Abstract
Dabigatran is a novel direct oral anticoagulant agent, whose plasma concentration is closely related to bleeding risk. Genetic polymorphisms can affect the level of plasma dabigatran. The purpose of this study was to understand the relationship between dabigatran-related genes and the plasma level of dabigatran in healthy Chinese subjects after taking a single oral dose. This study was performed with a single-center, single-dose, randomized, open-label, and four-period crossover trial design under both fasting and fed conditions. A total of 106 eligible healthy subjects were enrolled in the study and 104 were genotyped. One-way analysis of variance (ANOVA) was used to compare pharmacokinetic parameters among different genotypes and linear regression was applied to explore the multiplicative interaction between variables. In this study, we found that the genotype frequencies of CES1 rs2244613 and CES1 rs8192935 were significantly different between Chinese and Caucasians, but the genotype frequencies of ABCB1 rs1045642 and ABCB1 rs4148738 were similar in both populations. CES1 rs8192935 were associated with the peak concentration of dabigatran. There was no significant gender difference in the exposure level of dabigatran. Furthermore, food significantly delayed the absorption of dabigatran but had little effect on Cmax and AUC0-∞.
Keywords: anticoagulants, dabigatran, genetics, pharmacogenetics, CES1, ABCB1
Introduction
Dabigatran is a novel direct oral anticoagulant agent (NOAC),1 first marketed in Europe and then approved by the US Food and Drug Administration (FDA) in 2010 to prevent stroke risk in patients with non-valvular atrial fibrillation (AF).2 In 2014, an expanded dabigatran indication for preventing deep vein thrombosis and pulmonary thromboembolism was approved.3 As a specific inhibitor of thrombin, it can reversibly compete to inhibit both free and clot-bound thrombin and the thrombin-induced platelet activation. Dabigatran etexilate is an oral prodrug that can be quickly converted to dabigatran by esterases, especially liver esterase carboxylesterase (CES) 1. Dabigatran etexilate is a substrate of the P-glycoprotein (P-gp), encoded by the adenosine-triphosphate (ATP)-binding cassette sub-family B member (ABCB)1 genes. Potent P-gp inhibitors could increase the bioavailability of dabigatran by 12% to 23%.1,4,5 Studies indicated that dabigatran had a predictable pharmacokinetic profile, allowing for a fixed-dose regimen without the need for coagulation monitoring. However, wide inter-individual variability in pharmacokinetic and pharmacodynamic responses to dabigatran have recently been reported.6,7 Furthermore, dabigatran was also observed to cause clinical bleeding.8 It has been confirmed that genetic polymorphisms affect the plasma level of dabigatran,6,9,10 which is closely associated with bleeding risk.
The purpose of this study was to explore the relationship between single-nucleotide polymorphisms (SNPs) of dabigatran-related genes and the plasma level of dabigatran in healthy Chinese subjects after taking a single oral dose. We studied the relationship between the allele frequencies of single nucleotide polymorphisms in dabigatran-related genes and pharmacokinetic parameters in Chinese and compared the allele frequencies of these SNPs between Chinese and Caucasians. We hope that our findings could provide a basis for the safe use of dabigatran in future clinical practice.
Materials and Methods
Ethics
The study was designed following the Declaration of Helsinki, Good Clinical Practice (GCP) guidelines, and other related guiding principles. The study protocol was reviewed and approved by the independent ethics committee of Beijing Hospital China (Ethics approval number:2019BJYYEC-081–01). Informed consent was obtained from all individual subjects included in the study. According to the informed consent terms, all study results could be used for scientific purposes without uncovering personal identifiers. This study was registered at http://www.chinadrugtrials.org.cn/ (Identifier: CTR20191308). Before the screening, informed consent was obtained from all subjects.
Investigational Drug
The test product (T): dabigatran etexilate 150mg was manufactured by Chengdu Yuandong biopharmaceutical Co., Ltd.
The reference product (R): dabigatran etexilate 150mg (trade name: tibetaxel ®) was manufactured by Berlin Ingelheim, Germany.
Study Population
A total of 106 healthy Chinese subjects were recruited in this study. The subjects were selected according to the inclusion and exclusion criteria:males and females aged 18–45 years; body weight more than 50 kg and 45 kg for males and females, respectively; body mass index at 19–28 kg/m2; and assessed as healthy based on their medical history, physical examination, vital signs, electrocardiogram and laboratory tests at the screening.
Study Design
This study was performed with a single-center, single-dose, randomized, open-label, four-period crossover trial design, under both fasting and fed conditions. As a candidate gene approach based study, the relationship between SNPs of the CES1 and ABCB1 genes and the drug concentration in the plasma was explored. Five SNPs related to variability in dabigatran disposition were indexed in the Pharmacogenomics Knowledgebase (https://www.pharmgkb.org/): CES1 rs2244613, CES1 rs8192935, ABCB1 rs1045642 (C3435T changes in exon 26), rs2032582 (G2677T/A or Ala893Thr, located in exon 21) and ABCB1 rs4148738.
In the fasting study, 68 subjects were randomized into two groups (group A and group B), and in the fed study, 38 subjects were randomized into two groups (group C and group D). All of them underwent four study periods: taking a single oral dose of 150mg dabigatran etexilate (the test product or the reference product) with 240 mL water at each period. The administration sequence of group A and C was T-R-T-R whereas the administration sequence of group B and D was R-T-R-T. Each period was separated by a washout period of 7 days.
In each period, subjects were hospitalized one day before dosing. They were asked to fast for at least 10 hours before dosing and keep their body upright 4 hours after dosing. Drinking was also restricted 1 hour before dosing and 2 hours after dosing. Same standard meals were served to all the subjects during the study in the four periods. In the fed study, the subjects were given an extra standard high–fat breakfast which contained 800–1000 kcal (500–600 kcal fat, 150 kcal protein, and 250 kcal carbohydrates), 30 minutes before administration of the study drug.
Sample Collection and Analysis
In the fasting study, 4 mL of venous blood was collected at 0 h (before taking the drug) and 30 mins, 45 mins, 1 h, 1.5 h, 2 h, 2.5 h, 3 h, 4 h, 6 h, 8 h, 12 h, 24 h, 36 h, 48 h post-dose during each of the four periods. In the fed study, 4 mL of the venous blood was collected at 0 h (before taking the drug) and 30 min, 1.0 h, 1.5 h, 2 h, 2.5 h, 3 h, 3.5h, 4 h, 4.5h, 5h, 6 h, 8 h, 12 h, 24 h, 36 h, 48 h post-dose.
Plasma concentrations of unconjugated dabigatran and the total dabigatran were determined by a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method, at Fanweixi Pharmaceutical Technology Co., Ltd (Chengdu, China). LC-MS/CS is the gold standard test for measuring all DOAC concentrations.12 A sensitive and specific LC-MS/MS method for the determination of dabigatran in human plasma was established, and the method was systematically verified. The plasma samples were pretreated by the protein precipitation method, and dabigatran-13C6 was selected as the internal standard (IS). The analyte dabigatran and IS were separated on Chr.C-051 (GL Sciences-AQ-C18 (2.1 ×150mm, 3µm) with mobile phase of 15 mM ammonium formate + 0.1% formic acid aqueous solution (phase A), acetonitrile: water (containing 150 mM ammonium formate, 1% formic acid) = 9:1 (phase B) in gradient elution. The method exhibited good linearity over the concentration range of 1.000ng/mL~300.00ng/mL. The lower limit of quantification for dabigatran was 1.000ng/mL. The dabigatran in plasma was not interfered by other endogenous substances. Besides, a venous blood sample (2 mL) was collected for genotyping. Genotype determination and haplotype analysis of the samples were performed by Bestnovo (Beijing) Medical Laboratory. Genomic DNA was extracted from the peripheral blood using “FlexGen Blood DNA Kit” (CoWin Bioscience, China) according to the manufacturer’s protocol. Genotyping was performed with the ABI PRISM® Sequence Detector 3730 (Applied Biosystems) using a kangwei 2×Taq MasterMix and QIAquick® Gel Extraction Kit as per manufacturer’s instructions. The genotyping results were obtained through software chromas 2.4.1 (technelysium).
Pharmacokinetic Analyses
The pharmacokinetic parameters of each subject of the test and the reference product were described respectively, including the unconjugated dabigatran and the total dabigatran. The pharmacokinetic parameters included Cmax, Tmax, T1/2, AUC0-t, and AUC0-∞. Pharmacokinetic parameters were estimated with non‐compartmental methods using Phoenix WinNonlin software (Version 8.1).
Haplotyping and Statistical Analyses
The pharmacokinetic parameters of total dabigatran are used for statistical analysis. We used SPSS software (version 22.0) for analysis. We get Dose/weight (DW) ratio by dividing dose by weight. Then we divided AUC0-∞ and Cmax by DW ratio for (AUC/DW, Cmax/DW). In order to make the research more accurate, the fasting and fed groups were discussed separately. To explore whether there is an interaction among gender and genotyping, we used linear regression to explore the multiplicative interaction among variables. After clearance of interaction effect, we used Student’s t-test for binary variables (such as gender) and the ANOVA test for multi-categorical (more than two groups) respectively to test the difference in the continuous variables of AUC0-∞, Tmax, T1/2, AUC0-t, and AUC0-∞. Hardy–Weinberg’s equilibrium and haplotype frequency analyses were performed by Haploview 4.2 software. P<0.05 was defined as significance.
Results
Characteristics of Subjects
A total of 106 eligible healthy subjects were enrolled, 68 for the fasting and 38 for the fed study. Among them, 104 were genotyped with the above mentioned SNPs, 67 for the fasting and 37 for the fed study. 65 subjects completed the fasting study and 3 subjects withdrew form the study. Two of the three subjects withdrew from the study just before the fourth period due to adverse events while the other one voluntarily withdrew from the study after the first period. All other subjects completed the fed study. The adverse events were vasovagal reflexes (unrelated to the Investigational drug) and papular urticaria (may not relate to the Investigational drug). The demographic characteristics of subjects are presented in Table 1.
Table 1.
Demographic Characteristics
| Fasting | Fed | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age | Weight | Height | BMI | Number | Age | Weight | Height | BMI | |
| Sex | ||||||||||
| Women | 18 | 37.9(5.4) | 57.8(7.0) | 155.7(5.2) | 23.8(2.5) | 6 | 32.0(6.7) | 59.0(6.2) | 158.7(4.8) | 23.5(2.6) |
| Men | 49 | 29.9(6.3)* | 67.8(7.6)* | 168.7(5.0)* | 23.9(2.8) | 31 | 30.5(6.7) | 70.3(7.3)* | 168.4(6.2)* | 24.8(2.2) |
Notes: Data are shown as mean (SD). *p <0.05 after a t-test vs women.
Pharmacokinetics Parameters
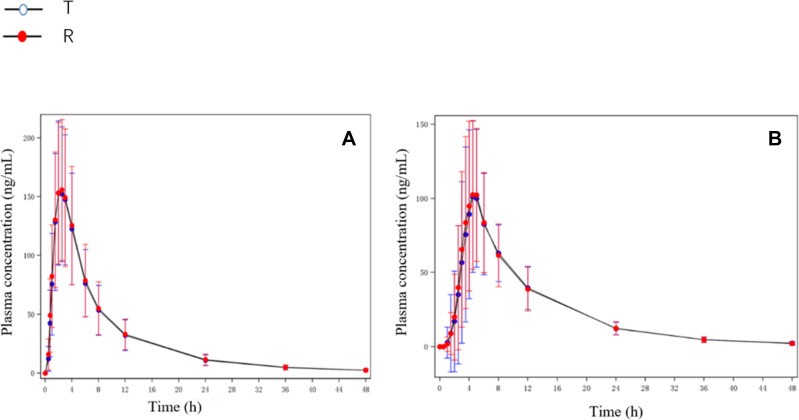
Mean plasma concentrations of the reference product and the test product at each timepoint in both the fasting and fed studies are shown in Figure 1.
Figure 1.
The mean plasma concentration–time curves of total dabigatran. (A) Total dabigatran concentration under fasting condition; (B) total dabigatran concentration under fed condition.
Genotyping Results and Analysis
Genotypes
Sixteen subjects carried the AA genotype of CES1 rs2244613, whereas 49 and 39 individuals carried the AC and CC genotype, respectively. CES1 rs8192935 AA was carried by 63 individuals, AG and GG were carried by 32 and 9 individuals, respectively. The genotype frequencies of the ABCB1 rs1045642 were shown as follows: GG genotype was carried by 35 (33.7%) individuals while AG and AA genotypes were carried by 47 (45.2%) and 22 (21.1%) individuals, respectively. On the other hand, the genotype frequencies of the ABCB1 rs4148738 were identified as follows: 34 individuals were TT genotype, 49 and 21 individuals were CT and CC genotype, respectively. The genotype frequencies of the rs2032582 in study group were: AA (21.2%), AC (29.8%), AT (16.3%), CC (19.2%), CT (9.6%) and TT (3.8%) (Table 2).
Table 2.
Allele Frequencies by Loci for the CES1 and ABCB1 in the Subjects
| Gene | SNP | Genotype | N (%) | Minor Allele | MAF (%) |
|---|---|---|---|---|---|
| CES1 | rs2244613 | CC | 39 (37.5) | A | 38.9 |
| AC | 49 (47.1) | ||||
| AA | 16 (15.4) | ||||
| rs8192935 | AA | 63 (60.6) | G | 24 | |
| AG | 32 (30.8) | ||||
| GG | 9 (8.6) | ||||
| ABCB1 | rs1045642 | GG | 35 (33.7) | A | 43.8 |
| AG | 47 (45.2) | ||||
| AA | 22 (21.1) | ||||
| rs4148738 | TT | 34 (32.7) | C | 43.8 | |
| CT | 49 (47.1) | ||||
| CC | 21 (20.2) | ||||
| rs2032582 | AA | 22 (21.2) | C | 39 | |
| AC | 31 (29.8) | ||||
| AT | 17 (16.3) | ||||
| CC | 20 (19.2) | T | 17 | ||
| CT | 10 (9.6) | ||||
| TT | 4 (3.8) |
Abbreviations: SNP, single-nucleotide polymorphism; MAF, minor allele frequency.
Pharmacokinetics
Gender and CES1 rs8192935 had an interaction effect on T1/2 in fasting condition. Gender and CES1 rs2244613 had an interaction effect on AUC in fed condition (Table 1 and 2 in Data Supplement). There was a higher recombination rate between ABCB1 rs1045642, rs2032582 and rs4148738. ABCB1 variants were merged into 7 haplotypes. None of ABCB1 haplotypes showed significant relationships with pharmacokinetic variability (Table 3 in Data Supplement).
Pharmacokinetic parameters were showed in Table 3 according to sex, mode of administration, and polymorphisms under fasting condition. Under fasting condition, sex and mode of administration were not associated with variability in drug exposure. Neither individual CES1 SNPs nor ABCB1 SNPs were significantly associated with variability on dabigatran exposure. The pharmacokinetic study to CES1 rs8192935 (A>G) allele showed that Cmax/DW of GG (83.29 kg*ng/mL*mg) carriers and AG (78.08 kg*ng/mL*mg) carriers were higher than that of AA (66.47 kg*ng/mL*mg) carriers.
Table 3.
Pharmacokinetic Parameters According to Sex, Mode of Administration and Polymorphisms in the Fasting Study
| Number | AUC0-∞/DW(kg*h*ng/mL*mg) | Cmax/DW(kg*ng/mL*mg) | Tmax(h) | t1/2(h) | Vd/F(L/kg) | CL/F(L/h*kg) | |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Female | 18 | 599.43(157.97) | 71.67(15.91) | 2.48(0.39) | 10.53(2.17) | 26.68(7.76) | 1.77(0.42) |
| Male | 49 | 581.64(136.65) | 71.25(21.45) | 2.27(0.39) | 11.06(2.05) | 29.43(10.58) | 1.83(0.51) |
| Mode of administration | |||||||
| A | 34 | 572.93(146.10) | 66.93(17.40) | 2.32(0.42) | 10.99(2.33) | 29.77(11.21) | 1.85(0.47) |
| B | 33 | 600.31(137.81) | 75.92(21.68) | 2.33(0.38) | 10.84(1.83) | 27.58(8.42) | 1.77(0.51) |
| CES1.rs2244613.c.1165.33C.A | |||||||
| GG | 23 | 568.17(136.90) | 66.94(15.69) | 2.39(0.39) | 10.88(2.11) | 29.50(10.74) | 1.87(0.51) |
| GT | 34 | 588.36(161.85) | 73.31(24.22) | 2.28(0.43) | 10.80(2.11) | 28.71(10.44) | 1.83(0.53) |
| TT | 10 | 621.78(55.46) | 75.15(9.82) | 2.34(0.31) | 11.42(2.04) | 26.77(5.90) | 1.62(0.16) |
| CES1.rs8192935.c.257.885T.C | |||||||
| AA | 41 | 560.26(138.88) | 66.47(17.78) | 2.36(0.40) | 10.74(2.20) | 29.82(11.31) | 1.90(0.52) |
| AG | 21 | 617.22(152.53) | 78.08(23.09) | 2.29(0.41) | 10.97(1.86) | 27.11(7.72) | 1.72(0.44) |
| GG | 5 | 671.53(30.54) | 83.29(10.44) | 2.25(0.32) | 12.14(1.94) | 26.00(3.38) | 1.49(0.06) |
| ABCB1.rs1045642.c.3435T.C | |||||||
| GG | 22 | 559.60(138.34) | 67.37(19.39) | 2.31(0.40) | 11.42(2.47) | 32.08(13.23) | 1.92(0.58) |
| AG | 31 | 612.81(163.77) | 76.08(22.75) | 2.35(0.39) | 10.59(1.72) | 26.43(7.02) | 1.75(0.50) |
| AA | 14 | 570.12(76.99) | 67.17(11.34) | 2.29(0.43) | 10.86(2.16) | 28.37(8.46) | 1.78(0.24) |
| ABCB1.rs4148738.c.2482.2236G.A | |||||||
| TT | 20 | 557.03(137.21) | 65.89(19.38) | 2.31(0.41) | 11.54(2.49) | 32.31(12.40) | 1.93(0.58) |
| CT | 31 | 610.05(166.35) | 76.23(22.81) | 2.35(0.40) | 10.44(1.62) | 26.40(8.25) | 1.77(0.52) |
| CC | 16 | 577.37(82.05) | 68.75(12.09) | 2.30(0.41) | 11.07(2.22) | 28.60(8.59) | 1.76(0.24) |
| ABCB1.rs2032582.c.2677T.A.G | |||||||
| AA | 16 | 557.37(82.05) | 68.75(12.09) | 2.30(0.41) | 11.07(2.22) | 28.60(8.59) | 1.76(0.24) |
| AC/AT | 31 | 610.05(166.35) | 76.23(22.81) | 2.35(0.40) | 10.44(1.62) | 26.40(8.25) | 1.77(0.52) |
| CC/CT/TT | 20 | 557.03(137.21) | 65.89(19.38) | 2.31(0.41) | 11.54(2.49) | 32.31(12.40) | 1.93(0.58) |
Notes: Data are shown as mean (SD).
Pharmacokinetic parameters were showed in Table 4 according to sex, mode of administration and polymorphisms under fed condition. Under fed condition, mode of administration were not related to variability in drug exposure. In contrast, males had higher Cmax and Tmax than females. The CES1 SNP rs8192935 was associated with Cmax and t1/2.Cmax/DW of GG (86.12 kg*ng/mL*mg) carriers was significantly higher than that of AA (50.96kg*ng/mL*mg) and AG (56.31kg*ng/mL*mg) carriers, while T1/2 of GG (11.05h) carriers was significantly higher than that of AA (9.18h) and AG (9.67h) carriers. ABCB1 rs1045642 had a significant effect on Tmax. Tmax of AG carriers was significantly higher than that of GG or AA carriers.
Table 4.
Pharmacokinetic Parameters According to Sex, Mode of Administration and Polymorphisms in the Fed Study
| Number | AUC0-∞/DW(kg*h*ng/mL*mg) | Cmax/DW(kg*ng/mL*mg) | Tmax(h) | t1/2(h) | Vd/F(L/kg) | CL/F(L/h*kg) | |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Female | 6 | 416.13(95.34) | 39.11(10.17) | 6.63(2.04) | 8.60(1.22) | 30.38(3.49) | 2.51(0.59) |
| Male | 31 | 555.76(173.21) | 59.69(21.05)* | 4.77(1.28)* | 9.71(1.33) | 28.16(12.81) | 2.01(0.85) |
| Mode of administration | |||||||
| C | 18 | 525.06(195.05) | 55.39(22.24) | 4.97(1.26) | 9.68(1.16) | 30.94(15.64) | 2.22(1.06) |
| D | 19 | 540.75(148.04) | 57.26(20.40) | 5.17(1.82) | 9.39(1.54) | 26.22(5.98) | 1.97(0.51) |
| CES1.rs2244613.c.1165.33C.A | |||||||
| GG | 16 | 508.14(131.22) | 54.01(16.73) | 5.30(1.78) | 9.28(1.22) | 27.90(7.70) | 2.09(0.52) |
| GT | 15 | 527.16(184.66) | 51.85(20.56) | 5.21(1.49) | 9.51(1.06) | 30.02(16.13) | 2.19(1.10) |
| TT | 6 | 614.61(226.94) | 73.83(26.80) | 4.15(0.67) | 10.26(2.19) | 26.39(9.02) | 1.85(0.78) |
| CES1.rs8192935.c.257.885T.C | |||||||
| AA | 22 | 496.51(177.32) | 50.96(18.93) | 5.30(1.56) | 9.18(1.22) | 30.36(14.31) | 2.30(0.98) |
| AG | 11 | 541.84(102.78) | 56.31(17.17) | 5.11(1.68) | 9.67(0.82) | 26.64(5.96) | 1.90(0.34) |
| GG | 4 | 710.44(201.27) | 86.12(20.61)* | 3.78(0.34) | 11.05(2.34)* | 23.52(6.75) | 1.50(0.46) |
| ABCB1.rs1045642.c.3435T.C | |||||||
| GG | 13 | 568.80(165.51) | 66.00(20.12) | 4.25(0.73) | 9.79(1.67) | 26.02(5.77) | 1.88(0.47) |
| AG | 16 | 492.45(183.68) | 47.37(19.48) | 5.66(1.95)* | 9.70(1.25) | 32.70(15.97) | 2.34(1.07) |
| AA | 8 | 556.46(151.83) | 58.62(20.14) | 5.25(1.15) | 8.78(0.73) | 24.21(6.28) | 1.94(0.63) |
| ABCB1.rs4148738.c.2482.2236G.A | |||||||
| TT | 14 | 568.68(159.39) | 62.13(21.00) | 4.41(0.84) | 9.77(1.63) | 25.85(5.32) | 1.87(0.45) |
| CT | 18 | 502.83(193.15) | 51.36(21.53) | 5.58(1.94) | 9.57(1.25) | 31.86(15.58) | 2.32(1.07) |
| CC | 5 | 542.59(105.28) | 58.11(18.65) | 5.10(0.91) | 8.72(0.49) | 23.95(5.19) | 1.90(0.39) |
| ABCB1.rs2032582.c.2677T.A.G | |||||||
| AA | 6 | 504.08(133.29) | 53.20(20.57) | 5.48(1.23) | 8.53(0.64) | 25.81(6.51) | 2.12(0.64) |
| AC/AT | 17 | 514.08(192.92) | 52.70(21.41) | 5.48(1.95) | 9.69(1.19) | 31.67(16.04) | 2.27(1.08) |
| CC/CT/TT | 14 | 568.68(159.39) | 62.13(21.00) | 4.41(0.84) | 9.77(1.63) | 25.85(5.32) | 1.87(0.45) |
Notes: Data are shown as mean (SD). *P< 0.05 after an ANOVA or t-test.
Discussion
There was a gene-dose effect between ABCB1 and CES1 genes and concentrations of dabigatran etexilate. To the best of our knowledge, this was the first study that exploring the correlation between allele pairs at different loci and drug metabolism of dabigatran etexilate in healthy Chinese subjects. This study adopted a repeated design and relied on validated mass spectrometry for analysis. The accuracy of the pharmacokinetic results was high. The subjects enrolled were all healthy subjects with no known diseases or other factors that affected drug metabolism. Therefore, our study could provide a good basis for elucidating the relationship between gene polymorphisms and pharmacokinetics.
Under fasting conditions, Tmax of dabigatran was 2.5h, shorter than the 4.5h under fed conditions. Cmax of dabigatran was slightly reduced under fed conditions. Whereas, AUC0-t, AUC0– ∞ and T1/2 of dabigatran were similar under both fasting and fed conditions (Table 4 in Data Supplement). Our findings suggested that food may delay the absorption of dabigatran etexilate. Although the Tmax obtained in the our study is consistent with those reported in the Caucasians, the Cmax and AUC0–∞ of total dabigatran under the fasting and fed conditions in our study were approximately 20% higher than those in Caucasians.13,14 Although males showed higher values of Cmax and Tmax compared to females under fed conditions (P<0.05), there was no gender difference in AUC0–∞ and T1/2. There was no significant gender difference in the exposure level of dabigatran. This was contrary to the conclusion that gender did not affect the exposure of dabigatran in a previous study.15
The genotypes of 104 healthy Chinese subjects were obtained in this study, CES1 rs2244613 allele C accounted for 61.1%, CES1 rs8192935 allele G accounted for 24%. ABCB1 rs1045642 allele A accounted for 43.8% and ABCB1 rs414738 allele C accounted for 43.8%. Previous studies have shown that CES1 rs2244613 allele C accounted for 15.3–28.3%,6,7,16–18 CES1 rs8192935 allele G accounted for 67–68.5%,6,7 ABCB1 rs1045642 allele A accounted for 50.8%18 and ABCB1 rs414738 allele C accounted for 52.7–54.2%7,18 among Caucasians. The allele frequencies of CES1 rs2244613 and rs8192935 in Chinese were significantly different from those in Caucasians, but the frequencies of ABCB1 rs1045642 and rs4148738 alleles were similar to those of Caucasians. In addition, the genotype frequency of Chinese in our study is very similar to that of Chinese in other study.19 In the present study the Cmax and AUC0–∞ of total dabigatran were approximately 20% higher than that in Caucasians. Allele C of CES1 rs2244613 was associated with decreased dabigatran trough concentration and decreased risk of bleeding, but not to the peak concentration.6,7,18 According to the RE-LY study in Chinese patients with non-valvular atrial fibrillation, dabigatran had a lower risk of bleeding than warfarin.11 Based on these findings, it was reasonable to suppose that the genetic difference between Chinese and Caucasians may contributed to the differences in bleeding risk.
In the present study, CES1 rs2244613 was not related to peak dabigatran concentration. Besides, the G allele of CES1 rs8192935 was related to the increase of the peak dabigatran concentration. Previous study showed that CES1 rs8192935 has been associated with variability of both peak and trough concentrations of dabigatran with no clinical impact on bleeding or ischemic events.6,10 In RE-LY study, the presence of CES1 SNP rs8192935 was associated with a 12% decrease in peak plasma concentration of the drug per allele (A) present in the genotype.6 In Dimatteo’s study, rs8192935 was associated with the trough concentration of dabigatran, rather than the peak concentration in patients with atrial fibrillation.7 Linkage disequilibrium of CES1 rs8192935 with unknown functional variants may contribute to alterations in plasma levels and regulate dabigatran anticoagulant efficacy.7 The CES1 gene encodes the hepatic CES1 while the CES2 gene encodes intestinal CES2. CES1 primarily converted dabigatran etexilate into its active metabolites in the liver. However, a small part of dabigatran etexilate could also be metabolized to M2 through CES2. This flexibility in dabigatran metabolism suggested that the deficiency of CES1 might be compensated by the activation of the intestinal CES2.15,20 It may explain the divergence between the conclusions of the different studies. More clinical trials are still needed to confirm further the role of the CES1 gene in the metabolism of Dabigatran.
In the present study, ABCB1 rs2032582, rs4148738, and rs1045642 occur as haplotype together, polymorphisms and haplotypes of ABCB1 genes were not linked with pharmacokinetic variability. Previous study showed that the allele A of ABCB1 rs1045642 was significantly associated with the peak dabigatran concentration and increased incidence of bleeding episodes.21 In RE-LY study,6 each allele (C) of the ABCB1 rs4148738 was associated with up to 12% decrease in peak concentration of dabigatran without significant impact on bleeding risk. The relationship between ABCB1 rs2032582 and the pharmacokinetics of dabigatran had only been reported in haplotype–based studies.15,22 Haplotype 2677–3435 was inferred by exclusively considering ABCB1 rs2032582, rs1045642 variants, and haplotype 1236-2677-34355 was inferred by exclusively considering ABCB1 rs1128503, rs2032582 and rs1045642. The two haplotypes were not significantly associated with variability on dabigatran exposure. According to previous studies, ABCB1 variants may alter dabigatran exposure. In this study, none ABCB1 haplotypes or individual SNPs had significant relationship with pharmacokinetic variability, probably because of their low impact on P-gp activity. ABCB1 genotype may not a significant determinant of interindividual variability in dabigatran.
The major limitation was the limited sample size of our study. Larger studies are needed to further confirm our observations. Furthermore, the subjects took dabigatran from two manufacturers, although the results showed that the two drugs were biologically equivalent (Table 5 in Data Supplement). This study is a single dose study in healthy subjects. Trough concentration was unable to obtain and therefore we were unable to evaluate the association between SNPs and the trough concentration of Dabigatran. Besides, long-term safety data are not available to assess the relationship of drug concentrations, genotypes, and safety events.
Conclusion
The allele frequencies of CES1 rs2244613 and CES1 rs8192935 were significantly different between Chinese and Caucasians, but the allele frequencies of ABCB1 rs1045642 and ABCB1 rs4148738 were similar in both populations. CES1 rs8192935 were associated with the peak concentration of dabigatran. There was no significant gender difference in the exposure level of dabigatran. Furthermore, food significantly delayed the absorption of dabigatran, but had little effect on Cmax and AUC0-∞.
Acknowledgment
We thank the Fanweixi Pharmaceutical Technology Co., Ltd and Bestnovo (Beijing) Medical Laboratory for the technical support.
Funding Statement
This study was supported by 135 National Science and Technology Major New Drug Creation Project NO: 2017ZX09304026, Capital Characteristics Research NO: Z161100000516053, Research projects on health development in the capital NO: BJ-2016-071.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Stangier J, Clemens A. Pharmacology, pharmacokinetics, and pharmacodynamics of dabigatran etexilate, an oral direct thrombin inhibitor. Clin Appl Thromb Hemost. 2009;15(Suppl 1):9s–16s. doi: 10.1177/1076029609343004 [DOI] [PubMed] [Google Scholar]
- 2.Ahmed S, Levin V, Malacoff R, Martinez MW. Dabigatran: a new chapter in anticoagulation. Cardiovasc Hematol Agents Med Chem. 2012;10(2):116–123. doi: 10.2174/187152512800388911 [DOI] [PubMed] [Google Scholar]
- 3.Armbruster AL, Buehler KS, Min SH, Riley M, Daly MW. Evaluation of dabigatran for appropriateness of use and bleeding events in a community hospital setting. Am Health Drug Benefits. 2014;7(7):376–384. [PMC free article] [PubMed] [Google Scholar]
- 4.Liesenfeld KH, Lehr T, Dansirikul C, Reilly PA, Connolly SJ, Ezekowitz MD. Population pharmacokinetic analysis of the oral thrombin inhibitor dabigatran etexilate in patients with non-valvular atrial fibrillation from the RE-LY trial. J Thromb Haemost. 2011;9(11):2168–2175. doi: 10.1111/j.1538-7836.2011.04498.x [DOI] [PubMed] [Google Scholar]
- 5.Stangier J, Sthle H, Rathgen K, Roth W, Reseski K, Körnicke T. Pharmacokinetics and pharmacodynamics of dabigatran etexilate, an oral direct thrombin inhibitor, with coadministration of digoxin. J Clin Pharmacol. 2012;52(2):243–250. doi: 10.1177/0091270010393342 [DOI] [PubMed] [Google Scholar]
- 6.Paré G, Eriksson N, Lehr T, et al. Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation. 2013;127(13):1404–1412. doi: 10.1161/CIRCULATIONAHA.112.001233 [DOI] [PubMed] [Google Scholar]
- 7.Dimatteo C, Dimatteo C, D’Andrea G, et al. Pharmacogenetics of dabigatran etexilate interindividual variability. Thromb Res. 2016;144(p):1–5. doi: 10.1016/j.thromres.2016.05.025 [DOI] [PubMed] [Google Scholar]
- 8.Connolly SJ, Ezekowitz MD, Yusuf S, Reilly PA, Wallentin L. Newly identified events in the RE-LY trial. N Engl J Med. 2010;363(19):1875–1876. doi: 10.1056/NEJMc1007378 [DOI] [PubMed] [Google Scholar]
- 9.Kanuri SH, Kreutz RP. Pharmacogenomics of novel direct oral anticoagulants: newly identified genes and genetic variants. J Pers Med. 2019;9(1):7. doi: 10.3390/jpm9010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross S, Pare G. Pharmacogenetics of antiplatelets and anticoagulants: a report on clopidogrel, warfarin and dabigatran. Pharmacogenomics. 2013;14(13):1565–1572. doi: 10.2217/pgs.13.149 [DOI] [PubMed] [Google Scholar]
- 11.Gao X, Yang YM, Zhu J, Dai Y, Tan HQ. [Dabigatran versus warfarin for the prevention of stroke in Chinese patients with nonvalvular atrial fibrillation: Chinese subpopulation analysis of RE-LY]. Zhonghua Xin Xue Guan Bing Za Zhi. 2016;44(11):929–934. Chinese. doi: 10.3760/cma.j.issn.0253-3758.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 12.Gosselin RC, Adcock DM, Bates SM, et al. International Council for Standardization in Haematology (ICSH) Recommendations for Laboratory Measurement of Direct Oral Anticoagulants. Thromb Haemost. 2018;118:437–450. doi: 10.1055/s-0038-1627480 [DOI] [PubMed] [Google Scholar]
- 13.Stangier J, Eriksson BI, Dahl OE, et al. Pharmacokinetic profile of the oral direct thrombin inhibitor dabigatran etexilate in healthy volunteers and patients undergoing total hip replacement. J Clin Pharmacol. 2005;45(5):555–563. doi: 10.1177/0091270005274550 [DOI] [PubMed] [Google Scholar]
- 14.Hartter S, Sennewald R, Nehmiz G, Reilly P. Oral bioavailability of dabigatran etexilate (Pradaxa(R)) after co-medication with verapamil in healthy subjects. Br J Clin Pharmacol. 2013;75:1053–1062. doi: 10.1111/j.1365-2125.2012.04453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pablo MSR, Dolores O, Marcos NG, et al. Effect of Sex, Use of Pantoprazole and Polymorphisms in SLC22A1, ABCB1, CES1, CYP3A5 and CYP2D6 on the Pharmacokinetics and Safety of Dabigatran. Adv Ther. 2020;37(8):3537–3550. doi: 10.1007/s12325-020-01414-x [DOI] [PubMed] [Google Scholar]
- 16.Chin PK, Wright DF, Zhang M, et al. Correlation between trough plasma dabigatran concentrations and estimates of glomerular filtration rate based on creatinine and cystatin C. Drugs R D. 2014;14(2):113–123. doi: 10.1007/s40268-014-0045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamzic S, Kummer D, Milesi S, et al. Novel genetic variants in carboxylesterase 1 predict severe early-onset capecitabine-related toxicity. Clin Pharmacol Ther. 2017;102(5):796–804. doi: 10.1002/cpt.641 [DOI] [PubMed] [Google Scholar]
- 18.Sychev D, Skripka A, Ryzhikova K, et al. Effect of CES1 and ABCB1 genotypes on the pharmacokinetics and clinical outcomes of dabigatran etexilate in patients with atrial fibrillation and chronic kidney disease. Drug Metab Pers Ther. 2020;35(1):254. [DOI] [PubMed] [Google Scholar]
- 19.Ji QY, Zhang CY, Xu Q, et al. The impact of ABCB1 and CES1 polymorphisms on dabigatran pharmacokinetics and pharmacodynamics in patients with atrial fibrillation. Br J Clin Pharmacol. 2020. doi: 10.1111/bcp.14646 [DOI] [PubMed] [Google Scholar]
- 20.Shi J, Wang X, Nguyen J-H, et al. Dabigatran etexilate activation is affected by the CES1 genetic polymorphism G143E (rs71647871) and gender. Biochem Pharmacol. 2016;119:76–84. doi: 10.1016/j.bcp.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sychev DA, Nikolaevich LA, Shelekhova TV, et al. The impact of ABCB1 (rs1045642 and rs4148738) and CES1 (rs2244613) gene polymorphisms on dabigatran equilibrium peak concentration in patients after total knee arthroplasty. Pharmgenomics Pers Med. 2018;11:127–137. doi: 10.2147/PGPM.S169277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouin-Thibault I, Delavenne X, Blanchard A, et al. Interindividual variability in dabigatran and rivaroxaban exposure: contribution of ABCB1 genetic polymorphisms and interaction with clarithromycin. J Thromb Haemost. 2017;15(2):273–283. doi: 10.1111/jth.13577 [DOI] [PubMed] [Google Scholar]