Quantitative assessments of microbial activity in crustal fluids yield potential strategies to supplement recalcitrant DOC.
Abstract
Biogeochemical processes occurring in fluids that permeate oceanic crust make measurable contributions to the marine carbon cycle, but quantitative assessments of microbial impacts on this vast, subsurface carbon pool are lacking. We provide bulk and single-cell estimates of microbial biomass production from carbon and nitrogen substrates in cool, oxic basement fluids from the western flank of the Mid-Atlantic Ridge. The wide range in carbon and nitrogen incorporation rates indicates a microbial community well poised for dynamic conditions, potentially anabolizing carbon and nitrogen at rates ranging from those observed in subsurface sediments to those found in on-axis hydrothermal vent environments. Bicarbonate incorporation rates were highest where fluids are most isolated from recharging bottom seawater, suggesting that anabolism of inorganic carbon may be a potential strategy for supplementing the ancient and recalcitrant dissolved organic carbon that is prevalent in the globally distributed subseafloor crustal environment.
INTRODUCTION
In the deep ocean, seawater is entrained into the rocky crust, chemically altered by abiotic and microbial processes, and discharged from the seafloor as hydrothermal fluid with a global flux that rivals riverine inputs (1, 2). More than 90% of this hydrothermal fluid discharge is from low-temperature fluids (5° to 20°C) circulating on the flanks of mid-ocean ridges (1), where these fluids are generally inaccessible and their microbial assemblages are largely unexplored (3). The biogeochemical influence of this cool, ridge-flank microbiome on net chemical fluxes, and particularly on the enormous, climate-sensitive reservoir of deep-ocean dissolved organic carbon (DOC), is potentially substantial, but poorly constrained. For this study, pristine, cool, basaltic subseafloor fluids (4) from 8–million year-old crust were recovered from ocean drilling borehole observatories of the North Pond site located at 22°N on the western flank of the Mid-Atlantic Ridge. Here, oxygenated crustal fluids are largely indistinguishable from bottom seawater and concentrations of ammonium, methane, hydrogen sulfide, and iron(II) are below detection, indicating an overall low redox energy potential in these subseafloor crustal fluids (5). While DOC sourced from the deep ocean is thought to be unreactive and resistant to microbial degradation (6, 7), previous isotopic data from these fluids suggest that selective removal of DOC via microbial oxidation does occur (8, 9) and DOC may, therefore, be the most abundant reduced substrate available for microbes to oxidize in cool crustal fluids (9).
While the abundance and diversity of microorganisms in the subseafloor have been explored for decades via scientific drilling programs (10), slow growth and often low biomass present challenges for demonstrating microbial activity under environmentally relevant conditions. Microbial activity via uptake of labeled substrates has been successfully observed in sedimentary (11–13) and diffuse flow hydrothermal vent fluids (14–16) using stable isotope probing (SIP) incubations coupled to single-cell measurements with nanoscale secondary ion mass spectrometry (NanoSIMS), providing constraints on the potential microbial contribution to primary production and organotrophy in these habitats. However, no such data exist from the crustal biome. Here, we determine single-cell and bulk estimates of microbial carbon and nitrogen incorporation from the ridge flank crustal habitat that represents the majority of fluid flux between the subsurface and the overlying ocean. Our extensive and quantitative assessments highlight a microbial population poised to incorporate fresh sources of labile organic carbon and a consistent, wide range of intercell incorporation rates across fluids and conditions. We also report significant bicarbonate incorporation, despite the absence of abundant inorganic sources of redox energy that could fuel chemolithotrophy, and suggest that this may be part of a metabolic strategy of supplementing anabolic carbon needs with bicarbonate to reduce the reliance on aged and recalcitrant deep-ocean DOC.
North Pond hosts two CORK (Circulation Obviation Retrofit Kit) seafloor borehole observatories installed in 2011 at Integrated Ocean Drilling Program (IODP) Sites U1382A and U1383C (Fig. 1A). The CORK at site U1382A accesses circulating fluids from one depth interval below the sediment in the rocky subseafloor [90 to 210 meters below seafloor (mbsf)], and the U1383C CORK accesses three depth ranges: Shallow (70 to 146 mbsf), Middle (146 to 200 mbsf), and Deep (200 to 332 mbsf). Although the geochemistry of fluids recovered from both CORK observatories is largely similar to overlying seawater, radiocarbon measurements and larger differences in dissolved oxygen and DOC concentrations indicate that fluids recovered from U1383C are more isolated from bottom water recharge than U1382A (5, 8, 9). Recent numerical simulations suggest that there is convective and oscillatory fluid movement through the rocky crust (17) rather than the simple linear flow along the north-south axis as had been hypothesized in earlier studies of North Pond (4).
Fig. 1. North Pond is located in young, cool crust (8 Ma) with a “sediment pond” roughly 13 km long by 7 km wide and up to 200 m deep.
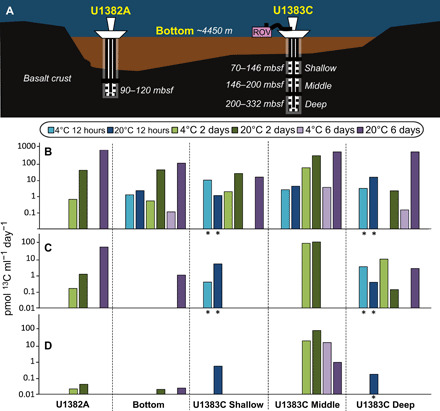
(A) Diagram of CORK observatories at North Pond with different sampling locations (U1382A, bottom seawater, and U1383C) and depths in meters below surface (m) or meters below seafloor (mbsf). U1383C is located about 6 km to the NE of U1382A. Bulk 13C-carbon isotope incorporation rates calculated from (B) 13C-acetate, (C) 13C-bicarbonate, and (D) 13C-methylamine SIP incubations of each fluid sample for 12-hour, 2-day, and 6-day incubation periods in order shown in (A). Asterisk indicates the value computed from SIP-NanoSIMS single-cell measurements (Table 3) instead of bulk elemental analysis. No bar indicates that the data are below detection (see Materials and Methods).
RESULTS AND DISCUSSION
Fluids described in this study were collected using a mobile pumping system (MPS) (18) in October 2017 as a part of the third sampling expedition to North Pond. Bottom seawater was also collected by Niskin bottles on a conductivity, temperature, depth (CTD) water sampling rosette. 16S ribosomal RNA (rRNA) gene sequencing of all samples (North Pond crustal fluids and bottom seawater) indicates that the microbial community in crustal fluids is distinct from those communities in bottom seawater in 2017 (figs. S1 and S2), as has been seen in previous years (8, 19). Within the crustal fluids, the microbial community at U1382A is more similar to bottom water than the U1383C fluid horizons, which is also consistent with geochemical data (5). Moreover, in a separate study of a diverse range of mineral chips incubated in the CORKs for 4 to 6 years (retrieved in 2017), the authors recovered similar taxa across all sample types (CORK fluids, bottom water, and mineral chips) and determined that the incubation fluid explained more of the microbial community composition colonizing minerals than the type of mineral surface (20). Together, these results indicate that the fluids collected in 2017, and used in the current study, capture the dominant microbial communities in the North Pond crustal fluids.
Cell counts from CORK fluids in 2017 ranged from 2.1 × 103 to 5.1 × 103 cells ml−1 (Table 1). These 2017 counts are lower than historical cell count data for previously collected fluids, which have ranged from high 103 to low 104 cells ml−1 of fluid (8, 19). Decreasing cell concentrations after drilling has also been observed in other CORKs [Juan de Fuca ridge flank; (21)] and groundwater well systems (22). Together with geochemical data [Table 1; (5)], low cell counts likely indicate that the North Pond system had recovered from drilling and that the 2017 fluids (and resulting data) are the best representation of microbial activity in the cold, oxic crustal subseafloor aquifer to date.
Table 1. North Pond CORK fluids and bottom water values for cell enumeration, dissolved inorganic carbon (DIC), and dissolved organic carbon (DOC) from 2017 samples collected for this study.
Additional geochemistry (oxygen, nitrate, and pH) reproduced from (5) also collected in 2017. Ammonium concentrations were all below detection (<0.1 μmol/kg) from 2017 (5).
| Sample | Depth (m) | Cells ×103 ml−1 | ±95% CI | pH | DIC (μmol/kg) | O2 (μM) | Nitrate (μmol/kg) | DOC (μmol/kg) |
| Bottom Water | 4397* | 9.0 | 0.4 | 7.92 | 2188 | 250 | 21.8 | 39 |
| U1382A | 90–210 | 5.1 | 0.2 | 7.91 | 2164 | 228 | 22.3 | 31 |
| U1383C Shallow | 70–146 | 3.7 | 0.3 | 8.07 | 2167 | 198 | 22.8 | 20 |
| U1383C Middle | 146–200 | 2.1 | 0.2 | 8.13 | 2189 | 205 | 22.9 | 22 |
| U1383C Deep | 200–332 | 3.7 | 0.3 | 8.06 | 2156 | 173 | 22.7 | 22 |
*Depth in meters below surface, remaining depths are in meters below seafloor (mbsf). Bicarbonate concentrations are ±5 μmol/kg.
Incubations of bottom seawater and crustal fluids were all amended with deuterated (2H2O) water, which can be used as a general tracer of microbial anabolic activity (23). Select combinations of 13C carbon (bicarbonate, acetate, methylamine, and diatom lysate) and 15N nitrogen (ammonium, methylamine, and diatom lysate) were provided as substrate-specific tracers of anabolic activity (Table 2). No substrates were added to CN controls. For diatom lysate, diatoms were grown in the presence of isotopically enriched 13C-bicarbonate and 15N-nitrate and then lysed before addition as a proxy for environmentally relevant complex organic matter. Incubations were conducted at temperatures bracketing the expected range in the North Pond aquifer (4° to 20°C), and cells were harvested from separate incubations prepared for three time points (12 hours, 2 days, and 6 days; Table 2). Isotope incorporation rates were calculated from bulk elemental analysis (Fig. 1, B to D) and single-cell NanoSIMS measurements (Figs. 2 and 3).
Table 2. Experimental conditions of stable isotope probing incubations for the five fluid sources collected at North Pond (bottom water, U1382A, U1383C Shallow, U1383C Middle, and U1383C Deep).
All incubations were conducted in triplicate separate bottles for each time point at 12 hours, 2 days, and 6 days. Condition identifiers used in figures referenced here, where C is control, D is diatom lysate, M is methylamine, A is acetate, and B is bicarbonate or bicarbonate, and the temperature is in subscript.
| Condition | 2H label | 13C label | 15N label | Temperature (°C) |
| C20 | Water | None | None | 20 |
| D20 | Water | Diatom lysate | Diatom lysate | 20 |
| M20 | Water | Methylamine | Methylamine | 20 |
| A20 | Water | Acetate | Ammonium | 20 |
| B20 | Water | Bicarbonate | Ammonium | 20 |
| C4 | Water | None | None | 4 |
| D4 | Water | Diatom lysate | Diatom lysate | 4 |
| M4 | Water | Methylamine | Methylamine | 4 |
| A4 | Water | Acetate | Ammonium | 4 |
| B4 | Water | Bicarbonate | Ammonium | 4 |
Fig. 2. Selected NanoSIMS ratio images for each stable isotope (2H, 15N, and 13C).
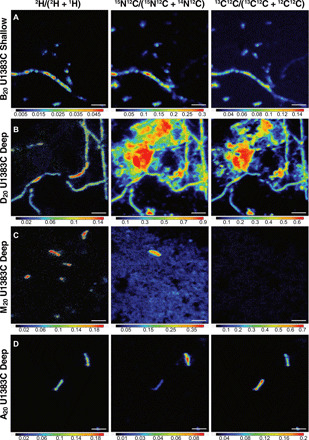
(A) The most active Shallow sample amended with 13C-DIC and 15N-ammonium (B20 U1383C Shallow), (B) U1383C Deep 13C15N–diatom lysate amendment, where the remaining diatom lysate is visible in 15N and 13C images, but 2H uptake is only seen in associated filaments (D20 U1383C Deep), (C) 13C15N-methylamine uptake in 2H and 15N without visible 13C incorporation (M20 U1383C Deep), and (D) most active Deep sample (A20 U1383C Deep) amended with 13C-acetate and 15N-ammonium. All incubations were amended with 2H2O. Scale bars, 3 μm. Ratio range scale differs between samples to show maximum values per image.
Fig. 3. Single-cell isotope incorporation rates calculated from U1383C Shallow and Deep incubations.
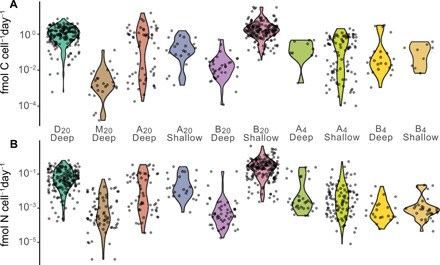
Computed fmol (A) C and (B) N cell−1 day−1 rates are based on NanoSIMS data and plotted as the kernel density distribution with true values overlain.
Microbial populations in North Pond fluids appear poised for dynamic and heterogeneous conditions. Cell counts from organic carbon–amended incubations (acetate and diatom lysate) had the highest cell densities in earlier time points (fig. S3). By 2 days, all incubations with acetate (1383C Middle) or diatom lysate (all other samples) had the highest cell densities. By the final time point of 6 days, most of these organic carbon incubations had a lower cell density than earlier time points or CN controls, suggesting an initial period of cell growth followed by death. This pattern of cell increase in the first few days followed by decrease has also been observed in groundwater wells with no amendments (22), which the authors attribute to necromass-induced growth from cells that die off at the onset of the incubation.
In bulk analysis performed on 13C-acetate–, 13C-methylamine–, and 13C-bicarbonate–amended incubations, acetate incorporation was widespread across fluids and conditions (Fig. 1B). Bicarbonate incorporation was not detected as consistently as acetate incorporation but was on par with acetate incorporation in some fluids (e.g., U1383C Middle and Deep; Fig. 1C). Methylamine incorporation rates were lower overall, compared to acetate and bicarbonate, but uniquely elevated in U1383C Middle at both 4° and 20°C (Fig. 1D).
Bulk rates of carbon incorporation from acetate were highest at 6 days for all 20°C fluids except U1383C Shallow, where the rate decreased between days 2 and 6 (Fig. 1B). Acetate 4°C incubations were generally lower than their 20°C counterparts (except for U1383C Shallow where the 12-hour rate was highest at 4°C; Fig. 1B). Examination of the single-cell rates for carbon incorporation (Fig. 3A) showed a bimodal distribution of rates for U1383C Deep (20°C) and Shallow (4°C) at their respective in situ temperatures. These data suggest that microbes in deeper crustal fluids may persist on recalcitrant carbon sources, with select members poised to quickly respond to an influx of more labile organic carbon.
Previous genomic studies of North Pond crustal fluid microbial communities collected in 2012 recovered genes for autotrophic CO2 fixation (8, 19), and much higher incorporation of 13C-bicarbonate than 13C-acetate (800 to 4300 pmol ml−1 day−1 versus no more than 104 pmol ml−1 day−1; table S4) from SIP incubations (8). However, the fluids collected in 2012, only 6 months after drilling, were particle-laden and geochemical data suggested that they represented a mixture of crustal fluids, bottom seawater, and surface seawater (5). 13C-bicarbonate incorporation rates from 2017 CORK fluids were much lower and ranged from below detection to 94 pmol ml−1 day−1. 2017 bottom seawater rates (0.95 pmol ml−1 day−1) were comparable to a study from the western branch of North Atlantic Deep Water, which overlays the North Pond site (0.24 pmol ml−1 day−1 from 14C-bicarbonate, 2537-m water depth; table S4) (24). Thus, it appears that bicarbonate incorporation may have been artificially elevated in these early studies.
Still, bicarbonate incorporation rates on par with acetate incorporation in some 2017 fluids is surprising because of the lack of abundant electron donors typically used for chemolithotrophy. Though oxygen was abundant, methane, hydrogen sulfide, ammonium, and reduced iron are all below detection limits (5, 8) and unlikely to fuel extensive autotrophic carbon fixation in North Pond crustal fluids.
Limited carbon fixation driven by the oxidation of reduced iron and sulfur minerals on basalt surfaces is still possible, although 13C-bicarbonate stable isotope incubations conducted with basalt samples and metagenomics investigations of these same rocks showed no conclusive evidence for carbon fixation (25, 26). However, global estimates indicate its importance in the first ~10 million-years (Ma) of crustal evolution (27), and chemolithotrophy in biofilms on rock surfaces may not have been captured by observations of bulk rates.
Given the lack of electron donors that could be paired with chemolithotrophy in our incubations, it is unlikely that rock-driven autotrophy is occurring in our experiments. Metagenomic and metatranscriptomic analysis of the same 2017 North Pond fluids used in our experiments showed more transcription of organotrophic genes than autotrophic genes across all sampling horizons, suggesting a microbial community utilizing organic carbon (28). Carbon fixation transcripts were most abundant in 1383C Middle and Deep where we also detected the highest rates of bicarbonate incorporation. Furthermore, metagenomic-assembled genomes (MAGs) from the fluids revealed that a number of MAGs contained carbon fixation pathways linked to oxidation of sulfide and thiosulfate, but these same MAGs also contained numerous extracellular protease and carbohydrate catabolism genes, consistent with a mixotrophic lifestyle.
These North Pond results are in accordance with the predominance of heterotrophic bacteria reported in ~33- and 104-Ma basalt crust cored beneath the South Pacific Gyre (29), as well as in subseafloor ultramafic and gabbroic rocks cored from the Atlantic Ocean (30, 31), where organotrophic microbial processes also dominate.
Because DOC is the reduced substrate with the highest concentration in the North Pond crustal fluids (20 to 31 μmol/kg DOC; Table 1), organotrophy coupled with anabolism of both organic and inorganic carbon is the more likely explanation for observed uptake of bicarbonate. While anabolic incorporation of bicarbonate is underexplored in environmental settings, it has been demonstrated in laboratory conditions with pure cultures. For example, Pseudomonas AM1, Hyphomicrobium vulgare, and Methylobacterium extorquens are able to use a combination of the ethylmalonyl–coenzyme A pathway and serine cycle, resulting in as much as 50% of biomass carbon derived from bicarbonate (32, 33).
We hypothesize that organotrophy coupled to anabolism of both organic and inorganic carbon reflects microbial communities optimizing the low potential redox energy conditions in the rocky subseafloor. Crustal fluid DOC likely represents the fraction of deep-ocean DOC that remains after the more bioavailable components are removed on short time scales after fluids are entrained in the crust (9). Our results indicate that this degraded DOC alone supports an active microbial population because cell growth was observed even when no carbon or nitrogen or other reduced substrates were added to incubations. DOC may be largely oxidized for energy by organotrophic pathways and bicarbonate may provide a supplementary, and less metabolically expensive, anabolic source of carbon.
Bicarbonate incorporation rates from 2017 were consistently high across time points in U1383C Deep fluids and highest with U1383C Middle fluids in 2-day incubations at 4° and 20°C (Fig. 1C). U1383C Middle and Deep fluids are also the fluids with the most “aged” or 14C-depleted DOC (9). DOC in fluids from U1383C has a radiocarbon age of 7300 to 9200 years and a higher aromaticity index and percent carboxyl-rich alicyclic molecules (CRAM) than bottom water or fluids from U1382A (9). Because DOC concentrations are lower at all subseafloor depths at U1383C compared to U1382A (Table 1), and compositional evidence indicates that the DOC is the most aged and degraded at U1383C (9), microbial communities at this site may have the most to gain from supplementing their anabolic needs with bicarbonate incorporation. Our results indicate that there is not a concentration limit below which natural DOC is unavailable to microbes (34), but rather, there could be a different strategy for accessing the aged and recalcitrant fraction of deep-ocean DOC by combining biomass production from both inorganic and organic carbon sources (fig. S8). This could be supplemented by other oligotrophic strategies to use diverse sources of organic carbon in low-DOC (<4 μM) conditions (35).
While dissolved nitrogen has been less studied at North Pond than DOC, ammonium loss through nitrification has been hypothesized in North Pond fluids based on geochemical fluxes (5). All of the provided nitrogen sources analyzed from U1383C Deep (15N-ammonium, 15N-methylamine, and 15N–diatom lysate) and U1383C Shallow fluids (15N-ammonium) were incorporated by cells based on NanoSIMS analysis (Fig. 3B). It was generally observed that when 15N-ammonium was paired with 13C-acetate, the nitrogen incorporation rate was higher than when paired with 13C-bicarbonate. It was also observed that 13C– and 15N–diatom lysate C and N incorporation rates were similar to the 15N-ammonium and 13C-acetate C and N rates for comparable 12-hour 20°C U1383C Deep incubations. Therefore, organic nitrogen anabolic rates may have been higher when organic carbon was also provided (Table 3 and fig. S8).
Table 3. Average per-cell and per-milliliter rates of carbon and nitrogen uptake from NanoSIMS data of 12-hour incubations.
Incubations that were conducted near in situ temperatures for each fluid source are shaded in gray.
| Condition | U1383C fluid | fmol C cell−1 day−1 | fmol N cell−1 day−1 | pmol C ml−1 day−1 | pmol N ml−1 day−1 | C:N |
| D20 | Deep | 1.3 | 0.096 | 86 | 6.2 | 14 |
| M20 | Deep | 0.0085 | 0.0087 | 0.16 | 0.17 | 0.98 |
| A20 | Deep | 1.15 | 0.097 | 15 | 1.3 | 12 |
| B20 | Deep | 0.042 | 0.021 | 0.4 | 0.17 | 2.0 |
| A4 | Deep | 0.21 | 0.12 | 2 | 1.5 | 1.7 |
| B4 | Deep | 0.46 | 0.0029 | 3 | 0.020 | 155 |
| A20 | Shallow | 0.30 | 0.054 | 1.1 | 0.2 | 5.5 |
| B20 | Shallow | 3.0 | 0.42 | 5 | 0.6 | 7.0 |
| A4 | Shallow | 0.33 | 0.025 | 10 | 0.8 | 13 |
| B4 | Shallow | 0.13 | 0.0085 | 0.4 | 0.023 | 16 |
Methylamine incorporation was observed in all fluids. The highest methylamine incorporation rate (72 pmol C ml−1 day−1) was from the same condition that had the highest bicarbonate incorporation rate, U1383C Middle 20°C at 2 days. This suggests that Middle fluids were the most amenable to different carbon sources, with the highest rates of bicarbonate and methylamine carbon incorporation and the third highest acetate incorporation. Bulk carbon incorporation of methylamine was comparable to carbon uptake rates of other simple organic N compounds from oxygen-deficient zones (ODZs) with similar oxygen concentrations to North Pond crustal fluids (36), such as urea and cyanate (5 to 12 pmol C ml−1 day−1 and 2 to 14 pmol C ml−1 day−1, respectively; table S4). However, nitrogen incorporation rates from the same compounds in ODZ incubations ranged from 2 to 265 pmol N ml−1 day−1, which were much higher than average nitrogen incorporation rates derived from single-cell analysis of North Pond microbial communities (Table 3), where the highest average rate from diatoms was 6 pmol N ml−1 day−1 and ammonium was 1 pmol ml−1 day−1. Overall, this suggests that both C and N can be incorporated from organic N sources in North Pond crustal fluids and, from these organic N sources, North Pond C incorporation rates were more similar to ODZ rates than the N incorporation rates.
Cool, crustal aquifer fluids appear to support a wider range of microbial activities than more stable subsurface sedimentary and diffuse flow hydrothermal vent counterparts. The average observed single-cell uptake rates (10−3 to 100 fmol C or N cell−1 day−1; Table 3) fall between SIP-NanoSIMS rates derived from deep, organic-rich marine sediments incubated with bicarbonate and ammonium (10−2 fmol C or N cell−1 day−1; table S5) and diffuse flow hydrothermal vent fluids of the East Pacific Rise incubated with bicarbonate (100 to 101 fmol C cell−1 day−1; table S5). The highest average C and N North Pond incorporation rates were observed in 20°C incubations (U1383C Deep with acetate and ammonium at 1 fmol C cell−1 day−1 and 0.1 fmol N cell−1 day−1; U1383C Shallow with bicarbonate and ammonium at 3 fmol C cell−1 day−1 and 0.4 fmol N cell−1 day−1; Table 3). While the upper end of this range may only be observed under incubation conditions, the highest average rates were from incubations where no additional DOC was provided (bicarbonate amendments), illustrating the potential for North Pond crustal microbes to exhibit high anabolic rates using aged DOC and little or no inorganic redox-active substrates for energy conservation. Higher bicarbonate uptake observed in the shallower fluids at 20°C may also be the result of supplemental bicarbonate anabolism, which allowed the colder in situ fluid community to more quickly take advantage of growth under warmer incubation temperatures, similar to deeper communities using bicarbonate to supplement available DOC. Therefore, in addition to implications for marine carbon and nitrogen cycling, the wide range in potential incorporation rates may have additional import for understanding evolutionary constraints in subseafloor ecosystems, which have been heretofore been based on sedimentary and diffuse flow “end-member” subseafloor biomes.
A global first-order estimate approximated that 1011 to 1012 mol organic carbon could be incorporated per year in the cool subseafloor crustal aquifer fluids. This is derived from an estimated habitable pore volume for North Pond–age crust (<10 Ma) and the average single-cell uptake rates from U1383C Shallow and Deep fluid incubations amended with organic carbon at the temperatures most relevant to in situ conditions (Table 4). This average estimate for fluid biomass production from organic carbon agrees with the estimated annual removal of DOC from cool crust at 1011 mol DOC year−1 (9) and suggests that the majority of DOC loss could be associated with organotrophic and anabolic processes rather than catabolism. We also find that organic nitrogen may be anabolized in this system at average rates of 1010 to 1011 mol N year−1.
Table 4. Estimated average, maximum, and minimum moles of C or N year−1 produced in young (<10 Ma) crustal fluids for incubations that were conducted near in situ temperatures for each fluid.
Estimates are based on single-cell average, maximum, and minimum carbon and nitrogen isotope incorporation rates from NanoSIMS analysis for U1383C Shallow and Deep samples.
| Fluid | Condition | mol C year−1 | mol N year−1 | ||||
| Avg | Max | Min | Avg | Max | Min | ||
| U1383C Deep | D20 | 2 × 1012 | 1 × 1012 | 8 × 108 | 1 × 1011 | 8 × 1011 | 2 × 108 |
| M20 | 1 × 1010 | 2 × 1011 | 2 × 107 | 1 × 1010 | 7 × 1010 | 2 × 107 | |
| A20 | 2 × 1012 | 2 × 1013 | 2 × 108 | 1 × 1011 | 5 × 1011 | 5 × 107 | |
| B20 | 6 × 1010 | 7 × 1011 | 2 × 108 | 3 × 1010 | 2 × 1010 | 9 × 107 | |
| U1383C Shallow | A4 | 4 × 1011 | 3 × 1012 | 1 × 108 | 3 × 1010 | 7 × 1011 | 1 × 107 |
| B4 | 2 × 1011 | 5 × 1011 | 9 × 109 | 1 × 1010 | 3 × 1010 | 6 × 107 | |
Estimates for bicarbonate incorporation were approximately 1010 to 1011 mol C year−1 (Table 4) and provide a potential mechanism for adding more labile, organic carbon to the crustal aquifer (fig. S8). This estimate from young crustal fluids is higher than estimates of annual primary productivity from hydrothermal systems at 109 mol C year−1 (24), suggesting that cool crustal fluid bicarbonate utilization may be equally or more significant than on-axis hydrothermal vent fluids on a global scale.
MATERIALS AND METHODS
Sample collection
Fluids were collected from the basaltic aquifer at North Pond (22°45′N and 46°05′W) in October 2017 during cruise AT39-01 of the R/V Atlantis using ROV JASON II during expedition AT39-01. Fluids were sampled from two CORK installations, each with umbilicals that are accessible at the seafloor and terminate in a subsurface hydrologic zone: U1382A (includes one umbilical) and U1383C (includes three umbilicals: Shallow, Middle, and Deep; Fig. 1 and Table 1). Filtered samples were collected using the MPS (8, 18). Sample purity was evaluated by monitoring geochemical parameters. Temperature and oxygen concentrations of the fluids were continuously monitored using an inline optode oxygen/temperature sensor (Aanderaa, Bergen, Norway). Before filtering, umbilical lines were flushed until oxygen concentrations were stable and reached the expected values previously observed in each horizon. Then, crustal fluid was pumped at a rate of ~0.5 LPM (liters per minute) for 80 min through a 0.22-μm, 47-mm GSWP filter (MilliporeSigma, Burlington, MA, USA). MPS sample purity has been demonstrated at other CORK installations where the geochemistry of the crustal fluids is more distinct from seawater (21). Fluids were also minimally disturbed during recovery and kept in a narrow temperature range between sampling and incubations. In addition, bottom seawater was filtered and collected at 4397-m water depth by holding the MPS pump inlet ~5 m off the seafloor and pumping for 80 min. All filters collected on the seafloor had a reservoir of RNAlater (Ambion, Austin, TX, USA) that could weep into the filter for in situ fixation as described in (18). Upon recovery, filters were removed from the reservoirs and placed in fresh RNAlater, incubated at 4°C for 18 hours and subsequently stored at −80°C shipboard.
Aquifer fluid was also collected by the MPS (~0.5 LPM for ~20 min) into multiple acid-washed custom 15-liter Tedlar bags for onboard SIP incubations and shipboard filtration. Approximately 10 liters was filtered in series to capture “particle-attached” (5 μm) and “free-living” (0.22 μm) fractions (47-mm GSWP, MilliporeSigma). Tedlar bag fluids were transferred to SIP microcosms using a peristaltic pump, sterile acid-washed tubing, and sterile needles (for incubations). Bottom seawater for microcosms was collected by Niskin bottles via a CTD water sampling rosette to the same bottom seawater depth as the in situ MPS filter. These fluids were collected by bringing Niskin bottles into the laboratory and processed in the same manner as MPS bag fluids. Filtering on deck was carried out using the same filter holders with in situ fixation as those used on the seafloor.
DNA extraction, 16S rRNA gene amplification, sequencing, and analyses
DNA was extracted using a phenol chloroform protocol (16) from in situ and onboard filters from all four CORK horizons (1382A and U1383C Shallow, Middle, and Deep) and the bottom seawater samples (MPS and CTD) and both filter size fractions. 16S rRNA gene amplicon libraries were prepared and sequenced by the UConn Microbial Analysis, Resources, and Services using modified Earth Microbiome Project primers (37–40). 16S rRNA gene reads were processed using mothur (v.1.39.5; 38), and 97% similarity operational taxonomic units (OTUs) were classified with the SILVA v128 database (41). A distance matrix of these OTUs was computed using Bray-Curtis dissimilarity with vegan (42). This matrix was then ordinated using classical multidimensional scaling with the R function cmdscale and clustered with the maximum allowable number of clusters (5) and membership exponent (1.5). For taxonomy figure generation, only OTUs greater than or equal to 0.5% of an individual sample were retained and R ggplot tools were used for display (43).
SIP incubations
Incubations were prepared in 50-ml serum vials with labeled substrates added before the cruise in acid-washed and combusted glassware with crimped autoclaved butyl rubber stoppers. All serum vials contained 5 ml of 99.99% 2H2O and carbon and nitrogen isotopes were added to a final concentration of 0.2 mM 13C–sodium bicarbonate, 1.5 μM 13C–sodium acetate (dual carbon label), 10 nM 15N–ammonium chloride, 4 μM 15N13C-methylamine HCl, and estimated 2 μM 13C and 4 μM 15N from diatom lysate. Sample fluid (~45 ml) was added to each vial for a total volume of 50 ml. All isotopes were from Cambridge Isotopes (Tewksbury, MA, USA). Controls with no C and N label were also included. After fluid addition to prelabeled bottles, incubations were carried out for 12 hours, 2 days, or 6 days at either room temperature (~20°C, monitored during the cruise and remained within ±1°C of 19°C) or 4°C in a walk-in refrigerator. We chose 20°C as the upper temperature limit because early temperature measurements from North Pond suggested basement temperatures up to 20°C at approximately 600 mbsf (44). We chose 4°C as the lower temperature limit to represent the shallowest fluids at bottom water temperatures.
While shipboard incubations inherently induce a variety of disturbances to the in situ microbial community, these changes may be similar to the spectrum of potential microenvironments crustal fluid microbes might experience between fluid flow paths or different thermal regimes beneath the seafloor [e.g., (5, 45)]. Samples remained within a 10° to 15°C range of in situ temperatures during the entire sampling and preparation process. A subset of large-scale (1 liter) incubations were attempted near in situ pressures with fluids that were repressurized onboard; however, they formed a cloudy, “rust colored” precipitate, and so it was determined that the geochemical conditions of those incubations differed too much from the unpressurized samples to be comparable in this study and were not included.
We compare our results to SIP-NanoSIMS incubations from hydrothermal vent fluids that remained pressurized throughout sampling and experimentation (table S5) but experienced marked temperature changes during the collection and incubation preparation process (dropping from hydrothermal vent fluid temperatures to bottom water temperatures of ~4°C at times). The sediment SIP-NanoSIMS incubations used for comparison (table S5) also underwent significant alteration as isotope labels were added by making sediment slurries that alter the in situ environment of compacted sediments. Therefore, all SIP-NanoSIMS subsurface sediment or fluid incubations have had to make some alterations from in situ conditions for the practicalities of undertaking these difficult experiments, and, as such, results from any of these systems should be interpreted as potential rates of activity rather than direct field measurements.
While oxygen was not monitored during the incubations, microcosms should have remained oxygenated during the full length of all incubations. A separate study of oxygen from bottled North Pond fluids collected in 2017 showed almost no change (<4 μM variation) in concentration (~300 μM) after 553 days of incubation across sampling horizons (5). The amount of organic carbon provided (<5 μM) was much lower than in situ oxygen concentrations (>100 μM) and, thus, should not be enough to stoichiometrically deplete oxygen during the incubation.
Cell enumeration
For in situ cell counts, fluids were preserved in labeled scintillation vials, 2 × 18-ml sample, with 1.8 ml of 37% formaldehyde. Vials were mixed via shaking after adding fixative, sealed with electrical tape, and stored at 4°C. Cells were quantified using 4′,6-diamidino-2-phenylindole (DAPI) DNA stain and 100× objective on an epifluorescence microscope (Table 1).
For incubation cell counts, incubations were ended by removing ~2 ml of the fluid, followed by the addition of 1.9 ml of 40% paraformaldehyde (PFA). Each bottle was shaken gently and stored at 4°C until filtration on shore. Cell density was quantified in each incubation using the DAPI DNA stain on a 0.22-μm polycarbonate membrane (MilliporeSigma) and visualized under 100× objective on an epifluorescence microscope (46). Replicates were pooled for a given incubation condition, with 3 ml from each replicate filtered together for a total of 9 ml.
Diatom lysate
For the diatom lysate amendment, Thalassiosira sp. were grown to a density of 106 ml−1 in f/2 Medium with 8.82 × 10−4 M 15N–sodium nitrate (100% of added nitrate) and of 1 M 13C–sodium bicarbonate (50% of added bicarbonate). All isotopes were from Cambridge Isotopes (Tewksbury, MA, USA). The diatom culture was then concentrated 100× via centrifugation, processed with three freeze-thaw cycles to lyse the cells, and 200 μl of this concentrated lysate was added to each diatom lysate–amended microcosm. Assuming no loss during concentration steps, this would be the equivalent of the lysed complex organics of 105 diatoms ml−1 incubation or 4 μM total C (10−12 mol C/diatom) and 2 μM 13C-diatom. Using the Redfield ratio, this would yield 0.6 μM nitrogen for that amount of carbon. Therefore, we estimated that 4 μM diatom concentrate carbon was equivalent to 2 μM 13C, and 4 μM 15N was added to each diatom lysate condition.
DIC measurement
Whole fluids were sampled for dissolved inorganic carbon (DIC) measurement by peristaltic pump transfer from Tedlar fluid bags through acid-cleaned Masterflex Biopharm tubing into 100-ml glass bottles sealed with ground glass stoppers and Apiezon M grease. Bottles and tubing were precleaned with 10% HCl, and glass bottles and stoppers were subsequently cleaned of organic carbon residues by muffling at 450°C. DIC samples were preserved with 20 μl of saturated HgCl2 solution and stored at room temperature in the dark until analysis. DIC concentrations were measured on an AS-C3 analyzer (Apollo Scitech, Newark, DE, USA) after acidification with phosphoric acid and quantification as CO2 by a LI-COR infrared gas analyzer. DIC concentrations were calibrated against certified reference material produced by A.G. Dickson (Scripps Institution of Oceanography, San Diego, CA, USA), and each sample was measured in duplicate with repeat concentrations within 0.1% and measurement uncertainty of 2 μmol/kg. Overall uncertainty is likely somewhat higher due to fluid handling during recovery and transfer to sample bottles. We assign a conservative estimate of ±5 μmol/kg for DIC concentrations.
DOC measurement
Whole fluids were sampled for DOC measurement similarly to DIC samples and collected in 1000-ml amber glass bottles sealed with Polytetrafluoroethylene-lined lids. Concentrations from unfiltered samples are presented here. DOC concentrations were measured at the National Ocean Sciences Accelerator Mass Spectrometry facility as the CO2 yield after ultraviolet photo-oxidation of total DOC. CO2 concentrations were determined by manometric quantification with uncertainty of ±2 μM. DOC concentrations reported here for subsurface fluids and bottom water are similar to those reported previously (6).
Elemental analysis and isotope mass balance calculations
Precombusted 1.0-μm AP15 glass fiber filters (MilliporeSigma) were washed three times with artificial seawater and then dried in a 50°C oven overnight in individual sterile petri plates. Filters were analyzed at the Marine Biological Laboratory Stable Isotope Laboratory for δ15N and δ13C using a Europa 20-20 continuous-flow isotope ratio mass spectrometer interfaced with a Europa ANCA-SL elemental analyzer (Sercon Ltd., Cheshire, UK). The analytical precision based on replicate analyses of isotopically homogeneous international standards was ±0.1‰ for both δ15N and δ13C measurements and about 1% relative to the %N and %C. While δ15N was measured, concentrations were too low to accurately quantify. 13C-carbon incorporation was calculated for 2-day and 6-day incubations by determination of the total particulate C with a background correction using blank filters (combusted and washed with artificial seawater only). Across all runs, blank filters had an average C concentration of 0.48 μmol C with an SD of 0.11 μmol. The average value plus 1 SD for the blank filters measured on each run was chosen as the conservative estimate of total background C (0.65 μmol C to 0.58 μmol C across runs; table S1). We then solved for the total moles of 13C that was incorporated into biomass (nlabel) from the labeled substrate using the estimated 13C fractional abundances of the isotope labels used (flabel) and average 13C fractional abundance of from filters where no C or N label was added that had greater than 14 μg C (finitial; 0.0185; table S2). This resulted in the following equation: [nfinal(finitial − ffinal)]/[finitial − flabel] = nlabel, where nfinal = nlabel + ninitial and initial refers to the initial conditions of the experiment, final refers to the final conditions, and label refers to the isotope label incorporated into biomass. The resulting moles of 13C (nfinal) was then divided by the total volume filtered (144 ml per filter from all replicate incubations) and the length of the respective incubation. Diatom samples were not included in elemental analysis because remaining 13C/15N-labeled diatom particles were included on the filters. Samples were listed as below detection when either (i) the total C was below the blank filter estimate or (ii) the 13C fractional abundance was below the estimate for unlabeled biomass, either of which would result in a negative value for nlabel.
Using 2017 fluid geochemistry, in situ 13C-DIC was 8.4% 13C. However, taking into account the dilution of the in situ DIC concentration from the addition of 5 ml 2H2O, the atom % 13C would be between 9 and 10 atom % 13C. Therefore, an estimate of 10 atom % 13C was used for the 13C fractional abundance for DIC-amended incubations, which would err on the side of underestimating rate of uptake. As there were no measurements of acetate, methylamine, or diatom lysate, we used 1.5 μM in situ concentration as an estimate for 13C isotope dilution calculations based on an estimated 15% of low–molecular weight DOM (~10 μM). This yielded an estimated 50 atom % 13C-acetate, 73 atom % 13C-methylamine, and 57 atom % 13C–diatom lysate. Ammonium concentrations were all below detection from 2017, but, using 2014 data, they ranged from <0.02 to 0.15 μM, which would yield enrichment ranging from 6.3 to 33 atom %. However, enrichment as high as 60 atom % was observed in ammonium incubations (fig. S4), suggesting that ammonium concentrations may have been even lower in 2017, possibly as low as 0.0055 μM or 5.5 nM (64.5 atom % enrichment). We also used 5.5 nM for the remaining organic nitrogen concentrations, yielding 99.9 atom % 15N-methylamine and 99.1 atom % 15N–diatom lysate. Final estimates for each amendment are in table S3. These estimates were rounded to one significant figure given the combined uncertainties of measurement errors and estimation of in situ concentrations of C and N compounds. Estimated values were below the maximal fractional abundance value of 13C measured for each condition, where DIC-amended incubations do not exceed 0.1 f13C and the remaining incubations do not exceed 0.5 f13C or f15N.
For the NanoSIMS-derived average rates plotting with bulk EA data, average single-cell measurements were multiplied by the cell density for each incubation. Cell counts and NanoSIMS were performed on 0.22-μm polyethersulfone filters, but elemental analysis was carried out on 1.0-μm glass fiber filters; therefore, some biomass that was analyzed by the first two techniques may have been missed by the elemental analysis. PFA fixation has also been shown to underestimate biomass incorporation rates [~10% loss in carbon; (47)]. Elemental analysis therefore likely represents a lower estimate of the total incubation biomass production.
NanoSIMS analysis and single-cell rate calculations
A subset of 12-hour U1383C Shallow and Deep incubations were analyzed via NanoSIMS. Twelve-hour incubations were selected for minimal incubation time, with samples from Shallow and Deep U1383C as crustal fluid depth endmembers. For this analysis, the remaining sample after cell counts was pooled from each replicate (~144 ml), filtered (0.22-μm polycarbonate membrane; MilliporeSigma), washed with artificial seawater, dehydrated with ethanol, and gold-coated (10 nm) before analysis. Masses 1 (1H), 2 (2H), 24 (12C2), 25 (13C12C), 26 (14N12C), and 27 (15N12C) were collected for all samples along with the secondary electrons using a NanoSIMS-50L (Cameca, Gennevilliers Cedex, France) at the Caltech Microanalysis Center. A focused primary Cs+ beam was used for data collection after presputtering to stabilize ion counts. Three frames of 512 × 512 pixels were collected for each image, collected in a randomized grid pattern per sample.
The data were processed using Look@NanoSIMS software (48). Individual ion image frames were merged and aligned using the 12C14N ion image to correct for drift during acquisition. Cell-based regions of interest (ROIs) were determined by “interactive thresholding” with the 12C14N ion and 2H images. Final ion images and counts per ROI were calculated by summation of ion counts for each pixel over all scans. For NanoSIMS isotope incorporation plots, only ROIs with counts greater than 2× the Poisson error of the NanoSIMS measurement were displayed. ROIs were also visually inspected to look for potential charging effects or edge effects that may affect isotopic measurements. A one-point correction was also applied to the C and N isotope values based on methods in (12) using EA measurements of the polycarbonate filter (table S1).
Single-cell incorporation rates were calculated using the same method as the bulk rates except the amount of C and N per cell was calculated using ROI size information from NanoSIMS images. First, cell volume was estimated based on the area of an ROI (fig. S5). Then, per-cell volume was converted to fg C and fg N per cell based on the formulas from (49). For comparison, estimates of 86 fg C and 20 fg N per cell have been used to represent biomass estimates from subsurface sedimentary environments (11). The range of C (21 to 443 fg cell−1) and N (6 to 76 fg cell−1) across all ROIs is plotted in fig. S6. The largest ROIs consist of aggregates of cells that could not be visually divided. Data analysis and visualization was done using R (50) with the “ggplot2” (43), “dplyr” (51), “gridExtra” (52), and “RColorBrewer” (53) packages. Cell counts for each incubation were used to convert from femtomoles per cell per day to picomoles per milliliter per day (Table 3).
Global rate calculations
Global rate calculations were computed by multiplying the 12-hour single-cell NanoSIMS rates by the in situ cell concentration (3700 cells/ml for both U1383C Shallow and Deep) to get femtomoles per milliliter per day. This rate was then converted to moles per cubic meter per year by estimating the integral from 1 to 10 of a function for crustal volume versus age (54). This estimate for young (<10 Ma) crust that could be occupied by the suboceanic biosphere (1 × 1017 m3) was then multiplied by an estimated crustal porosity of 1% (55), resulting in an estimated habitable pore volume for young ridge flank crust of 1015 m3. This estimated crustal fluid volume was multiplied by the single-cell rates to calculate moles per year production (Table 3).
Acknowledgments
We thank the captain and crew of the R/V Atlantis, the pilots and engineers of the ROV JASON II, and B. Orcutt, C. G. Wheat, M. Mullis, K. Yoshimura, O. Nigro, B. Tully, G. Stewart, K. Freel, C. Sullivan, and M. Rappé for assistance in accomplishing the field program. We thank M. Otter for analytical assistance and resources for POC isotopic and mass measurements; N. Hussain and W.-J. Cai for analytical assistance and resources for bicarbonate measurements; M. Johnson for growing and providing the diatom culture; V. Orphan and Y. Guan for access and assistance with NanoSIMS analysis; and M. Serres, J. Amend, and D. LaRowe for discussions regarding these data. Funding: The Gordon and Betty Moore Foundation sponsored most of the observatory components at North Pond through grant GBMF1609. This work was supported by the National Science Foundation through grants NSF OCE-1745589, OCE-1635208, and OCE-1062006 to J.A.H. and NSF OCE-1635365 to P.R.G. and S.R.S.W.; NASA Postdoctoral Fellowship with the NASA Astrobiology Institute to E.T.-R.; L’Oréal USA For Women in Science Fellowship to E.T.-R.; and Woods Hole Partnership Education Program, sponsored by the Woods Hole Diversity Initiative to M.A.F.O. The Center for Dark Energy Biosphere Investigations (C-DEBI OCE-0939564) also supported the participation of J.A.H. and P.D.C. This is C-DEBI contribution number 564. Author contributions: Conceptualization: E.T.-R., J.A.H., S.R.S.W., and P.R.G. Investigation: E.T.-R., M.A.F.O., P.D.C., and S.R.S.W. Writing—original draft: E.T.-R. and J.A.H. Writing—review and editing: E.T.-R., J.A.H., S.R.S.W., and P.R.G. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Raw sequencing data are available through the NCBI Short Read Archive under project number: PRJNA603242. All remaining data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/18/eabg0153/DC1
REFERENCES AND NOTES
- 1.Fisher A., Wheat C. G., Seamounts as conduits for massive fluid, heat, and solute fluxes on ridge flanks. Oceanography 23, 74–87 (2010). [Google Scholar]
- 2.Wheat C. G., Fisher A. T., McManus J., Hulme S. M., Orcutt B. N., Cool seafloor hydrothermal springs reveal global geochemical fluxes. Earth Planet. Sci. Lett. 476, 179–188 (2017). [Google Scholar]
- 3.Johnson H. P., Pruis M. J., Fluxes of fluid and heat from the oceanic crustal reservoir. Earth Planet. Sci. Lett. 216, 565–574 (2003). [Google Scholar]
- 4.Langseth M. G., Becker K., Herzen R. P. V., Schultheiss P., Heat and fluid flux through sediment on the western flank of the Mid-Atlantic Ridge: A hydrogeological study of North Pond. Geophys. Res. Lett. 19, 517–520 (1992). [Google Scholar]
- 5.Wheat C. G., Becker K., Villinger H., Orcutt B. N., Fournier T., Hartwell A., Paul C., Geochem. Geophys. Geosyst. 21, e2019GC008804 (2020). [Google Scholar]
- 6.Jiao N., Herndl G. J., Hansell D. A., Benner R., Kattner G., Wilhelm S. W., Kirchman D. L., Weinbauer M. G., Luo T., Chen F., Azam F., Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599 (2010). [DOI] [PubMed] [Google Scholar]
- 7.D. A. Hansell, C. A. Carlson, Biogeochemistry of Marine Dissolved Organic Matter (Elsevier, 2015). [Google Scholar]
- 8.Meyer J. L., Jaekel U., Tully B. J., Glazer B. T., Wheat C. G., Lin H.-T., Hsieh C.-C., Cowen J. P., Hulme S. M., Girguis P. R., Huber J. A., A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci. Rep. 6, 22541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah Walter S. R., Jaekel U., Osterholz H., Fisher A. T., Huber J. A., Pearson A., Dittmar T., Girguis P. R., Microbial decomposition of marine dissolved organic matter in cool oceanic crust. Nat. Geosci. 11, 334–339 (2018). [Google Scholar]
- 10.D’Hondt S., Inagaki F., Orcutt B., Hinrichs K.-U., IODP advances in the understanding of subseafloor life. Oceanography 32, 198–207 (2019). [Google Scholar]
- 11.Morono Y., Terada T., Nishizawa M., Ito M., Hillion F., Takahata N., Sano Y., Inagaki F., Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18295–18300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trembath-Reichert E., Morono Y., Ijiri A., Hoshino T., Dawson K. S., Inagaki F., Orphan V. J., Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc. Natl. Acad. Sci. U.S.A. 114, E9206–E9215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morono Y., Ito M., Hoshino T., Terada T., Hori T., Ikehara M., D’Hondt S., Inagaki F., Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years. Nat. Commun. 11, 3626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNichol J., Stryhanyuk H., Sylva S. P., Thomas F., Musat N., Seewald J. S., Sievert S. M., Primary productivity below the seafloor at deep-sea hot springs. Proc. Natl. Acad. Sci. U.S.A. 115, 6756–6761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkel M., Pjevac P., Kleiner M., Littmann S., Meyerdierks A., Amann R., Mußmann M., Identification and activity of acetate-assimilating bacteria in diffuse fluids venting from two deep-sea hydrothermal systems. FEMS Microbiol. Ecol. 90, 731–746 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Trembath-Reichert E., Butterfield D. A., Huber J. A., Active subseafloor microbial communities from Mariana back-arc venting fluids share metabolic strategies across different thermal niches and taxa. ISME J. 13, 2264–2279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price A. N., Fisher A. T., Weathers T. S., Gable C. W., Stauffer P. H., Two- and three-dimensional numerical simulations of coupled fluid and heat flow processes beneath a marine sediment pond. AGU Fall Meeting Abstracts 33, (2019). [Google Scholar]
- 18.Cowen J. P., Copson D. A., Jolly J., Hsieh C.-C., Lin H.-T., Glazer B. T., Wheat C. G., Advanced instrument system for real-time and time-series microbial geochemical sampling of the deep (basaltic) crustal biosphere. Deep-Sea Res. I Oceanogr. Res. Pap. 61, 43–56 (2012). [Google Scholar]
- 19.Tully B. J., Wheat C. G., Glazer B. T., Huber J. A., A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 12, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. N. Orcutt, T. D’Angelo, C. G. Wheat, E. Trembath-Reichert, Microbe-mineral biogeography from multi-year incubations in oceanic crust at North Pond, Mid-Atlantic Ridge. Environmental Microbiology (2021); 10.1111/1462-2920.15366. [DOI] [PubMed]
- 21.Jungbluth S. P., Grote J., Lin H.-T., Cowen J. P., Rappé M. S., Microbial diversity within basement fluids of the sediment-buried Juan de Fuca Ridge flank. ISME J. 7, 161–172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch P., Rades-Rohkohl E., Some special problems in the determination of viable counts of groundwater microorganisms. Microb. Ecol. 16, 99–113 (1988). [DOI] [PubMed] [Google Scholar]
- 23.Kopf S. H., McGlynn S. E., Green-Saxena A., Guan Y., Newman D. K., Orphan V. J., Heavy water and 15N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats. Environ. Microbiol. 17, 2542–2556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinthaler T., van Aken H. M., Herndl G. J., Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic’s interior. Deep-Sea Res. II Top. Stud. Oceanogr. 57, 1572–1580 (2010). [Google Scholar]
- 25.Orcutt B. N., Sylvan J. B., Rogers D. R., Delaney J., Lee R. W., Girguis P. R., Carbon fixation by basalt-hosted microbial communities. Front. Microbiol. 6, 904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Feng X., Wang F., Diversity and metabolic potentials of subsurface crustal microorganisms from the western flank of the mid-Atlantic ridge. Front. Microbiol. 7, 363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bach W., Edwards K. J., Iron and sulfide oxidation within the basaltic ocean crust: Implications for chemolithoautotrophic microbial biomass production. Geochim. Cosmochim. Acta 67, 3871–3887 (2003). [Google Scholar]
- 28.Seyler L. M., Trembath-Reichert E., Tully B. J., Huber J. A., Time-series transcriptomics from cold, oxic subseafloor crustal fluids reveals a motile, mixotrophic microbial community. ISME J. 15, 1192–1206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki Y., Yamashita S., Kouduka M., Ao Y., Mukai H., Mitsunobu S., Kagi H., D’Hondt S., Inagaki F., Morono Y., Hoshino T., Tomioka N., Ito M., Deep microbial proliferation at the basalt interface in 33.5–104 million-year-old oceanic crust. Commun. Biol. 3, 136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason O. U., Nakagawa T., Rosner M., Van Nostrand J. D., Zhou J., Maruyama A., Fisk M. R., Giovannoni S. J., First investigation of the microbiology of the deepest layer of ocean crust. PLOS ONE 5, e15399 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Mara P., Schubotz F., Sylvan J. B., Burgaud G., Klein F., Beaudoin D., Wee S. Y., Dick H. J. B., Lott S., Cox R., Meyer L. A. E., Quémener M., Blackman D. K., Edgcomb V. P., Recycling and metabolic flexibility dictate life in the lower oceanic crust. Nature 579, 250–255 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Large P. J., Peel D., Quayle J. R., Microbial growth on C1 compounds. 2. Synthesis of cell constituents by methanol- and formate-grown Pseudomonas AM1, and methanol-grown Hyphomicrobium vulgare. Biochem. J. 81, 470–480 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peyraud R., Schneider K., Kiefer P., Massou S., Vorholt J. A., Portais J.-C., Genome-scale reconstruction and system level investigation of the metabolic network of Methylobacterium extorquens AM1. BMC Syst. Biol. 5, 189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arrieta J. M., Mayol E., Hansman R. L., Herndl G. J., Dittmar T., Duarte C. M., Dilution limits dissolved organic carbon utilization in the deep ocean. Science 348, 331–333 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Schwedt A., Seidel M., Dittmar T., Simon M., Bondarev V., Romano S., Lavik G., Schulz-Vogt H. N., Substrate use of Pseudovibrio sp. growing in ultra-oligotrophic seawater. PLOS ONE 10, e0121675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widner B., Fuchsman C. A., Chang B. X., Rocap G., Mulholland M. R., Utilization of urea and cyanate in waters overlying and within the eastern tropical north Pacific oxygen deficient zone. FEMS Microbiol. Ecol. 94, (2018). [DOI] [PubMed] [Google Scholar]
- 37.Apprill A., McNally S., Parsons R., Weber L., Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137 (2015). [Google Scholar]
- 38.Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J. A., Smith G., Knight R., Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., Knight R., Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parada A. E., Needham D. M., Fuhrman J. A., Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F. O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs, H. Wagner, vegan: Community Ecology Package (2017); https://cran.r-project.org/package=vegan.
- 43.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2016); https://ggplot2.tidyverse.org.
- 44.Becker K., Bartetzko A., Davis E. E., Leg 174B synopsis: Revisiting Hole 395A for logging and long-term monitoring of off-axis hydrothermal processes in young oceanic crust. Proc. ODP, Sci. Results B. 174, 1–13 (2001). [Google Scholar]
- 45.Fisher A. T., Permeability within basaltic oceanic crust. Rev. Geophys. 36, 143–182 (1998). [Google Scholar]
- 46.Porter K. G., Feig Y. S., The use of DAPI for identifying and counting aquatic microflora1. Limnol. Oceanogr. 25, 943–948 (1980). [Google Scholar]
- 47.Musat N., Stryhanyuk H., Bombach P., Adrian L., Audinot J.-N., Richnow H. H., The effect of FISH and CARD-FISH on the isotopic composition of 13C- and 15N-labeled Pseudomonas putida cells measured by nanoSIMS. Syst. Appl. Microbiol. 37, 267–276 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Polerecky L., Adam B., Milucka J., Musat N., Vagner T., Kuypers M. M. M., Look@NanoSIMS—A tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol. 14, 1009–1023 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Khachikyan A., Milucka J., Littmann S., Ahmerkamp S., Meador T., Könneke M., Burg T., Kuypers M. M. M., Direct cell mass measurements expand the role of small microorganisms in nature. Appl. Environ. Microbiol. 85, e00493-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2017); https://www.r-project.org/.
- 51.H. Wickham, R. François, L. Henry, K. Müller, RStudio, dplyr: A Grammar of Data Manipulation (2020); https://CRAN.R-project.org/package=dplyr.
- 52.B. Auguie, A. Antonov, gridExtra: Miscellaneous Functions for “Grid” Graphics (2017); https://CRAN.R-project.org/package=gridExtra.
- 53.E. Neuwirth, RColorBrewer: ColorBrewer Palettes (2014; https://CRAN.R-project.org/package=RColorBrewer).
- 54.Heberling C., Lowell R. P., Liu L., Fisk M. R., Extent of the microbial biosphere in the oceanic crust. Geochem. Geophys. Geosyst. 11, 2010Q08003 (2010). [Google Scholar]
- 55.Orcutt B. N., Wheat C. G., Rouxel O., Hulme S., Edwards K. J., Bach W., Oxygen consumption rates in subseafloor basaltic crust derived from a reaction transport model. Nat. Commun. 4, 2539 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/18/eabg0153/DC1


