Abstract
High metastasis of clear cell renal cell carcinoma (ccRCC) significantly influenced survival rate of ccRCC patients. Here, we intended to investigate the impacts of circular RNA ITCH (circ-ITCH) on the metastasis of ccRCC. The expression of circ-ITCH in ccRCC tissues and cells was evaluated utilizing qRT-PCR. Transwell assay and wound healing were applied to investigate migration and invasion of ccRCC cells. Target gene prediction and screening and luciferase reporter gene assays were utilized to assess downstream target genes of circ-ITCH. Western blot was utilized to detect metastasis-related protein expression. A xenograft tumor model was established to evaluate the role of circ-ITCH in vivo. Results showed that circ-ITCH was low expressed in ccRCC tissues and cells. Downregulation circ-ITCH promoted cell migration, but overexpressing circ-ITCH inhibited cell migration and invasion in OSRC-2 and SW839 cells. Mechanism investigations claimed that circ-ITCH exerted its metastasis-inhibitory activity via sponging miR-106b-5p and regulating the expression of PDCD4. Conclusively, circ-ITCH suppresses ccRCC metastasis by enforcing PDCD4 expression through binding miR-106b-5p. circ-ITCH may function as a novel diagnostic target to suppress ccRCC metastasis.
1. Introduction
Clear cell renal cell carcinoma (ccRCC), the most common renal cell carcinoma (RCC) [1], characterizes with high metastasis [2, 3]. Therefore, the 5-year survival rate of RCC patients who are diagnosed at metastatic stage only have 12% [4]. At present, it lacks targeted and effective management marker for ccRCC metastasis in clinic. There is an urgent need to search for novel diagnosis target for ccRCC metastasis.
Circular RNAs (circRNAs) are particular type of noncoding RNA molecule that featured with closed circular structure and stability [5]. circRNAs are associated with proliferation, invasion, and metastasis in cancers including colorectal cancer, lung cancer, bladder cancer, pancreatic cancer, non-small-cell lung cancer, and endometrial carcinoma [6–9]. In RCC, previous studies showed that circ-AKT3 [10], cRAPGEF5 [11], circ_000926 [12], hsa-circ-0072309 [13], circ-EGLN3 [14], circ-ZNF652 [15], has_circ_0054537 [16], circTLK1 [17], circMTO1 [18], circ-0001368 [19], circPRRC2A [20], and has_circ_0035483 [21] were involved in cancer progression. However, there is no substantive progress in the RCC treatment yet. So it is necessary for RCC to find new circRNAs target. Among these circRNAs, circular RNA ITCH (circ-ITCH) acts as a tumor suppressor in the occurrence of several tumors, such as hepatocellular carcinoma [22], ovarian cancer [23], oral squamous cell carcinoma [24], cervical cancer [25], osteosarcoma [26], and prostate cancer [27]. But the role of circ-ITCH in ccRCC is not clarified now.
Programmed cell death 4 (PDCD4), a tumor suppressor, is firstly clarified as a gene that induces apoptosis in murine cell line [28]. It is frequently downregulated in cancers and inhibits tumor promotion, development, proliferation, invasion, and metastasis [29, 30]. Studies showed that PDCD4 was downregulated in RCC patients and strongly associated with tumor stage, tumor grade, tumor metastasis, and tumor-related death [31]. However, the corresponding mechanism in RCC is not understood clearly. Hao et al. [30] found that circular RNA ITCH could regulate PDCD4 to inhibit cell proliferation and promote cell apoptosis in oral squamous cell carcinoma. Whether circ-ITCH and PDCD4 control ccRCC development is not reported.
In this study, we examined the level of circ-ITCH in ccRCC tissues and cells and found that circ-ITCH was downexpressed in ccRCC. Further studies were conducted to explore the function of circ-ITCH in ccRCC by utilizing the ccRCC cell lines and xenograft model. Systematic studies revealed that circ-ITCH hindered ccRCC migration and invasion by sponging miR-106b-5p to regulate PDCD4. Consequently, circ-ITCH might become a novel target to inhibit ccRCC metastasis.
2. Materials and Methods
2.1. Tissue Samples
Fifty-four pairs of ccRCC tissues and paired adjacent normal kidney tissues were collected for research in this work; the characteristic information of samples were listed in supplementary file (Table S1). The tissue samples were quickly frozen in liquid nitrogen after surgery and stored until use. Histological and pathological diagnoses of the specimens were confirmed according to the 2016 World Health Organization Consensus Classification and Staging System of Renal Tumor and Fuhrman grade by two experienced pathologists. All specimens were obtained with informed consent of the patients and approved by the Ethics Committee of the Affiliated Huai'an Hospital of Xuzhou Medical University.
2.2. Cell Lines and Culture
Human normal kidney cell line HK-2 and ccRCC cell lines OSRC-2, A498, SW839, 786-O, Caki-1, and GRC-1 were purchased from the American Type Culture Collection (Shanghai, China). All cell lines were cultured in RPMI-1640 medium, which contained 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco, USA). Cells were cultured in CO2 incubator (37°C, 5% CO2).
2.3. Cell Transfection
Overexpression vector or downexpression vector of circ-ITCH was acquired by cloning the sequence of circ-ITCH into the pcDNA vector (Shanghai, China). miR-106b-5p mimic, miR-106b-5p inhibitor, si-PDCD4, OE-PDCD4, and their respective controls were purchased from RiboBio (Guangzhou, China). OSRC-2 and SW839 cells were transfected using lipofectamine 2000 (Invitrogen, USA).
2.4. Transwell Assay
Cell migration and invasion were performed by Transwell chambers (BD Biosciences, USA). After 48 h transfection of plasmid, OSRC-2 and SW839 cells were resuspended in 200 μL serum-free RPMI-1640 medium with the density of 3 × 105, then were placed in the upper chamber with 8 μm pore filters (Millipore, Germany). And RPMI-1640 medium with 10% FBS was added to the lower chambers. After incubation, cells were washed three times with PBS and handled by utilizing the methanol to fix and crystal violet to stain (Gibco, USA). Finally, cell photos were graphed under the microscope (Canon, Japan) at ×200 magnification.
2.5. Wound Healing
Cell migration was also tested by wound healing assay. OSRC-2 and SW839 cells (3 × 105 cells per well) separately were seeded into six-well plates. And lines were drawn on the back of the plate via marker pens. When cells covered the plate surface, the cells were treated overnight with 1% FBS. Then, 200 mL sterile pipette tips were used to draw a straight line on the cell surface. The fallen cells were washed with 2 mL of PBS for three times. After that, the cells were cultured with serum-free medium in CO2 incubator (37°C, 5% CO2). After 24 hours, the cells were photographed with the ImageJ software.
2.6. Luciferase Report Assay
The luciferase reporter vector pmirGLO was used to clone the full length of 3′-UTR of circ-ITCH and PDCD4. Then, the recombination plasmid (circ-ITCH-WT or circ-ITCH-MUT, PDCD4-WT or PDCD4-MUT) was transfected with miR-106b-5p mimic or miR-106b-5p NC into OSRC-2 and SW839 cells separately. And cells were incubated for 48 h. Then, the luciferase activities were analyzed via dual-luciferase reporter assay (Promega, USA).
2.7. Quantitative Real-Time PCR
Total RNA was isolated by using TRIzol reagent (Invitrogen, Canada). Reverse transcription was conducted by Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Switzerland). Real-time PCR was operated using SYBR ® Green PCR Kit (Qiagen). The expression levels of the fold changes of the genes were calculated by 2−ΔΔCt method and normalized with GAPDH (5′GGAGCGAGATCCCTCCAAAAT3′, 5′GGCTGTTGTCATACTTCTCATGG3′).
2.8. Western Blot
RIPA Lysis Buffer (HY-K1001, MedChem Express) was used to separate total protein in cells. The protein was transferred onto PVDF membranes (Millipore, MA), which were then blocked with skim milk for 1 h at room temperature. Else, the primary antibodies (anti-GAPDH, anti-E-cadherin, anti-N-cadherin, anti-PDCD4) were added for overnight incubation, followed by HRP-conjugated secondary antibody. Protein bands were analyzed by the ImageJ software.
2.9. In Vivo Studies
12 eight-week old male BALB/c mice were used to establish ccRCC-bearing xenograft models and were randomly divided into the NC group and OE-circ-ITCH group. Almost 3 × 105 OE-NC or OE-circ-ITCH RCC cells were injected into the armpits of nude mice. Mice were sacrificed after 8 weeks, and tumor width and length were measured by digital calipers. The tumor volumes were calculated using the formula: volume = (length × width2)/2. Tumor weights were collected after mice were sacrificed.
2.10. Statistical Analysis
Data analysis was carried out by the OriginPro 9 and the GraphPrism software. The data is presented as the mean ± SD. Group differences were tested for statistical significance using Student's t-test. p < 0.05 represents a significant difference. For each group, there were at least three replicates.
3. Results
3.1. circ-ITCH Expression Is Downregulated in ccRCC
We firstly detected the circ-ITCH expression in ccRCC tissues and cells by qRT-PCR. The results are shown in Figures 1(a) and 1(b); circ-ITCH levels were significantly reduced in ccRCC tissues than those in nearby normal tissues. And circ-ITCH levels in ccRCC cell lines were significantly downregulated comparing with normal kidney cell line. Above results show that circ-ITCH is downregulated in ccRCC.
Figure 1.
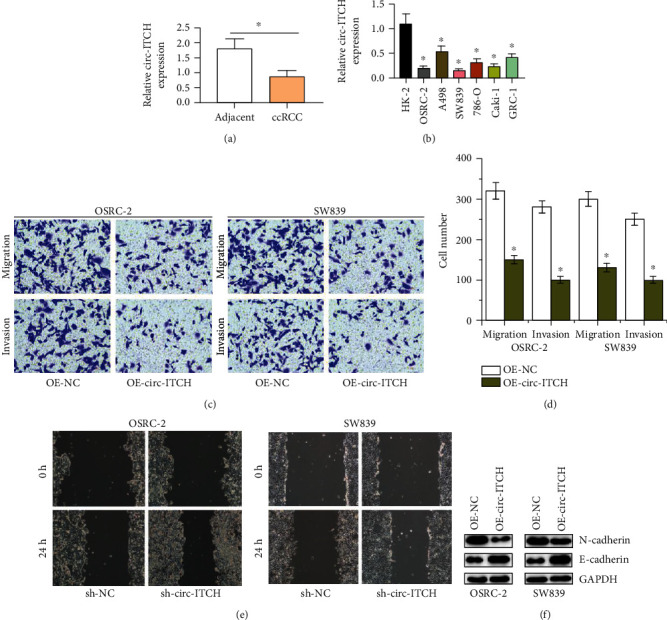
circ-ITCH inhibits ccRCC cell migration and invasion. (a, b) The expression of circ-ITCH in ccRCC tissues and cells by qT-PCR. (c, d) Transwell assay determines the role of circ-ITCH in migration and invasion of OSRC-2 and SW839 cells. (e) Wound healing detects the effect of circ-ITCH downregulation on the migration abilities of OSRC-2 and SW839 cells. (f) The expressions of E-cadherin and N-cadherin in OSRC-2 and SW839 cells transfected with OE-circ-ITCH.
3.2. circ-ITCH Inhibits Migration and Invasion of ccRCC Cells In Vitro
Then, to explore the effects of the circ-ITCH dysregulation on the ccRCC metastasis, we constructed circ-ITCH-overexpressing OSRC-2 and SW839 cell lines. Transwell assay showed that the overexpression of circ-ITCH inhibited the abilities of migration and invasion in OSRC-2 and SW839 cells, as shown in Figures 1(c) and 1(d). Wound healing assay showed that circ-ITCH knockdown promoted cell migration in RCC (Figure 1(e)). And the metastasis associated protein E-cadherin expressions were upregulated, and the N-cadherin expressions were downregulated in circ-ITCH-overexpressing ccRCC cells in Figure 1(f). These data suggest that circ-ITCH could inhibit migration and invasion of ccRCC cells.
3.3. circ-ITCH Acts as a Sponge for miR-106b-5p in ccRCC Cells
To study the mechanism of circ-ITCH mediated the migration and invasion of ccRCC cells, we applied the miRNA target prediction tool (StarBase Database, http://starbase.sysu.edu.cn/). Firstly, we identified the potential binding sites for circ-ITCH and miR-106b-5p in Figure 2(a). Then, we found that luciferase activity was significantly reduced in ccRCC cells stimulated with circ-ITCH-WT and miR-106b-5p, but this effect did not exist in ccRCC cells stimulated with circ-ITCH-MUT and miR-106b-5p, as shown in Figure 2(b). Furthermore, we measured the expression of miR-106b-5p in ccRCC tissues and noticed that miR-106b-5p was significantly upregulated in ccRCC tissues in Figure 2(c). Subsequently, functional studied showed that inducing the expression of miR-106b-5p would reverse the inhibitory effect of circ-ITCH overexpression on the migration and invasion abilities of ccRCC cells in Figures 2(d) and 2(e). Wound healing in Figure 2(f) showed that circ-ITCH knockdown promoted RCC cells migration, but extra miR-106b-5p inhibitor supplement in RCC cells transfected with sh-circ-ITCH would inhibited cell migration compared with RCC cells transfected with sh-circ-ITCH. The expressions of E-cadherin and N-cadherin were reversely regulated by promoting the miR-106b-5p expression in circ-ITCH overexpressing OSRC-2 and SW839 cells than that of circ-ITCH overexpressing OSRC-2 and SW839 cells, as shown in Figure 2(g). These findings indicate that circ-ITCH sponges miR-106b-5p in ccRCC cells to regulate cell metastasis.
Figure 2.
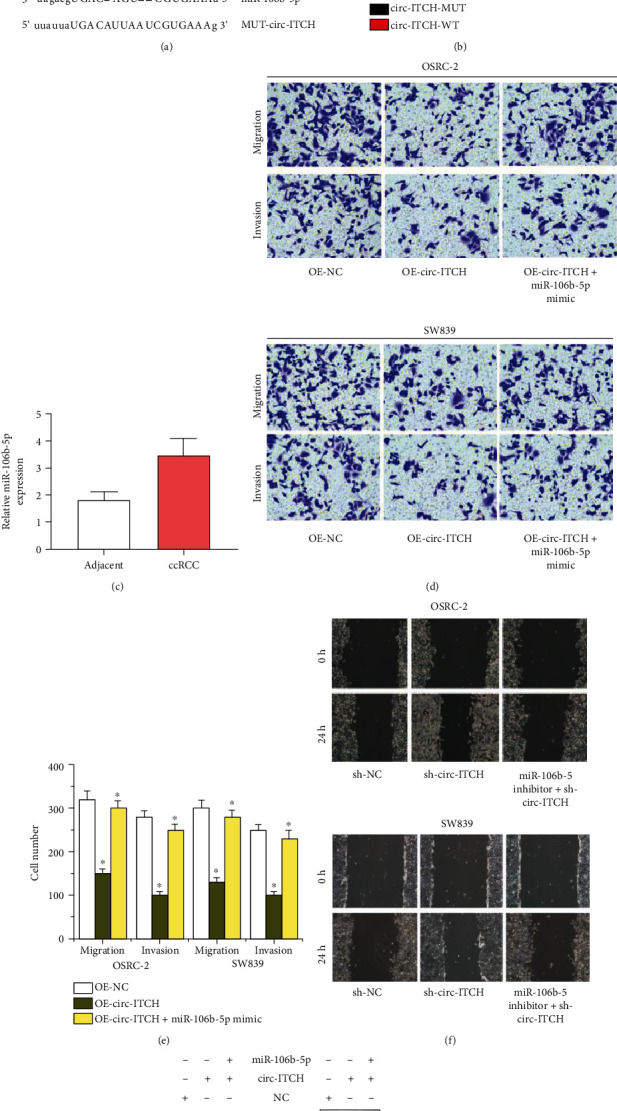
circ-ITCH inhibits ccRCC cell metastasis via sponging miR-106b-5p. (a) The predicted binding sites between circ-ITCH and miR-106b-5p. (b) The dual luciferase reporter assay detects the luciferase activity of circ-ITCH-WT/circ-ITCH-MUT in OSRC-2 and SW839 cells cotransfected with miR-106b-5p mimics. (c) The expression of miR-106b-5p in ccRCC tissues. (d, e) Transwell assay detects migration and invasion abilities of OSRC-2 and SW839 cells transfected with OE-NC, OE-circ-ITCH, OE-circ-ITCH, and miR-106b-5p mimic. (f) Wound healing detects migration abilities of OSRC-2 and SW839 cells transfected with sh-NC, sh-circ-ITCH, and sh-circ-ITCH + miR-106b-5p inhibitor. (g) Western blot analyzes the expressions of E-cadherin, N-cadherin in OSRC-2, SW839 cells transfected with NC, OE-circ-ITCH, OE-circ-ITCH, and miR-106b-5p mimic.
3.4. miR-106b-5p Directly Targets PDCD4 in ccRCC Cells
According to the StarBase database, PDCD4 was predicted as a target for miR-106b-5p in Figure 3(a). By dual luciferase reporting test, we found that luciferase activity was significantly reduced in ccRCC cells stimulated with PDCD4-WT and miR-106b-5p, but there were no evident differences in ccRCC cells transfected PDCD4-MUT and miR-106b-5p, as shown in Figure 3(b). In renal cancer patients, it was negatively related between the PDCD4 expressions and the miR-106b-5p expressions (Figure 3(c)). Western blot also showed that ccRCC cells transfected with miR-106b-5p mimic inhibited the PDCD4 expression, but ccRCC cells transfected with miR-106b-5p inhibitor induced the PDCD4 expression, as shown in Figure 3(d). Furthermore, Transwell assay suggests that the inhibition of miR-106b-5p would inhibit the migration and invasion of ccRCC cells. However, PDCD4 knockout in ccRCC cells transfected with miR-106b-5p inhibitor could induce the migration and invasion of ccRCC cells compared with ccRCC cells transfected with miR-106b-5p inhibitor, as shown in Figures 3(e) and 3(f). Wound healing showed that miR-106b-5p overexpression promoted RCC cells migration, but extra PDCD4 overexpression supplement in RCC cells transfected with miR-106b-5p mimic would inhibited cell migration compared with RCC cells transfected with miR-106b-5p mimic, as shown in Figure 3(g). Else, the miR-106b-5p inhibition induced the E-cadherin expression and inhibited the N-cadherin expression in ccRCC cells. However, PDCD4 knockout in ccRCC cells transfected with miR-106b-5p inhibitor could exerted an opposite effect on the E-cadherin and N-cadherin expressions compared with ccRCC cells transfected with miR-106b-5p inhibitor in Figure 3(h). Above results show that miR-106b-5p targets PDCD4 to mediate ccRCC cell migration and invasion.
Figure 3.
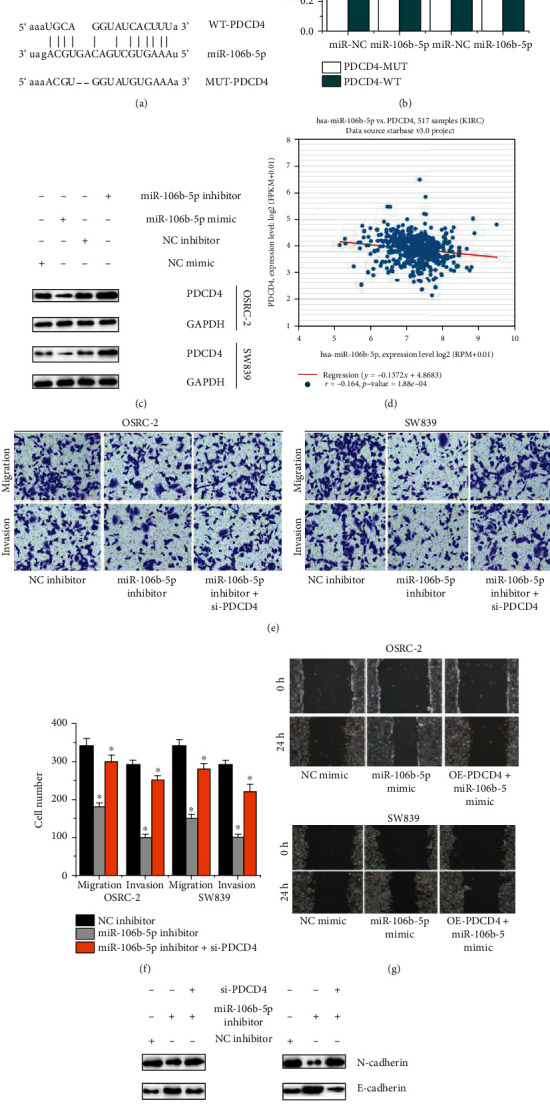
PDCD4 mediates miR-106b-5p-dependent ccRCC cell migration and invasion. (a) The predicted binding sites between miR-106b-5p and PDCD4. (b) The dual luciferase reporter assay detects the luciferase activity of PDCD4-WT/PDCD4-MUT in OSRC-2 and SW839 cells cotransfected with miR-106b-5p mimics. (c) The relation of the miR-106b-5p expressions and PDCD4 expressions in renal cancer. (d) Western blot determines the PDCD4 expressions in OSRC-2 and SW839 cells transfected with miR-106b-5p mimics and miR-106b-5p inhibitor. (e, f) Transwell assay detects migration and invasion abilities OSRC-2 and SW839 cells transfected with NC inhibitor, miR-106b-5p inhibitor, si-PDCD4, and miR-106b-5p inhibitor. (g) Wound healing detects migration abilities of OSRC-2 and SW839 cells transfected with NC mimic, miR-106b-5p mimic, OE-PDCD4, and miR-106b-5p mimic. (h) Western blot analyzes the expressions of E-cadherin, N-cadherin in OSRC-2, SW839 cells transfected with NC inhibitor, miR-106b-5p inhibitor, si-PDCD4, and miR-106b-5p inhibitor.
3.5. circ-ITCH Overexpression Restrained the Growth of ccRCC Cells In Vivo
To verify the role of circ-ITCH in vivo, we established a xenograft tumor mouse model of ccRCC. As shown in Figures 4(a)–4(c), the overexpression of circ-ITCH would significantly decrease the tumor volume and tumor weight. The data provide that circ-ITCH overexpression could inhibit the development of ccRCC.
Figure 4.
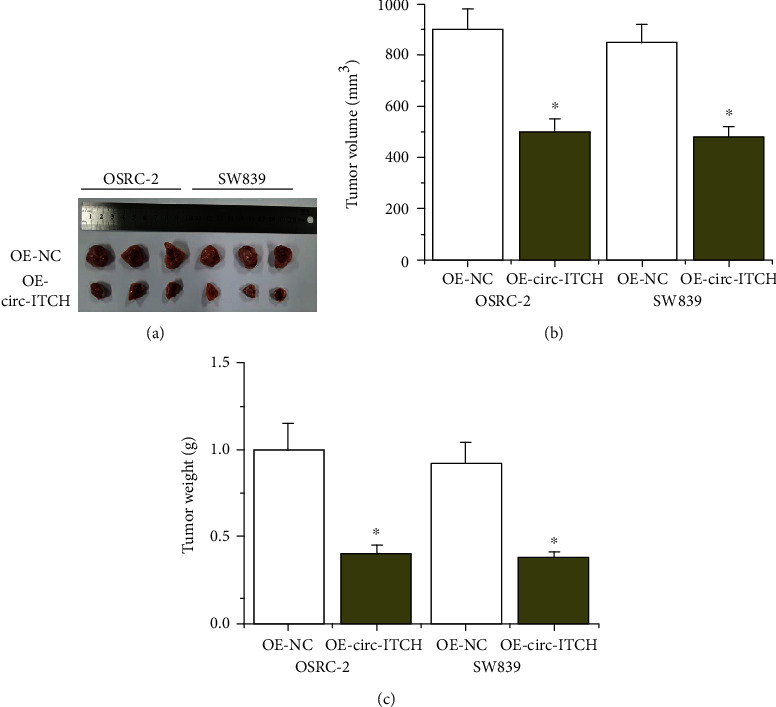
circ-ITCH exerts oncogenic effects of ccRCC cells in vivo. (a–c) Tumor specimens, volume, and weight in nude mice that injected with OE-circ-ITCH cells.
4. Discussion
circRNAs are reported to play important roles in taking part in tumorigenesis. As a common tumor suppressor in multiple cancers [22–27], the function of circ-ITCH in ccRCC is not clear. In this research, we firstly found that circ-ITCH levels were reduced in ccRCC tissues and ccRCC cell lines, which was similar with the fact that it existed the abnormal expression of circ-ITCH in other cancers [32]. And we observed that the downregulation of circ-ITCH promoted RCC cell migration. However, the upregulation of circ-ITCH suppressed the migration and invasion of ccRCC cell lines, decreased the expression of N-cadherin, and boosted the expression of E-cadherin.
To explore how circ-ITCH functioned in ccRCC, the deep investigations and research were operated. circRNAs can sponge miRNAs to regulate the mRNA expression in cancers, which is involved in the occurrence of cancers [33, 34]. We achieved the predicting binding sites between circ-ITCH and miR-106b-5p. By dual luciferase report assay, it is verified that miR-106b-5p coculture with ccRCC cells decreased the luciferase activity of circ-ITCH-WT not circ-ITCH-MUT. Different with the findings that circ-ITCH was downregulated in ccRCC tissues, miR-106b-5p was upregulated in ccRCC tissues. And circ-ITCH could mediate ccRCC cell migration and invasion via regulating miR-106b-5p. In addition, we found that miR-106b-5p negatively regulated PDCD4 in ccRCC cells and renal cancer patients. miR-106b-5p regulated ccRCC cell migration and invasion by targeting PDCD4. In previous works, PDCD4 was reported to be reduced in human renal cell carcinoma patients [27]. miR-21 downregulated PDCD4 to contribute to the RCC progression [35–37], which was similar with our findings that PDCD4 targeted miR-106b-5p to promote the ccRCC development. In vivo, we overexpressed circ-ITCH in BALB/c mice stimulated with ccRCC cells and noticed that the growth of tumors was then inhibited.
In this work, it reports the role of circ-ITCH in ccRCC migration and invasion. However, it still exists some limitations, such as small sample size, the unclear clinical diagnose effect of circ-ITCH in ccRCC metastasis. Subsequently, the samples of ccRCC patients will be increased to study the expression of circ-ITCH. It is worth exploring the relationship between circ-ITCH expression and tumor metastasis levels in ccRCC patients.
In conclusion, we demonstrated that circ-ITCH was a tumor suppressor in ccRCC via miR-106b-5p/PDCD4 pathway. It announced the novel molecular basis of circRNAs in the research of ccRCC metastasis.
Acknowledgments
We thank the Affiliated Huai'an Hospital of Xuzhou Medical University for providing ccRCC tissues and mice in this research.
Data Availability
I confirm that I have included a citation for available data in my references section.
Conflicts of Interest
No conflicts of interest.
Authors' Contributions
PG, YH, and YH analyzed data and wrote manuscript. QL designed the manuscript. HW revised the manuscript. All authors agreed with final manuscript. Ping Gao, Yong Huang, and Yanmei Hou contributed equally and are co-first authors.
Supplementary Materials
Clinical characteristics of 44 ccRCC patients in this work was listed in Table S1 in uploaded supplementary file.
References
- 1.Padala S. A., Barsouk A., Thandra K. C., et al. Epidemiology of renal cell carcinoma. World Journal of Oncology. 2020;11(3):79–87. doi: 10.14740/wjon1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobczuk P., Brodziak A., Khan M. I., et al. Choosing the right animal model for renal cancer research. Translational Oncology. 2020;13(3, article 100745) doi: 10.1016/j.tranon.2020.100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevrier S., Levine J. H., Zanotelli V. R. T., et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169(4):736–749.e18. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.SEER Explorer. March 2020, https://seer.cancer.gov/explorer/index.html.
- 5.Memczak S., Jens M., Elefsinioti A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z., Huang J., Feng Y., Li Z., Jiang Y. Screening and bioinformatics analysis of a ceRNA network based on the circular RNAs, miRNAs, and mRNAs in pan-cancer. Cancer Medicine. 2020;9(19):7279–7292. doi: 10.1002/cam4.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang P. C., Bu S. R. Clinical value of circular RNAs and autophagy-related miRNAs in the diagnosis and treatment of pancreatic cancer. Hepatobiliary & Pancreatic Diseases International. 2019;18(6):511–516. doi: 10.1016/j.hbpd.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y., Zheng R., Chen J., Ning D. CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC) Cancer Cell International. 2020;20(1):p. 289. doi: 10.1186/s12935-020-01390-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Q., He T., Yuan H. Hsa_circ_0002577 promotes endometrial carcinoma progression via regulating miR-197/CTNND1 axis and activating Wnt/β-catenin pathway. Cell Cycle. 2019;18(11):1229–1240. doi: 10.1080/15384101.2019.1617004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue D., Wang H., Chen Y., et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Molecular Cancer. 2019;18(1):p. 151. doi: 10.1186/s12943-019-1072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q., Liu T., Bao Y., et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Letters. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Zhang D., Yang X. J., Luo Q. D., et al. Down-regulation of circular RNA_000926 attenuates renal cell carcinoma progression through miRNA-411-dependent CDH2 inhibition. The American Journal of Pathology. 2019;189(12):2469–2486. doi: 10.1016/j.ajpath.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Chen T., Shao S., Li W., Liu Y., Cao Y. The circular RNA hsa-circ-0072309 plays anti-tumour roles by sponging miR-100 through the deactivation of PI3K/AKT and mTOR pathways in the renal carcinoma cell lines. Artificial Cells, Blood Substitutes, and Biotechnology. 2019;47(1):3638–3648. doi: 10.1080/21691401.2019.1657873. [DOI] [PubMed] [Google Scholar]
- 14.Lin L., Cai J. Circular RNA circ-EGLN3 promotes renal cell carcinoma proliferation and aggressiveness via miR-1299-mediated IRF7 activation. Journal of Cellular Biochemistry. 2020;121(11):4377–4385. doi: 10.1002/jcb.29620. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Guo Y. Retracted article: silencing circular RNA-ZNF652 represses proliferation and EMT process of renal carcinoma cells via raising miR-205. Artificial Cells, Blood Substitutes, and Biotechnology. 2020;48(1):648–655. doi: 10.1080/21691401.2020.1725532. [DOI] [PubMed] [Google Scholar]
- 16.Li R., Luo S., Zhang D. Circular RNA hsa_circ_0054537 sponges miR-130a-3p to promote the progression of renal cell carcinoma through regulating cMet pathway. Gene. 2020;754, article 144811 doi: 10.1016/j.gene.2020.144811. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Huang C., Zou Y., Ye J., Yu J., Gui Y. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Molecular Cancer. 2020;19(1):p. 103. doi: 10.1186/s12943-020-01225-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Li K., Wan C. L., Guo Y. Circular RNA circMTO1 suppresses RCC cancer cell progression via miR9/LMX1A axis. Technology in Cancer Research & Treatment. 2020;19, article 153303382091428 doi: 10.1177/1533033820914286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Wu D., Ding T. Circular RNA circ_0001368 inhibited growth and invasion in renal cell carcinoma by sponging miR-492 and targeting LATS2. Gene. 2020;753, article 144781 doi: 10.1016/j.gene.2020.144781. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Yang F. Q., Sun C. M., et al. circPRRC2A promotes angiogenesis and metastasis through epithelial-mesenchymal transition and upregulates TRPM3 in renal cell carcinoma. Theranostics. 2020;10(10):4395–4409. doi: 10.7150/thno.43239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan L., Liu G., Cao H., Zhang H., Shao F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochemical and Biophysical Research Communications. 2019;519(1):172–178. doi: 10.1016/j.bbrc.2019.08.093. [DOI] [PubMed] [Google Scholar]
- 22.Wu M., Deng X., Zhong Y., et al. MafF is regulated via the circ-ITCH/miR-224-5p axis and acts as a tumor suppressor in hepatocellular carcinoma. Oncology Research. 2020;28(3):299–309. doi: 10.3727/096504020X15796890809840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C., Xu X., Yang Q., Liang L., Qiao S. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell International. 2020;20(1):p. 336. doi: 10.1186/s12935-020-01420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao C., Wangzhou K., Liang Z., et al. Circular RNA ITCH suppresses cell proliferation but induces apoptosis in oral squamous cell carcinoma by regulating miR-421/PDCD4 axis. Cancer Management and Research. 2020;12:5651–5658. doi: 10.2147/CMAR.S258887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Guo R., Liu Q., Sun J., Wang H. Circular RNA Circ-ITCH inhibits the malignant behaviors of cervical cancer by microRNA-93-5p/FOXK2 axis. Reproductive Sciences. 2020;27(3):860–868. doi: 10.1007/s43032-020-00140-7. [DOI] [PubMed] [Google Scholar]
- 26.Ren C., Liu J., Zheng B., Yan P., Sun Y., Yue B. The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artificial Cells, Blood Substitutes, and Biotechnology. 2019;47(1):3359–3367. doi: 10.1080/21691401.2019.1649273. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Wang R., Wu Z., Bai P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell International. 2019;19(1):p. 328. doi: 10.1186/s12935-019-0994-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Shibahara K., Asano M., Ishida Y., Aoki T., Koike T., Honjo T. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene. 1995;166(2):297–301. doi: 10.1016/0378-1119(95)00607-9. [DOI] [PubMed] [Google Scholar]
- 29.Hu X., Wang Y., Liang H., et al. miR-23a/b promote tumor growth and suppress apoptosis by targeting PDCD4 in gastric cancer. Cell Death & Disease. 2017;8(10, article e3059) doi: 10.1038/cddis.2017.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuhashi S., Manirujjaman M., Hamajima H., Ozaki I. Control mechanisms of the tumor suppressor PDCD4: expression and functions. International Journal of Molecular Sciences. 2019;20(9):p. 2304. doi: 10.3390/ijms20092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X., Xin S., Yang D., et al. Down-regulation of PDCD4 expression is an independent predictor of poor prognosis in human renal cell carcinoma patients. Journal of Cancer Research and Clinical Oncology. 2012;138(3):529–535. doi: 10.1007/s00432-011-1121-y. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Ge Y. Z., Xu L., Jia R. Circular RNA ITCH: a novel tumor suppressor in multiple cancers. Life Sciences. 2020;254, article 117176 doi: 10.1016/j.lfs.2019.117176. [DOI] [PubMed] [Google Scholar]
- 33.Verduci L., Strano S., Yarden Y., Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Molecular Oncology. 2019;13(4):669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panda A. C. Circular RNAs act as miRNA sponges. Advances in Experimental Medicine and Biology. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 35.Yuan H., Xin S., Huang Y., et al. Downregulation of PDCD4 by miR-21 suppresses tumor transformation and proliferation in a nude mouse renal cancer model. Oncology Letters. 2017;14(3):3371–3378. doi: 10.3892/ol.2017.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Xin S., He Z., et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cellular Physiology and Biochemistry. 2014;33(6):1631–1642. doi: 10.1159/000362946. [DOI] [PubMed] [Google Scholar]
- 37.Bera A., Das F., Ghosh-Choudhury N., Kasinath B. S., Abboud H. E., Choudhury G. G. microRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKβ- mTORC1 axis to regulate renal cancer cell invasion. Experimental Cell Research. 2014;328(1):99–117. doi: 10.1016/j.yexcr.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of 44 ccRCC patients in this work was listed in Table S1 in uploaded supplementary file.
Data Availability Statement
I confirm that I have included a citation for available data in my references section.


