Abstract
The scientific community identified non stool-based biomarkers as the way forward to support soil-transmitted helminth (STH; Ascaris lumbricoides, Trichuris trichiura and the hookworms Ancylostoma duodenale and Necator americanus) and schistosome (S. mansoni and S. haematobium) deworming programs. This support is needed in making the decision of whether or not to stop preventive chemotherapy intervention efforts and to ultimately transition towards a post-intervention surveillance phase. We applied a two-step micro-array approach to identify antigenic linear epitopes in the STH and S. mansoni proteomes. In a first experiment, we identified antigenic peptides by applying sera from 24 STH and/or S. mansoni infected Ethiopian children on a high-density peptide microarray containing 3.3 million peptides derived from the complete STH and S. mansoni proteomes. A second array experiment with 170,185 peptides that were recognized in the first array was designed to identify non-specific antibody reactivity by applying sera from 24 healthy individuals from Belgium (a non-endemic country). From this array testing cascade, several peptides were identified for STH but none of them appeared to be unique for one species. We therefore concluded that for STH, none of the peptides revealed to be sufficiently sensitive or species specific. For S. mansoni, some promising peptides were identified prompting future investigation. Based on these results, it is unlikely that linear epitopes would be highly useful in detecting species-specific antibody responses to STH in endemic communities. For S. mansoni, one particular peptide of the micro-exon gene 12 (MEG-12) protein deserves further research. In addition, this study emphasizes the need of well-characterized biobanks for biomarker discovery, particularly when the integration of multiple disease programs is envisioned.
Author summary
Today, infections with intestinal (roundworms, whipworm and hookworms) and blood-dwelling worms (schistosomes) are traditionally diagnosed by demonstrating worm eggs in stool. This current practice comes with some important challenges, including but not limited to the low-throughput and the need of skilled operators. Especially in the context of monitoring and evaluation, there is a need for alternative tools. Therefore, one of the recommendations of the scientific community was to develop tests that are based on the detection of antibodies in blood, which reflect the natural immune response of the host to worm-specific components or antigens. In the present study, we screened the antigenicity of all the peptides that build up the proteome of intestinal and blood-dwelling worms. For intestinal worms, our results revealed that the antibody response to these peptides was either not unique for the worm species or absent in infected subjects. For blood-dwelling worms, the findings were less sobering, with a number of peptides inducing an antibody response that was only observed in infected subjects.
Introduction
Nearly two decades ago, during the 54th World Health Assembly, resolution WHA54.19 was approved. This resolution urged member states to increase efforts to control two of the most impactful neglected tropical diseases (NTDs), namely soil-transmitted helminthiasis (caused by Ascaris lumbricoides, Trichuris trichiura and the hookworms Ancylostoma duodenale and Necator americanus) and schistosomiasis (predominantly caused by Schistosoma mansoni and S. haematobium) by ensuring access to essential drugs. Combining their burden of disease, they affect an estimated 1 billion individuals worldwide, resulting in a health toll of over 3.3 million daily adjusted life years [1,2].
Periodic administration of anthelmintic drugs to at-risk populations has long been a cornerstone in the control of both soil-transmitted helminth (STH) and Schistosoma mansoni (SCH) infections. In 2012, continued support for control efforts and drug availability was stimulated and safeguarded with the development of the NTD roadmap for STH and SCH infections and the London declaration on NTDs [3]. For example, treatment coverage in school-aged children (SAC) for STH and SCH infections has since then increased from approximately 29% and 23% to 60% and 61% in 2018, respectively [4]. The recently published NTD roadmap for 2021–2030 reflects the continued ambition to continue the fight against these NTDs [5], with the specific aim of reaching elimination of soil-transmitted helminthiasis and schistosomiasis as a public health problem in 96% and 100% of the endemic countries, respectively. However, in order to monitor whether programs have reached these ambitious targets, the development and availability of sensitive and specific diagnostics is critical.
Today, standard diagnosis of STH and SCH infections is made by examining stool for the presence of helminth eggs using the Kato-Katz method (STH and S. mansoni infections) or by urine filtration (S. haematobium) [6,7]. Although both these methods are relatively simple and cheap, they are labor intensive, depend on human interpretation of the results (i.e. skills to recognize eggs in sample), have a limited sensitivity to detect low-intensity infections [8,9] and come with significant logistical challenges associated with sample collection and turnaround time (sample should be examined as soon as possible after collection). Therefore, as programs move towards a verification of elimination as a public health problem and examine the prospects of breaking disease transmission [10,11], the usefulness of these traditional methods is questionable. The use of other, sensitive and specific, high-throughput diagnostic options will be needed to measure progress towards WHO goals.
In 2018, the STH community produced target product profiles (TPPs) for each of the four different phases of a control program (use-case 1: determine disease transmission and identify type of mass drug administration (MDA), use-case 2: assess progress against program goals, use-case 3: confirm a decision to stop intervention and transition into surveillance, and use-case 4: verify a sustained break in transmission [12]). It was concluded that a diagnostic biomarker for implementation toward the later stages of a control program (use case 3&4) should be present and detectable in a readily accessible body fluid, excluding stool [12,13]. Moving away from stool as a diagnostic matrix provides some advantages including (i) increased sample throughput (will be necessary as sample sizes increase), (ii) improved compliance rates, especially in populations older than SAC and woman of child-bearing age, (iii) higher confidence regarding sample origin (collection on the spot, no stool exchanges or contaminations possible), (iv) no personnel and environmental contamination during field collection, lab analysis and discarding the leftover stool samples, (v) use of methods with higher sensitivity to detect parasite exposure compared to the detection of parasite eggs or DNA in the stool (which does not always accurately reflect exposure status [14], especially in low endemic settings) and (vi) finally, it would pave the way to a more integrated NTD control program, combining serological markers for multiple other co-endemic NTDs (e.g. lymphatic filariasis, onchocerciasis, and strongyloidiasis) for which serum-based diagnostic solutions already exist and are being used [15–17]. To date, the community lacks appropriate tools to confirm a break in transmission both for STH infections. For SCH the introduction of lateral flow tests detecting Schistosoma gut-associated polysaccharides (POC-CCA and POC-CAA) has shown great promise but especially in low endemic regions its accuracy has been questioned [18–20].
With the availability of parasite genomes and transcriptomes and the advances in high throughput technologies (e.g. microarray), it is now possible to screen for epitopes in numerous antigenic targets at once or even in complete proteomes [21–23]. In recent years, immunomics-based studies have identified novel antibody targets for schistosomiasis [24–25], hookworm [26] and other NTDs [21] by screening a pre-selected number of target proteins. However, scanning the complete proteome of any STH or SCH species has so far not been performed. The aim of this study was therefore to identify antigenic linear epitopes in the complete STH and S. mansoni proteome by performing two consecutive microarray experiments. The identified peptides could eventually support the development of novel diagnostic assays to be implemented in the different stages of STH and SCH control programs.
Materials and methods
Ethics statement
Human serum samples from Ethiopia were collected as part of a serological and parasitological screening in SAC and adults from Jimma, Ethiopia [27]. For the collection of stool and blood samples, ethical approval was obtained from the Institutional Review Board of both Ghent University, Belgium (reference number: 2015/0801 and study registration number: B670201526293) and Jimma University, Ethiopia (reference number: RPGC/181 and IHRPGD/680). Written informed consent was obtained from the parent/guardian of each participant under 18 years of age.
Plasma samples from Belgian healthy controls were collected as part of a biobank study in Beerse, Belgium [28–32]. The Ethics Committee of “ZiekenhuisNetwerk Antwerpen” and the ethics committee of the Antwerp University Hospital approved the protocol. Written informed consent was obtained from all individuals, and all samples were decoded and de-identified before they were provided for research purposes.
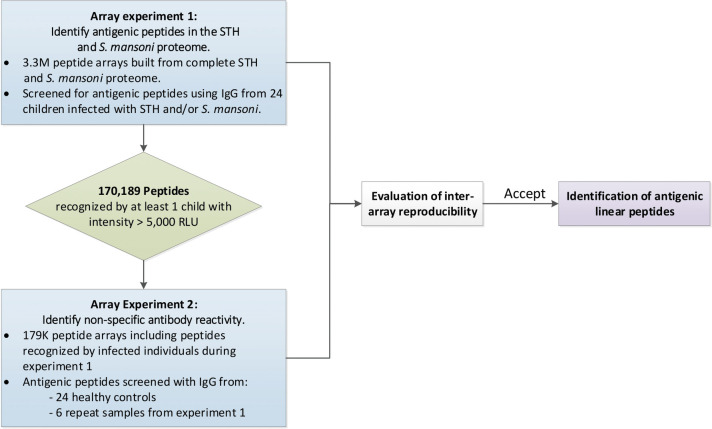
In this manuscript we describe the results of a two-step micro-array approach to identify antigenic linear epitopes in the complete STH and SCH proteomes. Fig 1 provides a schematic overview of the experiments described in this manuscript. In a first array experiment (array experiment 1), we identified antigenic STH and SCH peptides by applying sera from 24 STH and/or SCH infected children on a high-density peptide microarray containing 3.3 million peptides derived from the complete STH and SCH proteomes. In a second array experiment (array experiment 2), we identified any non-specific antibody reactivity. For this, we applied sera from 24 healthy individuals from Belgium (a non-endemic country for STH/SCH) on an array containing a subset of 179 thousand peptides that were detected in the array experiment 1 by at least one individual. To assess the inter-array reproducibility, six samples were included in both experiments. Following the assessment of the reproducibility across arrays, we developed a final list of antigenic linear peptides for each of the different helminth species.
Fig 1. Overview of the different operational steps.
(STH = soil-transmitted helminth, RLU = relative light units).
Array experiment 1: Identify antigenic peptides in the STH and S. mansoni proteome
Array design and synthesis
A total of 110,208 predicted protein sequences were used for array design. These data were obtained from WormBase ParaSite [33]. Table 1 provides more details on the used protein databases. A list of 2,832,723 peptides (13- to 16-mers) was generated from these proteins, by tiling all proteins using a 5-residue offset. A total of 165,854 peptide sequences from the worm proteomes were considered redundant as they were either repeated within the same organism or found across different organisms.
Table 1. Overview of STH and S. mansoni proteome databases used for array design.
Databases were obtained from the Wormbase Parasite repository in March 2017 [33].
| Organism | Reference genome | Number of predicted proteins | Total protein length (amino acids) |
|---|---|---|---|
| Ascaris lumbricoides | PRJEB4950 v1.5.4 | 23,604 | 7,164,414 |
| Ascaris suum | PRJNA80881 v1.0 | 18,542 | 6,058,746 |
| Trichuris trichiura | PRJEB535 v2.1 | 9,650 | 4,186,051 |
| Ancylostoma duodenale | PRJNA72581 v2.2 | 27,485 | 5,832,435 |
| Necator americanus | PRJNA72135 v1 | 19,153 | 5,136,657 |
| Schistosoma mansoni | PRJEA36577 v5.2 | 11,774 | 5,608,274 |
| Total | 110,208 | 33,986,577 |
The array also included control peptides, including a scramble peptide (WTKTPDGNFQLGGTEP) [34], one John Cunningham virus peptide (JCV: NLVRDDLPALTSQEI) [31] and two Epstein-Barr viral peptides (EBV1: PGRRPFFHPVGEADY and EBV2: FHPVGEADYFEYHQE) in 135-flold. The scramble peptide was included to confirm that the arrays did not detect a response for such a non-relevant peptide (i.e. average signal for this scramble peptide <5,000 relative light units (RLU) for all samples). Viral peptides were included to determine whether the arrays could detect an immune response against these viral peptides (i.e. average signal for this viral peptide >5,000 RLU) at a frequency similar to the seroprevalence described in literature for these viruses (82–94% for EBV and 64% for JCV [31,35]). Additionally, 2,220 peptides that originated from the Onchocerca volvulus proteome were included in 4-fold. Half of these peptides (1,110) were not recognized by O. volvulus infected individuals. The other 1,110 peptides included those against which the highest reactivity in O. volvulus patients was measured on a similar immune-array previously performed [23]. These O. volvulus peptides were included to assess the antigenicity of these peptides in a tropical, but non-Onchocerca endemic population.
Each peptide microarray slide (Roche Nimblegen) represented an entire set of peptides and one array per sample was used for screening. Peptides were synthesized in situ from C- to N-terminus and Serine was added N-terminally for improved solubility to conduct serum IgG profiling (NxT-Dx, Vienna, Austria). A list of all peptides, including their sequence and position on the arrays is available at https://osf.io/q5s9y/?view_only=8a464d9eafe6444b83b967a11f3211cd.
Selection of serum samples
For the first array experiment, a panel of 24 serum samples from Ethiopian SAC with known STH and S. mansoni infections were selected. Table 2 provides an overview of the samples and their parasitological measurements. Donated stool samples were analyzed by three copromicroscopic methods to reduce the chance of false negative samples. Each sample was analyzed by single Kato-Katz, McMaster and Mini-FLOTAC method according to standard protocols [6,36,37]. Additionally, serum IgG4 response to Ascaris lung stage L3 larval extract was analyzed as previously published [27]. Thus, only Ascaris EPG positive children that showed an Ascaris ELISA optical density ratio (ODr) over the cut-off for positivity of 0.08 were considered for inclusion as Ascaris positive samples. Samples that were copropositive for T. trichiura and hookworm or S. mansoni had to show negative Ascaris ELISA values (<0.08 ODr) before being included in the sample set. Antibody responses to other helminth antigens were not determined. A total of 10 samples were positive for A. lumbricoides, 11 for T. trichiura, 9 for hookworm and 7 for S. mansoni.
Table 2. Characteristics of the sera used to screen peptide microarrays.
Fecal egg counts (FEC; expressed as eggs per gram of stool (EPG) were based on a single stool sample. For STH, they represent the average FECs across single Kato-Katz, McMaster and Mini-FLOTAC method, whereas for S. mansoni, they represent the FEC based on a single Kato-Katz. For the Ascaris LungL3 IgG4 ELISA test, samples with an optical density ratio (ODr) of at least 0.08 were considered positive [27]. The grey cells indicate a positive test result for a given helminth or diagnostic test. Samples with an asterix (*) were included in both array experiments.
| Fecal egg count (EPG) | Ascaris LungL3 IgG4 ELISA (ODr) | Group | ||||||
|---|---|---|---|---|---|---|---|---|
| Sample ID | Age | Sex | A. lumbricoides | T. trichiura | hookworm | S. mansoni | ||
| 2015_287 | 6 | F | 1,626 | 0 | 0 | 0 | 0.147 | Al |
| 2015_330* | 14 | F | 545 | 0 | 0 | 0 | 0.428 | Al |
| 2015_129 | 8 | M | 16 | 0 | 0 | 0 | 0.613 | Al |
| 2015_218 | 14 | F | 34,699 | 1,523 | 0 | 0 | 1.063 | Al+Tt |
| 2015_268 | 14 | F | 1,715 | 496 | 0 | 0 | 0.664 | Al+Tt |
| 2015_229 | 17 | F | 120 | 0 | 1,773 | 0 | 1.266 | Al+Hw |
| 2015_293 | 9 | M | 3,565 | 0 | 403 | 0 | 0.102 | Al+Hw |
| 2015_234 | 15 | M | 331 | 0 | 53 | 0 | 0.409 | Al+Hw |
| 2015_132 | 9 | M | 24 | 0 | 0 | 480 | 0.433 | Al+Sm |
| 2015_499* | 15 | M | 17,500 | 0 | 0 | 360 | 0.045 | Al+Sm |
| 2015_587* | 5 | M | 0 | 5,389 | 0 | 0 | -0.057 | Tt |
| 2015_579 | 6 | M | 0 | 680 | 0 | 0 | -0.126 | Tt |
| 2015_285 | 9 | F | 0 | 582 | 0 | 0 | -0.106 | Tt |
| 2015_334 | 9 | F | 0 | 436 | 357 | 0 | -0.055 | Tt+Hw |
| 2015_537 | 6 | F | 0 | 119 | 1,005 | 0 | -0.109 | Tt+Hw |
| 2015_447 | 18 | F | 0 | 236 | 177 | 0 | -0.079 | Tt+Hw |
| 2015_173 | 14 | M | 0 | 69 | 0 | 1,296 | -0.030 | Tt+Sm |
| 2015_180 | 15 | M | 0 | 93 | 0 | 144 | -0.041 | Tt+Sm |
| 2015_549* | 16 | M | 0 | 643 | 0 | 528 | -0.133 | Tt+Sm |
| 2015_335 | 8 | F | 0 | 0 | 556 | 0 | -0.057 | Hw |
| 2015_368 | 18 | M | 0 | 0 | 122 | 0 | -0.010 | Hw |
| 2015_48* | 14 | M | 0 | 0 | 268 | 0 | -0.050 | Hw |
| 2015_338* | 8 | F | 0 | 0 | 0 | 1,248 | -0.098 | Sm |
| 2015_341 | 9 | M | 0 | 0 | 0 | 1,128 | -0.006 | Sm |
Serum sample processing
IgG from the serum samples was prepared by a simple spin-column purification procedure using Melon gel (Pierce, cat nr: 45206). Briefly, 15 μL of serum was diluted in 95 μL of Melon gel purification buffer, loaded onto Melon gel spin columns and centrifuged. The flow-through, containing pure IgG was collected and IgG concentrations were measured. Standardized IgG concentrations (0.025 mg/mL; final dilution in BLOTTO based binding buffer) of purified IgG were used for microarray analyses.
IgG microarray analysis
Microarray experiments were performed by NXT-Dx (NxT-Dx, Vienna, Austria). In brief, microarrays were washed once in TBST (Tris-Buffered Saline, 0.05% Tween 20), four times in TBS (Tris-Buffered Saline) and once in H2O. Microarrays were incubated at 4°C overnight with 5 μL of purified IgGs (final concentration 0.1 mg/mL) in 1% Alkali-soluble Casein in TBST. The microarrays were washed three times with TBST and incubated with secondary antibody (Alexa Fluor 647-AffiniPure Goat Anti-Human IgG, Fcγ Fragment Specific, Jackson 109-605-098) diluted 1:10,000 in 1% Alkali-soluble Casein TBST for 3 hours at room temperature. Finally, microarrays were washed three times in TBST, once in H2O and dried. Microarray fluorescent-signals were extracted from microarray images, and data sets were subjected to further statistical analysis in order to identify immunoreactive peptides.
Array experiment 2: Identify non-specific antibody reactivity
Based on the results obtained from array experiment 1, a second, 179K peptide array was designed. A total of 170,185 peptides (13- to 16-mers) that presented signals of at least 5,000 RLU in at least one of the 24 samples screened on array experiment 1 were included in a smaller, follow-up array (array experiment 2). The threshold of 5,000 RLU was chosen based on previous work using the same type of peptide arrays [23]. A conservative selection (only 1 positive sample was sufficient to retain the peptide) was used because of the limited sample size and to ensure inclusion of all possibly antigenic peptides.
Array design and synthesis
Except for the absence of the two influenza hemagglutinin peptides, other control peptides were the same as described in array experiment 1. Peptide microarray slides (Roche Nimblegen) were divided into 12 subarrays, each presenting the entire set of peptides. One subarray per sample was subsequently used for screening. A list of all peptides, including their sequence and position on the arrays is available at https://osf.io/q5s9y/?view_only=8a464d9eafe6444b83b967a11f3211cd.
Selection of serum samples
A total of 30 samples were run on this second array. These included a panel of 24 plasma samples from Belgian healthy controls and six samples that were also used in the previous array (array experiment 1) in order to evaluate inter-array reproducibility of the results (these six samples are indicated in Table 2). Since Belgium is a non-endemic country for STH and SCH, it was assumed Belgian healthy controls were not exposed to STH or SCH. Serum sample processing and IgG microarray analysis were performed identical as during array experiment 1.
Evaluation of inter-array reproducibility
To compare antibody responses measured in the 3.3M array and the 179K array, the data from both arrays were normalized together. Quantile normalization on base-2 log transformed data was used to correct for technical differences between arrays. Reproducibility between the two experiments was assessed for each of the six samples that were included on both arrays by correlating peptide specific antibody reactivity measured on the arrays. Correlations were performed using Spearman correlation. Statistical analysis was performed in R [38]. Both arrays were considered to be moderately or highly correlated in case Spearman’s correlation coefficient is >0.5 or >0.7, respectively [39,40].
Identification of antigenic linear peptides
In order to identify antigenic parasite-specific peptides, for each peptide the number of positive samples (“positive” is defined as signal >5,000 RLU) was determined. Peptides were considered to be antigenic when (i) at least one of the specific helminth infected individuals was positive, and when (ii) not more than 1 out of 24 (i.e. <4.2%) healthy controls was positive. For peptides that fulfilled these two criteria, we calculated the average response of the infected group minus the average response of the healthy controls expressed in RLUs (= the Δ-value). Finally, the set of peptides was ranked based on this Δ-value to identify promising linear peptides.
Peptide ELISA
Peptide ELISA’s were performed as described before [23,41–43].
Results
Array experiment 1: Identify antigenic peptides in the STH and S. mansoni proteome
We produced an array that contained 2,832,723 peptides representing a total of 110,208 protein sequences across the six different helminth species (Table 1) (A. lumbricoides, A. suum, T. trichiura, A. duodenale, N. americanus, and S. mansoni). These arrays were used to assess the IgG responses in 24 STH and/or S. mansoni infected individuals (Table 2).
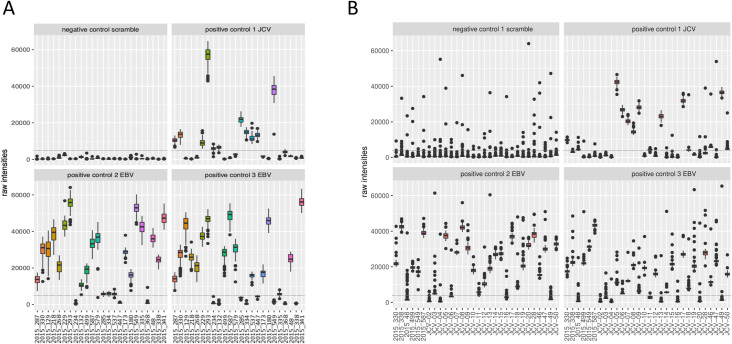
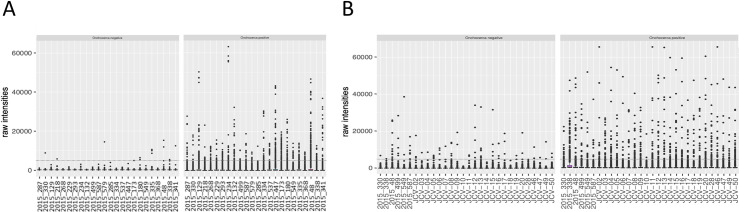
Even though there was some variability between sample reactivity to the negative scramble peptide, none of the samples showed an outlying signal above the threshold (5,000 RLU). Most of the subjects had high antibody responses against the two Epstein-Barr viral peptides (20 out of 24 for EBV1 and 16 out of 24 for EBV2). Fewer subjects reacted to the one John Cunningham virus peptide (JVC; 11 out of 24). Both the EBV and JCV peptides were comparable to the described prevalence of 82–94% and 64%, respectively (Fig 2A and S1 Info)). Non-reactive O. volvulus peptides showed very low reactivity, with only 1 peptide reacting consistently in 1 sample (OVOC5335; 523), indicating that these non-reactive O. volvulus peptides are also non-reactive in this non-Onchocerca endemic population. The highly reactive O. volvulus peptides were mostly negative, but some peptides were consistently found to be positive in one or more samples (Fig 3A and S1 Info).
Fig 2. Box plots of raw intensities measured against control peptides in both microarray experiments.
This figure represents the raw output values of IgG reactivity (y-axis) of the different samples (x-axis) to the different control peptides during array experiment 1 (A) and array experiment 2 (B). Both arrays included a scramble peptide (negative control), one John Cunningham virus (JCV) peptide and two Epstein-Barr viral peptides (EBV1 and EBV2) in 135-flold. Dotted line indicates the cut-off for positivity (5,000 relative light units (RLU)).
Fig 3. Box plots of raw intensities measured against Onchocerca volvulus peptides in both microarray experiments.
This figure represents the raw output values of IgG reactivity (y-axis) of the different samples (x-axis) to the 1,110 negative and 1,110 positive O. volvulus peptides included on the microarray during both array experiment 1 (A) and array experiment 2 (B). Dotted line indicates the cut-off for positivity (5,000 relative light units (RLU)).
Based on the data obtained from the 3.3M peptide arrays, a set of antigenic linear peptides was selected by filtering all peptides for which at least one sample had a signal above a threshold of 5,000 RLU. This resulted in a list of 170,185 peptides, comprising a total of 33,890 peptides that were selected from the proteome of A. lumbricoides; 29,424 from A. suum; 20,021 from T. trichiura; 28,669 from A. duodenale; 25,765 from N. americanus and 32,416 from S. mansoni. Of these 170,185 peptides, 150,451 peptides were unique, while 19,734 were present in more than one proteome.
Array experiment 2: Identify non-specific antibody reactivity
A second array was produced with the set of 170,185 peptides that were recognized (produced a signal >5,000 RLU) by at least one of the STH and/or S. mansoni infected children included in array experiment 1. On this second array, the IgG responses of 24 healthy control samples from Belgium to this set of peptides were assessed. Additionally, six samples were included that were also included in array experiment 1 to assess the reproducibility across both arrays (samples included in both arrays are indicated in Table 2). Similar to the 3.3M arrays, none of the analyzed samples showed an average signal for the negative control peptides above the threshold of 5,000 RLU, but some outliers were observed in the replicates (76 out of 2,880, i.e. 0.026%), indicating slightly increased variability on detection of non-antigenic peptides in the second array. Most of the subjects showed high IgG reactivity against the EBV peptides (25 out of 30 for EBV1 and 21 out of 30 for EBV2) and 13 out of 30 subjects reacted to the JCV peptide. (Fig 2B and S2 Info). These data for the positive control peptides of this second array experiment are comparable to those of the first array and to the described prevalence. Non-reactive O. volvulus peptides showed again very low reactivity, except for some outliers, indicating that these non-reactive O. volvulus peptides are also non-reactive in this healthy control population. The highly reactive O. volvulus peptides were mostly negative, but some peptides were found to be positive in one or more samples (Fig 3B and S2 Info).
Evaluating inter-array reproducibility
The inter-array reproducibility between the 3.3M array and the 179K array was assessed by comparing the normalized profiles obtained in both experiments for six samples. Spearman correlation analysis revealed a high positive linear correlation between repeatedly analyzed peptides in five samples and moderate correlation in one sample (correlation coefficient range between 0.67 and 0.81 with p-value <0.001, S1 Fig).
Identifying infection-specific antigenic peptides
For all peptides that were included in the 179K array, the number of positive samples (defined as signal >5,000 RLU) were determined both for subjects with a specific helminth infection (data from array experiment 1: A. lumbricoides: 10, T. trichiura: 11, hookworm: 9 and S. mansoni: 8) and healthy subjects (data from array experiment 2: 24) (All data available at https://osf.io/q5s9y/?view_only=8a464d9eafe6444b83b967a11f3211cd). Peptides for which at least one infected individual and not more than 1 out of 24 (i.e. ≤ 4.2%) healthy controls were positive (signal >5,000 RLU) were selected and the resulting set of peptides was sorted based on the Δ-value (= average response in the infected group—average response in healthy controls). For each infection, the 20 peptides with the highest Δ-value are listed in Table 3. Several peptides in this list were found to be strongly antigenic in several infection groups (e.g. ALUE_0000851001_mRNA_1;551). It is however not clear from the current data whether these peptides are truly pan-helminth markers or whether this is caused by the fact that several of the investigated individuals had multiple infections or had experienced prior exposure to the other STHs but without a patent infection at the moment of sample collection.
Table 3. Top 20 peptides identified for each infection.
This table reports the 20 peptides with the highest Δ-values for each infection. The Δ-values were calculated by subtracting the average intensity (in relative light units (RLU)) in healthy controls (HC) from the average intensity (in RLU) in the infected group. The number of positive samples (i.e. signal > 5,000 RLU) for both the infected group (n = 10 for A. lumbricoides, n = 11 for T. trichiura, n = 9 for hookworm and n = 7 for S. mansoni) and the healthy control group (n = 24) is indicated for each of the peptides listed.
| Species | Peptide ID | Uniprot Protein name | Peptide sequence | Number of positive samples (>5,000 RLU) | RLU Range Infected | RLU Range Healthy Controls | Δ (Avg. Infected–Avg. HC) | |
|---|---|---|---|---|---|---|---|---|
| Infected | Healthy controls | |||||||
| A. lumbricoides | ALUE_0000851001_mRNA_1;551 | Unknown | AKQDSFISNESTKLH | 8 | 1 | 26–46837 | 101–5111 | 21908 |
| Smp_034080.1;353 | Putative erythrocyte membrane protein | GIEAIKQNDESEFPNN | 8 | 1 | 294–48867 | 31–8289 | 18297 | |
| NECAME_06710;144 | HEAT repeat protein | INESSMKQASLISNVM | 7 | 1 | 19–49175 | 23–5893 | 15963 | |
| ALUE_0001246601_mRNA_1;617 | DNA-directed RNA polymerase III subunit RPC3 | ERQLKMESIIANIEAD | 6 | 0 | 37–44595 | 41–3581 | 15640 | |
| ANCDUO_02559;123 | Zinc finger, C3HC4 type | GIDEAVLRRTVDDLLT | 9 | 0 | 2951–47823 | 30–4345 | 14554 | |
| GS_23998;441 | Unknown | LIERQLKMESIIANIE | 6 | 0 | 20–41166 | 22–3109 | 14321 | |
| Smp_152630.1;1 | MEG-12 | GENYEQQLQQPKAYGI | 4 | 0 | 10–62200 | 22–1453 | 13232 | |
| TTRE_0000850001_mRNA_1;188 | 205 kDa Pk1(B+)1+ SICAvar antigen | GVTANDLRTAEAMVRS | 6 | 0 | 8–46446 | 19–571 | 13109 | |
| NECAME_05279;46 | Uncharacterized protein | MAKEDGLISNLRSYPP | 7 | 1 | 49–41397 | 22–5999 | 13088 | |
| Smp_063110.2;144 | Putative zinc finger protein | GIEQQVILQPAAPVAM | 6 | 0 | 240–40053 | 22–3087 | 12608 | |
| NECAME_07428;155 | PAP2_C domain-containing protein | GIEQLRPRELCGDLIV | 7 | 0 | 962–34212 | 26–4284 | 12265 | |
| GS_23620;782 | Unknown | GIEEMTPSKKANNTAG | 8 | 1 | 68–45073 | 28–8035 | 12252 | |
| Smp_103170.1;78 | Unknown | PPNYTPERYLEMGNRN | 4 | 1 | 431–51442 | 188–8920 | 12195 | |
| Smp_151380.1;507 | Anaphase-promoting complex subunit 1 | KTESLISNSPSTKKSK | 6 | 0 | 9–40354 | 22–2015 | 12078 | |
| ALUE_0000793101_mRNA_1;694 | Unknown | GIEKIREKLNPFVVEK | 5 | 1 | 182–51442 | 50–10302 | 11861 | |
| NECAME_00764;3928 | Uncharacterized protein | LKKIIGGHLQMPTAIV | 5 | 0 | 6–57516 | 20–386 | 11605 | |
| ANCDUO_18503;111 | Uncharacterized protein | SKFDSVILNGYNIVGD | 6 | 0 | 5–50886 | 20–895 | 11425 | |
| TTRE_0000749301_mRNA_1;12 | Unknown | KTVPPRKKDSEISNSV | 6 | 0 | 91–32314 | 26–2676 | 11359 | |
| GS_00707;166 | Unknown | GIDRVIDAFRNAYTQV | 4 | 0 | 19–33437 | 23–1963 | 11106 | |
| NECAME_01953;144 | Uncharacterized protein | KNLISAQLNQTATGQP | 6 | 0 | 1131–39427 | 36–4993 | 10934 | |
| T. trichiura | ALUE_0000851001_mRNA_1;551 | Unknown | AKQDSFISNESTKLH | 10 | 1 | 4645–48005 | 101–5111 | 21749 |
| ANCDUO_02559;123 | Zinc finger, C3HC4 type | GIDEAVLRRTVDDLLT | 11 | 0 | 5567–47823 | 30–4345 | 21584 | |
| Smp_034080.1;353 | Putative erythrocyte membrane protein | GIEAIKQNDESEFPNN | 10 | 1 | 294–38372 | 31–8289 | 21424 | |
| NECAME_01953;144 | Uncharacterized protein | KNLISAQLNQTATGQP | 6 | 0 | 44–53181 | 18–2211 | 16910 | |
| GS_00707;166 | Unknown | GIDRVIDAFRNAYTQV | 8 | 0 | 1356–39427 | 36–4993 | 15994 | |
| NECAME_06710;144 | HEAT repeat protein | INESSMKQASLISNVM | 8 | 1 | 733–43011 | 23–5893 | 15854 | |
| Smp_152630.1;1 | MEG-12 | GENYEQQLQQPKAYGI | 5 | 0 | 8–61569 | 22–1453 | 15837 | |
| ALUE_0001246601_mRNA_1;617 | DNA-directed RNA polymerase III subunit RPC3 | ERQLKMESIIANIEAD | 7 | 0 | 646–45073 | 41–3581 | 14147 | |
| GS_23620;782 | Unknown | GIEEMTPSKKANNTAG | 8 | 1 | 68–49508 | 28–8035 | 13550 | |
| ANCDUO_21963;35 | Uncharacterized protein | GVEHLARVNCVLWQND | 7 | 0 | 485–46611 | 52–4998 | 13079 | |
| TTRE_0000257301_mRNA_1;122 | Unknown | GIDERLAHVWQKTTNA | 6 | 1 | 1231–45116 | 59–10335 | 12956 | |
| GS_23998;441 | Unknown | LIERQLKMESIIANIE | 7 | 0 | 440–40911 | 22–3109 | 12878 | |
| Smp_063110.2;144 | Putative zinc finger protein | GIEQQVILQPAAPVAM | 9 | 0 | 240–34765 | 22–3087 | 12661 | |
| ANCDUO_10263;496 | Putative polyribonucleotide nucleotidyltransferase | GIDHVLDKMAVMLDRP | 8 | 1 | 2039–40107 | 25–6745 | 12588 | |
| ANCDUO_05567;1 | Uncharacterized protein | MEIANGKEWKSDVRET | 5 | 0 | 20–51748 | 23–2684 | 12323 | |
| GS_00126;243 | Unknown | GVLEMIKKYLINYEHM | 5 | 1 | 7–50732 | 32–5119 | 12294 | |
| NECAME_02686;166 | Unknown | DKIVSEASRYKDMLQF | 5 | 1 | 366–54645 | 60–7441 | 12170 | |
| GS_08916;309 | Unknown | GIDDIDEDELECILAN | 7 | 1 | 3027–40053 | 33–8529 | 12008 | |
| TTRE_0000898701_mRNA_1;67 | Unknown | GTVYKLQEMKPSVVKE | 4 | 0 | 4–51748 | 20–895 | 11972 | |
| ALUE_0001542901_mRNA_1;23 | Unknown | ITCKVTGRYTSALENL | 6 | 0 | 26–60526 | 19–941 | 11783 | |
| Hookworm | Smp_034080.1;353 | Putative erythrocyte membrane protein | GIEAIKQNDESEFPNN | 8 | 1 | 3894–36306 | 31–8289 | 18206 |
| ANCDUO_02559;123 | Zinc finger, C3HC4 type | GIDEAVLRRTVDDLLT | 8 | 0 | 1097–37428 | 30–4345 | 17623 | |
| ALUE_0000851001_mRNA_1;551 | Unknown | AKQDSFISNESTKLH | 5 | 1 | 26–44812 | 101–5111 | 13798 | |
| GS_23620;782 | Unknown | GIEEMTPSKKANNTAG | 7 | 1 | 1542–34806 | 28–8035 | 11960 | |
| ALUE_0002154501_mRNA_1;221 | Uncharacterized protein | FKELANKLLNKQQLAE | 6 | 1 | 932–52374 | 44–5966 | 11891 | |
| ALUE_0001868101_mRNA_1;34 | Unknown | SLAKKLFGSSAGLNDD | 4 | 0 | 183–43591 | 30–1890 | 11671 | |
| ANCDUO_21963;35 | Uncharacterized protein | GVEHLARVNCVLWQND | 7 | 0 | 485–30867 | 52–4998 | 11594 | |
| GS_15548;34 | Unknown | SLAKKLFGSSAGLNDD | 4 | 0 | 183–43591 | 32–2488 | 11582 | |
| GS_00707;166 | Unknown | GIDRVIDAFRNAYTQV | 5 | 0 | 1942–31701 | 36–4993 | 11247 | |
| ALUE_0000733401_mRNA_1;298 | Unknown | AKNGRQLYDNYIEARK | 4 | 0 | 126–32809 | 27–3427 | 11236 | |
| GS_08916;309 | Unknown | GIDDIDEDELECILAN | 7 | 1 | 1710–40053 | 33–8529 | 11231 | |
| GS_01502;573 | Unknown | KNGRQLYDNYIEARKR | 4 | 0 | 78–40862 | 28–1451 | 10892 | |
| ANCDUO_10263;496 | Putative polyribonucleotide nucleotidyltransferase | GIDHVLDKMAVMLDRP | 6 | 1 | 1227–40107 | 25–6745 | 10755 | |
| ANCDUO_06401;122 | Uncharacterized protein | GPWHQKLTAKKVAKSM | 4 | 1 | 15–38687 | 20–5099 | 10624 | |
| ANCDUO_05567;1 | Uncharacterized protein | MEIANGKEWKSDVRET | 3 | 0 | 9–51748 | 23–2684 | 10587 | |
| ALUE_0001434301_mRNA_1;111 | Unknown | QDILNNKALLGDEKAR | 4 | 0 | 13–57516 | 19–941 | 10349 | |
| TTRE_0000259701_mRNA_1;67 | Unknown | ARAGLQGDLLGLNVLC | 4 | 0 | 6–38452 | 19–2095 | 10332 | |
| NECAME_02954;232 | Beta-lactamase | VATKKLIGDQMLNRLQ | 4 | 0 | 159–33821 | 23–863 | 10284 | |
| TTRE_0000798101_mRNA_1;166 | Unknown | ILKKHDKVLQNTAGNE | 3 | 0 | 43–43168 | 19–2709 | 10235 | |
| ANCDUO_24637;1 | COesterase domain-containing protein | MTKPVKKTRWHQELSA | 4 | 0 | 7–39427 | 21–3782 | 10169 | |
| S. mansoni | Smp_152630.1;1 | MEG-12 | GENYEQQLQQPKAYGI | 6 | 0 | 4443–62200 | 22–1453 | 33784 |
| NECAME_01953;144 | Uncharacterized protein | KNLISAQLNQTATGQP | 5 | 0 | 262–53181 | 18–2211 | 32222 | |
| TTRE_0000898701_mRNA_1;67 | Unknown | GTVYKLQEMKPSVVKE | 6 | 0 | 2496–51748 | 20–895 | 27482 | |
| ALUE_0000851001_mRNA_1;551 | Unknown | AKQDSFISNESTKLH | 6 | 1 | 676–48005 | 101–5111 | 25208 | |
| ANCDUO_19104;23 | Sas10 domain-containing protein | GTVRKEMQKYSGESRG | 6 | 0 | 1313–44286 | 27–2391 | 21927 | |
| NECAME_06710;144 | HEAT repeat protein | INESSMKQASLISNVM | 5 | 1 | 180–43011 | 23–5893 | 20615 | |
| NECAME_10074;1464 | Uncharacterized protein | IELDPSAATSNTLHDV | 4 | 0 | 85–62200 | 18–2359 | 20462 | |
| Smp_162000.1;6582 | UBR-type domain-containing protein | QKIAISEELNAKQINS | 5 | 0 | 383–51442 | 28–1930 | 20002 | |
| NECAME_00764;3928 | Uncharacterized protein | LKKIIGGHLQMPTAIV | 5 | 0 | 9–57516 | 20–386 | 19697 | |
| ALUE_0001246601_mRNA_1;617 | DNA-directed RNA polymerase III subunit RPC3 | ERQLKMESIIANIEAD | 4 | 0 | 146–45073 | 41–3581 | 19667 | |
| TTRE_0000164301_mRNA_1;56 | Unknown | FVVIENLSNDEKQIFT | 5 | 0 | 176–41166 | 28–2467 | 19404 | |
| TTRE_0000806301_mRNA_1;342 | Unknown | ELELDPAKTNVNGGAI | 5 | 0 | 179–60104 | 18–3029 | 19162 | |
| ALUE_0001256701_mRNA_1;1255 | Unknown | KKLISEALNSETRLQN | 5 | 0 | 466–39540 | 27–3464 | 18956 | |
| Smp_034080.1;353 | Putative erythrocyte membrane protein | GIEAIKQNDESEFPNN | 4 | 1 | 2764–39968 | 31–8289 | 17679 | |
| ALUE_0000089701_mRNA_1;89 | Uncharacterized protein | GTVLKFPSLSPFANAA | 5 | 1 | 66–40220 | 26–6676 | 17505 | |
| GS_00094;969 | Unknown | FVVKEKRNANQFDVNN | 4 | 1 | 445–42092 | 136–13481 | 17494 | |
| TTRE_0000788501_mRNA_1;287 | Unknown | YKHVIGIDLNKEAIEN | 4 | 0 | 306–39209 | 26–2762 | 16791 | |
| TTRE_0000483901_mRNA_1;89 | Unknown | VSKQTISAHLKPLGSN | 4 | 1 | 28–59047 | 22–6293 | 16727 | |
| TTRE_0000134801_mRNA_1;243 | Unknown | SPLIGKEIVSLSLNGD | 4 | 0 | 31–46525 | 22–687 | 16646 | |
| ANCDUO_07179;45 | REX1 DNA Repair | IDREIVRILEEKLDHL | 5 | 0 | 606–42756 | 27–1848 | 16589 | |
Conversion to peptide ELISA for MEG-12 peptide
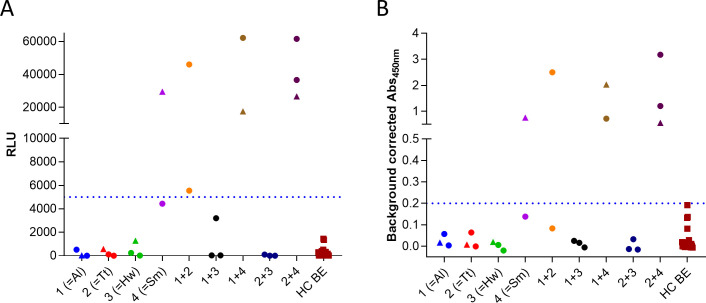
The data presented in Table 3 indicated that one particular peptide in the S. mansoni protein MEG-12 (Smp_152630.1;1) appeared to be a promising candidate to detect infection with S. mansoni. In order to confirm these findings, a peptide ELISA was setup for this peptide and the same samples as used in the arrays were tested on this assay (Fig 4B). The data obtained on the peptide ELISA were comparable to the data from the arrays (Fig 4A). One S. mansoni infected individual was found to be negative, but just below the cut-off on both the array and the ELISA. One other individual that did not have a patent infection with S. mansoni (but with A. lumbricoides and T. trichiura) was found to have a strong immune response against this MEG-12 peptide in both formats. This latter individual was a 14-year old girl for which it can’t be excluded that she had ever been infected with S. mansoni.
Fig 4.
Assessment of immune response against Smp_152630.1;1 using peptide array (A) and peptide ELISA (B). Immunoreactivity against the MEG-12 peptide was determined in peptide ELISA on the same samples that were used in the arrays. Samples that were only measured in array 1 are indicated by circles, samples that were measured in both array 1 and 2 by triangles, and samples that were measured only in array 2 by squares. Dotted line indicates the cut-off for positivity. For the peptide ELISA this was defined as the signal that corresponds to 100% specificity.
Discussion
The STH community identified non stool-based (e.g. serum and urine) biomarkers as the way forward to support programs in making the decision of whether or not to stop intervention efforts and to ultimately transit towards a sustainable post-intervention surveillance phase. In the current study, we aimed to identify a subset of linear antigenic peptides in the complete STH and S. mansoni proteome by performing two consecutive microarray experiments that could serve as potential biomarkers for helminth exposure in serum samples.
Identification of antigenic linear peptides for specific STH species revealed to be sobering
Our approach existed of two subsequent peptide microarray experiments. The first experiment was developed to identify STH and S. mansoni peptides that are recognized by antibodies from children infected with one or more of these helminth infections. This allowed us to reduce the number of target peptides from 2,832,723 to 170,185. In the second part of the experiment, we were able to eliminate 27,034 peptides which elicited a detectable response in more than one out of the 24 healthy control sera. Eventually, the resulting set of peptides was sorted based on the Δ-value (average response in the infected group—average response in healthy controls) and the 20 peptides with the highest Δ-value were reported for each helminth species. However, results of both peptide arrays indicate that the identification of antigenic linear peptides for each STH species separately is not straightforward. A huge amount of overlap is seen between the peptides detected by the children identified to be infected with one or more STHs. The major reason for this is likely the fact that it is impossible to have information on the infection history of endemic individuals. We did have a high level of certainty on the type and level of the patent infections the children were carrying at the time of sampling by performing stool analysis using multiple different diagnostic methods (Kato-Katz, McMaster and Mini-FLOTAC). However, it remains impossible to tell what infections these children have been exposed to over their lifetime. In this respect, it has been described before that seroprevalence for STH appears to be higher than coproprevalence, further supporting this hypothesis [27]. It is very likely that these children have been exposed to a number of different endemic helminth species in the past. Even some helminth species which were not investigated or for which diagnosis was not performed (e.g. Enterobius vermicularis and Strongyloides stercoralis).
Linear peptide of MEG-12 protein highly antigenic for S. mansoni infections
Although several peptides were found to be antigenic in multiple infection groups, the signal intensities of a particular peptide derived from the S. mansoni protein MEG-12 (for micro-exon gene 12) was notably higher in the S. mansoni infected group (Δ-value = 33,784; min. RLU = 4,443; max. RLU = 62,200), indicating a possible link with S. mansoni infection. The findings from the array were also confirmed in peptide ELISA. Interestingly, MEG-12 has been suggested as diagnostic marker for schistosome infection [44,45] and many of these MEGs have been shown to be immunogenic [46,47]. They have even been suggested as new group of targets that might be exploited for vaccine development [48]. Of interest, on position 100 of the top performing peptides in the array for S. mansoni, a peptide was found from MEG-2 (Smp_159800.1). Like MEG-12, also its N-terminal tail was found to be immunogenic in the array. MEG-2 is a protein that was found to be present in Schistosoma egg secretions, making it readily accessible for the immune system [49]. A more targeted evaluation of the serodiagnostic potential of these MEG proteins, either as recombinant proteins or as overlapping peptides, might be of interest for future studies. Recently, an epitope mapping study of a set of secreted or exposed proteins from the alimentary tract and tegument of S. mansoni identified exactly the same MEG-12 peptide as was detected in this study as one of the most highly immunogenic peptides and suggested it to be included in a multi-epitope vaccine construct [50]. The use of this epitope in a vaccine construct would however preclude the use of the same epitope in a serodiagnostic test. Increased emphasis on the coordination of both diagnostic and vaccine efforts would be essential to avoid vaccine-induced seroreactivity.
There is a need for well-characterized biobanks for biomarker discovery
For further downstream analysis of the usefulness of the identified peptides as well as for future studies aimed at identifying serological biomarkers, serum samples are needed which are much better defined. This could be achieved through (i) using longitudinal samples from endemic subjects with changing infection status; (ii) using samples from young children living in areas which are endemic for only a single STH (e.g. Ascaris-only positive children from region A, Trichuris only positive children from region B, etc.); (iii) using sera from experimentally infected non-endemic individuals (e.g. experimental infections in man) [51,52] or (iv) by using sera from experimentally infected animals using the original or a closely related parasite species (e.g. hookworm infections in gerbils/dogs, Schistosoma spp. or Trichuris muris infections in mice, A. suum or T. suis infections in pigs [53–56]).
The importance of well-characterized biobanks will become even more wanted in the near future as countries will attempt to integrate control or monitoring programs of multiple NTDs [57]. Clear regulations on sample sharing and the design of well-formulated informed consent forms that allow for broader sample use would be very useful for future trials. A similar discussion on the need for well-characterized biobanks has also been made for O. volvulus. Several peptides were identified that were assumed to be linked to O. volvulus infection but were later found to be linked–at least partly—to other immunogens, possibly from parasitic origin [58].
Limitations of linear peptide microarrays
Microarray results show that a great number of the peptides detected by the different infected populations actually originate form proteomes of other helminths. We can think of two explanations for this. First, some proteins are identical or have high degree of homology between the included helminth species. When constructing the 15-mer peptides to include onto the microarray, it is possible that for example a peptide that originates from the A. lumbricoides proteome also exists in identical or near identical form in the proteome of any of the other helminths. Second, it is so that, even though we measure the binding of antibodies to the different peptides on the microarray, this antibody affinity to these targets is not necessarily induced by that respective peptide or mother protein. Peptides could be recognized by antibodies that were initially targeting much different proteins carrying similar epitopes or mimotopes. The respective peptide thus acquires the structure of an (conformational) epitope that mimics the epitope that originally stimulated the immune system and antibody production. Our microarray approach focusses on linear epitopes. This has proven a successful approach for viral diseases like HIV and HCV [59–61]. However, our current data seem to indicate that it will not be as straightforward for helminth parasites [58]. It seems difficult to use antibody detection against linear epitopes as a good measure of helminth or parasite exposure. Maybe the use of other antibody Isotypes (IgG4) could reduce the level of cross-reactivity or be more indicative of active or recent infection (IgM). Alternatively, a set of peptides, either individually as a classifier, or as a multi-epitope construct, might be useful to cover the multitude of individual immune responses against these helminths.
Conclusion
Based on the data presented here, it is unlikely that linear epitopes will serve to be highly useful in detecting species-specific antibody responses to STH in endemic communities. This study also emphasis the need of well-characterized biobanks for serological biomarker discovery, this is in particular when integration of multiple NTD programs is envisioned. The fact that our experiments resulted in limited sensitive and specific peptides that can act in multiplex technology with performance characteristics as described for use case 4 casts a doubt over the approach [12]. The developing timelines for STH and SCH peptide-based point-of-care diagnostics will most likely become cost-prohibitive. Instead, other (non-immune based) approaches might be more useful in this respect.
Supporting information
(PNG)
(XLSX)
(XLSX)
Acknowledgments
The authors like to thank Janssen Biobank for logistic support and Benny Baeten and Marc Engelen from Janssen Global Public Health for programmatic support. We thank all study participants for providing samples and supporting the study.
Data Availability
Large data files are available at https://osf.io/q5s9y/?view_only=8a464d9eafe6444b83b967a11f3211cd, as indicated in the manuscript.
Funding Statement
The study was funded in part by the Bill and Melinda Gates Foundation (Grant no OPP1174629, https://www.gatesfoundation.org/). This grant was awarded to LJS and OL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.collaborators GDaH. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1859–922. 10.1016/S0140-6736(18)32335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789–858. Epub 2018/11/30. 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uniting to Combat Neglected Tropical Diseases. Resource. The London Declaration [18-Jan-2021]. Available from: http://unitingtocombatntds.org/resource/london-declaration.
- 4.WHO. Schistosomiasis and soil-transmitted helminthiasis: numbers of people treated in 2018. Weekly epidemiological record. 2019;50(94):601–12. [Google Scholar]
- 5.WHO. Ending the neglect to attain the sustainable development goals: A road map for neglected tropical diseases 2021–2030. Geneva, Switserland: World Health Organization, 2020. [Google Scholar]
- 6.WHO. Basic laboratory methods in medical parasitology . Geneva: World Health Organization, 1991. [Google Scholar]
- 7.WHO. Helminth control in school-age children: a guide for managers of control programmes. Geneva, Switserland: World Healt Organisation, 2011. [Google Scholar]
- 8.Nikolay B, Brooker SJ, Pullan RL. Sensitivity of diagnostic tests for human soil-transmitted helminth infections: a meta-analysis in the absence of a true gold standard. Int J Parasitol. 2014;44(11):765–74. 10.1016/j.ijpara.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mwinzi PN, Kittur N, Ochola E, Cooper PJ, Campbell CH Jr., King CH, et al. Additional Evaluation of the Point-of-Contact Circulating Cathodic Antigen Assay for Schistosoma mansoni Infection. Frontiers in public health. 2015;3:48. 10.3389/fpubh.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asbjornsdottir KH, Ajjampur SSR, Anderson RM, Bailey R, Gardiner I, Halliday KE, et al. Assessing the feasibility of interrupting the transmission of soil-transmitted helminths through mass drug administration: The DeWorm3 cluster randomized trial protocol. PLoS Negl Trop Dis. 2018;12(1):e0006166. 10.1371/journal.pntd.0006166 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jourdan PM, Montresor A, Walson JL. Building on the success of soil-transmitted helminth control—The future of deworming. PLoS Negl Trop Dis. 2017;11(4):e0005497. 10.1371/journal.pntd.0005497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim MD, Brooker SJ, Belizario VY Jr., Gay-Andrieu F, Gilleard J, Levecke B, et al. Diagnostic tools for soil-transmitted helminths control and elimination programs: A pathway for diagnostic product development. PLoS neglected tropical diseases. 2018;12(3):e0006213. Epub 2018/03/02. 10.1371/journal.pntd.0006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solomon AW, Engels D, Bailey RL, Blake IM, Brooker S, Chen JX, et al. A Diagnostics Platform for the Integrated Mapping, Monitoring, and Surveillance of Neglected Tropical Diseases: Rationale and Target Product Profiles. PLoS Negl Trop Dis. 2012;6(7). ARTN e1746 10.1371/journal.pntd.0001746 WOS:000307101900038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truscott JE, Turner HC, Farrell SH, Anderson RM. Soil-Transmitted Helminths: Mathematical Models of Transmission, the Impact of Mass Drug Administration and Transmission Elimination Criteria. Adv Parasitol. 2016;94:133–98. Epub 2016/10/21. 10.1016/bs.apar.2016.08.002 . [DOI] [PubMed] [Google Scholar]
- 15.Won KY, Sambou S, Barry A, Robinson K, Jaye M, Sanneh B, et al. Use of Antibody Tools to Provide Serologic Evidence of Elimination of Lymphatic Filariasis in The Gambia. Am J Trop Med Hyg. 2017. 10.4269/ajtmh.17-0371 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vargas P, Krolewiecki AJ, Echazu A, Juarez M, Cajal P, Gil JF, et al. Serologic Monitoring of Public Health Interventions against Strongyloides stercoralis. Am J Trop Med Hyg. 2017;97(1):166–72. 10.4269/ajtmh.16-0857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unnasch TR, Golden A, Cama V, Cantey PT. Diagnostics for onchocerciasis in the era of elimination. Int Health. 2018;10(suppl_1):i20–i6. 10.1093/inthealth/ihx047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haggag AA, Rabiee A, Abd Elaziz KM, Campbell CH, Colley DG, Ramzy RMR. Thirty-Day Daily Comparisons of Kato-Katz and CCA Assays of 45 Egyptian Children in Areas with Very Low Prevalence of Schistosoma mansoni. The American journal of tropical medicine and hygiene. 2019;100(3):578–83. Epub 2019/01/05. 10.4269/ajtmh.18-0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danso-Appiah A, Minton J, Boamah D, Otchere J, Asmah RH, Rodgers M, et al. Accuracy of point-of-care testing for circulatory cathodic antigen in the detection of schistosome infection: systematic review and meta-analysis. Bulletin of the World Health Organization. 2016;94(7):522–33A. Epub 2016/07/20. 10.2471/BLT.15.158741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulibaly JT, N’Gbesso YK, Knopp S, N’Guessan NA, Silue KD, van Dam GJ, et al. Accuracy of urine circulating cathodic antigen test for the diagnosis of Schistosoma mansoni in preschool-aged children before and after treatment. PLoS neglected tropical diseases. 2013;7(3):e2109. Epub 2013/04/05. 10.1371/journal.pntd.0002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmona SJ, Nielsen M, Schafer-Nielsen C, Mucci J, Altcheh J, Balouz V, et al. Towards High-throughput Immunomics for Infectious Diseases: Use of Next-generation Peptide Microarrays for Rapid Discovery and Mapping of Antigenic Determinants. Molecular & cellular proteomics: MCP. 2015;14(7):1871–84. 10.1074/mcp.M114.045906 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassegne K, Abe EM, Chen JH, Zhou XN. Immunomic approaches for antigen discovery of human parasites. Expert Rev Proteomics. 2016;13(12):1091–101. Epub 2016/10/25. 10.1080/14789450.2016.1252675 . [DOI] [PubMed] [Google Scholar]
- 23.Lagatie O, Van Dorst B, Stuyver LJ. Identification of three immunodominant motifs with atypical isotype profile scattered over the Onchocerca volvulus proteome. PLoS Negl Trop Dis. 2017;11(1):e0005330. 10.1371/journal.pntd.0005330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Assis RR, Ludolf F, Nakajima R, Jasinskas A, Oliveira GC, Felgner PL, et al. A next-generation proteome array for Schistosoma mansoni. Int J Parasitol. 2016;46(7):411–5. Epub 2016/05/02. 10.1016/j.ijpara.2016.04.001 . [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Zhang Y, Lin D, Zhang J, Xu J, Liu YM, et al. Serodiagnosis of Schistosoma japonicum infection: genome-wide identification of a protein marker, and assessment of its diagnostic validity in a field study in China. Lancet Infect Dis. 2014;14(6):489–97. Epub 2014/03/25. 10.1016/S1473-3099(14)70067-2 . [DOI] [PubMed] [Google Scholar]
- 26.Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the human hookworm Necator americanus. Nat Genet. 2014;46(3):261–9. Epub 2014/01/21. 10.1038/ng.2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dana D, Vlaminck J, Ayana M, Tadege B, Mekonnen Z, Geldhof P, et al. Evaluation of copromicroscopy and serology to measure the exposure to Ascaris infections across age groups and to assess the impact of 3 years of biannual mass drug administration in Jimma Town, Ethiopia. PLoS neglected tropical diseases. 2020;14(4):e0008037. Epub 2020/04/14. 10.1371/journal.pntd.0008037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuyver LJ, Verbeke T, Van Loy T, Van Gulck E, Tritsmans L. An antibody response to human polyomavirus 15-mer peptides is highly abundant in healthy human subjects. Virology journal. 2013;10:192. 10.1186/1743-422X-10-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Loy T, Thys K, Tritsmans L, Stuyver LJ. Quasispecies analysis of JC virus DNA present in urine of healthy subjects. PLoS One. 2013;8(8):e70950. 10.1371/journal.pone.0070950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagatie O, Van Loy T, Tritsmans L, Stuyver LJ. Circulating human microRNAs are not linked to JC polyomavirus serology or urinary viral load in healthy subjects. Virology journal. 2014;11(1):41. 10.1186/1743-422X-11-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagatie O, Van Loy T, Tritsmans L, Stuyver LJ. Antibodies reacting with JCPyV_VP2 _167-15mer as a novel serological marker for JC polyomavirus infection. Virology journal. 2014;11:174. 10.1186/1743-422X-11-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagatie O, Van Loy T, Tritsmans L, Stuyver LJ. Viral miRNAs in plasma and urine divulge JC polyomavirus infection. Virology journal. 2014;11:158. 10.1186/1743-422X-11-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parasite W. 2017 [cited 2017 March 28th]. Available from: https://parasite.wormbase.org/ftp.html.
- 34.Somers K, Geusens P, Elewaut D, De Keyser F, Rummens JL, Coenen M, et al. Novel autoantibody markers for early and seronegative rheumatoid arthritis. J Autoimmun. 2011;36(1):33–46. Epub 2010/11/13. 10.1016/j.jaut.2010.10.003 . [DOI] [PubMed] [Google Scholar]
- 35.Loebel M, Eckey M, Sotzny F, Hahn E, Bauer S, Grabowski P, et al. Serological profiling of the EBV immune response in Chronic Fatigue Syndrome using a peptide microarray. PLoS One. 2017;12(6):e0179124. Epub 2017/06/13. 10.1371/journal.pone.0179124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cringoli G, Maurelli MP, Levecke B, Bosco A, Vercruysse J, Utzinger J, et al. The Mini-FLOTAC technique for the diagnosis of helminth and protozoan infections in humans and animals. Nature protocols. 2017;12(9):1723–32. Epub 2017/08/05. 10.1038/nprot.2017.067 . [DOI] [PubMed] [Google Scholar]
- 37.Barda B, Cajal P, Villagran E, Cimino R, Juarez M, Krolewiecki A, et al. Mini-FLOTAC, Kato-Katz and McMaster: three methods, one goal; highlights from north Argentina. Parasit Vectors. 2014;7:271. 10.1186/1756-3305-7-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Team RC. R: A language and environment for statistical computing. Vienna, Australia: R foundation for statistical computing; 2019. [Google Scholar]
- 39.Schober P, Boer C, Schwarte LA. Correlation Coefficients: Appropriate Use and Interpretation. Anesth Analg. 2018;126(5):1763–8. Epub 2018/02/27. 10.1213/ANE.0000000000002864 . [DOI] [PubMed] [Google Scholar]
- 40.Mukaka MM. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24(3):69–71. Epub 2013/05/03. [PMC free article] [PubMed] [Google Scholar]
- 41.Lagatie O, Verheyen A, Nijs E, Batsa Debrah L, Debrah YA, Stuyver LJ. Performance evaluation of 3 serodiagnostic peptide epitopes and the derived multi-epitope peptide OvNMP-48 for detection of Onchocerca volvulus infection. Parasitology research. 2019. Epub 2019/05/16. 10.1007/s00436-019-06345-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagatie O, Verheyen A, Nijs E, Van Dorst B, Batsa Debrah L, Debrah A, et al. Evaluation of the Diagnostic Performance of Onchocerca volvulus Linear Epitopes in a Peptide Enzyme-Linked Immunosorbent Assay. The American journal of tropical medicine and hygiene. 2018;98(3):779–85. Epub 2018/01/10. 10.4269/ajtmh.17-0756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlaminck J, Lagatie O, Verheyen A, Dana D, Van Dorst B, Mekonnen Z, et al. Patent infections with soil-transmitted helminths and Schistosoma mansoni are not associated with increased prevalence of antibodies to the Onchocerca volvulus peptide epitopes OvMP-1 and OvMP-23. Parasites & vectors. 2019;12(1):63. Epub 2019/01/30. 10.1186/s13071-019-3308-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–8. Epub 2009/07/17. 10.1038/nature08160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeMarco R, Mathieson W, Manuel SJ, Dillon GP, Curwen RS, Ashton PD, et al. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 2010;20(8):1112–21. Epub 2010/07/08. 10.1101/gr.100099.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson RA, Li XH, MacDonald S, Neves LX, Vitoriano-Souza J, Leite LC, et al. The Schistosome Esophagus Is a ’Hotspot’ for Microexon and Lysosomal Hydrolase Gene Expression: Implications for Blood Processing. PLoS Negl Trop Dis. 2015;9(12):e0004272. Epub 2015/12/08. 10.1371/journal.pntd.0004272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li XH, de Castro-Borges W, Parker-Manuel S, Vance GM, Demarco R, Neves LX, et al. The schistosome oesophageal gland: initiator of blood processing. PLoS Negl Trop Dis. 2013;7(7):e2337. Epub 2013/08/13. 10.1371/journal.pntd.0002337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XH, Vance GM, Cartwright J, Cao JP, Wilson RA, Castro-Borges W. Mapping the epitopes of Schistosoma japonicum esophageal gland proteins for incorporation into vaccine constructs. PLoS One. 2020;15(2):e0229542. Epub 2020/02/29. 10.1371/journal.pone.0229542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathieson W, Wilson RA. A comparative proteomic study of the undeveloped and developed Schistosoma mansoni egg and its contents: the miracidium, hatch fluid and secretions. International journal for parasitology. 2010;40(5):617–28. 10.1016/j.ijpara.2009.10.014 . [DOI] [PubMed] [Google Scholar]
- 50.Farias LP, Vance GM, Coulson PS, Vitoriano-Souza J, Neto APdS, Wangwiwatsin A, et al. Epitope Mapping of Exposed Tegument and Alimentary Tract Proteins Identifies Putative Antigenic Targets of the Attenuated Schistosome Vaccine. Frontiers in immunology. 2021;11(3671). 10.3389/fimmu.2020.624613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crosnier C, Hokke CH, Protasio AV, Brandt C, Rinaldi G, Langenberg MCC, et al. Screening of a library of recombinant Schistosoma mansoni proteins with sera from murine and human controlled infections identifies early serological markers. J Infect Dis. 2020. Epub 2020/06/12. 10.1093/infdis/jiaa329 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce D, Merone L, Lewis C, Rahman T, Croese J, Loukas A, et al. Safety and tolerability of experimental hookworm infection in humans with metabolic disease: study protocol for a phase 1b randomised controlled clinical trial. BMC Endocr Disord. 2019;19(1):136. Epub 2019/12/13. 10.1186/s12902-019-0461-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farah IO, Kariuki TM, King CL, Hau J. An overview of animal models in experimental schistosomiasis and refinements in the use of non-human primates. Lab Anim. 2001;35(3):205–12. Epub 2001/07/21. 10.1258/0023677011911570 . [DOI] [PubMed] [Google Scholar]
- 54.Shepherd C, Wangchuk P, Loukas A. Of dogs and hookworms: man’s best friend and his parasites as a model for translational biomedical research. Parasites & vectors. 2018;11(1):59. Epub 2018/01/27. 10.1186/s13071-018-2621-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kringel H, Thamsborg SM, Petersen HH, Goring HH, Skallerup P, Nejsum P. Serum antibody responses in pigs trickle-infected with Ascaris and Trichuris: Heritabilities and associations with parasitological findings. Veterinary parasitology. 2015;211(3–4):306–11. Epub 2015/06/23. 10.1016/j.vetpar.2015.06.008 . [DOI] [PubMed] [Google Scholar]
- 56.Fujiwara RT, Geiger SM, Bethony J, Mendez S. Comparative immunology of human and animal models of hookworm infection. Parasite immunology. 2006;28(7):285–93. Epub 2006/07/18. 10.1111/j.1365-3024.2006.00821.x . [DOI] [PubMed] [Google Scholar]
- 57.Arnold BF, Scobie HM, Priest JW, Lammie PJ. Integrated Serologic Surveillance of Population Immunity and Disease Transmission. Emerg Infect Dis. 2018;24(7):1188–94. Epub 2018/06/19. 10.3201/eid2407.171928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagatie O, Granjon E, Odiere MR, Zrein M, Stuyver LJ. Assessment of multiplex Onchocerca volvulus peptide ELISA in non-endemic tropical regions. Parasites & vectors. 2019;12(1):570. Epub 2019/12/01. 10.1186/s13071-019-3824-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcaro MC, Peroni E, Rovero P, Papini AM. Synthetic peptides in the diagnosis of HIV infection. Curr Protein Pept Sci. 2003;4(4):285–90. Epub 2003/10/08. 10.2174/1389203033487117 . [DOI] [PubMed] [Google Scholar]
- 60.Kotwal GJ, Baroudy BM, Kuramoto IK, McDonald FF, Schiff GM, Holland PV, et al. Detection of acute hepatitis C virus infection by ELISA using a synthetic peptide comprising a structural epitope. Proc Natl Acad Sci U S A. 1992;89(10):4486–9. Epub 1992/05/15. 10.1073/pnas.89.10.4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stuyver L, Van Arnhem W, Wyseur A, DeLeys R, Maertens G. Analysis of the putative E1 envelope and NS4a epitope regions of HCV type 3. Biochemical and biophysical research communications. 1993;192(2):635–41. Epub 1993/04/30. 10.1006/bbrc.1993.1462 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG)
(XLSX)
(XLSX)
Data Availability Statement
Large data files are available at https://osf.io/q5s9y/?view_only=8a464d9eafe6444b83b967a11f3211cd, as indicated in the manuscript.