Abstract
As part of the safety assessment process, all industrial sectors employ genotoxicity test batteries, starting with well-established in vitro assays. However, these batteries have limited predictive capacity for the in vivo situation, which may result in unnecessary follow-up in vivo testing or the loss of promising substances where animal tests are prohibited or not desired. To address this, a project involving regulators, academia and industry was established to develop and validate in vitro human skin-based genotoxicity assays for topically exposed substances, such as cosmetics ingredients. Here, we describe the validation of the 3D reconstructed skin (RS) Comet assay. In this multicenter study, chemicals were applied topically three times to the skin over 48 h. Isolated keratinocytes and fibroblasts were transferred to slides before electrophoresis and the resulting comet formation was recorded as % tail DNA. Before decoding, results of the validation exercise for 32 substances were evaluated by an independent statistician. There was a high predictive capacity of this assay when compared to in vivo outcomes, with a sensitivity of 77 (80)%, a specificity of 88 (97)% and an overall accuracy of 83 (92)%. The numbers reflect the calls of the performing laboratories in the coded phase, whereas those in parenthesis reflect calls according to the agreed evaluation criteria. Intra- and inter-laboratory reproducibility was also very good, with a concordance of 93 and 88%, respectively. These results generated with the Phenion® Full-Thickness skin model demonstrate its suitability for this assay, with reproducibly low background DNA damage and sufficient metabolic capacity to activate pro-mutagens. The validation outcome supports the use of the RS Comet assay to follow up positive results from standard in vitro genotoxicity assays when the expected route of exposure is dermal. Based on the available data, the assay was accepted recently into the OECD test guideline development program.
Introduction
Testing for genotoxic properties is a core pillar of ensuring the safe use of substances. The genotoxicity hazard is generally explored by using so-called test batteries as no in vitro or in vivo assay can individually consider all three types of DNA damage which need to be addressed for regulatory purposes. State-of-the-art genotoxicity testing always starts with a battery of in vitro assays covering gene mutations, structural and numerical chromosome aberrations, including the Ames assay (1) and the in vitro micronucleus test (2). If a chemical substance is without any genotoxic effect in vitro it might be classified as non-genotoxic in chemicals legislations (e.g. REACH, CLP, Cosmetics Directive), while others (i.e. legislations for pharmaceuticals or veterinary drugs) always require follow-up testing in animals. There have, however, been reports of reduced specificity of the standard in vitro test battery, meaning that in vivo non-genotoxic chemicals are often mistakenly identified as genotoxic (3–5). Positives from the standard battery, including such ‘misleading positive’ findings, were traditionally investigated further by follow-up testing using animals. Testing of chemical substances in animals has been increasingly questioned in the last decades and a paradigm shift away from animal studies is on the way in multiple countries, driven by animal welfare concerns. As a result, the European Union launched the 7th Amendment to the Cosmetics Directive of the European Commission which prohibits the use of in vivo follow-up assays for cosmetic ingredients, starting in 2009 (6). In the above ‘test battery’ context that means a positive result from an in vitro standard genotoxicity assay would result in the loss of this ingredient for use in cosmetic products. In non-cosmetic sector industries with regulations that still allow in vivo testing, ‘misleading positive’ findings would trigger potentially unnecessary in vivo follow-up testing and animal usage. When follow-up testing is performed, increasing focus is now being placed on ensuring an appropriate selection of the route of exposure, as shown by recently developed and/or revised OECD genotoxicity testing guidelines (TGs). These now emphasise that the intended/expected route of human exposure should be taken into account (e.g. OECD TGs 474, 488 and 489) (2,7,8), as well as the need for testing the so-called site-of-contact (OECD TGs 488 and 489) (7,8). For ingredients in cosmetics, household products or pesticides, agrochemicals, etc., in most instances, that is the skin.
There are currently no validated higher-tier in vitro genotoxicity assays in the standard testing toolbox of mutagenicity and genotoxicity assays to specially address potential genotoxicity via the dermal exposure route. To close this gap, Cosmetics Europe initiated and led a project aiming at addressing the lack of adequate alternatives to traditional in vivo genotoxicity tests and supporting the development and validation of skin-based genotoxicity assays. The idea was to combine classical genotoxicity read-out parameters with existing 3D skin technology utilising Reconstructed Human Skin (RHS, referred to as reconstructed skin (RS) if used in conjunction with the assays) (9–12), resulting in the development of the “3D” RS Comet assay (13,14), described here in more detail, and an RS micronucleus assay (RSMN) (15,16). The combination of these assays is thought to be adequate for follow-up of positive results from the standard 2D in vitro genotoxicity assays. Depending on the outcome from the standard in vitro battery, which generally consists of assays that cover all genotoxicity endpoints (gene mutation, clastogenicity and aneugenicity), a skin Comet assay or skin micronucleus assay (or both) would be chosen (17,18). The results of the RS Comet assay are described here, while the results of the validation of the RSMN assay are shown in Pfuhler et al. (19) and the overall strategy for the use of RHS model-based genotoxicity assays is presented in Pfuhler et al. (18).
The Comet assay detects a broad spectrum of DNA damage, including modifications that lead to gene mutation (20). This is underlined by the high sensitivity of the in vivo Comet assay for carcinogens that show gene mutation activity, which this assay detects as efficiently as the transgenic rodent (TGR) mutation assays (21). A more recent comprehensive analysis again confirmed that the Comet assay shows similar sensitivity in detecting DNA reactive (Ames positive) rodent carcinogens to the TGR mutation assays (22). Its increased recognition and demand for regulatory testing has led to the implementation of the In vivo Mammalian Alkaline Comet Assay OECD TG (OECD TG 489) (8).
The RS Comet assay, therefore, seems well suited as a complement to the RSMN test, which can detect clastogenic and aneugenic properties of a substance. Research efforts from others using in vitro and in vivo models have helped to shape the protocol and validation set-up for the current RS Comet assay. The first approaches to assessing DNA strand breaks in ‘in vitro skin’ employed 2D monolayer cultures of primary human keratinocytes (23), or the HaCaT keratinocyte cell line (24,25). The first study evaluating DNA strand breaks via the Comet assay in an RHS model after topical application was published in 2006 (26). DNA damage, however, was not measured in the keratinocytes but in dendritic cells which were cultured in the medium below the skin model. The first published reports measuring DNA damage in keratinocytes isolated from RHS models stem from studies that were designed to measure photoprotective effect of UV filters, i.e. the protection from reduction of DNA damage caused by irradiation with UV light (26,27). While it had already been shown for a few model chemicals that the Comet assay can be applied to rodent skin in vivo, Reus et al. (28) successfully demonstrated that ex vivo human skin can also be utilised to predict DNA damage after topical exposures in vitro. In this study, 20 known in vivo genotoxins and non-genotoxins were applied to the surface of fresh human skin obtained after breast or abdominal surgery, resulting in an excellent sensitivity, specificity and accuracy of 89, 90 and 89%, respectively (28). The observed high predictivity of the ex vivo human skin Comet assay supports the usefulness of a human skin-based follow-up testing concept, although the test chemicals in this study were not tested in a blinded manner.
The ‘RS Comet project’ started with the establishment of the assay using the EpiDerm™ skin model (MatTek, Ashland, MA), which was intended to measure DNA damage by chemicals in the same skin model as that used for the RSMN assay. Initial experiments showed that the method could be transferred to different laboratories, with good intra- and inter-laboratory reproducibility (13). However, high background levels of DNA damage in this skin tissue model caused a significant number of invalid experiments. Therefore, two independent project teams collaborated to explore the suitability of alternative test systems, i.e. RHS models comprised of both epidermis and dermis [full thickness (FT) skin models]. One of these projects was funded by Cosmetics Europe and the other by the German Federal Ministry for Education and Research (‘Bundesministerium für Bildung und Forschung’, BMBF). The data presented here are a result of a merger of these two projects, which enabled more efficient testing of a larger number of validation chemicals. At this stage, as a measure to improve the sensitivity of the RS Comet assay by accumulating strand breaks, aphidicolin (APC), an inhibitor of DNA polymerases α and δ, was added into the protocol (29) when the standard protocol did not show any effects. Inhibiting the DNA repair function of the polymerases by APC amplifies single strand breaks generated during excision repair which leads to increased comet formation (30). This approach has been used successfully also by others (31,32), although when used to measure cellular DNA incision and repair capacity in human whole blood cultures considerable intra- and inter-individual variability was observed (33). In the context of this validation, the suitability of this approach was supported by the results obtained in earlier phases of this project; adding APC to the solvent control (SC) did not significantly increase the background DNA damage level but did increase the sensitivity of the RS Comet to detect pro-mutagens, while the high predictivity of non-genotoxins was not impacted (14,29).
The project followed a modular validation approach (34) and was performed in three phases: Phase 0 – transferability, optimisation and within-laboratory reproducibility with model genotoxins, Phase 1 – between-laboratory reproducibility with eight coded chemicals (14) and Phase 2 – predictive capacity to increase the domain of chemicals tested which included additional chemicals that were added to broaden the overlap of substances also tested in the RSMN. In Phases 0 and 1, Phenion® (Henkel, Germany) FT skin tissues were demonstrated to be useful for the 3D Skin Comet Assay methodology (14). Moreover, FT skin models were shown to be a better choice for this genotoxic endpoint than simpler reconstructed human epidermal (RHE) models, because of lower and less variable background DNA strand breaks than in RHE (13, 14).
3D tissue constructs are logical follow-up tools for standard ‘2D’ genotoxicity assays because they allow for more natural cell–cell and cell–matrix interactions, and show ‘in vivo-like’ behavior for key parameters, such as cell proliferation, differentiation, morphology, gene and protein expression, and function (35–39). Specifically, the FT models used for the 3D Skin Comet assay are composed of primary cells of human origin and are p53 competent. The latter feature was suggested to be important to avoid the generation of misleading positive results during a workshop hosted by the European Union Reference Laboratory for alternatives to animal testing (EURL ECVAM) (40). The use of primary human cells not only eliminates concerns due to rodent-specific effects but also preserves normal cell cycle control in addition to DNA repair competence (29). The similarity of RHS to native human skin was also shown regarding the activity of enzymes involved in xenobiotic metabolism (41–46), negating the need for external metabolising systems such as rat liver S9 for the given (dermal) route of exposure. Importantly, while the skin has a low background level of cytochrome P450 (CYP) phase 1 activities, these can be induced during the course of the experiment when using a repeated treatment protocol, as described by Götz et al. (47), Wiegand et al. (41) and recently confirmed by Downs et al. (48).
The goal of the work described here is to confirm intra-laboratory reproducibility observed in Phases 0 and 1 and to evaluate the predictive capacity of the RS Comet assay. Ultimately, we aimed to establish its usefulness as a ‘tier 2’ assay that can be used to follow up on positive results from the standard in vitro test battery. An experimental dataset was established by testing a series of chemicals in a double-blind fashion. Chemicals were selected, coded, decoded and analysed by independent experts, as detailed in the Materials and Methods section.
Material and Methods
The RS Comet assay was conducted according to the protocol previously described in detail (14) and outlined below.
Characterisation and selection of coded test chemicals
Test chemicals were selected by external experts [Raffaella Corvi (EC, Joint Research Centre) and David Kirkland (Kirkland Consulting)] from a master list prepared for Cosmetics Europe by a larger group of external experts. These chemicals had previously been tested in in vivo genotoxicity and/or carcinogenicity studies with dermal exposure and were grouped according to three categories: true negative (TN) and true positive (TP) chemicals, with concordant in vitro and in vivo data, as well as misleading positives (MP) for which positive in vitro findings were reported, but not confirmed in in vivo studies. Originally, a total of 30 chemicals were selected with a balanced dataset of 15 genotoxicants (TP) with various modes of action, and 15 chemicals with an expected negative outcome (TN and MP), all covering different chemical classes. For Phase 1 of validation reported previously (14), eight of the 30 chemicals were tested in three laboratories each to demonstrate intra- and inter-laboratory reproducibility.
For the predictivity phase reported here (Phase 2), the remaining 22 chemicals were each tested in one laboratory only. An additional four chemicals were later chosen for testing to close a data gap with chemicals tested in the RSMN assay, bringing the number of chemicals in Phase 2 of the RS Comet assay validation to 26 and increasing the overall total number of coded chemicals tested to 34. However, after decoding the chemicals in this second phase of the validation exercise, technical issues relating to the concentration of the stock chemical and the solvent used (described in detail later) were discovered with two of the test chemicals, dimethylnitrosamine (DMNA), and potassium bromate (KBr), and these were excluded from the validation data set by the Steering Committee after consultation with the external advisory board (see Results and discussion). Literature information on the in vitro and in vivo genotoxicity of the 32 chemicals included in the final evaluation are shown in supplementary Table 1, available at Mutagenesis Online. The test chemicals were purchased from Sigma–Aldrich (≥95% purity), assigned a unique code and shipped to each laboratory by independent co-workers at the German Federal Institute for Risk Assessment (BfR) and BioTeSys (Esslingen, Germany). Each laboratory received chemicals from all subcategories and all chemicals were tested under blinded conditions.
Comet assay reagents
The following reagents were standardised between laboratories: low melting temperature agarose (LMA; SeaPlaque® GTG® Agarose) from Lonza (Basel, Switzerland), agarose (NEEO Ultra-Quality) from Carl Roth (Karlsruhe, Germany), Aphidicolin (APC), DMSO (>99.7% purity), thermolysin, methyl methane sulfonate (MMS), benzo[a]pyrene (BaP) from Sigma–Aldrich (Saint Louis, MO) and SYBR Gold from Life Technologies (Carlsbad, CA). All other reagents were obtained from local suppliers and were not harmonised among laboratories.
Experimental design
For each coded chemical, testing included a solubility study (assessed by visual inspection), a dose range-finding (DRF) experiment (up to a maximum dose of 10% w/v, 1600 µg/cm2), and at least two valid main experiments (14). Chemicals were preferably dissolved in acetone at 10%; however, if they were not soluble at this concentration, they were carefully diluted to determine their maximum solubility. If the solubility in acetone was below 1% (w/v), 70% (v/v) ethanol was used instead. The DRF experiment defined the maximum dose for the main experiments, depending on (a) solubility (if <10%), (b) precipitation of the chemical on the skin upon visual inspection at the end of the experiment, or (c) strong cytotoxicity (>50% relative to the SC). A clear positive finding in the first main experiment was confirmed in a second experiment. Depending on the outcome of the first main experiment, the dose spacing of the second experiment was adjusted, usually by using tighter spacing between doses. If the test chemical provided negative or equivocal results (see ‘Evaluation and interpretation of results’ section for a definition of equivocal), an experimental approach with improved sensitivity was applied which introduced the DNA repair inhibitor APC into the protocol before cell harvest to allow for accumulation of unrepaired DNA lesions.
RS Comet assay
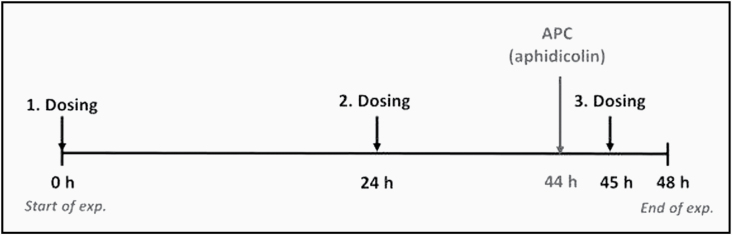
The RS Comet assay was conducted according to the protocol previously described in detail (14). RHS models (Phenion®FT Skin Models, Henkel, Düsseldorf, Germany; www.phenion.com) were cultured individually in 35-mm Petri dishes filled with 5-ml warm air–liquid-interface (ALI) medium (supplied by the manufacturer) overnight at 37°C and 5% CO2 before use. As with the previous validation phases, a main experiment comprised at least three dose groups of a test chemical, as well as the SC and a positive control [PC; methyl methane sulfonate (MMS) at 5 µg/cm2 in acetone]. In case of negative or equivocal findings (please refer to ‘Evaluation and interpretation of results’ section for details), another experiment was performed in which the DNA inhibitor aphidicolin (APC) was added. In these experiments benzo(a)pyrene (BaP) was added as PC. All groups were tested in triplicate. Untreated negative control tissues were omitted since it had been previously demonstrated that the solvents used in these studies (acetone or 70% ethanol) did not affect background DNA damage (14). Chemicals were applied topically in a 25 µl volume (equivalent to 16 μl/cm2 skin surface area) for a total of 48 h to ensure possible metabolic transformation (Figure 1). The chemical was reapplied to the same tissues after 24 h and 45 h. A negative or equivocal outcome in the first main experiment was followed by an APC experiment that involved the addition of APC at 4 h prior to the end of the experiment (Figure 1). SC + APC was then used as SC and BaP (12.5 µg/cm2) plus APC as PC. Solutions of the test chemicals were prepared fresh daily just prior to each dosing. Keratinocytes and fibroblasts were isolated separately and prepared on slides for electrophoresis, which was carried out for 30 min at 39 V and 450 ± 10 mA with fresh buffer in an electrophoresis chamber harmonised among laboratories (Carl Roth, Karlsruhe, Germany, Cat.# N610.1). After electrophoresis, slides were neutralised in 0.4 M Tris–HCl, pH 7.5, for at least 5 min, dehydrated in absolute ethanol, and allowed to dry (14).
Figure 1.
Treatment schedule of skin models. Tissues were exposed with test chemicals for a total of 48 h. A maximum of 100 mg/mL in either acetone or 70% (v/v) ethanol was applied three times. In case of negative or equivocal findings, APC was added 4 h before the end of experiment. Reproduced from Reisinger et al., 2018; by permission of Oxford University Press.
Slide analysis
Analysis of slides was harmonised among laboratories based on published standards (13). Two laboratories used Comet Assay IV software (Perceptive Instruments, Suffolk, UK) and one used CometImager (MetaSystems, Altlußheim, Germany). DNA was stained for 15 min with a 1:10 000 dilution of SYBR Gold in Tris–EDTA buffer pH 7.2. Tail intensity (% tail DNA) was chosen as parameter to assess genotoxicity. For each tissue compartment, two slides were analysed, and a third slide was stored as back-up. Fifty cells per slide were analysed, i.e. 100 cells per tissue compartment (epidermis or dermis). For three tissue replicates, this resulted in 300 cells per compartment and 600 cells total per dose group. Sample size and number of analysed cells were in line with published recommendations (49).
Cytotoxicity assessment
Cytotoxicity was measured according to the relative intracellular adenosine triphosphate (ATP) content (50) and the activity of adenylate kinase (AK) released into the culture medium due to cell membrane damage (51). For determining intracellular ATP content, frozen tissue samples (epidermis plus dermis) were homogenised in 1 ml of precooled PBS w/o Ca2+/Mg2+ in a TissueLyzer II (Qiagen, Hilden, Germany) for 5 min and 30 Hz using a 5-mm stainless steel bead. Homogenates were then heated for 5 min at 105°C, cooled on ice and centrifuged (2 min, 10 000 × g). ATP levels were determined in the supernatants using the ATPlite kit (Perkin Elmer, Waltham, MA) and normalised to protein content (µg ATP/mg protein) in the supernatant measured using the Bradford assay (BioRad, Hercules, CA). AK levels in the culture media were quantified using the ToxiLight bioassay kit (Lonza, Basel, Switzerland). The cytotoxicity markers in the treated tissue groups were expressed relative to those in the SC and were used to assess the validity of each experiment. If cytotoxicity was observed with both measurements, the more sensitive parameter was used (14).
Data evaluation
Raw data (% tail DNA) of single experiments were submitted in Excel spreadsheets to the independent statistician for further analysis as previously (14). The raw data were aggregated as follows: For each slide, the median was calculated from the 50 comet scores and the median values were then arcsine square-root transformed to approach normality and variance homogeneity. For each compartment, the transformed medians of the two slides were averaged. This procedure resulted in n = 3 values (three tissues) per control or treatment group for the epidermis and the dermis compartments, respectively, which were used for statistical testing. In addition, the laboratories provided information on the validity of dose groups, which could be compromised due to strong cytotoxicity or precipitations visible at the end of experiments on top of the skin tissues (please refer to next sections for details).
Validity criteria
Prior to statistical analysis, the validity of each experiment was determined (14). First, the experiment needed to follow the predefined experimental design: SC, PC and at least three dose groups of the test chemical, each represented by three valid tissues and two slides per compartment, with 50 comets scored per slide. Second, the validity criteria for the control groups were applied. In the standard main experiment, the SC had to display ≤20% tail DNA and the PC (MMS) had to show at least a two-fold increase in % tail DNA compared to SC, as well as an absolute increase in % tail DNA by ≥15% points above the SC. In the APC experiment, the SC + APC had to also display ≤ 20% tail DNA; whereas, the PC (BaP + APC) had to be above the SC + APC reference range and had to show an increase in % tail DNA by ≥5% points above the SC + APC. Third, a dose group was considered valid when the thresholds set for strong cytotoxicity (i.e. 2-fold increase in AK leakage compared to SC and/or 50% reduction in normalised ATP content compared to SC) were not exceeded. If excessive cytotoxicity in a dose group or precipitation on top of a tissue at the end of experiments was observed, the dose group was not considered for the evaluation of genotoxicity and was not included in the statistical analysis. In rare cases of treatments triggering neither of the above-mentioned cytotoxicity thresholds, a clear decrease in the number of comets observed on slides was used as indication to not consider these high dose groups for genotoxicity assessment.
Evaluation and interpretation of results
Data interpretation included the statistical analysis of the datasets as well as judgment of the biological relevance of the results and was performed in collaboration with the Steering Committee and external experts but also prior to decoding by the performing laboratory, which is described in more detail further below. A detailed description of the statistical analysis can be found in Reisinger et al. (14). Specifically, an experiment was identified as positive if at least one of the two statistical prediction models indicated a significant and dose-related increase in tail intensity for doses that did not induce cytotoxicity exceeding the cut-off. In case only one dose group produced a statistically significant increase in % tail DNA without dose-dependency, the effect had to be reproduced in a second main experiment to trigger a positive call. In both scenarios, at least one dose group needed to be outside the historical control range (laboratory specific: mean of the historical SCs plus 2 SD). In any case, a positive result in one cell type/compartment was sufficient to consider a main or an APC experiment as positive. In case of negative or equivocal findings where some, but not all, criteria were fulfilled for a positive call, an additional APC experiment was considered positive if the test chemical caused a statistically significant increase in % tail DNA in the presence of APC compared to SC + APC and this value exceeded the historical control range for APC experiments. For the final conclusion, criteria based on both the statistical significance and the biological relevance were taken into account. These criteria followed the standards of the OECD Test Guideline of the ‘In Vivo Mammalian Alkaline Comet Assay‘ (8).
In addition to the above, the performing laboratories were asked to provide their expert judgement regarding the study outcome overall, prior to decoding. Information on the outcome of the two statistical prediction models was available at that time to assist the laboratories in their judgement. To note: Historical background ranges were calculated at the end of the experimental phase (after decoding) and, together with the statistical predictions, formed the core of the (later established) evaluation criteria. The predictive capacity of the RS comet assay was calculated and presented two ways, as a function of the outcome of the evaluation and interpretation of results as described above, and as a function of the outcome of the laboratories’ expert judgements.
Results and Discussion
During Phase 1 of the validation each chemical was investigated in three laboratories to obtain information on the intra- and inter-laboratory reproductivity (14). As reproducibility was found to be sufficient, for Phase 2 described here, the Steering Committee decided to test each chemical in one laboratory to expand the range of chemicals tested in the RS Comet assay.
Study results of validation Phase 2
Chemicals with an expected positive outcome
2-Acetylaminofluorene
In Experiment 1, 2-acetylaminofluorene (2-AAF) was tested up to its maximum solubility (precipitation was evident at doses at and above 128 µg/cm2) in Lab B, in which no increase in DNA migration was observed (supplementary Figure 1A, available at Mutagenesis Online; see Table 1 and Supplementary Table 2 for an overview). In the second experiment in the presence of APC, 2-AAF did not cause any significant increases in % tail DNA in keratinocytes, except at 80 µg/cm2, which slightly exceeded the cytotoxicity limit (supplementary Figure 1B, available at Mutagenesis Online). However, the SC for fibroblasts was also outside the historical control range in this experiment; therefore, a second experiment with APC was conducted. In this experiment, 2-AAF induced a significant increase in DNA migration at all doses tested (supplementary Figure 1C, available at Mutagenesis Online). The level of cytotoxicity was slightly below the threshold limits in this experiment; however, based on the clear increase in DNA damage in both cell types, 2-AAF was correctly classified as positive. This result is supported by recent data published by Downs et al. (48), which showed that a reactive metabolite (as demonstrated by DNA adduct formation) accumulates with the multiple treatment protocol utilised in this validation study, leading to positive responses in the RS Comet assay. Importantly, 2-AAF is a pro-mutagen, the known metabolic pathways of which involves NAT and CYP1A2 and leads to the formation of genotoxic metabolites (52,53).
Table 1.
(a and b) Overview of validation outcome
| Chemical | CAS No | Cat | Phase | Lab A | Lab B | Lab C | Lab D | Lab E | |
|---|---|---|---|---|---|---|---|---|---|
| (a) | |||||||||
| Chemicals with an expected positive outcome | |||||||||
| 1 | 2-AAF | 53-96-3 | TP | 2 | Pos | ||||
| 2 | IQ | 76180-96-6 | TP | 2 | Neg | ||||
| 3 | AZT | 30516-87-1 | TP | 2 | Pos | ||||
| 4 | BaP | 50-32-8 | TP | 2 | Pos | ||||
| 5 | 4-Chloroaniline | 106-47-8 | TP | 2 | Neg | ||||
| 6 | Cadmium chloride | 10108-64-2 | TP | 1 | Pos | Pos | Neg | ||
| 7 | CPPE | 27208-37-3 | TP | 2 | Pos | ||||
| 8 | CP | 6055-19-2 | TP | 2 | Pos | ||||
| 9 | 2,4-DAT | 95-80-7 | TP | 2 | Neg | ||||
| 10 | DMBA | 57-97-6 | TP | 1 | Pos | Pos | Pos | ||
| 11 | EMS | 62-50-0 | TP | 2 | Pos | ||||
| 12 | ENU | 759-73-9 | TP | 1 | Pos | Pos | Pos | ||
| 13 | Etoposide | 33419-42-0 | TP | 2 | Pos | ||||
| 14 | MNNG | 70-25-7 | TP | 2 | Pos | ||||
| 15 | Mitomycin C | 50-07-7 | TP | 1 | Pos | Equiv | Pos. | ||
| Chemicals with an expected negative outcome | |||||||||
| 16 | Amitrole | 61-82-5 | TN | 2 | Neg | ||||
| 17 | Ampicillin sodium salt | 69-52-3 | TN | 2 | Neg | ||||
| 18 | N-butyl chloride | 109-69-3 | TN | 2 | Neg | ||||
| 19 | Curcumin | 458-37-7 | MP | 2 | Neg | ||||
| 20 | Cyclohexanone | 108-94-1 | TN | 1 | Neg | Neg | Neg | ||
| 21 | 2,6-DAT | 823-40-5 | MP | 2 | Neg | ||||
| 22 | N,N-dicyclohexyl thiourea | 1212-29-9 | TN | 2 | Neg | ||||
| 23 | Ethionamide | 536-33-4 | MP | 2 | Equiv | ||||
| 24 | DEHP | 117-81-7 | TN | 1 | Neg | Neg | Neg | ||
| 25 | Eugenol | 97-53-0 | MP | 1 | Neg | Neg | Neg | ||
| 26 | Glyoxal | 107-22-2 | MP | 2 | Neg | ||||
| 27 | d-Mannitol | 69-65-8 | TN | 2 | Neg | ||||
| 28 | 4-Nitrophenol | 100-02-7 | MP | 2 | Neg | ||||
| 29 | Propyl gallate | 121-79-9 | MP | 1 | Neg | Neg | Neg | ||
| 30 | Resorcinol | 108-46-3 | MP | 2 | Pos | ||||
| 31 | Sodium xylene sulfonate | 1300-72-7 | MP | 2 | Neg | ||||
| 32 | TBHQ | 1948-33-0 | MP | 2 | Equiv | ||||
| (b) | |||||||||
| Chemicals with an expected positive outcome | |||||||||
| 15 | Mitomycin C | 50-07-7 | TP | 1 | Pos | . | |||
| Chemicals with an expected negative outcome | |||||||||
| 23 | Ethionamide | 536-33-4 | MP | 2 | Neg | ||||
| 30 | Resorcinol | 108-46-3 | MP | 2 | Equiv | ||||
| 32 | TBHQ | 1948-33-0 | MP | 2 | Neg |
In Phase 1, each chemical was tested in three laboratories (14). Subsequently, in Phase 2, chemicals were each tested in one laboratory only to expand the chemical space tested in the RS comet assay or to expand the overlap with chemicals tested in the RSMN. Cat – Category, i.e., expected outcome based on historical in vitro and in vivo genotoxicity or carcinogenicity data as provided in supplementary Table 1, available at Mutagenesis Online; MP = misleading positive; TN = true negative; TP = true positive; Pos = positive study outcome, DNA damage detected; Neg = negative; Equiv = equivocal. (a) Outcome as per expert judgement of the performing laboratory before decoding; (b) discordant calls as per application of the (later established) evaluation criteria agreed by the Steering Committee and other external experts
2-Amino-3-methylimidazo[4,5-f]quinolone
2-Amino-3-methylimidazo[4,5-f]quinolone (IQ) was tested up to its solubility limit in the initial experiment in Lab A but showed strong cytotoxicity at all doses tested (33.3–300 µg/cm2), resulting in a non-valid experiment (supplementary Figure 2A, available at Mutagenesis Online). In the second experiment, in which the dose range was modified according to cytotoxicity observed in the first experiment, the two lowest doses were within acceptable cytotoxicity limits, but a moderate increase in % tail DNA was detected in one mid-dose group that was slightly outside the cytotoxicity limits (supplementary Figure 2B, available at Mutagenesis Online). A third main experiment using the APC protocol also did not show a statistically significant increase in DNA damage at any dose, possibly due to a high variability in the SC (supplementary Figure 2D, available at Mutagenesis Online). However, the APC experiment was repeated, and a significant increase in % tail DNA was only detected in keratinocytes in one dose group that exceeded the cytotoxicity cut-off (supplementary Figure 2D, available at Mutagenesis Online). Overall, despite some borderline increases, IQ was incorrectly detected as negative.
IQ has been shown to be an in vivo genotoxicant and is listed as an IARC class 2A carcinogen (i.e. reasonably anticipated to be a human carcinogen). This chemical requires metabolic activation to cause genotoxicity. Evidence suggests that the chemical is activated by N-hydroxylation via CYP1A2 and O-acetylation by NAT (54), followed by spontaneous degradation of N-acetoxy-IQ to an unstable nitrenium ion capable of binding to DNA (55). The NAT isoform responsible for O-acetylation could be organ-specific since NAT1 was shown to O-acetylate IQ in human mammary glands (56) but studies using salmonella expressing human NAT1 or NAT2 showed that NAT2 was mainly involved in the activation (57). Studies investigating the metabolism of IQ in ex vivo pig or human skin (over 24 h) or by EpiSkin™ S9 (over 4 h), indicate that it is not metabolised in either skin model, not even to the O-acetylated metabolite (58)]. These finding suggest that while IQ can be metabolised to a genotoxic metabolite in vivo in the rat (59,60), this does not occur in human ex vivo skin and RHS models.
Azidothymidine
Azidothymidine (AZT) was tested up to concentrations producing signs of strong cytotoxicity (700 µg/cm2) in Lab B, based on the results of the DRF experiment. AZT caused a strong dose-dependent increase in % tail DNA in keratinocytes and fibroblasts in both main experiments at non-cytotoxic doses ≤700 µg/cm2 (supplementary Figure 3A and B, available at Mutagenesis Online) without APC. Due to the clear increase in DNA damage at doses below the cytotoxicity limit, AZT was correctly classified as positive.
Benzo(a)pyrene
In Lab A, the top dose for BaP (160 µg/cm2) was limited by its solubility in acetone (10 mg/ml). BaP was not cytotoxic at this dose but failed to increase % tail DNA in the first main experiment (supplementary Figure 4A, available at Mutagenesis Online). However, in the follow-up experiment with APC, BaP clearly and significantly increased DNA migration at all doses in both cell types (supplementary Figure 4B, available at Mutagenesis Online). The response reached a maximum level at the lowest BaP dose tested (10 µg/cm2) and thus was not dose-related within the dose range used. Notably, BaP is a promutagen that requires metabolic activation by CYP1 isoforms, primarily CYP1A1 and/or CYP1B1, in order to interact with DNA (61). This suggests that while CYPs are known to be of low abundance in the skin compared with the liver (62), there was sufficient activity of these enzymes to be able to metabolise BaP to reactive metabolite(s). Indeed, several CYP-mediated genotoxic metabolites, including the highly mutagenic (+)-anti-BP-7,8-diol-9,10-epoxide (BPDE), have been shown to be formed in human skin and 3D skin tissue models (29). The 10 µg/cm2 dose is also similar to that used for the BaP PC (12.5 µg/cm2) in the APC experiments and the results observed with the coded BaP sample were consistent with the reproducible effects seen from its regular use in that context. Taken together, BaP was correctly classified as positive.
4-Chloroaniline
The top dose for 4-chloroaniline (5 µg/cm2) was limited by strong cytotoxicity, as determined in the DRF experiment in Lab B. In Experiment 1, 4-chloroaniline induced strong cytotoxicity at this top dose but did not increase % tail DNA in either cell type (supplementary Figure 5A, available at Mutagenesis Online). A second experiment with APC, the cytotoxicity effects were reproduced at doses ≥10 µg/cm2 4-chloroaniline but this dose did not induce an increase in DNA migration (supplementary Figure 5B, available at Mutagenesis Online). These negative results were reproduced in another experiment with APC (supplementary Figure 5C, available at Mutagenesis Online). As a result, 4-chloroaniline was incorrectly classified as negative.
4-Chloroaniline is classified as an in vivo genotoxic (63,64) and carcinogenic (65) chemical. N-Acetylation has been shown to be a detoxification pathway for aromatic amines and there is evidence that the skin provides a ‘first pass’ detoxification capability for this class of compounds, as discussed by Zeller and Pfuhler (24). However, 4-chloroaniline was not metabolised in ex vivo human skin over 24 h, but was N-acetylated to N-acetyl-4-chloroaniline by skin S9 subcellular fractions (58). The authors hypothesised that its lack of metabolism in ex vivo skin may be due to its high reactivity to extracellular proteins preventing it from entering the keratinocytes and being metabolised, unlike S9, which was homogenously mixed with test chemical, the preparation being free of functional membranes. This finding may help explain why a negative response was obtained for the dermal exposure route.
Cyclopenta[c,d]pyrene
Cyclopenta[c,d]pyrene (CPPE) was investigated in Lab A up to concentrations producing signs of strong cytotoxicity (75 µg/cm2), as observed in the DRF experiment. In Experiment 1, cytotoxicity did not exceed the cut-off limit and DNA migration was not increased in either cell type (supplementary Figure 6A, available at Mutagenesis Online). In the APC experiment, a significant increase in % tail DNA was observed in the absence of limiting cytotoxicity at four of the top five doses tested in the keratinocytes (supplementary Figure 6B, available at Mutagenesis Online) and CPPE was correctly classified as positive. As with BaP, this result was significant because CPPE is metabolised by CYPs (CYP1A1, CYP1A2 and CYP3A4) to form cyclopenta[c,d]pyrene-3,4-epoxide, trans-3,4-dihydrocyclopenta[c,d]pyrene-3,4-diol and other DNA reactive metabolites which form DNA adducts (66).
Cyclophosphamide
Cyclophosphamide (CP) is genotoxic in vivo (67,68) and is often used as a PC in genotoxicity assays. This chemical was tested up to concentrations producing signs of strong cytotoxicity (1200 µg/cm2) in Lab B. CP induced a dose-dependent increase in % tail DNA in both cell types in both main experiments at non-cytotoxic doses ≤1000 µg/cm2 (supplementary Figure 7A and B, available at Mutagenesis Online) without APC. Due to the clear increase in DNA damage at doses below the cytotoxicity limit, CP was correctly classified as positive. CP is metabolised by human hepatic CYP enzymes, mainly by CYP2B6 but also by CYP3A4 and CYP2C8/9 (69). Of these, CYP3A4 and CYP2C9 have been shown to be present in native human skin and 3D skin models, albeit at very low levels (42). This CYP-mediated hydroxylation results in the formation of 4-hydroperoxy-cyclophosphamide which is subsequently converted to the genotoxic metabolites, acrolein and the potent alkylating agent, phosphoramide mustard (70,71).
2,4-Diaminotoluene
The maximum dose of 2,4-diaminotoluene (2,4-DAT; 1280 µg/cm2) was limited by strong cytotoxicity observed in the DRF experiment observed in Lab B. In Experiment 1, 2,4-DAT did not increase the % tail DNA of either cell type (supplementary Figure 8A, available at Mutagenesis Online). In the second experiment with APC, DNA migration was statistically significantly induced by 2,4-DAT at 400 and 800 µg/cm2 in keratinocytes only (supplementary Figure 8B, available at Mutagenesis Online). As these responses remained within the SC + APC historical control ranges and an increase was not observed at a higher dose (1200 µg/cm2) at which cytotoxicity levels were marginally exceeded 2,4-DAT was incorrectly considered negative.
Ethyl methanesulfonate
Ethyl methanesulfonate (EMS) was tested up to concentrations producing signs of strong cytotoxicity, based on the results of the DRF experiment, i.e. 200 µg/cm2 in Lab B. EMS induced a clear and dose-dependent increase in % tail DNA in both cell types in the absence of APC in both main experiments (supplementary Figure 9A and B, available at Mutagenesis Online). As a result of the clear increase in DNA damage at doses below the cytotoxicity limit, EMS was correctly classified as positive.
Etoposide
The top dose of etoposide was limited to 120 µg/cm2 due to its cytotoxicity at higher doses observed in the DRF experiment in Lab C. Etoposide induced a small dose-dependent increase in % tail DNA in both cell types at non-cytotoxic doses ≤120 µg/cm2 without APC in the first main experiment (supplementary Figure 10B, available at Mutagenesis Online). However, since none of the % tail DNA values were outside of the historical ranges of the SC, a second experiment with APC was conducted. In the presence of APC, a stronger dose-dependent increase in DNA damage was observed at doses below the cytotoxicity limit (supplementary Figure 10B, available at Mutagenesis Online), and etoposide was correctly classified as positive.
N-methyl-N’-nitro-N-nitrosoguanidine
The top dose of N-methyl-N’-nitro-N-nitrosoguanidine (MNNG; 2 µg/cm2) was limited by strong cytotoxicity in Lab B. In the first main experiment, MNNG was tested up to this cytotoxicity limit and was shown to cause a dose-dependent increase in % tail DNA in keratinocytes only (supplementary Figure 11A, available at Mutagenesis Online). In a second standard experiment, MNNG reproduced the effects in keratinocytes. MNNG also caused significant increase in DNA damage in fibroblasts in Experiment 2. The responses were dose-dependent and exhibited little variability but did not exceed the historical range of the SC for fibroblasts (supplementary Figure 11B, available at Mutagenesis Online). However, due to the reproducible effect on DNA damage in keratinocytes at doses below the limit of cytotoxicity, MNNG was correctly classified as positive.
Chemicals with an expected negative outcome
This section comprises chemicals considered as ‘true negative’, i.e. those for which concordant negative results in historical in vitro and in vivo experiments were obtained, as well as ‘misleading positive’ chemicals, which showed positive in vitro findings, which were not confirmed in vivo.
Amitrole
In Lab A, in the first main experiment, amitrole was investigated up to the maximum dose of 1600 µg/cm2 in Lab A, with no observed cytotoxicity outside the cut-off limits or increases in DNA damage (supplementary Figure 12A, available at Mutagenesis Online). In Experiment 2 with APC, a small but statistically significant increase in DNA migration was observed at several doses but this was not dose-dependent, and all values remained well within the respective historical SC range (supplementary Figure 12B, available at Mutagenesis Online). The observed small increases in % tail DNA in the second experiment were therefore considered not of biological relevance. Amitrole was correctly classified as negative.
Ampicillin sodium salt
Ampicillin sodium salt was tested up to the maximum dose of 1600 µg/cm2 in Experiment 1. In this experiment, none of the doses caused cytotoxicity that exceeded the limit in Lab B, except at one of the mid-doses. However, it was noted that precipitation that remained on the surface of the tissues at the time of the cell harvest was present at doses of ≥400 µg/cm2 (supplementary Figure 13A, available at Mutagenesis Online). Two experiments with APC were conducted using different dose ranges. In these, ampicillin sodium salt did not increase DNA migration except at one mid-dose (800 µg/cm2) in the second experiment that exceeded the historical SC range in keratinocytes (supplementary Figure 13B and C, available at Mutagenesis Online). However, this increase was only observed in the presence of precipitation, which occurred at doses ≥200–400 µg/cm2 in both experiments. The performing laboratory decided to perform an additional experiment without APC, using a modified dose range that did not result in precipitation. A small but statistically significant increase in DNA migration was seen in fibroblasts only at the high dose. The respective value, however, was well within the historical SC range (supplementary Figure 13D, available at Mutagenesis Online). Therefore, the observed increase in % tail DNA in the initial experiment with APC was considered not of biological relevance and ampicillin sodium salt was correctly classified as negative.
N-Butyl chloride
In the first main experiment, N-butyl chloride was tested up to the maximum dose of 1600 µg/cm2 in Lab A without any strong cytotoxicity being observed. A small but statistically significant increase in DNA migration was observed at the lowest dose (400 µg/cm2) only in keratinocytes but the response did not exceed the historical SC range (supplementary Figure 14A, available at Mutagenesis Online). A second experiment with APC was performed using the same dose range. This reproduced the lack of strong cytotoxicity and did not show any significant increases in DNA migration (supplementary Figure 14B, available at Mutagenesis Online). Taken together, the increase observed in the initial experiment was considered not to be of biological relevance and N-butyl chloride was correctly classified as negative.
Curcumin
The top dose of curcumin tested (50 µg/cm2) was limited by strong cytotoxicity, as well as by precipitation that remained on the surface of the tissues at the time of the cell harvest in Lab B. In Experiment 1, a marked increase in DNA damage (being well above the historical SC range) was observed in keratinocytes and fibroblasts at all doses. However, this chemical exhibited excessive cytotoxicity at a dose of 50 µg/cm2, and precipitation was present at all the tested doses, thus invalidating the experiment (supplementary Figure 15A, available at Mutagenesis Online). This experiment was repeated using a lower dose range and similar results were observed (supplementary Figure 15B, available at Mutagenesis Online). Two experiments with and without APC were conducted using different dose ranges in which curcumin did not increase DNA migration in either experiment, except some marked increases in keratinocytes at several doses that either exceeded the cytotoxicity limits or were limited by precipitation (supplementary Figure 15C and D, available at Mutagenesis Online). Therefore, the increases in DNA damage in these experiments were considered not of biological relevance and curcumin was correctly classified as negative.
2,6-Diaminotoluene
In the first experiment 2,6-diaminotoluene (2,6-DAT) was tested up to a maximum dose of 800 µg/cm2 in Lab B, which was limited by strong cytotoxicity, evident in the DRF experiment. 2,6-DAT did not increase the DNA migration under these conditions (supplementary Figure 16A, available at Mutagenesis Online). In the second experiment in the presence of APC, 2,6-DAT caused a moderate increase in % tail DNA in keratinocytes at a dose of 160 µg/cm2. However, the limit of cytotoxicity was exceeded at this and higher doses, resulting in only one valid dose group in this experiment (supplementary Figure 16B, available at Mutagenesis Online). Therefore, a second experiment with APC was conducted using a modified dose range. This showed that 2,6-DAT caused an increase in DNA migration in keratinocytes at one dose (140 µg/cm2), which was not statistically significant and inside the historical control range for the SC (supplementary Figure 16D, available at Mutagenesis Online). Since there was some inconsistency in the cytotoxicity values amongst experiments, a third experiment with APC was run using higher doses of 2,6-DAT. Two doses caused statically significant increases in % tail DNA in keratinocytes but these responses were well within the historical control range for the SC and were flagged as statistically significant because of the very low values for the corresponding SC (supplementary Figure 16D, available at Mutagenesis Online). 2,6-DAT was therefore correctly predicted as negative.
N,N-dicyclohexyl thiourea
N,N-dicyclohexyl thiourea was tested up to concentrations producing signs of strong cytotoxicity (200 µg/cm2) in Lab B but did not induce an increase in % tail DNA in either cell type in the first main experiment (supplementary Figure 17A, available at Mutagenesis Online). In Experiment 2 with APC, N,N-dicyclohexyl thiourea caused signs of strong cytotoxicity at ≥160 µg/cm2 but did not cause any significant increases in % tail DNA (supplementary Figure 17B, available at Mutagenesis Online).N,N-dicyclohexyl thiourea was correctly classified as negative. It was also not carcinogenic in rats and mice (72).
Ethionamide
The top dose for ethionamide (320 µg/cm2) was limited by its solubility in acetone. However, the cytotoxicity at this dose exceeded the cut-off limit in the first main experiment in Lab A. A statistically significant increase in DNA migration was observed in fibroblasts at this (toxic) dose and the next lowest dose (160 µg/cm2), which showed acceptable toxicity; however, neither response exceeded the historical SC range (supplementary Figure 18A, available at Mutagenesis Online). A second experiment was performed using a top dose of 240 µg/cm2, based on the cytotoxicity observed in Experiment 1. Here, ethionamide was not cytotoxic and failed to increase % tail DNA (supplementary Figure 18B, available at Mutagenesis Online). In the follow-up experiment with APC, ethionamide significantly increased
DNA migration at the mid-dose only in fibroblasts but this response was again within the historical SC range and exceeded the cytotoxicity limit at the highest dose tested (supplementary Figure 18C, available at Mutagenesis Online). To verify the negative result, a second experiment with APC was conducted. As before, no significant increases in % tail DNA or cytotoxicity were observed (supplementary Figure 18D, available at Mutagenesis Online). Overall, based on the lack of reproducible, biologically relevant increases in DNA migration, ethionamide was classified correctly as negative. To note, the expert call of the performing laboratory was ‘equivocal’ (Table 1).
Glyoxal
Lab B tested glyoxal up to concentrations producing signs of strong cytotoxicity (25 µg/cm2), as observed in the DRF experiment. In Experiment 1, cytotoxicity did not exceed the cut-off limit and DNA migration was not increased in either cell type (supplementary Figure 19A, available at Mutagenesis Online). Despite of an increase in % tail DNA at the highest dose tested in fibroblasts in the initial APC experiment, the experiment was invalid due to strong cytotoxicity at all doses tested (supplementary Figure 19B, available at Mutagenesis Online). The cytotoxicity measurements were quite variable in Experiment 2 and were not considered reliable. Therefore, a second APC experiment was conducted in which a statistically significant increase in DNA migration was observed in keratinocytes at the highest dose tested (supplementary Figure 19C, available at Mutagenesis Online). While prediction models 1 and 2 identified it as being positive, expert judgement predicted it as negative since these values were clearly within historical control range. Although positive in several in vitro genotoxicity assays, glyoxal tested negative for genotoxicity in vivo and was non-carcinogenic; therefore, based on the overall weight of evidence from in vivo studies (supplementary Table 1, available at Mutagenesis Online), was correctly considered as negative.
Glyoxal has been shown to cause mutations in vitro by forming DNA adducts, indicated as a positive response in vitro using a standard cell line (TK6) in the comet assay (73). In addition, it also possesses DNA crosslinking activity in vitro (74–76). In general, chemicals with crosslinking activities are not easy to detect with the standard comet assay protocol since the crosslinking of DNA with DNA or protein can inhibit the migration of single strand DNA and thus prevent the detection of tail DNA (8,77,78). It is important to mention, though, that these types of chemicals are more easily identified using clastogenicity assays when investigated as part of a standard genotoxicity testing battery. Indeed, in vitro chromosomal aberration studies using a Chinese hamster V79 fibroblast cell line have all shown clear positive results for glyoxal (79), but such an effect did not replicate in vivo. Multiple in vivo micronucleus tests in mice treated with glyoxal by either intraperitoneal injection or oral administration have all shown negative results, supporting the conclusion of independent expert panels that glyoxal is non-genotoxic in vivo (80,81).
d-Mannitol
The top dose for d-mannitol (320 µg/cm2) was limited by its solubility in acetone. In Experiment 1, cytotoxicity did not exceed the limits in Lab C and no increase in DNA migration was observed at any dose (supplementary Figure 20A, available at Mutagenesis Online). In two follow-up experiments with APC, d-mannitol also did not significantly increase DNA migration (supplementary Figure 20B and C, available at Mutagenesis Online). d-Mannitol was classified correctly as negative.
4-Nitrophenol
4-Nitrophenol was tested up to concentrations producing signs of strong cytotoxicity (48 µg/cm2) in Lab C. In the first main experiment, a statistically significant increase in % tail DNA was observed in keratinocytes only at the two lowest doses (supplementary Figure 21A, available at Mutagenesis Online), in the absence of a dose-response, which were slightly outside the historical control range. In Experiment 2 with APC, 4-nitrophenol induced a small but non-significant increase in DNA migration at the highest non-cytotoxic dose (32 µg/cm2) in keratinocytes (supplementary Figure 21B, available at Mutagenesis Online). A second experiment with APC was conducted in which a small but statistically significant increase in DNA migration at two out of 5 doses (10 and 35 µg/cm2) was observed only in fibroblasts (supplementary Figure 21C, available at Mutagenesis Online). The respective % tail DNA values were well within the corresponding historical control range data and the statistical significance versus the control likely was caused by a very low value for the corresponding SC. Based on this, as well as the lack of reproducibility of the observed (marginal) increases in DNA migration, 4-nitrophenol was correctly classified as negative.
Resorcinol
Resorcinol was tested up to concentrations producing signs of strong cytotoxicity (250 µg/cm2) in Lab B, as observed in the DRF experiment. In the first main experiment, a small but statistically significant increase in DNA migration was observed in keratinocytes, supported by a low value of the corresponding SC, while not exceeding the historical SC range (supplementary Figure 22A, available at Mutagenesis Online). In a second experiment with APC, a statistically significant increase in % tail DNA was observed in fibroblasts at the lowest dose again aided by low values of the SC (supplementary Figure 22B, available at Mutagenesis Online). This increase exceeded the self-calculated historical SC range of the performing laboratory at the time of the testing, resulting in a positive expert call by the laboratory in the coded phase. Later on, when the final HC range for APC experiments was calculated after decoding and applied to the dataset, the respective dose group was within the HC, i.e. the criteria for a positive call were no longer fulfilled. At the same time, the two highest dose groups tested in the presence of APC showed increases that were clearly outside the HC and only marginally exceeded the cytotoxicity cut-off limits. Since such borderline results were not suitable to support a negative call either, the overall call for resorcinol was equivocal (Table 1).
While positive in most other in vitro genotoxicity assays (supplementary Table 1, available at Mutagenesis Online), resorcinol led to somewhat controversial results in vivo. Resorcinol was tested positive in an in vivo micronucleus test in male B6C3F1 mice (82) but showed no effects in male and female Crl:CD (SD)BR rats (83) and was non-carcinogenic in rats and mice (82). Overall, both the European Food Safety Authority (EFSA) and the Scientific Committee on Consumer Safety (SCCS), the independent expert panel mandated by the European Commission, classified resorcinol as in vivo non-genotoxic (83,84).
Sodium xylene sulfonate
Sodium xylene sulfonate was tested up to concentrations producing signs of strong cytotoxicity (e.g., 512 µg/cm2 in the DRF) in Lab C and did not induce an increase in % tail DNA in either cell type in the first main experiment (supplementary Figure 23A, available at Mutagenesis Online). In Experiment 2 with APC, sodium xylene sulfonate caused signs of strong cytotoxicity at 640 µg/cm2 but no significant increases in % tail DNA (supplementary Figure 23B, available at Mutagenesis Online). The experiment with APC was repeated with a modified dose range (supplementary Figure 23C, available at Mutagenesis Online) in which similar results were observed, with no increase in DNA damage. Sodium xylene sulfonate was therefore correctly classified as negative and is also negative for carcinogenicity after topical application to rats and mice (85).
Tert-butylhydroquinone
Tert-butylhydroquinone (TBHQ) demonstrated a steep cytotoxicity response in the DRF experiment, limiting the top dose tested in Experiment 1 to 40 µg/cm2 in Lab A. Moderate increases in % tail DNA were observed in this experiment but only at the two highest doses that both exceeded the cytotoxicity limit (supplementary Figure 24A, available at Mutagenesis Online). The dose range for TBHQ was adjusted according to the observed cytotoxicity and the second experiment used a reduced top dose of 20 µg/cm2. TBHQ was still cytotoxic at the top dose but did not lead to an increased % tail DNA. By contrast, in fibroblasts only, the next lower dose of 16 µg/cm2 did cause a statistically significant increase in % tail DNA (supplementary Figure 24B, available at Mutagenesis Online); however, the value remained within the respective historical control range. In a follow-up experiment with APC, TBHQ caused a small but statistically significant increase in DNA migration at the two highest doses in fibroblasts only but, as previously observed, the respective value remained well within the historical SC range (supplementary Figure 24C, available at Mutagenesis Online). The performing laboratory decided to conduct a third experiment without APC to help resolve uncertainty of the substance’s profile. In this experiment, the highest two doses exceeded the cytotoxicity limit, but no significant increases in % tail DNA were observed (supplementary Figure 24D, available at Mutagenesis Online). Based on the overall dataset, TBHQ was classified correctly as negative while the original expert call of the performing laboratory was ‘equivocal’ (Table 1).
Excluded chemicals due to technical reasons
There were two chemicals that were tested but the results of which were excluded from the validation dataset, namely, DMNA and KBr. DMNA was one of the chemicals selected by the external experts, sent to the laboratories as a coded chemical and tested negative (data not shown). However, post decoding, it became clear that DMNA was not supplied pure but dissolved in methanol as a very dilute (0.5%) solution. Since the laboratory treated the 0.5% solution as if it was a pure test compound, the resulting top dose in the assay was 200× lower than the desired top dose. After consultation with the external advisory panel it was decided that this chemical will be excluded from the validation dataset.
Due to the extremely limited solubility in either acetone or 70% ethanol (<0.1 mg/ml), Lab A decided to test KBr using a vehicle (water) that was not approved by the Standard Operating Procedure and had not been previously validated in the Phenion®FT skin model (data not shown). After decoding and consultation with the external advisory panel, it was decided to exclude the KBr data from the validation dataset due to the use of a non-authorised solvent.
Historical database with and without APC
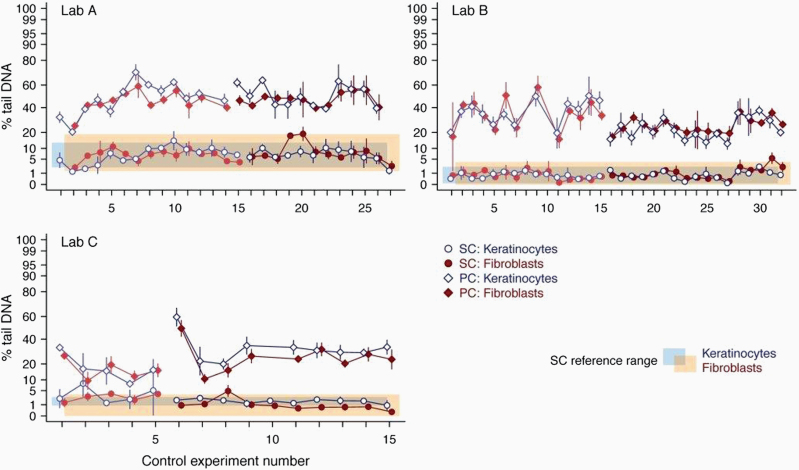
For all laboratories participating in Phase 2, descriptive statistics were performed displaying the percentage of tail DNA for both SC and PC of both cell types, namely keratinocytes and fibroblasts, in relation to the experiment number (Figure 2). These data were used to obtain information on average levels and on the variability of background DNA damage across experiments, which were then used to derive laboratory-specific reference ranges (calculated as mean ± 2 SDs).
Figure 2.
Historical control data of standard experiments. Percentage of tail DNA in the solvent control (SC) and positive control (PC, i.e., MMS) of individual experiments as obtained with the Phenion®FT during Phase 1 and 2 of coded testing are shown. The SC values (circles) and PC values (diamonds) for the keratinocytes (dark blue symbols and lines) and fibroblasts (red symbols and lines) are given as mean ± SD (N = 3 samples each). Faint symbols indicate values obtained in Phase 1 and dark symbols indicate values from Phase 2 of the validation. The light blue-shaded and orange-shaded areas indicate the reference range (mean ± 2 SD) for the SC, i.e. historical control, for keratinocytes and fibroblasts, respectively. The reference ranges were derived from the control data of Phase 2. The y-axis is on the arcsine square-root transformed scale, but the tick labels are expressed in units of the percentage scale.
While variable among laboratories, each one (A, B, C) showed a low and reproducible level of background DNA damage for the SC in the absence of APC, as well as a clear induction of DNA migration by the PC (MMS) for both cell types. When compared to the values observed for an earlier validation phase (Figure 2, faint symbols), similar results were obtained in terms of reproducibility, as well as the level of DNA damage observed, demonstrating the robustness of the method over time. Application of PC produced clear, significant increases in % tail DNA in all experiments performed (Figure 2). The first acceptance criterion for a PC (a 2-fold increase in tail % DNA above SC) was fulfilled in all main experiments in Phase 2; the second criterion (an absolute increase in % tail DNA of 15 percentage points above the SC) was fulfilled in all main experiments except four in Lab B.
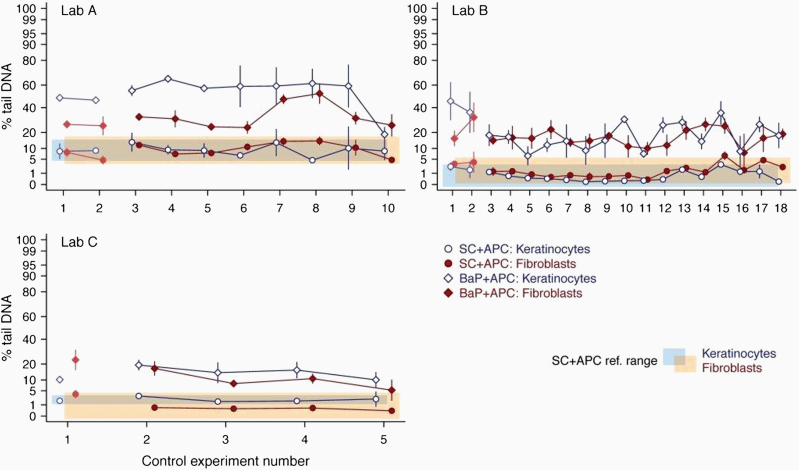
In Phase 1, a negligible impact of APC on the background DNA level was indicated (14). To further investigate this aspect on a broader data set, the historical control data for the experiments of Phase 2 were added to the dataset (Figure 3). SC + APC data (Figure 3) indicated that all laboratories achieved a low and reproducible level of background DNA damage that was similar to the levels observed for the standard protocol for the respective laboratory. This demonstrates that the addition of the DNA repair inhibitor, APC, had no major impact on the background DNA damage in the RS Comet assay. In parallel, positive control values (BaP + APC) induced an increase above the BaP + APC reference range and an absolute increase in % tail DNA by ≥5% points above the SC + APC in all experiments performed in Phase 2, except one experiment in Lab C (Figure 3).
Fig. 3.
Historical control data of APC experiments. Percentage of tail DNA in the solvent control (SC) and positive control (PC, i.e., BaP) both with APC of individual experiments as obtained with the Phenion®FT during Phases 1 and 2 of coded testing are shown. The SC + APC values (circles) and BaP + APC values (diamonds) for the keratinocytes (dark blue symbols and lines) and fibroblasts (red symbols and lines) are given as mean ± SD (N = 3 samples each). Faint symbols indicate values obtained in Phase 1 and dark symbols indicate values from Phase 2 of the validation. The light blue-shaded and orange-shaded areas indicate the reference range (mean ± 2 SD) for the SC, i.e., historical control, for keratinocytes and fibroblasts, respectively. The reference ranges were derived from the control data of Phase 2. The y-axis is on the arcsine square-root transformed scale, but the tick labels are expressed in units of the percentage scale.
Assessment of intra-and inter-laboratory reproducibility
In order to assess intra- and inter-laboratory reproducibility, all data generated within the coded validation effort were tabulated and also included data calls from Phase 1 of the validation study published previously (14) and from Phase 2 of the present study (see Tables 2 and 3, supplementary Figures 1–24 and Table 2, available at Mutagenesis Online).
Table 2.
Reproducibility within one laboratory over time (within-laboratory concordance) in Phases 1–2
| Discordant | Concordant | Total | % | |
|---|---|---|---|---|
| Lab A | 0 | 8 | 8 | 100 |
| Lab B | 2 | 13 | 15 | 87 |
| Lab C | 0 | 8 | 8 | 100 |
| Lab D | 1 | 4 | 5 | 80 |
| Lab E | 0 | 4 | 4 | 100 |
| All labs | 3 | 37 | 40 | 93 |
Table 3.
Reproducibility between laboratories (between-laboratory concordance) in Phase 1
| Discordant | Concordant | Total | % |
|---|---|---|---|
| 1 | 7 | 8 | 88 |
The reproducibility of the RS Comet assay within a laboratory over time was assessed by comparing the outcome of individual experiments when more than one experiment was performed under identical conditions (without or with APC) in the same laboratory. Across all five laboratories, 44 double and triple experiments could be identified and counted towards assessing the concordance of classification (see Table 2, supplementary Table 2, available at Mutagenesis Online). The overall within-laboratory reproducibility for the validation exercise was 93% (Table 2), with values between 80 and 100% for the individual laboratories that participated in the validation.
Reproducibility between laboratories was calculated based on the final overall call (from each laboratory) for each chemical obtained for the ‘reproducibility’ phase of the validation which evaluated eight coded chemicals, each tested in three laboratories (14). Of these, six chemicals obtained 100% concordant calls and two led to partially discordant calls, as described by Reisinger et al. (14), leading to a between-laboratory reproducibility of 88% (see Table 3, supplementary Table 2, available at Mutagenesis Online). Both the within- and the between-laboratory reproducibility are found to be in a similar range to other in vitro genotoxicity assays when testing was done in a coded fashion and were therefore considered acceptable. For example, the intra-laboratory reproducibility of the in vitro micronucleus test was reported to vary between 83% and 100% (86) and the inter-laboratory reproducibility of the Ames assay is ~85% (87–89).
Predictive capacity of the RS Comet assay
The predictive capacity of the RS Comet assay was calculated using all available data from all phases of the validation exercise that were performed with coded chemicals. Where the call for a chemical unequivocally agreed with the expected classification, it was assigned a value of 1.0 when applied to the calculation, and if it unequivocally disagreed with the expected classification, that chemical was assigned a value of 0. Equivocal calls counted as 0.5. Discordant calls for one chemical among laboratories went in according to their weight, e.g. if a chemical was tested in three labs and two found the expected results and one gave an unexpected result it would be assigned a value of 0.66. Applying these principles reveals an overall sensitivity of the RS Comet assay of 77%, and an overall specificity of 88% (see Table 4). The respective overall accuracy is calculated to be 83%. These numbers reflect the calls of the performing laboratories in the coded phase which improved when expert calls were based on the earlier described evaluation criteria agreed by the Steering Committee and external experts. Taking these criteria into account, RS Comet assay showed a sensitivity of 80%, a specificity of 97%, and an overall accuracy of 92%. Importantly, when the RS Comet assay is used as intended, i.e. in combination with the RSMN assay, as an endpoint-specific follow-up to positive results from the standard in vitro testing battery, the sensitivity increases to about 90%, while the specificity remains high. A manuscript with a detailed analysis of the predictive capacity of these two assays in a ‘battery’ approach is in preparation.
Table 4.
Predictive capacity: (a) calculated based on the expert calls by the performing laboratories before decoding and (b) calculated based on the (later established) evaluation criteria agreed on by the Steering Committee and other external experts
| Parameter | Lab A | Lab B | Lab C | Lab D | Lab E | Overall (mean) |
|---|---|---|---|---|---|---|
| (a) | ||||||
| Sensitivity (%) | 80 | 80 | 100 | 25 | 100 | 77 |
| Specificity (%) | 86 | 87 | 100 | 100 | 100 | 88 |
| Accuracy (%) | 83 | 83 | 100 | 70 | 100 | 83 |
| (b) | ||||||
| Sensitivity (%) | 80 | 80 | 100 | 50 | 100 | 80 |
| Specificity (%) | 100 | 94 | 100 | 100 | 100 | 97 |
| Accuracy (%) | 92 | 86 | 100 | 80 | 100 | 92 |
Please note that for the representation of the assay predictivity via the two prediction models only one value was shown, since both models performed equally well (data not shown).
Requirements for measurements of cytotoxicity
Two different cytotoxicity measurements were used with the RS Comet assay to capture general signs of cellular damage, (i) during the entire treatment period, i.e. adenylate kinase (AK), which is released upon damage that leads to permeable cell membranes and accumulates in the culture medium and (ii) the intracellular ATP concentration, which is determined at harvest, representing the energy status of the cells 3 h after the last treatment. These measurements are not dependent on the cells undergoing proliferation, as cell division is not necessary to manifest the genotoxicity read-out (the types of DNA damage detected with comet assay), nor for monitoring cell cycle. Throughout the validation exercise AK was shown to be more reliable in terms reproducibility between experiments when testing the same compound and in terms of lower standard deviations when compared to ATP measurements; however, not all cytotoxic chemicals cause plasma membrane leakage. Therefore, for the current validation set, the recommendation introduced in the Standard Operating Procedure after Phase 1 of the validation, i.e. to use both measurement with each of the experiments, remained in place until the end of the validation exercise. For future studies, AK may be the preferred cytotoxicity measurement due to its reliability and only in cases in which AK leakage does not show a relevant increase in the DRF, the main experiment should still include the measurement of cellular ATP. After the validation exercise, the thresholds established for AK leakage and cellular ATP concentration to indicate strong cytotoxicity were re-evaluated by, e.g. introducing alterative cut-offs (i.e. 300% increase for AK instead of 200%, and/or a decrease of ATP to 33% instead of 50%). This approach, however, clearly decreased the predictivity of the RS Comet assay (data not shown) and it was, therefore, concluded that the original thresholds used during the validation exercise are suitable to adequately monitor toxicity in the low proliferating cells of full-thickness skin tissues.
Measurement of DNA damage in keratinocytes versus fibroblasts
In Phase 1 studies, in most experiments that were judged to be positive, both cell types exhibited an increase in % tail DNA. In the rare cases in which only one cell type showed genotoxic effects, results were confirmed in a subsequent experiment in which both cell types were positive. This suggested that testing in one cell type may be sufficient, which would reduce the overall workload of the assay but needed to be confirmed in Phase 2. Results of these phases showed that DNA migration increased in both cell types when tissues were treated with TP chemicals of different mode of actions: DNA reactive chemicals (EMS, MNNG), pro-mutagens with different requirements for phase 1 and 2 enzymes (2-AAF, BaP, CP) or chemicals which either interfere with the nucleotide pool of cells (AZT) or with topoisomerase (etoposide). Cyclopenta(c,d)pyrene was the only chemical that induced an increase in DNA migration in keratinocytes only. Laboratories which observe a dose-dependent increase in % tail DNA in keratinocytes or fibroblasts in a DRF experiment may want to proceed with the cell type that gave the positive result only for the main experiments. However, at present, a reliable negative call should be based on both cell types. We assume that extended use of the assay will help clarify this point.
Impact of the use of aphidicolin
As expected from the data generated in the RS Comet assay previously (14, 29), the introduction of APC into the protocol helped increase the sensitivity of the assay. Five ‘true positive’ chemicals were exclusively, or predominantly, detected in the presence of APC only: CCPE, BaP, 2-AAF, DMBA (in two out of three labs) and MMC (in two out of three laboratories). This clearly shows that the accumulation of strand breaks in the last hour before cell harvest is important to boost the overall sensitivity of the RS comet assay to detect in vivo genotoxic agents. As was speculated earlier (29), it seems especially critical in the context of pro-mutagens that require metabolic activation, a category that all of the aforementioned except MMC falls into. It is likely that the Phase I activation capacity of the skin models is not sufficient to produce enough DNA reactive metabolite(s) to cause an increase in strand breaks strong enough for a significant effect versus control levels. This assumption is also supported by the APC-induced accumulation of DNA adducts in 2-AAF treated skin models (48). While the sensitivity was greatly enhanced, the specificity of the RS comet assay was reduced slightly. Two expected negative compounds produced positive or equivocal results in the presence of APC only; cyclohexanone (in one out of two labs), and resorcinol which led to an equivocal result, as discussed in more detail in the Results section.
Strategic use of the RS Comet assay
The comet assay in general is considered an indicator test since the DNA damage detected could be repaired or may be lethal to the cell resulting in non-persistent effects, but strand breaks could also be fixed into mutations or chromosomal damage both resulting in heritable DNA damage of viable cells (20). The alkaline comet assay identifies double strand breaks, as well as single breaks, which may result from direct interaction of the test chemical with the DNA or which are related to incomplete excision repair and alkali labile sites. Consequently, the alkaline comet assay not only detects clastogenic DNA damage but also lesions which could be precursors of gene mutation events. The suitability of the comet assay to detect chemical mutagens was shown in an analysis of rodent carcinogens giving negative or equivocal results in the in vivo micronucleus test. The in vivo comet assay was positive for ~90% of these chemicals and was negative for nearly 80% of the non-carcinogens (20). This has been confirmed by Kirkland and colleagues, who recently published a comparison of in vivo mammalian alkaline comet assay data with those obtained with the transgenic rodent mutation (TGR) assay (90). The authors showed that ‘the comet assay can detect potent genotoxicants which would otherwise be detected using the TGR in liver and GI tract’. After further analysis of in vivo mammalian alkaline comet assay data, the authors concluded that the ‘Results from TGR and in vivo comet assays for 91 chemicals showed they have similar ability to detect in vivo genotoxicity per se with bacterial mutagens and Ames-positive carcinogens’ (91).
In line with these findings, the RS Comet assay is suggested as higher tier test method for dermally exposed substances to follow up positive Ames results. The results from Phase 2 testing and the final evaluation of all the data from Phases 1 and 2 provide confidence that the RS Comet assay can be used as a predictive tier 2 assay to follow up positive Ames results from the standard in vitro testing battery. In line with our conclusions, a SWOT analyses (Strengths, Weaknesses, Opportunities, Threads) published by the ILSI-HESI (37) noted that ‘3-D constructs have the potential to serve as follow-up assays based on results in the traditional 2-D tests, especially when 2-D models cannot be used’. Likewise, experts at two International Working Group on Genetic Toxicology (IWGT) Working Group meetings in 2009 (40) and 2017 (92) accepted the proposed endpoint-triggered follow-up strategy and concluded that the combination of both assays, the RS Comet assay, as well as the RSMN assay, resulted in a ‘highly predictive’ testing of genotoxicity. Moreover, the IWGT 2017 Working Group believed that both assays are sufficiently validated to move towards the development of individual OECD test guidelines. Two separate genotoxicity OECD TGs based on 3D skin models have also been proposed as a result of these validation studies and OECD accepted both assays into their test guideline development program in April of 2019. Compared with the TG based on testing in rodents, these proposed TGs are of increased human relevance as they use reconstituted human skin models with metabolic capacity similar to human skin (41,42,46,93).
The SCCS, the independent expert panel mandated by the European Commission to ensure safe use of consumer products, foresees a role for the RS genotoxicity assays since 2014 (5). The committee has recently updated their Notes of Guidance to include a toolbox of ‘tier 2’ assays to follow up on unfavorable results from the standard in vitro testing battery in a weight-of-evidence approach. This ‘tier 2’ toolbox includes the RS-based assays in the context of a dermal exposure scenario (94). In fact, the SCCS has already applied data from the RS Comet assay in support of safe use of a hair dye, Basic Brown 17 (95). Initial experiments with this dye in standard genotoxicity assays provided positive findings with the Ames test but negative findings in the standard in vitro micronucleus test with human lymphocytes. Due to the ban of animal studies in Europe for testing of cosmetic ingredients, in vivo data could not be generated to address the positive findings for gene mutation in the Ames test. Therefore, the assessment was exclusively based on in vitro data, including the RS Comet assay which showed a lack of DNA damaging properties after application of the dye to the skin. The SCCS accepted these data in a weight-of-evidence approach as evidence that it does not have a genotoxic potential for the given dermal exposure scenario (95). Meanwhile, the toxicological safety assessments of two additional hair dye ingredients have successfully been supported by negative findings of the RS Comet assay using the Phenion® FT skin equivalents (96,97).
Conclusions
The dataset for the validation of the RS Comet assay is complete.
The intra-laboratory reproducibility on the experimental level was very good, as was the inter-laboratory reproducibility, which was based on the final call.
The Phenion® Full-Thickness Skin model was shown to be suitable for the use in the RS Comet assay, with reproducibly low background DNA damage and sufficient metabolic capacity to metabolise pro-mutagens.
The capacity of the assay to predict expected genotoxic effects of a difficult set of coded chemicals was very good, providing a sensitivity of 77 (80)% and a specificity of 88 (97)%. The overall accuracy was calculated to be 83 (92)%. The numbers reflect the expert calls in the coded phase, while those in parenthesis reflect the calls in accordance to the agreed evaluation criteria.
The RS Comet assay is considered sufficiently validated for use as a tier 2 assay for dermally exposed compounds and was accepted into the OECD’s test guideline development program.
The RS Comet assay has gained scientific acceptance from expert panels and from the SCCS, who now use data from this assay as a follow-up assay to help address positive findings from the Ames test.
The RS Comet assay is applicable as a follow-up tool for positive results from standard in vitro genotoxicity assays when the expected route of exposure is dermal, as is the case, e.g. for the majority of cosmetics.
Supplementary Material
Acknowledgements
The presented work is the result of a broad joint research project. Due to limited space, not all colleagues that contributed to this study could be added to the author list and their excellent contributions are therefore acknowledged here. These are for the BfR Joep Brinkmann, Manfred Liebsch, Frank Henkler, Stefanie Jeschke and Nils Dommershausen, for P&G/ATL Ryan Posgai, Andreas Zeller (now F. Hoffmann-La Roche), Brenda Barnett, Demetria Thomas and Amanda Isom, as well as for Henkel Jürgen Kreutz, Bita Tarazouei-Bahrami and Lars Vierkotten. Julian Tharmann (BfR) supported the ordering, coding and shipment of chemicals. The Cosmetics Europe Genotoxicity Task Force has provided support to this project all along the way.
A special thank goes to Raffaella Corvi (EURL-ECVAM) and David Kirkland (Kirkland Consulting) for selecting the test chemicals and for in-depth discussion of data and processes and their important contributions to the steering of the project. The authors finally like to thank all other experts that have contributed to the project over the years: Guenter Speit, Rodger Curren, Tom Slaga, Peter Kasper, Jan van Benthem, Ralph Pirow, Johannes Doehmer, Sebastian Hoffmann, Jochen Kühnl.
The work was financially supported by the Federal Ministry of Education and Research (German: Bundesministerium für Bildung und Forschung) (grant Nos. 0315226, 0316008) and by Cosmetics Europe – The Personal Care Association.
Conflict of interest statement: Kerstin Reisinger is an employee of the company which developed and commercializes the Phenion® Full-Thickness Skin Tissue. None of the other authors report a conflict of interest.
References
- 1. OECD (1997) OECD guideline for testing of chemicals. Bacterial reverse mutation test. OECD TG 471, Adopted 21 July 1997. Retrieved from https://www.oecd.org/chemicalsafety/risk-assessment/1948418.pdf.
- 2. OECD (2016) Test No. 474: Mammalian Erythrocyte Micronucleus Test, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. Retrieved from doi: 10.1787/9789264264762-en. [DOI] [Google Scholar]
- 3. EU (2006) Regulation EC No. 1907/2006 OF The European Parliament and of the Councilof concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC. Retrieved from https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2007:136:0003:0280:en:PDF.
- 4. ICH (2011) ICH Guideline, Guidance on Genotoxicity Testing and Data Interpretation for Pharmaceuticals Intended for Human Use s2(r1), Approval by the Steering Committee of S2(R1) under Step 4 and recommendation for adoption to the three ICH regulatory bodies (9 November 2011). Retrieved from http://www.fda.gov/downloads/Drugs/Guidances/ucm074931.pdf.
- 5. SCCS (2014) Addendum to the SCCS’s Notes of Guidance (NoG) for the Testing of Cosmetic Ingredients and Their Safety Evaluation 8th Revision, SCCS/1501/12. Retrieved from: http://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_156.pdf.
- 6. EU (2009) European Union 2009. Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on cosmetic products (recast). Official Journal L342, 22.12.2009. Retrieved from https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=uriserv:OJ.L_.2009.342.01.0059.01.ENG. pp. 59.
- 7. OECD (2011) Test No. 488: Transgenic Rodent Somatic and Germ Cell Gene Mutation Assays. OECD Publishing, Paris, France. Retrieved from doi:10.1787/9789264122819-en. [Google Scholar]
- 8. OECD (2016) Test No. 489: In Vivo Mammalian Alkaline Comet Assay, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. Retrieved from doi: 10.1787/9789264264885-en. [DOI] [Google Scholar]
- 9. OECD (2015) Test No. 430: In Vitro Skin Corrosion: Transcutaneous Electrical Resistance Test Method (TER), OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. Retrieved from doi: 10.1787/9789264242739-en. [DOI] [Google Scholar]
- 10. OECD (2015) Test No. 435: In Vitro Membrane Barrier Test Method for Skin Corrosion, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. Retrieved from doi: 10.1787/9789264242791-en. [DOI] [Google Scholar]
- 11. OECD (2019) Test No. 439: In Vitro Skin Irritation: Reconstructed Human Epidermis Test Method, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. Retrieved from doi: 10.1787/9789264242845-en. [DOI] [Google Scholar]
- 12. OECD (2019) Test No. 431: In Vitro Skin Corrosion: Reconstructed Human Epidermis (RHE) Test Method, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. Retrieved from doi: 10.1787/9789264264618-en. [DOI] [Google Scholar]
- 13. Reus, A. A., Reisinger, K., Downs, T. R., Carr, G. J., Zeller, A., Corvi, R., Krul, C. A. and Pfuhler, S. (2013) Comet assay in reconstructed 3D human epidermal skin models–investigation of intra- and inter-laboratory reproducibility with coded chemicals. Mutagenesis, 28, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reisinger, K., Blatz, V., Brinkmann, J., et al. (2018) Validation of the 3D Skin Comet assay using full thickness skin models: Transferability and reproducibility. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 827, 27–41. [DOI] [PubMed] [Google Scholar]
- 15. Curren, R. D., Mun, G. C., Gibson, D. P. and Aardema, M. J. (2006) Development of a method for assessing micronucleus induction in a 3D human skin model (EpiDerm). Mutat. Res., 607, 192–204. [DOI] [PubMed] [Google Scholar]
- 16. Dahl, E. L., Curren, R., Barnett, B. C., et al. (2011) The reconstructed skin micronucleus assay (RSMN) in EpiDerm™: detailed protocol and harmonized scoring atlas. Mutat. Res., 720, 42–52. [DOI] [PubMed] [Google Scholar]
- 17. Pfuhler, S., Kirst, A., Aardema, M., et al. (2010) A tiered approach to the use of alternatives to animal testing for the safety assessment of cosmetics: genotoxicity. A COLIPA analysis. Regul. Toxicol. Pharmacol., 57, 315–324. [DOI] [PubMed] [Google Scholar]
- 18. Pfuhler, S., Fautz, R., Ouedraogo, G., et al. (2014) The Cosmetics Europe strategy for animal-free genotoxicity testing: project status up-date. Toxicol. In Vitro, 28, 18–23. [DOI] [PubMed] [Google Scholar]
- 19. Pfuhler, S., Barnett, B. C., Downs, T. R., Hewitt, N. J., Hoffman, S., Mun, G. C., Roy, S., Curren, R. D., Aardema, M. J. (2019) Validation of the 3D reconstructed human skin micronucleus (RSMN) assay, an animal-free alternative for following-up positive results from standard in vitro genotoxicity assays Mutagenesis, in this issue. [DOI] [PMC free article] [PubMed]
- 20. Hartmann, A., Agurell, E., Beevers, C., et al. ; 4th International Comet Assay Workshop . (2003) Recommendations for conducting the in vivo alkaline Comet assay. 4th International Comet Assay Workshop. Mutagenesis, 18, 45–51. [DOI] [PubMed] [Google Scholar]
- 21. Kirkland, D. and Speit, G. (2008) Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens III. Appropriate follow-up testing in vivo. Mutat. Res., 654, 114–132. [DOI] [PubMed] [Google Scholar]
- 22. Kirkland, D., Levy, D. D., LeBaron, M. J., et al. (2019) A comparison of transgenic rodent mutation and in vivo comet assay responses for 91 chemicals. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 839, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tzung, T. Y. and Rünger, T. M. (1998) Assessment of DNA damage induced by broadband and narrowband UVB in cultured lymphoblasts and keratinocytes using the comet assay. Photochem. Photobiol., 67, 647–650. [PubMed] [Google Scholar]
- 24. Zeller, A. and Pfuhler, S. (2014) N-acetylation of three aromatic amine hair dye precursor molecules eliminates their genotoxic potential. Mutagenesis, 29, 37–48. [DOI] [PubMed] [Google Scholar]
- 25. Wischermann, K., Popp, S., Moshir, S., et al. (2008) UVA radiation causes DNA strand breaks, chromosomal aberrations and tumorigenic transformation in HaCaT skin keratinocytes. Oncogene, 27, 4269–4280. [DOI] [PubMed] [Google Scholar]
- 26. Flamand, N., Marrot, L., Belaidi, J. P., Bourouf, L., Dourille, E., Feltes, M. and Meunier, J. R. (2006) Development of genotoxicity test procedures with Episkin, a reconstructed human skin model: towards new tools for in vitro risk assessment of dermally applied compounds? Mutat. Res., 606, 39–51. [DOI] [PubMed] [Google Scholar]
- 27. Marrot, L., Belaïdi, J. P., Lejeune, F., Meunier, J. R., Asselineau, D. and Bernerd, F. (2004) Photostability of sunscreen products influences the efficiency of protection with regard to UV-induced genotoxic or photoageing-related endpoints. Br. J. Dermatol., 151, 1234–1244. [DOI] [PubMed] [Google Scholar]
- 28. Reus, A. A., Usta, M. and Krul, C. A. (2012) The use of ex vivo human skin tissue for genotoxicity testing. Toxicol. Appl. Pharmacol., 261, 154–163. [DOI] [PubMed] [Google Scholar]
- 29. Brinkmann, J., Stolpmann, K., Trappe, S., et al. (2013) Metabolically competent human skin models: activation and genotoxicity of benzo[a]pyrene. Toxicol. Sci., 131, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Speit, G., Schütz, P. and Hoffmann, H. (2004) Enhancement of genotoxic effects in the comet assay with human blood samples by aphidicolin. Toxicol. Lett., 153, 303–310. [DOI] [PubMed] [Google Scholar]
- 31. Vande Loock, K., Decordier, I., Ciardelli, R., Haumont, D. and Kirsch-Volders, M. (2010) An aphidicolin-block nucleotide excision repair assay measuring DNA incision and repair capacity. Mutagenesis, 25, 25–32. [DOI] [PubMed] [Google Scholar]
- 32. Odongo, G. A., Skatchkov, I., Herz, C. and Lamy, E. (2019) Optimization of the alkaline comet assay for easy repair capacity quantification of oxidative DNA damage in PBMC from human volunteers using aphidicolin block. DNA Repair (Amst)., 77, 58–64. [DOI] [PubMed] [Google Scholar]
- 33. Speit, G., Leibiger, C., Kuehner, S. and Högel, J. (2013) Further investigations on the modified comet assay for measuring aphidicolin-block nucleotide excision repair. Mutagenesis, 28, 145–151. [DOI] [PubMed] [Google Scholar]
- 34. Hartung, T., Bremer, S., Casati, S., et al. (2004) A modular approach to the ECVAM principles on test validity. Altern. Lab. Anim., 32, 467–472. [DOI] [PubMed] [Google Scholar]
- 35. Kubilus, J., Hayden, P. J., Ayehunie, S., Lamore, S. D., Servattalab, C., Bellavance, K. L., Sheasgreen, J. E. and Klausner, M. (2004) Full Thickness EpiDerm: a dermal-epidermal skin model to study epithelial-mesenchymal interactions. Altern. Lab. Anim., 32 (Suppl 1A), 75–82. [DOI] [PubMed] [Google Scholar]
- 36. Mewes, K. R., Raus, M., Bernd, A., Zöller, N. N., Sättler, A. and Graf, R. (2007) Elastin expression in a newly developed full-thickness skin equivalent. Skin Pharmacol. Physiol., 20, 85–95. [DOI] [PubMed] [Google Scholar]
- 37. Zeiger, E., Gollapudi, B., Aardema, M. J., et al. (2015) Opportunities to integrate new approaches in genetic toxicology: an ILSI-HESI workshop report. Environ. Mol. Mutagen., 56, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Balansin Rigon, R., Kaessmeyer, S., Wolff, C., et al. (2018) Ultrastructural and molecular analysis of ribose-induced glycated reconstructed human skin. Int. J. Mol. Sci, 19, 3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hausmann, C., Zoschke, C., Wolff, C., et al. (2019) Fibroblast origin shapes tissue homeostasis, epidermal differentiation, and drug uptake. Sci. Rep., 9, 2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pfuhler, S., Fellows, M., van Benthem, J., et al. (2011) In vitro genotoxicity test approaches with better predictivity: summary of an IWGT workshop. Mutat. Res., 723, 101–107. [DOI] [PubMed] [Google Scholar]
- 41. Wiegand, C., Hewitt, N. J., Merk, H. F. and Reisinger, K. (2014) Dermal xenobiotic metabolism: a comparison between native human skin, four in vitro skin test systems and a liver system. Skin Pharmacol. Physiol., 27, 263–275. [DOI] [PubMed] [Google Scholar]
- 42. Hewitt, N. J., Edwards, R. J., Fritsche, E., et al. (2013) Use of human in vitro skin models for accurate and ethical risk assessment: metabolic considerations. Toxicol. Sci., 133, 209–217. [DOI] [PubMed] [Google Scholar]
- 43. Jäckh, C., Blatz, V., Fabian, E., Guth, K., van Ravenzwaay, B., Reisinger, K. and Landsiedel, R. (2011) Characterization of enzyme activities of Cytochrome P450 enzymes, Flavin-dependent monooxygenases, N-acetyltransferases and UDP-glucuronyltransferases in human reconstructed epidermis and full-thickness skin models. Toxicol. In Vitro, 25, 1209–1214. [DOI] [PubMed] [Google Scholar]
- 44. Hu, T., Khambatta, Z. S., Hayden, P. J., et al. (2010) Xenobiotic metabolism gene expression in the EpiDermin vitro 3D human epidermis model compared to human skin. Toxicol. In Vitro, 24, 1450–1463. [DOI] [PubMed] [Google Scholar]
- 45. Bätz, F. M., Klipper, W., Korting, H. C., Henkler, F., Landsiedel, R., Luch, A., von Fritschen, U., Weindl, G. and Schäfer-Korting, M. (2013) Esterase activity in excised and reconstructed human skin–biotransformation of prednicarbate and the model dye fluorescein diacetate. Eur. J. Pharm. Biopharm., 84, 374–385. [DOI] [PubMed] [Google Scholar]
- 46. Grohmann, L., Becker, D., Rademann, J., Ma, N., Schäfer-Korting, M. and Weindl, G. (2017) Biotransformation of 2,4-toluenediamine in human skin and reconstructed tissues. Arch. Toxicol., 91, 3307–3316. [DOI] [PubMed] [Google Scholar]
- 47. Götz, C., Hewitt, N. J., Jermann, E., Tigges, J., Kohne, Z., Hübenthal, U., Krutmann, J., Merk, H. F. and Fritsche, E. (2012) Effects of the genotoxic compounds, benzo[a]pyrene and cyclophosphamide on phase 1 and 2 activities in EpiDerm™ models. Xenobiotica., 42, 526–537. [DOI] [PubMed] [Google Scholar]
- 48. Downs, T. R., Arlt, V. M., Barnett, B. C., Posgai, R., and Pfuhler, S. (2019) Effect of 2-acetylaminofluorene and its genotoxic metabolites on DNA adduct formation and DNA damage in 3D reconstructed human skin tissue models. Mutagenesis [Epub ahead of print], doi: 10.1093/mutage/gez044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lovell, D. P. and Omori, T. (2008) Statistical issues in the use of the comet assay. Mutagenesis, 23, 171–182. [DOI] [PubMed] [Google Scholar]
- 50. Kangas, L., Grönroos, M. and Nieminen, A. L. (1984) Bioluminescence of cellular ATP: a new method for evaluating cytotoxic agents in vitro. Med. Biol., 62, 338–343. [PubMed] [Google Scholar]
- 51. Olsson, T., Gulliksson, H., Palmeborn, M., Bergström, K. and Thore, A. (1983) Leakage of adenylate kinase from stored blood cells. J. Appl. Biochem., 5, 437–445. [PubMed] [Google Scholar]
- 52. Rudo, K., Meyers, W. C., Dauterman, W. and Langenbach, R. (1987) Comparison of human and rat hepatocyte metabolism and mutagenic activation of 2-acetylaminofluorene. Cancer Res., 47, 5861–5867. [PubMed] [Google Scholar]
- 53. Rodrigues, A. S., Silva, I. D., Caria, M. H., Laires, A., Chaveca, T., Glatt, H. R. and Rueff, J. (1994) Genotoxicity assessment of aromatic amines and amides in genetically engineered V79 cells. Mutat. Res., 341, 93–100. [DOI] [PubMed] [Google Scholar]
- 54. Aryal, P., Terashita, T., Guengerich, F. P., Shimada, T. and Oda, Y. (2000) Use of genetically engineered Salmonella typhimurium OY1002/1A2 strain coexpressing human cytochrome P450 1A2 and NADPH-cytochrome P450 reductase and bacterial O-acetyltransferase in SOS/umu assay. Environ. Mol. Mutagen., 36, 121–126. [DOI] [PubMed] [Google Scholar]
- 55. Probst, M. R., Blum, M., Fasshauer, I., D’Orazio, D., Meyer, U. A. and Wild, D. (1992) The role of the human acetylation polymorphism in the metabolic activation of the food carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline (IQ). Carcinogenesis, 13, 1713–1717. [DOI] [PubMed] [Google Scholar]
- 56. Sadrieh, N., Davis, C. D. and Snyderwine, E. G. (1996) N-acetyltransferase expression and metabolic activation of the food-derived heterocyclic amines in the human mammary gland. Cancer Res., 56, 2683–2687. [PubMed] [Google Scholar]
- 57. Muckel, E., Frandsen, H. and Glatt, H. R. (2002) Heterologous expression of human N-acetyltransferases 1 and 2 and sulfotransferase 1A1 in Salmonella typhimurium for mutagenicity testing of heterocyclic amines. Food Chem. Toxicol., 40, 1063–1068. [DOI] [PubMed] [Google Scholar]
- 58. Géniès, C., Jamin, E. L., Debrauwer, L., et al. (2019) Comparison of the metabolism of 10 chemicals in human and pig skin explants. J. Appl. Toxicol., 39, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Snyderwine, E. G., Nouso, K. and Schut, H. A. (1993) Effect of 3-methylcholanthrene induction on the distribution and DNA adduction of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) in F344 rats. Food Chem. Toxicol., 31, 415–423. [DOI] [PubMed] [Google Scholar]
- 60. Davis, C. D., Schut, H. A. and Snyderwine, E. G. (1993) Enzymatic phase II activation of the N-hydroxylamines of IQ, MeIQx and PhIP by various organs of monkeys and rats. Carcinogenesis, 14, 2091–2096. [DOI] [PubMed] [Google Scholar]
- 61. Harrigan, J. A., McGarrigle, B. P., Sutter, T. R. and Olson, J. R. (2006) Tissue specific induction of cytochrome P450 (CYP) 1A1 and 1B1 in rat liver and lung following in vitro (tissue slice) and in vivo exposure to benzo(a)pyrene. Toxicol. In Vitro, 20, 426–438. [DOI] [PubMed] [Google Scholar]
- 62. van Eijl, S., Zhu, Z., Cupitt, J., Gierula, M., Götz, C., Fritsche, E. and Edwards, R. J. (2012) Elucidation of xenobiotic metabolism pathways in human skin and human skin models by proteomic profiling. PLoS One, 7, e41721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. NTP (1998) National toxicology program comparative toxicity studies of o-, m-, and p-Chloroanilines (CAS Nos. 95-51-2; 108-42-9; and 106-47-8) administered by Gavage to F344/N Rats and B6C3F1 Mice. Toxic Rep Ser, 43, 1–f20. [PubMed] [Google Scholar]
- 64. Barfield, W. and Burlinson, B. (2015) p-Chloroaniline, t-butylhydroquinone, and methyl carbamate: Rat in vivo comet test, JaCVAM trial phase 4.2. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 786-788, 98–103. [DOI] [PubMed] [Google Scholar]
- 65. IARC (1993) para-Chloroaniline. IARC Working Group on the Evaluation of Carcinogenic Risk to Humans. Occupational Exposures of Hairdressers and Barbers and Personal Use of Hair Colourants; Some Hair Dyes, Cosmetic Colourants, Industrial Dyestuffs and Aromatic Amines, https://monographs.iarc.fr/wp-content/uploads/2018/06/mono57-21.pdf, pp. 305–321.
- 66. Kwon, H., Sahali, Y., Skipper, P. L. and Tannenbaum, S. R. (1992) Oxidation of cyclopenta[cd]pyrene by human and mouse liver microsomes and selected cytochrome P450 enzymes. Chem. Res. Toxicol., 5, 760–764. [DOI] [PubMed] [Google Scholar]
- 67. Ashby, J. and Beije, B. (1985) Concomitant observations of UDS in the liver and micronuclei in the bone marrow of rats exposed to cyclophosphamide or 2-acetylaminofluorene. Mutat. Res., 150, 383–392. [DOI] [PubMed] [Google Scholar]
- 68. Doherty, A. T., Hayes, J., Holme, P. and O’Donovan, M. (2012) Chromosome aberration frequency in rat peripheral lymphocytes increases with repeated dosing with hexamethylphosphoramide or cyclophosphamide. Mutagenesis, 27, 533–539. [DOI] [PubMed] [Google Scholar]
- 69. Gervot, L., Rochat, B., Gautier, J. C., Bohnenstengel, F., Kroemer, H., de Berardinis, V., Martin, H., Beaune, P. and de Waziers, I. (1999) Human CYP2B6: expression, inducibility and catalytic activities. Pharmacogenetics, 9, 295–306. [PubMed] [Google Scholar]
- 70. Dearfield, K. L., Jacobson-Kram, D., Huber, B. E. and Williams, J. R. (1986) Induction of sister chromatid exchanges in human and rat hepatoma cell lines by cyclophosphamide and phosphoramide mustard and the effects of cytochrome P-450 inhibitors. Biochem. Pharmacol., 35, 2199–2205. [DOI] [PubMed] [Google Scholar]
- 71. Goldberg, M. T. and Josephy, P. D. (1987) Studies on the mechanism of action of diallyl sulfide, an inhibitor of the genotoxic effects of cyclophosphamide. Can. J. Physiol. Pharmacol., 65, 467–471. [DOI] [PubMed] [Google Scholar]
- 72. NTP (1978) Bioassay of N,N’-Dicyclohexylthiourea for Possible Carcinogenicity, [web site], National Toxicology Program [web site], https://ntp.niehs.nih.gov/ntp/htdocs/lt_rpts/tr056.pdf (accessed July 1, 2019).
- 73. Henderson, L., Wolfreys, A., Fedyk, J., Bourner, C. and Windebank, S. (1998) The ability of the Comet assay to discriminate between genotoxins and cytotoxins. Mutagenesis, 13, 89–94. [DOI] [PubMed] [Google Scholar]
- 74. Kasai, H., Iwamoto-Tanaka, N. and Fukada, S. (1998) DNA modifications by the mutagen glyoxal: adduction to G and C, deamination of C and GC and GA cross-linking. Carcinogenesis, 19, 1459–1465. [DOI] [PubMed] [Google Scholar]
- 75. Kuchenmeister, F., Schmezer, P. and Engelhardt, G. (1998) Genotoxic bifunctional aldehydes produce specific images in the comet assay. Mutat. Res., 419, 69–78. [DOI] [PubMed] [Google Scholar]
- 76. Vilanova, B., Fernández, D., Casasnovas, R., et al. (2017) Formation mechanism of glyoxal-DNA adduct, a DNA cross-link precursor. Int. J. Biol. Macromol., 98, 664–675. [DOI] [PubMed] [Google Scholar]
- 77. Pfuhler, S. and Wolf, H. U. (1996) Detection of DNA-crosslinking agents with the alkaline comet assay. Environ. Mol. Mutagen., 27, 196–201. [DOI] [PubMed] [Google Scholar]
- 78. Merk, O. and Speit, G. (1999) Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ. Mol. Mutagen., 33, 167–172. [DOI] [PubMed] [Google Scholar]
- 79. Nishi, Y., Miyakawa, Y. and Kato, K. (1989) Chromosome aberrations induced by pyrolysates of carbohydrates in Chinese hamster V79 cells. Mutat. Res., 227, 117–123. [DOI] [PubMed] [Google Scholar]
- 80. SCCP (2005) Scientific Committee on Consumer Products, SCCP Opinion on Glyoxal, Adopted on 21 June 2005, SCCP/0881/05. Retrieved from: https://ec.europa.eu/health/ph_risk/committees/04_sccp/docs/sccp_o_023.pdf.
- 81. MAK (1999) Glyoxal [MAK Value Documentation in German language]. The MAK‐Collection for Occupational Health and Safety. Documents and methods. ISBN 9783527600410. Wiley-VCH Verlag GmbH & Co. KGaA, pp. 1–5. [Google Scholar]
- 82. NTP Testing Status of Resorcinol 10163-Y. National Toxicology Program [web site], https://ntp.niehs.nih.gov/testing/status/agents/ts-10163-y.html (accessed 1 July 2019).
- 83. EFSA (2010) Panel on food additives: scientific opinion on the use of resorcinol as a food additive. EFSA Journal, 8, 1411. [Google Scholar]
- 84. SCCS (2010) Scientific Committee on Consumer Safety Opinion on Resorcinol, SCCS/1270/09. The SCCS adopted this opinion at its 6th plenary meeting of 23 March 2010 [web site], http://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_015.pdf (accessed July 1, 2019).
- 85. NTP (1998) National toxicology program toxicology and carcinogenesis studies of technical grade sodium xylenesulfonate (CAS No. 1300-72-7) in F344/N rats and B6C3F1 mice (Dermal Studies). Natl. Toxicol. Program. Tech. Rep. Ser., 464, 1–272. [PubMed] [Google Scholar]
- 86. Corvi, R., Albertini, S., Hartung, T., Hoffmann, S., Maurici, D., Pfuhler, S., van Benthem, J. and Vanparys, P. (2008) ECVAM retrospective validation of in vitro micronucleus test (MNT). Mutagenesis, 23, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. McCann, J., Horn, L. and Kaldor, J. (1984) An evaluation of Salmonella (Ames) test data in the published literature: application of statistical procedures and analysis of mutagenic potency. Mutat. Res., 134, 1–47. [DOI] [PubMed] [Google Scholar]
- 88. Kamber, M., Flückiger-Isler, S., Engelhardt, G., Jaeckh, R. and Zeiger, E. (2009) Comparison of the Ames II and traditional Ames test responses with respect to mutagenicity, strain specificities, need for metabolism and correlation with rodent carcinogenicity. Mutagenesis, 24, 359–366. [DOI] [PubMed] [Google Scholar]
- 89. Piegorsch, W. W., and Zeiger, E. (1991) Measuring intra-assay agreement for the Ames Salmonella assay. In Hothorn, L. (ed.), Statistical Methods in Toxicology. Springer, Berlin Germany, pp. 35–41. [Google Scholar]
- 90. Kirkland, D., Levy, D. D., LeBaron, M. J., et al. (2019) A comparison of transgenic rodent mutation and in vivo comet assay responses for 91 chemicals. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 839, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kirkland, D., Uno, Y., Luijten, M., et al. (2019) In vivo genotoxicity testing strategies: report from the 7th International workshop on genotoxicity testing (IWGT). Mutat. Res., 847, 403035. [DOI] [PubMed] [Google Scholar]
- 92. Pfuhler, S., van Benthem, J., Curren, R., et al. (2019) Use of 3D Tissues in Genotoxicity testing: strategic fit, validation status and way forward report of the IWGT working group. Report from the 7th International workshop on genotoxicity testing (IWGT). Mutat. Res./Genetic Toxicol. Environ. Mutag, 850-851, 503135. [DOI] [PubMed] [Google Scholar]
- 93. Oesch, F., Fabian, E. and Landsiedel, R. (2018) Xenobiotica-metabolizing enzymes in the skin of rat, mouse, pig, guinea pig, man, and in human skin models. Arch. Toxicol., 92, 2411–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. SCCS (2018) SCCS’s Notes of Guidance (NoG) for the Testing of Cosmetic Ingredients and Their Safety Evaluation 10th Revision, SCCS/1602/18. Retrieved from https://ec.europa.eu/health/sites/health/files/scientific_committees/consumer_safety/docs/sccs_o_224.pdf (accessed July 1, 2019).
- 95. SCCS (2014) Scientific Committee on Consumer Safety Opinion on Basic Brown 17, COLIPA n° B007, revision of 18 June 2014, SCCS/1531/14. Retrieved from: https://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_152.pdf (accessed July 1, 2019).
- 96. SCCS (2016) Scientific Committee on Consumer Safety. Opinion on N,N’-Bis-(2-hydroxyethyl)-2-nitro-p-phenylenediamine (B34). Adopted on 16 September 2016, SCCS/1572/16. Retrieved from: https://ec.europa.eu/health/sites/health/files/scientific_committees/consumer_safety/docs/sccs_o_196.pdf (accessed July 1, 2019).
- 97. SCCS (2015) Scientific Committee on Consumer Safety. Opinion on 2,6-Dihydroxyethylaminotoluene COLIPA no A138. Adopted on 25 June 2015., SCCS/1563/15. Retrieved from: http://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_188.pdf (accessed July 1, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.