RH6, RH8, and RH12 are previously unidentified D-body components and drive the formation of D-bodies through LLPSs.
Abstract
Eukaryotic cells contain numerous membraneless organelles that are made from liquid droplets of proteins and nucleic acids and that provide spatiotemporal control of various cellular processes. However, the molecular mechanisms underlying the formation and rapid stress-induced alterations of these organelles are relatively uncharacterized. Here, we investigated the roles of DEAD-box helicases in the formation and alteration of membraneless nuclear dicing bodies (D-bodies) in Arabidopsis thaliana. We uncovered that RNA helicase 6 (RH6), RH8, and RH12 are previously unidentified D-body components. These helicases interact with and promote the phase separation of SERRATE, a key component of D-bodies, and drive the formation of D-bodies through liquid-liquid phase separations (LLPSs). The accumulation of these helicases in the nuclei decreases upon Turnip mosaic virus infections, which couples with the decrease of D-bodies. Our results thus reveal the key roles of RH6, RH8, and RH12 in modulating D-body formation via LLPSs.
INTRODUCTION
In addition to membrane-bound organelles, such as nuclei, mitochondria, chloroplasts, and lysosomes, eukaryotic cells contain numerous membraneless organelles, such as nucleoli, Cajal bodies, processing bodies (P-bodies), and stress granules (SGs). It has been proposed that the components of membraneless organelles with intrinsically disordered regions (IDRs) control the formation of these membraneless organelles through liquid-liquid phase separations (LLPSs) (1, 2). For example, yeast DEAD-box RNA helicases Dhh1 and Ded1 drive the formation and turnover of P-bodies and SGs through LLPS in yeast (3–5). Lacking a lipid-rich barrier, membraneless organelles are highly dynamic macromolecular assemblages that can quickly form, undergo turnover, and exchange contents with the cellular milieu or other membraneless organelles in response to stress (1). However, the molecular mechanisms underlying and functional consequences of the rapid formation and turnover of most membraneless organelles remain elusive, especially those that occur in response to various biotic or abiotic stresses.
Viruses are obligate intercellular pathogens that produce a massive amount of viral RNAs and proteins in a short time, which causes severe cellular stresses to cells that they invaded. Viral RNAs and proteins are targeted by host membraneless organelles to attenuate virus proliferation. Triggered by viral infection and other environmental stresses, SG, a translationally silent membraneless organelle, deactivates the translation of the majority of mRNAs to conserve nutrients and energy for cell survival (6–9). Upon viral infection, SGs also sequester proteins that are critical for viral replication and translation (6). Likewise, P-bodies are cytosolic membraneless organelles that are normally required for mRNA storage and translation repression (10) and degrade viral RNAs to confer virus resistance upon viral infections (6–8). To achieve a robust and productive replication, viruses have evolved different strategies to repress the formation and function of membraneless organelles (6–9, 11). For example, some viruses encode proteins that interact with nucleator of SGs, such as Ras-GAP SHB domain-binding proteins (G3BPs), through the Phe-Gly-Asp-Phe (FGDF) motif to block SG assembly (12). Viruses can also inhibit P-body assembly by degrading or dispersing P-body components (8, 9, 11). However, for most membraneless organelles, the nature of their dynamics upon viral infections and the corresponding functions of the virus-induced alterations have not been determined.
An important nuclear membraneless organelle in plants is the dicing body (D-body), in which microRNAs (miRNAs) are processed (13). D-bodies contain at least three key components: the ribonuclease (RNase) III enzyme DICER-LIKE 1 (DCL1), the double-stranded RNA binding protein HYPONASTIC LEAVES 1 (HYL1), and the zinc-finger protein SERRATE (SE). Increasing amounts of evidence show that, in addition to their crucial roles in regulating plant growth and development, miRNAs are involved in plant antiviral defense (14). For counterdefense, viruses have evolved RNA silencing suppressors that inhibit host small interfering RNA and/or miRNA biogenesis and functional machinery, including D-body components (14–16). The constitutive expression of some RNA silencing suppressors in transgenic plants leads to developmental aberrations resembling mutants with hypomorphic mutations in D-body components (17, 18). These observations indicate that viruses may generate phenotypes resembling D-body mutants by interfering with D-body accumulation. However, the molecular mechanism of D-body formation and the dynamics of D-body accumulation upon stress treatments are still poorly understood.
In this study, we explored the new components of D-bodies and analyzed their roles in D-body formation. We found DEAD-box helicases RH6 (RNA helicase 6), RH8, and RH12 as previously unidentified components of D-bodies that modulate the formation of D-bodies and demonstrated that D-body accumulation decreases upon viral infection. Our results reveal the key roles of DEAD-box RNA helicases in modulating the formation and stress-induced dynamics of D-bodies and potentially other membraneless organelles.
RESULTS
Plant homologs of Dhh1/DDX6 are novel components of D-bodies
To identify novel components controlling the assembly of D-bodies, we analyzed D-body–associated proteins via immunoprecipitation (IP) coupled with mass spectrometry (MS) (IP-MS) in transgenic Arabidopsis thaliana plants expressing yellow fluorescent protein (YFP)–tagged HYL1 (HYL1-YFP) (13). In addition to HYL1, a total of 81 unique proteins were detected in the IP fractions of HYL1-YFP samples but were absent in the fractions of green fluorescent protein (GFP)–tagged hemagglutinin (HA) controls (table S1). Among these proteins, RH6, RH8, and RH12 were particularly interesting, as they are homologs of yeast Dhh1 and human DEAD-Box Helicase 6 (DDX6) (fig. S1 and tables S1 and S2) and Dhh1 is known to control P-body assembly and turnover in yeast (3, 5).
To test whether RH6, RH8, and RH12 are novel components of D-bodies, we first verified the physical associations of these three DEAD-box helicases with D-bodies. Bimolecular fluorescence complementation (BiFC) assays between RH6, RH8, or RH12 with the three key known D-body components HYL1, DCL1, or SE were first performed. Strong granular YFP signals were detected in the nuclei of Nicotiana benthamiana epidermal cells coexpressing RH6-cYFP, RH8-cYFP, or RH12-cYFP with DCL1-nYFP, HYL-nYFP, or nYFP-SE but were absent in epidermal cells coexpressing these helicases together with histone 2A (H2A) control proteins (Fig. 1A and fig. S2, A to B). Notably, these fluorescent granules resemble D-bodies (13). We also analyzed the colocalization of RH6 and RH8 together with HYL1 in the root tip elongation cells of 1-week-old A. thaliana transgenic seedlings stably expressing pRH6::RH6-3FLAG/pHYL1::HYL1-YFP or pRH8::RH8-3FLAG/pHYL1::HYL1-YFP. Immunofluorescence assays with FLAG and HYL1 antibodies showed that both RH6 and RH8 colocalized with HYL1 in the nucleus (Fig. 1B). In N. benthamiana epidermal cells, transiently expressed red fluorescent protein (RFP)–SE also colocalized with RH6-HA–cyan fluorescent protein (CFP), RH8-HA-CFP, and RH12-CFP as punctate structures in the nucleus (Fig. 1C). The interactions of RH6, RH8, and RH12 with DCL1, SE, and HYL1 were then validated by co-IP assays in N. benthamiana leaves. When RH12-FLAG was coexpressed with DCL1-YFP or 3HA-GFP, RH12 specifically coprecipitated with DCL1-YFP but not with 3HA-GFP (Fig. 1D). When IP with anti-FLAG antibodies RH12-CFP, RH6-HA-CFP, and RH8-HA-CFP was detected in the SE-FLAG and HYL1-FLAG fractions but not in the control Membrin 12 (MEMB 12)-FLAG fractions (Fig. 1, E to F, and fig. S2, C to F). These interactions were further validated in the leaves of 2-week-old A. thaliana transgenic plants stably expressing pRH12::RH12-3FLAG, which showed that RH12-3FLAG coprecipitated both SE and HYL1 (Fig. 1G). Direct interaction between RH12 and SE or HYL1 was evaluated by a pull-down assay using glutathione S-transferase (GST)– and 6×His-tagged recombinant proteins. A strong RH12 signal was detected from the GST-SE beads but not from the GST control beads, and a weak RH12 signal was detected from the GST-HYL1 beads (Fig. 1H). Together, these results suggest that RH6, RH8, and RH12 are previously unidentified components of D-bodies that physically associate with known D-body components.
Fig. 1. Plant homologs of Dhh1/DDX6 are novel components of D-bodies.
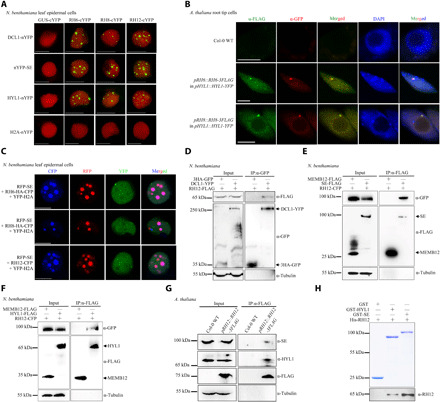
(A) BiFC analyses of N. benthamiana cells coexpressed with NLS-mCherry. YFP signals (green) and mCherry fluorescent signals (red) were observed at 72 hours post-inoculation (hpi). Scale bars, 10 μm. (B) Immunofluorescence analyses performed in A. thaliana root tip cells in the elongation region of stable transgenic plants. The immunofluorescence of RH6-3FLAG/RH8-3FLAG appears green, and the immunofluorescence of HYL1-YFP appears red. The positive cell percentages in the RH6-3FLAG and RH8-3FLAG lines were 74% (n = 87) and 80% (n = 70), respectively. The blue signals from 4′,6-diamidino-2-phenylindole (DAPI) represent nuclei. Scale bars, 5 μm. (C) Colocalization of RFP-SE together with RNA helicases in N. benthamiana. Fluorescence was analyzed at 72 hpi. Scale bars, 5 μm. YFP-H2A was used as nuclear marker. (D to F) Co-IP assays in N. benthamiana showing interactions between RH12 and DCL1 (D), SE (E), or HYL1 (F). (G) Co-IP assays in pRH12::RH12-3FLAG transgenic A. thaliana plants showing interactions between RH12 with SE and HYL1. (H) Detection of the direct interactions between RH12 with SE and HYL1 by pull-down assays. GST, GST-HYL1, and GST-SE (top) were used as matrix-bound bait, where RH12 served as prey. WT, wild type.
RH6, RH8, and RH12 are required for D-body formation
Yeast Dhh1 and human DDX6, which are homologs of RH6/RH8/RH12, drive P-body assembly via LLPS (3, 5). This prompted us to investigate whether RH6, RH8, and RH12 also influence D-body formation. To this end, we first examined D-body dynamics on the basis of fluorescence recovery after photobleaching (FRAP) by bleaching the fluorescence of a punctate structure with a laser. In both N. benthamiana leaves that transiently expressed DCL1-YFP or RFP-SE and stable transgenic A. thaliana plants expressing HYL1-YFP, fluorescence signals of the bleached punctata rapidly recovered after photobleaching (Fig. 2A), suggesting that D-bodies are membraneless organelles that can quickly exchange contents with their surrounding cellular environment.
Fig. 2. RH6/RH8/RH12 is required for D-body formation.
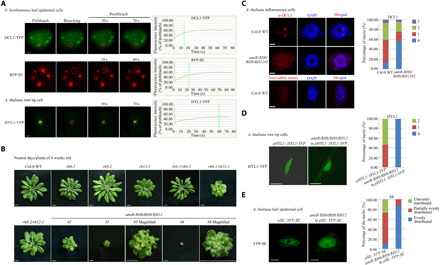
(A) Representative images and fluorescence recovery curves of D-body components. Leaf epidermal cells of N. benthamiana (inoculated with DCL1-YFP/RFP-SE for 72 hours) and root tip cells (elongation region) of 1-week-old A. thaliana (pHYL1::HYL1-YFP) plants were analyzed. The region of interest is highlighted with a red circle. Scale bars, 2 μm. The intensity was normalized against the average prebleach fluorescence. n = 3 for each group. The green line indicates the half-recovery time. (B) Whole-plant images of 6-week-old plants. Scale bars, 0.5 cm. (C) Detection of DCL1 subcellular localization with anti-DCL1 antibodies. Left: One set of representative images. Right: The quantification analysis. The numbers 0 to 3 represent the number of DCL1 granules in each nucleus. In total, 150 to 200 nuclei were analyzed for each genotype. Scale bars, 1 μm. (D and E) Subcellular localization of HYL-YFP (D) and YFP-SE (E) in 3-week-old A. thaliana plants. Fifteen root tip cells (elongation region) for HYL-YFP and 115 to 125 leaf epidermal cells for YFP-SE were analyzed for each genotype. Scale bars, 5 μm. Photo credit: Ningkun Liu, Institute of Zoology, Chinese Academy of Sciences.
To determine the possible functions of RH6, RH8, and RH12 in the formation of D-bodies, A. thaliana transferred DNA (T-DNA) insertion mutants of these helicases were obtained (fig. S3, A and B). Of the three single mutants, only rh12-1 exhibited obvious developmental abnormalities, e.g., smaller plant size and rosette leaf, under neutral-day conditions (12-hour light and 12-hour dark photoperiod; Fig. 2B and fig. S3C). The rh6-1/rh8-2 double mutant exhibited no obvious phenotypic changes, whereas rh6-1/rh12-1 and rh8-2/rh12-1 produced relatively small leaves. To verify that the observed phenotypes are due to loss of function of these helicases, the rh6-1/rh12-1 double mutant was transformed with either pRH6::RH6-3FLAG or pRH12::RH12-3FLAG. Both RH6 and RH12 were able to reverse the developmental defects of rh6-1/rh12-1 (fig. S3, D to E). Likely because of embryonic lethality, we were unable to obtain homozygous rh6-1/rh8-2/rh12-1 triple mutant. Therefore, we generated transgenic plants expressing an artificial miRNA (amiR-RH6/RH8/RH12) that can simultaneously knock down all three genes (fig. S3, A to B). The resulting transgenic lines were substantial smaller than the double mutants and produced severely wrinkled and downward-curling leaves (Fig. 2B and fig. S3C) under neutral-day conditions. Under long-day conditions (16-hour light and 8-hour light photoperiod), the amiR-RH6/RH8/RH12 transgenic plants also exhibited strong developmental phenotypes, but the single and double mutants exhibited no obvious phenotypes (fig. S3, F to G). The accumulation of amiR-RH6/RH8/RH12 in 4-week-old transgenic plants under neutral-day conditions was higher than that under long-day conditions, while the transcript levels of RH6, RH8, and RH12 and the protein level of RH12 are in opposite trends (fig. S3, H to J), suggesting that the enhanced phenotypes of amiR-RH6/RH8/RH12 transgenic plants at neutral-day conditions are likely due to higher level of amiR-RH6/RH8/RH12 expression.
The roles of RH6, RH8, and RH12 in the formation of D-bodies were then determined in amiR-RH6/RH8/RH12 transgenic plants. Immunofluorescence assays with DCL1 antibodies showed that nuclear D-bodies in the inflorescence cells were less frequently formed in the amiR-RH6/RH8/RH12 transgenic plants compared with the Col-0 wild-type (WT) plants (Fig. 2C). Similarly, nuclear D-bodies were eliminated in the root tip elongation cells of pHYL1::HYL1-YFP plants transformed with the amiR-RH6/RH8/RH12 construct (Fig. 2D and fig. S4A). In addition, YFP-SE signals became uniformly distributed in the leaf epidermal cells of amiR-RH6/RH8/RH12/pSE::YFP-SE transgenic plants, whereas they were unevenly distributed in the control plants (Fig. 2E and fig. S4B). These results suggest that the RH6/RH8/RH12 helicases are required for D-body accumulation. Since the transcript and protein levels of other D-body components (i.e., DCL1, HYL1, and SE) were not decreased in the amiR-RH6/RH8/RH12 plants (fig. S4, C to F), it is unlikely that these helicases facilitate D-body formation by regulating the abundance of other D-body components. Therefore, we reasoned that the RH6/RH8/RH12 helicases may play a more direct role in D-body formation.
RH6, RH8, and RH12 promote the formation of D-body via LLPS
We then tried to determine the molecular mechanisms underlying RH6/RH8/RH12-mediated D-body formation. An in silico prediction using IUPred2A (19) indicates that RH6, RH8, and RH12 contain an IDR at their N termini (fig. S5A). FRAP assays were conducted in N. benthamiana plants transiently expressing RH6-HA-CFP, RH8-HA-CFP, or RH12-CFP. Both RH6 and RH12 are conspicuously localized in nuclear speckles, whereas RH8 displayed less puncta (Fig. 3A and fig. S5B). The RH6-HA-CFP, RH8-HA-CFP, and RH12-CFP signals quickly recovered after photobleaching, indicating that RH6, RH8, and RH12 proteins rapidly redistribute between membraneless speckles and the surrounding protein pools. To further determine the phase separation characteristics of these helicases, A. thaliana transgenic plants expressing RH12-eGFP under its native promoter (pRH12::RH12-eGFP) were generated (fig. S5C). The GFP signal of the RH12-eGFP (enhanced GFP) granules in the nuclei of root tip meristematic cells of 1-week-old pRH12::RH12-eGFP plants also quickly recovered after photobleaching (Fig. 3A and fig. S5D). Like Dhh1 and DDX6 (3), the RH6, RH8, and RH12 proteins purified from Escherichia coli cells phase-separated into liquid-like droplets following the addition of single-stranded RNA (ssRNA) (polyU) and adenosine 5′-triphosphate (ATP) (Fig. 3B and fig. S5, E to F). In contrast, RH12, RH6, and RH8 remained dispersed in the absence of RNA or ATP, indicating that these helicases at high concentrations can undergo phase separation in an RNA- and ATP-dependent manner in vitro.
Fig. 3. RH6/RH8/RH12 drives D-body formation through LLPS.
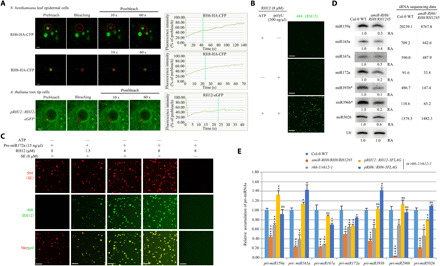
(A) Representative images and fluorescence recovery curves of the RNA helicase FRAP in N. benthamiana leaf epidemical cells or root tip meristematic cells of 1-week-old A. thaliana. Scale bars, 2 μm. The fluorescence intensity was normalized against the average prebleach fluorescence. n = 3 for each protein. The green dashed line indicates the half-recovery time. (B) In vitro phase separation of RH12 proteins with or without polyU (300 ng/μl) and/or 5 mM ATP. Scale bars, 20 μm. (C) In vitro phase separation of SE with/without RH12 under pre-miR172a (15 ng/μl). Scale bars, 20 μm. (D) Northern blot (left panels) and small RNA (sRNA) sequencing (right) analysis of miRNAs. U6 was used as a loading control. The relative abundance (RA) of miRNAs in amiR-RH6/RH8/RH12 transgenic plants compared to Col-0 WT plants is indicated. The sequencing data are shown as reads per million mapped reads. (E) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of selected pri-miRNAs in 3-week-old plants. Elongation factor 1-α (EF-1α) was used as an internal control. Each error bar represents 1 SD of three biological replicates. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; for the others, P ≥ 0.05. eGFP, enhanced GFP; ns, not significant.
In silico prediction via IUPred2A suggests that SE also contains long IDRs (fig. S5G). A recent study showed that SE contains an intrinsically unfolded region and undergoes degradation via the 20S proteasome to secure the functionality of folded and functional SE (20). Hence, we suspected that RH6, RH8, and RH12 may facilitate the phase separation of SE during D-body assembly. We purified prokaryotically expressed SE protein (fig. S5H) and tested its phase separation capacity in vitro. The SE proteins underwent phase separation in the absence of ATP and RNA (fig. S5I), and miRNA precursors (pre-miRNA) promoted their phase separation in vitro (fig. S5J). A study published while this study was under review also reported the phase separation property of SE (21). In contrast, phase separation of RH12 is ATP and RNA dependent (Fig. 3B). The purified SE protein was then mixed with varied concentrations of RH12 protein in the absence of ATP. As expected, RH12 remained dispersed in the absence of ATP (Fig. 3C), whereas the size and number of SE droplets increased along with the amount of RH12, suggesting that RH12 promotes the phase separation of SE in the absence of ATP in vitro.
RH6, RH8, and RH12 promote the accumulation of miRNAs
Given the important role of RH6, RH8, and RH12 in D-body formation and that D-bodies are where miRNA processing occur (13, 22), we hypothesized that these helicases may contribute to miRNA biogenesis. Therefore, we compared small RNA (sRNA) expression profiles between the aerial parts of 4-week-old amiR-RH6/RH8/RH12 plants and Col-0 WT plants with two replicates each. After removing adapter sequences and low-quality reads, we obtained 13.5 million to 18.5 million clean sRNA reads for each of the four samples (fig. S6A). In total, 13,155,146, 11,955,793, 13,598,138, and 15,264,650 reads were mapped to the A. thaliana genome. After removing the reads that mapped to structural RNAs [ribosomal RNAs (rRNAs), tRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs)], 8,196,598, 7,350,497, 8,146,914, and 9,117,670 perfectly matched reads remained, representing 1,644,096, 1,449,140, 1,488,037, and 1,684,795 unique sequences, respectively (fig. S6A). The accumulation of sRNAs from each library was normalized to the number of reads per million. Both 21- and 24-nucleotide (nt) sRNAs accumulated to lower levels in the amiR-RH6/RH8/RH12 plants compared with the Col-0 WT plants (fig. S6B). Plant miRNAs typically start with a uracil (U) at their 5′ termini (23–25). sRNAs (21 nt) with a 5′-U were less abundant in the amiR-RH6/RH8/RH12 plants than in the Col-0 WT plants (fig. S6C). The results of the miRNA profiling analysis (fig. S6D and table S3) and Northern blot of several well-studied miRNAs (Fig. 3D) further confirmed that malfunction of RH6, RH8, and RH12 inhibit miRNA accumulation. In addition, compared to Col-0 WT, the tested miRNA/miRNA* (the passenger strand of miRNAs) levels were also lower in rh6-1/rh12-1 and rh8-2/rh12-1 double mutants but not in single mutants or rh6-1/rh8-2 double mutants (fig. S6E). As expected, the transcriptional and translational levels of the miRNA target genes increased in the rh6-1/rh12-1, rh8-2/rh12-1, and amiR-RH6/RH8/RH12 plants (fig. S6, F to G).
miRNA genes are typically transcribed by RNA polymerase II (Pol II) as hairpin-containing primary transcripts (pri-miRNAs), which are subsequently processed by dicing complexes into mature and functional forms (22). The transcription and processing of plant pri-miRNAs are tightly coupled (22, 26, 27). To elucidate the mechanism through which RH6, RH8, and RH12 regulate miRNA biogenesis, the accumulation of pri-miRNAs in different genotypes was measured. The transcripts of examined pri-miRNAs were less abundant in rh6-1/rh12-1, rh8-2/rh12-1, and amiR-RH6/RH8/RH12 plants than in Col-0 WT plants but were not obviously altered in single mutants or rescued lines (Fig. 3E and fig. S6H). Chromatin IP (ChIP) coupled with quantitative polymerase chain reaction (PCR) assays were then performed with Pol II–specific antibodies in 2-week-old Col-0 WT and rh6-1/rh12-1 seedling plants. The association of Pol II with miRNA-encoding genes was attenuated in rh6-1/rh12-1 mutant plants (fig. S6I), suggesting that these helicases may facilitate the binding of Pol II to miRNA-encoding genes and promote the transcription of pri-miRNAs.
Accumulations of D-bodies and nuclear helicases decrease upon Turnip mosaic virus infection
Viral infection often results in alterations of membraneless organelles, such as P-bodies, SGs, and small interfering RNA bodies (6–9, 11, 28). Therefore, viral infection provides an excellent opportunity to study the dynamics of D-bodies upon stress treatment. Turnip mosaic virus (TuMV) is one of the most extensively studied members of the family Potyviridae, the largest family of plant RNA viruses (29). When N. benthamiana leaves were coinfiltrated with pDCL1::DCL1-YFP, p35S::RFP-SE, p35S::RH6-HA-CFP, or p35S::RH12-CFP together with TuMV (30), nuclear punctate structures labeled by DCL1-YFP, RFP-SE, RH6-HA-CFP, or RH12-CFP were significantly decreased (Fig. 4, A to C, and fig. S7, A to E). We also inoculated pHYL1::HYL1-YFP and pRH12::RH12-eGFP transgenic A. thaliana plants with TuMV or buffer. Nuclear punctate structures labeled by HYL1-YFP in A. thaliana root tip meristematic cells also significantly decreased in TuMV-infected plants (Fig. 4, D to E, and fig. S7F). In contrast to the localization patterns of RH12-eGFP in cells of 1-week-old A. thaliana roots (Fig. 3A and fig. S5D), RH12-eGFP in nuclei of 4-week-old A. thaliana roots were unevenly distributed, but punctate structures were rarely observed (Fig. 4D). The uneven distribution of RH12-eGFP in the nuclei of A. thaliana root tip meristematic cells decreased upon TuMV infection (Fig. 4, D and F). A time course further confirmed the decrease of D-bodies labeled by RFP-SE in epidermal cells of N. benthamiana leaves during TuMV infection (fig. S7, G to H). No significant alteration of the nuclear and perinuclear punctate structures was observed upon the infection of TuMV-GDD (31), a replication-defective clone of TuMV (fig. S8), suggesting that massive replication of TuMV per se is required for reduced D-body accumulation. Moreover, the infection of Cucumber mosaic virus (CMV; another well-studied plant virus) did not significantly alter the punctate structures of these D-body components in nuclei, suggesting that the decrease of D-body accumulation is not an artifact of the viral infection process (fig. S9).
Fig. 4. TuMV infection leads to decreased accumulation of D-bodies.
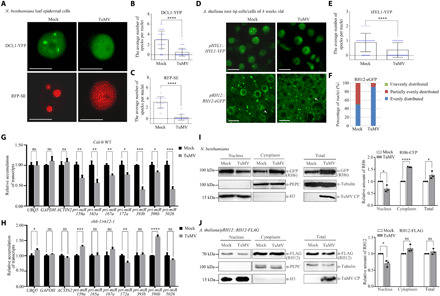
(A to F) Diffusion of the punctate signals of D-body components in the nucleus. (A) N. benthamiana plants were coinoculated with buffer (Mock)/TuMV with DCL1-YFP/RFP-SE. Fluorescence was observed at 72 hpi. (D) Three-week-old A. thaliana transgenic plants were inoculated with buffer (Mock)/TuMV. Fluorescence was observed at 10 days post-inoculation (dpi). The white arrowheads represent nucleus. Scale bars, 10 μm. (B, C, E, and F) Quantification analysis of the studies in (A) and (D). A total of 100 nuclei of root tip cells (elongation region) of five A. thaliana plants and 30 to 50 nuclei of N. benthamiana leaf epidermal cells of three leaves (one sample per leaf) were analyzed. (G and H) qRT-PCR analysis of selected pri-miRNAs in 6-week-old A. thaliana plants treated with Mock/TuMV. Systemic leaves were collected at 14 dpi. EF-1α was used as an internal control. (I and J) Quantitative detection of RH6/RH12 in nuclei after TuMV infection. H3 and PEPC: nuclear and cytoplasmic markers. Each error bar represents 1 SD of three biological replicates. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; for the others, P ≥ 0.05.
The decreased D-body accumulation upon TuMV infection intrigued us to study the function of TuMV on miRNA biogenesis. Similar to that in rh6/rh8/rh12 mutant plants (Fig. 3E and fig. S6H), pri-miRNA transcript levels but not control mRNA levels were decreased in the Col-0 WT plants after TuMV infection (Fig. 4G). In contrast, the transcript levels of pri-miRNAs did not further decrease in response to viral infection in the rh6-1/rh12-1 mutant plants (Fig. 4H), which indicated that these helicases play important roles in pri-miRNA decrease in Col-0 WT plants upon TuMV infection. The accumulation of mature miRNAs did not significantly decrease during TuMV infection (fig. S10A), which is likely due to P1/HC-Pro, a well-studied TuMV-encoded RNA silencing suppressor that promotes the accumulation of miRNAs (32).
The observations that RH6, RH8, and RH12 control the formation of D-bodies and play important roles in mediating pri-miRNA decrease upon TuMV infection implied that TuMV infection may reduce the accumulation of these helicases to a level below the threshold insufficient to trigger LLPS to form D-bodies. However, we found no significant decrease in the abundance of the three RNA helicases or the major D-body components DCL1, SE, and HYL1 in TuMV-infected plants compared to control plants (fig. S10, B to J). The accumulation of these helicase even increased in the TuMV-infected N. benthamiana plants (fig. S10, H to J), likely because of the RNA silencing suppressor function of TuMV P1/HC-Pro proteins (32). However, although the total abundance of RH6, RH8, and RH12 did not decrease upon TuMV infection, the accumulation of these proteins in the nuclei may differentially drop to a low level. We found that protein levels of RH6 and RH12 significantly decreased in nuclei and increased in the cytoplasm of TuMV-infected N. benthamiana plants compared to control plants (Fig. 4I and fig. S10K). The accumulation of RH12 protein also decreased in the nuclei but not in the cytoplasm of TuMV-infected A. thaliana plants (Fig. 4J). TuMV-induced decrease of RH6 and RH12 accumulation in the nucleus may lead to insufficient amount to trigger LLPS and formation of D-bodies. Therefore, viral infection most likely decreases the accumulation of D-bodies by reducing their formation efficiency. Moreover, under TuMV infection, punctate structures labeled by RFP-SE disappeared quickly compared to those inoculated with buffer (fig. S10L), which implies that TuMV infection may also promote the turnover of D-bodies.
RH6, RH8, and RH12 form virus bodies with viral replication proteins
Upon TuMV infection, RH6 and RH12 were enriched as punctate structures in perinuclear regions (figs. S7A and S8A). The 6-kDa viral protein (6K2), a small membrane protein encoded by potyviruses, induces the formation of viral vesicles where the replication of potyvirus takes place. These vesicles can gradually aggregate into large irregular inclusions in a similar perinuclear area (33–36). We thus hypothesized that these RH6/RH12-enriched puncta might be membraneless organelles containing viral proteins, and we termed them virus bodies (V-bodies).
To test this hypothesis, the colocalization of RH6, RH8, and RH12 granules with 6K2-induced perinuclear inclusions was first determined. TuMV::6K2-mCherry, a TuMV infectious clone expressing an N-terminal mCherry-tagged 6K2 protein between the P1 and HC-Pro cistrons, allows the direct monitoring of virus replication sites (37). In TuMV::6K2-mCherry-infected N. benthamiana and A. thaliana cells, many punctate structures of RH6-HA-CFP, RH8-HA-CFP, RH12-CFP, and RH12-eGFP were observed in 6K2-induced perinuclear inclusions and on chloroplast periphery (Fig. 5, A and B). In contrast, there were no perinuclear globular structures and few signals on the chloroplast periphery in the Mock-treated plants (fig. S11A). Next, we determined the dynamics of these structures with FRAP assay. The fluorescence signals of bleached puncta quickly recovered after bleaching (fig. S11B), which indicates that these compartments are membraneless organelles.
Fig. 5. RH6/RH8/RH12 promotes the phase separation of VPg.
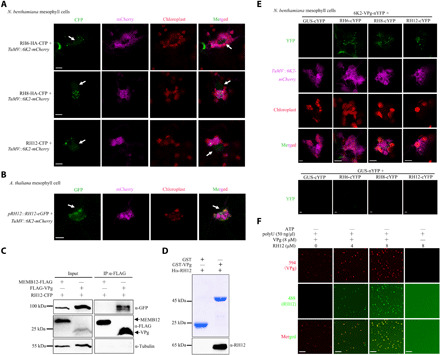
(A) Confocal fluorescence microscopy images showing the localization change of RH6-HA-CFP, RH8-HA-CFP, and RH12-CFP upon viral infection. RH6-HA-CFP, RH8-HA-CFP, or RH12-CFP was coinoculated together with TuMV in N. benthamiana plants. 6K2-mCherry represents the localization of virus protein 6K2 in cells. Fluorescence signals were observed at 72 hpi. The white arrows indicate nuclei. Scale bars, 10 μm. (B) Localization change of RH12-eGFP in pRH12::RH12-eGFP transgenic A. thaliana plants upon viral infection. Scale bar, 10 μm. (C) Co-IP analysis of the interaction between RH12 and VPg in N. benthamiana leaves. (D) GST pull-down assays confirming the direct interaction between RH12 and VPg. GST and GST-VPg (top) were used as matrix-bound bait, whereas RH12 served as prey. (E) BiFC analysis of the interactions of RH6/RH8/RH12 with 6K2-VPg in N. benthamiana plants infected with TuMV. Reconstituted YFP fluorescence (green), mCherry-tagged viral protein 6K2 (purple), and chloroplast autofluorescence (red) were observed at 72 hpi. Scale bars, 5 μm. (F) In vitro phase separation of 8 μM VPg proteins with or without RH12 under polyU (50 ng/μl; without ATP). Fluorescence microscopy images present liquid droplets formed by phase-separated proteins. Scale bars, 20 μm.
V-bodies may contain viral proteins. A previous study showed that viral protein linked to the genome (VPg) of Plum pox virus (another virus in the family Potyviridae) interacts with RH8 for robust replication (38). The high sequence similarity among RH6, RH8, and RH12 suggests that these helicases may also interact with TuMV VPg. To test this possibility, RH6-HA-CFP, RH8-HA-CFP, or RH12-CFP was coexpressed with FLAG-VPg or MEMB12-FLAG in N. benthamiana plants. The HA or CFP signal was detected in the FLAG-VPg fraction but not in the MEMB12-FLAG fractions (Fig. 5C and fig. S11, C to D). Similar results were obtained in TuMV-infected N. benthamiana plants (fig. S11, E to G). To test for direct interaction between VPg and RH12, a GST pull-down assay was performed with purified His-RH12 and GST-VPg proteins. Strong His-RH12 signals were detected from GST-VPg beads but not from control GST beads (Fig. 5D), confirming direct interaction between RH12 and VPg.
The possible localization of the VPg-RH6/8/12 protein aggregates in V-bodies was then studied in TuMV-infected plants using BiFC assays. Besides the mature form, VPg is also present as precursors, such as 6K2-VPg, 6K2-VPg-Pro, and VPg-Pro, during TuMV infection (39). We therefore determined the interaction between 6K2-VPg and these helicases by BiFC assay. Granules with strong YFP signals were readily detected in TuMV-infected plants coexpressing RH6-cYFP, RH8-cYFP, or RH12-cYFP with 6K2-VPg-nYFP but not in control plants (Fig. 5E and fig. S11H), which further confirmed the interactions between these helicases and VPg in vivo. Similar to the localization of RH6, RH8, and RH12 in TuMV-infected plants (Fig. 5A), the granules predominately located near 6K2-labeled membrane structures in TuMV-induced perinuclear globular structures or on the chloroplast periphery (Fig. 5E). These results showed that these membraneless V-bodies contain both viral and host proteins.
Since RH6, RH8, and RH12 play key roles in the formation of endogenous membraneless organelles, we speculated that they also contribute to the formation of V-bodies via promoting phase separation of viral proteins. An in silico prediction suggests that VPg also contains IDRs (fig. S12A). Using purified VPg protein from E. coli (fig. S12B), we found that VPg protein undergoes LLPS in the absence of RNA, and ssRNA (polyU) decreases its phase separation (fig. S12, C to E). VPg directly interacts with RH12 in vitro (Fig. 5D). Although high concentration of RNA inhibits the phase separation of VPg, phase separation of RH12 is dependent on RNA (Fig. 3B). Therefore, VPg may recruit RH12 to facilitate its phase separation. We tested whether these helicases promote VPg phase separation under high concentration of RNA. As phase separations of these helicases are ATP dependent (Fig. 3B), these helicase proteins remain dispersed without ATP (Fig. 5F and fig. S12F). As expected, the droplet number and size of VPg proteins increased with the addition of RH8 or RH12 proteins under high concentration of RNAs (Fig. 5F and fig. S12F). Moreover, we measured the fluorescence intensity to characterize the strength of phase separation. We found that phase separation of VPg in reaction with RH8 was slightly stronger than with RH12 (fig. S12G). Together, these results indicated that these helicases facilitate the formation of V-bodies via promoting the phase separation of VPg in the presence of RNA. To evaluate whether VPg directly inhibits RH12 phase separation, we mixed VPg and RH12 under high concentration of RNA. VPg proteins remain dispersed at polyU (300 ng/μl; fig. S12H). However, RH12 underwent LLPS similarly when mixed with VPg, which argues against an inhibition effect of VPg on the phase separation of these helicases. Together, these results suggest that these helicases facilitate the formation of V-bodies via promoting the phase separation of VPg.
Because of the close localization of these V-bodies and the 6K2-induced vesicles that conduct virus replication, we hypothesized that these V-bodies may facilitate virus proliferation. To evaluate this possibility, we compared viral accumulation in different genotypes. Arabidopsis plants (n = 8) were inoculated with TuMV::GFP, a TuMV infectious clone expressing a free GFP between P1 and HC-Pro cistrons for directly visualizing virus infection (30, 37). We found that TuMV::GFP infection was delayed in all single mutants and was further attenuated in double mutants. For instance, TuMV::GFP had invaded all rosette leaves of Col-0 WT plants but infected only half of rosette leaves in single mutants and was restricted to the middle of the rosette in double mutants at 14 days post-inoculation (dpi; Fig. 6A). These results were confirmed by the Plant Computer Vision (PlantCV) platform, a quantitative phenotyping protocol that has been used to analyze viral disease progress (Fig. 6, A to C) (40). The data suggested that mutation in these genes could significantly decrease the accumulation of TuMV, and the virus-resistance phenotype seems to be additive between these helicase genes. TuMV still infects amiR-RH6/RH8/RH12 transgenic plants, although the infecting area was restricted to the base of the rosette leaves (Fig. 6D). In the later stage after viral infection (28 dpi), Col-0 WT plants infected by TuMV were more stunted than TuMV-infected mutants (fig. S13). Consistently, the accumulation of TuMV coat protein (CP), HC-Pro, and VPg proteins in the systemic leaves of the mutant plants were much lower than that in the control plants (Fig. 6E). Together, these results suggest that these helicases play an important role in TuMV proliferation.
Fig. 6. RH6/RH8/RH12 facilitates viral proliferation.
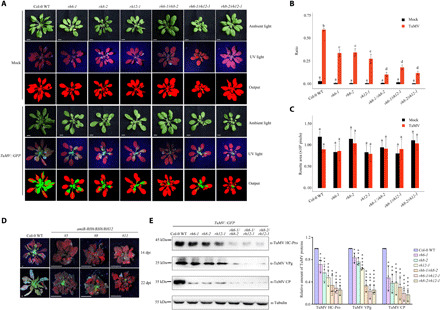
(A) Visualization of rosette leaves of plants inoculated with TuMV::GFP or buffer (Mock). Images were taken at 14 dpi. In the pseudocolor images, green color refers to infected plant pixels, while red color refers to noninfected plant pixels. Scale bars, 0.5 cm. (B) Bar plot showing the ratio of infected area to the total area of Col-0 WT and mutant plants at 14 dpi. (C) Bar plot showing the rosette size at 14 dpi. Statistical analysis was performed between treatments in each genotype. The boxes with different letters are significantly different (n = 8, Tukey post hoc test, with α = 0.05). (D) TuMV-infected amiR-RH6/RH8/RH12 plants. Four-week-old A. thaliana plants with TuMV::GFP were examined, and the plants were imaged under ultraviolet (UV) light at 14 and 22 dpi. Scale bars, 0.5 cm. (E) Western blot analysis of the accumulation of CP, VPg, and HC-Pro proteins. Representative images and quantitative analysis are shown on the left and right, respectively. Photo credit: Ningkun Liu, Institute of Zoology, Chinese Academy of Sciences.
DISCUSSION
In this study, we identified three DEAD-box helicases—RH6, RH8, and RH12—as previously unidentified D-body components. These helicases interact with SE, DCL1, and HYL1 and drive the formation of D-bodies by promoting LLPS (Fig. 7). Upon viral infection, these helicase proteins are decreased in nuclei and enriched near cytoplasmic viral replication sites as membraneless V-bodies, which correlate with D-body reduction and enhanced viral proliferation. Therefore, RH6, RH8, and RH12 play key roles in the formation of D-bodies and possibly other endogenous and exogenous membraneless organelles in plants through LLPS.
Fig. 7. Model of the formation of D-bodies driven by RH6/RH8/RH12.
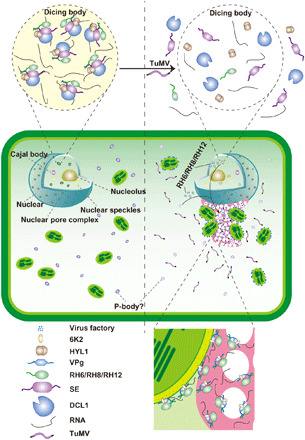
RH6, RH8, and RH12 interact with SE, the key D-body component, and drive the formation of D-bodies (left) under normal conditions. Upon infection with TuMV, RH6, RH8, and RH12 may be retained or translocated to the perinuclear globular structure and on chloroplast periphery, coupling with the formation of V-bodies and decreased accumulation of D-bodies (right).
A very recent study showed that RH6, RH8, and RH12 associate with and contribute to the formation of P-bodies and SGs to limit overaccumulation and translation of stress-responsive mRNAs, and malfunction of these helicases leads to a constitutive immune response in plants (41). It would be interesting to determine the dynamics of P-bodies and SGs upon the infections of TuMV. As VPg proteins do not harbor an FGDF motif (12), they may inhibit the assembly of SGs via their interactions with these helicases. Both SE and VPg contain IDRs, and their direct interaction with RH6, RH8, and RH12 suggests that intrinsically disordered proteins are likely to interact preferentially with other intrinsically disordered proteins to facilitate the formation of membraneless organelles. Many DEAD-box helicases contain IDRs, including human DDX39B and yeast Ded1 and UAP56, and regulate the phase separation of nuclear speckles, SGs, and nuclear foci (5). Therefore, DEAD-box helicases may serve as a common scaffold for the assembly of many membraneless organelles. While this study was under review, another related work revealed that SEs can undergo phase separation and control the assembly of D-bodies (21), which supports our conclusion that scaffold RNA helicase proteins indeed interact with unique IDR-containing proteins of membraneless organelles and coordinately drive the formation of distinct membraneless organelles.
All DEAD-box helicases contain a highly conserved core structure responsible for RNA binding, ATP binding, and hydrolysis. The phase separations of Dhh1, Ded1 (3, 5), RH6, RH8, and RH12 are both RNA and ATP dependent (Fig. 3B and fig. S5F). Moreover, the RNA-binding core of Dhh1 is required for LLPS and P-body assembly (5), and the adenosine triphosphatase activity of Dhh1 controls RNA partitioning from P-bodies to SGs. Chromatin remodelling factors 2 (CHR2), a core component of the switch/sucrose nonfermenting (SWI/SNF) chromatin-remodeling complex, also directly interacts with SE (42). CHR2 binds pri-miRNAs through SE and remodels the folding of pri-miRNAs, which prevents the processing of DCL1 and HYL1. As RH6, RH8, and RH12 also bind SE, these helicases may remodel the secondary structure of pri-miRNAs. All these observations point to a key role of RNAs in membraneless organelle assembly controlled by DEAD-box helicases. However, these RNA helicases interact primarily with the sugar-phosphate backbone of RNAs, but they lack the ability to recognize specific RNAs and require specific cofactors to help them perform spatial and temporal activities (43). The central core of many DEAD-box helicases is surrounded by a variable tail at one or both ends; this tail provides a platform for a variety of protein-protein interactions and is critical for the diverse functions of different helicases (43–46). RH6, RH8, and RH12 also contain a tail at their N termini and can directly interact with the D-body component SE and the viral protein VPg (Figs. 1H and 5D). In yeast, only DEAD-box helicases with IDRs at their tails, not those without IDRs, can undergo phase separations (5). Therefore, these helicases may target both RNAs and proteins, and this characteristic makes them key regulators of the formation of membraneless organelles. A recent study showed that RH27 associates with both major D-body proteins and pri-miRNA components and facilitates the processing of pri-miRNAs (47), which supports our conclusion.
Because of the important roles of RNA membraneless organelles in host resistance to viruses, viruses have evolved various strategies to interfere with these organelles and even explore key components for their own benefit (6–9, 11). For instance, the translation-to-replication transition of Brome mosaic virus occurs in P-bodies (48, 49); the BV1 protein of Cabbage leaf curl virus recruits ASYMMETRIC LEAVES 2 to P-bodies for reducing RNA interference (RNAi) and promoting virus infection through asymmetric leaves 2 (AS2)–activated, decapping 2 (DCP2)–mediated mRNA decapping (50). Some viruses also antagonize the formation/accumulation or functions of these immune organelles (6, 7). miRNAs regulate plant resistance to viruses by finely tuning the activation of plant resistance genes (R genes), RNA silencing components, hormone signaling pathways, and other resistance mechanisms (51). Our results showed that RH6, RH8, and RH12 were retained in cytoplasm and formed V-bodies during TuMV infection (Figs. 4, I and J, and 5, A and E, and figs. S7A and S10K), which correlates with the reduction of nuclear D-bodies and enhanced virus proliferation. Therefore, the enhanced virus replication may be associated with the decrease of D-bodies. Emerging evidence has shown that the infection of some animal viruses also modulates P-bodies and SGs by redistributing P-body and SG components (9). The DEAD-box RNA helicase components may play important roles in these processes. For example, DDX6 and DDX3 are recruited from P-bodies and SGs by Flaviviridae West Nile virus (52) and Hepatitis C virus (53–56) to viral replication factories, resulting in the disassembly of P-bodies and SGs. Dengue virus recruits DDX6 to the 3′ untranslated region of viral genomic RNAs, resulting in compromised P-body formation (57). Considering the regulatory roles of Dhh1 and Ded1 in yeast P-body and SG dynamics, the turnover of these membraneless organelles may also be due to the redistribution of DDX6 and DDX3 upon viral infection.
Although our results indicate that TuMV may target RH6, RH8, and RH12 to regulate D-body alteration, TuMV may also target other D-body components for function. TuMV may inhibit the phase separation and alter the localization or modification of other D-body components, leading to decrease in D-body accumulation. However, the accumulation of pri-miRNAs decreased in TuMV-infected Col-0 WT plants but not in TuMV-infected rh6-1/rh12-1 mutant plants, supporting the role of these helicases instead of other D-body components in D-body decrease upon TuMV infections. At the same time, although our results indicate that the localization change of RH6, RH8, and RH12 may trigger the decreased accumulation of D-bodies in the nucleus, TuMV may also inhibit the phase separation or cause posttranslational modification of these DEAD-box helicases. However, these hypotheses are inconsistent with the observation that these helicases can still undergo phase separation in perinuclear punctate structures (figs. S7A and S8A). Alternatively, RH6, RH8, and RH12 proteins may be protected by viruses from being posttranslationally modified in virus factories, which leads to the phase separation of them at viral replication sites but not in D-bodies. VPg interacts with RH6, RH8, and RH12 (Fig. 5, C to E, and fig. S11, C to G) (38), suggesting that VPg proteins may sequester these helicases into V-bodies. However, other TuMV proteins may interact with RH6, RH8, and RH12, and TuMV may indirectly alter the localization of these helicases. How TuMV targets RH6, RH8, and RH12 and alters their localization and whether TuMV alters their modifications need to be further investigated.
Viruses also organize viral and host proteins and nucleic acids into exogenous compartments in host cells. These virus-induced membranous and membraneless compartments not only concentrate molecules for their rapid proliferation but also shield viruses from host surveillance systems, such as RNA silencing and innate defenses (36, 58, 59). For example, the replication of negative-sense ssRNA viruses takes place in membraneless inclusion bodies that are recognized as exogenous membraneless organelles formed through phase separation (60–62). The HC-Pro encoded by Potato virus A and Potato virus Y, two other potyviruses, also induces the assembly of viral RNA granules that contain host varicose and viral VPg (63, 64). Although these viral RNA granules induced by HC-Pro are not enriched in the perinuclear area, it has been reported that these RNA granules may overcome the RNAi and stimulate the viral protein translation to achieve optimal viral gene expression and virus accumulation (63, 65). Our results indicate that TuMV generates membraneless V-bodies adjacent to virus factories in the perinuclear area to facilitate viral proliferation (Fig. 5E). V-bodies may also facilitate viral proliferation by stimulating viral protein translation or releasing viral RNA from host translational repression. Whether V-bodies and the viral RNA granules induced by HC-Pro share similar components and are functionally related are interesting questions for further investigation.
In summary, the membraneless characteristic enables membraneless organelles to respond quickly to stress by altering components or assembly/disassembly. It would be interesting to investigate the general roles of DEAD-box RNA helicases in the dynamics of other endogenous and exogenous membraneless organelles. Because of the key role of DEAD-box RNA helicases in the formation of different stress-related membraneless organelles, DEAD-box RNA helicases may be one of the key regulators of host-virus/pathogen interactions in both plants and animals.
MATERIALS AND METHODS
Plant materials and growth conditions
N. benthamiana and A. thaliana plants were grown in a green house at 22°C under 12-hour light/12-hour dark photoperiod (neutral days) or 16-hour light/8-hour dark photoperiod (long days). A. thaliana T-DNA insertion lines rh6-1 (Sail_111_H08), rh8-2 (GK_447_H07), and rh12-1 (Salk_016921) were obtained from the Nottingham Arabidopsis Stock Centre. Primers for genotyping analyses are listed in table S4. rh6-1/rh8-2, rh6-1/rh12-1, and rh8-2/rh12-1 double mutants were generated by crossing. Rescue lines pRH6::RH6-3FLAG/rh6-1, pRH8::RH8-3FLAG/rh8-2, pRH12::RH12-3FLAG/rh12-1, pRH6::RH6-3FLAG/rh6-1/rh12-1, and pRH12::RH12-3FLAG/rh6-1/rh12-1 were obtained by floral dip transformation using rh6-1, rh8-2, and rh12-1 single mutants and rh6-1/rh12-1 double mutants. p35S::amiR-RH6/RH8/RH12 was transformed into Col-0 WT to obtain amiR-RH6/RH8/RH12 transgenic plants. pRH12::RH12-eGFP/rh12-1 were generated by transformation pRH12::RH12-eGFP into rh12-1 mutant plants.
Transgenic plants pHYL1::HYL1-YFP and pSE::YFP-SE were described previously (13). p35S::amiR-RH6/RH8/RH12 were transformed into pHYL1::HYL1-YFP transgenic plants and pSE::YFP-SE transgenic plants to obtain amiR-RH6/RH8/RH12/pHYL1::HYL1-YFP and amiR-RH6/RH8/RH12/pSE::YFP-SE transgenic plants, respectively. pEarleyGate201-3HA-GFP was transformed into Col-0 WT to generate p35S::3HA-GFP transgenic plants.
Viral infection
The inoculations of TuMV, TuMV::GFP (TuMV-P1-GFP), TuMV::GFP-GDD, and TuMV::6K2-mCherry, were performed as described (30, 37, 66). Briefly, Agrobacterium tumefaciens GV3101 carrying TuMV constructs were resuspended with buffer (10 mM MgCl2, 0.15 mM AS, and 10 mM MES) to a final concentration of OD600 (optical density at 600 nm) = 0.5. Three- to 4-week-old A. thaliana plants were infiltrated with the agrobacterial solution. Fluorescence images of plants were taken at 14 or 22 dpi under an ultraviolet (UV) lamp or white light. Systemic tissues were harvested at 14 dpi for protein and RNA analyses. Four-week-old N. benthamiana leaves were infiltrated with the agrobacterial solution. Fluorescence microscopy analysis was performed at 3 dpi.
Plasmid constructions
To generate pENTR constructs, the full-length CDS (coding sequence) of RH6, RH8, RH12, and H2A (AT5G27670) and 4.2-kb genomic DNA fragment containing RH12 promoter and gene were amplified from Col-0 WT cDNA and genomic DNA. The TuMV VPg fragment was amplified from TuMV infectious clone. 2HA-GFP fragment was amplified from pEarleyGate103 with PrimeSTAR HS DNA Polymerase (Takara) and subsequently cloned into pENTR vector with a pENTR/SD/D-TOPO cloning kit (Invitrogen), which was cloned into pEarleyGate201 with Gateway LR Clonase II Enzyme Mix (Invitrogen) to form pEarleyGate201-3HA-GFP plasmid. BiFC vectors DCL1-nYFP, HYL1-nYFP, and nYFP-SE were constructed as described previously (26). After amplification, fragments containing CDS of RH6, RH8, RH12, and H2A were digested with Sal I and Spe I and then cloned into pCAMBIA1300-cYFP or pCAMBIA1300-nYFP to generate RH6-cYFP, RH8-cYFP, RH12-cYFP, and H2A-nYFP. 6K2-VPg fragment was amplified from TuMV infectious clone and cloned into pCAMBIA1300-nYFP to generate 6K2-VPg-nYFP. GUS-cYFP and GUS-nYFP were described previously (26).
For co-IP assays and transient expression assay, RH12 gene fragment was inserted into pCAMBIA1300-CFP by using the ClonExpress II One Step Cloning Kit (Vazyme, C112) to generate pCAMBIA1300-RH12-CFP. pEarleyGate102-RH6-HA-CFP, pEarleyGate102-RH8-HA-CFP, pEarleyGate102-FLAG-VPg, and pFH-RH12 were generated by the Gateway LR Clonase II Enzyme Mix (Invitrogen) with corresponding pENTR constructs and pEarleyGate102 or p35S:RP-FH (23). p35S::HYL1-FLAG, p35S::SE-FLAG, pDCL1::DCL1-YFP, and p35S::MEMB12-FLAG were described previously (13, 23, 67).
For transient expression assay, SE CDS fragment was amplified and digested with Sal I and Bam HI and inserted into pGD-dsRed2 though T4 ligation to generate pGD-dsRed2-SE. To generate the nuclear marker vector for colocalization, pEarleyGate104-YFP-H2A was generated by the Gateway LR Clonase II Enzyme Mix (Invitrogen) with pENTR-H2A.
To generate the pRH6::RH6-3FLAG, pRH8::RH8-3FLAG, and pRH12::RH12-3FLAG rescue plasmid, the 4.3-, 5.9-, and 4.2-kb genomic DNA fragments containing promoters and genes of RH6, RH8, and RH12, respectively, were amplified from Col-0 WT genomic DNA and cloned into pCAMBIA1305-3FLAG binary vector (68) by using the ClonExpress II One Step Cloning Kit (Vazyme, C112). To generate p35S::amiR-RH6/RH8/RH12 vector, the amiR-RH6/RH8/RH12 precursor was generated by overlapping PCR using pre-miR319a backbone and cloned into pEarleyGate100 as described previously (23). For IP-MS assay, p35S::3HA-GFP was generated with pENTR-2HA-GFP and pEarleyGate201 by recombination reactions using a Gateway LR Clonase II Enzyme Mix (Invitrogen). For observation of the localization of RH12 in Arabidopsis, pRH12::RH12-eGFP rescue plasmid was generated with pENTR-RH12 and pK7FWG0 by recombination LR reactions using Gateway LR Clonase II Enzyme Mix (Invitrogen).
For prokaryotic expression, pET-28a-6×His-RH6, pET-28a-6×His-RH8, and pET-28a-6×His-RH12 were constructed by inserting RH6 CDS, RH8 CDS, or RH12 CDS into pET-28a digested with Nde I and Sal I. Codon-optimized SE was synthesized (GenScript) and inserted into pET-28a that digested with Bam HI and Xho I. PreScission Protease (PSP) recognition sequences were added to the 5′ terminal of VPg by PCR; then, pGEX4T-1-GST-VPg was generated by inserting PSP-VPg into pGEX4T-1 that digested with Eco RI and Xho I. Prokaryotic expression vectors GST-SE and GST-HYL1 were constructed as described previously (69). All primers used were listed in table S4.
RNA extraction
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions and a previous study (70).
Northern blot analysis
A total of 15 to 120 μg of total RNAs isolated by TRIzol reagent (Thermo Fisher Scientific) was separated on 14% polyacrylamide-denaturing gel. RNA was cross-linked to membrane by chemical as described previously (70). Oligo was labeled with γ-32P ATP for probing. U6 was probed as internal control. The sRNA probes were listed in table S4. Image quantification was performed with ImageJ.
Reverse transcription PCR analysis
To perform mRNA reverse transcription PCR (RT-PCR), 10 μg of total RNA isolated by TRIzol reagent (Thermo Fisher Scientific) was firstly treated with deoxyribonuclease I (DNase I) [New England Biolabs (NEB)]. After reextraction, 1 μg of total RNA was converted to cDNA with the PrimeScript RT Reagent Kit (Takara). Semiquantitative RT-PCR was performed with Taq enzyme, and quantitative RT-PCR (qRT-PCR) was performed with a TB Green Premix Ex Taq kit (Tli RNase H Plus) (Takara). Elongation factor 1-α (EF-1α) or ACTIN2 was used as internal control, and primers used were listed in table S4.
To perform sRNA qRT-PCR analysis, 1 μg of total RNA isolated by TRIzol reagent was treated with DNase I (NEB). cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (NEB) primed by miRNA/U6-specific primers. qRT-PCR was performed with a TB Green Premix Ex Taq kit (Tli RNase H Plus) (Takara). U6 was used as internal control, and primers used were listed in table S4.
sRNA sequencing and analysis
sRNA sequencing and analysis were performed as described before (70). Briefly, 20 μg of total RNA isolated from 4-week-old plants by TRIzol reagent was used for sRNA library construction. sRNA libraries were constructed with the Illumina TruSeq sRNA Library Preparation Kit (Illumina) according to the manufacturer’s instructions. Two libraries for each sample were sequenced on Illumina HiSeq 2500 platform. The adaptors of sequencing reads were removed using cutadapt version 2.8. Reads that aligned to rRNAs/tRNAs/snoRNAs/snRNAs were removed using bowtie. The rest of the 18- to 30-nt reads that perfectly matchs the A. thaliana genome (TAIR10 version) were used for further analysis. For miRNA profiling, known mature miRNA sequences were downloaded from miRBase release 22.1. Reads with multiple matches were weighted by dividing the read count and equally assigning to each miRNA. For each sample, reads were normalized to per million mapped reads (RPM). miRNAs with average RPM ≥ 1 were used for differential analysis.
ChIP analysis
ChIP assay was performed as described previously (68). Briefly, nuclei were purified from 2 g of 14-day-old (neutral days) seedlings of Col-0 WT and rh6-1/rh12-1 plants. The Pol II antibody (Abcam, ab817) was used for ChIP assays, and rabbit immunoglobulin G (IgG) was used as negative control. The genomic fragments were enriched and eluted into 100 μl of TE (tris-EDTA) buffer. A total of 0.5 μl of sample was used for each qPCR reaction. Primers used were listed in table S4.
IP and MS
The IP was performed as described previously (23). Briefly, 8 g of 3-week-old seedlings of p35S::3HA-GFP and pHYL1::HYL1-YFP transgenic plants was collected and ground in liquid nitrogen. The plant powder was suspended in 40 ml of lysis buffer [50 mM tris-HCl (pH 8.0), 200 mM NaCl, 5% glycerol, 0.12% NP-40, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 protease inhibitor cocktail tablet (Roche)] and incubated for 10 min at 4°C. After 15 min of 18,300g centrifugation at 4°C, the supernatant was incubated with 80 μl of GFP-Trap beads (ChromoTek) overnight at 4°C. Followed by five times washing with lysis buffer, proteins copurified with 3HA-GFP or HYL1-YFP were extracted by boiling the beads in 1× SDS sample buffer. After electrophoresis and staining, each sample was cut from gel into five portions for further study.
The MS was done as described previously (68). For protein identification in A. thaliana, Proteome Discoverer (Thermo Fisher Scientific) and UniProt were used. The proteins with score >0 and normalized spectral abundance factor (NSAF) > 0.0007 and that are absent in negative control were considered as positive candidates.
Co-IP in N. benthamiana
For the co-IP between SE and HYL1 with RH12, RH6, or RH8, A. tumefaciens expressing p35S::SE-FLAG or p35S::HYL1-FLAG was coinoculated with A. tumefaciens containing pCAMBIA1300-RH12-CFP, pEarleyGate102-RH6-HA-CFP, or pEarleyGate102-RH8-HA-CFP in N. benthamiana, respectively. A. tumefaciens containing p35S::MEMB12-FLAG was coinoculated with A. tumefaciens containing pCAMBIA1300-RH12-CFP, pEarleyGate102-RH6-HA-CFP, or pEarleyGate102-RH8-HA-CFP as negative controls. A total of 2 g of leaves was collected and grounded in liquid nitrogen. The procedure of IP was the same as described above, and copurified proteins were immunoprecipitated by using 20 μl of anti-FLAG beads (Sigma-Aldrich, A2220).
For the co-IP assay between DCL1 with RH12, A. tumefaciens containing pDCL1::DCL1-YFP (13) was coinoculated with A. tumefaciens containing pFH-RH12 in N. benthamiana. A. tumefaciens containing p35S::3HA-GFP was coinoculated with A. tumefaciens containing pFH-RH12 as negative control. After grounding in liquid nitrogen, 3 g of plant power was suspended in 15 ml of lysis buffer [50 mM tris-HCl (pH 8.0), 250 mM NaCl, 5% glycerol, 0.1% NP-40, 2 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail tablet (Roche)]. After centrifugation, protein extract was incubated with 45 μl of GFP-Trap beads (ChromoTek) overnight.
For co-IP between VPg and RH6, RH8, or RH12, A. tumefaciens containing pEarleyGate102-FLAG-VPg and pEarleyGate102-RH6-HA-CFP, pEarleyGate102-RH8-HA-CFP, or pCAMBIA1300-RH12-CFP was coinoculated in N. benthamiana leaves. A. tumefaciens containing p35S::MEMB12-FLAG was used as negative control. Leaves (2 g) were grounded in liquid nitrogen and suspended in 10 ml of lysis buffer [50 mM tris-HCl (pH 8.0), 200 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail]. IP was performed by using 15 μl of anti-FLAG beads (Sigma-Aldrich, A2220). All the beads for Co-IP were suspended and boiled in 50 μl of SDS loading buffer and then resolved with 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel.
Co-IP in A. thaliana
Two-week-old seedlings of pRH12::RH12-3FLAG/rh12-1 transgenic plants (3 g) were collected and grounded in liquid nitrogen. The samples were then suspended in 15 ml of lysis buffer [50 mM tris-HCl (pH 8.0), 250 mM NaCl, 5% glycerol, 0.1% NP-40, 0.2 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail]. Anti-FLAG beads (20 μl; Sigma-Aldrich, A2220) were used to immunoprecipitate RH12-3FLAG overnight at 4°C. Col-0 WT was used as negative control. The beads were boiled in 50 μl of SDS loading buffer and then resolved with 10% SDS-PAGE gel.
Immunofluorescence
Immunofluorescence was performed as described previously (71). For DCL1 immunofluorescence, inflorescence tissue of Col-0 WT and amiR-RH6/RH8/RH12 were fixed in 3% paraformaldehyde under vacuum for 15 min at room temperature. DCL1 antibody (72) was used as the primary antibody (1:150 dilution). Goat anti-rabbit antibody labeled with Dylight 594 (Abbkine, A23420) was used as secondary antibody (1:300 dilution). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at 1 μg/ml. Col-0 WT inflorescence was used as negative control.
Protein extraction and Western blot
Plant samples were ground in liquid nitrogen, and total proteins were isolated by extraction buffer [0.2 M NaCl, 5 mM MgCl2, 5 mM DTT, 20 mM tris-HCl (pH 7.5), 0.03% Tween 20 (Ameresco), and 0.5 tablets of protease inhibitor (Roche)]. The supernatant was collected by centrifuging at 12,000 rpm for 15 min. Total proteins were examined by Western blot analysis using α-tubulin (1:5000; EASYBIO, BE0031) as a loading control.
Proteins in the study were also probed with α-RH12 (1:2000; GenScript, GTGRGAPPNPDYHQC), α-SE (1:2000; Agrisera, AS09532A), α-HYL1 (1:2000; Agrisera, AS06136), α-GFP (1:2000; EASYBIO, BE2001), α-FLAG (1:2000; Sigma-Aldrich, F7425), α-HA (1:2000; EASYBIO, BE7002), α-TuMV-VPg (1:200; GenScript, KGKSKGRTRGIGHKC), α-TuMV-CP (1:5000; GenScript, AGETLDAGLTDEQKC), α-TuMV-HC-Pro (1:2000), α-CMV-CP (1:3000), and α-MEMB12 (1:2000) (23). Secondary antibodies were goat anti-rabbit IgG (1:5000; EASYBIO, BE0101) and goat anti-mouse IgG (1:5000; EASYBIO, BE0102).
Phylogenetic analysis and sequence alignment
The DEAD-box RNA helicases from A. thaliana (73) and Homo sapiens (74) as well as the DDX6 ortholog in Saccharomyces cerevisiae, Xenopus laevis, Mus musculus, Caenorhabditis elegans, and Drosophila melanogaster were included in the phylogenetic analysis. Multiple sequence alignment was performed using MUSCLE (www.ebi.ac.uk/Tools/msa/muscle/). Conserved regions of the alignment were selected for subsequent phylogenetic analysis. A neighbor-joining tree was constructed using PAUP_dev_icc (http://phylosolutions.com/paup-test/). Clade support was estimated by 500 bootstrap replicates. The tree was rooted by midpoint rooting.
Transient expression in N. benthamiana
A. tumefaciens GV3101 containing overexpression vector was cultivated overnight and then suspended in inoculation buffer [150 μM acetosyringone, 10 mM MgCl2, and 10 mM MES (pH 5.7)] at an OD600 of 0.5. A. tumefaciens was injected into 4-week-old N. benthamiana plants with a needleless syringe, and further studies were performed at 3 dpi. For BiFC and co-IP assays, A. tumefaciens GV3101 was suspended in inoculation buffer at OD600 of 1.2.
Bimolecular fluorescence complementation
To determine the associations of helicases with DCL1, SE, and HYL1, A. tumefaciens containing DCL1-nYFP, HYL1-nYFP, or nYFP-SE was coinoculated with RH6-cYFP, RH8-cYFP, or RH12-cYFP in N. benthamiana (26). A. tumefaciens containing GUS-cYFP or H2A-nYFP was used as negative control. NLS-mCherry was coinoculated as nuclear marker (75).
To study the association of these helicases with 6K2-VPg, A. tumefaciens containing TuMV::6K2-mCherry or 6K2-VPg-nYFP was coinoculated with A. tumefaciens containing RH6-cYFP, RH8-cYFP, or RH12-cYFP in N. benthamiana. A. tumefaciens containing GUS-nYFP and GUS-cYFP was used as the negative controls. The fluorescence was determined at 3 dpi.
Protein colocalization in plants
To determine the colocalization of SE with RH6, RH8, and RH12, A. tumefaciens containing pGD-dsRed2-SE was coinoculated with A. tumefaciens containing pEarleyGate102-RH6-HA-CFP, pEarleyGate102-RH8-HA-CFP, or pCAMBIA1300-RH12-CFP in N. benthamiana, respectively. The fluorescence was determined at 3 dpi. The images were taken under confocal fluorescence microscopy with the same snap parameters.
Dual immunolocalization of RH6 or RH8 with HYL1 in A. thaliana was performed to detect their colocalization. Root tissue of F1 hybrids of pRH6::RH6-3FLAG, pRH8::RH8-3FLAG, and pHYL1::HYL1-YFP was fixed in 3% paraformaldehyde for 15 min. FLAG antibody (Sigma-Aldrich, F3165) and GFP antibody (Invitrogen, A-11122) were used as primary antibody (1:100 dilution) and incubated 3 hours in room temperature. Goat anti-mouse coupled with Dylight 488 (Abbkine, A23210) and goat anti-rabbit coupled with Dylight 594 (Abbkine, A23420) were used as secondary antibody (1:300 dilution). Nuclei were stained with DAPI at 1 μg/ml. The Col-0 WT root tissue was as negative control.
To determine the localization of RH6, RH8, and RH12 in TuMV-induced perinuclear globular structures in N. benthamiana, A. tumefaciens containing TuMV::6K2-mCherry was coinoculated with A. tumefaciens containing pEarleyGate102-RH6-HA-CFP, pEarleyGate102-RH8-HA-CFP, or pCAMBIA1300-RH12-CFP, respectively. The fluorescence was determined at 3 dpi.
To determine the localization of RH12 in TuMV-induced perinuclear globular structures in A. thaliana, 3-week-old pRH12::RH12-eGFP/rh12-1 transgenic plants were infiltrated with TuMV::6K2-mCherry. The fluorescence was determined in infiltrated leaf at 10 dpi.
Prediction of IDRs
The IDRs were predicted with the online tool IUPred2A (https://iupred2a.elte.hu) with default parameters.
Protein expression and purification
Recombinant proteins were expressed in E. coli BL21 (DE3) cells. Transformed cells were grown in LB at 37°C until OD600 of 0.6. E. coli strain expressing GST-SE, GST-HYL1, 6×His-RH6, 6×His-RH8, 6×His-RH12, or 6×His-SE protein was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C overnight. E. coli strain expressing GST or GST-VPg protein was induced with 0.2 mM IPTG at 37°C for 3 hours. Protein purification was performed at 4°C.
For 6×His-RH12 and 6×His-SE purifications, bacterial cells collected were resuspended in lysis buffer [50 mM phosphate buffer (pH 8.0), 300 mM NaCl, 1% Triton X-100, and 1 pellet per 50 ml of cOmplete EDTA-free protease inhibitor (Roche)] and lysis by sonication. After centrifugation, the supernatants were affinity-extracted with Ni–nitrilotriacetic acid (NTA) resin and dialyzed into protein storage buffer [20 mM Hepes (pH 7.5), 200 mM NaCl, and 1 mM DTT]. After that, proteins were further purified by size exclusion with a Superdex 200 column (GE Healthcare) in protein storage buffer. The peak fractions containing the proteins were collected and concentrated by 10-kDa molecular weight cutoff (MWCO) Centricon (Millipore).
For 6×His-RH6 and 6×His-RH8 purifications, bacterial cells collected were resuspended in lysis buffer [50 mM phosphate buffer (pH 7.5), 300 mM NaCl, 1% Triton X-100, and 1 pellet per 50 ml of cOmplete EDTA-free protease inhibitor (Roche)] and lysis by sonication. After centrifugation, the supernatants were affinity-extracted with Ni-NTA resin and dialyzed into protein storage buffer 2 [20 mM Hepes (pH 7.5), 200 mM NaCl, 5% glycerol, and 2 mM DTT]. After that, proteins were further purified by size exclusion with a Superdex 200 column (GE Healthcare) in protein storage buffer 2. The peak fractions containing the proteins were collected and concentrated by 10-kDa MWCO Centricon (Millipore).
To purify the GST-VPg protein, bacterial cells collected were resuspended in lysis buffer [50 mM tris-HCl (pH 8.0), 200 mM NaCl, 1% Triton X-100, and 1 pellet per 50 ml of cOmplete EDTA-free protease inhibitor] and lysis by sonication. After centrifugation, the supernatants were affinity-extracted with Glutathione Sepharose 4B. For phase separation assay, VPg was cleaved with PSP from the beads at 4°C overnight. For pull-down assay, GST-VPg was eluted by glutathione. Then, proteins were dialyzed into storage buffer [20 mM Hepes (pH 7.5), 200 mM NaCl, and 1 mM DTT]. After that, proteins were further purified by size exclusion with a Superdex 75 column (GE Healthcare) in storage buffer. The peak fractions containing VPg were collected and concentrated by 10-kDa MWCO Centricon (Millipore).
For GST, GST-HYL1, and GST-SE protein purifications, bacterial cells collected were resuspended in lysis buffer [50 mM tris-HCl (pH 8.0), 200 mM NaCl, 1% Triton X-100, and 1 pellet per 50 ml of cOmplete EDTA-free protease inhibitor] and lysis by sonication. After centrifugation, the supernatants were affinity-extracted with Glutathione Sepharose 4B and dialyzed into protein storage buffer [20 mM Hepes (pH 7.5), 200 mM NaCl, and 1 mM DTT]. After that, the supernatants were concentrated by 10-kDa MWCO Centricon (Millipore). The final purified protein was quantified by Bradford assay and SDS-PAGE, then snap-frozen in small aliquots by liquid nitrogen, and stored at −80°C.
GST pull-down assay
Recombinant GST, GST-HYL1, GST-SE, or GST-VPg protein (10 μg) was incubated with 30 μg of 6×His-RH12 and GST beads, respectively. Mixer were rotated at 4°C for 2 hours, and GST beads were washed six times with wash buffer [100 mM NaCl, 20 mM tris-HCl (pH 7.5), and 0.05% NP-40]. Proteins were eluted by boiling in 5× SDS sample buffer for 5 min. After that, proteins were separated on a 10% acrylamide gel and stained with Coomassie brilliant blue R-250. The RH12 prey protein was determined by Western blot with α-RH12 antibody.
Protein labeling
6×His-RH6, 6×His-RH8, and 6×His-RH12 proteins were added to Alexa Fluor 488 NHS Ester (succinimidyl ester) (Thermo Fisher Scientific, A20000) solution at a 1:1 molar ratio and rotated at room temperature for 1 hour. 6×His-SE and VPg proteins were added to Alexa Fluor 594 NHS Ester (succinimidyl ester) (Thermo Fisher Scientific, A37572) solution at a 1:1 molar ratio and rotated at room temperature for 1 hour. After incubation, free dyes were removed with Sephadex G-25 column (GE Healthcare). The labeled proteins were snap-frozen in liquid nitrogen and stored at −80°C for further study.
In vitro liquid droplet reconstitution assay
Proteins were diluted to required concentration using storage buffer and added into a 20-μl reaction. ATP [from a 100 mM stock in 0.5 M Hepes-KOH (pH 7.5)], ssRNA (polyU; Sigma-Aldrich), or pre-miR172a was added to reactions. With 1 M KCl and 1 M Hepes-KOH with different pH, reaction mixers were adjusted to appropriate salt concentration and pH.
For the phase separation of RH6, RH8, or RH12, a master mix was prepared with 1 μl of Hepes-KOH (pH 6.6); 3 μl of polyU (2 mg/ml); 1 μl of ATP (100 mM ATP together with 200 mM MgCl2); 5 μl of RH6, RH8 (diluted to appropriate concentration with protein storage buffer 2), or RH12 (diluted to appropriate concentration with protein storage buffer); and 2 μl of 50% PEG8000 (polyethylene glycol, molecular weight 800) and added up to 20 μl with nuclease-free water and mixed by pipetting. For the phase separation of SE, a master mix was prepared with 1 μl of Hepes-KOH (pH 7.0), 2 μl of 1 M KCl, and 5 μl of SE (diluted to appropriate concentration with protein storage buffer) and added up to 20 μl with nuclease-free water; additional RNA and KCl were added as described. For the phase separation of VPg, a master mix was prepared with 1 μl of Hepes-KOH (pH 6.6), 1 μl of 200 mM MgCl2, and 5 μl of VPg (diluted to appropriate concentration with protein storage buffer) and added up to 20 μl with nuclease-free water; additional RNA and KCl were added as described.
For the phase separation of SE with RH12, a master mix was prepared with 1 μl of Hepes-KOH (pH 6.6), 2 μl of 1 M KCl, 1 μl of pre-miR172a (0.3 mg/ml), 7 μl of SE and/or RH12 (diluted to appropriate concentration with protein storage buffer) and added up to 20 μl with nuclease-free water. For the phase separation of VPg with RH12 in Fig. 5F, a master mix was prepared with 1 μl of Hepes-KOH (pH 6.6), 1 μl of 200 mM MgCl2, 1 μl of polyU (1 mg/ml), 8 μl of VPg, and/or RH12 (diluted to appropriate concentration with protein storage buffer) and added up to 20 μl with nuclease-free water. For the phase separation of VPg with RH8/RH12 in fig. S12F, a master mix was prepared with 1 μl of Hepes-KOH (pH 6.6), 1 μl of 200 mM MgCl2, 1 μl of polyU (1 mg/ml), 8 μl of VPg, and/or RH8/RH12 (diluted to appropriate concentration with protein storage buffer 2) and added up to 20 μl with nuclease-free water. For the phase separation of RH12 with VPg in fig. S12H, 1 μl of Hepes-KOH (pH 6.6), 3 μl of polyU (2 mg/ml), 1 μl of ATP (100 mM ATP together with 200 mM MgCl2), 8 μl of RH12 and/or VPg (diluted to appropriate concentration with protein storage buffer), and 2 μl of 50% PEG8000 and added up to 20 μl with nuclease-free water. Microscopy was performed with a Zeiss LSM 710 laser scanning confocal microscope, and the Zen Black software was used to collect and process data.
Microscopy
Fluorescence microscopy pictures were taken under Zeiss LSM 710 confocal microscope with Zeiss Zen 2010 software. CFP, YFP or eGFP, RFP/mCherry fluorescence, and chlorophyll autofluorescence were excited/visualized under 405 nm/470 to 530 nm, 514 nm/510 to 520 nm, 543 nm/620 to 630 nm, and 633 nm/633 to 645 nm, respectively. The fluorescence of Dylight 488 and Dylight 594 that were used in colocalization experiments were excited under 488 and 594 nm, respectively. Alexa Fluor 488 and Alexa Fluor 594 used in protein labeling experiments were excited under 488 and 594 nm, respectively. Sequential model was used when more than one fluorescent signals were recorded.
Fluorescence recovery after photobleaching
To bleach the fluorescence of RH6-HA-CFP, RH8-HA-CFP, or RH12-CFP, a punctate structure was selected by a red circle with Zeiss Zen 2010 software and photobleached with a 405-nm laser at speed of 10. The recovery images were taken at equal time intervals (about 1 to 2 s) under 470- to 530-nm absorption spectrum, and the signal intensity was calculated by Zeiss Zen 2010 software. DCL1-YFP, HYL1-YFP, and RH12-eGFP signals were bleached with a 488-nm laser. The recovery images were taken at equal time intervals (about 1 to 2 s) under 510- to 520-nm absorption spectrum. RFP-SE signal was bleached under a 543-nm laser. The recovery images were taken at equal time intervals (about 1 to 2 s) under 620- to 630-nm absorption spectrum. Number of each sample ≥ 3. The intensity of photobleaching is about 70 to 80% for each region of interest.
Statistical analysis of microscopy
A. tumefaciens containing pEarleyGate102-RH6-HA-CFP, pCAMBIA1300-RH12-CFP, pDCL1::DCL1-YFP, or pGD-dsRed2-SE was inoculated in N. benthamiana with or without A. tumefaciens containing TuMV. The number of punctate structures of CFP, YFP, or RFP in the nucleus was calculated under fluorescence microscope with the same settings. The signals close to perinuclear membranes were excluded. Three-week-old pHYL1::HYL1-YFP and pRH12::RH12-eGFP Arabidopsis plants were inoculated with or without A. tumefaciens containing TuMV. The root cells of these infected plants were observed at 10 dpi, and the punctate structures of YFP and eGFP in the nucleus were calculated.
Nuclear-cytoplasmic fractionation
The aerial part of 4-week-old A. thaliana under neutral days was collected. Nuclear-cytoplasmic separation was performed as described previously (76). α-H3 (EASYBIO, BE7004) and α-PEPC phosphoenolpyruvate carboxylase (Agrisera, AS09458) were used as nuclear and cytoplasmic controls, respectively.
PlantCV image analysis
Color images of individual Arabidopsis plants infected with virus or mock solution under UV light were processed using the PlantCV package to quantify the severity of disease symptoms (77, 78). PlantCV version 3.9 was used (https://plantcv.danforthcenter.org). Briefly, the naïve Bayes approach was applied to segment image pixels into three classes: (i) green plant pixels (“infected”), (ii) red plant pixels (“noninfected”), (iii) and background pixels (40). By using these three classifiers, plant tissues with GFP signals and other part of the rosette were highlighted in green and red. Then, the number of infected and noninfected plant pixels was summed and used to calculate the total area of rosette and the percentage of infection, as an indicator for disease severity. The raw input images, the analyzed output images, and analysis results are available at Figshare (https://doi.org/10.6084/m9.figshare.12923981.v1). The Tukey post hoc test in one-way analysis of variance (ANOVA) was applied to determine whether any statistically significant difference exist between the group means under different treatment in one genotype. As mentioned in the figure legends, boxes with different letters are statistically different (at α = 0.05). The statistical analysis was performed in R version 3.4.4.
Acknowledgments
We thank C.-e. A. Chang and K. Y. Wu for modulating the interaction site between VPg with RH12. We also thank J. C. Carrington, H. Jin, Y. Lii, and J. Chen for constructive suggestions. We are grateful to X. Chen, X. Cao, Y. Qi, J.-F. Laliberté, F. Ponz, G. Qin, B. Zheng, W. Qian, X. Tao, D. Xi, and M. Shi for sharing materials. Funding: This work is supported by the National Natural Science Foundation of China (NSFC 32090012, 91954105, 32090012, 31622046, 31900224, and 31872302), Ministry of Agriculture transgenic major projects (2016ZX08010001), the National Key Plan for Scientific Research and Development of China (2017YFD0200904 and 2019YFC1200504), Beijing Municipal Natural Science Foundation (5202017), Program of CAS (ZDBS-LY-SM027), and the Strategic Priority Research Program of the CAS (XDB11050700). Author contributions: X.Zha. initiated the project. X.Zha., Q. Li, Y.-W.Y., G.R., X.C., X.Zhe., and M.Y. designed the experiments. Q. Li, N.L., Q. Liu, X.Zhe., W.G., Yang Liu, Yan Liu, S.Z., Q.W., J.P., C.C., Y.M., and X.Zha. conducted the experiments. L.L., X.Zhe., and Y.-W.Y. performed the bioinformatic analyses. X.Zha., Y.-W.Y., G.R., X.C., X.Zhe., Q. Li, N.L., L.L., and Q. Liu wrote the paper. All authors have read and approved the manuscript for publication. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The dataset of sRNA deep sequencing has been deposited to NCBI under the accession number PRJNA603736. Additional data related to this paper may be requested from the authors. All materials must be obtained through a material transfer agreement.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/18/eabc6266/DC1
REFERENCES AND NOTES
- 1.Mitrea D. M., Kriwacki R. W., Phase separation in biology; functional organization of a higher order. Cell Commun. Sig. 14, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., Fuxreiter M., Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mugler C. F., Hondele M., Heinrich S., Sachdev R., Vallotton P., Koek A. Y., Chan L. Y., Weis K., ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife 5, e18746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomes E., Shorter J., The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hondele M., Sachdev R., Heinrich S., Wang J., Vallotton P., Fontoura B. M. A., Weis K., DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019). [DOI] [PubMed] [Google Scholar]
- 6.White J. P., Lloyd R. E., Regulation of stress granules in virus systems. Trends Microbiol. 20, 175–183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lloyd R. E., How do viruses interact with stress-associated RNA granules? PLOS Pathog. 8, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mäkinen K., Lõhmus A., Pollari M., Plant RNA regulatory network and RNA granules in virus infection. Front. Plant Sci. 8, 2093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaete-Argel A., Marquez C. L., Barriga G. P., Soto-Rifo R., Valiente-Echeverria F., Strategies for success. Viral infections and membraneless organelles. Front. Cell. Infect. Microbiol. 9, 336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang G.-J., Jang J.-C., Wu S.-H., Dynamics and functions of stress granules and processing bodies in plants. Plants 9, 1122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q., Sharma N. R., Zheng Z.-M., Chen M., Viral regulation of RNA granules in infected cells. Virol. Sin. 34, 175–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panas M. D., Schulte T., Thaa B., Sandalova T., Kedersha N., Achour A., McInerney G. M., Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLOS Pathog. 11, e1004659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y., Spector D. L., Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z., Li Y., Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 32, 88–99 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Csorba T., Kontra L., Burgyan J., Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 479, 85–103 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Zheng L., Zhang C., Shi C., Yang Z., Wang Y., Zhou T., Sun F., Wang H., Zhao S., Qin Q., Qiao R., Ding Z., Wei C., Xie L., Wu J., Li Y., Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice. PLOS Pathog. 13, e1006662 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mlotshwa S., Schauer S. E., Smith T. H., Mallory A. C., Herr J. M., Roth B., Merchant D. S., Ray A., Bowman L. H., Vance V. B., Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17, 2873–2885 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jay F., Wang Y., Yu A., Taconnat L., Pelletier S., Colot V., Renou J.-P., Voinnet O., Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLOS Pathog. 7, e1002035 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meszaros B., Erdos G., Dosztanyi Z., IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Sun D., Ma Z., Yamaguchi K., Wang L., Zhong S., Yan X., Shang B., Nagashima Y., Koiwa H., Han J., Xie Q., Zhou M., Wang Z., Zhang X., Degradation of SERRATE via ubiquitin-independent 20S proteasome to survey RNA metabolism. Nat. Plants 6, 970–982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie D., Chen M., Niu J., Wang L., Li Y., Fang X., Li P., Qi Y., Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 23, 32–39 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Mei J., Ren G., Plant microRNAs: Biogenesis, homeostasis and degradation. Front. Plant Sci. 10, 360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Zhao H., Gao S., Wang W.-C., Katiyar-Agarwal S., Huang H.-D., Raikhel N., Jin H., Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42, 356–366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mi S., Cai T., Hu Y. G., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C., Chen S., Hannon G. J., Qi Y., Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery T. A., Howell M. D., Cuperus J. T., Li D., Hansen J. E., Alexander A. L., Chapman E. J., Fahlgren N., Allen E., Carrington J. C., Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Fang X., Cui Y., Li Y., Qi Y., Transcription and processing of primary microRNAs are coupled by elongator complex in Arabidopsis. Nat. Plants 1, 15075 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Zhang S., Xie M., Ren G., Yu B., CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 110, 17588–17593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Contu L., Steiner S., Thiel V., Muhlemann O., The role of stress granules and the nonsense-mediated mRNA decay pathway in antiviral defence. Chimia 73, 374–379 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Riechmann J. L., Laín S., García J. A., Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73, 1–16 (1992). [DOI] [PubMed] [Google Scholar]
- 30.Sánchez F., Matínez-Herrera D., Aguilar I., Ponz F., Infectivity of Turnip mosaic potyvirus cDNA clones and transcripts on the systemic host Arabidopsis thaliana and local lesion hosts. Virus Res. 55, 207–219 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Cui H., Wang A., Plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early stage of infection. J. Virol. 90, 5119–5131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasschau K. D., Xie Z. X., Allen E., Llave C., Chapman E. J., Krizan K. A., Carrington J. C., P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Wan J., Basu K., Mui J., Vali H., Zheng H., Laliberté J.-F., Ultrastructural characterization of turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J. Virol. 89, 12441–12456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grangeon R., Agbeci M., Chen J., Grondin G., Zheng H., Laliberté J.-F., Impact on the endoplasmic reticulum and Golgi apparatus of turnip mosaic virus infection. J. Virol. 86, 9255–9265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei T., Huang T.-S., McNeil J., Laliberté J.-F., Hong J., Nelson R. S., Wang A., Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novoa R. R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C., Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97, 147–172 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cotton S., Grangeon R., Thivierge K., Mathieu I., Ide C., Wei T., Wang A., Laliberte J.-F., Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 83, 10460–10471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang T.-S., Wei T., Laliberté J.-F., Wang A., A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 152, 255–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beauchemin C., Boutet N., Laliberté J.-F., Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81, 775–782 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X., Fahlgren N., Abbasi A., Berry J. C., Carrington J. C., Antiviral ARGONAUTEs against turnip crinkle virus revealed by image-based trait analysis. Plant Physiol. 180, 1418–1435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chantarachot T., Sorenson R. S., Hummel M., Ke H., Kettenburg A. T., Chen D., Aiyetiwa K., Dehesh K., Eulgem T., Sieburth L. E., Bailey-Serres J., DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat. Plants 6, 675–685 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Wang Z., Ma Z., Castillo-Gonzalez C., Sun D., Li Y., Yu B., Zhao B., Li P., Zhang X., SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557, 516–521 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Sloan K. E., Bohnsack M. T., Unravelling the mechanisms of RNA helicase regulation. Trends Biochem. Sci. 43, 237–250 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Linder P., Jankowsky E., From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Jarmoskaite I., Russell R., RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 83, 697–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourgeois C. F., Mortreux F., Auboeuf D., The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17, 426–438 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Hou X.-L., Chen W.-Q., Hou Y., Gong H.-Q., Sun J., Wang Z., Zhao H., Cao X., Song X.-F., Liu C.-M., DEAD-BOX RNA HELICASE 27 regulates MicroRNA biogenesis, zygote division and stem cell homeostasis. Plant Cell 33, 66–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beckham C. J., Light H. R., Nissan T. A., Ahlquist P., Parker R., Noueiry A., Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 81, 9759–9768 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedro Galao R., Chari A., Alves-Rodrigues I., Lobao D., Mas A., Kambach C., Fischer U., Diez J., LSm1-7 complexes bind to specific sites in viral RNA genomes and regulate their translation and replication. RNA 16, 817–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye J., Yang J., Sun Y., Zhao P., Gao S., Jung C., Qu J., Fang R., Chua N.-H., Geminivirus activates ASYMMETRIC LEAVES 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLOS Pathog. 11, e1005196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S.-R., Zhou J.-J., Hu C.-G., Wei C.-L., Zhang J.-Z., MicroRNA-mediated gene silencing in plant defense and viral counter-defense. Front. Microbiol. 8, 1801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chahar H. S., Chen S., Manjunath N., P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 436, 1–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pager C. T., Schütz S., Abraham T. M., Luo G., Sarnow P., Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology 435, 472–484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ariumi Y., Kuroki M., Kushima Y., Osugi K., Hijikata M., Maki M., Ikeda M., Kato N., Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 85, 6882–6892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ariumi Y., Kuroki M., Abe K.-i., Dansako H., Ikeda M., Wakita T., Kato N., DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 81, 13922–13926 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pène V., Li Q., Sodroski C., Hsu C.-S., Liang T. J., Dynamic interaction of stress granules, DDX3X, and IKK-α mediates multiple functions in hepatitis C virus infection. J. Virol. 89, 5462–5477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward A. M., Bidet K., Yinglin A., Ler S. G., Hogue K., Blackstock W., Gunaratne J., Garcia-Blanco M. A., Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3’ UTR structures. RNA Biol. 8, 1173–1186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laliberté J.-F., Sanfaçon H., Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48, 69–91 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Den Boon J. A., Diaz A., Ahlquist P., Cytoplasmic viral replication complexes. Cell Host Microbe 8, 77–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinrich B. S., Maliga Z., Stein D. A., Hyman A. A., Whelan S. P. J., Phase transitions drive the formation of vesicular stomatitis virus replication compartments. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nikolic J., Le Bars R., Lama Z., Scrima N., Lagaudriere-Gesbert C., Gaudin Y., Blondel D., Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y., Su J. M., Samuel C. E., Ma D., Measles virus forms inclusion bodies with properties of liquid organelles. J. Virol. 93, e00948-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hafren A., Lohmus A., Makinen K., Formation of Potato virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLOS Pathog. 11, e1005314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hafren A., Eskelin K., Makinen K., Ribosomal protein P0 promotes Potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J. Virol. 87, 4302–4312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eskelin K., Hafren A., Rantalainen K. I., Makinen K., Potyviral VPg enhances viral RNA translation and inhibits reporter mRNA translation in planta. J. Virol. 85, 9210–9221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu G., Cui X., Dai Z., He R., Li Y., Yu K., Bernards M., Chen X., Wang A., A plant RNA virus hijacks endocytic proteins to establish its infection in plants. Plant J. 101, 384–400 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Su C., Li Z., Cheng J., Li L., Zhong S., Liu L., Zheng Y., Zheng B., The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to Promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev. Cell 41, 527–539.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Li Q., Wang X., Sun H., Zeng J., Cao Z., Li Y., Qian W., Regulation of active DNA demethylation by a methyl-CpG-binding domain protein in Arabidopsis thaliana. PLOS Genet. 11, e1005210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang X., Ren W., Zhao Q., Zhang P., Wu F., He Y., Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res. 42, 12224–12236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y., Gao W., Wu S., Lu L., Chen Y., Guo J., Men S., Zhang X., AtXRN4 affects the turnover of chosen miRNA*s in Arabidopsis. Plants 9, 362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tao J., Zhang L., Chong K., Wang T., OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. Plant J. 51, 919–930 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Zhang B., You C., Zhang Y., Zeng L., Hu J., Zhao M., Chen X., Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis. Nat. Plants 6, 956–969 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mingam A., Toffano-Nioche C., Brunaud V., Boudet N., Kreis M., Lecharny A., DEAD-box RNA helicases in Arabidopsis thaliana: Establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol. J. 2, 401–415 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Ostareck D. H., Naarmann-de Vries I. S., Ostareck-Lederer A., DDX6 and its orthologs as modulators of cellular and viral RNA expression. Wiley Interdis. Rev. 5, 659–678 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Ye R., Wang W., Iki T., Liu C., Wu Y., Ishikawa M., Zhou X., Qi Y., Cytoplasmic assembly and selective nuclear import of Arabidopsis ARGONAUTE4/siRNA complexes. Mol. Cell 46, 859–870 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Wang W., Ye R., Xin Y., Fang X., Li C., Shi H., Zhou X., Qi Y., An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23, 3565–3576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gehan M. A., Fahlgren N., Abbasi A., Berry J. C., Callen S. T., Chavez L., Doust A. N., Feldman M. J., Gilbert K. B., Hodge J. G., Hoyer J. S., Lin A., Liu S., Lizarraga C., Lorence A., Miller M., Platon E., Tessman M., Sax T., PlantCV v2: Image analysis software for high-throughput plant phenotyping. Peerj 5, e4088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fahlgren N., Feldman M., Gehan M. A., Wilson M. S., Shyu C., Bryant D. W., Hill S. T., McEntee C. J., Warnasooriya S. N., Kumar I., Ficor T., Turnipseed S., Gilbert K. B., Brutnell T. P., Carrington J. C., Mockler T. C., Baxter I., A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in setaria. Mol. Plant 8, 1520–1535 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/18/eabc6266/DC1


