Abstract
Preterm birth (PTB) is associated with increased risk of type 2 diabetes and neurocognitive impairment later in life. We analyzed for the first time the associations of PTB with blood miRNA levels in adulthood. We also investigated the relationship of PTB associated miRNAs and adulthood phenotypes previously linked with premature birth. Blood MicroRNA profiling, genome-wide gene expression analysis, computer-based cognitive testing battery (CANTAB) and serum NMR metabolomics were performed for Young Finns Study subjects (aged 34–49 years, full-term n = 682, preterm n = 84). Preterm birth (vs. full-term) was associated with adulthood levels of hsa-miR-29b-3p in a fully adjusted regression model (p = 1.90 × 10–4, FDR = 0.046). The levels of hsa-miR-29b-3p were down-regulated in subjects with PTB with appropriate birthweight for gestational age (p = 0.002, fold change [FC] = − 1.20) and specifically in PTB subjects with small birthweight for gestational age (p = 0.095, FC = − 1.39) in comparison to individuals born full term. Hsa-miR-29b-3p levels correlated with the expressions of its target-mRNAs BCL11A and CS and the gene set analysis results indicated a target-mRNA driven association between hsa-miR-29b-3p levels and Alzheimer's disease, Parkinson's disease, Insulin signaling and Regulation of Actin Cytoskeleton pathway expression. The level of hsa-miR-29b-3p was directly associated with visual processing and sustained attention in CANTAB test and inversely associated with serum levels of VLDL subclass component and triglyceride levels. In conlcusion, adult blood levels of hsa-miR-29b-3p were lower in subjects born preterm. Hsa-miR-29b-3p associated with cognitive function and may be linked with adulthood morbidities in subjects born preterm, possibly through regulation of gene sets related to neurodegenerative diseases and insulin signaling as well as VLDL and triglyceride metabolism.
Subject terms: Gene expression, Gene regulation, miRNAs, Type 2 diabetes
Introduction
Preterm birth (PTB), defined as birth of an infant at less than 37 weeks’ gestational age associates with increased risk of developmental defects and mortality in infants as well as multiple diseases later in life1. In addition to PTB, growth restriction and being born small for gestational age have been shown to increase the risk of multiple maladies2. Altogether, PTB subjects are at greater risk of neurodevelopmental disorders, have worse academic performance at school and poorer performance in childhood cognitive tests3–5. Cohort studies from Nordic countries have also indicated that late PTB (34 weeks 0 days to 36 weeks 6 days of gestation) is associated with poorer episodic memory performance in adulthood, increased risk of neurodegenerative diseases6 and risk of accelerated age related cognitive dysfunction7. Subjects born preterm have been shown to have increased prevalence of chronic diseases, such as asthma8, type 1 and type 2 diabetes (T2D)9, and cardiovascular risk factors and decreased arterial health10–12. The effect of PTB on later life health is conveyed by malformations associated with PTB13; altered pattern of growth, increased whole body adiposity, and partitioning of adipose tissue14,15; and subtle abnormalities of central nervous system function16. It has also been suggested that intrauterine conditions and PTB affect individuals’ health through changes in epigenetic factors, namely, histone modifications17, DNA-methylation profiles18,19, and non-coding RNAs20. Prenatal insults and stress can affect individuals’ epigenome and long-term gene expression regulation, which in turn can convey the increased risk of chronic diseases later in life21.
MicroRNAs (miRNAs, miRs) are small non-coding RNAs that primarily regulate gene expression by binding to target mRNAs and interfering with the translation22,23. In addition to translational repression, miRNAs have been shown to function as translational24 and transcriptional activators, the latter of which may happen via epigenetic modulation25. MicroRNAs can also be transported between cells and tissues via circulation. These circulating miRNAs have been shown to participate in cell-to-cell communication26, potentially contributing to disease progression, but also have been shown to act as indicators for pathological processes elsewhere in the body.
Changes in miRNA profiles have been associated with many chronic diseases with partly developmental origins, such as asthma27, T2D28 and cardiovascular diseases29. Additionally, miRNA expression has been shown to be altered in complications of pregnancy, such as miscarriages30 and preeclampsia31,32. In animal studies, the nutritional and stress status of the mother has been shown to alter the offspring’s miRNA profile in various tissues and affect the gestational length and insulin sensitivity of the offspring20,33. It has also been shown that the miRNA profiles of the placenta31, amniotic fluid34, and maternal serum32, whole blood and monocytes35 differ between preterm and full term births. Although the lasting effect on PTB on DNA methylation pattern19,36 and subjects’ health37 have been studied, only a few studies analyzing the associations of PTB with the subjects’ circulatory miRNA profile have been conducted on newborns38–41 and one case–control study (n = 50) with 4 selected miRNAs in young adults42.
Our hypothesis is that PTB is associated with the blood levels of miRNAs in adulthood and that this altered miRNA expression may associate with increased risk of chronic diseases in prematurely born subjects later in their lives. More specifically, we aimed to (1) analyze the differences in blood miRNA levels in adulthood between subjects born full term (FTB) and preterm or alternatively PTB with appropriate birth weight for gestational age (AGA) or PTB with small birth weight for gestational age (SGA), (2) discover blood target mRNAs affected by these dysregulated miRNAs, (3) research the biological gene sets over the whole transcriptome that are affected by these miRNAs (4) and to investigate the possible association of these dysregulated miRNAs and adulthood indices of those conditions previously associated with PTB, such as T2D, dyslipidemias and lower cognitive function.
Methods
More detailed materials and methods can be found in the Additional File 1.
The Young Finns Study (YFS)
YFS is a multicenter follow-up study on cardiovascular risk factors from childhood to adulthood in Finland. The YFS was launched in 1980, when 3,596 children and adolescents (3–18 years old) participated in the baseline study43. Thereafter, the subjects have been followed up with several examinations including comprehensive risk factor assessments. The 30-year follow-up was conducted in 2011, with 2,063 adults, aged 34–49 years, participating in the study. The examinations included physical measurements, blood tests, and questionnaires. All measurements utilized in this study, excluding birthweight, prematurity at birth and parental education level at baseline, are derived from the 2011 follow-up study and samples collected during it. The miRNA profiling population comprises 871 subjects from the 2011 follow-up study. When the population in the miRNA analysis was compared to the whole 2011 follow-up study population, the only difference was observed in the prevalence of T2D, which in the whole population is 6.0% and in the miRNA profiling cohort only 3.0%. Birth weight and preterm status were available for 761 subjects in the miRNA profiling sub-population. The demographics are presented in Supplementary Table S1.
Fetal growth and PTB
PTB status was defined as birth before 37 weeks’ gestation, and the number of weeks preterm was ascertained in those who reported PTB. Birthweight was self-reported during the follow-up studies in 1983 and 1986. Subjects born preterm were categorized as either AGA or with SGA using a cut point of − 1 SD z score (corresponding to the 15th percentile) based on Finnish sex and gestational age-stratified birthweight percentiles12. AGA describes a newborn infant whose size is within the normal range for his or her gestational age, while newborns with SGA were smaller than expected for their gestational weeks. The study population did not include any subjects born preterm and large for their gestational age.
RNA isolation, miRNA and gene expression profiling
RNA was isolated and miRNA and gene expression profiling was performed as described before44. Briefly, whole blood was collected in PaXgene Blood RNA Tubes (PreAnalytix) and isolated with a PAXgene Blood microRNA Kit (Qiagen) including the DNase Set using the QiaCube according to the manufacturer’s instructions.
As described before in44, microRNA expression profiling was performed with the TaqMan MicroRNA Panel (Applied Biosystems) containing 758 microRNAs using the standard protocol. MicroRNA panels were loaded using the AccuFill System and run with the QuantStudio 12K Flex (Applied Biosystems). Primary data analysis was performed with Expression Suite Software version 1.0.1. U6 snRNA, RNU44, and RNU48 were used as housekeeping small RNAs. 243 miRNAs that were expressed in at least 2/3 of the samples were included in further analysis. After quality control and removal of outlier miRNA, profiling was successful on 871 samples. To correct for batch effects, principal components analysis was performed for miRNA expression data. The data was adjusted for 10 of the first 20 principal components from the principal component analysis.
As described in44, the expression levels were analyzed with an Illumina HumanHT-12 version 4 Expression BeadChip (Illumina Inc.). The BeadChips were scanned with the Illumina iScan system. The expression data was processed using nonparametric background correction, followed by quantile normalization with control and expression probes, using the neqc function in the limma package and log2 transformation. The expression analysis was successful in 743 of the 871 samples with a miRNA expression profile.
Clinical and biochemical measurements
As previously described44, weight, height, waist circumference, and blood pressure were measured, and body mass index (BMI) was calculated. Blood cell parameters were measured with flow cytometric particle counting and photometry. The serum triglyceride, glucose, and total cholesterol concentrations were analyzed using the enzymatic methods. High density lipoprotein (HDL) cholesterol levels were estimated after the precipitation of apolipoprotein B-containing lipoproteins. Low density lipoprotein cholesterol was calculated indirectly using the Friedewald formula. For glycated hemoglobin (HbA1c) fraction measurement, the concentration of total hemoglobin was determined colorimetrically, after which the concentration of HbA1c was measured immunoturbidimetrically. These two concentrations were used to calculate the HbA1c percentage (HbA1c%). Insulin levels were measured by a microparticle enzyme immunoassay kit. Subjects were categorized into the normoglycemic, impaired fasting glucose, and T2D groups, based on fasting serum glucose and HbA1c according to the criteria of the WHO45 and self-reported diagnosis of T2D by a physician. Subjects with type 1 diabetes were excluded from the analysis.
NMR metabolomics
A high-throughput serum NMR metabolomics platform was used for absolute quantification of serum lipids and metabolites, including lipoprotein subclass distributions, fatty acids, and various small molecules such as amino acids and glycolysis precursors46,47. The analyzed 14 lipoprotein subclasses were defined based on particle size. The details of the experimentation have been described46–48. Data were available from all the subjects with successful miRNA profiling.
Cognitive function
A test battery developed by the Cambridge Cognition (CANTAB) was used to assess cognitive function among the subjects in the follow-up study in 2011. The CANTAB test is a computerized, predominantly non-linguistic and culturally neutral test focusing on a wide range of cognitive domains. The test was performed using a validated touch-screen computer system and was successful on all the 871 subjects with miRNA expression data. In YFS, the test battery was compiled and performed as described earlier49. During cognitive testing the participants conducted a Motor screening test, Paired associates learning (PAL) test, Spatial working memory (SWM) test, Reaction time (RTI) test and Rapid visual information processing (RVP) test. Each test produced several variables. Principal component analysis was conducted for each test to identify components accounting for the majority of the variation within the dataset. The first principal component was selected to represent the performance in each separate test. After distribution analyses, the Motor screening test component was excluded from further analyses due to ceiling effect. Other components were normalized using rank order normalization procedure resulting in four variables (mean 0, standard deviation (SD) 1)49.
Statistical analysis
Association between PTB status (full term birth vs. preterm birth) and adulthood miRNA levels was analyzed with linear regression using age, sex, smoking (yes/no), serum insulin and glucose levels, leucocyte, erythrocyte and thrombocyte count and liver status (fatty liver yes/no) as covariates (MODEL 1). To account for multiple testing, only miRNAs with FDR (Benjamini-Hochberg) < 0.05 were further investigated. Median fold changes (FC) between different birth status groups (FTB, PTB with AGA or PTB with SGA) were calculated and pairwise statistical significance was evaluated with Mann–Whitney U test and over all groups using Kruskal–Wallis test for trend.
The flow and results of transcriptomic and gene set analysis are presented in the Supplementary Fig. S1. The predicted mRNA targets of miRNAs of interest were included in the correlation analysis if they were recognized by one or more in silico miRNA target prediction programs in miRGator50. Correlations between miRNAs of interest and their predicted targets were calculated with Spearman’s correlation and individual correlations with FDR < 0.05 (both positive and negative correlations) were considered significant. Predicted target mRNAs with correlation at the level of FDR < 0.25 with the levels miRNA of interest were selected in further analysis. The overlaps between the selected target mRNAs and gene set in KEGG51 and BIOCARTA were analyzed in molecular signature database. Gene sets containing at least 5 of the selected target mRNAs and with FDR < 0.05 for the enrichment were included in gene set enrichment analysis (GSEA). In GSEA (adjusted for age and sex), the association between the expression of all genes in the identified gene sets and blood levels of the miRNA of interest were evaluated and FDR < 0.25 was considered significant.
Association between the miRNAs of interest and the metabolite levels as well as physiological features previously associated with metabolic dysfunction (listed in44) were analyzed one by one with linear regression analysis using age, sex, smoking, birth status, leucocyte, erythrocyte and thrombocyte count, fatty liver status and polygenic risk score for metabolic syndrome52 as covariates (MODEL 2). Associations between the miRNA and diagnosis of hypertension, T2D and impaired fasting glucose were analyzed separately with binominal regression models using the covariates according to the MODEL 2.
The associations between the miRNA of interest and each studied cognitive domain were analyzed separately with linear regression with age, sex, smoking, birth status, leucocyte, erythrocyte and thrombocyte count and fatty liver status as well as parental education years (less than 9 years, 9–12 years or more than 12 years), subject’s own education level (participated in higher academic education yes/no) and polygenic risk score for cognitive function as covariates (MODEL 3). If the miRNA of interest associated significantly (FDR < 0.05) with any of the cognitive domains, its association with the individual variables produced by the specific cognitive test for that particular cognitive domain was also analyzed one by one utilizing the adjustments according to the MODEL 3. FDR < 0.05 was considered significant when utilizing models 1, 2 and 3.
Ethics approval and consent to participate
The present study has been approved by the 1st ethical committee of the Hospital District of Southwest Finland on September 21st, 2010 and by local ethical committees. All study subjects gave an informed consent, and the study was conducted according to the principles of the Declaration of Helsinki.
Results
Insulin levels and prevalence of impaired fasting glucose are higher in subjects with PTB
Our data comprises of 681 subjects with FTB, 67 with PTB and AGA and 17 with PTB and SGA (Supplementary Table S1). The prevalence of impaired fasting glucose was higher in subjects with PTB vs FTB (chi square test, p = 0.039). Difference was detected also when comparing only the subjects with PTB and SGA to those with FTB (p = 0.046). In line with these results, insulin levels were also higher in the subjects with PTB in comparison to those with reported FTB (p = 0.042). In addition to these indicators of impaired glucose metabolism, we observed higher thrombocyte count on the subjects with PTB and AGA in comparison to those with FTB (p = 0.043). The median BMI, and the prevalence of fatty liver and female sex were higher in the subjects with PTB and SGA than in the FTB subjects, but most likely due to the small number of subjects with PTB and SGA, these differences were statistically non-significant.
Adulthood hsa-miR-29b-3p levels are lower in PTB subjects than in subjects born full term
Of the analyzed 243 miRNAs, adult blood levels of hsa-miR-29b-3p, -409-3p and -21-5p were nominally associated with birth status (FTB vs. PTB) in MODEL 1 (Supplementary Table S2). Only blood levels of hsa-miR-29b-3p were associated with PTB after multiple testing correction (p = 1.90 × 10–4, FDR = 0.046, β = − 0.521 95% CI − 0.792 to − 0.249) in this model. When comparing non-adjusted miRNA FC, there was a trend over birth status groups (p = 0.002, Fig. 1). Subjects born preterm with AGA had lower levels of hsa-miR-29b-3p than subjects born full-term (FC = − 1.20, p = 0.002) and this down-regulation was quantitatively more substantial (FC = − 1.39, p = 0.095), in subjects born preterm with SGA, compared to subjects born full-term (Fig. 1). Trend of expression was similar for males and females separately, but the association was statistically significant only in women (Supplementary Fig. S2). The levels of hsa-miR-29b-3p were not associated with birthweight in subjects with FTB.
Figure 1.
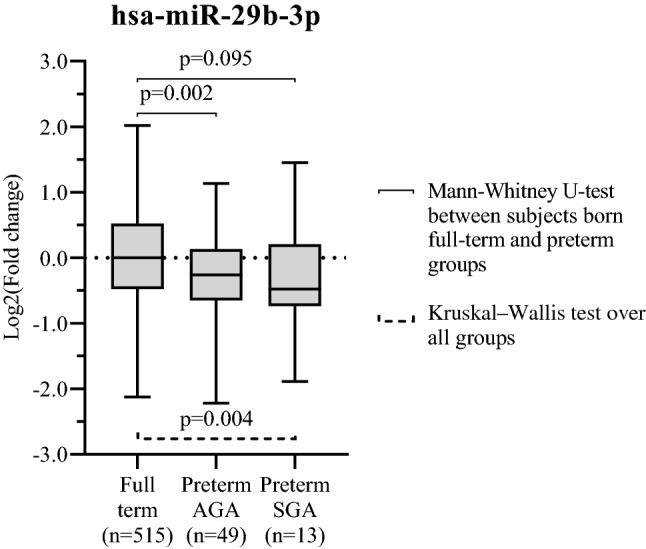
Blood levels of hsa-miR-29b-3p in subjects born preterm in comparison to those born full term. Blood levels of hsa-miR-29b-3p of subjects born preterm and either appropriate birth weight for gestational age (AGA) or small birth weight for gestational age (SGA) described by the Log2(fold changes) in comparison to subjects born full-term. MicroRNA-29b-3p expression was detected in 551 participants with birth weight and birth status information.
Levels of hsa-miR-29n-3p in subjects with FTB or PTB in infancy
As we did not have miRNA data from birth from our study subjects, we retrieved data from Rager et al.53 from Gene Expression Omnibus (GSE48353), to address the question whether miR-29b-3p levels were lower in the blood of PTB infants at birth in comparison to those with FTB. Even though their data set of miRNA profiling performed for 40 cord blood samples collected from the infants immediately following delivery, contained only two subjects who were born before gestational week 37, their median hsa-miR-29b-3p levels were lower than the median of those who were born at or after gestational week 37 (FC = − 1.82, p = 0.023) (Supplementary Fig. S3).
Hsa-miR-29b-3p blood levels correlate negatively with BAF chromatin remodeling complex subunit BCL11A (BCL11A) and positively with citrate synthase (CS) mRNA levels
MiRGator v. 3.0 gives a total of 6102 predicted targets for hsa-miR-29b-3p. In our whole blood transcriptomics data hsa-miR-29b-3p levels correlated at the level of FDR < 0.25 with 269 of these predicted targets (Supplementary Table S3 in Additional file 2). These mRNAs were included in the subsequent gene set analysis. After appropriate multiple testing correction, only BCL11A mRNA levels correlated inversely with hsa-miR-29b-3p levels in blood (p = 1.31 × 10–5, FDR = 0.035, r = − 0.183). In addition, hsa-miR-28b-3p levels correlated positively with CS mRNA levels (p = 1.90 × 10–5, FDR = 0.035, r = 0.180).
Target mRNA driven whole transcriptome wide gene set analysis for hsa-miR-29b-3p levels pinpoints pathways associated with neurodegenerative diseases and insulin signaling
The 269 correlated targets of hsa-miR-29b-3p were enriched in 13 biological gene sets (Table 1). When performing GSEA for these selected 13 pathways and hsa-miR-29b-3p levels, the expression of Alzheimer’s and Parkinson’s disease pathways were negatively associated with hsa-miR-29b-3p levels and the expression of Insulin signaling pathway and Regulation of actin cytoskeleton were associated positively with hsa-miR-29b-3p levels (Table 1).
Table 1.
Target-mRNA driven gene set enrichment analysis for hsa-miR-29b-3p levels.
| Gene set | Target genes enriched in gene sets | Gene sets association with hsa-miR-29b-3p levels | ||||
|---|---|---|---|---|---|---|
| p-value | FDR | Genes in overlap | FWER | FDR | Direction of association | |
| Alzheimer's disease | 3.83 × 10–4 | 0.014 | ADAM10, COX7A2L, COX7C, ITPR1, NDUFA10, UQCRQ | 0.237 | 0.084 | Negative |
| Insulin signaling pathway | 0.001 | 0.026 | FASN, PPP1CB, PRKX, PYGB, RPS6KB2 | 0.080 | 0.145 | Positive |
| Parkinson's disease | 1.05 × 10–4 | 0.008 | COX7A2L, COX7C, NDUFA10, SEPT5, UBE2L3, UQCRQ | 0.234 | 0.165 | Negative |
| Regulation of actin cytoskeleton | 0.001 | 0.033 | ITGA5, ITGB2, ITGB5, PPP1CB, RAC2, VCL | 0.202 | 0.234 | Positive |
| Gap junction | 1.51 × 10–4 | 0.008 | GNAS, ITPR1, PRKX, TUBA1B, TUBB1 | 0.511 | 0.353 | Positive |
| MAPKinase Signaling Pathwaya | 1.29 × 10–4 | 0.008 | MAP2K3, MAP2K4, RPS6KA1, RPS6KA3, RPS6KB2 | 0.560 | 0.361 | Positive |
| Hematopoietic cell lineage | 1.36 × 10–4 | 0.008 | CD4, CD44, IL11RA, ITGA5, MS4A1 | 0.629 | 0.366 | Positive |
| Cell adhesion molecules (CAMs) | 0.001 | 0.025 | ALCAM, CD4, CD58, ITGB2, SELP | 0.599 | 0.369 | Positive |
| GnRH signaling pathway | 2.60 × 10–4 | 0.012 | GNAS, ITPR1, MAP2K3, MAP2K4, PRKX | 0.492 | 0.416 | Positive |
| Vascular smooth muscle contraction | 4.72 × 10–4 | 0.015 | GNAS, ITPR1, KCNMB2, PPP1CB, PRKX | 0.411 | 0.429 | Positive |
| MAPK signaling pathway | 1.35 × 10–4 | 0.008 | MAP2K3, MAP2K4, PPM1B, PRKX, PTPN7, RAC2, RPS6KA1, RPS6KA3 | 0.760 | 0.545 | Positive |
| Long-term potentiation | 4.56 × 10–5 | 0.008 | ITPR1, PPP1CB, PRKX, RPS6KA1, RPS6KA3 | 0.823 | 0.639 | Positive |
| Oocyte meiosis | 4.54 × 10–4 | 0.015 | ITPR1, PPP1CB, PRKX, RPS6KA1, RPS6KA3 | 0.856 | 0.674 | Negative |
Thirteen gene sets were enriched with predicted and correlating (FDR < 0.25) targets of hsa-miR-29b-3p. When performing GSEA for these 13 gene sets and hsa-miR-29b-3p levels, four were associated (FDR < 0.25, in bold) with the expression of hsa-miR-29b-3p. This indicates a possible hsa-miR-29b-3p target mRNA driven regulation of these gene sets. The flow and results of transcriptomics and gene set analysis are presented in Supplementary Fig. S1
Notes All gene sets except MAPKinase Signaling Pathway (BIOCARTAa) are from KEGG collection. Both gene-expression data and hsa-miR-29b-3p levels were available from 559 study subjects.
GSEA gene set enrichment analysis; FDR False discovery rate.
Hsa-miR-29b-3p levels associated with visual processing and sustained attention
From the 4 cognitive domains analyzed, hsa-miR-29b-3p levels were associated directly and independently with visual processing and sustained attention (Rapid visual information processing test; p = 0.005, FDR = 0.020; β = 0.115, 95%CI = 0.035–0.195) (Table 2, MODEL 2). When analyzing the individual variables within the test measuring this cognitive domain, the association was strongest with the total of correct rejections (p = 0.008, FDR = 0.025; β = 0.106, 95%CI = 0.028–0.184).
Table 2.
Association between hsa-miR-29b-3p levels and cognitive domains and variables indicating visual processing and sustained attention.
| p-value | FDR | β | 95% CI | |
|---|---|---|---|---|
| Cognitive domains | ||||
| Visual processing and sustained attention (RVP test) | 0.005 | 0.020 | 0.115 | 0.035–0.195 |
| Episodic memory and associative learning (PAL test) | 0.280 | 0.547 | 0.043 | − 0.035 to 0.121 |
| Short term working memory (SWM test) | 0.410 | 0.547 | 0.034 | − 0.047 to 0.116 |
| Reaction and movement time (RTI test) | 0.569 | 0.569 | 0.025 | − 0.112 to 0.061 |
| Variables indicating visual processing and sustained attention | ||||
| Total correct rejections | 0.005 | 0.025 | 0.112 | 0.034–0.191 |
| Signal detection measure of sensitivity to the target (A’) | 0.008 | 0.025 | 0.105 | 0.028–0.184 |
| Total misses | 0.013 | 0.025 | − 0.099 | − 0.177 to − 0.021 |
| Probability of hit | 0.013 | 0.025 | 0.098 | 0.021–0176 |
| Total hits | 0.014 | 0.025 | 0.098 | 0.020–0.175 |
| Probability of false alarm | 0.041 | 0.061 | − 0.078 | − 0.152 to 0.003 |
| Total false alarms | 0.070 | 0.089 | − 0.070 | − 0.143 to 0.005 |
| Signal detection measure of the strength of trace required to elicit a response (B’) | 0.091 | 0.102 | 0.068 | − 0.011 to 0.146 |
| Mean latency | 0.362 | 0.362 | − 0.044 | − 0.126 to 0.046 |
The association between hsa-miR-29b-3p levels and the cognitive domains was analyzed one by one with linear regression with age, sex, smoking, birth status, leucocyte, erythrocyte and thrombocyte count and liver status as well as parental education years, participants own education level and and polygenic risk score for cognitive function as covariates (MODEL 3). All variables were available from 513 subjects. As the principal component of Rapid visual information processing test was associated with hsa-miR-29b-3p levels the association to the individual variables produced by the CANTAB test battery were also analyzed one by one utilizing the same MODEL 3.
Note The CANTAB test battery used to determine the cognitive domains including four separate tests: (1) Paired associates learning (PAL) test, (2) Spatial working memory (PWM) test, (3) Reaction time (RTI) test and (4) Rapid visual information processing (RVP) test.FDR, False discovery rate.
Hsa-miR-29b-3p levels associated with levels of serum very low-density lipoprotein (VLDL) subclasses lipid and triglyceride levels
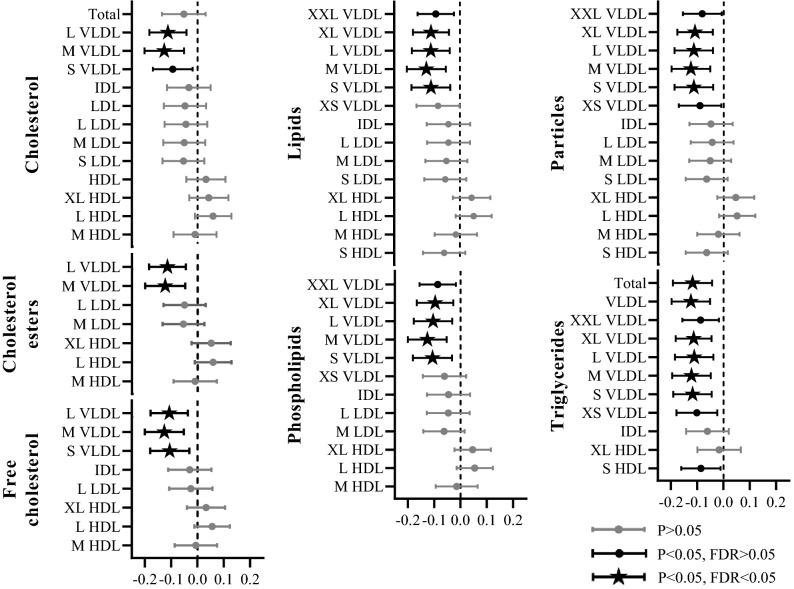
Hsa-miR-29b-3p levels inversely and independently associated with the levels of different subclasses of VLDL in MODEL 3, most frequently with those of medium VLDL. From different types of lipoprotein components, the association was most systematically observed with triglyceride levels. In addition to above NMR-metabolic measures, hsa-miR-29b-3p levels were associated with the conventionally measured triglyceride levels (p = 0.003, FDR = 0.023, β = − 0.111, 95%CI = − 0.185 to − 0.037) (Fig. 2, Supplementary Table S4). There was no association between the diagnosis of hypertension, impaired fasting glucose or T2D and hsa-miR-29b-3p levels (p > 0.05).
Figure 2.
Associations of blood hsa-miR-29b-3p levels with serum lipoprotein subclass particle size and lipid components. The serum lipoprotein subclass particle size and lipid components measured by NMR-spectroscopy. Values are standard deviation (SD) increment in metabolite measure and 95% confidence intervals (95% CI) per one SD change of miRNA of interest are presented. Associations have been analyzed with a linear regression model adjusted with age, sex, smoking, birth status, liver status, polygenic risk score for metabolic syndrome and blood cell counts (MODEL 2). n for the model is 551. XL extra large, L large, M medium, S small, XS extra small, HDL high-density lipoprotein, LDL low-density lipoprotein, IDL intermediate-density lipoprotein, VLDL very-low-density lipoprotein.
Discussion
The association of PTB and adulthood blood miRNA levels was for the first time studied here in the population based YFS data. Lower levels of hsa-miR-29b-3p were identified in subjects born preterm with either AGA or SGA. Hsa-miR-29b-3p levels were shown to correlate negatively with the mRNA levels of BCL11A and positively with CS mRNA levels and the predicted targets of this specific miRNA were enriched in 13 biological gene sets. The expression of Alzheimer’s and Parkinson’s disease pathways were negatively associated and the expression of the Insulin signaling pathway and Regulation of actin cytoskeleton associated positively with hsa-miR-29b-3p levels. We also observed a direct association between the levels of this miRNA and visual processing and sustained attention as well as inverse association with extra-large, large and medium VLDL subclasses and their lipids and triglyceride levels.
Adult (aged 34–49 years) blood levels of hsa-miR-29b-3p, -409-3p and -21-5p were shown here to be associated with individuals birth status. Of these, only the association between hsa-miR-29b-3p levels and birth status remained significant after multiple testing correction. The levels of hsa-miR-29b-3p were shown to be lower in subjects born preterm when comparing to those born full-term and even more so in those born preterm and SGA. This finding is supported by the data from Rager et al.53 showing that the two individuals born before the week of 37 had lower miR-29b-3p levels than the median of those who were born at full term. In addition, Pavlek et al. have reported lower levels of miR-29b umbilical cord blood in infants born with spontaneous PTB in comparison to those whose PTB was medically induced. Intriguingly in their data, the 4 individuals born preterm and SGA had higher hsa-miR-29b-3p levels in comparison to those with PTB and AGA38. The levels of hsa-miR-29b-3p have been also associated with complications in PTB infants such as bronchopulmonary dysplasia39 and adverse events during pregnancy, like preeclampsia31,32. All-in-all, the literature describing the associations between circulatory miRNAs and complicated pregnancies or PTB are scare and recent studies have either not analyzed miR-29b-3p or don’t report association between this miRNA and PTB40,41.
MicroRNA expression has often been thought to be short-termed and responsive to stimulus. The levels and effect of miRNA in the cytoplasm hindering transcription are seen to be transient. In this light, the association between preterm birth and adulthood miRNA levels could be doubted. Our association does not imply causality and transient hsa-miR-29b-3p levels can be affected by many confounding factors. Hsa-miR-29b-3p is nonetheless a credible candidate for more sustained regulation. Instead of the cytoplasm, miR-29b has been shown to be transported to the nucleus where it can act as a transcriptional or splicing factor54. The whole microRNA-29b family has been named epi-miRNAs, as they are capable of controlling epigenetic landscape by targeting key epigenetic effectors, such as DNA methyltransferases and histone deasetylases55,56 having longer lasting effects on gene expression. It is also plausible that hsa-miR-29b-3p levels themselves are regulated by pre- and perinatal conditions and/or their consequences as this miRNA has been suggested to have an extensive regulatory role in embryonic development57,58. Furthermore, Pavlek et al. reported an association between miR-29b-3p levels and inflammation on preterm infants, suggesting thus a possibility of a link between this miRNA, prenatal life and adulthood health38.
The predicted targets of hsa-miR-29b-3p that correlated with its expression were enriched in Insulin signaling pathway and the expression of this gene set as a whole was positively associated with hsa-miR-29b-3p levels. Association between miR-29b and T2D is supported by literature59. Levels of hsa-miR-29b have been shown to be down-regulated in plasma of individuals who will later develop T2D60 and in blood fractions of individuals with either impaired glucose tolerance61 or impaired fasting glucose62. Additionally, this miRNA has been associated with type 1 and gestational diabetes28, and it has been linked to several complications of T2D in small arteries, such as diabetic retinopathy and nephropathy63–65. Like us, others have not seen a correlation between fasting glucose60 or other indications of sugar metabolism62 and hsa-miR-29b-3p levels. Instead, this miRNA has been shown to contribute to insulin resistance and repression of insulin-stimulated glucose uptake in animal-models66–68. Interestingly, Dooley et al. have also described the effects of miR-29 family members on both increased insulin signaling in the pancreas and truncating the duration of insulin signaling in the liver69, while Jacovetti et al. emphasized the effect of miR-29b-3p on maturation of pancreatic islet cells in newborn rats and the effect of this on later manifestation of metabolic diseases70. Both of the predicted target genes of hsa-miR-29b-3p, which levels correlated statistically significantly with the miRNA levels in our data, have also been associated with T2D. BCL11A, which had a negative correlation with hsa-miR-29b-3p levels in our study, has been associated with T2D in genome-wide association studies66,67 and in consequence functional studies73,74. Alike, CS, which had a positive correlation with hsa-miR-29b-3p levels in our data, has been shown to be down-regulated in the muscle of T2D patients, possibly as a consequence of impaired response to insulin75,76.
Our results show a negative association between hsa-miR-29b-3p and VLDL and triglyceride levels. Insulin resistance at the level of the fat cells leads to increased intracellular hydrolysis of triglycerides and release of fatty acids into the circulation. The subsequent excessive flow of fatty acids to the liver results in increased fatty deposition within the hepatocytes and promotes enhanced large VLDL production77. This increased secretion of triglyceride-enriched large VLDL particles and elevated plasma triglycerides is commonly seen in patients with insulin resistance and T2D78. Dyslipidemia including hypertriglyceridemia is an early event accompanying insulin resistance and precedes beta cell failure79. In addition, elevated triglyceride levels have been shown to predict future incidence of T2D independently of traditional risk factors80. The miR-29 family has been previously shown to control hepatic lipogenic programs81 and dampen de novo synthesis of triglycerides through targeting of Sirtuin-1 in the mouse liver82. In cell culture experiments, miR-29b has been shown to target lipoprotein lipase, and thus, to possibly regulate triglyceride production83. However, to our knowledge, this is the first study reporting an association between miR-29b and triglyceride levels in humans.
Targets of hsa-miR-29b-3p, which correlated with the miRNA levels in blood, were also enriched in gene sets related to memory (Long-term potentiation pathway) and neurodegenerative diseases (Alzheimer's disease and Parkinson’s disease pathways). Of these, Alzheimer's disease and Parkinson’s disease pathways expression as a whole were negatively associated with hsa-miR-29b-3p levels. Supporting our findings, down-regulation of hsa-miR-29b-3p has been previously shown in the peripheral blood mononuclear cells84, plasma85 and brain of subjects86 with AD whilst up-regulation of this miRNA has been detected in the cerebrospinal fluid of AD patients87. Positive correlations have also been reported between serum mir-29b levels and human cortical thickness and cortical glucose metabolism88, which both decrease in AD. Interestingly, pre-mir-29b has even been studied as a potential therapeutic agent targeted against AD89.
The expression of Regulation of acting cytoskeleton pathway was directly associated with the levels of hsa-miR-29b-3p. Dysregulation of gene sets governing actin cytoskeleton stability leading to synapse loss has been thought to be an early insult in AD90. This can be a response to insulin resistance in the brain, possibly being one of the many shared molecular, biochemical and mechanistic abnormalities in T2D and AD91. However, even if these two pathologies are known to act in concert, and simultaneously and diabetes is suggested to exacerbate AD pathology, the complex interactions are still not fully understood92.
We found a direct association between hsa-miR-29b-3p levels and visual processing and sustained attention (RVP-test), with the strongest associations being observed between total correct rejection and signal detection measure of sensitivity to the target (RVP A´). A significant impairment in this cognitive domain or its individual variables have been previously reported in subjects with AD93 or mild cognitive impairment94. Though the RVP-test primarily measures processing speed and sustained attention, its successful execution requires also selective attention and working memory i.e. two cognitive subdomains in which impairment has been well documented along with the progression of AD95. Importantly, our results are the first to connect hsa-miR-29b-3p to cognitive function in young and healthy adult population without clinically detectable signs of neurodegenerative diseases.
The strength of this study is the birth records and versatile phenotyping of the YFS. As the miRNA profiling has been performed as a cross-sectional study, we cannot infer causality or assess the dynamics of hsa-miR-29b-3p levels during subjects’ lifespans. Previous reports and our analysis show associations between miR-29b-3p levels and PTB38. However, as miR-29b levels have been shown to decrease with aging96, the differences in the pace of this decrease could also affect the levels in adulthood. Profiling miRNAs from blood poses a challenge for identifying the origin of the miRNAs as blood contains miRNAs from circulatory cells but also circulatory miRNAs originating from various tissues44. Whole blood was selected to enable gene-expression analysis from the same sample. Oral glucose tolerance tests had not been performed for the study population, hence we are unable to reflect upon the association between hsa-miR-29b-3p and impaired glucose tolerance. In addition, our analysis does not take account for all occupation-related or other lifestyle factors which affect the later risk of metabolic diseases and dementia or the effects of early life nurture on cognition. Larger replication cohorts are needed to verify the findings. Due to the age of the study populations, there was a low frequency of subjects with T2D and none with cognitive deficits and/or neurodegenerative diseases. Thus, associations between miRNA levels and clinical outcomes were not achievable. However, the observed associations with VLDL levels and cognitive function indicate a possible link to later morbidities.
Conclusions
Within this study we showed for the first time in humans that adulthood blood levels of hsa-miR-29b-3p were lower in subjects born preterm compared to subjects born full term, and that hsa-miR-29b-3p may be linked in the association between PTB and poor adulthood cognitive function and dyslipidemia. The association of this dysregulated miRNA with cognitive function and metabolic dysfunction could be mediated through its role in regulating gene sets related with neurodegenerative diseases and insulin signaling. We also pointed out that hsa-miR-29b-3p levels were associated with serum VLDL lipid and triglyceride levels, possibly reflecting impaired glucose tolerance which in theory could be related to prediabetes. Similarly, the association between this miRNA and visual processing and sustained attention is linked to the previous findings connecting miR-29b in the pathology of AD. In summary, our results reveal hsa-miR-29b-3p being associated with preterm birth and adulthood metabolic and cognitive health and open new perspectives for pre-clinical investigations.
Supplementary Information
Abbreviations
- AD
Alzheimer's disease
- AGA
Appropriate birth weight for gestational age
- BCL1A
BAF chromatin remodeling complex subunit BCL11A
- CANTAB
Cambridge Neuropsychological Test Automated Battery
- CS
Citrate synthase
- FC
Fold change
- FDR
False discovery rate
- FTB
Full term birth
- GSEA
Gene set enrichment analysis
- HbA1c
Glycated hemoglobin
- HDL
High density lipoprotein
- hsa
Homo Sapiens
- LDL
Low density lipoprotein
- miR
MicroRNA
- miRNA
MicroRNA
- mRNA
Messenger RNA
- NMR
Nuclear magnetic resonance
- PAL
Paired associates learning
- PTB
Preterm birth
- RTI
Reaction time
- RVP
Rapid visual information processing
- SD
Standard deviation
- SGA
Small birth weight for gestational age
- SWM
Spatial working memory
- T2D
Type 2 diabetes
- WHO
World Health Organization
- VLDL
Very low-density lipoprotein
- YFS
The Young Finns Study
Author contributions
S.M. and E.R. participated in the writing of the manuscript. S.R., M.J., M.W., N.O., M.A.-K., E.H., N.H.-K., O.R. and T.L. contributed to the discussion and edited the manuscript. P.P.M., I.L., L.-P.L. and E.R. performed statistical analysis. S.R., M.J., M.A.-K., N.H.-K., M.K., O.R., T.L. and E.R. participated in cohort collection/acquired data. N.O., O.R., T.L. and E.R. handled funding and supervision. All authors reviewed and approved the final manuscript.
Funding
The Young Finns Study has been financially supported by the Academy of Finland: Grants 286284 and 322098 (T.L.), 285902 and 330809 (E.R.), 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi); the Social Insurance Institution of Finland; the Kuopio, Tampere, and Turku University Hospital Medical Funds (Grant X51001 for T.L and 9X047 and 9S054 for E.R); the Juho Vainio Foundation; the Paavo Nurmi Foundation; the Finnish Foundation of Cardiovascular Research (T.L.); the Finnish Cultural Foundation; the Tampere Tuberculosis Foundation (T.L. and N.O.); the Emil Aaltonen Foundation (T.L. and N.O.); and the Yrjö Jahnsson Foundation (T.L., N.O. and E.R.); Signe and Ane Gyllenberg Foundation; Diabetes Research Foundation of Finnish Diabetes Association; EU Horizon 2020 (Grant 755320 for TAXINOMISIS; Grant 848146 for To_Aition); European Research Council (Grant 742927 for MULTIEPIGEN project). M. A-K. is also holding a research Grant from the Sigrid Juselius Foundation, Finland. This work was supported by the Foundation of Clinical Chemistry, Laboratoriolääketieteen edistämissäätiö sr., the Orion-Farmos Research Foundation and the Paulo Foundation.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available. Due to legal restrictions, the Ethics Committee of the Hospital District of Southwest Finland has in 2016 stated that individual level data cannot be stored in public repositories or otherwise made publicly available. However, data are available upon request from YFS Project Application. Data requests can be submitted by email and are subject to approval by the YFS Board.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88465-4.
References
- 1.Behrman RE, Butler AS. Preterm Birth: Causes, Consequences, and Prevention Causes. Washington, DC: National Academic Press; 2007. [PubMed] [Google Scholar]
- 2.Chernausek SD. Update: Consequences of abnormal fetal growth. J. Clin. Endocrinol. Metab. 2012;97:689–695. doi: 10.1210/jc.2011-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson S, Evans TA, Draper ES, Field DJ, Manktelow BN, Marlow N, et al. Neurodevelopmental outcomes following late and moderate prematurity: A population-based cohort study. Arch. Dis. Child. 2015;100(4):F301–F308. doi: 10.1136/archdischild-2014-307684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipkind HS, Slopen ME, Pfeiffer MR, McVeigh KH. School-age outcomes of late preterm infants in New York City. Am J Obstet Gynecol. 2012;206(3):222.e1–222.e6. doi: 10.1016/j.ajog.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Petrini JR, Dias T, McCormick MC, Massolo ML, Green NS, Escobar GJ. Increased risk of adverse neurological development for late preterm infants. J. Pediatr. 2009;154(2):169–176. doi: 10.1016/j.jpeds.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Heinonen K, Eriksson JG, Lahti J, Kajantie E, Pesonen AK, Tuovinen S, et al. Late preterm birth and neurocognitive performance in late adulthood: A birth cohort study. Pediatrics. 2015;135(4):e818–e825. doi: 10.1542/peds.2014-3556. [DOI] [PubMed] [Google Scholar]
- 7.Mosing MA, Lundholm C, Cnattingius S, Gatz M, Pedersen NL. Associations between birth characteristics and age-related cognitive impairment and dementia: A registry-based cohort study. PLoS Med. 2018;15:e1002609. doi: 10.1371/journal.pmed.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaakkola JJ, Ahmed P, Ieromnimon A, Goepfert P, Laiou E, Quansah R, et al. Preterm delivery and asthma: A systematic review and meta-analysis. J. Allergy Clin. Immunol. 2006;118(4):823–830. doi: 10.1016/j.jaci.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Zhang M, Tian H, Liu Z, Yin X, Xi B. Preterm birth and risk of type 1 and type 2 diabetes: Systematic review and meta-analysis. Obes. Rev. 2014;15(10):804–811. doi: 10.1111/obr.12214. [DOI] [PubMed] [Google Scholar]
- 10.Kerkhof GF, Breukhoven PE, Leunissen RW, Willemsen RH, Hokken-Koelega AC. Does preterm birth influence cardiovascular risk in early adulthood? J. Pediatr. 2012;161(3):390–396.e1. doi: 10.1016/j.jpeds.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 11.Juonala M, Cheung MM, Sabin MA, Burgner D, Skilton MR, Kahonen M, et al. Effect of birth weight on life-course blood pressure levels among children born premature: The Cardiovascular Risk in Young Finns Study. J. Hypertens. 2015;33(8):1542–1548. doi: 10.1097/HJH.0000000000000612. [DOI] [PubMed] [Google Scholar]
- 12.Skilton MR, Viikari JS, Juonala M, Laitinen T, Lehtimaki T, Taittonen L, et al. Fetal growth and preterm birth influence cardiovascular risk factors and arterial health in young adults: The Cardiovascular Risk in Young Finns Study. Arterioscler. Thromb. Vasc. Biol. 2011;31(12):2975–2981. doi: 10.1161/ATVBAHA.111.234757. [DOI] [PubMed] [Google Scholar]
- 13.Laas E, Lelong N, Thieulin AC, Houyel L, Bonnet D, Ancel PY, et al. Preterm birth and congenital heart defects: A population-based study. Pediatrics. 2012;130(4):e829–e837. doi: 10.1542/peds.2011-3279. [DOI] [PubMed] [Google Scholar]
- 14.Finken MJ, Inderson A, Van Montfoort N, Keijzer-Veen MG, van Weert AW, Carfil N, et al. Lipid profile and carotid intima-media thickness in a prospective cohort of very preterm subjects at age 19 years: Effects of early growth and current body composition. Pediatr. Res. 2006;59(4 Pt 1):604–609. doi: 10.1203/01.pdr.0000203096.13266.eb. [DOI] [PubMed] [Google Scholar]
- 15.Thomas EL, Parkinson JR, Hyde MJ, Yap IK, Holmes E, Dore CJ, et al. Aberrant adiposity and ectopic lipid deposition characterize the adult phenotype of the preterm infant. Pediatr. Res. 2011;70(5):507–512. doi: 10.1203/PDR.0b013e31822d7860. [DOI] [PubMed] [Google Scholar]
- 16.Ment LR, Vohr BR. Preterm birth and the developing brain. Lancet Neurol. 2008;7:378. doi: 10.1016/S1474-4422(08)70073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joss-Moore LA, Albertine KH, Lane RH. Epigenetics and the developmental origins of lung disease. Mol. Genet. Metab. 2011;104(1–2):61–66. doi: 10.1016/j.ymgme.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruickshank MN, Oshlack A, Theda C, Davis PG, Martino D, Sheehan P, et al. Analysis of epigenetic changes in survivors of preterm birth reveals the effect of gestational age and evidence for a long term legacy. Genome Med. 2013;5(10):96. doi: 10.1186/gm500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholas LM, Rattanatray L, MacLaughlin SM, Ozanne SE, Kleemann DO, Walker SK, et al. Differential effects of maternal obesity and weight loss in the periconceptional period on the epigenetic regulation of hepatic insulin-signaling pathways in the offspring. FASEB J. 2013;27(9):3786–3796. doi: 10.1096/fj.13-227918. [DOI] [PubMed] [Google Scholar]
- 21.Vickers MH. Early life nutrition, epigenetics and programming of later life disease. Nutrients. 2014;6(6):2165–2178. doi: 10.3390/nu6062165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvestri P, Di Russo C, Rigattieri S, Fedele S, Todaro D, Ferraiuolo G, et al. MicroRNAs and ischemic heart disease: Towards a better comprehension of pathogenesis, new diagnostic tools and new therapeutic targets. Recent Pat. Cardiovasc. Drug Discov. 2009;4(2):109–118. doi: 10.2174/157489009788452977. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Amodio N, Bellizzi D, Leotta M, Raimondi L, Biamonte L, D’Aquila P, et al. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle. 2013;12(23):3650–3662. doi: 10.4161/cc.26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.Larner-Svensson HM, Williams AE, Tsitsiou E, Perry MM, Jiang X, Chung KF, et al. Pharmacological studies of the mechanism and function of interleukin-1beta-induced miRNA-146a expression in primary human airway smooth muscle. Respir. Res. 2010;11:68. doi: 10.1186/1465-9921-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collares CV, Evangelista AF, Xavier DJ, Rassi DM, Arns T, Foss-Freitas MC, et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res. Notes. 2013;6:491. doi: 10.1186/1756-0500-6-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raitoharju E, Oksala N, Lehtimaki T. MicroRNAs in the atheroclerotic plaque. Clin. Chem. 2013;59:1708–1721. doi: 10.1373/clinchem.2013.204917. [DOI] [PubMed] [Google Scholar]
- 30.Winger EE, Reed JL, Ji X. First-trimester maternal cell microRNA is a superior pregnancy marker to immunological testing for predicting adverse pregnancy outcome. J. Reprod. Immunol. 2015;110:22–35. doi: 10.1016/j.jri.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Li P, Hao S, Liu L, Zhao J, Hou Y. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin. Chem. Lab. Med. 2009;47(8):923–929. doi: 10.1515/CCLM.2009.228. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MS, Casselman RC, Tayade C, Smith GN. Differential expression of plasma microRNA in preeclamptic patients at delivery and 1 year postpartum. Am. J. Obstet. Gynecol. 2015;213(3):367.e1–367.e9. doi: 10.1016/j.ajog.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Yao Y, Robinson AM, Zucchi FC, Robbins JC, Babenko O, Kovalchuk O, et al. Ancestral exposure to stress epigenetically programs preterm birth risk and adverse maternal and newborn outcomes. BMC Med. 2014;12:121–126. doi: 10.1186/s12916-014-0121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enquobahrie DA, Hensley M, Qiu C, Abetew DF, Hevner K, Tadesse MG, et al. Candidate gene and MicroRNA expression in fetal membranes and preterm delivery risk. Reprod. Sci. 2016;23:731–737. doi: 10.1177/1933719115612925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paquette AG, Shynlova O, Wu X, Kibschull M, Wang K, Price ND, et al. MicroRNA-transcriptome networks in whole blood and monocytes of women undergoing preterm labour. J. Cell. Mol. Med. 2019;23:6835–3845. doi: 10.1111/jcmm.14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Jaffe AE, Feinberg JI, Tryggvadottir R, Brown S, Montano C, et al. DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. Int. J. Epidemiol. 2012;41(1):188–199. doi: 10.1093/ije/dyr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Outcomes I of M (US) C on UPB and AH. No Title. (2007).
- 38.Pavlek LR, Vudatala S, Bartlet CW, Buhimschi IA, Buhimschi CS, Rogers LK. MiR-29b is associated with perinatal inflammation in extremely preterm infants. Pediatr. Res. 2020 doi: 10.1038/s41390-020-0943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durrani-Kolarik S, Pool CA, Gray A, Heyob KM, Cismowski MJ, Pryhuber G, et al. miR-29b supplementation decreases expression of matrix proteins and improves alveolarization in mice exposed to maternal inflammation and neonatal hyperoxia. Am. J. Physiol. 2017;313:339–349. doi: 10.1152/ajplung.00273.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hromadnikova I, Kotlabova K, Krofta L, Sirc J. Association analysis in children born from normal and complicated pregnancies—cardiovascular disease associated micrornas and the incidence of prehypertension/hypertension, overweight/obesity, valve problems and heart defects. Int. J. Mol. Sci. 2020;21:8413. doi: 10.3390/ijms21218413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gródecka-Szwajkiewicz D, Ulańczyk Z, Zagrodnik E, Łuczkowska K, Rogińska D, Kawa MP, et al. Differential secretion of angiopoietic factors and expression of microRNA in umbilical cord blood from healthy appropriate-for-gestational-age preterm and term newborns—in search of Biomarkers of Angiogenesis-Related Processes in Preterm Birth. Int. J. Mol. Sci. 2020;21:1305. doi: 10.3390/ijms21041305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inzaghi E, Kistner A, Germani D, Deodati A, Vanpee M, Legnevall L, et al. A prospective case-control study on miRNA circulating levels in subjects born small for gestational age (SGA) evaluated from childhood into young adulthood. PLoS ONE. 2020;15:e0228075. doi: 10.1371/journal.pone.0228075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raitakari OT, Juonala M, Ronnemaa T, Keltikangas-Jarvinen L, Rasanen L, Pietikainen M, et al. Cohort profile: The cardiovascular risk in Young Finns Study. Int. J. Epidemiol. 2008;37(6):1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 44.Raitoharju E, Seppala I, Lyytikainen LP, Viikari J, Ala-Korpela M, Soininen P, et al. Blood hsa-miR-122-5p and hsa-miR-885-5p levels associate with fatty liver and related lipoprotein metabolism-The Young Finns Study. Sci. Rep. 2016;6:38262. doi: 10.1038/srep38262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Organization WHO. Global Report on Diabetes. Geneva: WHO Organization; 2016. [Google Scholar]
- 46.Soininen P, Kangas AJ, Wurtz P, Tukiainen T, Tynkkynen T, Laatikainen R, et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134(9):1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 47.Soininen P, Kangas AJ, Wurtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ. Genet. 2015;8(1):192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- 48.Inouye M, Kettunen J, Soininen P, Silander K, Ripatti S, Kumpula LS, et al. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol. Syst. Biol. 2010;6:441. doi: 10.1038/msb.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rovio SP, Pahkala K, Nevalainen J, Juonala M, Salo P, Kahonen M, et al. Cognitive performance in young adulthood and midlife: Relations with age, sex, and education-the Cardiovascular risk in young Finns study. Neuropsychology. 2015;30:532. doi: 10.1037/neu0000239. [DOI] [PubMed] [Google Scholar]
- 50.Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, et al. MiRGator v3.0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41:D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS, et al. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ. Genet. 2012;5(2):242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, et al. Prenatal arsenic exposure and the epigenome: Altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 2014;55:196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 55.Amodio N, Rossi M, Raimondi L, Pitari MR, Botta C, Tagliaferri P, et al. miR-29s: A family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget. 2015;6:12837. doi: 10.18632/oncotarget.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugar SS, Heyob KM, Cheng X, Lee RJ, Rogers LK. Perinatal inflammation alters histone 3 and histone 4 methylation patterns: Effects of MiR-29b supplementation. Redox. Biol. 2021;38:101783. doi: 10.1016/j.redox.2020.101783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh S, Shyam S, Sah S, Singh MK, Palta P. Treatment of buffalo (Bubalus bubalis) somatic cell nuclear transfer embryos with MicroRNA-29b mimic improves their quality, reduces DNA methylation, and changes gene expression without affecting their developmental competence. Cell. Reprogram. 2019;21:210–219. doi: 10.1089/cell.2019.0007. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Zhou T, Wan J, Yang Y, Chen X, Wang J, et al. Comparative transcriptome analysis reveals a regulatory network of microRNA-29b during mouse early embryonic development. Oncotarget. 2016;7:53772. doi: 10.18632/oncotarget.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang YZ, Li JJH, Xiao HB, He Y, Zhang L, Yan YX. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: A systematic review and meta-analysis. J. Diabetes. 2018;12:633–644. doi: 10.1111/1753-0407.12643. [DOI] [PubMed] [Google Scholar]
- 60.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 61.Razny U, Polus A, Goralska J, Zdzienicka A, Gruca A, Kapusta M, et al. Effect of insulin resistance on whole blood mRNA and microRNA expression affecting bone turnover. Eur. J. Endocrinol. 2019;181:525–537. doi: 10.1530/EJE-19-0542. [DOI] [PubMed] [Google Scholar]
- 62.Nunez Lopez YO, Garufi G, Seyhan AA. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016;13(1):106–121. doi: 10.1039/C6MB00596A. [DOI] [PubMed] [Google Scholar]
- 63.Silva VA, Polesskaya A, Sousa TA, Correa VM, Andre ND, Reis RI, et al. Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol. Vis. 2011;17:2228–2240. [PMC free article] [PubMed] [Google Scholar]
- 64.Chen HY, Zhong X, Huang XR, Meng XM, You Y, Chung AC, et al. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 2014;22(4):842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chien HY, Chen CY, Chiu YH, Lin YC, Li WC. Differential microRNA profiles predict diabetic nephropathy progression in Taiwan. Int. J. Med. Sci. 2016;13:457. doi: 10.7150/ijms.15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 2007;21(11):2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 67.Liu J, Ye C, Liu W, Zhao W, Zhang YJ, Zhang H, et al. AICAR enhances insulin signaling via downregulation of miR-29. Can. J. Physiol. Pharmacol. 2015;94:199–205. doi: 10.1139/cjpp-2015-0159. [DOI] [PubMed] [Google Scholar]
- 68.Liang J, Liu C, Qiao A, Cui Y, Zhang H, Cui A, et al. MicroRNA-29a-c decrease fasting blood glucose levels by negatively regulating hepatic gluconeogenesis. J Hepatol. 2013;58(3):535–542. doi: 10.1016/j.jhep.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 69.Dooley J, Garcia-Perez JE, Sreenivasan J, Schlenner SM, Vangoitsenhoven R, Papadopoulou AS, et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes. 2016;65:53–61. doi: 10.2337/db15-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacovetti C, Rodriguez-Trejo A, Guay C, Sobel J, Gattesco S, Petrenko V, et al. MicroRNAs modulate core-clock gene expression in pancreatic islets during early postnatal life in rats. Diabetologia. 2017;60:2011–2020. doi: 10.1007/s00125-017-4348-6. [DOI] [PubMed] [Google Scholar]
- 71.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keaton JM, Cooke Bailey JN, Palmer ND, Freedman BI, Langefeld CD, Ng MC, et al. A comparison of type 2 diabetes risk allele load between African Americans and European Americans. Hum Genet. 2014;133(12):1487–1495. doi: 10.1007/s00439-014-1486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jonsson A, Ladenvall C, Ahluwalia TS, Kravic J, Krus U, Taneera J, et al. Effects of common genetic variants associated with type 2 diabetes and glycemic traits on alpha- and beta-cell function and insulin action in humans. Diabetes. 2013;62(8):2978–2983. doi: 10.2337/db12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simonis-Bik AM, Nijpels G, van Haeften TW, Houwing-Duistermaat JJ, Boomsma DI, Reiling E, et al. Gene variants in the novel type 2 diabetes loci CDC123/CAMK1D, THADA, ADAMTS9, BCL11A, and MTNR1B affect different aspects of pancreatic beta-cell function. Diabetes. 2010;59(1):293–301. doi: 10.2337/db09-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asmann YW, Stump CS, Short KR, Coenen-Schimke JM, Guo Z, Bigelow ML, et al. Skeletal muscle mitochondrial functions, mitochondrial DNA copy numbers, and gene transcript profiles in type 2 diabetic and nondiabetic subjects at equal levels of low or high insulin and euglycemia. Diabetes. 2006;55(12):3309–3319. doi: 10.2337/db05-1230. [DOI] [PubMed] [Google Scholar]
- 76.Ortenblad N, Mogensen M, Petersen I, Hojlund K, Levin K, Sahlin K, et al. Reduced insulin-mediated citrate synthase activity in cultured skeletal muscle cells from patients with type 2 diabetes: Evidence for an intrinsic oxidative enzyme defect. Biochim. Biophys. Acta. 2005;1741(1–2):206–214. doi: 10.1016/j.bbadis.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Ginsberg HN. Insulin resistance and cardiovascular disease. J. Clin. Investig. 2000;106:453–458. doi: 10.1172/JCI10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malmström R, Packard CJ, Caslake M, Bedford D, Stewart P, Yki-Järvinen H, et al. Defective regulation of triglyceride metabolism by insulin in the liver in NIDDM. Diabetologia. 1997;40:454–464. doi: 10.1007/s001250050700. [DOI] [PubMed] [Google Scholar]
- 79.Adiels M, Borén J, Caslake MJ, Stewart P, Soro A, Westerbacka J, et al. Overproduction of VLDL1 driven by hyperglycemia is a dominant feature of diabetic dyslipidemia. Arterioscler. Thromb. Vasc. Biol. 2005;25:1697–1703. doi: 10.1161/01.ATV.0000172689.53992.25. [DOI] [PubMed] [Google Scholar]
- 80.Tirosh A, Shai I, Bitzur R, Kochba I, Tekes-Manova D, Israeli E, et al. Changes in triglyceride levels over time and risk of type 2 diabetes in young men. Diabetes Care. 2008;31:2032–2037. doi: 10.2337/dc08-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurtz CL, Fannin EE, Toth CL, Pearson DS, Vickers KC, Sethupathy P. Inhibition of miR-29 has a significant lipid-lowering benefit through suppression of lipogenic programs in liver. Sci. Rep. 2015;5:1–13. doi: 10.1038/srep12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hung YH, Kanke M, Kurtz CL, Cubitt RL, Bunaciu RP, Zhou L, et al. MiR-29 regulates de novo lipogenesis in the liver and circulating triglyceride Levels in a Sirt1-dependent manner. Front. Physiol. 2019;10:1367. doi: 10.3389/fphys.2019.01367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Y, Pan Q, Sun B, Yang R, Fang X, Liu X, et al. MiR-29b targets LPL and TDG genes and regulates apoptosis and triglyceride production in MECs. DNA Cell Biol. 2016;35:758–765. doi: 10.1089/dna.2016.3443. [DOI] [PubMed] [Google Scholar]
- 84.Villa C, Ridolfi E, Fenoglio C, Ghezzi L, Vimercati R, Clerici F, et al. Expression of the transcription factor Sp1 and its regulatory hsa-miR-29b in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J. Alzheimers Dis. 2013;35(3):487–494. doi: 10.3233/JAD-122263. [DOI] [PubMed] [Google Scholar]
- 85.Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, et al. Plasma exosomal miRNAs in persons with and without Alzheimer disease: Altered expression and prospects for biomarkers. PLoS ONE. 2015 doi: 10.1371/journal.pone.0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. U S A. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiko T, Nakagawa K, Tsuduki T, Furukawa K, Arai H, Miyazawa T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 2014;39(2):253–259. doi: 10.3233/JAD-130932. [DOI] [PubMed] [Google Scholar]
- 88.Maldonado-Lasuncion I, Atienza M, Sanchez-Espinosa MP, Cantero JL. Aging-related changes in cognition and cortical integrity are associated with serum expression of candidate MicroRNAs for Alzheimer disease. Cereb Cortex. 2019;29:4426–4437. doi: 10.1093/cercor/bhy323. [DOI] [PubMed] [Google Scholar]
- 89.Pereira PA, Tomás JF, Queiroz JA, Figueiras AR, Sousa F. Recombinant pre-miR-29b for Alzheimer’s disease therapeutics. Sci. Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Penzes P, Vanleeuwen JE. Impaired regulation of synaptic actin cytoskeleton in Alzheimer’s disease. Brain Res. Rev. 2011;67(1–2):184–192. doi: 10.1016/j.brainresrev.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moroz N, Tong M, Longato L, Xu H, de la Monte SM. Limited Alzheimer-type neurodegeneration in experimental obesity and type 2 diabetes mellitus. J. Alzheimers Dis. 2008;15(1):29–44. doi: 10.3233/JAD-2008-15103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sims-Robinson C, Kim B, Rosko A, Feldman EL. How does diabetes accelerate Alzheimer disease pathology? Nat. Rev. Neurol. 2010;6:551. doi: 10.1038/nrneurol.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matos Gonçalves M, Pinho MS, Simões MR. Construct and concurrent validity of the Cambridge neuropsychological automated tests in Portuguese older adults without neuropsychiatric diagnoses and with Alzheimer’s disease dementia. Aging. Neuropsychol Cogn. 2018;25:290–317. doi: 10.1080/13825585.2017.1294651. [DOI] [PubMed] [Google Scholar]
- 94.Égerházi A, Berecz R, Bartók E, Degrell I. Automated Neuropsychological Test Battery (CANTAB) in mild cognitive impairment and in Alzheimer’s disease. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2007;31:746–751. doi: 10.1016/j.pnpbp.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 95.Rizzo M, Anderson SW, Dawson J, Myers R, Ball K. Visual attention impairments in Alzheimer’s disease. Neurology. 2000;54:1954–1959. doi: 10.1212/WNL.54.10.1954. [DOI] [PubMed] [Google Scholar]
- 96.Zhang H, Yang H, Zhang C, Jing Y, Wang C, Liu C, et al. Investigation of microRNA expression in human serum during the aging process. J. Gerontol. A. 2015;70(1):102–109. doi: 10.1093/gerona/glu145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available. Due to legal restrictions, the Ethics Committee of the Hospital District of Southwest Finland has in 2016 stated that individual level data cannot be stored in public repositories or otherwise made publicly available. However, data are available upon request from YFS Project Application. Data requests can be submitted by email and are subject to approval by the YFS Board.