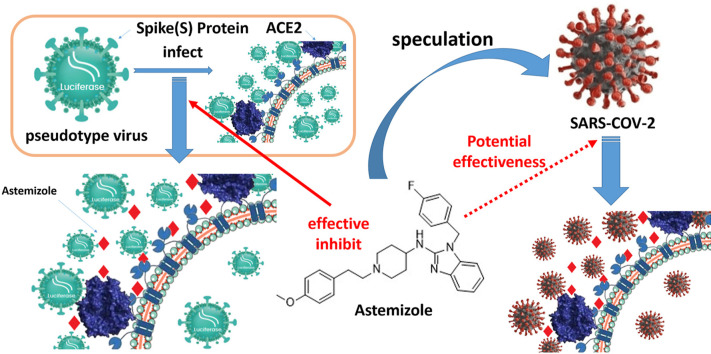
Abstract
Since the beginning of December 2019, a novel Coronavirus severe respiratory disease, caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) which also been termed 2019-new CoV (2019-nCoV), has continued to spread worldwide. As of August 27, 2020, a total of 24,232,429 people have been infected and 826,518 people have died. In our study, we found that astemizole can antagonize ACE2 and inhibit the entry of SARS-COV-2 spike pseudovirus into ACE2-expressed HEK293T cells (ACE2hi cells). We analysied the binding character of astemizole to ACE2 by molecular docking and surface plasmon resonance (SPR) assays and molecule docking, SARS-COV-2 spike pseudotype virus was also taken to investigate the suppression viropexis effect of astemizole. The results showed that astemizole can bind to the ACE2 receptor and inhibit the invasion of SARS-COV-2 Spike pseudoviruses. Thus astemizole represent potential drug candidates that can be re-used in anti-coronavirus therapies.
Keywords: Astemizole, Drug re-use, SARS-COV-2, ACE2
Graphical abstract
1. Introduction
Since the beginning of December 2019, a novel Coronavirus severe respiratory disease, caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) which also been termed 2019-new CoV (2019-nCoV), has continued to spread worldwide. The gene virus is very closed to bat coronavirus Bat-CovratG13 and Bat-SL-CovzC45 CoVs, which can cause diseases similar to Middle East respiratory syndrome (MERS)-CoV and SARS-CoV [1], with higher infectibility and lower fatality [2]. Angiotensin-converting enzyme II (ACE2), as the SARS-CoV receptor, can also bind to SARS-CoV-2 [3]. SARS-CoV-2 binds to the pockets of the ACE2 receptor via the N-terminal part of viral protein unit S1, thereby enters the cell [[4], [5], [6], [7], [8], [9], [10], [11]].Then, the body fluid and cellular immune system are turned on by antigen presentation and inflammatory factor aggregation [[9], [10], [11]].
As of August 27, 2020, a total of 24,232,429 people have been infected and 826,518 people have died. Unfortunately, no special antiviral drugs have been used for clinical treatment of this viral infection, and vaccines are still in the development stage. Therefore, it is crucial to find a treatment, and the scheme of reusing existing drugs is widely proposed [[12], [13], [14]]. In the traditional drug development process, from target identification, target verification, lead optimization to preclinical and clinical trials, it generally takes more than 10 years [15] and would cost huge amounts of money. However, facing the current urgent situation of SARS-CoV-2, there is hardly enough time to develop new drugs. Therefore, the most feasible method is to find potential treatment methods from existing clinical drugs through the re-use of old drugs (also known as drug rediscovery). Drug re-use can quickly obtain data on the human pharmacokinetics, safety and preclinical results of related drugs [16], while saving time and money.
Benzimidazole is a special substructure that exists in many basic cell components and biologically active compounds, used in a variety of important drugs in different therapeutic fields, such as proton pump inhibitors, angiotensin receptor antagonists, Antihistamines and antiparasitic drugs [17]. In previous studies, benzimidazoles have been found to play an important role in anti-HIV-1, HCV and RSV and other viral compounds, and a variety of benzimidazoles have been shown to be effective against other viruses (influenza B virus, Rhinovirus, poliovirus, Coxsackie B3, cytomegalovirus, etc.) also have an impact [18].The SARS-CoV-2 virus enters the body through the ACE2 [19]. Blocking or antagonizing the ACE2 may help co-treatment of SARS-CoV-2 virus infection. Benzimidazole is the parent nucleus structure of some angiotensin receptor antagonists. Therefore, we hypothesized that the benzimidazole structure may bind to the ACE2 receptor to block the virus from entering the body.
In this study, we found that a histamine receptor antagonist with the structure of benzimidazole parent nucleus, astemizole, can antagonize ACE2 and inhibit the entry of SARS-COV-2 spike pseudovirus into ACE2-expressed HEK293T cells (ACE2hi cells).
2. Materials and methods
2.1. Drugs and reagents
Dulbecco's Modification of Eagle's Medium (DMEM) high glucose (Cat. No. SH30022.01) and fetal bovine serum (FBS) (Cat. No. 16140071) was obtained from HyClone (Logan, UT, USA). Penicillin–streptomycin solution was purchased from Xi'an Hat Biotechnology Co., Ltd (Xi'an, Shaanxi, China). Cell Counting Kit was purchased from 7Sea Pharmatech Co., Ltd (Shanghai, China).
2.2. Cell lines
ACE2 high expressing HEK293T cells (ACE2hi cells) were kept in DMEM high glucose medium containing 10% FBS, 1% penicillin-streptomycin, 4 μg/mL puromycin and cultured at 37 °C in a 5% CO2 incubator.
2.3. Cytotoxicity assay
5 × 103 ACE2 high expression cells per well were seeded in 96-well plate and the repetitive wells were set as 3. Then treated with different concentrations of Astemizole (0, 0.1, 1, 5, 10, 20 and 40 μM) for 24 h. An abbkine-Cell Counting Kit assay (7 Sea, China) was used to determine the Cytotoxicity assay Cell viability. Next, using a microplate reader (Bio‐Rad, Carlsbad, CA, USA) to assess the relative cell viability by the detection of the absorbance at 450 nm. All the steps were finished followed the instructions strictly. The survival rate of ACE2hi cells was calculated as the following formula:
| [(ODTreated − ODBlank) / (ODControl − ODBlank)] × 100% |
2.4. The detection of SARS-COV-2 spike pseudotype virus entry into ACE2 cells
5 × 104 of ACE2hi cells in 50 μL DMEM per well were seeded into white 96‐well plates. The cells were cultured in a 37 °C incubator containing 5% CO2 for 2 h 25 μL medium was aspirated carefully from 96 wells, 25 μL medium containing corresponding dose of the medicine was added and incubated for 2 h. Then 5 μL of SARS-COV-2 Spike pseudotype virus was added (Sino Biological, PSC001), the virus titer is 104.4TCID50/mL and 860 ng SARS-CoV-2 spike S1 protein was contained in 1 mL. The copy number is 1010 virus copies per mL. So, there was 152.59TCID50 in 5 μL coronavirus spike pseudotype virus. Then incubated in the 37 °C incubator containing 5% CO2 for 4 h followed with adding 100 μL of complemented DMEM per well. After 6–8 h of further infection, the culture medium containing the virus was sucked away and replaced by 200 μL of fresh DMEM, and incubated continuously at 37 °C for 48 h, the culture medium was aspirated and 20 μL of cell lysate was add from the Luciferase Assay System (Promega, E1500) to each well, then 100 μL of luminescence solution was added to wells before the luciferase luminescence detection, chemiluminescence was detected by a microplate reader under 560 nm, the exposure time was 1 s.
2.5. Docking studies
Molecular docking studies were conducted using the Sulflex-Dock Mode of Sybyl-X program package (New Tripos International, St. Louis, USA). The docking model of ACE2 is download from PDB. Bank, and the PDB code is 6M0J. The docking model of H1R is download from PDB. Bank, and the PDB code is 3RZE. Astemizole and the protein structure were both optimized by minimizing the Hückel charge.
2.6. Surface plasmon resonance assay
For the analysis of surface plasmon resonance (SPR), ACE2 protein with a 6-his tag (30 μg/mL) was fixed on the COOH sensor chip by capture-coupling, the interaction of Astemizole (0, 0.1, 1, 5, 10, 20 and 40 μM) with ACE2 fixed was detected by OpenSPRTM (Nicoya Lifesciences, Waterloo, Canada) at 25 °C. The binding time and disassociation time were both 250 s, the flow rate was 20 μl/s. A one to one diffusion corrected model was fitted to the wavelength shifts corresponding to the varied drug concentration. The data was retrieved and analyzed with TraceDrawer software.
2.7. Statistical analysis
Data were presented as the mean ± standard error of the mean (S.D.). Significant differences were determined by one-way ANOVA test. Two-tailed unpaired Student's t-test was used to conduct all analyses between two groups. Differences were deemed statistically significant at P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
3. Results
3.1. The effect of astemizole on ACE2-293T cell viability
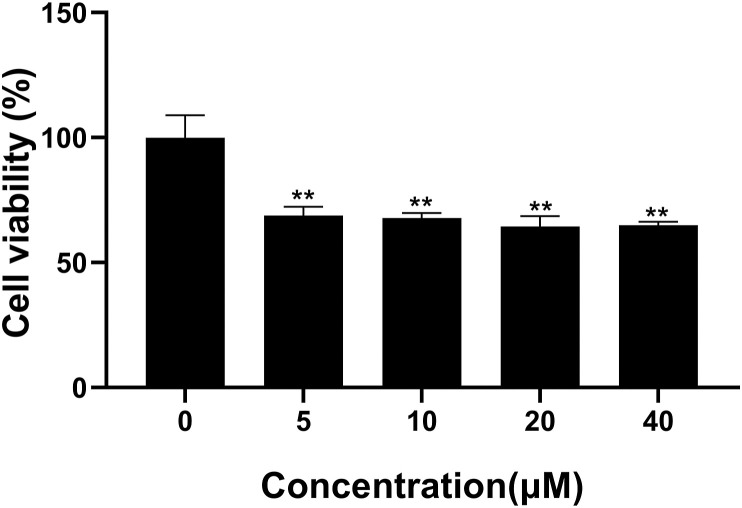
The potential cytotoxicity of Astemizole was evaluated with Cell Counting kit assay. Astemizole was tested in different concentrations range from 0.1 to 40 μM. As it shows in Fig. 1 , Astemizole had no significant effect on the activity of ACE2hi cells when the concentration was than 1 μM, and the survival rate of ACE2hi cells could be reduced in a slight dose-dependent manner when the concentration was above 5 μM. This result showed that Astemizole didn't influence the viability of ACE2 cells significantly.
Fig. 1.
The effect of cell viability on ACE2 cell by Astemizole. ACE2 cells were pretreated with different doses of Astemizole. Viability of ACE2 cells treated with Astemizole for 24 h. The samples were tested in triplicate. Data are expressed as mean standard deviation (SD) of triplicate assays. Astemizole showed no effect on apoptosis in ACE2 cells.
3.2. Astemizole suppressed the entrance of SARS-COV-2 spike pseudotype virus into ACEhi cells
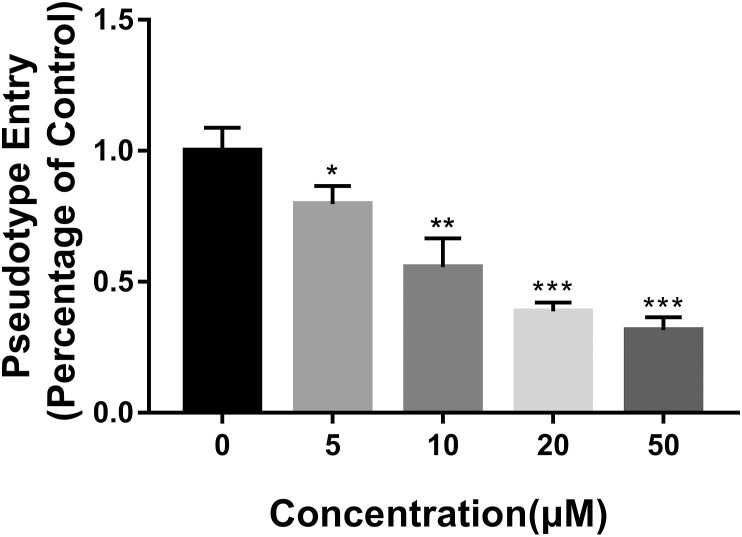
ACEhi cells infected only by SARS-COV-2 Spike pseudotype virus were considered as control, and the luciferase luminescence value was defined as 1. Under the treatment of 5, 10, 20, and 50 μM Astemizole, the SARS-COV-2 Spike pseudotype virus entrance ratio were reduced to 0.80 ± 0.05, 0.56 ± 0.09, 0.39 ± 0.03, and 0.14 ± 0.01 (Fig. 2 ). The ability of the SARS-COV-2 Spike pseudotype virus to enter the ACE2 cells were reduced significantly.
Fig. 2.
The effect of Astemizole on the entrance of SARS-COV-2 Spike pseudotype virus into ACE2 cells. Data are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control.
3.3. The binding character of astemizole with ACE2
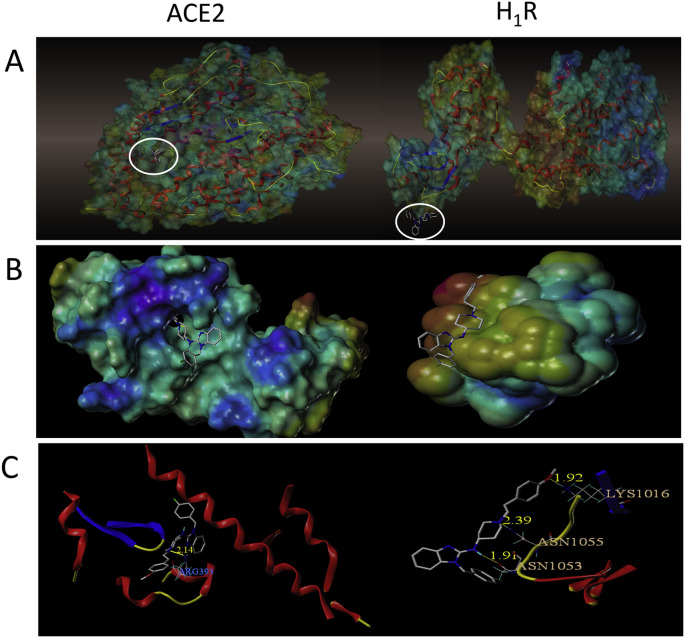
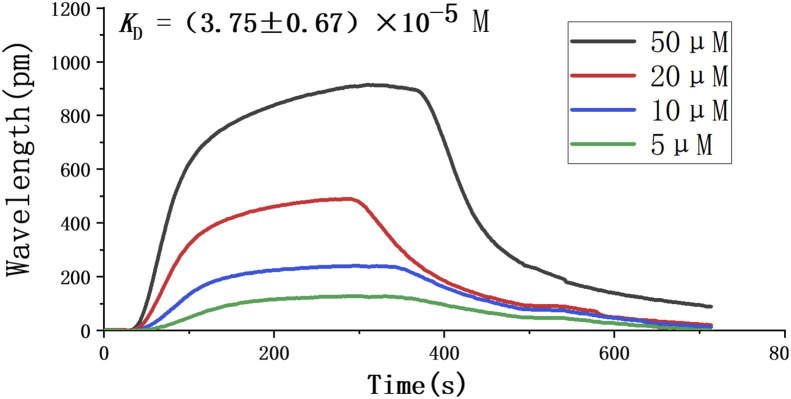
Since it is believed that the SARS-COV-2 virus enters the host cell by binding to the ACE2 receptor, we focused on whether Astemizole could bind with ACE2. The virtual molecular docking test was performed to investigate the binding character of Astemizole with ACE2 and H1R. Fig. 3 show that Astemizole formed one hydrogen bond with ACE2, while three hydrogen bonds with H1R. Nitrogen on the hexahydropyridine ring of astemizole form hydrogen bonds with ARG393 of ACE2 with distances of 2.14 Å. Asmidazole forms hydrogen bonds with LYS1016, ANS1055, and ASN1053 of H1R with distances of 1.92 Å, 2.39 Å, and 1.91 Å, respectively. We further used SPR to confirm the binding between Astemizole and ACE2. The binding constant K D of this compounds and ACE2 protein was (3.75 ± 0.67) × 10−5 M (Fig. 4 ).
Fig. 3.
The binding character of Astemizole with ACE2 and H1R. (A) Docked pose of Astemizole with ACE2 and H1R. (B) Surface representation of the best ranked docking pose of Astemizole in ACE2 and H1R. binding pocket. (C) Residues involved in binding are displayed as sticks, with hydrogen bonds shown as yellow lines.
Fig. 4.
Binding response curves andKDof Astemizole with ACE2 by SPR.
4. Discussion
Current efforts to control the SARS-COV-2 pandemic are mainly focused on improving sanitation, quarantining infected people, maintaining social distance to limit transmission, and developing vaccines [12]. Despite the initial success of vaccine development, there are still hundreds of thousands of patients in urgent need of therapeutic intervention [20]. Previous studies reported that ACE2 is the main receptor for the new coronavirus to invade the human body [19]. Therefore, it is very important to find chemical drugs that directly target the ACE2, which may be able to prevent its interaction with SARS-CoV-2. After analyzing a large number of marketed vascular converting enzyme inhibitors, it was found that most drugs can only bind to ACE2 and regulate the number of ACE2 receptors, without any reports of antiviral activity. However, we found that some angiogenic inhibitors use benzimidazole as the core structure. Considering that the benzimidazole may bind to the ACE2, we searched for drugs that use benzimidazole as the core structure and with antiviral effects.
In this study, we found asitemizole, with benzimidazole as the parent nucleus, used for many years as an H1-histamine receptor antagonist, is a long-acting, non-sedating, second-generation anti-histamine, which is currently used in certain countries to treat allergy symptoms [21]. Using pharmacokinetics and pharmacodynamics (PK/PD) data from previous clinical trials, we assessed the possibility of the second-generation antihistamines being reused without modification. A typical adult astemizole dosage regimen is 100 mg/day. In animal experiments with beagle dogs and cynomolgus monkeys, after oral administration of 10 mg/kg, the maximum plasma concentrations measured are 27.0 ± 30.5 and 22.6 ± 5.44 ng/mL, respectively [23]. In this study, it can be seen that a concentration of 20 μM can significantly inhibit the invasion of pseudoviruses. Therefore, at normal dosages, these compounds are unlikely to provide the desired antiviral effect without modification. Therefore, the focus of future work is to modify these antihistamine drugs to increase their effectiveness to the nM range.
Secondly, surface plasma resonance assay and pseudovirus experiments also show that astemizole can bind to the ACE2 receptor and inhibit the invasion of pseudoviruses. However, the mechanism by which astemizole inhibits virus invasion is not yet clear. Previous research has reported that histamine receptor inhibitors have anti-filoviruses properties, and are mainly aimed at an intranuclear step of filovirus entry, similar to cathepsin inhibitor CA-074, which is known to block filoviruses in the nucleus proteolysis in the body [22].Thus, it can be considered that astemizole inhibits the virus from invading cells through the same method, but it requires more research.
In the experiment, we virtually docked the molecular structure of astemizole with the ACE2 structure and found that astemizole can bind to the ACE2 receptor. However, according to the results of the molecular docking model, the binding site of astemizole and ACE2 is the nitrogen atom on the hexahydropyridine ring, not the benzimidazole. Perhaps the benzimidazole ring can bind to ACE2 with other forces. We also docked astemizole with the H1R receptor. From the docking results, it was found that the N bound to ACE2 is also the site where astemizole binds to H1R. However, astemizole can form three hydrogen bonds with the H1R receptor, so the binding force is stronger. Not only that, it can be seen from the results of virtual docking that the binding pocket of astemizole with ACE2 is located in the center of ACE2, which is different from the binding pocket of H1R receptor located at the edge. This difference may cause different steric hindrances when astemizole binds to the proteins. This may be the reason that makes the combination of astemizole and ACE2 more difficult. This may explain the fact that astemizole cannot antagonize the invasion of SARS-CoV-2 at the same clinical dose that inhibit allergic reactions.
In conclusion, the results of this study show that antihistamines represent a huge library of potential drug candidates that can be re-used in anti-coronavirus therapies and modified to improve their effectiveness. Because their anti-coronavirus properties depend on the ACE2 and not the classic GPCR, these compounds will become ideal targets for further optimization. The focus is to strengthen antiviral properties and reduce side effects by enhancing the interaction between the drug and the ACE2 while reducing the interaction with other receptors. Efforts can be made to optimize the binding affinity of these compounds to ACE2 in the future to improve the efficacy and drug-like properties of these antihistamines.
Author statement
Xiangjun Wang: Writing – original draft, Experiment and Writing Manuscript, Jiayu Lu: Experiment and Review Manuscript, Shuai Ge: Experimental Collaboration, Yajing Hou: Intellectual Collaboration and Experimental Design, Tian Hu: Intellectual Collaboration and Experimental Design, Yuexin Lv: Experimental Collaboration, Cheng Wang: Orientation of Experiment and Review Manuscript, Huaizhen He: Orientation of Experiment and Review Manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81930096, 81903573); the Postdoctoral Research Foundation of China (No. 2018M643682); and the Natural Science Basic Research Plan in Shaanxi Province of China (No. 2020JQ-089).
Declaration of competing interest
The authors declare no competing financial interest.
References
- 1.Bimonte S., Crispo A., Amore A., Celentano E., Cuomo A., Cascella M. vol. 34. 2020. Potential antiviral drugs for SARS-cov-2 treatment: preclinical findings and ongoing clinical research; pp. 1597–1602. (Vivo). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madabhavi I., Sarkar M., Kadakol N. COVID-19. A review. Monaldi Arch. Chest Dis. 2020;90 doi: 10.4081/monaldi.2020.1298. [DOI] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y., Shang J., Graham R., Baric R.S., Li F., Gallagher T. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Hao G. The role of angiotensin-converting enzyme 2 in coronaviruses/influenza viruses and cardiovascular disease. Cardiovasc. Res. 2020;116:1932–1936. doi: 10.1093/cvr/cvaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Touyz R.M., Li H., Delles C. ACE2 the Janus-faced protein - from cardiovascular protection to severe acute respiratory syndrome-coronavirus and COVID-19. Clin. Sci. (Lond.) 2020;134:747–750. doi: 10.1042/CS20200363. [DOI] [PubMed] [Google Scholar]
- 9.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., et al. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z., Wang J., Qin Y., Zhang X., Yan X., et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the Perspectives of clinical immunologists from China. Clin. Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 12.Kruse R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020;9:72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaitin K.I. Deconstructing the drug development process: the new face of innovation. Clin. Pharmacol. Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha Y., Erez T., Reynolds I.J., Kumar D., Ross J., Koytiger G., Kusko R., Zeskind B., Risso S., Kagan E., et al. Drug repurposing from the perspective of pharmaceutical companies. Br. J. Pharmacol. 2018;175:168–180. doi: 10.1111/bph.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bansal Y., Silakari O. The therapeutic journey of benzimidazoles: a review. Bioorg. Med. Chem. 2012;20:6208–6236. doi: 10.1016/j.bmc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Tonelli M., Novelli F., Tasso B., Vazzana I., Sparatore A., Boido V., Sparatore F., La Colla P., Sanna G., Giliberti G., et al. Antiviral activity of benzimidazole derivatives. III. Novel anti-CVB-5, anti-RSV and anti-Sb-1 agents. Bioorg. Med. Chem. 2014;22:4893–4909. doi: 10.1016/j.bmc.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 19.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell M., García-Quiroz J., García-Becerra R., Barrera D., Santos N., Avila E., Ordaz-Rosado D., Rivas-Suárez M., Halhali A., Rodríguez P., et al. Astemizole synergizes calcitriol antiproliferative activity by inhibiting CYP24A1 and upregulating vdr: a novel approach for breast cancer therapy. PloS One. 2012;7 doi: 10.1371/journal.pone.0045063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misasi J., Chandran K., Yang J.Y., Considine B., Filone C.M., Cote M., Sullivan N., Fabozzi G., Hensley L., Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J. Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Back H.M., Lee J.H., Chae J.W., Song B., Seo J.W., Yun H.Y., Kwon K.I. A novel HPLC-MS/MS method for the simultaneous determination of astemizole and its major metabolite in dog or monkey plasma and application to pharmacokinetics. J. Pharmaceut. Biomed. Anal. 2015;114:121–126. doi: 10.1016/j.jpba.2015.04.036. [DOI] [PubMed] [Google Scholar]