Abstract
Among the hallmarks of melanoma, are impaired proteostasis and the rapid development of resistance to targeted therapy, that represent a major clinical challenge. However, the molecular machinery that links these processes is unknown. Here we describe that by stabilizing key melanoma oncoproteins the ubiquitin ligase RNF4 promotes tumorigenesis and confers resistance to targeted therapy in melanoma cells, xenograft mouse models and patient samples. In patients, RNF4 protein and mRNA levels correlate with poor prognosis and with resistance to MAPK inhibitors. Remarkably, RNF4 tumorigenic properties including therapy resistance require the translation initiation factor eIF2α. RNF4 binds, ubiquitinates and stabilizes the phosphorylated eIF2α (p-eIF2α), but not ATF4 or CHOP that mediate eIF2α–dependent integrated stress response. In accordance, p-eIF2α level was significantly elevated in high-RNF4 patient derived melanomas. Thus, RNF4 and p-eIF2α establish a positive feed-forward loop connecting oncogenic translation and ubiquitin-dependent protein stabilization in melanoma.
Keywords: STUbL, RNF4, Ubiquitin, Translation, p-eIF2α, Gene-regulation, Melanoma, Cancer
Introduction
Despite major advances in targeted and immune therapy for metastatic melanoma, a substantial percentage of patients fail to respond (Tsao et al., 2012; Merlino et al., 2016; The & Alpin, 2018). Among the hallmarks of melanoma progression and unresponsiveness to therapy, is impaired ubiquitin-dependent degradation of short lived oncoproteins (Fuchs, 2005; Qi et al., 2008; Kim et al., 2015; Senft et al., 2018). Like ubiquitination, SUMOylation is also implicated in melanomagenesis (Moschos et al., 2007; Tsao et al., 2012; Bertolotto et al., 2011).
Ubiquitination and SUMOylation are connected via SUMO-targeted ubiquitin ligases (STUbLs). The human genome encodes for two STUbL proteins, RNF111 and RNF4, that are both implicated in cancer (Briones-Orta et al., 2013; Sriramachandran et al., 2014; Kumar et al., 2017). RNF4 effects on tumorigenesis are context dependent. For example, in promyelocytic leukemia (PML), RNF4 regulates the SUMO-dependent degradation of the oncogenic fusion protein PML-RAR, exhibiting a tumor suppressive function (Lallemand-Breitenbach et al., 2007; Gärtner A et al., 2014; Hands et al., 2014). In contrast, in epithelial cancer cells, RNF4 has pro-tumorigenic effects (Thomas et al., 2016). These oncogenic effects stem from RNF4-dependent stabilization of selected short-lived phosphorylated oncoproteins including p-c-Myc, p-β-catenin, p-c-Jun. RNF4 binds to these oncoproteins via its short arginine-rich-motif (ARM), independent of its SUMO-interacting motifs (SIM) or of substrate SUMOylation. RNF4 binding requires site-specific phosphorylation, which is often dependent on the MAPK signaling pathway that is hyper-activated in melanoma (Thomas et al., 2016; Amaral et al., 2017). The roles of RNF4 in melanoma, and in conferring resistance to molecular therapy, are currently unknown.
We report that RNF4 promotes tumorigenesis and confers resistance to targeted therapy in melanoma cells, xenograft mouse models and patient samples. In patients, high levels of RNF4 mRNA coincide with resistance to BRAF inhibitors and with poor prognosis. Mechanistically, RNF4 contributes to melanoma progression by increasing p-eIF2α levels, connecting oncogenic translation and protein stabilization in melanoma.
RESULTS:
High RNF4 expression is associated with poor prognosis in melanoma patients.
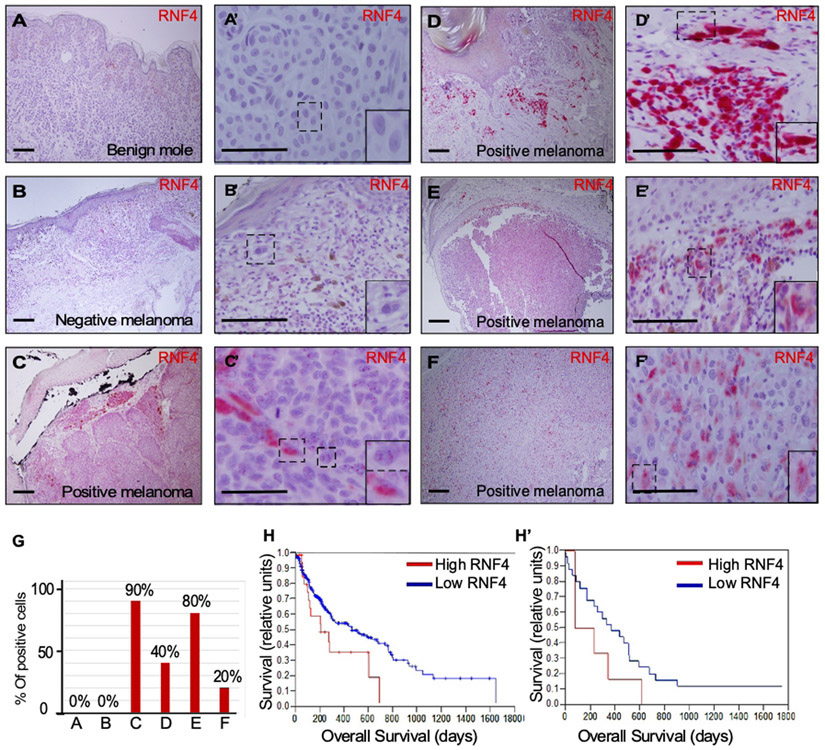
We analyzed the protein expression of RNF4 in biopsies of melanoma patients as well as benign nevi. RNF4 protein was not detected in benign nevi (Fig 1A-A’) but was expressed in 40% of primary melanoma samples (Fig. 1B-F’; 4/10 n=10). In positive biopsies, RNF4 was nuclear as well as cytoplasmatic. An average of 57% of melanoma cells expressed RNF4 in the positive biopsies (Fig. 1G). Assessment of RNF4 in the TCGA database (n=330), revealed that high levels of RNF4 mRNA correlate with poor prognosis (Fig. 1H). Employing a tissue microarray of 28 samples from metastatic melanoma patients revealed that overall survival tended to be shorter in patients with high RNF4 protein expression (Fig 1H’). Collectively, these data point to an inverse correlation between RNF4 expression and melanoma prognosis.
Figure 1: RNF4 mRNA and protein levels are elevated in human melanoma and correlate with poor survival.
(A-F’) Immunohistochemistry of patient-derived biopsies of nevi (A, A’), and melanoma tumors (B-F’) (H&E x40). RNF4 was identified using 810D mAb (red). (B, B’) A negative melanoma biopsy, and (C-F’) positive biopsies. A’, B’, C’, D’, E’, F’, are higher magnification (H&E x40). Insets are higher magnification of the regions in the dashed squares. Scale bar is 50μm. (G) Quantification of positive cells in each biopsy. (H, H’) Kaplan-Meier overall survival curves of melanoma patients, stratified according to RNF4 mRNA, (H; n=330, p< 0.05) and protein (H’; n= 28 p; ns, likely due to small size of the TMA) levels. In both panels Low RNF4 is shown in blue and High RNF4 in red.
RNF4 is essential for the tumorigenicity of melanoma cells.
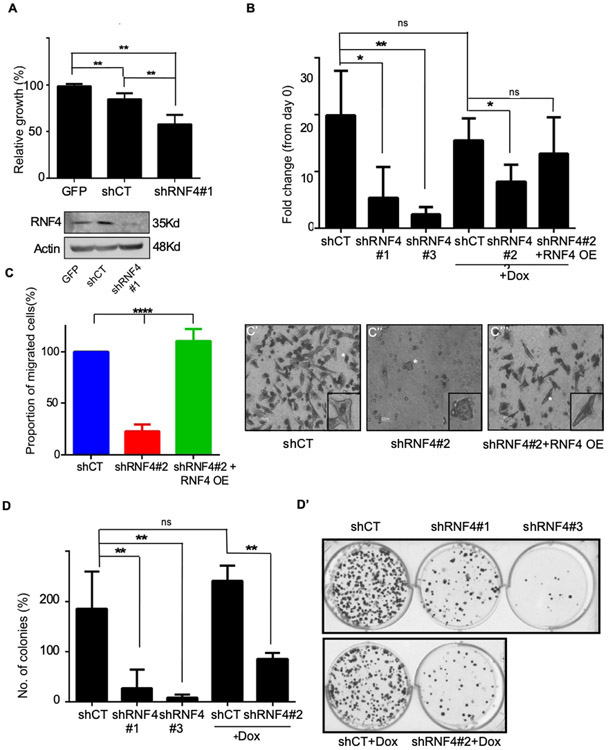
To determine possible role(s) for RNF4 in melanoma, we monitored changes in key melanoma phenotypes, upon altered RNF4 expression in A375 (BRAF mutated) and MeWo (BRAF WT, p53 mutated) human melanoma cell lines. To reduce RNF4 level we generated three independent shRNA RNF4 vectors. Expression of shRNF4s, but not scrambled control shRNA (shCT) resulted in reduced mitochondrial activity and cell viability as measured by MTT and ATP-Lite respectively (Figure. 2A, B, Supplemental Figure. S1A). Reduced RNF4 expression also impaired the migration of A375 cells (Figure 2C-C’”, Supplemental Figure. S1B). shRNF4-mediated reduction of RNF4 via these shRNF4s but not shCT inhibited colony formation (Figure 2D, D’). The reduced viability and attenuated cell migration were partially restored upon co-expression of RNF4 that was not sensitive to shRNF4#2 (Figure 2B, C, Supplemental. Figure. 1C-D). These findings suggest that RNF4 is essential for the tumorigenic properties of melanoma cells in culture.
Figure 2: RNF4 is essential for proliferation, migration, and clonogenicity of human melanoma cells.
(A) Upper panel: Mitochondrial activity of A375 melanoma cells as determined by MTT assay. Cells were infected with GFP or the indicated lentiviral shRNA vectors, **=p<0.01; n=3. Lower panel: Western blot analysis of cells used in A. Actin serves as loading control. (B) Viability of A375 cells as determined by ATP-lite® assay **=p<0.01; *=p<0.05; ns: non-significant; n=3. (C-C’”) Trans-well migration assay of A375 human melanoma cells infected with the indicated vectors. Quantification is shown in (C), and (C’-C’”) are representative images. White stars indicate cells shown in the insets. Where indicated in (B), and in (C), the activation of shRNF4#2 and RNF4 over-expression (RNF4 OE) are induced by the addition of Dox. The RNF4 OE vector is not sensitive to the shRNF4#2. (D-D’) Colony formation assay of A375 cells. Cells were infected with the indicated shRNA vectors. (D) Quantification of three biological repeats **= P<0.01; n=3 (D’) representative wells. shCT denoted scrambled RNF4 control; shRNF4#1-3 are shRNAs targeting RNF4.
RNF4 contributes to melanoma tumorigenesis in vivo.
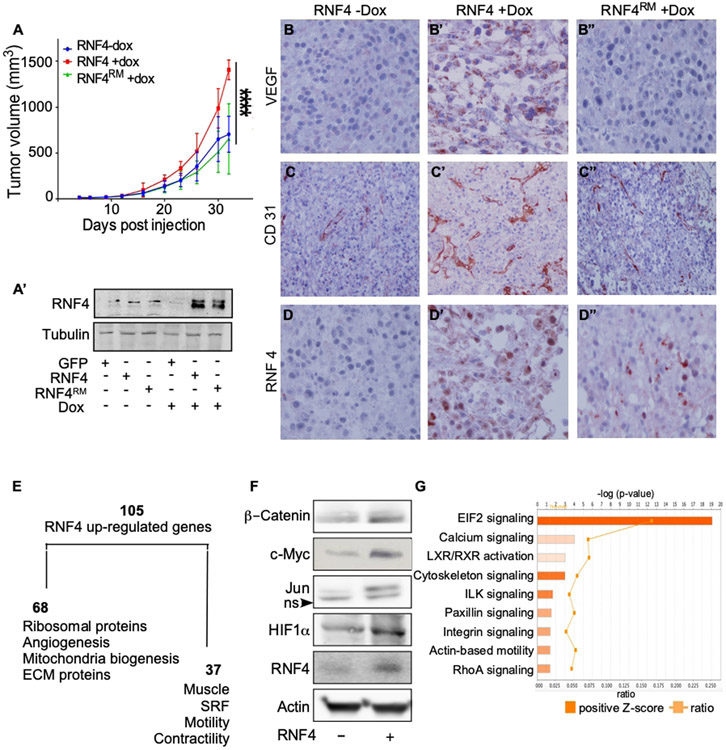
To investigate the role of RNF4 in melanomagenesis in vivo, we injected human A375 cells stably expressing either GFP or Dox-induced WT-RNF4, or a Dox-induced catalytic inactive RING mutant RNF4C159A (termed RNF4RM) into immunocompromised nude mice (n=7 in each group, Figure 3A). Expression of either RNF4, or RNF4RM, was induced 3 days post injection by the addition of Dox to drinking water and validated by western blot analysis (Figure 3A’). Dox-induced expression of RNF4, but not of RNF4RM, accelerated tumor growth compared with GFP-expressing tumors (Figures. 3A, Supplemental Figures 1E, E’). RNF4-expressing tumors exhibited increased levels of the endothelial CD31 marker, and VEGF protein (Figure 3B-D’). These changes were not observed in tumors expressing RNF4RM (Figure 3B”-D”). Notably, the increase in VEGFA levels coincides with TCGA data of high VEGFA mRNA levels in tumors expressing high RNF4 (p=2.19e−07).
Figure 3: RNF4 potentiates the growth of melanoma cells after xenografting in immunodeficient mice.
(A) Tumor size of A375-expressing Dox-inducible RNF4 or RNF4C159A catalytic inactive RING mutant (RNF4RM) injected subcutaneously to xenograft nude mice in the absence or presence of Dox, as indicated (−/+Dox; n=7 per group; ****=p<0.0001). (A’) Western blot analysis of RNF4 protein level in transplanted A375 cells at time of transplantation. (B-D”) Representative immuno-histochemistry images with indicated antibodies (immunoperoxidase, x200). Dox-induced the expression of RNF4 (B’-D’) or RNF4RM (B”-D”) in indicated tumors. (E) A diagram of RNF4-upregulated genes identified by RNA-seq of RNF4-expressing tumors and respective gene ontology. (F) Western blot analysis of the indicated proteins in A375 cells expressing GFP control or RNF4. (G) Ingenuity upstream pathway analysis (IPA) of RNF4-expressing tumors. Positive-z scores indicated in orange.
The tumorigenic activity of RNF4 was previously attributed to regulation of gene expression, requiring its association with nucleosomes (Thomas et al. 2016). To discover RNF4 and melanoma-related gene signature, we performed RNA-seq analysis of the xenograft-derived tumors (Figures. 3E, Supplemental Table 1). Using a cut-off of 2 (log2(FC), p< 0.05), we identified 105 genes that were significantly upregulated upon RNF4 overexpression, compared to control GFP expressing tumors. The expression of two genes was downregulated. By Gene Ontology analysis 64% of these upregulated genes were related to the regulation of cell growth, angiogenesis and ribosomal protein network, and 36% corresponded to contractility, motility, and calcium signaling (Figure. 3E). These results are consistent with the observed effect of RNF4 on migration (Figure 2C). To validate the RNA-seq data, representative genes were subjected to qPCR analysis (Supplemental Figure. S2).
We used Qiagen Upstream Regulator Analysis to identify transcription factors that may regulate this signature. Among these regulators we identified transcription factors that were previously reported as RNF4-stabilized oncoproteins, including c-Myc, β-catenin, c-Jun, and a novel one, HIF1α. Indeed, the expression of these oncoproteins and their activity increased upon expression of RNF4 in A375 cells (Figure. 3F, Supplemental Figure S3).
RNF4 regulates p-eIF2α.
To identify regulatory pathways enriched in the group of 105 upregulated genes, we used the ingenuity pathway analysis (IPA), which identified eIF2 as the most prominent node (Fig. 3G. p<10−19). The trimeric translation initiation factor eIF2 complex conveys the initiator tRNA to the ribosomes. In non-transformed cells, phosphorylation of the eIF2α subunit on Ser51 in humans and Ser52 in mice (p-eIF2α) is an essential regulator of the integrated cellular stress response and attenuates canonical translation. Remarkably in human squamous skin cancer and rodent cells the eIF2 complex was shown to be essential for promoting tumorigenesis (Sendoel et al., 2017; Robichaud et al., 2018;Koromilas, 2015; Rosenwald et al., 2009). In accordance increased levels of p-eIF2α were reported in several melanoma cell lines (Ferretta et al., 2016).
To study the molecular connection between RNF4 and p-eIF2α, we tested whether RNF4 affects p-eIF2α stability. We observed that A375 cells express low levels of both endogenous RNF4 and p-eIF2α (Fig. 4A-C, Supp. Fig. 4A-G). Expression of RNF4, but not of the catalytic inactive mutant RNF4RM, resulted in an increased steady state level of endogenous p-eIF2α protein in melanoma cell lines (Figure 4A, Supp. Figure S4A, B). Similarly, reduction of RNF4 protein levels using shRNF4, but not control, reduced p-eIF2α levels (Figure 4B). The activity of RNF4 is directed towards p-eIF2α as the levels of other translational initiation factors were not altered by RNF4 expression (Supplemental Figure. S4C). Moreover, expression or knockdown of RNF4 in A375 cells did not affect global translation activity as evident by the surface sensing of translation (SUnSET) assay (Supplemental Figures. S4D, E; Schmidt et al., 2009).
Figure 4: RNF4 binds, ubiquitinates, and increases the level of p-Ser51-eIF2α.
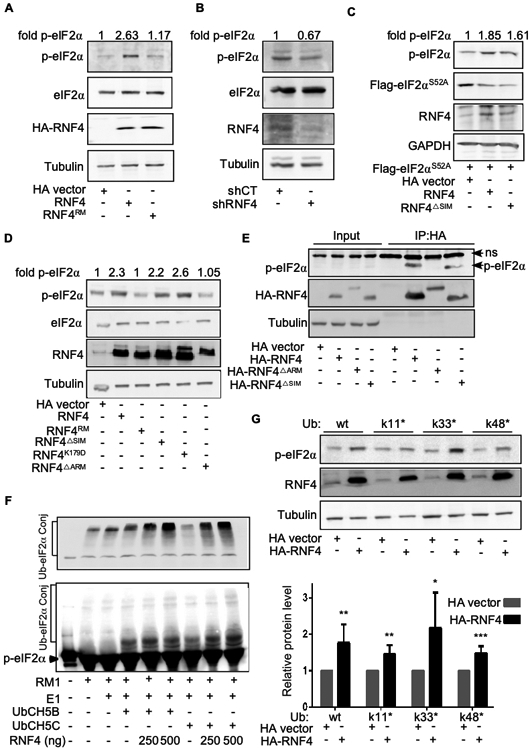
(A-G) Western blot analysis with the indicated antibodies. Tubulin or GAPDH serve as loading controls. HA vector indicates HA-only-expressing vector compared with HA-RNF4-expressing vectors. Fold change represents p-eIF2α/Tubulin relative ratio (A). Expression of RNF4, but not of catalytic inactive RING mutant C159A (RNF4RM), in A375 melanoma cells results in elevated p-Ser51-eIF2α (p-eIF2α) level. (B) Expression of shRNF4, but not of scrambled control (shCT), results in reduced p-eIF2α levels in A375 melanoma cells . (C) A375 melanoma cells were transfected with a plasmid coding for FLAG-tagged eIF2αS51A vector, or HA-empty vector, along with RNF4 or RNF4ΔSIM. Endogenous p-eIF2α and the transfected non-phosphorylatable FLAG--eIF2αS51A mutant level were monitored concomitantly using α–p-eIF2α and α-FLAG antibodies respectively. (D) HEK293T cells were transfected with the indicated HA-RNF4 coding plasmids and the protein level of p-eIF2α was determined. (E) RNF4 but not RNF4ΔARM binds to p-eIF2α in a co-IP assay. HEK293T cells were transfected with the indicated HA-RNF4 coding plasmids and the ability of RNF4 or RNF4 mutants to bind endogenous p-eIF2α in a co-immunoprecipitation assay was determined. 5% input is shown; ns, non-specific band. F) RNF4 ubiquitinates immuno-precipitated FLAG-eIF2α together with either UbC5B or UbC5C in vitro. Upper panel eIF2α~ubiquitin conjugates are detected using α-ubiquitin antibody. Lower panel, p-eIF2α~ubiquitin conjugates are detected using α-p-eIF2α antibody (G) Upper panel p-eIF2α protein level in HEK293 cells expressing either HA control vector or RNF4, as well as wide-type ubiquitin, or ubiquitin harboring a single internal lysine as indicated. Lower panel, quantification of six independent biological repeats *** =p<0.001. **= p<0.01. *=<0.05.
RNF4 stabilized endogenous p-eIF2α, but failed to increase the level of a non-phosphorylatable FLAG-tagged eIF2αS51A mutant (Figure 4C), suggesting that the activity of RNF4 is directed towards Ser51-phosphorylated form of eIF2α (p-eIF2α). In accordance, the arginine-rich motif (ARM) of RNF4, which mediates the recognition of phosphorylated oncoproteins (Thomas et al., 2016; Kuo et al., 2014), was essential for increasing p-Ser51-eIF2α levels. It was also required for binding to p-eIF2α, in a co-immunoprecipitation assay (Figures. 4D, E, Supplemental Figure 4F). In contrast, the SIM domains, which mediate the recognition of SUMOylated proteins by RNF4, were dispensable for the increased level or binding to p-eIF2α (Figure. 4D, E). Similarly, expression of the RNF4K179D mutant, which cannot bind to nucleosomes, also increased the level of p-eIF2α (Figure. 4D, and see discussion).
In agreement, RNF4 ubiquitinated in vitro a FLAG-tagged p-eIF2α that was purified from A375 melanoma cells (Figure 4F). RNF4 ubiquitination and protein stabilization were shown to involve the assembly of heterotypic ubiquitin chains containing internal links, using K11 and K33 within ubiquitin (Thomas et al. 2018). Likewise, the increase in p-eIF2α required the formation of heterotypic ubiquitin chains, and predominantly internal linkage of K33 (Figure 4G). In all, the increase in p-eIF2α requires recognition of p-eIF2α by the ARM domain of RNF4, and catalysis of ubiquitin chains with heterotypic topology.
RNF4 tumorigenic properties in melanoma require eIF2α .
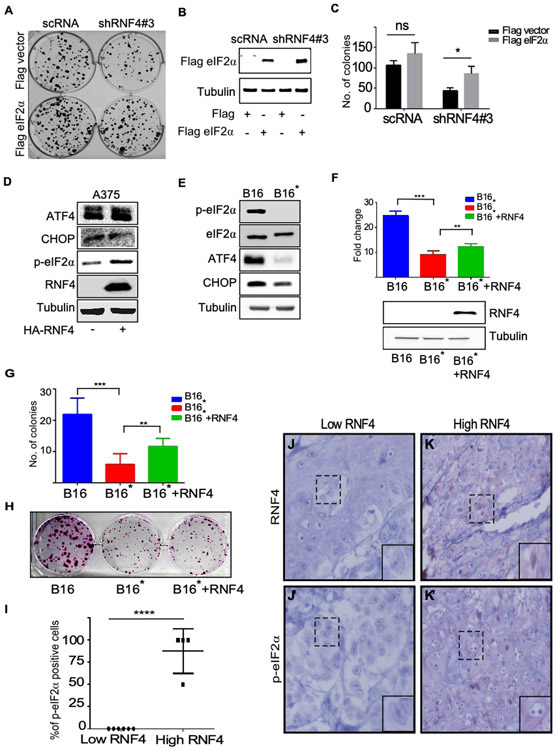
We examined whether eIF2α is required for RNF4’s tumorigenic activity. We transduced a plasmid coding for eIF2α to A375 melanoma cells that were subjected to shRNA-mediated knockdown of RNF4 (shRNF4). Indeed, the attenuated ability of these cells to form colonies was partially restored upon expression of eIF2α , but not of a control vector (Figure 5A-C). This suggests that eIF2α can partially compensate for the loss of RNF4, and that an increase in eIF2α is essential for the tumorigenic properties of melanoma.
Figure 5: p-eIF2α is critical for RNF4 tumorigenic activity.
(A) Colony formation of A375 cells infected with the indicated vectors. Expression of eIF2α restores the ability of A375 cells lacking RNF4 to form foci. A375 cells were infected with the indicated shRNA vectors and co-transfected with control (FLAG vector) or FLAG-eIF2α-coding vectors. (B) Western-blot example of eIF2α protein level in extract derived fromA375 cells at time of seeding colonies. (C ) Quantification of foci formation of three independent biological repeats. (D) Western blot analysis with the indicated antibodies, tubulin serves as a loading control. Expression of RNF4 in A375 human cells does not stabilize ATF4 and CHOP. (E) Western blot analysis of the indicated proteins in B16 and B16* mouse melanoma cells. (F) Upper panel, viability analysis using Celltiter-Glo assay of mouse melanoma B16 cells expressing HA-control vector, B16* cells expressing HA-control vector, or HA-RNF4, n=3. Lower panel; Western blot analysis of RNF4 protein in one of the experiments shown in (F). (G) Quantification of colony formation assay of mouse melanoma B16 cells expressing HA-control vector, B16* cells expressing HA-control vector or HA-RNF4. (H) Representative foci in one of the experiments quantitated in (G). (I) Immunohistochemistry analysis of p-eIF2α positive cells in melanoma patient biopsies stratified by low- and high-RNF4, n=10, **** p<0.0001. (J-K’) Representative immunohistochemistry of RNF4 protein and p-eIF2α in patient samples quantified in “(I)”. Dashed square indicates magnified area; (immunoperoxidase, x400).
Phosphorylation of eIF2α is required for the activation of the ER integrated stress response, and its downstream transcription factors ATF4 and CHOP (Rozpedek et al., 2016). However, the expression of ATF4 and CHOP did not change upon expression of RNF4, albeit its ability to increase the protein levels of p-eIF2α (Figure 5D). Moreover, the gene signature of RNF4 expressing tumors did not correlate with ATF4 or the integrated stress response signature.
To further study the linkage between the tumorigenic activity of RNF4 and p-eIF2α , we generated B16 mouse melanoma cell line where the endogenous eIF2α gene was deleted using CRISPER/CAS9 genome editing. Concomitantly, we expressed a mutant mouse FLAG-eIF2αS52A , that cannot be phosphorylated. These cells, B16*, exhibited similar level of total eIF2α, but no p-eIF2α (Figure 5E). As expected, the endogenous levels of ATF4 and CHOP were reduced in B16* compared to B16 control cells. We found that B16* proliferated less than B16 cells, and that the expression of RNF4 only minimally restored proliferation of B16* cells (Figure 5F, p<0.001). In accordance, B16* cells formed less colonies, that also exhibited smaller size than B16 cells. The expression of RNF4 in B16* cells only minimally restored the number or size of the colonies (Figure 5G, H).
To establish a clinical relevance, we determined the levels of RNF4 and p-eIF2α in patient-derived melanoma samples (Figures 5I-K’). Elevated levels of p-eIF2α were observed only in high-RNF4-expressing tumors (Figure 5I, n= 10, p<0.001) Taken together these data suggest that p-eIF2α is part of the RNF4-dependent pathway in melanoma.
RNF4 confers resistance to BRAF inhibitors.
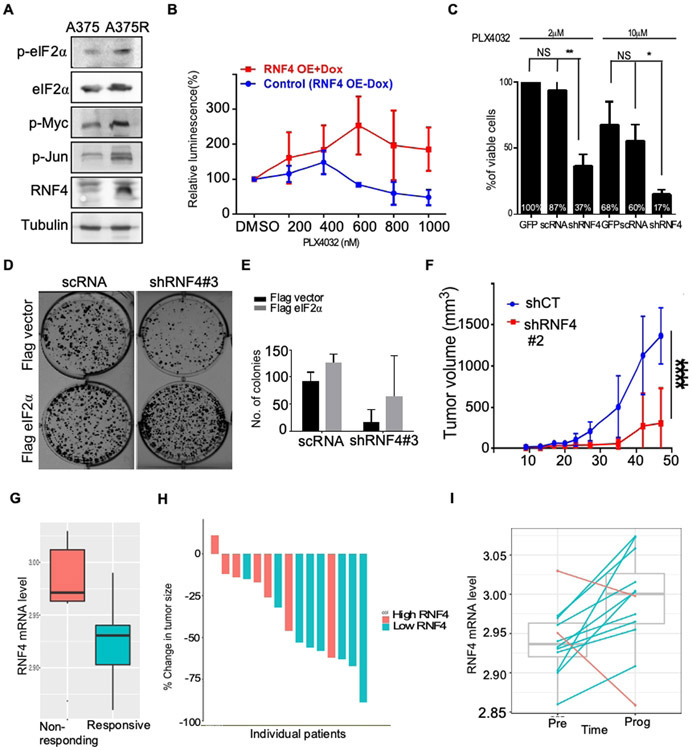
One characteristic of melanoma aggressiveness is the development of resistance to targeted therapy. Resistance to PLX4032 (Vemurafenib®), an inhibitor of BRAFV600E, develops rapidly, and represents a clinical challenge, even in the era of immunotherapies (Flaherty et al., 2010; Nazarian et al., 2010; Luke et al., 2017; Wahid et al., 2018). Given the role of RNF4 in melanoma tumorigenesis and the related MAPK signaling components, we studied the involvement of RNF4 in resistance to BRAF inhibition. Indeed, higher RNF4 protein levels as well as its stabilized substrates were elevated in PLX4032-resistant cell lines, such as A375R, LU1205R, UACC1113 (Figure. 6A, Supplemental Figure 4G; Kim et al. 2015). Both gain- and loss-of function experiments established a critical role for RNF4 in conferring resistance to PLX4032. Expression of RNF4 in A375 PLX4032-sensitive cells induced PLX-4032 resistance (Figure 6B), and PLX4032-resistant A375 cells (A375R) expressing shRNF4 re-gained sensitivity to treatment (Figure 6C). In accordance, A375R expressed higher RNF4 as well as higher p-eIF2α protein levels. Moreover, over-expression of eIF2α in A375R cells subjected to RNF4 knockdown, partially restored resistance to PLX4032 (Figure 6D, E).
Figure 6: RNF4 confers resistance to RTK inhibitors.
(A) Western blot analysis of the indicated proteins derived from PLX4032-sensitive (A375) or PLX4032-resistant human A375R cells. (B) Expression of RNF4 in A375 cells induces resistance to PLX4032 (Vemurafenib). A375 Cells were infected with Dox-inducible RNF4-coding viruses. Cell viability was monitored in the absences or presence of Dox and treatment with PLX4032 at the indicated concentrations for six days, n=3. (C) shRNF4-medeiated knockdown of RNF4 resulted in reduced viability of A375R cells as measured by MTT assay. A375R cells were infected with the indicated shRNA and PLX4032 concentrations. Cell viability was determined six days post-infection. (D) Colony formation of A375R cells infected with the indicated vectors, in the presence of PLX4032 at 5μM. Expression of eIF2α, but not of FLAG vector, in shRNF4-expressing A375R cells partially restores sensitivity to PLX4032. (E) Quantification of colonies number shown in (D), n=3. (F) Size of A375R xenograft mouse tumors expressing either scRNA control or shRNF4 in the presence of PLX4072. ****= P<0.0001 (G-I) High RNF4 mRNA level correlates with resistance to RTK therapy (see methods). (G) Boxplot depict RNF4 mRNA expression in melanoma samples at time of pre-treatment comparing responders to none responders, n=15. (H) Reduction in tumor size upon treatment for individual patients shown in (G). (I) comparison of RNF4 mRNA levels between pre-treatment and upon disease progression. Blue lines denote increase of RNF4 expression in upon progression while red lines denote decrease (n=12, paired Wilcoxon Ranksum P<0.02).
To substantiate observations made in melanoma cells, we tested the effects of RNF4 knockdown on PLX4032-resistant xenografts in vivo. We injected subcutaneously to nude mice, A375R cells expressing Dox-inducible shRNF4 or control (shRenilla). Control shRNA expressing A375R cells rapidly developed tumors (8/8) that resisted PLX4720 treatment. However, only 3/8 animals in the group injected with RNF4-knockdown cells developed tumors, that were notably smaller (p<0.0001, Figure 6F).
Lastly, we analyzed patient samples that were either sensitive or resistant to BRAF/MEK inhibition (Figure 6G, H; Kakavand et al. 2017). Higher RNF4 mRNA levels correlated with smaller reduction in tumor size (Mann whitney test p <0.04) and with failure to respond to treatment (Cohen's D = 0.85, within top 1% of all protein coding genes). Moreover, upon progression, resistant tumors expressed higher RNF4 level (Figure. 6I; paired Wilcoxon ranksum p<0.02). This correlation suggests that RNF4 is likely to play an important role in the resistance to MAPK inhibition in melanoma patients. In summary, we established in vitro, in vivo and in patient-samples, that RNF4 promotes melanoma progression and resistance to targeted therapy which is partly mediated via p-eIF2α.
Discussion
The ubiquitin ligase RNF4 regulates diverse nuclear processes (Sriramachandran & Dohmen, 2014). Our study suggests a cytoplasmic role for RNF4 in melanoma, as a positive regulator of the largely cytoplasmic p-eIF2α. During early Drosophila embryogenesis, the localization of the fly RNF4 ortholog, Dgrn, alternates between the cytoplasm and nucleus (Barry et al., 2011). In human bronchial epithelial cells, RNF4 was shown to target the cytoplasmic cystic fibrosis transmembrane regulator for degradation (Ahner et al., 2013). In patient derived tumor samples, RNF4 protein accumulates in the cytoplasm of melanoma and colon cancer patient samples (Figure 1; Thomas et al., 2016). However, the cytoplasmic substrates of RNF4 in cancer cells were unknown. Here we show that RNF4 binds, ubiquitinates and increases the level of p-eIF2α. This is further supported by the observation that the RNF4K179D mutant, which cannot bind to nucleosomes, efficiently increased p-eIF2α levels. In addition, RNF4 also stabilized nuclear melanoma-related oncoproteins such as c-Myc, c-Jun and HIF1α. Thus, RNF4 emerges as a nuclear and cytoplasmic regulator of melanoma tumorigenesis.
Phosphorylation of Ser51 within human eIF2α is required for the RNF4-mediated increase in p-eIF2α protein level. This phosphorylation is mediated by several protein kinases, including HRI, PERK, PKR, and GCN2 (Leprivier et al., 2015; Wek, 2018), and at least one of these kinases would be required for RNF4 activity. Regardless of the kinase involved, the potentiating activity of RNF4 on p-eIF2α takes place in wild-type, as well as in BRAF mutated cells. This phosphorylation is known to induce the ER stress response. However, RNF4 expression did not increase ATF4 or CHOP mRNA or protein levels in RNF4 expressing melanoma cells or tumors, suggesting that RNF4~p-eIF2α axis is part of a non-canonical oncogenic translation pathway. Possibly, RNF4-stabilizes a subpopulation of p-eIF2α complexes that induces a translation shift by using alternative initiation sites, without inhibiting global translation (Sendoel et al., 2017; Robichaud et al, 2018).
While we have focused on the role of RNF4 in the human melanoma cancer cell itself, our data suggest a potential role for RNF4 in promoting metastasis and effecting the tumor microenvironment. RNF4 was required for migration of melanoma cells, and a third (37/105) of RNF4-upregulated genes are involved in cell migration and motility. Moreover, tumors expressing RNF4 are more vascular and express high level of VEGF. These results hint that RNF4 may potentiate metastasis and impact the tumor microenvironment. However, the experimental settings that we used (e.g. human A375 cells and immune-deficient nude mice) are not optimal for studying metastasis. Therefore, the effects of RNF4 on metastasis require studies using mouse melanoma cells and immune competent hosts.
An important role of RNF4 in melanoma progression, is its role in conferring resistance to BRAF-inhibitors. The ability of eIF2α expression to restore resistance to BRAF-inhibition in RNF4 knockdown cells, presents a unique mechanism involving RNF4~ eIF2α–dependent resistance and promoting tumorigenesis.
The prognostic value of RNF4 expression in predicting response to targeted therapies and the novel role of RNF4-eIF2α in conferring drug resistance, may have direct clinical implications. Analysis of RNF4 mRNA and protein levels in patient samples may assist in tailoring treatment among the variety of current therapies. Moreover, pharmacological inhibition of RNF4 offers an opportunity to target a melanoma-related oncogenic translation pathway in a selective manner, resulting in collapse of resistant melanoma tumors.
Material and Methods:
Detailed material and methods are under supplemental data file.
Cell viability assay:
Cell viability was assessed using MTT (Sigma-Aldrich) and ATP-lite (Perkin Elmer) assays according to the manufacture instructions. In assays evaluating resistance to PLX4032, PLX4032-treated cells were cultured for six days before analysis.
Colony formation assay:
Cells were suspended in DMEM medium and seeded at a density of 1000 cells/well. Plates were maintained at 37°C for six days, fixed overnight with 4% paraformaldehyde (PFA), stained with 0.05% crystal violet.
Trans-well migration assay:
Cells were grown in serum-free medium for five hours and 1x105 cells/well were seeded in trans-well culture chambers in 100 μl of serum-free DMEM medium (BD Falcon). Bottom chamber contained medium with 10% fetal calf serum and allowed to migrate overnight.
Determination of global translation activity using SUnSET assay:
Surface sensing of translation (SUnSET) assay, was as described previously (Schmidt et al. 2009). Briefly, A375 cells were treated with Dox 500ng/ml for 48-96 hours. Puromycin (final concentration 5μg/ml), was then added to the medium for 30 min. Cell extracts were subjected to western blot analysis using α-puromycin antibody.
Immunoprecipitation:
Cell extracts were prepared using cell lysis buffer [10% glycerol, 1% Triton, 3% NaCl 5M, 0.2% EDTA 0.5M, 20mM (2%) Hepes pH7.4, and protease, phosphatase and proteasome inhibitors). Protein extract (1mg) was incubated overnight with pre-washed HA beads (Pierce™ #88836). Beads were then washed 5 times with lysis buffer, and proteins resolved over SDS-PAGE.
Protein levels and stability and ubiquitination:
Protein stability was determined in steady state and dynamic CHX chase experiments as described (Trausch-Azar et al. 2015). Proteins resolved over SDS-PAGE, using the indicated antibodies, and visualized using chemiluminescence (Image-Quan LAS4000).
In vitro ubiquitination was performed as previously described (Thomas et al. 2016); Briefly, FLAG-tagged eIF2α was immunoprecipitated using FLAG antibody coupled protein G- Sepharose (1:100). IP material was washed 3 times with IP-buffer and one time with 20mM Tris pH 7.2. Ubiquitination was reconstituted in vitro using 10μl beads as substrates and the indicated, purified, bacterially expressed enzymes for 1h incubation at 37°C. Proteins resolved using SDS-PAGE and visualized using the indicated antibody.
Generation of CAS9-edited A375p-eIF2αAS52A cells:
A 19-nt guide sequence (GCATCGTAGGCACCGTATCC) targeting exon 3 of the mouse eIF2α gene was ligated into a pX330 hSpCas9 plasmid (Addgene # 42230). The resulting construct was co-transfected into B16 melanoma cells together with a bi-cistronic construct encoding from its first open reading frame (ORF) a FLAG-tagged eIF2αS52A. This vector contains silent mutations that make it refractory to the guide RNA and puromycin resistance from its second ORF. Foci established following puromycin selection and tested by western blot analysis using eIF2α antibodies. Foci displaying FLAG-tagged eIF2α protein instead of the endogenous protein were selected and subjected to single cell cloning. In parallel total genomic DNA was purified and sequenced ensuring that the only coding DNA is that of the FLAG- eIF2αS52A vector.
In-vivo xenograft experiments:
Melanoma cells (1×106 cells/100 μl DMEM) were injected subcutaneously into 6-week-old female nude mice. RNF4 or shRNF4 vectors were induced by adding doxycycline (2 mg/ml) to drinking water 3 days post-injection. When relevant, a Vemurafenib analog, PLX4720, was administered with food according to manufacturer instructions (PLX4720, 417mg/Kg in chow, Research Diets). Tumor growth was monitored twice a week for 4-6 weeks, at which point the mice were sacrificed and tumors were analyzed.
Immunohistochemistry of tumors and patient samples:
Xenograft-tumors were fixed in formalin 5%, embedded in paraffin and immunostained with indicated antibodies. Patient samples were obtained from the Institute of Pathology at the Rambam Health Care Campus, Haifa, Israel. Experiments involving human tissues were according to Helsinki approvals (# 0239-12-RMB, RMB-0634-16131539).
RNA extraction, cDNA synthesis, and qPCR analysis:
RNA was extracted using GenElute Mammalian Total RNA Miniprep Kit (Sigma Aldrich). cDNA was synthesized from 1μg of total RNA using qScript cDNA Synthesis Kit (Quantabio). Real-time PCR was performed with Essential SYBR Green Master Mix® (Roche).
RNA-seq and data analyses:
RNA-sequencing and bio-informatic analysis are detailed in the supplemental data file.
Correlation between RNF4 mRNA levels and patient survival:
RNF4 expression (RSEM) and survival data for all TCGA SKCM samples were from cBioPortal (Cerami et al. 2012). We compared two groups of samples that represent the 10% highest and lowest RNF4 expression. Kaplan-Meier survival curve comparison, adjusted for patient age and sex, was conducted between these two groups using the logrank test, and a P-value of ≤0.05 was considered statistically significant.
Tissue microarray (TMA)
Tissue microarray were constructed as described (Jilaveanu et al. 2009). Tumors were stained using α-RNF4 mAb8D10 antibody and assessed for RNF4 level. Clinical data regarding response to treatment, disease-free and overall survival were collected and analyzed by HK.
Correlation between RNF4 mRNA levels and resistance to therapy in patient samples:
We analyzed a melanoma cohort treated with MAPK inhibitors; Post-treatment biopsies were taken when the tumor progressed. Fifteen patient samples that were analyzed for RNF4 mRNA level; 9 responders, and 6 non-responders and set a threshold of 30% reduction in tumor size. Additionally, we analyzed 12 patient samples; 3 patients were treated with Dabtrafenib and Trametinib, 7 patients with Dabrafenib, and 2 patients with Vemurafenib (Kakavand et al. 2017). RNF4 mRNA expression was compared between pre-treatment and post-treatment samples using paired Wilcoxon Ranksum test.
Statistical analysis.
SEM, and t-test comparisons were performed using GraphPad Prism and ANOVAs software. In all experiments, significance is as follows: ****=p<0.0001, ***= p<0.001, ** = p<0.01, *=p<0.05.
Supplementary Material
Acknowledgments:
We are grateful to A.C.O. Vertegaal, Israel Vlodavsky and Amit Meller for generous reagents and antibodies, to Eliya Lotan-Bitman for her assistance in bioinformatic analysis. EAH was supported by Fine and Wolf Fellowships. YZ was supported by an Atidim research grant. ZR was supported by NCI Grants R35CA197465 and R01CA202021, and AO was supported by ICRF 2017 and GIF I-1431-412.3 grants.
Footnotes
Conflict of Interest: The authors declare that they have no conflicts of interests.
Data Availability Statement:
Datasets related to this article can be found at [URL/s linked to dataset/s], hosted at GSE124064
References:
- Ahner A, Gong X, Schmidt BZ, Peters KW, Rabeh WM, Thibodeau PH, Lukacs GL, Frizzell RA. Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier-dependent pathway. Mol Biol Cell. 2013; 24:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral T, Sinnberg T, Meier F, Krepler C, Levesque M, Niessner H, Garbe C. The mitogen-activated protein kinase pathway in melanoma part I - Activation and primary resistance mechanisms to BRAF inhibition. Eur J Cancer 2017; 73:85–92. [DOI] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Bressac-de Paillerets B. et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011; 480:94–98. [DOI] [PubMed] [Google Scholar]
- Barry KC, Abed M, Kenyagin D, Werwie TR, Boico O, Orian A, Parkhurst SM. The Drosophila STUbL protein Degringolade limits HES functions during embryogenesis. Development. 2011; 138:1759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones-Orta MA, Levy L, Madsen CD, Das D, Erker Y, Sahai E, Hill CS. Arkadia regulates tumor metastasis by modulation of the TGF-β pathway. Cancer Res. 2013; 73:1800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretta A, Maida I, Guida S, Azzariti A, Porcelli L, Tommasi S, Zanna P, Cocco T, Guida M, Guida G. New insight into the role of metabolic reprogramming in melanoma cells harboring BRAF mutations. Biochim Biophys Acta. 2016; 1863:2710–2718. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010; 363:809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY. De-regulation of ubiquitin-dependent proteolysis and the pathogenesis of malignant melanoma. Cancer Metastasis Rev. 2005; 24:329–38. [DOI] [PubMed] [Google Scholar]
- Gärtner A, Muller S. PML, SUMO, and RNF4: guardians of nuclear protein quality. Mol Cell. 2014; 55:1–3. [DOI] [PubMed] [Google Scholar]
- Hands KJ, Cuchet-Lourenco D, Everett RD, Hay RT. PML isoforms in response to arsenic: high-resolution analysis of PML body structure and degradation. J Cell Sci. 2014; 127:365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Frederick DT, Levesque MP, Cooper ZA, Feng Y, Krepler C, Brill L, Samuels Y, Hayward NK, Perlina A, Piris A, Zhang T, Halaban R, Herlyn MM, Brown KM, Wargo JA, Dummer R, Flaherty KT, Ronai ZA. (2015) Downregulation of the Ubiquitin Ligase RNF125 Underlies Resistance of Melanoma Cells to BRAF Inhibitors via JAK1 Deregulation. Cell Rep. 2015:11:1458–1473. doi: 10.1016/j.celrep.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakavand H, Rawson RV, Pupo GM, Yang JYH, Menzies AM, Carlino MS, Kefford RF, Howle JR, Saw RPM, Thompson JF, et al. (2017). PD-L1 Expression and Immune Escape in Melanoma Resistance to MAPK Inhibitors. Clinical cancer research: an official journal of the American Association for Cancer Research 23, 6054–6061. [DOI] [PubMed] [Google Scholar]
- Koromilas AE. Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim Biophys Acta. 2015; 1849:871–880. [DOI] [PubMed] [Google Scholar]
- Kumar R, González-Prieto R, Xiao Z, Verlaan-de Vries M, Vertegaal ACO. The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat Commun. 2017; 8:1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CY, Li X, Kong XQ, Luo C, Chang CC, Chung Y, Shih HM, Li KK, Ann DK. An arginine-rich motif of ring finger protein 4 (RNF4) oversees the recruitment and degradation of the phosphorylated and SUMOylated Krüppel-associated box domain-associated protein 1 (KAP1)/TRIM28 protein during genotoxic stress. J Biol Chem. 2014; 289:20757–20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–55 [DOI] [PubMed] [Google Scholar]
- Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochim Biophys Acta. 2015; 1849:845–860. [DOI] [PubMed] [Google Scholar]
- Luke JJ, Flaherty KT, Ribas A, Long GV (2017). Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 14:463–482. [DOI] [PubMed] [Google Scholar]
- Merlino G, Herlyn M, Fisher DE, Bastian BC, Flaherty KT, Davies MA, Wargo JA, Curiel-Lewandrowski C, Weber MJ, Ronai ZA et al. The state of melanoma: challenges and opportunities. Pigment Cell Melanoma Res. 2016; 29: 404–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschos SJ, Smith AP, Mandic M, Athanassiou C, Watson-Hurst K, Jukic DM, Edington HD, Kirkwood JM, Becker D. SAGE and antibody array analysis of melanoma-infiltrated lymph nodes: identification of Ubc9 as an important molecule in advanced-stage melanomas. Oncogene 2007; 26:4216–4225 [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010; 468:973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Nakayama K, Gaitonde S, Goydos JS, Krajewski S, Eroshkin A, Bar-Sagi D, Bowtell D, Ronai Z. The ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by HIF-dependent and -independent pathways. Proc Natl Acad Sci USA. 2008. 105:16713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud N, Sonenberg N, Ruggero D, Schneider RJ. Translational Control in Cancer. Cold Spring Harb Perspect Biol. 2018; pii: a032896. doi: 10.1101/cshperspect.a032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald I, Wang S, Savas L, Woda B, Pullman J. Expression of translation initiation factor eIF-2alpha is increased in benign and malignant melanocytic and colonic epithelial neoplasms. Cancer. 2009; 8:1080–88. [DOI] [PubMed] [Google Scholar]
- Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med. 2016; 16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009; 4:275–7. [DOI] [PubMed] [Google Scholar]
- Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018. 18: 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, Weissman JS, Fuchs E. Translation from unconventional 5' start sites drives tumour initiation. Nature 2017; 541:494–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta. 2014; 1843:75–85 [DOI] [PubMed] [Google Scholar]
- Teh JLF, Aplin AE. Playing the melanoma endgame. Clin Cancer Res 2018; pii: clincanres.0989.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JJ, Abed M, Heuberger J, Novak R, Zohar Y, Beltran Lopez AP, Trausch-Azar J5, Ilagan MXG, Benhamou D, Dittmar G, Kopan R, Birchmeier W, Schwartz AL, Orian A. RNF4-Dependent Oncogene Activation by Protein Stabilization. Cell Rep. 2016;16:3388–400. doi: 10.1016/j.celrep.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WB, Aiba I, Long Y, Lin HK, Feun L, Savaraj N, Kuo MT. Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res. 2012; 72:2622–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes & Dev. 2012; 26:1131–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid M, Jawed A, Mandal RK, Dar SA, Akhter N, Somvanshi P, Khan F, Lohani M, Areeshi MY, Haque S. (2018) Recent developments and obstacles in the treatment of melanoma with BRAF and MEK inhibitors. Crit Rev Oncol Hematol. 2018; 125:84–88. [DOI] [PubMed] [Google Scholar]
- Wang JY, Liu GZ, Wilmott JS, La T, Feng YC, Yari H, Yan XG, Thorne RF, Scolyer RA, Zhang XD, Jin L. Skp2-Mediated Stabilization of MTH1 Promotes Survival of Melanoma Cells upon Oxidative Stress. Cancer Res. 2017; 77:6226–6239. [DOI] [PubMed] [Google Scholar]
- Wek RC. Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb Perspect Biol. 2018; pii: a032870. doi: 10.1101/cshperspect.a032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets related to this article can be found at [URL/s linked to dataset/s], hosted at GSE124064