixtures of microarray-synthesized oligonucleotides. Nucleic Acids Res. 38, e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JY, Chen HH, Kao YS, Kao WC, Peck K, 2002. High throughput parallel synthesis of oligonucleotides with 1536 channel synthesizer. Nucleic Acids Res. 30, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb RE, Sun N, Zhao H, 2013. Directed evolution as a powerful synthetic biology tool. Methods 60, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H, 2007. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl. Acad. Sci. U. S. A. 104, 9387–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Mulligan J, Hennessy K, Yelton MA, Allen E, Araujo R, Aviles E, Berno A, Brennan T, Carpenter J, et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome V. Nature 387 (6632 Suppl.), 78–81. [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P, 1999. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19, 353–361. [DOI] [PubMed] [Google Scholar]
- Gan CF, 1990. Compact synthesizers let small labs make their own genes. Scientist 4 (6), 30–31. [Google Scholar]
- Gao X, Gulari E, Zhou X, 2004. In situ synthesis of oligonucleotide microarrays. Biopolymers 73, 579–596. [DOI] [PubMed] [Google Scholar]
- Goforth S, 2002. The core of DNA synthesis. Scientist 16 (12), 43. [Google Scholar]
- Hager KM, Fox JW, Gunthorpe M, Lilley KS, Yeung A, 1999. Survey of current trends in DNA synthesis core facilities. J. Biomol. Tech. 10 (4), 187–193. [PMC free article] [PubMed] [Google Scholar]
- Hardenbol P, Baner J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, et al. , 2003. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21, 673–678. [DOI] [PubMed] [Google Scholar]
- Jensen MA, Anderson KM, Davis RW, 2010a. Gas-phase cleavage and dephosphorylation of universal linker-bound oligodeoxynucleotides. Nucleosides Nucleotides Nucleic Acids 29, 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Fukushima M, Davis RW, 2010b. DMSO and betaine greatly improve amplification of GC-rich constructs in de novo synthesis. PLoS ONE 5, e11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Jauregui L, Davis RW, 2012. A rapid cost-effective method of assembly and purification of synthetic DNA probes > 100 bp. PLoS ONE 7 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Han H, Ahn J, Lee J, Cho N, Jang H, Kwon S, Bang D, 2012. ‘Shotgun DNA synthesis’ for the high-throughput construction of large DNA molecules. Nucleic Acids Res. 40, e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong DS, Carr PA, Chen L, Zhang S, Jacobson JM, 2007. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 35, e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921. [DOI] [PubMed] [Google Scholar]
- Lashkari DA, Hunicke-Smith SP, Norgren RM, Davis RW, Brennan T, 1995. An automated multiplex oligonucleotide synthesizer: development of high-throughput, low-cost DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 92, 7912–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebl M, Pistek C, Hachmann J, Mudra P, Pesek V, Pokorny V, Poncar P, Zenisek L, 2007. Economical parallel oligonucleotide and peptide synthesizer – pet oligator. Int. J. Pept. Res. Ther. 13, 367–375. [Google Scholar]
- Lee CC, Snyder TM, Quake SR, 2010. A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 38, 2514–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesay EA, Liu YH, Luebke KJ, Irick J, Belosludtsev Y, Rayner S, Balog R, Johnston SA, 2002. A scalable high-throughput chemical synthesizer. Genome Res. 12, 1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Saaem I, Tian J, 2012a. Error correction in gene synthesis technology. Trends Biotechnol. 30, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Tang N, Tian J, 2012b. DNA synthesis, assembly and applications in synthetic biology. Curr. Opin. Chem. Biol. 16 (3–4), 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Coleman JR, Wimmer E, 2009. Putting synthesis into biology: a viral view of genetic engineering through de novo gene and genome synthesis. Chem. Biol. 16 (3), 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan JY, Saaem I, Tang N, Ma SM, Negre N, Gong H, White KP, Tian JD, 2011. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat. Biotechnol. 29, 449. [DOI] [PubMed] [Google Scholar]
- Ravikumar VT, Kumar RK, Olsen P, Moore MN, Carty RL, Andrade M, Gorman D, Zhu X, Cedillo I, Wang Z, et al. , 2008. UnyLinker: an efficient and scaleable synthesis of oligonucleotides utilizing a universal linker molecule: a novel approach to enhance the purity of drugs. Org. Process Res. Dev. 12, 399–410. [Google Scholar]
- Rayner S, Brignac S, Bumeister R, Belosludtsev Y, Ward T, O’dell G, O’Brien K, Evans GA, Garner HR, 1998. MerMade: an oligodeoxyribonucleotide synthesizer for high throughput oligonucleotide production in dual 96-well plates. Genome Res. 8, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MP, Hanna NB, Farooqui F, 1994. Fast cleavage and deprotection of oligonucleotides. Tetrahedron Lett. 35, 4311–4314. [Google Scholar]
- Weber W, Fussenegger M, 2012. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 13, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Wilson WJ, Czajka J, Luo J, Barany F, Batt CA, 1994. Ligase chain reaction (LCR) – overview and applications. PCR Methods Appl. 3, S51–S64. [DOI] [PubMed] [Google Scholar]
- Yasgar A, Furdas SD, Maloney DJ, Jadhav A, Jung M, et al. , 2012. High-throughput 1,536-well fluorescence polarization assays for α1-acid glycoprotein and human serum albumin binding. PLoS ONE 7 (9), e45594, 10.1371/journal.pone.0045594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.