Abstract
Wastewater based epidemiology is a potential early warning tool for the detection of COVID-19 outbreak. Sewage surveillance for SARS-CoV-2 RNA was introduced in Hungary after the successful containment of the first wave of the pandemic to forecast the resurge of infections. Three wastewater treatment plants servicing the entire population (1.8 million) of the capital, Budapest were sampled weekly. 24 h composite (n = 44) and grab samples (n = 21) were concentrated by an in-house flat sheet membrane ultrafiltration method. The efficiency and reproducibility of the method was comparable to those previously published. SARS-CoV-2 RNA was quantified using RT-qPCR of the N gene. The first positive signal in sewage was detected 2 weeks before the rise in case numbers. Viral concentration and volume-adjusted viral load correlated to the weekly new cases from the same week and the rolling 7-day average of active cases in the subsequent week. The correlation was more pronounced in the ascending phase of the outbreak, data was divergent once case numbers plateaued. Wastewater surveillance was found to be effective in predicting the second wave of the outbreak in Hungary. Data indicated that even relatively low frequency (weekly) sampling is useful and at the same time, cost effective tool in outbreak detection.
Keywords: COVID-19, SARS-CoV-2, Wastewater-based epidemiology, Environmental surveillance, Early warning
Graphical abstract
1. Introduction
COVID-19 pandemic was announced by WHO on March 11th, 2020 (WHO, 2020a). It was at that time already present in 55 countries and spread to all continents since. Most European countries exercised severe control measures, including complete lockdown in the spring (March–May), successfully limiting the rate of transmission (ECDC, 2020a). The second, larger wave of the outbreak started in late July or early August in European countries (ECDC, 2020b). RT-PCR analysis of nasopharyngeal swab samples is commonly used to detect new cases (ECDC, 2020c). However, the efficiency of surveillance based on clinical testing depends on the local testing strategy, the availability and accessibility of clinical tests, and the turnover time of test results (WHO, 2020b). SARS-CoV-2 was shown to be shed in nasal fluids and other respiratory secretions, faeces and (rarely) in urine by both symptomatic and asymptomatic individuals (Walsh et al., 2020). According to the meta-analysis by Walsh et al. (2020), shedding may start before the clinical symptoms, and can extend to several weeks, especially in faeces. Respiratory secretions and human excreta are collected in sewage, offering a possibility to obtain information on community transmission through wastewater sampling and analysis (Ahmed et al., 2020; Gonçalves et al., 2021; Kocamemi et al., 2020; Sherchan et al., 2020).
Wastewater based epidemiology (WBE) was suggested early on in the COVID-19 pandemic as a potential tool for community screening and trend analysis (Medema et al., 2020b). Several countries around the world reported presence/absence or quantitative data for SARS-CoV-2 RNA in sewage (e.g. Ahmed et al., 2020; Haramoto et al., 2020; La Rosa et al., 2020; Lodder and de Roda Husman, 2020; Randazzo et al., 2020; Wurtzer et al., 2020). Epidemiological studies confirmed the correlation of viral titres detected in raw sewage or primary sewage sludge and clinical data with a delay of 4–10 days (Wu et al., 2020; Peccia et al., 2020). Correlation to new cases, cumulated number of active cases, hospital admissions and deaths were investigated (Larsen and Wigginton, 2020; Medema et al., 2020c; Nemudryi et al., 2020). Apparently, the strength of WBE lies in applications where relative trend of viral titres are sufficient, such as early warning, rather than direct estimation of infected individuals (Medema et al., 2020a). Faecal shedding varies widely (between 103 and 107 genome copies (GC)/g faeces) in association with the stage of infection, starting before the symptoms appear and extending well into the post-symptomatic phase (Foladori et al., 2020). Studies failed to identify significant difference in the viral load from symptomatic and asymptomatic cases (Walsh et al., 2020).
Concentration is a key step in the detection of SARS-CoV-2 RNA in sewage. Ultrafiltration by 10–100 kDa molecular weight cut-off filters (Kocamemi et al., 2020, Medema et al., 2020b, Nemudryi et al., 2020) and flocculation with PEG or other chemicals (Randazzo et al., 2020; Wu et al., 2020) are the most frequently used methods, but filtration on electronegative membranes (Ahmed et al., 2020) and ultracentrifugation (Wurtzer et al., 2020) were also suggested. The efficiency of RNA extraction is less likely to be affected by the choice of the method. A number of commercial RNA extraction kits were applied by different laboratories (Ahmed et al., 2020, Kocamemi et al., 2020, La Rosa et al., 2020, Medema et al., 2020b, Trottier et al., 2020, Wu et al., 2020, etc.). Detection currently unanimously relies on quantitative RT-PCR, usually using the same protocols (approved by CDC and WHO) as for clinical samples. The most frequently used targets of the RT-PCR are N, ORF1ab and E genes (Kitajima et al., 2020). If the wastewater is collected in a combined system, volume of precipitation can greatly influence the concentration of the viral RNA. To overcome this bias, it is suggested to normalise the data to the flow volume of the wastewater, or parameters related to the ratio of sewage within the discharge, such as microbial faecal indicators or chemical parameters (e.g. conductivity, or endogenous or exogenous biomarkers) (Medema et al., 2020a; Polo et al., 2020).
Hungary was successful in controlling the first wave of the pandemic. The first cases were identified on March 4th and were followed by control measures within a week. Schools were closed on March 16th, and lockdown was introduced on March 27th. Starting on May 4th, public constraints were gradually raised. The peak number of new cases in the first wave was 2.1/100,000 inhabitants, amounting to a maximum of 21.0 active cases and 10.5 hospitalised patients/100,000 inhabitants on May 4th. The total number of deaths was below 600 in the first wave (6.1/100,000 inhabitants). Approximately 60% of all cases and deaths in the first wave were connected to Budapest, which is much higher than its proportion within the population (18%). The second wave was more severe: as of November 1st, the number of active cases and hospitalised patients was 586.5 and 43.0/100,000 inhabitants (25% of the total Hungarian cases). Regular sewage surveillance for SARS-CoV-2 started in early June in Budapest and was extended to all counties by the end of the month with the aim of signalling a potential resurge of infections. The current paper presents sewage surveillance results from Budapest and the association to the second wave of infections.
2. Materials and methods
2.1. Sample collection
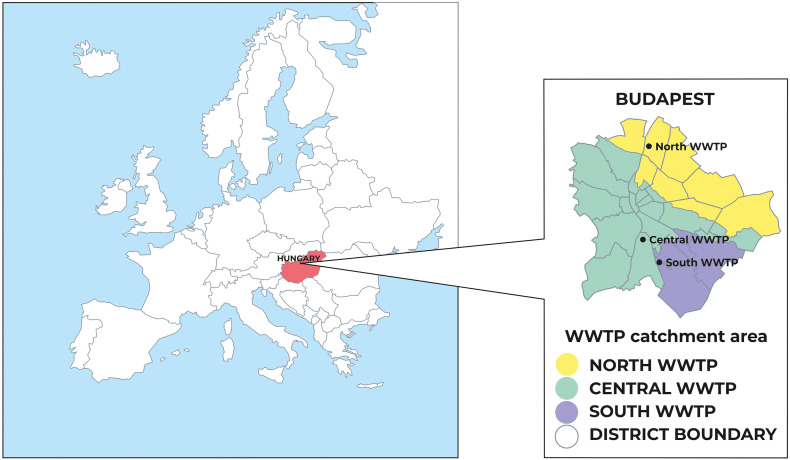
Raw sewage samples (44 composite and 21 grab samples in total) were collected in the three wastewater treatment plants (WWTPs) of Budapest (North, South and Central WWTP) (Fig. 1 ). In the exploratory phase of sewage surveillance, two grab samples were taken from raw sewage of the main hospital dedicated to COVID-19 patients in the service area of the Central WWTP. Weekly samples were taken from the beginning of June 2020 by accredited personnel of the WWTPs on the same day of the week, between 8 and 10 AM. 250 mL grab samples were collected in sterile glass containers in all three WWTPs; 24 h automated composite samples were only available in the Central WWTP. Samples were transported at 4 °C to the laboratory and processed within 6 h.
Fig. 1.
Location and service area of the sampled WWTPs in Budapest.
2.2. Sample preparation and concentration
Four concentration methods based on flocculation and ultrafiltration were compared for method development. Skimmed milk flocculation (SMF) method was performed as published previously (Rusiñol et al., 2014), using 50 mL initial sample volume (n = 2). Before ultrafiltration, samples were sedimented by centrifugation (4500 g, 30 min, 4 °C) and the supernatant was used for further concentration. Pierce™ Protein Concentrator PES 10 kDa molecular weight cut-off 5–20 mL ultrafilter (Thermo Fisher Scientific, Waltham, MA, USA) (n = 3) and Centriprep 50 kDa nominal molecular weight limit 15 mL centrifugal filter unit (Merck KGaA, Darmstadt, Germany) (n = 14) were used according to the manufacturers' instructions. Briefly, on Pierce™ Protein Concentrator units, 50 mL samples were concentrated in multiple aliquots by centrifugation at 4700 g, room temperature, until the volume of the concentrate was below 1 mL. Using Centriprep columns, samples were centrifuged at 1500g, room temperature until the volume of the concentrate was below 1 mL or the unit clogged (0.6–12.6 mL final volume). The processed sample volume was 36–50 mL.
Due to availability issues of most previously used filter units, an in-house method was developed using a custom-developed flat-sheet polyvinylidene-difluoride-based ultrafiltration membrane of 30 nm average pore size and 270 kDa cut-off (Suez Water Technologies & Solutions, Membrane Research Center, Tatabánya, Hungary). The filtration membranes were shipped in aqueous sodium hypochlorite solution as a preservative. Membranes were washed in phosphate buffer at 37 °C to remove the preservative and used without drying. 50 mL samples were processed by vacuum membrane filtration. Membranes were transferred to centrifuge tubes and washed by vortexing for 1 min with 1 mL virus transport medium (VTM) to recover viral particles. Supernatant was removed and stored at −20 °C until RNA extraction.
Recovery of the concentration methods was calculated using sewage samples spiked prior to the centrifugation with heat inactivated SARS-CoV-2 virus (quantity: 1.72 × 105 GC/L) cultured from a clinical isolate, obtained from the National Safety Laboratory. Virus concentration was compared to direct RNA extraction from 1 mL VTM spiked with the same amount of inactivated virus by RT-qPCR.
Positive external process control was prepared using heat inactivated SARS-CoV-2 virus as described above and was used to confirm the efficiency of the concentration method. The results of the process controls are shown in Supplementary material, Fig. S1. Negative process control was 50 mL sterile tap water. Controls were prepared fresh daily and processed with every batch of samples.
Enterococcus count of the sewage samples was determined according to ISO 7899-1 standard method. MUD/SF plates were obtained from Bio-mnRad (CA, USA). Plates were incubated 44 °C for 48 h and read under UV light.
2.3. RNA extraction
RNA was isolated using a commercial kit (QIAamp Viral RNA Mini Kit 250, Qiagen, Germany) in accordance with the manufacturer's instructions. Briefly, 140 μL concentrate was lysed and loaded on centrifuge extraction columns. After 2 washing steps, RNA was eluted in 30 μL final volume to obtain more concentrated RNA. This elution volume still yields acceptable results according to the manufacturer's specification. Before RT-PCR, the extracted RNA was stored at −20 °C for maximum 48 h.
2.4. Detection of SARS-CoV-2
For the detection of SARS-CoV-2 RNA, quantitative RT-PCR was conducted on the LightCycler 480 Instrument II platform using the LightCycler Multiplex RNA Virus Master kit (Hoffmann-La Roche, Basel, Switzerland). The applied primers and probes were specific for the nucleocapsid 1 (N1) gene of the virus (2019-nCoV-N1-F, GAC CCC AAA ATC AGC GAA AT; 2019-nCoV-N1-R: TCT GGT TAC TGC CAG TTG AAT CTG; 2019-nCoV-N1-P: FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1) (CDC, Atlanta, https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html). Virus copy number was calculated based on the standard curve of the commercially available copy control (EURM-019) obtained from the European Commission Joint Research Center, Geel, measured with an automatic threshold. To detect any possible RT-PCR inhibition, a spike-in internal control (LightMix Modular EAV RNA Extraction Control, Roche, Basel) was added to the extracted RNA samples. The 70 bp long fragment from the Equine Arteritis Virus genome was amplified with specific primers included in the kit and detected by an Atto647 labelled hydrolysis probe. Limit of detection of the RT-PCR was measured using log10 serial dilution of the EURM-19 copy control.
2.5. Sewage quality data
Regular monitoring data on flow volumes and conductivity, where available, were provided by the WWTP operators (data shown in Supplementary material, Table S1).
2.6. Epidemiological data
Daily epidemiological data of newly identified, active and hospitalised COVID-19 cases in Budapest were obtained from the official records of the National Public Health Centre.
2.7. Statistical analysis
For the concentration method comparison, paired sample t-test was performed using Microsoft Excel.
Epidemiological and viral genome copy number data collected in weeks 23–44 (from June to November) were used for the analysis. As the virus genome copy numbers were available on a weekly basis, daily epidemiological data were converted to rolling 7-day cumulative data in case of new cases and deaths. For active and hospitalised cases rolling 7-day average data were calculated for each week. Viral genome copy numbers were log10 transformed. Correlation among the datasets was analysed by linear regression using R version 3.6.2 (released on 12.12. 2019), (The R Foundation for Statistical Computing, https://www.r-project.org/). For new and active cases, daily shifts (i.e. rolling average 1 to 12 days after sampling) were also analysed.
3. Results and discussion
3.1. Comparison of concentration methods
Concentration methods were compared on a limited number of samples due to the limited availability of materials for several previously published methods. Skim milk flocculation (SMF) showed low recovery on spiked samples (n = 2, 0.8 and 12% recovery). It was also more time consuming as other ultrafiltration methods, as it required 8 h of mixing plus 8 h of sedimentation. SMF was used successfully for simultaneous concentration of various pathogens, including viruses, bacteria and protozoa (Gonzales-Gustavson et al., 2017), with recoveries ranging 15–66% for different viruses. A recent study detected naturally occurring SARS-CoV-2 in sewage samples using SMF (Rusiñol et al., 2020). Recoveries (23–37%) – calculated for phage MS2 as an internal process control – were higher and more reliable than in the present study. However, based on considerations of time demand and poor initial data, this method was abandoned in spite of its low cost and sustainable availability of materials.
Of the tested commercially available ultrafiltration columns, SARS-CoV-2 was not recovered from spiked sewage samples using the Pierce™ Protein Concentrator (n = 3). Mean recovery of Centriprep filter units was 80% (n = 3) compared to direct extraction of the spiked virus quantity. The in-house method using flat ultrafilter membranes showed 96% mean concentration recovery (n = 7) under the same test conditions.
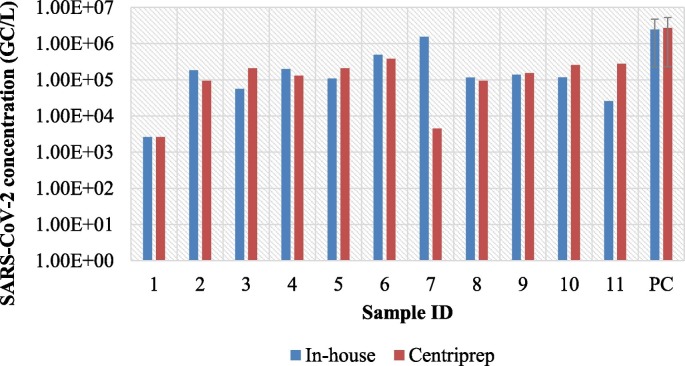
Based on the initial results, Centriprep filter units and the in-house method were tested further on spiked and non-spiked sewage samples. The SARS-CoV-2 concentration was within one order of magnitude for almost all (10/11) non-spiked sewage samples, mean concentration and standard deviation of spiked samples were also similar (Fig. 2 ). The two methods were statistically not different (paired t-test, t value: 0.073, critical two tailed, 2.16, p < 0.05).
Fig. 2.
Comparison of concentration efficiency of the in-house flat membrane ultrafiltration method and Centriprep 50 kDa molecular weight cut-off filter units. Viral concentration of sewage samples (samples 1–11) and spiked positive control samples (PC, average of 3 samples; negative result (ID: 1) was substituted by LOD = 2640 GC/L for data visualisation.)
Several methods are being used for the concentration and detection of SARS-CoV-2 RNA in sewage by various research groups (Ahmed et al., 2020; Haramoto et al., 2020; Kocamemi et al., 2020; La Rosa et al., 2020; Medema et al., 2020b; Wurtzer et al., 2020, among others). An interlaboratory comparison of 36 different standard operation procedures indicated that various methods can be applied successfully, but the same procedure should be used in a single laboratory (Pecson et al., 2020). In this study, the in-house method proved to be at least as efficient as the commercial filtration units, and filters were more readily available. To obtain comparable results, this method was used in the subsequent analysis.
3.2. Detection of SARS-CoV-2 RNA in WWTPs
Three WWTPs service the entire area of Budapest and some of its agglomeration (total population: 1.8 million) (Fig. 1). The nominal capacity of the North, South and Central WWTPs is 1.33, 0.29 and 1.63 million person equivalents, and the served population is 700,000, 300,000 and 800,000, respectively. Daily commuters may increase daytime population of the city up to 2.0–2.1 million. The number of commuters varied in a wide range in the study period due to different lockdown measures affecting the number of remote workers. Some of the suburbs where people commute from are also serviced by the WWTPs of Budapest. Due to the above difficulties in estimating the number of commuters, this variation was not considered in data analysis.
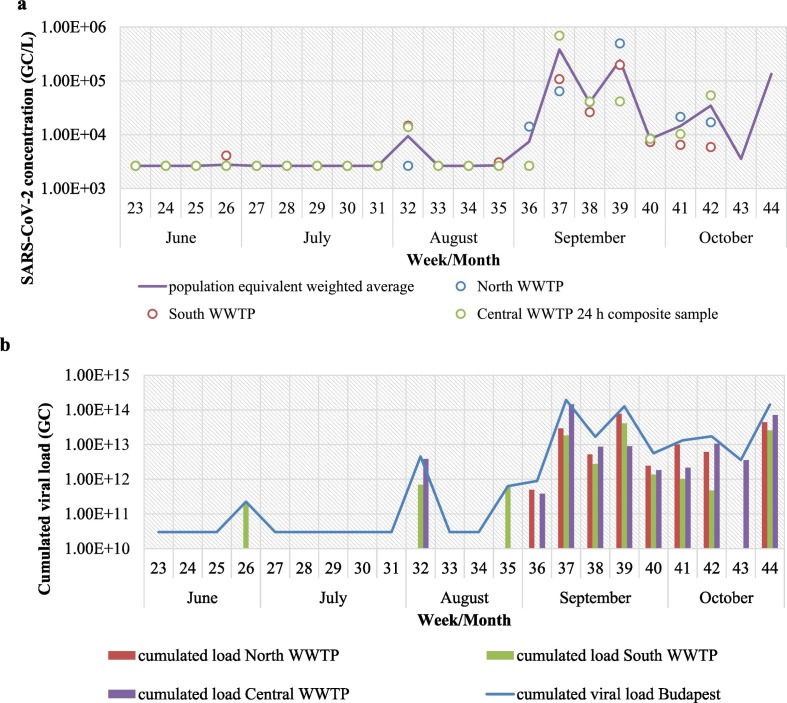
In the first 9 weeks of regular monitoring, all samples were below LOD with a single weak positive in South WWTP on week 26. The experimentally determined LOD of the RT-PCR was 35.8 Ct value corresponding to approximately 1.54 copies/reaction and 2640 GC/L in the initial sewage sample. Positive samples were detected on week 32, and increasing copy numbers were seen in the subsequent period (Fig. 3 ). The observed concentration range (up to 7.14 × 105 GC/L) in the sewage samples was similar to those seen in metropolitan areas in France, the Netherlands, Massachusetts and Spain, among others (Chavarria-Miró et al., 2021; Medema et al., 2020b; Wu et al., 2020; Wurtzer et al., 2020). Higher values (up to 108 GC/L) were reported from Brazil at the peak of the outbreak (Prado et al., 2021).
Fig. 3.
Presence of SARS-CoV-2 viral RNA in the Budapest WWTPs between June and October 2020 (weeks 23–44). (a) Viral concentrations normalised to 10,000 MPN/mL Enterococcus counts and the calculated average weighed by capacity (person equivalent) of the WWTPs. Negative results were substituted by LOD = 2640 GC/L for data visualisation. (b) Viral load by WWTP and cumulated for the entire service area. Negative results are substituted by 1010 GC/L for data visualisation.
Viral concentrations in the three WWTPs were generally within one order of magnitude, larger discrepancies were mostly seen at low concentrations, where some samples were below LOD. The results of 24 h composite samples and grab samples from the Central WWTP were also different on several sampling occasion. Therefore it is likely that the variations observed among the three WWTPs are due to the heterogeneity of the sewage itself rather than actual differences of the infected population in the service area. No trends were observed, e.g. either of the WWTPs yielding consistently higher viral titres. Similar variability was seen between WWTPs serving the Frankfurt metropolitan area (Agrawal et al., 2021). No data is available on the distribution of infected persons within Budapest. The hospital sampled in the exploratory phase was among the first ones designated to COVID-19 positive patients, so it was expected that first positive results would be detected there. One building was positive for SARS-CoV-2 (3.27 × 105 GC/L), but the other, housing high risk patients, was negative. Enterococcus count was also unusually low (300 MPN/mL), implying that wastewater was disinfected, and thus not contributing to the viral load. Disinfection of hospital sewage is not a general practice in Hungary and there is no information on its application at these specific sampling sites. Occasional disinfection might have been applied after the surge of COVID-19 cases. WBE in hospital sewage was suggested to be a potentially useful complimentary tool for monitoring the spread of SARS-CoV-2 in the population (Gonçalves et al., 2021). However, in smaller communities or single institutions the low number of shedders results in high temporal variability, thus the chance of false negative is higher, especially in grab samples. In the present study, hospital samples served as a confirmation of the method applied. Larger sample numbers and more detailed information on hospital sewage composition would be necessary to draw further conclusions.
24 h composite sampling is deemed more representative of the actual viral concentration as it corrects for the variability resulting from the different sewage loads during the day (Medema et al., 2020a). In the present study, morning samples as a peak load were used in the WWTPs where 24 h sampling was not available. The variability of the data from samples taken at different sites may also be partially attributed to the method of sampling. However, since all sampled WWTPs serve very large number of people and their service area is extensive, samples are still likely to be representative of the served population.
3.3. Estimation of viral load
Majority of the sewerage in Budapest is a combined system, receiving precipitation as well as sewage. Wastewater can be significantly diluted during rain events, causing a bias in the measured viral concentrations. To obtain comparable results over space and time, it is therefore necessary to correct for dilution. Different approaches have been suggested for data normalisation (Medema et al., 2020a). Flow volume provides direct information on the dilution of the sewage and can be used to calculate viral load, which was suggested to be a better predictor of COVID-19 cases than viral concentration (Westhaus et al., 2021). The other approach is to use faecal indicators to estimate the faecal fraction within the wastewater sample. The advantage of this method is that faecal indicators can be analysed from the same sample as the viral concentration.
The daily volume of wastewater varied between 118–241, 52.8–82.7 and 196–489 thousand m3 in the North, South and Central WWTP, respectively. During the study period, there were one major and three minor rain events coinciding with the sampling dates, causing approximately 124% and 30% increase in the volume of wastewater, respectively (Supplementary material, Table S1). Conductivity (where available) correlated negatively to the flow volume (Supplementary material, Table S1). Enterococcus counts varied between 570 and 84,000 MPN/mL (Supplementary material, Table S1), but the detected titres did not reflect the effect of dilution in case of rain events. As discussed above, the main driver of the observed variability between WWTPs was most probably the inherent inhomogeneity of sewage samples, associated with different shedding patterns. However, sewage composition can also affect the detection methods. Dilution leads to less efficient concentration of SARS-CoV-2 RNA while more concentrated sewage might contain higher amount of PCR inhibitor substances.
Total viral load was calculated by multiplying the viral concentration by the volume of wastewater on the day of sampling (Fig. 3b). Daily calculated viral load of Budapest was 8 × 1011–1.93 × 1014 genome copies, similar to Frankfurt metropolitan area in the same period (Agrawal et al., 2021). Normalisation by flow volume decreased week-to-week and site-to-site variations, but did not modify the observed trends. Viral faecal indicators such as F-specific RNA phages indicated by Medema et al. (2020b) were not measured. The faecal fraction of the sewage sample was estimated based on counts of intestinal Enterococci, which is generally considered a more conservative indicator of faecal pollution in the environment than E. coli. Weighted average of viral concentrations was calculated for the three WWTPs, taking into account their service area (person equivalent) (Fig. 3a).
3.4. Correlation to epidemiological data
Viral load values (cumulated viral load calculated from the viral concentration and the daily flow volume of each WWTP), representative of the entire population of Budapest were used for the analysis. Negative results were substituted by half of LOD for regression analysis. Viral data were compared to the 7-day rolling average of epidemiological data on the same week and next week. The number of new COVID-19 infections in Budapest and the cumulated viral loads between June and November 2020 are shown in Fig. 4 a.
Fig. 4.
Weekly cumulated numbers of new (a) and active (b) COVID-19 infections (cases/100,000 inhabitants) and cumulated viral loads in Budapest between June–November 2020 (a: weekly cumulated data, b: 7-day averages). Reference population is 1.8 million.
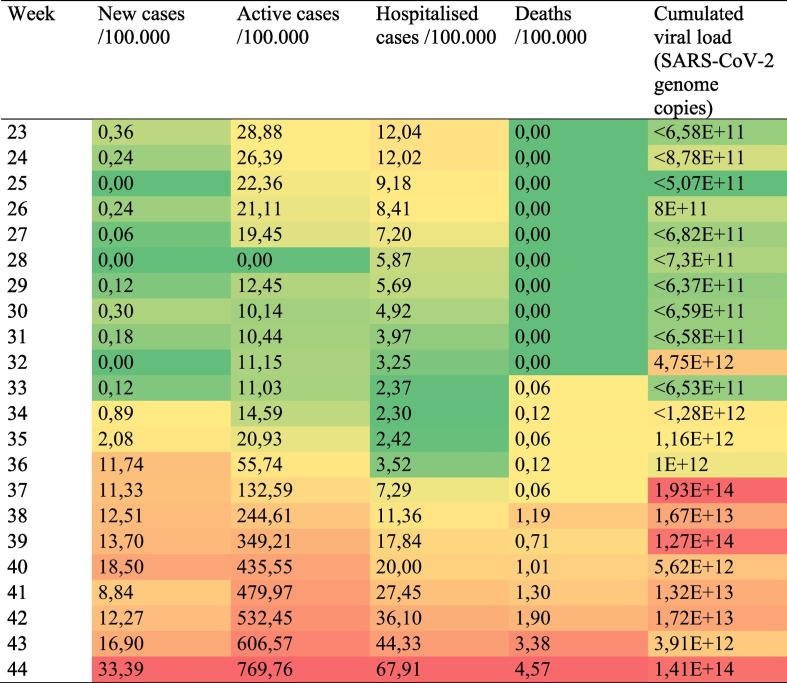
Epidemiological data per 100,000 population and cumulated viral loads are shown in Table 1 .
Table 1.
Epidemiological data per 100,000 population and cumulated viral loads (showing aggregated data for the three WWTPs) in Budapest in the study period. Reference population is 1.8 million.
First positive signals of SARS-CoV-2 in sewage were detected 2 weeks before case numbers started to rise. On that week (week 32), 51 new cases were detected (2.8 cases/100,000), and the number of active cases was 11/100,000. Similar lag was observed between the rise in viral titres and the increase in case numbers in other metropolitan areas, such as Barcelona (Chavarria-Miró et al., 2021).
From week 34, new cases doubled every week (Fig. 4a), and the viral titres also increased exponentially. Between weeks 40–42, the number of new cases plateaued, then increased again, while viral load was also variable in this period. Based on regression analysis, viral load correlated to the number of new cases on the week of sampling and the active cases on the next week (Table 2 ). Correlation to hospitalisations and deaths were low. Analysing only the rapidly ascending phase (up to week 40) of the second wave, higher r2 values were observed, though trends remained the same.
Table 2.
Correlation of normalised viral concentration/viral load to epidemiological outcomes (7-day rolling average) for the entire study period (weeks 23–44) and the rising phase of the second wave (weeks 23–40).
| Weighted average |
Viral load |
|||
|---|---|---|---|---|
| Same week r2 |
Next week r2 | Same week r2 |
Next week r2 | |
| 23–44 weeks | ||||
| Active cases | 0,421⁎⁎ | 0,462⁎⁎⁎ | 0,555⁎⁎⁎ | 0,589⁎⁎⁎ |
| Daily new cases | 0,575⁎⁎⁎ | 0,433⁎⁎⁎ | 0,670⁎⁎⁎ | 0,522⁎⁎⁎ |
| Hospitalised cases | 0,242⁎ | 0,250⁎ | 0,351⁎⁎ | 0,363⁎⁎ |
| Deaths | 0,076 | 0,119 | 0,187⁎ | 0,235⁎ |
| 23–40 weeks | ||||
| Active cases | 0,514⁎⁎⁎ | 0,645⁎⁎⁎ | 0,589⁎⁎⁎ | 0,697⁎⁎⁎ |
| Daily new cases | 0,720⁎⁎⁎ | 0,693⁎⁎⁎ | 0,716⁎⁎⁎ | 0,665⁎⁎⁎ |
| Hospitalised cases | 0,222 | 0,347⁎ | 0,278⁎ | 0,397⁎⁎ |
| Deaths | 0,145 | 0,560⁎ | 0,246 | 0,653⁎ |
p < 0.05.
p < 0.01.
p < 0.001.
Correlation of new and cumulated active cases to viral load data was also calculated in daily shifts for the day of sampling and subsequent days (day+1, +2… to day+12) (Table S2). Highest correlation was seen at +3 and + 11 days for new cases and active cases, respectively. Association of epidemiological data to population-equivalent weighted average of viral concentrations from the three WWTPs was also analysed yielding similar result, but lower r2 values than viral load data.
Linking environmental surveillance data to disease cases is the key challenge of WBE. Viral RNA concentrations in sewage cannot be directly converted into case numbers due to extensive variations in the excreted viral load both person-to-person and in time as the infection progresses (Foladori et al., 2020; Hart and Halden, 2020). According to the theoretical model of Medema et al. (2020a), the concentration range which most samples in this study fall into (104–106 GC/L SARS-CoV-2 RNA) translates to a median of 50–500 infected persons/100,000, but the confidence interval of the model expands to more than an order of magnitude at these data points. The number of recognised active cases in Budapest reached 50/100,000 on week 36 and exceeded 500/100,000 on week 41, but the number of shedders is expected to be much higher, due to pre-symptomatic and asymptomatic shedding and undiagnosed individuals. Positive results were consistently detected accordingly from week 32 onward. Sporadic occurrence was observed even earlier, similar to the findings of Trottier et al. (2020), who found an increase in viral titres several weeks before the re-emergence in case numbers.
Different approaches were used previously for correlating environmental and epidemiological data. Several studies report spatial variability by sampling several WWTPs in parallel. Medema et al. (2020a) found correlation between SARS-CoV-2 concentrations and 4 weeks cumulated COVID-19 case numbers in 7 Dutch cities in the ascending phase of the first wave. Westhaus et al. (2021) correlated viral load with both acute and cumulative prevalence data in various cities of North Rhine-Westphalia, but did not observe correlation to concentration, not even after normalising to creatinine, a human biomarker. Other studies looked at trends over time: Weidhaas et al. (2020) used viral load data normalised to population size to compare trends in different WWTPs in France, and found correlation to daily and weekly case numbers, though not in every site. Correlation improved if viral load was compared to next week's prevalence data. Wu et al. (2020) found the strongest correlation to new daily cases in Massachusetts with 4 days off-set in the ascending phase of the outbreak, while Wurtzer et al. (2020) estimated an 8-day lag phase in the Parisien urban area. According to Nemudryi et al. (2020) SARS-CoV-2 RNA levels in wastewater followed symptom onset by 5–8 days, and preceded clinical PCR test results by 2–4 days.
Similar results were observed for sewage sludge: D'Aoust et al. (2020) found correlation to daily cases, active cases and the percentage of positive samples using SARS-CoV-2 concentration normalised to pepper mild mottle virus in two WWTPs. Peccia et al. (2020) also confirmed the correlation of virus concentrations in sewage sludge and epidemiological data, but with a lag time of 0–2 days and 6–8 days for new cases by sampling date and reporting date, respectively, and 1–4 days for the number of hospitalised cases.
The present study indicated similar off-set between viral concentrations and epidemiological data in the ascending phase of the second wave. Of the two normalisation methods, viral load yielded higher r2 values, but the correlation patterns with the epidemiological outcomes were almost identical. The 7-day cumulated number of daily new cases followed the rise of viral load with an off-set of 3 days. Since cumulated new cases were calculated on a weekly basis, this is the sum of cases reported on 2–8 days after the sampling. Taking the turn-over time of clinical samples into account (which was 24–48 h in the study period), it is likely that samples corresponding to these results were collected 0–6 days after sewage sampling. The correlation to cumulative number of active cases increased with the number of shifted days, reaching the highest r2 value at 10–12 days (Table S2).
However, when looking at the entire data series, including the weeks, when the number of new cases plateaued (see Fig. 4a), associations were weaker, though still statistically significant (Table 2). Several reasons may lie behind divergence of environmental and epidemiological data as the outbreak advances. Increasing case numbers may overburden the testing capacity, leading to lower recognition rates. This effect is reflected the ratio of positive samples, which first exceeded 10% in week 39 and was consistently above 10% from week 42 in Hungary. The strain on testing facilities can also lead to longer result turn-over times increasing the gap between sampling dates and reporting (though in the study period, no significant difference was observed in the laboratory reporting time). Number of active cases might be overestimated in later phase of outbreaks as administrative capacities are becoming restricted while the healthcare system is under prolonged pressure. On the other hand, extended virus shedding in the post-symptomatic phase introduces a bias in the viral concentrations. A combination of these factors can lead to less reliable prediction. Fitting the data to the theoretical model on viral RNA shedding by Medema et al. (2020a) exhibited similar divergence as the epidemiological data: at lower infection rates (active cases up to 400/100,000 inhabitants) RNA concentrations were within the expected range of the model, but at higher number of shedders fell below it. Laboratory bias cannot be ruled out, but currently we are not aware of any factor or inhibition mechanism that would hinder the detection of RNA concentrations above 106 GC/L.
A limiting factor of the present study is the issue of non-resident shedders. The resident population of Budapest is 1.8 million. In the study period, international and national tourism in Budapest was limited and unlikely to have a considerable impact on the results. However, daily commuters from the agglomeration (approximately 225,000 people) may also contribute to the viral load, but will not be represented in the counts of new and active cases. The number of hospitalised cases also captures commuters, as majority of the agglomeration is serviced by Budapest hospitals.
Most previously published studies were conducted in the first wave of the COVID-19 pandemic, mostly when case numbers were rapidly rising. Two studies looking at the descending phase after lockdowns arrived to conflicting conclusions: Wu et al. (2020) observed a sharp drop of viral titres which was only slowly followed by the reduction of case numbers, while in the study of Wurtzer et al. (2020) virus concentrations plateaued while cases were decreasing. Lockdown was introduced after the study period in Hungary, thus its effect could not yet be evaluated. The ongoing weekly data collection by sewage sampling will clarify association between the environmental surveillance data and epidemiological outcomes in the more advanced phases of the infection waves. The future aim is to develop a predictive model that can further support outbreak management decisions.
4. Conclusions
-
•
Surveillance of sewage in Budapest for SARS-CoV-2 confirmed the usefulness of WBE as an early warning tool. The method provides screening data within 24 h, in case of Budapest representing approximately 1.8 million people through sampling three WWTPs. The first positive signal in sewage preceded the resurge of the outbreak by 2 weeks.
-
•
Normalised SARS-CoV-2 concentration in sewage correlated to the number of new cases on the week of sampling and the number of active cases on the following week.
-
•
Viral load was a better predictor of clinical data than population-weighted average of SARS-CoV-2 RNA concentration.
-
•
The correlation was more pronounced in the uprising phase of infection. No clear association was seen when the number of cases plateaued.
-
•
The efficiency and reproducibility of the in-house concentration method using flat sheet membrane ultrafiltration was comparable or superior to commercial ultrafiltration units.
CRediT authorship contribution statement
Eszter Róka: Conceptualization, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Bernadett Khayer: Conceptualization, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Zoltán Kis: Methodology. Luca Bella Kovács: Investigation. Eszter Schuler: Investigation. Nóra Magyar: Methodology. Tibor Málnási: Formal analysis. Orsolya Oravecz: Writing – original draft, Writing – review & editing, Visualization. Bernadett Pályi: Methodology. Tamás Pándics: Conceptualization. Márta Vargha: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The support of Budapest Waterworks and the Budapest Sewage Works Pte Ltd. in sampling and sample transportation is highly appreciated.
We thank dr. Kristóf Vízvárdi and Dorottya Zverger (Suez Water Technologies & Solutions, Hungary) for their cooperation in the development of the ultrafiltration method and for the preparation of the custom-designed membrane.
We are also grateful to Peter Pollner (MTA-ELTE Statistical and Biological Physics Research Group) for his help in the planning of sampling activities.
Editor: Yolando Picó
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.147398.
Appendix A. Supplementary data
Supplementary material
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11:5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona, Spain. Appl. Environ. Microbiol. 2021;87, 7 doi: 10.1128/aem.02750-20. e02750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust P.M., Mercier E., Montpetit D., Jia J.-J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2020;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . Coronavirus disease 2019 (COVID-19) in the EU/EEA and the UK – ninth update, 23 April 2020. 2020. European Centre for disease prevention and control.https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-rapid-risk-assessment-coronavirus-disease-2019-ninth-update-23-april-2020.pdf Available from: (accessed on 13.11.2020) [Google Scholar]
- ECDC . Increased transmission of COVID-19 in the EU/EEA and the UK – 24 September 2020. 2020. European Centre for disease prevention and control.https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-risk-assessment-increased-transmission-12th-update-september-2020.pdf Available from: (accessed on 13.11.2020) [Google Scholar]
- ECDC . Diagnostic testing and screening for SARS-CoV. 2020. European centre for disease prevention and control.https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing Available from: (accessed on 13.11.2020) [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J., Koritnik T., Mioč V., Trkov M., Bolješič M., Berginc N., Prosenc K., Kotar T., Paragi M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Gustavson E., Cárdenas-Youngs Y., Calvo M., da Silva M.F.M., Hundesa A., Amorós I., Moreno Y., Moreno-Mesonero L., Rosell R., Ganges L., Araujo R., Girones R. Characterization of the efficiency and uncertainty of skimmed milk flocculation for the simultaneous concentration and quantification of water-borne viruses, bacteria and protozoa. J. Microbiol. Methods. 2017;134:46–53. doi: 10.1016/j.mimet.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacıoglu S., Yaralı C., Saatci A.M., Pakdemirli B. Infectious Diseases (except HIV/AIDS) 2020. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey (preprint) [DOI] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. The Lancet Gastroenterology & Hepatology. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. 2020. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Medicine. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y. medRxiv. 2020. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. 2020.11.02.20221622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Vale V.H.C., Braz R.M.S., Andrade J.S.R., Maranhão A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Research. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Fernandez-Cassi X., Hundesa A., Vieira C., Kern A., Eriksson I., Ziros P., Kay D., Miagostovich M., Vargha M., Allard A., Vantarakis A., Wyn-Jones P., Bofill-Mas S., Girones R. Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res. 2014;59:119–129. doi: 10.1016/j.watres.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020;10 doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., Ahern S., Carty P.G., O’Brien K.K., O’Murchu E., O’Neill M., Smith S.M., Ryan M., Harrington P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Inf. Secur. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z., Roper D., VanDerslice J., Gaddis E., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. In Review. 2020. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO Director-General's Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 Available from: (accessed on 13.11.2020)
- WHO Public Health Surveillance for COVID-19 Interim Guidance 7 August 2020. 2020. https://www.who.int/publications/i/item/who-2019-nCoV-surveillanceguidance-2020.7 Available from: (accessed on 13.11.2020)
- Wu F., Zhang J., Xiao A., Bonneau R., Brown M.A., Bushman M., Gu X., Huang K.H., Xiaoqiong G., Lee W.L., Armas F., Kauffman K., Hanage W.P., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K.S., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4) doi: 10.1128/2FmSystems.00614-20. 2020. 2020. e00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters (preprint) Epidemiology. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material