Abstract
Harmful algal blooms (HABs) are increasing in frequency, magnitude, and duration around the world. Prymnesium parvum is a HAB species known to cause massive fish kills, but the toxin(s) it produces contributing to this acute toxicity has not been confirmed. In the present study, a 2 X 2 factorial design was employed to examine influences of salinity (2.4 or 5 ppt) and nutrient limitation (f/2 or f/8) on P. parvum acute toxicity to fish and produced molecules. Acute toxicity (LC50) of these cultures, following a 48-hour mortality assay, ranged from 10,213 to 96,816 cells mL−1. Non-targeted analysis was performed using liquid chromatography high-resolution mass spectrometry (LC-HRMS) to investigate compounds contributing to the differential toxicological responses. When P. parvum elicited toxicity to fish, suspect screening confirmed the presence of several prymnesins, and the peak area of PRM-A (3 Cl; prymnesin2aglycone) was significantly (p < 0.05) and positively related to acute toxicity. In addition, a non-targeted approach to highlighting peaks that differ between two chemical fingerprints was developed, termed a relative difference plot, and used to search for peaks co-varying with P. parvum acute toxicity. Several peaks were highlighted along with the prymnesins identified through suspect screening when acute toxicity to fish was observed.
Keywords: harmful algal blooms, prymnesin, relative difference, differential analysis
Graphical Abstract
1. Introduction
Harmful algal blooms (HABs) are an increasing threat to global water supplies, affecting local economies, public health, and aquatic ecosystems. HAB occurrence has routinely been examined in coastal areas and estuaries, but HAB incidence in freshwater bodies continues to increase in frequency, duration, and magnitude (Preece et al., 2017). Increased watershed development (causing eutrophication) and atmospheric temperatures (among other factors) have provided the necessary environments for inland HABs to proliferate, producing and releasing secondary metabolites, some of which are toxins (Paerl and Huisman, 2009). Human exposure to these toxins occurs through recreation (dermal absorption via swimming), ingestion (drinking/eating contaminated water or seafood), and inhalation (through aerosols). HAB toxins cause health problems ranging from rashes and gastrointestinal distress to death in humans and other animals (Carmichael, 2001). To minimize human exposure, monitoring programs (if they exist) routinely measure total phytoplankton biomass or species composition which oftentimes is not correlated to toxin(s) production. Implementation of more advanced surveillance systems that examine toxins is needed to support management decisions (Brooks et al., 2017, 2016).
Prymnesium parvum, a unicellular haptophyte that causes fish kills around the world, was first documented in the United States in 1985 when a massive fish kill occurred at the Pecos River in Texas (James and De La Cruz, 1989). However, it was not until 2001 that P. parvum gained widespread attention when another large-scale fish kill in Texas caused significant economic losses (Southard et al., 2010). Four compound classes (prymnesins (Igarashi et al., 1999, 1996; Rasmussen et al., 2016), fatty acids (Henrikson et al., 2010), GATs (Henrikson et al., 2010), and fatty acid amides (Bertin et al., 2012a, 2012b)) have been isolated from P. parvum strains showing some level of toxicity; however, recent research indicates fatty acids, fatty acid amides, and GATs are not responsible for observed toxicity to aquatic life (Blossom et al., 2014). This suggests prymnesins and prymnesin-type molecules produced by P. parvum are likely contributing to fish kills resulting from P. parvum blooms. However, only recently have prymnesins been isolated from a toxic strain for the first time since 1995 (Manning and La Claire, 2013). This could be due to nonstandard growth procedures, inadequate extraction, insufficient analytical methodologies, lack of analytical standards, or limited prymnesin production when other compound classes were detected (Roelke et al., 2015). Historical studies of P. parvum toxicity have routinely employed non-standardized assays, which limit robust comparative studies among laboratories, species and model systems (Brooks et al., 2011,2010). Further, different strains of P. parvum have been reported to exert differential levels of aquatic toxicity (Blossom et al., 2014). Most importantly, the major molecule(s) contributing to fish toxicity during P. parvum HAB events is (are) yet to be confirmed.
The identification of novel toxins is difficult. Despite improvements to instruments and software, the most common approach to identifying toxins within a complex mixture still utilizes a bottom-up method where entire mixtures are analyzed, and an attempt is made to identify each component. Alternatively, response-directed approaches with bioassays are used to identify toxic fractions of a sample, and then specific fractions are selected for subsequent analysis and identification (Blossom et al., 2014). Both options are valuable, but can be resource intensive, which suggests a top-down approach to examining differences in toxic samples could be advantageous over the current methodology.
A top-down approach in high-resolution mass spectrometry (HRMS) has recently been made possible due to new advancements including soft ionization sources that allow detection of the molecular ion, high-resolution mass spectrometers that can be coupled to high-performance liquid chromatography (HPLC) to monitor a wide mass-to-charge (m/z) range with high resolution and mass accuracy (i.e. FTICR and Orbitrap), and the development of readily available mass spectral databases consisting of thousands of compounds (Hollender et al., 2017). Analysis is possible with a number of software packages that can be tailored to the analytes of interest for non-targeted methods, such as proteins and metabolites in biological samples (Chen et al., 2018; Yi et al., 2016) or emerging contaminants in environmental samples (Bader et al., 2017; Fu et al., 2018; Nürenberg et al., 2015). In the present study, we (1) examined the extent to which specific nutrient and salinity conditions influenced aquatic toxicity associated with P. parvum; (2) investigated whether prymnesins were differentially produced among these environmental conditions; and (3) determined if any relationships existed between prymnesin and acute toxicity to fish. We also employed non-target HPLC-HRMS analysis to develop a novel top-down approach for visualizing differences in chemical composition of complex mixtures.
2. Methods
2.1. Chemicals and Reagents
The internal standard clarithromycin-N-methyl-d3 (CLA-d3) was purchased from Toronto Research Chemicals (North York, Ontario, Canada). For heat map visualization studies (described below), we selected known algal toxins for study and acquired nodularin (NOD) and microcystin (MC-) LA, LR, LY, LF, YR, and RR from Abraxis (Warminster, PA, USA). Ammonium formate was purchased from Sigma Aldrich (St. Louis, MO, USA). All chemicals were used as received. Formic acid and LCMS-grade acetonitrile (ACN) were obtained from Fisher Scientific (Fair Lawn, NJ, USA). A Thermo Scientific Barnstead™ Nanopure™ (Marietta, OH, USA) water purification system was used to deliver 18Ω nanopure water for all samples and HPLC solvent preparation.
2.2. Prymnesium parvum experiment
A 2 x 2 factorial design with duplication was applied with the Texas strain (UTEX 2797) of P. parvum to examine nutrient sufficient (f/2) and deficient (f/8) conditions at two salinity levels (2.4 and 5 ppt). Temperature was maintained at 15 °C with a 12:12 light:dark cycle (Franco et al., 2019; Hill et al., n.d.). Once late stationary growth was observed (day 60 or 67), an aliquot was taken for acute toxicity bioassays. The remaining volume for each experimental unit was frozen at −80 °C, thawed, and then 500 mL was filtered with a 0.45 μm filter. The analyzed sample should contain, in theory, both intra and extracellular toxins. Because this was an initial exploration of P. parvum toxins coupled with acute mortality, a freeze/thaw cycle of both components was used to capture as much as possible and not rule anything out. Ten mL of cell-free filtrate was concentrated 10-fold by drying under N2 gas and reconstituted in 1 mL of 95:5 H2O:ACN containing 0.1% formic acid for HPLC-HRMS analysis. An additional bioassay experiment was conducted with the freeze/thawed units to mimic the samples analyzed with HPLC-HRMS and ensure the relative toxicity levels for the experimental units remained the same.
Acute toxicity of each P. parvum experimental unit was identified using a 48-hour fish mortality bioassay with larval fathead minnows (Pimephales promelas), following standard methods previously reported from our laboratory and approved by the Institutional Animal Care and Use Committee at Baylor University (Hill et al., n.d.; Prosser et al., 2012; Theodore W. Valenti et al., 2010). Briefly, five dilutions of each culture (100, 50, 25, 12.5, and 6.25%) were prepared using corresponding media conditions, which also served as controls (e.g., media without algae). These dilutions were made using unfiltered media from experimental units containing in-tact cells with pH titrated to 8.5 using 1.0 N hydrochloric acid or 1.0 N sodium hydroxide, because increasing pH increases acute toxicity (Valenti et al., 2010). A second bioassay (mentioned above) was completed three months later with the same cultures that had been frozen at −80 °C, thawed and filtered. The experimental unit dilution to cause 50% organism lethality (LC50) were estimated using the EPA Toxicity Relationship Analysis Program, which fits a sigmoidal curve to the data set. Nominal cell concentrations were calculated by multiplying the LC50 dilution by the 100% culture cell density. All sample preparation and toxicity analysis were conducted in the dark as substances produced by P. parvum that elicit acute toxicity to fish are known to degrade when exposed to light (James et al., 2011).
2.3. HPLC-HRMS Instrumentation and Method
2.3.1. Instrumentation
A Dionex UltiMate 3000 RS LC system coupled to a Q Exactive Focus (Thermo Scientific, Waltham, MA, USA) was utilized for HPLC-HRMS analysis. Chromatographic separation was achieved with a 2.7 μm, 2.1 x 100 mm Agilent Poroshell 120 SB-C18 column equipped with a 2.7 μm, 2.1 x 5 mm Poroshell 120 SB-C18 guard column (Santa Clara, CA, USA). An injection volume of 10 μL was used for all samples, and the solvent systems were delivered at a flow rate of 0.5 mL/min. Eluent was diverted to waste for the first minute of every run to avoid the introduction of salts and highly polar compounds into the mass spectrometer.
The Q Exactive Focus was equipped with a heated electrospray ionization source (HESI) using the following MS parameters: spray voltage of 4000 V, capillary temperature at 350 °C, sheath gas pressure 40 (arbitrary units), auxiliary gas pressure 20 (arbitrary units), probe heater temperature 30 °C, and the S-lens RF set to 50. The spectrometer was operated in full-scan positive ionization mode scanning 200-1200 m/z with FWHM resolution set at 70,000 and AGC at 1 x 106. All spectra were collected in centroid mode to minimize the size of data files. This was necessary to create relative difference plots on a standard computer operating system.
2.3.2. Liquid Chromatography Separation
For P. parvum samples, a gradient elution program was used with (A) H2O and (B) ACN both containing 0.1% formic acid to promote ionization. The gradient began at 5% B (held for 1 min) and was increased to 100% B over 29 minutes (held for 2 min) before returning to 5% B and equilibrating for 5 minutes (total run time of 37.5 min). Column temperature was held at 35 °C. Microcystin spiked samples for workflow optimization were analyzed using a previously published separation method (Haddad et al., 2019). Briefly, a gradient elution program was used with (A) 95:5 H2O:ACN and (B) ACN both containing 5 mM ammonium formate and 3.6 mM formic acid. The gradient program began at 30% B, increased to 50% B over 5 minutes, increased to 100% B over 0.1 min (held for 1 min) before equilibrating at 30% B for 4 minutes (a total run time of 10 min). Column temperature was held at 60 °C.
2.4. Peak List Generation
Compound Discoverer 3.0 (Thermo Scientific, Waltham, MA, USA) was used to generate peak lists for the algal HPLC-HRMS data files. An automatic workflow was utilized with the following settings. Any peaks below a 1.5 S/N threshold were discarded for a given file, and no total ion intensity minimum was selected. Blank solvent runs were used to identify background peaks. If the peak area ratio was less than 5:1 sample/blank, the peak was removed. Retention time alignment between injections was performed using the adaptive curve function with a mass tolerance of 5 ppm and maximum shift time of 2 minutes. The following parameters were chosen for extracted ion chromatogram creation: minimum peak intensity of 50,000, mass tolerance of 10 ppm, and S/N threshold of 2. Further grouping of detected compounds between samples was performed when retention times and masses were within 0.1 minutes and 5 ppm of each other, respectively. Linear regression analysis between peak areas and LC50 values was performed in SigmaPlot 14.0.
2.5. Sample Preparation for Method of Visualizing Differences in a MS Heatmap
We developed an LCMS heatmap for visualizing algal toxins, but because toxins responsible for P. parvum toxicity remain under study, we selected known cyanobacterial toxins (nodularin, microcystins) for this exercise. These cyanotoxins samples were prepared from 1 mg/L stock solutions of each standard and diluted to final concentrations of 1, 10, and 100 μg/L in 95:5 H2O:ACN containing 5 mM ammonium formate and 3.6 mM formic acid. Two “unknown” samples were also prepared with concentrations of each cyanotoxin varying between the two solutions. “Unknown one” and “unknown two” had the following concentrations of MC-LA, MC-LF, MC-LR, MC-LY, MC-YR, MC-RR, NOD, and CLA-d3: 100, 10, 100, 50, 100, 50, 100, 10 μg/L and 50, 100, 10, 100, 10, 100, 10, 10 μg/L, respectively. Two additional “unknowns” (unknown three and unknown four) were prepared in filtered water taken from Lake Waco to mimic real matrix conditions. They had the following concentrations of MC-LR, MC-LY, and CLA-d3: 66.7, 6.67, 6.67 μg/L and 33.3, 66.7, 6.67 μg/L, respectively. Between 2 and 20 μL of spiked standards were diluted to a final volume of 300 μL using lake water in order to provide the above concentrations and prevent unwanted dilution of the environmental matrix.
2.6. HPLC-HRMS Heatmap and Relative Difference Plot Workflow
Raw data files were converted to netCDFs using Xcalibur’s File Converter (Thermo Scientific). All netCDFs were automatically loaded into MATLAB R2017a (The MathWorks, Inc., Natick, MA, USA) using a loop function within the Bioinformatics package that separates each file into an intensity matrix and respective retention time and m/z vectors. MATLAB automatically uses zeros to fill columns where a m/z value is not detected for a given scan. When a zero occurs in a reference scan, it can cause the relative difference in Equation (1) to return a value of infinity. To account for this, the standard deviation of the intensity matrices for all blanks was used as the background threshold to remove all zeros from sample matrices (i.e., all intensity values less than or equal to the standard deviation of the blanks was set equal to the standard deviation of the blanks). The standard deviation of blanks varied from day to day but was typically between 1000 and 3000 arbitrary units. This is a reasonable noise threshold and below a peak intensity that would be considered relevant (below the limit of detection of 4200 cps calculated from 3× standard deviation of the blanks).
After this initial normalization, each file was subjected to the same processing scheme adapted from the MATLAB Bioinformatics package in the following order: alignment to a common m/z grid, resampling to a fixed number of m/z values, Gaussian smoothing along the retention time axis, LOWESS smoothing in both time and m/z axis, and time alignment to a reference ion map using the Correlation Optimized Warping (COW) algorithm (Tomasi et al., 2004). Intensity matrices for each triplicate measurement were then averaged. This workflow provided intensity matrices for each sample that were aligned to a single retention time and m/z vector. The script used for this procedure is provided in supplemental information (Figure S1), and alignment scripts are freely available at http://www.models.life.ku.dk/dtw_cow. A detailed explanation and examples of workflow optimization on spiked samples are also provided in supplemental information (Figures S3–S9).
To create a relative difference plot, a relative difference intensity matrix (ZRD) is created between two averaged intensity matrices using Equation 1:
| (1) |
where intensity matrix Zsample is expected to have the chemicals of interest at higher concentrations (e.g., toxic) than intensity matrix Zref (e.g., non-toxic). Relative difference plots were graphed using the retention time and m/z vector of the reference sample.
3. Results and Discussion
3.1. Prymnesium Parvum Toxicity
To examine whether prymnesins are associated with fish toxicity commonly associated with P. parvum blooms, P. parvum were grown under a 2 X 2 factorial design varying salinity and nutrient conditions. These conditions are known to promote a magnitude gradient of acute toxicity based on our previous work showing non-optimal conditions for growth increase acute toxicity per cell (Baker et al., 2009, 2007; Brooks et al., 2010; James et al., 2011; Theodore W Valenti et al., 2010). LC50 values calculated from the 48-hour acute mortality bioassay on larval fathead minnows are shown in Table 1. All duplicate cultures had essentially the same LC50 values except for the low salinity (2.4 ppt) x nutrient limited (f/8) condition. When bioassays were completed immediately after the growth study with in-tact cells, replicate #2 of the low salinity (2.4 ppt) x nutrient limited (f/8) treatment combination indicated that replicate #2 was acutely toxic with an LC50 value similar to the higher salinity (5 ppt) treatment. To investigate this observation, a second bioassay experiment was performed several months later using the frozen, filtered cultures. A general decrease in the acute toxicity of all algal cultures during the second bioassay experiment was observed; therefore, relative toxicity was compared within a given bioassay. The duplicate low salinity (2.4 ppt) x nutrient limited (f/8) treatment demonstrated similar toxicity (non-toxic) for the second bioassay. The reason for the differences between toxicity measurements in the first bioassay of low salinity (2.4 ppt) x nutrient limited (f/8) replicates is unknown; however, it is likely that the fish mortality difference was influenced by several factors. The overall decrease in toxicity for the second bioassay is potentially due to lack of active cells excreting toxins in this second study, degradation of toxins while stored frozen for several months, and/or brief exposures to light while preparing solutions.
Table 1.
Toxicity results (LC50) for 48-hour acute mortality bioassays on larval fathead minnow conducted with duplicate (1 or 2) P. parvum cultures are shown.a
| 2.4 ppt x f/2 | 2.4 ppt x f/8 | 5 ppt x f/2 | 5 ppt x f/8 | |||||
|---|---|---|---|---|---|---|---|---|
| (1) | (2) | (1) | (2) | (1) | (2) | (1) | (2) | |
| First Bioassayb | NT | NT | 96,816 (67,899-125,735) | 30,661 (20,240-41,083) | 32,438 (NA) | 32,174 (18,582-45,767) | 13,029 (NA) | 10,213 (6,785-27,211) |
| Second Bioassayc | NT | NT | NT | NT | 118,986 (91,944-146,034) | 208,195 (155,033-261,350) | 182,616 (87,903-278,510) | 136,335 (107,535-165,139) |
LC50 values (cells mL−1) are estimated using a sigmoidal curve where values estimated above the 100% culture concentration are denoted as non-toxic (NT), and 95% confidence intervals are shown in parentheses.
Bioassay conducted with live cells.
Bioassay conducted with frozen, thawed and filtered culture.
During both toxicity bioassays, higher salinity (5 ppt) conditions exhibited greater fathead minnow mortality than the low salinity (2.4 ppt) treatment, regardless of nutrient treatment level, suggesting a higher salinity environment promoted increased toxins production in the current study. In fact, P. parvum reached higher cell densities in these elevated salinity conditions (Franco et al., 2019; Hill et al., n.d.). However, LC50 values could not be estimated in the low salinity (2.4 ppt) x nutrient sufficient (f/2) experimental units and are denoted as non-toxic hereafter, along with both replicates of the low salinity (2.4 ppt) x nutrient deficient (f/8) treatment combination in the second bioassay. These differences may have resulted from differences in toxin concentration and/or bioavailability of toxins among experimental conditions. Since the salinity was optimized using Instant Ocean®, a mixture containing mostly calcium and magnesium ions shown to increase the potency of toxic P. parvum (Ulitzur and Shilo, 1963), it is possible that the ion concentrations led to differences in toxicity. Ulitzer et al. (1963) showed toxicity to minnows increased with increasing Ca2+ concentration to 200 mg/L. The concentrations of Ca2+ and Mg2+ in this study were approximately 57 and 189 mg L−1 or 27 and 91 mg L−1 for the 5 ppt and 2.4 ppt conditions, respectively. We specifically investigated the hypothesis that acute toxicity differences were associated by differential toxin production.
3.2. Prymnesin analysis
The same experimental units used for acute mortality bioassays were frozen, thawed, filtered and preconcentrated for HPLC-HRMS analysis such that each chemical mixture analyzed by mass spectrometry (MS) was the same mixture used in a corresponding acute bioassay. A suspect screening approach was used to search for known prymnesin type molecules in the chromatograms of each sample. Using masses reported elsewhere for known prymnesin and aglycone (loss of all sugar moieties) molecules, a database was created to search the peak lists generated by Compound Discoverer (Table S1). It is important to note that several compounds have the same molecular weight in the database. For example, prymnesin1 minus a hexose and pentose has the same mass as prymnesin2; therefore, these two compounds cannot be differentiated using MS unless they are resolved with chromatography.
After searching the prymnesin database, Compound Discoverer identified three prymnesin-type molecules. The most prominent prymnesin peak in all 8 cultures was prymnesin1-hexose-pentose-glycone (or prymnesin2-aglycone), which has an m/z of 918.8839 and elution time of 17.1 minutes (red curve in Figure 1). Prymnesin2 (or prymnesin1-hexose-pentose) and a recently reported A-type prymnesinaglycone (one less chlorine atom compared to Prymnesin1 or -2) are also present in replicate #1 of the higher salinity (5ppt) x nutrient deficient (f/8) treatment combination (black and teal curves in Figure 1). These prymnesin analogues were recently identified by another group analyzing the same UTEX 2797 P. parvum strain (Binzer et al., 2019). A systematic naming system was recently proposed (Binzer et al., 2019) and will be used for the remainder of this paper; the 91-carbon backbone prymnesin is abbreviated as PRM-A with the presence of pentose and hexose conjugates denoted with plus signs and the number of chlorines and extra double bonds added where necessary (e.g., PRM-A (3 Cl) + pentose corresponds to prymnesin-2). As shown in Figure 1, several other masses with isotope patterns characteristic of a prymnesin appear within a two-minute elution period around the 918.8839 m/z peak. Extracted ion chromatograms of these m/z values show peaks at two distinct elution times that are separated by 12 seconds (Figure 1).
Figure 1.
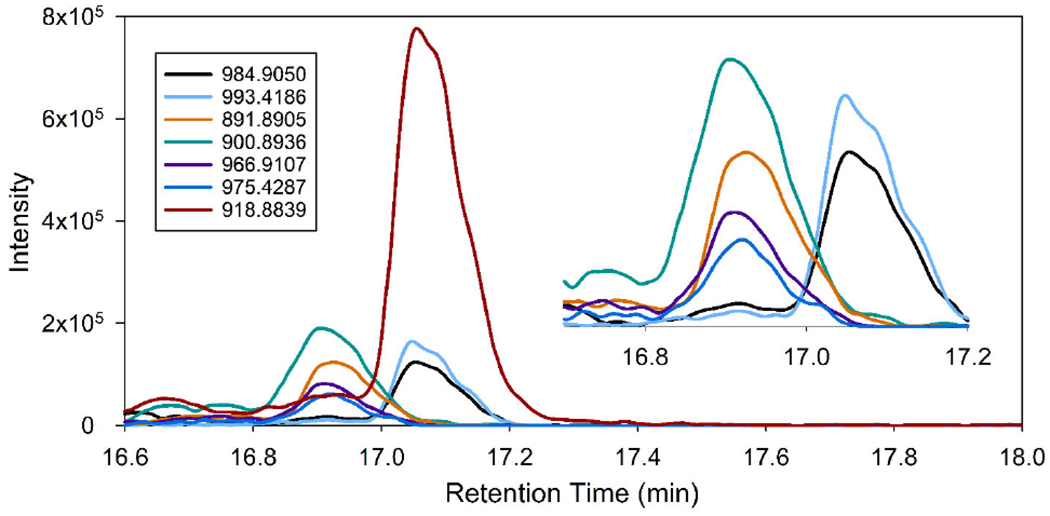
Suspect screening identified three prymnesin type molecules (red, black, and teal traces), and their extracted chromatograms are shown for the 5ppt f/8 replicate one culture. These were tentatively identified as prymnesin (1 or 2) aglycone, prymnesin-2, and prymnesin a-type (-Cl) aglycone. Manual screening found additional prymnesin-type molecules and/or adducts that suggest additional prymnesin analogues are present and elute in two distinct regions (magnification shown in inset).
A cumulative mass spectrum over the two-minute elution period shows at least ten potential prymnesins. Of these masses, four are tentatively assigned to previously published prymnesin-type congeners and adducts (Figure 2) (Binzer et al., 2019; Manning and La Claire, 2013). The congeners are all A-type prymnesins with varying numbers of double bonds and chlorine and pentose attachments. A comparison of an experimental mass spectrum against a simulated mass spectrum for PRM-A (3 Cl) is shown in Figure S2. Isotopic peak ratios and masses agree within 1 ppm of expected values, increasing confidence that prymnesins are indeed being produced by these cultures. Prymnesin standards are not available, so further confirmation and quantitation are not possible.
Figure 2.
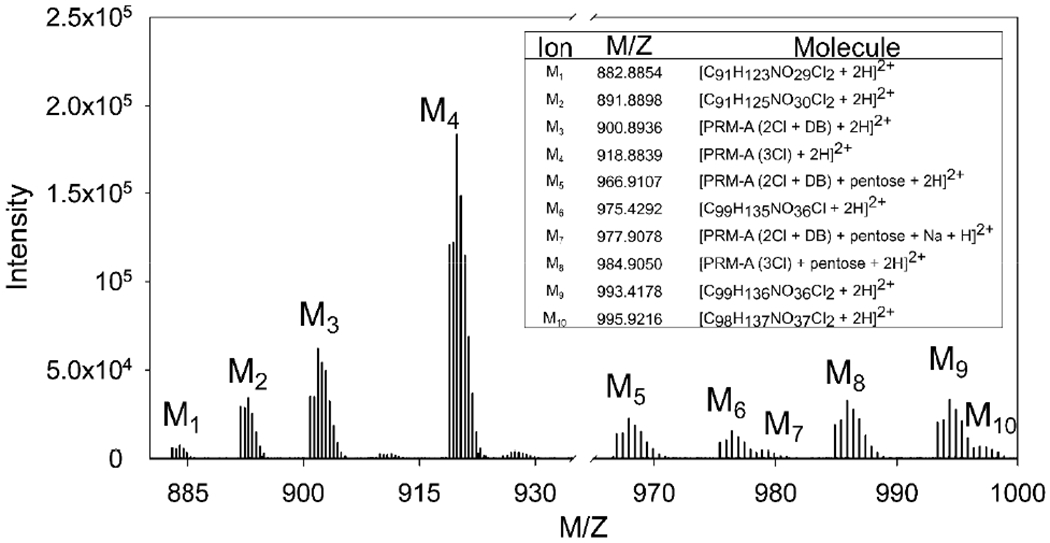
An extracted mass chromatogram of the prymnesin elution period showed several additional masses with isotope patterns characteristic of prymnesins corresponding to other adduct types or prymnesin types not detected by the software. Four prymnesin types were identified that have been previously reported in literature. Potential chemical formulas with the proton adducts (< 1 ppm mass error) are shown for masses with isotopic patterns similar to the prymnesins, but not previously reported.
There are five isotopic patterns in the mass spectrum that are indicative of prymnesins, but the molecular weight does not match those previously identified within a 2 ppm window. A chemical formula was generated for those molecular weights, assuming a doubly protonated ion. No more than five chemical formulas agreed with the masses within 2 ppm mass error using the following atom criteria: C = 85 – 110, H = 120 – 150, O = 28 – 45, N = 1, and Cl = 1 – 3. Predicted formulas were narrowed down by selecting those with degrees of unsaturation less than 32, which is typical of prymnesins. The potential chemical formulas for the unidentified masses are presented in Figure 2. In total, four identified prymnesins (PRM-A (2 Cl +DB), PRM-A (3 Cl), PRM-A (2 Cl + DB) + pentose, PRM-A (3 Cl) + pentose) and five unidentified potential analogues are present in the cultures.
3.3. Relationship between Acute Toxicity and Prymnesins Concentration
The peak area of 918.8839 m/z was used to investigate the relationship between concentration of prymnesins and identified acute toxicity since it was present in all cultures and likely includes contributions from congeners 1 and 2. If prymnesins’ concentration was potentially responsible for the measured toxicity, an inverse relationship between peak area and the LC50 value was expected. Indeed, a general trend between the concentration of prymnesins and LC50 values was observed (Figure 3A). Comparing replicate #1 toxicity measurements (bars) with prymnesins concentration, a smaller peak area (lower concentration of PRM-A (3 Cl)) corresponded to a larger LC50 value or less toxic culture. Figures 3B, C show the percent mortality at 50% and 25% culture dilutions against PRM-A (3 Cl) peak area. A significant (p ≤ 0.05) positive relationship between the concentration and fish mortality was present.
Figure 3.
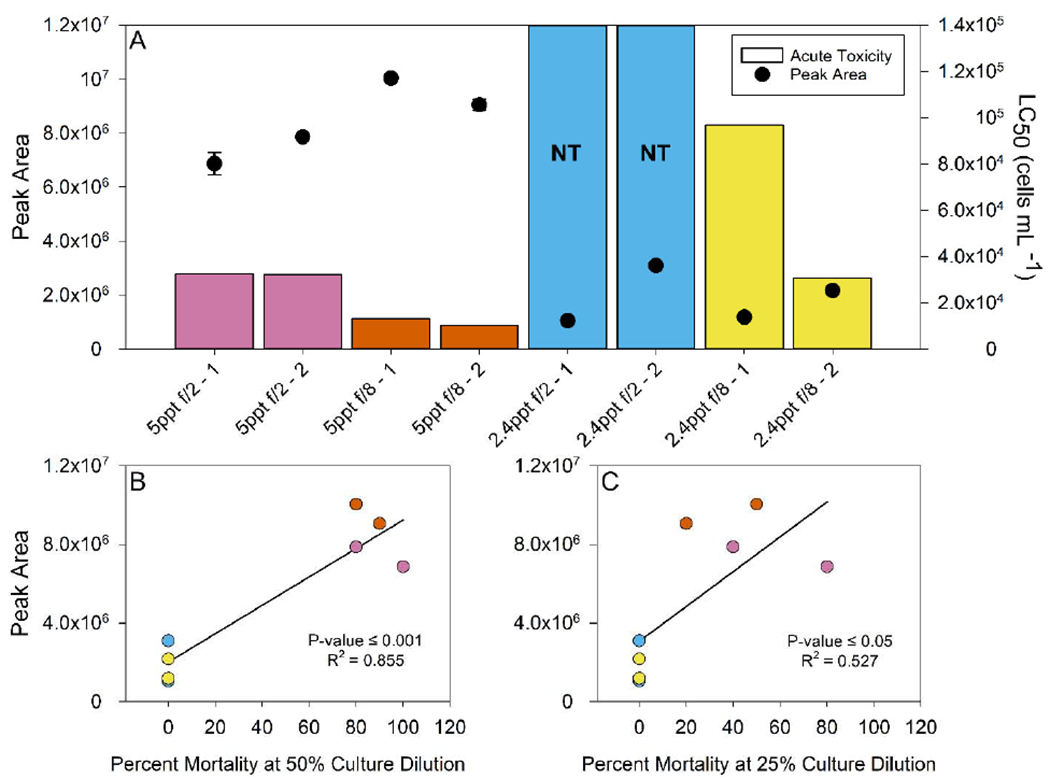
The peak area of PRM-A (3 Cl) (circles) present in the duplicate cultures of all four growth conditions is shown in (A) against the larval fathead minnow acute toxicity LC50 values (bars). Samples with a large LC50, considered non-toxic (NT), are shown with bars extended to the top of the graph (blue). An increase in prymnesin concentration shows an inverse relationship with toxicity (lower LC50). Linear regression analysis between percent mortality of cultures at 50% (B) and 25% (C) dilution and PRM-A (3 Cl) peak area show a positive relationship is present (p-value ≤ 0.05).
To the best of our knowledge, this is the first demonstration of a positive relationship between increasing prymnesin concentrations and elevated acute toxicity to fish from P. parvum. However, a gap remains to further define this dose - response gradient because we could only examine acutely and non-toxic conditions. Future work should focus on filling this gap with conditions of intermediate toxicity, accompanied by toxicity identification evaluation, to confirm this potentially causative relationship. Similarly, this analysis utilized a targeted approach and only considered previously identified suspect toxins within the ca. 2000 peaks recognized by the peak picking software. It is possible that other compounds in the complex mixture co-vary and influence acute toxicity. Whether these molecules are associated with sublethal toxicity remains unknown and similarly deserves future attention.
3.4. Visualizing Differences in the Chemical Fingerprint of Complex Mixtures
Current methods for locating peaks that change significantly between sample groups rely on software that identifies peaks within a given group of full-scan HRMS data and compares the intensity of each peak between samples. This approach provides a table of peaks that can be filtered by ANOVA p-values and log fold change (Hu et al., 2016; Nürenberg et al., 2015) but does not present a visual representation of all the peaks that significantly changed between sample groups in an intuitive heatmap similar to the original data. Utilizing relative difference plots from two HPLC-HRMS heatmaps allows for easy visualization of the fluctuating peaks between two sample groups. A relative difference approach is considered the most suitable method because it does not display bias towards high intensity peaks, since it is known that signals with high intensities are not always compounds of interest, especially for toxins where a slight change in concentration can cause a significant increase in toxicity.
A MATLAB script (Figure S1) was developed to align HPLC-HRMS files to a selected reference so that the heatmaps (intensity matrices) of various samples could be compared. The method was developed using spiked samples, and a description and demonstration of this process are given in supplemental information. Using the developed approach, relative difference plots were created with HPLC-HRMS heatmaps of each P. parvum experimental unit (replicate #2) to examine differences in the chemical composition. The heatmap of the non-toxic treatment combination of low salinity (2.4 ppt) and nutrient sufficient (f/2) conditions was used as a reference such that any peaks with an intensity higher than the reference would be highlighted, and peaks with an intensity below the reference would disappear. In theory, peaks highlighted in the relative difference plots could be contributing to the differences in the toxicological response.
Plots for P. parvum grown under higher (5 ppt) salinity, regardless of nutrient treatment level, highlighted numerous peaks and looked relatively similar (Figure 4A and 4B), whereas the relative difference plot of low salinity (2.4 ppt) and nutrient deficient (f/8) showed only a few highlighted peaks (Figure 4C). This is expected since the 5 ppt treatment had similar acute toxicity profiles that are higher than the reference sample, and the low salinity (2.4 ppt) and nutrient deficient (f/8) combination elicited low/no toxicity to fish, which is similar to the reference sample. Prymnesins are among the highlighted compounds for both higher salinity (5 ppt) and nutrient deficient (f/8) and higher salinity (5 ppt) and nutrient sufficient (f/2) conditions (Inset Figure 4A and 4B), which supports the suspect screening results. Both approaches showed a positive relationship between prymnesin concentration and acute toxicity that agrees with recent work suggesting prymnesins are most likely the toxins linked to P. parvum fish toxicity (Blossom et al., 2014). However, prymnesins are not the only peaks highlighted in the relative difference plots, suggesting other molecules might be contributing to the acute toxicity observed here. Future studies need to develop a robust statistical approach that pinpoints molecules displaying a strong correlation with diverse toxicological responses.
Figure 4.
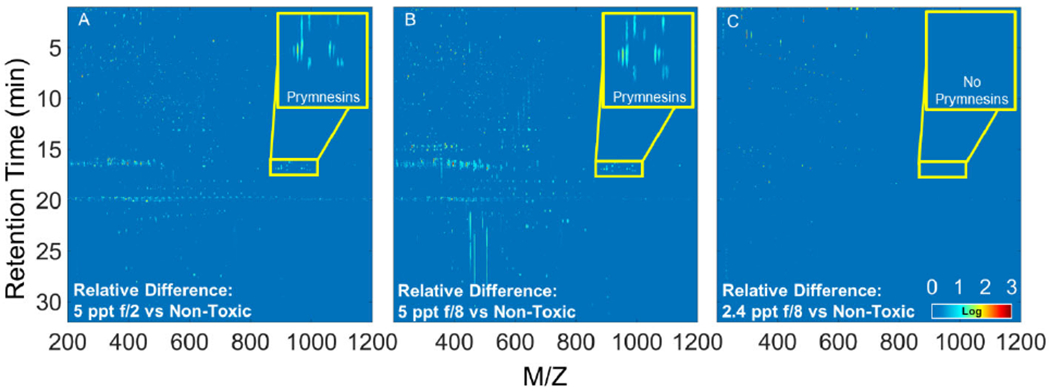
Relative difference plots for the second replicate of Prymnesium parvum cultures created with the non-toxic 2.4 ppt f/2 culture as the reference matrix. Plots of the 5 ppt f/2 (A) and f/8 (B) cultures are similar and highlight several peaks including prymnesins (inset), which are likely compounds responsible for the increased toxicity of these cultures. The relative difference plot for 2.4 ppt f/8 (C) shows fewer highlighted compounds, which is expected as the 2.4 ppt cultures exhibited similar acute toxicity.
4. Conclusion
In the present study, we report the first significantly (p < 0.05) positive relationship between levels of prymnesins and acute toxicity to fish elicited by P. parvum, suggesting that prymnesin molecules are the primary cause of fish kills during toxic golden algae blooms. However, while prymnesins displayed a positive relationship with fish acute toxicity, it is important to note that other molecules in the toxic P. parvum cultures co-varied with toxicity and are potential toxins contributing to the fish mortality observed in the current study. Future work should focus on investigating other molecules that have a positive relationship with acute toxicity before confirming prymnesins are indeed the responsible toxins for acute aquatic toxicity.
Evaluating differences in toxicity measurements using a statistical approach could alleviate the difficulties associated with the time and cost intensive fractionation/TIE approach. However, future experiments should utilize these traditional approaches and compare them against statistical methods to ensure results are consistent. To the extent additional work confirms prymnesins as the responsible toxin, it would be highly beneficial for surface water quality assessors and managers to develop a quick and easy prymnesins screening method that allow plans of action to be implemented based on toxins instead of biomass concentrations. Additional work should focus on treatment protocols and their effects on the biosynthesis and degradation of prymnesins.
Supplementary Material
Highlights:
Prymnesium parvum causes major fish kills but the toxins have not been confirmed
Nutrients and salinity altered P. parvum growth and acute toxicity to fish
Cultures were analyzed with LC-HRMS and screened for known prymnesins
Identified prymnesins were significantly related to the magnitude of acute toxicity
Novel relative difference plots using LC-HRMS heatmaps highlighted additional peaks
Acknowledgements:
The authors acknowledge the Baylor University Mass Spectrometry Center (BU-MSC) for support during this work.
Funding: This research was partially supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under award number 1P01ES028942 to BWB. Dr. Hering's work was supported in part by the King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) under award no: OSR-2015-CRG4-2582.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Bader T, Schulz W, Kümmerer K, Winzenbacher R, 2017. LC-HRMS Data Processing Strategy for Reliable Sample Comparison Exemplified by the Assessment of Water Treatment Processes. Anal. Chem 89, 13219–13226. 10.1021/acs.analchem.7b03037 [DOI] [PubMed] [Google Scholar]
- Baker JW, Grover JP, Brooks BW, Ureña-Boeck F, Roelke DL, Errera R, Kiesling RL, 2007. Growth and toxicity of Prymnesium parvum (Haptophyta) as a function of salinity, light, and temperature. J. Phycol 43, 219–227. 10.1111/j.1529-8817.2007.00323.x [DOI] [Google Scholar]
- Baker JW, Grover JP, Ramachandrannair R, Black C, Valenti TW, Brooks BW, Roelke DL, 2009. Growth at the edge of the niche: An experimental study of the harmful alga Prymnesium parvum. Limnol. Oceanogr 54, 1679–1687. 10.4319/lo.2009.54.5.1679 [DOI] [Google Scholar]
- Bertin MJ, Zimba PV, Beauchesne KR, Huncik KM, Moeller PDR, 2012a. Identification of toxic fatty acid amides isolated from the harmful alga Prymnesium parvum carter. Harmful Algae 20, 111–116. 10.1016/j.hal.2012.08.005 [DOI] [Google Scholar]
- Bertin MJ, Zimba PV, Beauchesne KR, Huncik KM, Moeller PDR, 2012b. The contribution of fatty acid amides to Prymnesium parvum Carter toxicity. Harmful Algae 20, 117–125. 10.1016/j.hal.2012.08.004 [DOI] [Google Scholar]
- Binzer SB, Svenssen DK, Daugbjerg N, Alves-de-Souza C, Pinto E, Hansen PJ, Larsen TO, Varga E, 2019. A-, B- and C-type prymnesins are clade specific compounds and chemotaxonomic markers in Prymnesium parvum. Harmful Algae 81, 10–17. 10.1016/j.hal.2018.11.010 [DOI] [PubMed] [Google Scholar]
- Blossom HE, Rasmussen SA, Andersen NG, Larsen TO, Nielsen KF, Hansen J, Hansen PJ, 2014. Prymnesium parvum revisited: Relationship between allelopathy, ichthyotoxicity, and chemical profiles in 5 strains. Aquat. Toxicol 157, 159–166. 10.1016/j.aquatox.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Brooks BW, Grover JP, Roelke DL, 2011. Prymnesium parvum: An emerging threat to inland waters. Environ. Toxicol. Chem 30, 1955–1964. 10.1002/etc.613 [DOI] [PubMed] [Google Scholar]
- Brooks BW, James SV, Valenti TW Jr., Urena-Boeck F, Serrano C, Berninger JP, Schwierzke L, Mydlarz LD, Grover JP, Roelke DL, 2010. Comparative Toxicity of Prymnesium parvum in Inland Waters. JAWRA J. Am. Water Resour. Assoc 46, 45–62. 10.1111/j.1752-1688.2009.00390.x [DOI] [Google Scholar]
- Brooks BW, Lazorchak JM, Howard MDA, Johnson M-VV, Morton SL, Perkins DAK, Reavie ED, Scott GI, Smith SA, Steevens JA, 2017. In some places, in some cases, and at some times, harmful algal blooms are the greatest threat to inland water quality. Environ. Toxicol. Chem 36, 1125–1127. 10.1002/etc.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BW, Lazorchak JM, Howard MDA, Johnson MVV, Morton SL, Perkins DAK, Reavie ED, Scott GI, Smith SA, Steevens JA, 2016. Are harmful algal blooms becoming the greatest inland water quality threat to public health and aquatic ecosystems? Environ. Toxicol. Chem 35, 6–13. 10.1002/etc.3220 [DOI] [PubMed] [Google Scholar]
- Carmichael WW, 2001. Health Effects of Toxin-Producing Cyanobacteria: “The CyanoHABs.” Hum. Ecol. Risk Assess. An Int. J 7, 1393–1407. 10.1080/20018091095087 [DOI] [Google Scholar]
- Chen B, Brown KA, Lin Z, Ge Y, 2018. Top-Down Proteomics: Ready for Prime Time? Anal. Chem 90, 110–127. 10.1021/acs.analchem.7b04747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco ME, Hill BN, Brooks BW, Lavado R, 2019. Prymnesium parvum differentially triggers sublethal fish antioxidant responses in vitro among salinity and nutrient conditions. Aquat. Toxicol 213, 105214. 10.1016/j.aquatox.2019.05.016 [DOI] [PubMed] [Google Scholar]
- Fu Y, Zhang Y, Zhou Z, Lu X, Lin X, Zhao C, Xu G, 2018. Screening and Determination of Potential Risk Substances Based on Liquid Chromatography-High-Resolution Mass Spectrometry. Anal. Chem 90, 8454–8461. 10.1021/acs.analchem.8b01153 [DOI] [PubMed] [Google Scholar]
- Haddad SP, Bobbitt JM, Taylor RB, Lovin LM, Conkle JL, Chambliss CK, Brooks BW, 2019. Determination of microcystins, nodularin, anatoxin-a, cylindrospermopsin, and saxitoxin in water and fish tissue using isotope dilution liquid chromatography tandem mass spectrometry. J. Chromatogr. A 1599, 66–74. 10.1016/j.chroma.2019.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson JC, Gharfeh MS, Easton AC, Easton JD, Glenn KL, Shadfan M, Mooberry SL, Hambright KD, Cichewicz RH, 2010. Reassessing the ichthyotoxin profile of cultured Prymnesium parvum (golden algae) and comparing it to samples collected from recent freshwater bloom and fish kill events in North America. Toxicon 55, 1396–1404. 10.1016/j.toxicon.2010.02.017 [DOI] [PubMed] [Google Scholar]
- Hill BN, Saari GN, Steele WB, Corrales J, Brooks BW, n.d. Influences of nutrients and salinity on Prymnesium parvum elicited sublethal toxicity in two common fish models. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender J, Schymanski EL, Singer H, Ferguson PL, 2017. Non-target screening with high resolution mass spectrometry in the environment: Ready to go? Environ. Sci. Technol 51, 11505–11512. 10.1021/acs.est.7b02184 [DOI] [PubMed] [Google Scholar]
- Hu M, Krauss M, Brack W, Schulze T, 2016. Optimization of LC-Orbitrap-HRMS acquisition and MZmine 2 data processing for nontarget screening of environmental samples using design of experiments. Anal. Bioanal. Chem 408, 7905–7915. 10.1007/s00216-016-9919-8 [DOI] [PubMed] [Google Scholar]
- Igarashi T, Satake M, Yasumoto T, 1999. Structures and partial stereochemical assignments for prymnesin-1 and prymnesin-2: Potent hemolytic and ichthyotoxic glycosides isolated from the red tide alga Prymnesium parvum. J. Am. Chem. Soc 121, 8499–8511. 10.1021/ja991740e [DOI] [Google Scholar]
- Igarashi T, Satake M, Yasumoto T, 1996. Prymnesin-2: A Potent Ichthyotoxic and Hemolytic Glycoside Isolated from the Red Tide Alga Prymnesium parvum. J. Am. Chem. Soc 118, 479–480. 10.1021/ja9534112 [DOI] [Google Scholar]
- James TL, De La Cruz A, 1989. Prymnesium-parvum Carter (Chrysophyceae) as a suspect of mass mortalities of fish and shellfish communities in western texas. Texas J. Sci 41, 429–430. [Google Scholar]
- James SV, Valenti TW, Prosser KN, Grover JP, Roelke DL, Brooks BW, 2011. Sunlight amelioration of Prymnesium parvum acute toxicity to fish. J. Plankton Res 33, 265–272. 10.1093/plankt/fbq082 [DOI] [Google Scholar]
- Manning SR, La Claire JW, 2013. Isolation of polyketides from Prymnesium parvum (Haptophyta) and their detection by liquid chromatography/mass spectrometry metabolic fingerprint analysis. Anal. Biochem 442, 189–195. 10.1016/j.ab.2013.07.034 [DOI] [PubMed] [Google Scholar]
- Nürenberg G, Schulz M, Kunkel U, Ternes TA, 2015. Development and validation of a generic nontarget method based on liquid chromatography - high resolution mass spectrometry analysis for the evaluation of different wastewater treatment options. J. Chromatogr. A 1426, 77–90. 10.1016/j.chroma.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Paerl HW, Huisman J, 2009. Climate change: A catalyst for global expansion of harmful cyanobacterial blooms. Environ. Microbiol. Rep 1, 27–37. 10.1111/j.1758-2229.2008.4.x [DOI] [PubMed] [Google Scholar]
- Preece EP, Hardy FJ, Moore BC, Bryan M, 2017. A review of microcystin detections in Estuarine and Marine waters: Environmental implications and human health risk. Harmful Algae 61, 31–45. 10.1016/j.hal.2016.11.006 [DOI] [Google Scholar]
- Prosser KN, Valenti TW, Hayden NJ, Neisch MT, Hewitt NC, Umphres GD, Gable GM, Grover JP, Roelke DL, Brooks BW, 2012. Low pH preempts bloom development of a toxic haptophyte. Harmful Algae 20, 156–164. 10.1016/j.hal.2012.10.002 [DOI] [Google Scholar]
- Rasmussen SA, Meier S, Andersen NG, Blossom HE, Duus JØ, Nielsen KF, Hansen PJ, Larsen TO, 2016. Chemodiversity of Ladder-Frame Prymnesin Polyethers in Prymnesium parvum. J. Nat. Prod 79, 2250–2256. 10.1021/acs.jnatprod.6b00345 [DOI] [PubMed] [Google Scholar]
- Roelke DL, Barkoh A, Brooks BW, Grover JP, Hambright KD, LaClaire JW, Moeller PDRR, Patino R, 2015. A chronicle of a killer alga in the west: ecology, assessment, and management of Prymnesium parvum blooms. Hydrobiologia 764, 29–50. 10.1007/s10750-015-2273-6 [DOI] [Google Scholar]
- Southard GM, Fries LT, Barkoh A, 2010. Prymnesium parvum: The texas experience. J. Am. Water Resour. Assoc 46, 14–23. 10.1111/j.1752-1688.2009.00387.x [DOI] [Google Scholar]
- Tomasi G, Van Den Berg F, Andersson C, 2004. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J. Chemom 18, 231–241. 10.1002/cem.859 [DOI] [Google Scholar]
- Ulitzur S, Shilo M, 1963. A Sensitive Assay System for Determination of the Ichthyotoxicity of Prymnesium parvum. J. Gen. Microbiol 36, 161–169. 10.1099/00221287-36-2-161 [DOI] [PubMed] [Google Scholar]
- Valenti Theodore W., James SV, Lahousse M, Schug KA, Roelke DL, Grover JP, Brooks BW, 2010. Influence of pH on amine toxicology and implications for harmful algal bloom ecology. Toxicon 55, 1038–1043. 10.1016/j.toxicon.2010.02.001 [DOI] [Google Scholar]
- Valenti Theodore W, James SV, Lahousse MJ, Schug KA, Roelke DL, Grover JP, Brooks BW, 2010. A mechanistic explanation for pH-dependent ambient aquatic toxicity of Prymnesium parvum carter. Toxicon 55, 990–998. 10.1016/j.toxicon.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Yi L, Dong N, Yun Y, Deng B, Ren D, Liu S, Liang Y, 2016. Chemometric methods in data processing of mass spectrometry-based metabolomics: A review. Anal. Chim. Acta 914, 17–34. https://doi.Org/10.1016/j.aca.2016.02.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


