An in-depth look at mammalian brain size evolution prompts a reevaluation of a traditional paradigm.
Abstract
Relative brain size has long been considered a reflection of cognitive capacities and has played a fundamental role in developing core theories in the life sciences. Yet, the notion that relative brain size validly represents selection on brain size relies on the untested assumptions that brain-body allometry is restrained to a stable scaling relationship across species and that any deviation from this slope is due to selection on brain size. Using the largest fossil and extant dataset yet assembled, we find that shifts in allometric slope underpin major transitions in mammalian evolution and are often primarily characterized by marked changes in body size. Our results reveal that the largest-brained mammals achieved large relative brain sizes by highly divergent paths. These findings prompt a reevaluation of the traditional paradigm of relative brain size and open new opportunities to improve our understanding of the genetic and developmental mechanisms that influence brain size.
INTRODUCTION
The brain is directly responsible for governing an animal’s interactions with its environment. As such, the brain is often considered to flexibly respond to selection in changing environments (1–3). Brain size is, however, also commonly accepted to be restrained by energetic requirements that are considered universal across all vertebrates (4, 5). This apparent paradox highlights that brain size is one of the most salient traits for understanding the fundamental balance between adaptability and constraint in evolution. Despite this importance, crucial aspects related to the timing, pattern, and drivers that underlie modern phenotypic diversity in brain size remain undescribed.
It has long been recognized that brain size scales with body size following a standard linear allometric power law (6). The scaling coefficient (slope) of this allometry is assumed to be relatively stable across vertebrate classes and orders (most often estimated as between 2/3 and 3/4) (7) and is thought to reflect universal energetic growth constraints (4, 5). Largely because of methodological limitations in phylogenetic comparative statistics, this working hypothesis has received little scrutiny. Previous studies have therefore mostly been limited to comparing residual variation along a stable slope [i.e., mean relative brain size or encephalization quotient (EQ), quantified through differences in the intercept of the evolutionary allometry] (7, 8). There is, however, evidence to suggest that changes in the slope (quantifying changes in brain-body covariation) may constitute an important additional source of comparative variation (9–12).
Identifying the points at which evolutionary shifts in brain-body covariation occur is of paramount importance to understanding the selection pressures that may be operating. Whereas shifts in intercept address changes in mean among traits, shifts in slope address changes in variation among traits (e.g., stabilizing selection restrains variation, divergent selection allows for variation) (13). Failing to account for the possibility that trait covariation may differ among groups of species could potentially hide crucial sources of variation that contribute to explaining phenotypic diversity. Moreover, evolutionary allometries (allometries quantified across species) are determined by ontogenetic and static allometries (across developmental time in the same individual and individuals of the same species, respectively) and thus are indicative of the genetic and developmental mechanisms that regulate growth (14). Consequently, more detailed information on the allometric patterns that characterize the brain-body relationship across evolutionary time will provide new opportunities to investigate the nature of the processes that shape those patterns. Last, the occurrence of shifts in slope would indicate that comparisons of relative brain size (and EQ) are only valid among groups with a similar slope. The ubiquitous approach of quantifying only residual variation along a stable slope may therefore lead to biased results and incorrect interpretations.
Birds and mammals are of particular interest in this context because they both independently evolved relatively larger brains than other vertebrate classes (7). This innovation was likely facilitated by an easing of the phenotypic integration between brain size and body size (15). Such decoupling leads to increased variation available to selection, which, in turn, is expected to heighten flexibility in response to selection (16). Recent work has shown that shifts in slope are paramount to explaining the brain’s evolutionary diversification in birds (12), demonstrating that the selective response to increased variation is not restricted solely to changes in mean relative brain size but may also play out in terms of changes in brain-body covariation. In mammals, shifts in brain-body covariation have been suggested to occur in primates (9), carnivorans (17), marsupials, (18), and among mammalian orders (11). However, it has remained unclear where and when such shifts occur throughout mammalian evolution and how they contribute to explaining variation in the brain-body relationship.
Several methodological innovations (19, 20), in conjunction with ever-increasing data availability, make it possible to test the widely held assumption that the slope of brain-body allometry is stable in mammals. Here, we use bivariate Bayesian multipeak Ornstein-Uhlenbeck (OU) modeling (19, 21) in combination with phylogenetic analysis of covariance (20) to identify changes in both slope and intercept of the evolutionary allometry of the brain-body relationship in mammals. We apply these methods to the largest taxonomic sampling to date, comprising 107 extinct species and 1311 extant species spanning 21 orders (table S1; we quantify size in terms of mass for both brain and body). Our approach allows us to identify where and when allometric shifts occur in mammalian evolution and whether these shifts are driven primarily by changes in brain or body size. Our results provide new insight into the types of selection that have shaped extant diversity and open new opportunities for research into the underlying mechanisms.
RESULTS
Allometric patterning through time
Across more than 1400 species, mammalian brain-body allometry comprises 30 distinct grades (F21,2 = 29.02, P < 0.001; L.Ratio = 525.68, P < 0.001, AICΔ = 488, AICѡ > 0.999; Figs. 1 and 2, Table 1, and table S2). The ancestral mammalian grade has a slope of 0.51 and is retained by several early radiating orders (golden moles and tenrecs, elephant shrews, dugongs and manatees, hyraxes, sloths, and armadillos), as well as by tree shrews, hares and rabbits, squirrels, flying lemurs, and tarsiers (Fig. 1). Shifts in slope are common (of 29 grade shifts, 16 include a shift in slope) and characterize both early and late diversification. Most early slope shifts occurred near the Cretaceous-Paleogene boundary (K-Pg; ~66 million years (Ma) ago; Fig. 1), and all indicate a shift toward a higher slope. This temporal clustering suggests that changes in the relative growth trajectory of brain and body size were fundamental for mammalian diversification in the wake of the K-Pg mass extinction. This aligns with a pattern recently observed in birds (12), suggesting that ecological radiation and subsequent niche expansion following the K-Pg mass extinction played a major role in shaping the trajectories by which both birds and mammals became the largest-brained vertebrate classes.
Fig. 1. Time-calibrated phylogeny of mammals with branch colors corresponding to the 30 significantly different allometric grades identified in this study (Table 1).
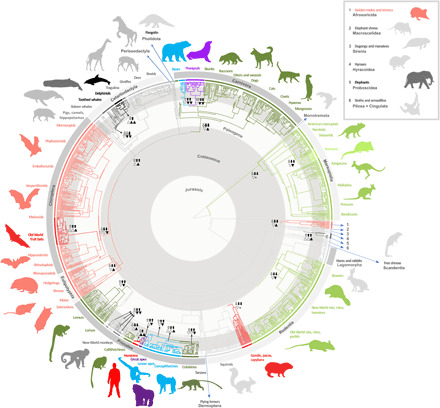
The ancestral mammalian grade is indicated in gray, with warmer colors (green and red) assigned to higher-slope grades, and colder colors (blue and purple) to lower-slope grades. For each grade, a lighter color hue indicates grades with a lower intercept, and a darker hue indicates grades with a higher intercept (Table 1). Arrows indicate changes in mean body size (white arrows) or mean brain size (black arrows) resulting in grade shifts, with double arrows indicating one of these variables is changing faster than the other after considering allometry. Triangles indicate changes in cross-species trait variance in body size (white triangles) or brain size (black triangles), with normal triangles indicating increase in mean variance and inverted triangles indicating decrease in mean variance (tables S3 to S5). The equality sign (=) indicates no discernible change in brain size variance. See data S2 for individual species labels. Illustration by J. Lázaro.
Fig. 2. pGLS regressions for each of the grades.
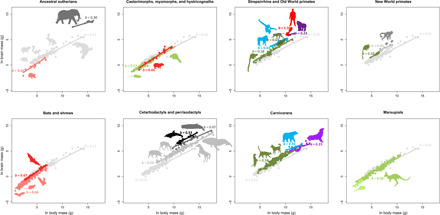
The ancestral mammalian grade is indicated in each display to provide an evolutionary context. Numbers indicate the value of the slope for each grade. Colors and silhouettes are as in Fig. 1.
Table 1. Phylogenetic generalized least-squares parameters for all grades identified in the analysis.
Grade numbers (1 to 6) indicate groups of clades with significantly different slopes (clades with the same grade number indicate slopes that are not significantly different from each other). Within each grade with a similar slope, grade letters indicate clades with a significantly different intercept. b and a refer to the values for the slope and intercept. “Lower” and “Upper” refer to the lower and upper bounds of the 95% confidence intervals. Note that some grades with low sample size are not listed here because they did not include a significant shift in slope (only a significant shift in intercept). These grades include elephants (n = 8), Cebus (n = 6), Atelinae (n = 8), Saki/Uakari (n = 4), Daubentonia (n = 1), Tragulina (n = 4), and pangolin (n = 1). These grades are, however, listed in table S3, which analyzes their mean brain and body sizes. NW, New World; OW, Old World.
| Slope | Intercept | ||||||||||
| Grade | Clade | n | b | SE | Lower | Upper | a | SE | Lower | Upper | |
| 1A | Pinnipeds | 32 | 0.226 | 0.080 | 0.064 | 0.389 | 3.001 | 1.045 | 0.872 | 5.130 | |
| 1B | Stem great apes | 7 | 0.229 | 0.061 | 0.085 | 0.373 | 3.520 | 0.657 | 1.967 | 5.073 | |
| 2A | Stem apes | 9 | 0.377 | 0.159 | 0.018 | 0.736 | 1.394 | 1.475 | −1.944 | 4.731 | |
| 2A | Ursids | 10 | 0.323 | 0.077 | 0.151 | 0.496 | 1.922 | 0.801 | 0.137 | 3.707 | |
| 2A | Cercopithecines | 56 | 0.426 | 0.033 | 0.360 | 0.493 | 0.675 | 0.312 | 0.050 | 1.301 | |
| 3A | Ancestral | 130 | 0.510 | 0.021 | 0.469 | 0.551 | −1.588 | 0.357 | −2.293 | −0.882 | |
| 3B | Stem NW monkeys | 16 | 0.402 | 0.051 | 0.294 | 0.510 | 0.309 | 0.383 | −0.503 | 1.121 | |
| 3B |
Stem fereuungulates |
100 | 0.542 | 0.020 | 0.501 | 0.582 | −1.028 | 0.295 | −1.612 | −0.443 | |
| 3D |
Stem-toothed whales* |
31 | 0.468 | 0.035 | 0.396 | 0.540 | 1.093 | 0.436 | 0.204 | 1.982 | |
| 3E | Delphinids | 16 | 0.533 | 0.046 | 0.435 | 0.631 | 0.847 | 0.550 | −0.320 | 2.013 | |
| 4A | Dunnarts | 15 | 0.545 | 0.092 | 0.349 | 0.741 | −2.645 | 0.251 | −3.180 | −2.111 | |
| 4B |
Castorimorphs/ myomorphs |
180 | 0.567 | 0.014 | 0.538 | 0.595 | −2.226 | 0.130 | −2.482 | −1.969 | |
| 4B | Stem marsupials | 150 | 0.580 | 0.018 | 0.544 | 0.615 | −2.140 | 0.174 | −2.484 | −1.797 | |
| 4C | Strepsirrhines | 56 | 0.577 | 0.031 | 0.514 | 0.639 | −1.403 | 0.267 | −1.938 | −0.869 | |
| 4C | Callitrichines | 16 | 0.578 | 0.059 | 0.452 | 0.704 | −1.279 | 0.361 | −2.044 | −0.515 | |
| 4C | Stem carnivorans | 178 | 0.577 | 0.019 | 0.539 | 0.614 | −1.666 | 0.204 | −2.068 | −1.264 | |
| 4D |
Stem cercopithecoids° |
29 | 0.671 | 0.074 | 0.520 | 0.822 | −1.676 | 0.673 | −3.052 | −0.300 | |
| 5A | Eulipotyphlans | 48 | 0.618 | 0.029 | 0.560 | 0.676 | −2.712 | 0.158 | −3.030 | −2.394 | |
| 5A | Stem bats | 217 | 0.672 | 0.015 | 0.642 | 0.701 | −2.834 | 0.068 | −2.968 | −2.700 | |
| 5A | Afrosoricids | 13 | 0.659 | 0.072 | 0.503 | 0.815 | −3.010 | 0.369 | −3.808 | −2.213 | |
| 5B | Hystricomorphs | 19 | 0.663 | 0.047 | 0.565 | 0.760 | −2.674 | 0.357 | −3.421 | −1.926 | |
| 5B | OW fruit bats | 47 | 0.665 | 0.016 | 0.633 | 0.697 | −2.366 | 0.071 | −2.510 | −2.223 | |
| 6A | Hominins | 11 | 1.097 | 0.164 | 0.736 | 1.458 | −5.304 | 1.722 | −9.095 | −1.514 | |
*“Stem toothed whales” refer to stem and crown nondelphinid toothed whales.
°“Stem cercopithecoids” consists of the extinct species Victoriapithecus and crown colobines.
Shifts in slope (both increases and decreases) were also crucial in later diversifications, with one prime example being anthropoid primates (Fig. 1). Stem and early crown anthropoids retained the ancestral mammalian condition until shortly after the Paleogene-Neogene boundary (~23 Ma ago), after which several significant shifts occurred rapidly (Table 1 and table S2). These shifts from the ancestral grade (slope: b = 0.51) resulted in new slopes for colobines (b = 0.67), cercopithecines (b = 0.43), lesser apes (b = 0.32), great apes (b = 0.23), hominins (b = 1.10), and callitrichines (b = 0.58). Other shifts in slope near the Paleogene-Neogene boundary occurred in bears and pinnipeds (Fig. 1; Table 1 and table S2), which are noteworthy for having the largest downward shifts in slope in the sample: from b = 0.58 in other carnivorans to b = 0.39 and b = 0.23, respectively. See the Supplementary Results for a complete description of allometric repatterning across clades.
DISCUSSION
Different evolutionary trajectories for the largest-brained mammals
Five mammalian groups (elephants, great apes, hominins, toothed whales, and delphinids) attained their position at the top of the bivariate brain-body space (Fig. 2) by following an unexpectedly diverse range of trajectories. Elephants represent the simplest case, as they evolved directly from the ancestral mammalian grade and achieved large relative brain size by vastly increasing their body size while increasing brain size even more rapidly (table S3). In toothed whales and delphinids, relative brain size increased in a stepwise manner. The first intercept shift occurred in the stem fereuungulates, which follow a trajectory similar to (although less pronounced than) that in elephants, by increasing brain size more than body size. This was followed by an intercept shift in stem toothed whales, which decreased brain and body size relative to stem cetaceans, with body size decreasing more rapidly than brain size (uncertainty surrounding this scenario exists and is a function of the interpretation of the fossil record; see the Supplementary Results). Delphinids show a third intercept shift driven by body size decrease and brain size increase relative to other toothed whales. The evolutionary trajectory of great apes and hominins was the most complex, starting with two consecutive downward shifts in slope (while increasing both brain and body size) in stem apes and stem great apes, followed by the most marked increase in slope observed in this study, in hominins (from b = 0.23 to b = 1.10). Delphinids and hominins, which converge at the apex of the brain-body space, are the only two grades with negative brain-body correlations, that is, brain size increased while body size decreased (table S3). All five of the mammal groups with the largest brain size may have originated in the Neogene. This remarkable diversity in evolutionary trajectories and late attainment of peak relative brain size parallels patterns in birds in which the largest-brained taxa (parrots and corvids) attained large brain sizes during the Neogene via very different trajectories (12). These parallel patterns between birds and mammals suggest that, similar to the K-Pg transition, the Paleogene-Neogene transition may have created conditions ripe for ecological radiations and niche expansions that affected brain size evolution.
Similar evolutionary trajectories for the smallest-brained mammals
In contrast to relatively large-brained clades, species that lie near the bottom of the brain-body space are consistently characterized by a shift toward a higher slope and a lower intercept. Such shifts occur in afrosoricids (b = 0.66), hystricomorph rodents (b = 0.66), myomorph and castorimorph rodents (b = 0.57), bats (b = 0.66), eulipotyphlans (b = 0.62), and marsupials (b = 0.58); all of which derive directly from the ancestral mammalian grade (b = 0.51) (Table 1). Bats, eulipotyphlans, and afrosoricids converge on the same grade (i.e., same intercept and slope) as the myomorph and castorimorph rodents and marsupials. These allometric changes are mostly explained by a disproportionally higher decrease in mean brain size relative to body size and a lower variance in body size relative to brain size (tables S3 to S5). This suggests that body size becomes more restrained than brain size at smaller body sizes, permitting smaller animals to evolve disproportionately small brain sizes.
The higher evolutionary flexibility of mean brain size relative to mean body size is apparent across all mammals, with stem toothed whales being a notable exception (table S3). This contrasts with birds, where pronounced changes in mean body size compared with mean brain size are common (12). This effect may be rooted in the scalability of mammalian neocortical architecture. While the mammalian neocortex (dorsal pallium) is organized as an outer layer of neurons surrounding scalable white matter, the bird dorsal pallium is organized in a nuclear manner that might limit its scalability (22).
Divergent versus stabilizing selection in brain and/or body size
A detailed account of changes in evolutionary allometry across deep time provides opportunities for understanding the types of selection operating in certain taxa. A crucial aspect in this regard are putative differences in the cross-species variance of a trait among groups. Although variance has long been considered crucial to understanding the types of selection operating in certain taxa (13) and to play a principal role in driving shifts in allometric slope between grades (fig. S1), it was undescribed by previous research. Several patterns revealed by our analyses (table S5) demonstrate how changes in the cross-species variance of a trait between groups affect shifts in slope and may reveal possible selective pressures.
Stem cercopithecoids derived from the ancestral mammalian grade by disproportionately increasing mean brain size relative to mean body size and disproportionately decreasing body size variance relative to brain size variance (tables S3 to S5). This pattern resulted in an increase in slope (from b = 0.51 to b = 0.67; Fig. 1 and Table 1). Within cercopithecoids, cercopithecines (baboons, macaques, and relatives) diverged toward a lower slope (b = 0.43) through a moderate increase in mean brain and body size (~1.28 times greater mean), a strong increase in body size variance (3.45 times greater), and a stabilization of brain size variance (1.16 times greater). Brain size of cercopithecines thus varies across a much wider range of body size than other cercopithecoids (i.e., colobines), suggesting more divergent selection on body size (scenario displayed in fig. S1B). Considering the effects of locomotion on body size (23), and the fact that cercopithecines include both arboreal and terrestrial species while colobines are predominantly arboreal, this allometric repatterning is likely associated with their differences in locomotor diversity.
A similar scenario plays out in pinnipeds, which underwent a significant decrease in slope compared with other carnivorans (from b = 0.58 in carnivorans to b = 0.23), primarily due to decreased brain size variance relative to body size variance (table S5). Body size in pinnipeds thus varies widely compared with other carnivorans given their range of brain sizes (table S5). This suggests divergent selection on body size, most likely influenced by the transition to a semiaquatic niche (24).
A contrasting scenario is provided by great apes and hominins, which have extremely different slopes. Great apes have the lowest slope in the sample (b = 0.23, which they share with pinnipeds), whereas hominins have the highest slope (b = 1.10). Although both mean brain size and mean body size are similar across these two nested grades, the variance in hominin brain size is 6.45 times greater than in great apes, while the variance in body size is only 1.58 times greater. In this case, a major shift in variance occurred in brain size while body size remained mostly static, suggesting divergent selection on brain size (scenario displayed in fig. S1C) and more stabilizing selection on body size (likely associated with the shift to bipedality in hominins).
Allometric shifts do not always represent shifts in cognition
Variation in relative brain size is traditionally associated with cognitive capacities and behavioral flexibility, but this notion has rested on several fundamental assumptions. First, it has been assumed that the slope of the brain-body relationship is stable across species and that deviations from this allometry reflect selection on brain size. As a result, numerous studies geared toward explaining the evolution of relative brain size have focused on the evolution of brain size and cognition (25, 26). Our results do not support this assumption. Evolutionary shifts in brain-body allometry commonly included changes in slope and were often driven by changes in body size. Rather than focusing on residual deviation from a common slope, the emphasis should be on the allometric shifts themselves. In addition, addressing factors that are not directly tied to brain size or cognition likely plays an important role. Some selective pressures that play a key role in species diversification are more directly tied to body size than brain size.
A prime example is locomotion. Known to influence body size and energetic expenditure [e.g., high cost of flying in bats (27)], major transitions in locomotion allow for a redistribution of energetic allocation to growth, thereby providing opportunities for allometric repatterning. This is also apparent in birds, where relative brain size is associated with flight style and complexity (28). Our analyses confirm the influence of locomotion on brain-body allometry in mammals by demonstrating that many major transitions in locomotion coincide with allometric repatterning events. For example, the evolution of flight in bats coincides with an allometric shift in intercept and slope (AICΔ = 69.339; AICѡ < 0.001) that is characterized by decreased body size variance, suggesting strong constraints on body size due to the highly specialized and energetically demanding locomotion in this group. The transition from the mammalian ancestral grade to terrestrial cursorial locomotion in fereuungulates (AICΔ = 46.051; AICѡ < 0.001) coincides with a disproportionate increase in mean brain size to body size. The transition to a semiaquatic niche in pinnipeds (AICΔ = 31.527; AICѡ < 0.001) coincides with disproportionate decrease in brain size variance relative to body size variance. Notably, the initial transition to an aquatic niche in cetaceans is not linked to a shift in brain-body allometry, although later shifts to higher mean brain size do occur in toothed whales and delphinids. This makes whales a compelling counter example, despite being largely freed from size constraints and reaching the largest size of any vertebrates, baleen whales retain the same brain-body allometry as their terrestrial cetartiodactylan relatives. Within primates, allometric shifts coincide with increased commitment to terrestriality in cercopithecines (AICΔ = 81.709; AICѡ < 0.001). The origins of the great ape grade coincide with a decreased slope (AICΔ = 14.632; AICѡ < 0.005) and is associated with a substantially larger increase in body size variance compared with brain size variance. This may similarly have been associated with increased reliance on orthograde behaviors related to terrestriality (29). The transition to bipedalism in hominins coincides with a marked increase in slope (AICΔ = 58.527, AICѡ < 0.001) that is characterized by higher brain size variance relative to body size variance.
Other factors that likely influence the brain-body allometry primarily through changes in body mass include, but are not limited to, sexual size dimorphism, diving depth in aquatic niches (30), antipredator defensive mechanisms (31), and energetic strategies to maintain homeostasis (possibly contributing to explaining the allometric shifts in the convergent eulipotyphlans and afrosoricids) (32). Overall, the general alignment of allometric shifts with major transitions in locomotion (table S2) and concomitant selection on body size (e.g., small body size in a volant niche, large body size in terrestrial and aquatic niches) suggests that the evolutionary repatterning of the brain-body relationship reflects an adaptive profile that extends beyond selection on brain size alone (33). The overwhelming focus on brain size and cognition in the literature therefore should be reconsidered.
A second fundamental assumption is that brain-body allometry reflects the maintenance of basic autonomic, and sensory functions and allometric deviations therefore reflect cognition (34). This in turn relies on the assumption that there is little variation in the relative volumes of brain regions and that larger brains are mostly scaled up small brains (35). If true, the only target for explanation would be the (allometrically adjusted) brain-to-body ratio, irrespective of how changes in body size alter allometric patterning. This assumption has, however, already been disproven by numerous multivariate studies of brain region sizes (36, 37). A poignant comparison in this context is that between the polar bear (Ursus maritimus) and the California sea lion (Zalophus californianus). Although these two species have a similar mean body size, the brain size of the polar bear is twice that of the California sea lion. Consequently, the polar bear exhibits a much higher (four times higher) relative brain size. However, the California sea lion has 3.6 times more volume devoted to brain regions that are associated with higher cognition relative to regions associated with basic autonomic and sensory functions (38). This is likely associated with vocal learning and other cognitive skills in the California sea lion (39). Although cognitive tests in polar bears are generally lacking, this example indicates that relative brain size alone may not be a sufficient proxy for the amount of brain tissue that is allocated to higher cognition (33).
This example further illustrates the potentially confounding effects of assuming a stable slope across all species. As mentioned, pinnipeds display the lowest slope among mammals, a pattern that is most likely driven by divergent selection on body size. Given that pinnipeds are large mammals, such a low slope inevitably results in low relative brain size when considering a common slope (the common slope is higher than that observed in pinnipeds alone). Attributing this low relative brain size to selection on brain size and cognition obscures the trajectory that resulted in their low relative brain size (namely, most predominantly selection on body size, not brain size). Overall, assuming a stable slope across all species implies that selection on body size is comparable across species—an assumption that is difficult to uphold given the wide variety of niches that mammals inhabit and the fundamental role that body size plays in ecological and evolutionary processes.
In general, these results indicate that the brain-body relationship reveals more than just selection on brain size. Therefore, relative brain size may not always be a valid proxy of cognition. The same argument applies to the widely used EQ measure (8), which is also quantified using deviations from a stable slope. A possible way to improve the comparative study of cognition is to compare different brain regions. Whereas comparisons among brain regions associated with different functions would reveal neurobehavioral specializations (36, 38), comparisons among brain regions from different developmental precursors would highlight changes in growth allocation (10). Such remapping factors ensure validity by using hypotheses that are based on established neuroanatomical and neuroscientific principles (40). Comparisons among brain regions also have the potential to reveal which patterns of brain region evolution explain brain size evolution (41) and whether such patterns of brain region evolution can be tied to cognition (38). Such analyses are essential because the evolution of brain size may not always be in line with the evolution of brain regions (or other neuroanatomical features) that are associated with higher cognition. The association between increased brain size and increased complexity is assuredly strong (42), but crucial exceptions to this trend suggest that much may be left to discover on this topic. For example, whereas some archaeocetes (fossil stem cetaceans) had brains that are larger than toothed whales (data S2), their brains have relatively smaller cerebral hemispheres compared with toothed whales (43). Early cetacean brain evolution may thus have comprised two different trajectories: increased brain size with low complexity in archaeocetes, and stable or decreased brain size with high complexity in toothed whales. In this example, complexity does not match absolute brain size, although it does match with relative brain size as toothed whales have a higher relative brain size. In the above described comparison of the polar bear and the California sea lion (38), however, complexity does not match either absolute or relative brain size (or EQ). Both these empirical cases confirm that deviations from the general association between brain size and complexity occur and may be a source of future discovery. Such trends are of paramount importance to the study of cognition and can only be revealed through comparisons among brain regions (or other neuroanatomical features).
Allometric shifts reveal comparative differences in adaptive profile
The primary importance of identifying evolutionary allometric shifts lies in the fact that they provide fundamental information on both the patterns and the processes that shape extant variation. Because evolutionary allometries are determined by ontogenetic and population-level allometries, they can be considered as macroevolutionary signatures of changes in the microevolutionary mechanisms that regulate growth (14). Accurate identification of macroevolutionary shifts thus provides crucial information on the mechanisms that shape comparative differences in adaptive profile.
The potential of this approach is arguably best exemplified by humans, where a shift to bipedality allowed for a redistribution of energy from locomotion to reproduction and brain growth (44). This redistribution of energy to the brain effected changes in the mechanisms of its growth, for example, delayed expression of genes associated with synaptic development (45) and neotenic changes in mRNA expression (46) for those brain regions that explain human brain size expansion (36, 41). In turn, these developmental changes caused an evolutionary allometric shift that is characterized by a significantly higher ratio of brain variance to body variance, which indicates divergent selection on brain size (causing an increase in slope; see Figs. 1 and 2 and table S5).
Humans represent an evolutionary allometric shift that is driven primarily by changes in brain size. We observed other allometric repatterning events driven primarily by brain size in elephants, Old World fruit bats, toothed whales, and delphinids. In other clades evolutionary allometric shifts may be primarily driven by selection for body size (e.g., cercopithecines, great apes, and pinnipeds). In all cases, these allometric shifts occur at transitions into a new niche and/or changes in energetic requirements or energetic availability and further involve redistribution of growth allocation, shifts in the genetic and developmental mechanisms that regulate growth, and allometric repatterning.
Although the mechanisms underlying different types of allometric shifts are not yet fully understood, we emphasize that accurate identification of these shifts is a necessary first step toward reaching this goal. Contrary to the traditional paradigm of relative brain size, we show that allometric shifts can be characterized by changes in slope and may be caused by changes in both brain and/or body size. This prompts a reevaluation of the conventional concept of a grade shift as only representing changes in intercept and reveals that a full understanding of the evolution of brain size relative to body size requires the consideration of effects that extend beyond selection on brain size alone.
Implications for the use of “relative brain size”
Relative brain size and the related EQ measure are one of the most widely used measures in comparative biology and have played a fundamental role in developing several core theories in the life sciences (4, 47). Here, we demonstrate that the way in which relative brain size and EQ are traditionally quantified (using deviations from a stable slope) may result in erroneous inferences on which taxa increased or decreased brain size and hampers a deeper understanding of the patterns and types of selection that explain changes in brain size (and body size). In other words, our results demonstrate that the traditional statistical measures of relative brain size and EQ do not always validly capture variation in brain size. We demonstrate that a more nuanced approach to quantifying variation of brain size relative to body size (quantifying changes in both the intercept and the slope of the evolutionary allometry, combined with investigating univariate patterns of change in brain and/or body size that underpin these bivariate changes in intercept and slope) provides new insights and opens new opportunities for improving our understanding of the patterns and processes that characterize brain size evolution.
In general, our results do not contradict the notion that variation in brain size is associated with cognition. Our results rather demonstrate that the traditional measures of relative brain size and EQ do not always validly capture variation in brain size. This result cautions against the unequivocal use of relative brain size or EQ to quantify or study cognition. We argue that the evolution of cognition is more validly represented by comparisons among brain regions (or other neuroanatomical features). Such comparisons have the potential to identify which patterns of brain region evolution explain brain size evolution and therefore reveal more precise and relevant information regarding the evolution of cognition (38, 41). This does not render the study of brain size relative to body size useless. On the contrary, it reframes this trait more broadly to represent comparative differences in adaptive profile, thereby accounting for the complexity and diversity of the underlying processes and ultimately encapsulating aspects beyond cognition and brain size.
MATERIALS AND METHODS
Data
Data on brain and body size (both quantified as mass) were gleaned from the literature (table S1 and data S2). Comparisons between techniques using brain tissue mass data and endocranial volume data from the skulls of fossil species have been validated in multiple published studies (48, 49), and endocranial volume has been proven a reliable proxy estimate of brain size of both mammalian and nonmammalian taxa (11). The phylogeny is a consensus tree derived by Smaers et al. (38) from the mammalian supertree compiled by Faurby et al. (50). Fossil placement was done according to the literature (table S1) and is detailed in data S2.
Identifying shifts in allometric patterning
Methods are similar to those reported in previous work (12). We estimated differences in slope and intercept of the brain-to-body relationship directly from the data using a Bayesian multiregime OU modeling approach (19). The OU model assumes that the evolution of a continuous trait “X” along a branch over time increment “t” is quantified as dX(t) = α[θ − X(t)]dt + σdB(t) (51). Relative to the standard Brownian motion (BM) model [dX(t) = σdB(t)], the OU model adds parameters that estimate mean trait value (θ) and the rate at which changes in mean values are observed (α). The inclusion of these additional parameters allows an appropriate differentiation between changes in the mean (θ and α) and variance (σ) of a trait over time and thus renders the OU model framework more appropriate than BM for modeling changes in the direction of trait evolution. Here, we used a bivariate implementation of OU modeling that is explicitly geared toward estimating shifts in slope and intercept of evolutionary allometries by using reversible-jump Markov chain Monte Carlo (MCMC) machinery (21) (“OUrjMCMC”). We implemented this approach by combining 10 parallel chains of 2 million iterations each with a burn-in proportion of 0.3. We allowed only one shift per branch, and the total number of shifts was constrained by means of a conditional Poisson prior with a mean equal to 2.5% of the total number of branches in the tree and a maximum number of shifts equal to 5%. Starting points for MCMC chains were set by randomly drawing a number of shifts from the prior distribution and assigning these shifts to branches randomly drawn from the phylogeny with a probability proportional to the size of the clade descended from that branch. The MCMC was initialized without any birth-death proposals for the first 10,000 generations to improve the fit of the model. The output of this procedure generates an estimate of a best-fit allometric model with posterior probabilities assigned to each shift in slope and/or intercept.
In part due to difficulties in parameter estimation intrinsic to OU modeling (52), the bivariate OUrjMCMC output may include false positives and/or false negatives (21). To identify false negatives, we ran a univariate OU model estimation procedure (19) on the residuals of each grade to detect shifts in mean. If such shifts in mean were detected, they were added as shifts in intercept to the allometric model (no such shifts were detected for these data). To identify false positives, the allometric model was translated to a least-squares framework and used in a confirmatory analysis using phylogenetic analysis of covariance (“pANCOVA”) (20). Although pANCOVA uses a different evolutionary process than OU modeling (i.e., BM instead of OU), it is expected that grade membership as estimated by the OU modeling is confirmed using least-squares analysis. Because BM assumes fewer statistical parameters, pANCOVA can be considered as a conservative confirmatory test of the significance of grade membership as estimated by OU modeling. All reported results are those that were confirmed by pANCOVA (Table 1 and table S2).
Assessing differential changes in mean brain and/or body size
To assess whether changes in the brain-to-body allometry were driven primarily by mean brain size or body size, phylogenetic means for both brain size and body size were calculated for each of the allometric grades identified by the allometric patterning analysis. Phylogenetic means were calculated following standard phylogenetic generalized least-squares procedures (20).
Patterns of mean brain/body size increase/decrease were evaluated by comparing mean differences in brain size and body size between ancestral and descendant grades (or “derived grades”; note that we, here, consider “descendant” and “derived” as equivalent terms) (table S3). The ratio of the difference in ancestral-to-descendant mean brain size to the difference in ancestral-to-descendant mean body size (; log scale) was considered as an indication of the proportionality of ancestral-to-descendant change in mean brain size relative to mean body size. The scaling coefficient of the brain-to-body relationship of the ancestral grade was used as the expected proportionality of this ratio. The upper bound of the 95% confidence interval of the scaling coefficient of the ancestral grade was used as the cutoff to infer that the change in mean size observed from ancestral-to-descendant grade is characterized by more change in mean brain size than body size. To account for the fact that generalized least-squares procedures minimize residual error for the dependent variable, we inverse this procedure when evaluating changes in mean body size. For body size, we thus considered the proportion relative to the scaling coefficient of the ancestral body-to-brain relationship (table S4). Although we consider the use of the body-to-brain relationship for evaluating body size to be more rigorous than also using the brain-to-body relationship for these purposes, we emphasize that the results are largely unaffected by this choice (i.e., the same results regarding disproportionate brain/body increase/decrease are attained when using the brain-to-body relationship to evaluate both brain size and body size).
More change in mean brain size than mean body size is inferred when is higher than the upper bound of the ancestral brain-to-body expectation and is lower than the upper bound body-to-brain expectation. In Fig. 1, this scenario is indicated as two arrows for mean brain size and one arrow for mean body size. More change in mean body size than mean brain size is inferred when is lower than the upper bound of the ancestral brain-to-body expectation and is higher than the upper bound body-to-brain expectation. In Fig. 1, this scenario is indicated as one arrow for mean brain size and two arrows for mean body size (which is the case only for toothed whales). In pinnipeds, the observed proportion lies above the upper bound of both the ancestral brain-to-body and body-to-brain relationship. This is therefore indicated as two arrows for brain size and two for body size (both indicating an increase in size).
For example, stem cercopithecoid primates (consisting of the fossil Victoriapithecus and extant colobines) derive directly from the mammalian ancestral grade (Fig. 1). This gives this grade an expected change in mean brain size relative to change in mean body size of 0.47 with a maximum expectation of 0.55 (table S3). The mammalian ancestral grade has a mean brain size of 1.92 and a mean body size of 6.92 (log scale). The stem cercopithecoid/colobine grade has a mean brain size of 4.40 and a mean body size of 9.04. The difference in mean brain size from the mammalian ancestral grade to the colobine grade is +2.48; that of body size is +2.12. The ratio is thus 1.17, which is 0.62 points above the upper bound brain-to-body expectation (table S3). The ratio is 0.86, which is 0.88 below the upper bound of the body-to-brain expectation (table S4). Colobines thus indicate more change in mean brain size relative to change in mean body size than expected from their ancestral grade.
Stem toothed whales are the only grade in the sample that indicates more change in mean body size than change in mean brain size. Relative to stem cetaceans (archaeocetes), stem toothed whales decrease in size (although note the uncertainties inherent in this inference discussed in the Supplementary Results). Archaeocetes have a mean brain size of 7.25 and a mean body size of 15.03. Stem toothed whales have a mean brain size of 6.67 and a mean body size of 11.90. The differences in mean brain and mean body size from archaeocetes to stem toothed whales are thus −0.58 and − 3.13, respectively. The confidence interval of the slope for the ancestral grade of stem toothed whales is 0.50:0.58. The ratio is 0.19, which 0.39 below the upper bound of the brain-to-body relationship. The ratio is 5.39, which 3.65 above the upper bound of the body-to-brain relationship. Therefore, it is inferred that stem toothed whales indicated more change in mean body size than change in mean brain size than allometrically expected given their ancestral grade (specifically, more decrease in mean body size than decrease in mean brain size).
It should be noted that this procedure is valid only in the case of a positive brain-to-body correlation. This assumption is not upheld in delphinids and hominins, who demonstrate a decrease in body size and an increase in brain size (table S3). It is, however, evident that selection favors increased brain size relative to body size in these cases.
Assessing differential changes in the variance of brain and/or body size
Patterns of changes in the variance of brain size and body size among grades were evaluated by comparing the differences in variances in brain size and body size between ancestral and descendant grades [phylogenetic variance was calculated following standard phylogenetic generalized least-squares procedures (20)]. The change in ancestral-to-descendant body size variance is expected to be 1:1 for all grades, as this would maintain the proportionality of scaling differences from ancestral-to-descendant grades. If this ratio is >1, then changes in body size variance are greater than changes in brain size variance. If this ratio is <1, then changes in brain size variance are greater than changes in body size variance. Results are presented in table S5.
Acknowledgments
We thank E. R. Seiffert for the useful comments and discussion, and K. W. S. Ashwell for sharing marsupial data. We further thank J. Lázaro for making Fig. 1. Funding: J.B.S. was funded by the National Science Foundation (grant 80692). A.M.B. was funded by the National Science Foundation (grant DEB 1801224). A.G. was funded by the European Research Council (H2020 ERC-Stg-637171). C.S.M. was supported by the Gerstner Fellowship and the Gerstner Family Foundation, the Kalbfleisch Fellowship, and the Richard Gilder Graduate School of the American Museum of Natural History. V.W. was funded by the Australian Research Council Discovery Grant (DP170103227). D.d.V. was supported by funds from the Natural Environment Research Council (NERC NE/T000341/1). Author contributions: J.B.S., D.K.N.D., K.S., R.S.R., and D.R.H. gleaned data from the literature. D.R.H., B.B., J.G.F., C.C.G., A.G., W.L.J., P.R.M., and C.S.M. assisted with phylogenetic placement of extinct species. J.B.S., R.S.R., and D.R.H. performed the statistical analyses. J.B.S. wrote the first draft. All authors read and edited the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/18/eabe2101/DC1
REFERENCES AND NOTES
- 1.Eisenberg J. F., Wilson D. E., Relative brain size and feeding strategies in the Chiroptera. Evolution 32, 740–751 (1978). [DOI] [PubMed] [Google Scholar]
- 2.Shultz S., Dunbar R., Both social and ecological factors predict ungulate brain size. Proc. R. Soc. Lond. B Biol. Sci. 273, 207–215 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sol D., Duncan R. P., Blackburn T. M., Cassey P., Lefebvre L., Big brains, enhanced cognition, and response of birds to novel environments. Proc. Natl. Acad. Sci. 102, 5460–5465 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin R. D., Relative brain size and basal metabolic rate in terrestrial vertebrates. Nature 293, 57–60 (1981). [DOI] [PubMed] [Google Scholar]
- 5.M. Kleiber, The Fire of Life: An Introduction to Animal Energetics (John Wiley&Sons, Inc., 1961). [Google Scholar]
- 6.Dubois E., Sur le rapport du poids de l’encéphale avec la grandeur du corps chez les mammifères. Bull. Mém. Soc. Anthropol. Paris 8, 337–376 (1897). [Google Scholar]
- 7.H. J. Jerison, Evolution of the Brain and Intelligence (Academic Press, New York, 1973). [Google Scholar]
- 8.Jerison H. J., Animal intelligence as encephalization. Philos. Trans. R. Soc. Lond B Biol. Sci. 308, 21–35 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Gilbert C. C., Jungers W. L., Comment on relative brain size in early primates and the use of encephalization quotients in primate evolution. J. H um. Evol. 109, 79–87 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Smaers J. B., Mongle C. S., Safi K., Dechmann D. K., Allometry, evolution and development of neocortex size in mammals. Prog. Brain Res. 250, 83–107 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Burger J. R., George M. A. Jr., Leadbetter C., Shaikh F., The allometry of brain size in mammals. J. Mammal. 100, 276–283 (2019). [Google Scholar]
- 12.Ksepka D. T., Balanoff A. M., Smith N. A., Bever G. S., Bhullar B. A. S., Bourdon E., Braun E. L., Burleigh J. G., Clarke J. A., Colbert M. W., Corfield J. R., Degrange F. J., de Pietri V. L., Early C. M., Field D. J., Gignac P. M., Gold M. E. L., Kimball R. T., Kawabe S., Lefebvre L., Marugán-Lobón J., Mongle C. S., Morhardt A., Norell M. A., Ridgely R. C., Rothman R. S., Scofield R. P., Tambussi C. P., Torres C. R., van Tuinen M., Walsh S. A., Watanabe A., Witmer L. M., Wright A. K., Zanno L. E., Jarvis E. D., Smaers J. B., Tempo and pattern of avian brain size evolution. Curr. Biol. 30, 2026–2036.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 13.G. G. Simpson, Tempo and Mode in Evolution (Columbia Univ. Press, 1944). [Google Scholar]
- 14.Pélabon C., Firmat C., Bolstad G. H., Voje K. L., Houle D., Cassara J., Rouzic A. L., Hansen T. F., Evolution of morphological allometry. Ann. N. Y. Acad. Sci. 1320, 58–75 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Tsuboi M., van der Bijl W., Kopperud B. T., Erritzøe J., Voje K. L., Kotrschal A., Yopak K. E., Collin S. P., Iwaniuk A. N., Kolm N., Breakdown of brain–body allometry and the encephalization of birds and mammals. Nat. Ecol. Evol. 2, 1492–1500 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Goswami A., Smaers J. B., Soligo C., Polly P. D., The macroevolutionary consequences of phenotypic integration: From development to deep time. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finarelli J. A., Flynn J. J., Brain-size evolution and sociality in Carnivora. Proc. Natl. Acad. Sci. U.S.A. 106, 9345–9349 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisbecker V., Goswami A., Marsupials indeed confirm an ancestral mammalian pattern: A reply to Isler. BioEssays 33, 358–361 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Uyeda J. C., Harmon L. J., A novel bayesian method for inferring and interpreting the dynamics of adaptive landscapes from phylogenetic comparative data. Syst. Biol. 63, 902–918 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Smaers J. B., Rohlf F. J., Testing species’ deviation from allometric predictions using the phylogenetic regression. Evolution 70, 1145–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Uyeda J. C., Pennell M. W., Miller E. T., Maia R., McClain C. R., The evolution of energetic scaling across the vertebrate tree of life. Am. Nat. 190, 185–199 (2017). [DOI] [PubMed] [Google Scholar]
- 22.G. F. Striedter, Principles of Brain Evolution (Sinauer Associates, 2005).
- 23.Fleagle J. G., Mittermeier R. A., Locomotor behavior, body size, and comparative ecology of seven Surinam monkeys. Am. J. Phys. Anthropol. 52, 301–314 (1980). [Google Scholar]
- 24.Gearty W., McClain C. R., Payne J. L., Energetic tradeoffs control the size distribution of aquatic mammals. Proc. Natl. Acad. Sci. U.S.A. 115, 4194–4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. Byrne, A. Whiten, Machiavellian Intelligence: Social Expertise and the Evolutoin of Intellect in Monkeys, Apes, and Humans (Oxford Univ. Press Inc., New York, 1988). [Google Scholar]
- 26.DeCasien A. R., Williams S. A., Higham J. P., Primate brain size is predicted by diet but not sociality. Nat. Ecol. Evol. 1, 0112 (2017). [DOI] [PubMed] [Google Scholar]
- 27.J. R. Speakman, D. W. Thomas, Physiological ecology and energetics of bats, in Bat Ecology, T. Kunz, M. Fenton, Eds. (University of Chicago Press, 2003), pp. 430–490. [Google Scholar]
- 28.Dial K. P., Evolution of avian locomotion: Correlates of flight style, locomotor modules, nesting biology, body size, development, and the origin of flapping flight. The Auk 120, 941–952 (2003). [Google Scholar]
- 29.Nakatsukasa M., Kunimatsu Y., Nacholapithecus and its importance for understanding hominoid evolution. Evol. Anthropol. Issues News Rev. 18, 103–119 (2009). [Google Scholar]
- 30.Marino L., Sol D., Toren K., Lefebvre L., Does diving limit brain size in cetaceans? Mar. Mamm. Sci. 22, 413–425 (2006). [Google Scholar]
- 31.Stankowich T., Romero A. N., The correlated evolution of antipredator defences and brain size in mammals. Proc. R. Soc. B Biol. Sci. 284, 20161857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genoud M., Energetic strategies of shrews: Ecological constraints and evolutionary implications. Mammal Rev. 18, 173–193 (1988). [Google Scholar]
- 33.Healy S. D., Rowe C., Costs and benefits of evolving a larger brain: Doubts over the evidence that large brains lead to better cognition. Anim. Behav. 86, e1–e3 (2013). [Google Scholar]
- 34.Jerison H. J., Brain to body ratios and the evolution of intelligence. Science 121, 447–449 (1955). [DOI] [PubMed] [Google Scholar]
- 35.Finlay B. L., Darlington R. B., Linked regularities in the development and evolution of mammalian brains. Science 268, 1578–1584 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Smaers J. B., Gómez-Robles A., Parks A. N., Sherwood C. C., Exceptional evolutionary expansion of prefrontal cortex in great apes and humans. Curr. Biol. 27, 714–720 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Barton R. A., Harvey P. H., Mosaic evolution of brain structure in mammals. Nature 405, 1055–1058 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Smaers J. B., Turner A. H., Gómez-Robles A., Sherwood C. C., A cerebellar substrate for cognition evolved multiple times independently in mammals. eLife 7, e35696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R. J. Schusterman, C. R. Kastak, D. Kastak, The cognitive sea lion: Meaning and memory in the laboratory and in nature, in The Cognitive Animal: Empirical and Theoretical Perspectives on Animal Cognition, M. Bekoff, C. Allen, G. M. Burghardt, Eds. (The MIT Press, Cambridge, MA, 2002), pp. 217–228. [Google Scholar]
- 40.Passingham R. E., Smaers J. B., Is the prefrontal cortex especially enlarged in the human brain? Allometric relations and remapping factors. Brain Behav. Evol. 84, 156–166 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Smaers J. B., Vanier D. R., Brain size expansion in primates and humans is explained by a selective modular expansion of the cortico-cerebellar system. Cortex 118, 292–305 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Marino L., Big brains do matter in new environments. Proc. Natl. Acad. Sci. U.S.A. 102, 5306–5307 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marino L., Cetacean brain evolution: Multiplication generates complexity. Int. J. Comp. Psychol. 17, 1–16 (2004). [Google Scholar]
- 44.Navarrete A., van Schaik C. P., Isler K., Energetics and the evolution of human brain size. Nature 480, 91–93 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Liu X., Somel M., Tang L., Yan Z., Jiang X., Guo S., Yuan Y., He L., Oleksiak A., Zhang Y., Li N., Hu Y., Chen W., Qiu Z., Paabo S., Khaitovich P., Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 22, 611–622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somel M., Franz H., Yan Z., Lorenc A., Guo S., Giger T., Kelso J., Nickel B., Dannemann M., Bahn S., Webster M. J., Weickert C. S., Lachmann M., Paabo S., Khaitovich P., Transcriptional neoteny in the human brain. Proc. Natl. Acad. Sci. U.S.A. 106, 5743–5748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunbar R. I. M., Shultz S., Evolution in the social brain. Science 317, 1344–1347 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Iwaniuk A. N., Nelson J. E., Can endocranial volume be used as an estimate of brain size in birds? Can. J. Zool. 80, 16–23 (2002). [Google Scholar]
- 49.Logan C. J., Clutton-Brock T. H., Validating methods for estimating endocranial volume in individual red deer (Cervus elaphus). Behav. Processes 92, 143–146 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Faurby S., Svenning J.-C., A species-level phylogeny of all extant and late quaternary extinct mammals using a novel heuristic-hierarchical Bayesian approach. Mol. Phylogenet. Evol. 84, 14–26 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Butler M. A., King A. A., Phylogenetic comparative analysis: A modeling approach for adaptive evolution. Am. Natural. 164, 683–695 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Ho L. S. T., Ané C., Intrinsic inference difficulties for trait evolution with Ornstein-Uhlenbeck models. Methods Ecol. Evol. 5, 1133–1146 (2014). [Google Scholar]
- 53.Montgomery S. H., Geisler J. H., McGowen M. R., Fox C., Marino L., Gatesy J., The evolutionary history of cetacean brain and body size. Evolution 67, 3339–3353 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Marino L., McShea D. W., Uhen M. D., Origin and evolution of large brains in toothed whales. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 281, 1247–1255 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Smaers J. B., Mongle C. S., On the accuracy and theoretical underpinnings of the multiple variance Brownian motion approach for estimating variable rates and inferring ancestral states. Biol. J. Linn. Soc. 121, 229–238 (2017). [Google Scholar]
- 56.Smaers J. B., Mongle C. S., Kandler A., A multiple variance Brownian motion framework for estimating variable rates and inferring ancestral states. Biol. J. Linn. Soc. 118, 78–94 (2016). [Google Scholar]
- 57.Khabbazian M., Kriebel R., Rohe K., Ané C., Fast and accurate detection of evolutionary shifts in Ornstein–Uhlenbeck models. Methods Ecol. Evol. 7, 811–824 (2016). [Google Scholar]
- 58.M. Archer, F. Jenkins Jr, S. Hand, P. Murray, H. Godthelp, Description of the skull and non-vestigial dentition of a Miocene Platypus (Obdurodon dicksoni, n. sp.) from Riversleigh, Australia, and the problem of monotreme origins, in Platypus and Echidnas, M. Augee, Ed. (Royal Zoological Society of New South Wales, Australia, 1993), pp. 15–27.
- 59.Macrini T. E., Rowe T., Archer M., Description of a cranial endocast from a fossil platypus, Obdurodon dicksoni (Monotremata, Ornithorhynchidae), and the relevance of endocranial characters to monotreme monophyly. J. Morphol. 267, 1000–1015 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Ashwell K. W. S., Encephalization of Australian and new guinean marsupials. Brain Behav. Evol. 71, 181–199 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Forasiepi A. M., Macphee R. D., del Pino S. H., Caudal Cranium of Thylacosmilus atrox (Mammalia, Metatheria, Sparassodonta), a South American Predaceous Sabertooth. Bull. Am. Museum Natural His. 2019, 1–66 (2019). [Google Scholar]
- 62.Quiroga J., Dozo M., The brain of Thylacosmilus atrox. Extinct South American saber-tooth carnivore marsupial. J. Hirnforsch. 29, 573–586 (1988). [PubMed] [Google Scholar]
- 63.Tambusso P. S., Fariña R. A., Digital cranial endocast of Pseudoplohophorus absolutus (Xenarthra, Cingulata) and its systematic and evolutionary implications. J. Vertebr. Paleontol. 35, e967853 (2015). [Google Scholar]
- 64.Kielan-Jaworowska Z., Evolution of the therian mammals in the Late Cretaceous of Asia. Part VI. Endocranial casts of eutherian mammals. Palaeontol. Polonica 46, 157–171 (1984). [Google Scholar]
- 65.Benoit J., Crumpton N., Mérigeaud S., Tabuce R., A memory already like an Elephant’s? The advanced brain morphology of the last common ancestor of afrotheria (Mammalia). Brain Behav. Evol. 81, 154–169 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Isler K., van Schaik C. P., Allomaternal care, life history and brain size evolution in mammals. J. Hum. Evol. 63, 52–63 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Benoit J., Legendre L. J., Tabuce R., Obada T., Mararescul V., Manger P., Brain evolution in Proboscidea (Mammalia, Afrotheria) across the Cenozoic. Sci. Rep. 9, 9323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D. P. Domning, Encyclopedia of Marine Mammals, B. Würsig, J. Thewissen, K. M. Kovacs, Eds. (Elsevier, London, United Kingdom, 2018). [Google Scholar]
- 69.Pirlot P., Kamiya T., Qualitative and quantitative brain morphology in the Sirenian Dugong dugong Erxl. J. Zoolog. Syst. Evol. Res. 23, 147–155 (1985). [Google Scholar]
- 70.Savage R., Domning D. P., Thewissen J., Fossil Sirenia of the West Atlantic and Caribbean region. V. The most primitive known sirenian,Prorastomus sirenoides Owen, 1855. J. Vert. Paleontol. 14, 427–449 (1994). [Google Scholar]
- 71.Shoshani J., Kupsky W. J., Marchant G. H., Elephant brain: Part I: Gross morphology, functions, comparative anatomy, and evolution. Brain Res. Bull. 70, 124–157 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Palkopoulou E., Lipson M., Mallick S., Nielsen S., Rohland N., Baleka S., Karpinski E., Ivancevic A. M., To T. H., Kortschak R. D., Raison J. M., Qu Z., Chin T. J., Alt K. W., Claesson S., Dalén L., MacPhee R. D. E., Meller H., Roca A. L., Ryder O. A., Heiman D., Young S., Breen M., Williams C., Aken B. L., Ruffier M., Karlsson E., Johnson J., di Palma F., Alfoldi J., Adelson D. L., Mailund T., Munch K., Lindblad-Toh K., Hofreiter M., Poinar H., Reich D., A comprehensive genomic history of extinct and living elephants. Proc. Natl. Acad. Sci. U.S.A 115, E2566–E2574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Delsuc F., Gibb G. C., Kuch M., Billet G., Hautier L., Southon J., Rouillard J. M., Fernicola J. C., Vizcaíno S. F., MacPhee R. D. E., Poinar H. N., The phylogenetic affinities of the extinct glyptodonts. Curr. Biol. 26, R155–R156 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Harel S., Watanabe K., Linke I., Schain R. J., Growth and development of the rabbit brain. Neonatology 21, 381–399 (1972). [DOI] [PubMed] [Google Scholar]
- 75.Herculano-Houzel S., Ribeiro P., Campos L., Valotta da Silva A., Torres L. B., Catania K. C., Kaas J. H., Updated neuronal scaling rules for the brains of Glires (rodents/lagomorphs). Brain Behav. Evol. 78, 302–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pirlot P., Kamiya T., Relative size of brain and brain components in three gliding placentals (Dermoptera: Rodentia). Can. J. Zool. 60, 565–572 (1982). [Google Scholar]
- 77.Beaudet A., Dumoncel J., de Beer F., Duployer B., Durrleman S., Gilissen E., Hoffman J., Tenailleau C., Thackeray J. F., Braga J., Morphoarchitectural variation in South African fossil cercopithecoid endocasts. J. Hum. Evol. 101, 65–78 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Bloch J. I., Silcox M. T., Boyer D. M., Sargis E. J., New Paleocene skeletons and the relationship of plesiadapiforms to crown-clade primates. Proc. Natl. Acad. Sci. U.S.A. 104, 1159–1164 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burney D. A., Burney L. P., Godfrey L. R., Jungers W. L., Goodman S. M., Wright H. T., Jull A. J. T., A chronology for late prehistoric Madagascar. J. Hum. Evol. 47, 25–63 (2004). [DOI] [PubMed] [Google Scholar]
- 80.E. Delson, C. J. Terranova, W. L. Jungers, E. J. Sargis, N. G. Jablonski, Body Mass in Cercopithecidae (Primates, Mammalia): Estimation and Scaling in Extinct and Extant Taxa, (Anthropological Papers, American Museum of Natural History, 2000), vol. 83.
- 81.Gómez J., Verdú M., Mutualism with plants drives primate diversification. Syst. Biol. 61, 567–577 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Gonzales L. A., Benefit B. R., McCrossin M. L., Spoor F., Cerebral complexity preceded enlarged brain size and reduced olfactory bulbs in Old World monkeys. Nat. Commun. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grabowski M., Hatala K. G., Jungers W. L., Richmond B. G., Body mass estimates of hominin fossils and the evolution of human body size. J. Hum. Evol. 85, 75–93 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Grabowski M., Jungers W. L., Evidence of a chimpanzee-sized ancestor of humans but a gibbon-sized ancestor of apes. Nat. Commun. 8, 880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halenar L. B., Cooke S. B., Rosenberger A. L., Rímoli R., New cranium of the endemic Caribbean platyrrhine, Antillothrix bernensis, from La Altagracia Province, Dominican Republic. J. Human Evol. 106, 133–153 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Isler K., Christopher Kirk E., Miller J. M. A., Albrecht G. A., Gelvin B. R., Martin R. D., Endocranial volumes of primate species: Scaling analyses using a comprehensive and reliable data set. J. Hum. Evol. 55, 967–978 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Kappelman J., The evolution of body mass and relative brain size in fossil hominids. J. Hum. Evol. 30, 243–276 (1996). [Google Scholar]
- 88.Kirk E. C., Simons E. L., Diets of fossil primates from the Fayum Depression of Egypt: A quantitative analysis of molar shearing. J. Hum. Evol. 40, 203–229 (2001). [DOI] [PubMed] [Google Scholar]
- 89.Kordos L., New results of Hominoid research in the Carpathian Basin. Acta Biol. Szegediensis 44, 71–74 (2000). [Google Scholar]
- 90.Marino L., A comparison of encephalization between odontocete cetaceans and anthropoid primates. Brain Behav. Evol. 51, 230–238 (1998). [DOI] [PubMed] [Google Scholar]
- 91.R. D. Martin, A.-E. Martin, Primate Origins and Evolution: A Phylogenetic Reconstruction (Princeton Univ. Press Princeton, 1990). [Google Scholar]
- 92.Montgomery S. H., Capellini I., Barton R. A., Mundy N. I., Reconstructing the ups and downs of primate brain evolution: Implications for adaptive hypotheses and Homo floresiensis. BMC Biol. 8, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parr W. C., Chatterjee H. J., Soligo C., Inter- and intra-specific scaling of articular surface areas in the hominoid talus. J. Anat. 218, 386–401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwartz G. T., Mahoney P., Godfrey L. R., Cuozzo F. P., Jungers W. L., Randria G. F. N., Dental development in Megaladapis edwardsi (Primates, Lemuriformes): Implications for understanding life history variation in subfossil lemurs. J. Hum. Evol. 49, 702–721 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Sears K. E., Finarelli J. A., Flynn J. J., Wyss A. R., Estimating body mass in New World “monkeys” (Platyrrhini, Primates), with a consideration of the Miocene platyrrhine, Chilecebus carrascoensis. Am. Museum Novitates 3617, 1–29 (2008). [Google Scholar]
- 96.Silcox M. T., Dalmyn C. K., Bloch J. I., Virtual endocast of Ignacius graybullianus (Paromomyidae, Primates) and brain evolution in early primates. Proc. Natl. Acad. Sci. U.S.A. 106, 10987–10992 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simons E. L., The cranium of Parapithecus grangeri, an Egyptian Oligocene anthropoidean primate. Proc. Natl. Acad. Sci. U.S.A. 98, 7892–7897 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Strait S. G., Dietary reconstruction of small-bodied omomyoid primates. J. Vert. Paleontol. 21, 322–334 (2001). [Google Scholar]
- 99.Orihuela J., Endocranial morphology of the extinct Antillean shrew Nesophontes (Lipotyphla: Nesophontidae) from natural and digital endocasts of Cuban taxa. Palaeontol. Electron. 17, 153 (2014). [Google Scholar]
- 100.G. Baron, H. Stephan, H. D. Frahm, Comparative Neurobiology in Chiroptera: Macromorphology, Brain Structures, Tables and Atlases (Birkhäuser, Basel, 1996), vol. 1.
- 101.U. Norberg, M. Fenton, P. Racey, J. Rayner (Cambridge Univ. Press Cambridge, 1987). [Google Scholar]
- 102.Simmons J. A., Comparative Neurobiology in Chiroptera. Science 273, 609–610 (1996). [Google Scholar]
- 103.Radinsky L., Evolution of brain size in carnivores and ungulates. Am. Nat. 112, 815–831 (1978). [Google Scholar]
- 104.Spaulding M., O’Leary M. A., Gatesy J., Relationships of Cetacea (Artiodactyla) among mammals: Increased taxon sampling alters interpretations of key fossils and character evolution. PloS ONE 4, e7062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zanazzi A., Kohn M. J., Terry D. O. Jr., Biostratigraphy and paleoclimatology of the Eocene-Oligocene boundary section at Toadstool Park, northwestern Nebraska, USA. Geol. Soc. Am. 452, 197–214 (2009). [Google Scholar]
- 106.Geisler J. H., McGowen M. R., Yang G., Gatesy J., A supermatrix analysis of genomic, morphological, and paleontological data from crown Cetacea. BMC Evol. Biol. 11, 112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lloyd G. T., Slater G. J., A total-group phylogenetic metatree of Cetacea and the importance of fossil data in diversification analyses. System. Biol. 2021, syab002 (2021). [DOI] [PubMed] [Google Scholar]
- 108.Manger P. R., An examination of cetacean brain structure with a novel hypothesis correlating thermogenesis to the evolution of a big brain. Biol. Rev. 81, 293–338 (2006). [DOI] [PubMed] [Google Scholar]
- 109.Marino L., Connor R. C., Fordyce R. E., Herman L. M., Hof P. R., Lefebvre L., Lusseau D., McCowan B., Nimchinsky E. A., Pack A. A., Rendell L., Reidenberg J. S., Reiss D., Uhen M. D., van der Gucht E., Whitehead H., Cetaceans have complex brains for complex cognition. PLOS Biol. 5, e139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ridgway S. H., Hanson A. C., Sperm whales and killer whales with the largest brains of all toothed whales show extreme differences in cerebellum. Brain Behav. Evol. 83, 266–274 (2014). [DOI] [PubMed] [Google Scholar]
- 111.M. D. Uhen, Form, Function, and Anatomy of Dorudon Atrox (Mammalia, Cetacea): An Archaeocete from the Middle to Late Eocene of Egypt (University of Michingan, 2004).
- 112.Radinsky L., Oldest horse brains: More advanced than previously realized. Science 194, 626–627 (1976). [DOI] [PubMed] [Google Scholar]
- 113.Dubied M., Solé F., Mennecart B., The cranium of Proviverra typica (Mammalia, Hyaenodonta) and its impact on hyaenodont phylogeny and endocranial evolution. Palaeontology 62, 983–1001 (2019). [Google Scholar]
- 114.R. Hunt, in The George Wright Forum (JSTOR, 1984), vol. 4, pp. 29–39.
- 115.Tomiya S., Tseng Z. J., Whence the beardogs? Reappraisal of the Middle to Late Eocene ‘Miacis’ from Texas, USA, and the origin of Amphicyonidae (Mammalia, Carnivora). R. Soc. Open Sci. 3, 160518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/18/eabe2101/DC1


