Abstract
Cancer Cachexia (CC) is a disease that changes various metabolisms in human body. Fat metabolism is significantly affected in CC, leading to fat loss. Non-coding RNAs (ncRNAs) in adipocytes and exosomes secreted by tumor play an important role in fat loss. However, there is no related reviews summarizing how ncRNAs contribute to fat loss during CC. This review screens recent articles to summarize how ncRNAs are packaged, transported in exosomes, and play the role in fat loss. Not only does this review summarize the mechanisms, we also point out the research orientations in the future.
Keywords: Non-coding RNAs, Cancer cachexia, Exosomes, Fat loss
1. Introduction
1.1. Cancer Cachexia
Cancer cachexia (CC) is a multifactorial, multiple organ syndrome with skeletal muscle mass loss caused by cancer (presence or absence of fat loss), while conventional nutritional support cannot be completely reversed [1]. Fat loss is one of the most prominent features of CC, and its mechanisms are mainly related to fat degradation, fatty acid oxidation, white adipocyte browning, lipid production and storage, and adipogenesis [2]. Studies have shown that the occurrence and development of fat loss during CC are a multi-factor, multi-step process, which are the result of mutual induction and joint effects of factors such as reducing food intake, systemic inflammatory response, and metabolic abnormalities [3,4].With the goal of more precise targeted therapy and good prognosis, researchers gradually reveal non-coding RNA-mediated fat loss during CC by exploring further mechanisms.
1.2. Non-coding RNAs
Non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into protein. It can be divided into two categories: housekeeping ncRNAs and regulatory ncRNAs. Among them, regulatory ncRNAs include long non‐coding RNAs (lncRNAs), microRNAs (miRNAs), circular RNA (circRNA), etc [5].
lncRNAs, the minimum size limit of 200 nt, can recruit regulatory complexes to affect gene expression through RNA-protein interactions [6], and three mechanisms have been summarized to explain lncRNAs targeting chromatin: 1) lnRNAs get targeted to chromatin by interacting with proteins having RNA or DNA binding capability. 2) lnRNAs form triple helix nucleic acid structures by interacting with double-stranded DNA in order to target specific complementary sequences directly in the genome to perform its regulatory functions. 3) lnRNAs form R-loop which can affect gene expression or interact with gene locus [7].
In short ncRNAs, miRNA is a short, non-coding RNA molecule of 19–25 nucleotides long, usually paired with incomplete complementary bases of the 3′-untranslated region. It regulates gene expression by inhibiting mRNA translation and reducing mRNA stability [8]. In the case of cancer, however, there are many miRNA expression disorders. In recent years, a large number of studies have concluded that the disorders of miRNAs in cancer are mainly related to the methylation status of miRNA promoters, the function and/or expression of enzymes associated with miRNA biogenesis, and changes in transcription factor activity [[9], [10], [11], [12]]. According to the different roles of miRNAs in regulating cancer, they are divided into tumor-suppressor miRNAs and carcinogenic miRNAs (oncomiRs). Generally, oncomiRs are over-expressed in cancer, while tumor suppressor miRNAs are under-expressed.
Covalently blocked circRNA is produced by reverse splicing of the precursor mRNA of exons of thousands of genes in eukaryotes [13]. circRNAs are known to function as miRNA sponges [14,15], regulating transcription and splicing [16], and producing peptides by translation [17].
The levels of ncRNAs are related to cancer development and metastasis. On the one hand, the dysregulation of ncRNAs regulates the proliferation, cycle, apoptosis, migration, and invasion of cancer cells through specific pathways [18,19]. On the other hand, it acts on the external factors of cancer cells [20,21]. Cancer cells actively communicate with the surrounding microenvironment to support malignant progress. Exosome, a class of extracellular vesicles between 40 nm and 100 nm, plays a key role in intercellular communication and regulate recipient cells by transmitting their contents, including proteins and nucleic acids [22]. Therefore, the dysregulation of ncRNA caused by cancer not only acts on the tumor microenvironment to promote or inhibit the development and metastasis of cancer under the transmission of exosomes [23,24], but also affects other cells including skeletal muscle cells, adipocytes, throughout the body to induce skeletal muscle mass loss and fat loss [25,26].
1.3. Fat metabolism
Adipose tissue can be divided into white adipose tissue (WAT), brown adipose tissue (BAT) and beige adipocyte. Commitment of mesenchymal stem cells (MSCs) to preadipocytes and terminal differentiation of preadipocytes toward mature adipocytes are the two phases of adipogenesis [27]. This process is caused by the sequential activation of a series of transcription factors. The essential ones are two families, CCAAT enhancer binding protein (C/EBPS) and peroxisome proliferation activated receptor (PPAR) [28].
WAT is composed of white adipocytes containing unilocular lipid droplet which stores triacylglycerol esterified from fatty acids (FAs). Many factors can promote the breakdown of WAT such as adipose triglyceride lipase (ATGL), hormone sensitive lipase (HSL), monoglyceride lipase, cell death induced DNA fragmentation-factor-α-like effector A(CIEDA) and lipolysis related genes [29]. Elevated levels of catecholamines and natriuretic peptides activate the protein kinase A (PKA) and protein kinase G (PKG) pathways when it comes to an energy demanding environment and tumor-derived factors such as IL-6 acting on downstream mediators function through PKA [29]. Thus, phosphorylating perilipin A promotes the translocation of HSL from cytoplasm to lipid droplet and it disassociates from comparative gene identification 58 (CGI-58), unbinding ATGL to G0G1 switch protein (GOS2) resulting in lipolysis [30].
BAT and beige adipocyte are rich in mitochondrion expressing uncoupling protein 1 (UCP1) in a high level which produces heat through adenosine triphosphate (ATP) oxygen-dephosphorylation. BAT is of the same lineage as myocytes, and its differentiation process can be regulated by PPARγ, PPARγ coactivator −1 alpha (PGC-1α) and the transcription factor PR domain containing 16 (PDRM16) interacting with C/EBPβ to promote the expression of UCP1 [31]. During CC, the levels of TNF-α, IL-6, IL-1, zinc-α2-glycoprotein (ZAG) are elevated, which upregulate oxidative genes in the mitochondria that promote the formation of brown adipose tissue [29]. The expression change of UCP1 is the main manifestation of WAT browning. When stimulated by the outside world, WAT expresses UCP1 and other thermogenic genes, and UCP1-positive multilocular cells appear in the area, which promotes energy consumption throughout the body and exerts a similar effect to BAT [32].
2. Changes of ncRNA expression in exosomes during cancers
Exosomes have multiple roles in cancer and CC. Wang S. et al. found that exosomes in lung cancer were internalized by human adipose tissue mesenchymal stem cells (hAD-MSCs) and significantly inhibited adipogenesis of hAD-MSCs through the transforming growth factor-β (TGF-β) signaling pathway [33]. However, if Lewis lung cancer cells are inhibited from producing and releasing exosomes, lipolysis and adipose tissue browning can be inhibited [34]. In some cases, the exosomes produced and released by cancer cells may play an important role in causing CC.
The levels of ncRNAs in exosomes change differently in patients with different cancers. For instance, researchers detected 79 exosomal RNAs whose levels were significantly higher than normal in stage I gastric carcinoma (GC) patients [35]. Researchers measured the RNA profiles of osteosarcoma cell lines and osteosarcoma-derived exosomes and found 10 down-regulated miRNAs and 11 up-regulated miRNAs in exosomes [36]. In addition, the expression of six exosomal lncRNAs were significantly up-regulated during colorectal cancer [37]. A lncRNA, LINC00161 was found significantly upregulated in hepatocellular carcinoma patients and showed excellent stability and specificity [38]. The researchers identified circRNA microarrays for frozen tumors of esophageal squamous cell carcinoma and two circRNAs could be found in exosome [39]. Another results of high-throughput sequencing data indicated that 105 exosomal circRNAs in plasma of lung adenocarcinoma patients were up-regulated and 78 were down-regulated [40].
Different types and degrees of ncRNA expression disorders are caused by different kinds of cancer. Disordered ncRNAs can affect the ncRNA content in other cells throughout the body under the transmission of exosomes, thereby inducing skeletal muscle mass loss and fat loss. A 2018 review innovatively summarized miRNAs that are dysregulated in various cachexia and cause skeletal muscle depletion and named ‘AtromiRs’ [41].So, we believe that there are also a series of ncRNAs that not only have dysregulated expression in a variety of cancers, but also cause fat loss.
3. Formation and sorting mechanism of exosomes
3.1. Formation of exosomes
Exosomes are a class of extracellular vesicles between 40 and 100 nm surrounded by lipid bilayer rich in sphingolipids which contain proteins and nucleic acids, and transmit information [22]. Exosomes are enveloped in multivesicular bodies formed in cells. After those multivesicular bodies are fused with the cell membrane, the intracapsular vesicles are released into the extracellular environment and become exosomes [42].
3.2. Cargo sorting mechanism of exosomes
Exosomes separate target proteins and RNAs mainly through three pathways, namely ESCRT, lipid raft and RNA-binding protein-related pathways [43]. ESCRT system includes 4 kinds of protein complexes named ESCRT-0, ESCRT-Ⅰ, SCRT-Ⅱ and ESCRT-Ⅲ, which have UBDs (ubiquitin combines domain) to combine with modified protein. ESCRT-0 has four UBDs to form a larger complex with low affinity and multiple binding domains [44].Tsg101 and sub UBAP1 are core subunits of ESCRT-Ⅰ and associated with ubiquitin directly. Studies show that ESCRT-Ⅱ is likely to be independent of others. Vps36, sub domain of ESCRT-Ⅱ, relies on membrane cholesterol improving efficiency and combine with mRNA 3 ‘UTR to involve in RNA sorting [45]. Effect of ESCRT-Ⅲ is not very clear and the interaction between complexes remains to be explored.
Lipid raft is found both in cell membrane and exosomes which provides new ideas. According to a research, neutral sphingomyelinase catalyze the formation of a large amount of ceramide, and ceramide can involve in the classification and selection [46].
RNA-binding proteins act as transporters by binding to specific RNA sequences. For example, HnRNPA2B1 binds with miRNAs or lncRNAs by identifying CCGA or CCCU [47]. YBP1 is a hnRNP that interacts with the hairpin ring of retrovirus RNA to keep the virus stable [48].
4. Transport and reception of exosomes
Exosomes transport contents to cells by means of justacrine secretion, paracrine secretion, and internal secretion. In internal secretion, exosomes enter the blood and lymph fluid to bind to specific cells in circulation. They either anchor ligands through the surface membrane, or act on cells through receptor-mediated endocytosis [49].
A large number of exosomes in the serum of patients with CC lead scientists to explore their role in the cell-cell communication. The reception of exosomes is a multiple process while the primary uptake is endocytic pathway which includes caveolin-mediated uptake, lipid raft-mediated internalization and so on. Exosomes can also fuse with cell membranes directly [50]. Chemotaxis of exosomes makes them concentrate on a certain tissue to regulate the growth, development and apoptosis of target cells including mesenchymal cells, tumor vascular associated endothelial cells, and adipocytes [51]. It suggests that cancer cells can act on fat cells in specific way such as endoplasmic reticulum stress to cause fat loss.
5. The role of lncRNAs in fat loss during CC
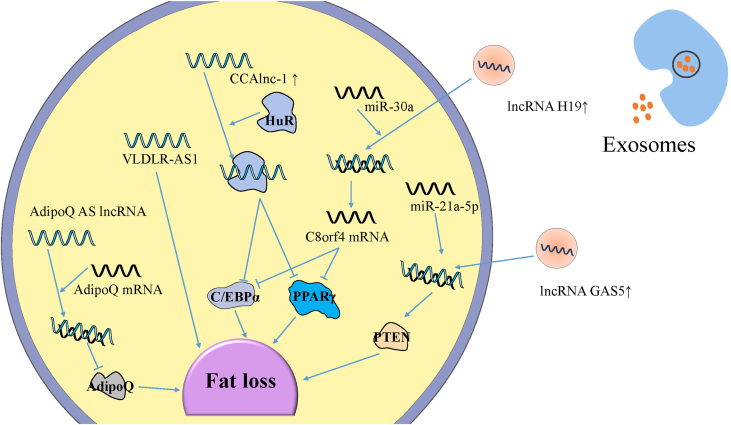
Lipid degradation, fatty acid oxidation and enhanced browning of WAT, lipid production, lipid storage, and adipocyte reduction are important factors that cause fat loss [52]. ncRNAs are encapsulated into exosomes and transported to target cells to perform functions. lncRNAs appear as a regulatory factor of adipose tissue including adipogenesis, browning of WAT and so on [53] (Fig. 1).
Fig. 1.
The changed expression level of exosomal and intracellular lncRNAs leads to fat loss during CC.
Exosomal lncRNAs from tumor cells always bind with specific microRNAs as a sponge to weaken their function. lncRNAs changed in adipose tissue can bind with a protein or a mRNA to promote fat loss. Some of these lncRNAs effect the genes related to adipogenic differentiation, while some down-regulate two vital proteins of adipogenesis, PPAR-γ and C/EBPα. An antisense lncRNA can suppress the expression of mRNA and then reduce the level of hormone about lipid metabolism.
5.1. Exosomal lncRNA-induced fat loss during CC
Research showed that the level of exosomal lncRNA H19 increased and the exosome was absorbed by the target cells in the colorectal cancer mouse model [54]. It has been reported that lncRNA H19 is associated with brown adipose tissue (BAT) activation in mice and humans to promote brown adipocyte differentiation, which enables the host to maintain insulin sensitivity and avoid excessive deposition of fat [55]. Kunli et al. found that lncRNA H19 could bind with miR-30a as a sponge or a ceRNA and inhibited the negative regulation of miR-30a on C8orf4 mRNA, thereby increasing C8orf4, reducing PPAR-γ and CEBP-α protein levels, and interfering with lipid accumulation [56].
In the patient of early-stage-non-small-cell lung cancer, the level of lncRNA GAS5 in cancer-derived exosomes is much higher than that in healthy humans and advanced-stage-non-small-cell lung cancer patients [57]. lncRNA GAS5 has been shown to bind to miR-21 in breast cancer cells [58]. Liu et al. [59] found that the overexpression of lncRNA GAS5 could reduce miR-21-5p level and inhibit the generation of 3T3-L1 cells. Further, miR-21-5p regulated the expression of phosphatase and tensin homolog (PTEN), and then the miR-21-5p/PTEN signaling pathway could suppress adipogenesis.
5.2. Intracellular lncRNA-induced fat loss during CC
Adiponectin (AdipoQ), a hormone has a positive effect on lipid metabolism. Dorota Diakowska et al. [60] reported that the serum levels of AdipoQ in cachexia patients with malignant tumors of the digestive tract were lower than normal. It indicates that the reduce of AdipoO may cause the fat loss during CC. AdipoQ antisense (AS) lncRNA is an antagonistic RNA of AdipoQ mRNA which forms an AdipoQ AS lncRNA/AdipoQ mRNA duplex. As a result, the transcription of AdipoO mRNA is weakened, which reduces the level of AdipoQ and inhibits fat deposition [61].
Lei Shen et al. found that cachexia-related anti-adipogenesis lncRNA 1 (CAAlnc1) was an inhibitory regulator of adipogenesis by means of preventing the binding of HuR antigen (HuR) and adipogenic transcription factors (TFs) required for adipogenesis [62]. The interaction between CCAlnc1 and HuR regulates the expression of C/EBP-α and PPAR-γ, therefore, the adipogenesis is cut down [63]. Another key lncRNA involved is VLDLR antisense RNA 1 (VLDLR-AS1) which can interact with 14 potential miRNAs and affect some pathways such as Wnt/β-catenin signaling pathway. However, vital target coding genes and specific functions are not explored which can be a promising orientation [64].
6. The role of miRNAs in fat loss during CC
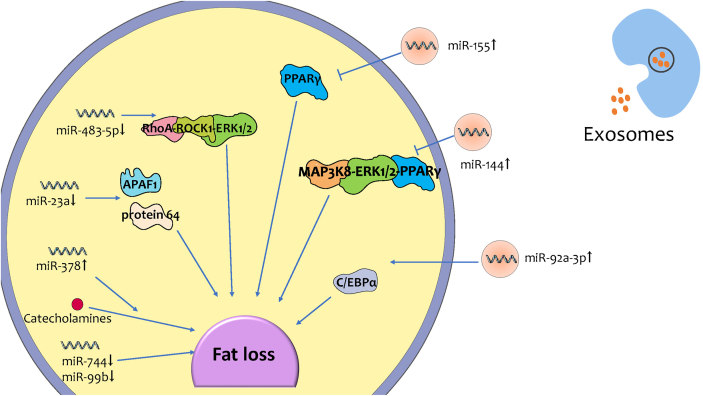
Recent studies on miRNA-mediated fat loss during CC can be roughly divided into two categories, some of which focus on the ncRNAs in exosomes acting on target cells, leading to fat loss; while some aim to explore the mechanism of intracellular miRNA inducing fat loss during CC (Fig. 2).
Fig. 2.
The changed expression level of exosomal and intracellular miRNAs leads to fat loss during CC.
The highly expressed exosomal miRNAs enter target cells. The expressions’ change of exsomal miRNAs and intracellular miRNAs lead to regulating adipogenesis and lipolysis, inducing adipose tissue inflammation, promoting beige/brown differentiation, etc. Finally, changes of exsomal miRNAs and intracellular miRNAs expression lead to fat loss during CC.
6.1. Exosomal miRNA-induced fat loss during CC
The expression of some exosomal miRNAs is changed in cancer cells during CC, and then they are packaged into exosomes. After they are transported to and received by target cells, they induce fat loss through multiple pathways. MiR-92a-3p is highly expressed in exosomes of chronic myelogenous leukemia cells and some other cachexia-causing cancers. The miRNA is transferred to adipose-derived mesenchymal stem cells (ADSCs) through exosomes, which then reduce C/EBPα expression to inhibit adipogenesis of ADSCs [65]. Another study found that in breast cancer exosomes, miRNA-144 induced beige/brown differentiation and enhanced the catabolism of adipocytes by down-regulating the MAP3K8/ERK1/2/PPARγ axis [66]. In addition, co-culture experiments of breast cancer cells and mature adipocytes showed that miR-155 in exosomes secreted by cancer cells could promote the beige/brown differentiation of adipocytes and metabolism by down-regulating PPARγ expression [26,67].
6.2. Intracellular miRNA-induced fat loss during CC
Currently, there are a few experimental researches on exosomal miRNA-induced fat loss during CC, while intracellular miRNAs have also received the attention of researchers. Researchers found that expression of miR-483–5p/-23a/-744/-99b was downregulated, whereas miR-378 was significantly upregulated in cancer subcutaneous adipose tissue in patients with CC. When miR-378 was overexpressed in adipocytes during CC, catecholamine was able to stimulate lipolysis more strongly. When the expression was inhibited, not only lipolysis, but also the expression of LIPE, PLIN1 and PNPLA2 which encoding important lipolytic regulators, was declined [68].
The expression of miR-23a and miR-483-5p was downregulated during CC [68]. There are no studies have discovered the mechanism of miR-23a and miR-483-5p on fat loss during CC. Nevertheless, other related studies on fat metabolism may predict the mechanism of miR-23a and miR-483-5p inducing fat loss. miR-23a played a critical role in conjugated linoleic acid-induced apoptosis in adipocytes via controlling APAF1 expression [69]. Furthermore, the up-regulated miR-483-5p promoted adipogenesis by inhibiting the RhoA/ROCK1/ERK1/2 pathway [70]. Therefore, it can be boldly conjectured that during CC, miR-23a expression is reduced and then induce adipocyte apoptosis through APAF1. On the other hand, the expression of miR-483-5p in adipose tissue is reduced, which may inhibit adipogenesis through the RhoA/ROCK1/ERK1/2 pathway.
7. CircRNAs mediating fat metabolism
Some circRNAs regulate the expression level of miRNAs in target cells through exosome transport and their own miRNA sponge action, then, resulting in fat loss. A recent research found that circNrxn2 could promote the browning of WAT by sponging miR-103 [71]. Hsa_circ_0010522, also known as ciRS-133, could interact with miR-133 in preadipocytes to reduce miR-133 levels [72]. As a result, the expression of PRDM16 was up-regulated, and UCP1 was also activated in preadipocytes, which promoted the differentiation of preadipocytes into brown-like cells. However, it has not been able to prove that these circRNAs change in exosomes during CC.
8. Summary and prospect
Generally, the role of fat loss during CC is gradually noticed and researched. We summarize the ncRNAs changed in exosomes and adipocytes (Table 1). Exosomal ncRNAs are absorbed into adipocytes and cause fat loss through inhibition of adipogenesis, lipolysis and beige/brown differentiation at the method of regulating the level of associated proteins. circRNAs and lncRNAs indirectly bind with specific miRNAs as a sponge to adjust their transcription. On the other hand, intracellular ncRNAs also play an important role in fat loss during CC.
Table 1.
ncRNAs that participate in fat loss.
| ncRNA | Target(s) | Function | |
|---|---|---|---|
| Exosomal | lncRNA H19 | miR-30a/C8orf4 | interferes lipid accumulation |
| lncRNA GAS5 | miR-21a-5p/PTEN | Inhibits adipogenesis and lipid accumulation | |
| miR-92a-3p | C/EBPα | inhibits adipogenesis | |
| miR-144 | MAP3K8/ERK1/2/PPARγ | beige/brown differentiation, catabolism of adipocytes | |
| miR-15 | PPARγ | promotes beige/brown differentiation of adipocytes and metabolism | |
| ciRS-133 | miR-133/PRDM16 | differentiation of preadipocytes into brown-like cells | |
| Intercellular | AdipoQ AS lncRNA | AdipoQ mRNA | inhibits adipogenesis |
| CAAlnc1 | HuR | inhibits adipogenesis | |
| VLDLR-AS1 | fat loss | ||
| miR-378 | lipolysis | ||
| miR-23a | APAF1 | adipocyte apoptosis | |
| miR-483-5p | RhoA/ROCK1/ERK1/2 | inhibits adipogenesis | |
| circNrxn2 | miR-103 | promotes browning of WAT |
However, researchers focused more on ncRNAs participating in skeletal muscle consumption, but few on those participating in fat loss during CC. As a result, ncRNAs causing fat loss during CC haven't been fully researched, and even the published researches in this field have flaws. On one hand, lncRNA H19 and lncGAS5 were reported to be high-level in exosomes of cancer patients. On the other hand, the expression of miR-23a and miR-483-5p was downregulated during CC. Although there are no studies have discovered the specific mechanism of them on fat loss during CC, related articles have discovered the ways they regulate lipid metabolism. Therefore, conjectures are made to summarize the mechanisms which still need to be proved. VLDLR-AS1 and miR-378 were screened by researchers to participate in fat loss during CC, specific targets of them inducing fat loss are not discovered though. In addition, ciRS-133 and circNrxn2 have been confirmed to act on miRNAs and cause fat loss, but there is no report that their contents are changed during CC. It seems that they may not receive the attention of researchers, or the biochemical detection technology in related fields still needs to be improved. Frankly speaking, there are still many unknown ncRNAs and mechanisms in fat loss during CC waiting to be discovered and studied.
It is expected that future studies can further clarify the mechanism of exosomal and intercellular ncRNAs in the occurrence and development of fat loss during CC. Researching on drugs targeting ncRNAs in cells or exosomes, and delivering drugs to target cells by encapsulating drugs alleviating fat loss into exosomes, may be new strategies and research directions for the treatment of fat loss during CC.
Role of the funding source
Prof. Yi Li et al. was sponsored by National Natural Science Foundation of China, China (Grant No. 81972546) in study concepts, study design, data acquisition, quality control, manuscript editing and review.
Declaration of competing interest
The author reports no conflicts of interest in this work.
Acknowledgments
We gratefully acknowledge the financial support from National Natural Science Foundation of China, China (Grant No. 81972546).
References
- 1.Fearon K., Strasser F., Anker S.D., Bosaeus I., Bruera E., Fainsinger R.L., Jatoi A., Loprinzi C., Macdonald N., Mantovani G., Davis M., Muscaritoli M., Ottery F., Radbruch L., Ravasco P., Walsh D., Wilcock A., Kaasa S., Baracos V.E. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12(5):489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 2.Daas S.I., Rizeq B.R., Nasrallah G.K. Adipose tissue dysfunction in cancer cachexia. J. Cell. Physiol. 2018;234(1):13–22. doi: 10.1002/jcp.26811. [DOI] [PubMed] [Google Scholar]
- 3.Han J., Meng Q., Shen L., Wu G. Interleukin-6 induces fat loss in cancer cachexia by promoting white adipose tissue lipolysis and browning. Lipids Health Dis. 2018;17(1):14. doi: 10.1186/s12944-018-0657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster J.M., Kempen L., Hardy R.S., Langen R.C.J. Inflammation and skeletal muscle wasting during cachexia. Front. Physiol. 2020;11:597675. doi: 10.3389/fphys.2020.597675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H., Xu Z., Liu D. Small non-coding RNA and colorectal cancer. J. Cell Mol. Med. 2019;23(5):3050–3057. doi: 10.1111/jcmm.14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., Wysocka J., Lei M., Dekker J., Helms J.A., Chang H.Y. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra K., Kanduri C. Understanding long noncoding RNA and chromatin interactions: what we know so far. Noncoding RNA. 2019;5(4):54. doi: 10.3390/ncrna5040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu T.X., Rothenberg M.E. MicroRNA. J. Allergy Clin. Immunol. 2018;141(4):1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Bao W., Liu Y., Wang S., Xu S., Li X., Li Y., Wu S. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018;9(5):447. doi: 10.1038/s41419-018-0390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuna M., Machado A.S., Calin G.A. Genetic and epigenetic alterations of microRNAs and implications for human cancers and other diseases. Genes Chromosomes Cancer. 2016;55(3):193–214. doi: 10.1002/gcc.22332. [DOI] [PubMed] [Google Scholar]
- 11.Acunzo M., Croce C.M. MicroRNA in cancer and cachexia--A mini-review. J. Infect. Dis. 2015;212(Suppl 1):S74–S77. doi: 10.1093/infdis/jiv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borzi C., Calzolari L., Ferretti A.M., Caleca L., Pastorino U., Sozzi G., Fortunato O. c-Myc shuttled by tumour-derived extracellular vesicles promotes lung bronchial cell proliferation through miR-19b and miR-92a. Cell Death Dis. 2019;10(10):759. doi: 10.1038/s41419-019-2003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8(1):1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R., Zhang Y., Han B., Bai Y., Zhou R., Gan G., Chao J., Hu G., Yao H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy. 2017;13(10):1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 17.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., Shenzis S., Samson M., Dittmar G., Landthaler M., Chekulaeva M., Rajewsky N., Kadener S. Translation of CircRNAs. Mol. Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M., Sha Y., Zhang X. MiR-22 functions as a biomarker and regulates cell proliferation, cycle, apoptosis, migration and invasion in renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2017;10(12):11425–11437. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Chen G., Zhang B., Liu C., Yu Y., Jin Y. miR-27b-3p suppresses cell proliferation, migration and invasion by targeting LIMK1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2017;10(9):9251–9261. [PMC free article] [PubMed] [Google Scholar]
- 20.Huffaker T.B., Lee S.H., Tang W.W., Wallace J.A., Alexander M., Runtsch M.C., Larsen D.K., Thompson J., Ramstead A.G., Voth W.P., Hu R., Round J.L., Williams M.A., O'connell R.M. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J. Biol. Chem. 2017;292(45):18530–18541. doi: 10.1074/jbc.M117.808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negrete-Garcia M.C., Ramirez-Rodriguez S.L., Rangel-Escareno C., Munoz-Montero S., Kelly-Garcia J., Vazquez-Manriquez M.E., Santillan P., Ramirez M.M., Ramirez-Martinez G., Ramirez-Venegas A., Ortiz-Quintero B. Deregulated MicroRNAs in cancer-associated fibroblasts from front tumor tissues of lung adenocarcinoma as potential predictors of tumor promotion. Tohoku J. Exp. Med. 2018;246(2):107–120. doi: 10.1620/tjem.246.107. [DOI] [PubMed] [Google Scholar]
- 22.Kowal J., Tkach M., Théry C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Hu J., Wang W., Lan X., Zeng Z., Liang Y., Yan Y., Song F., Wang F., Zhu X., Liao W. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Canc. 2019;18(1):91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu G., Zhang B., Ye J., Cao S., Shi J., Zhao Y., Wang Y., Sang J., Yao Y., Guan W. Exosomal miRNA-139 in cancer-associated fibroblasts inhibits gastric cancer progression by repressing MMP11 expression. Int. J. Biol. Sci. 2019;15(11):2320–2329. doi: 10.7150/ijbs.33750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan Z., Chen X., Gao X., Dong Y., Zhao Y., Wei M., Fan W., Yang G., Liu L. Chronic myeloid leukemia‐derived exosomes attenuate adipogenesis of adipose derived mesenchymal stem cells via transporting miR‐92a‐3p. J. Cell. Physiol. 2019;234(11):21274–21283. doi: 10.1002/jcp.28732. [DOI] [PubMed] [Google Scholar]
- 26.Wu Q., Sun S., Li Z., Yang Q., Li B., Zhu S., Wang L., Wu J., Yuan J., Wang C., Li J., Sun S. Breast cancer-released exosomes trigger cancer-associated cachexia to promote tumor progression. Adipocyte. 2019;8(1):31–45. doi: 10.1080/21623945.2018.1551688. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lee J.E., Schmidt H., Lai B., Ge K. Transcriptional and epigenomic regulation of adipogenesis. Mol. Cell Biol. 2019;39(11):e00601–e00618. doi: 10.1128/MCB.00601-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L.Y., Chen C.W., Chen L.K., Chou H.Y., Chang C.L., Juan C.C. Curcumin attenuates adipogenesis by inducing preadipocyte apoptosis and inhibiting adipocyte differentiation. Nutrients. 2019;11(10):2307. doi: 10.3390/nu11102307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalini D. Lipid metabolism in cancer cachexia. Ann. Palliat. Med. 2019;8(1):13–23. doi: 10.21037/apm.2018.10.01. [DOI] [PubMed] [Google Scholar]
- 30.Tsoli M., Swarbrick M.M., Robertson G.R. Lipolytic and thermogenic depletion of adipose tissue in cancer cachexia. Semin. Cell Dev. Biol. 2016;54:68–81. doi: 10.1016/j.semcdb.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 31.Kir S., White J.P., Kleiner S., Kazak L., Cohen P., Baracos V.E., Spiegelman B.M. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature: International weekly journal of science. 2014;513(7516):100–104. doi: 10.1038/nature13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townsend L.K., Wright D.C. Looking on the "brite" side exercise-induced browning of white adipose tissue. Pflügers Archiv. 2019;471(3):455–465. doi: 10.1007/s00424-018-2177-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang S., Li X., Xu M., Wang J., Zhao R.C. Reduced adipogenesis after lung tumor exosomes priming in human mesenchymal stem cells via TGFβ signaling pathway. Mol. Cell. Biochem. 2017;435(1–2):59–66. doi: 10.1007/s11010-017-3056-3. [DOI] [PubMed] [Google Scholar]
- 34.Hu W., Ru Z., Xiao W., Xiong Z., Wang C., Yuan C., Zhang X., Yang H. Adipose tissue browning in cancer-associated cachexia can be attenuated by inhibition of exosome generation. Biochem. Biophys. Res. Commun. 2018;506(1):122–129. doi: 10.1016/j.bbrc.2018.09.139. [DOI] [PubMed] [Google Scholar]
- 35.Lin L.Y., Yang L., Zeng Q., Wang L., Chen M.L., Zhao Z.H., Ye G.D., Luo Q.C., Lv P.Y., Guo Q.W., Li B.A., Cai J.C., Cai W.Y. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Canc. 2018;17(1):84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raimondi L., De Luca A., Gallo A., Costa V., Russelli G., Cuscino N., Manno M., Raccosta S., Carina V., Bellavia D., Conigliaro A., Alessandro R., Fini M., Conaldi P.G., Giavaresi G. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis. 2019;41(5):666–677. doi: 10.1093/carcin/bgz130. [DOI] [PubMed] [Google Scholar]
- 37.Hu D., Zhan Y., Zhu K., Bai M., Han J., Si Y., Zhang H., Kong D. Plasma exosomal long non-coding RNAs serve as biomarkers for early detection of colorectal cancer. Cell. Physiol. Biochem. 2018;51(6):2704–2715. doi: 10.1159/000495961. [DOI] [PubMed] [Google Scholar]
- 38.Sun L., Su Y., Liu X., Xu M., Chen X., Zhu Y., Guo Z., Bai T., Dong L., Wei C., Cai X., He B., Pan Y., Sun H., Wang S. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J. Canc. 2018;9(15):2631–2639. doi: 10.7150/jca.24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan L., Cao Q., Liu J., Zhang J., Li B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol. Canc. 2019;18(1):16. doi: 10.1186/s12943-018-0936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F., Huang C., Wu Q., Jiang L., Chen S., Chen L. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J. Cell. Biochem. 2020;121(3):2525–2533. doi: 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- 41.Van De Worp W.R., Theys J., Van Helvoort A., Langen R.C. Regulation of muscle atrophy by microRNAs:‘AtromiRs’ as potential target in cachexia. Curr. Opin. Clin. Nutr. Metab. Care. 2018;21(6):423–429. doi: 10.1097/MCO.0000000000000503. [DOI] [PubMed] [Google Scholar]
- 42.Chitti S.V., Fonseka P., Mathivanan S. Emerging role of extracellular vesicles in mediating cancer cachexia. Biochem. Soc. Trans. 2018;46(5):1129–1136. doi: 10.1042/BST20180213. [DOI] [PubMed] [Google Scholar]
- 43.Kerui W., Fei X., Shih-Ying W., Kounosuke W. Extracellular vesicles as emerging targets in cancer: recent development from bench to bedside. Biochim. Biophys. Acta. 2017;1868(2):538–563. doi: 10.1016/j.bbcan.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayers J.R., Fyfe I., Schuh A.L., Chapman E.R., Edwardson J.M., Audhya A. ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J. Biol. Chem. 2010;286(11):9636–9645. doi: 10.1074/jbc.M110.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uwe I., Daniel S.J. Bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445(7127):554–558. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F.…Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 47.Carolina V.-B., Cristina G.-V., Fátima S.-C., Daniel P.-H., Jesús V., Noa M.-C., Jorge M.-H.D., Alberto P.-M., María M., Francisco S.-M. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J S.M., M T.-D.M., V K.K., Sayaka R., Randy S. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5 doi: 10.7554/eLife.19276. e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S., Wang J., Wei W., Ma G. Exosomes: the indispensable messenger in tumor pathogenesis and the rising star in antitumor applications. Advanced Biosystems. 2019;3(5) doi: 10.1002/adbi.201900008. e1900008. [DOI] [PubMed] [Google Scholar]
- 50.Ann M.L., Charles P.R., Francisco C.D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014;3(1):24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholas S., Lingzhi W., Gautam S., Jean-Paul T., Boon-Cher G. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol. Sci. 2016;37(7):606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Ebadi M., Mazurak V.C. Evidence and mechanisms of fat depletion in cancer. Nutrients. 2014;6(11):5280–5297. doi: 10.3390/nu6115280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yongzhi Z., Kun R., Xiao Z., Zhi Z., Guanghui Y. Long noncoding RNAs: advances in lipid metabolism. Adv. Clin. Chem. 2018;87:1–36. doi: 10.1016/bs.acc.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Ren J., Ding L., Zhang D.Y., Shi G.P., Xu Q.Y., Shen S.N., Wang Y.P., Wang T.T., Hou Y.Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt E., Dhaouadi I., Gaziano I., Oliverio M., Klemm P., Awazawa M., Mitterer G., Fernandez-Rebollo E., Pradas-Juni M., Wagner W., Hammerschmidt P., Loureiro R., Kiefer C., Hansmeier N.R., Khani S., Bergami M., Heine M., Ntini E., Frommolt P., Zentis P., Orom U.A., Heeren J., Bluher M., Bilban M., Kornfeld J.W. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 2018;9(1):3622. doi: 10.1038/s41467-018-05933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li K., Wu Y., Yang H., Hong P., Fang X., Hu Y. H19/miR-30a/C8orf4 axis modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. J. Cell. Physiol. 2019;234(11):20925–20934. doi: 10.1002/jcp.28697. [DOI] [PubMed] [Google Scholar]
- 57.Chuling L., Yanling L., Chenye S., Cen C., Tianli Z., Yuqing W., Hang F., Tangfeng L., Hongbin L., Yong S. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell. Physiol. 2019;234(11):20721–20727. doi: 10.1002/jcp.28678. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z., Zhu Z., Watabe K., Zhang X., Bai C., Xu M., Wu F., Mo Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20(11):1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H., Li H., Jin L., Li G., Hu S., Ning C., Guo J., Shuai S., Li X., Li M. Long noncoding RNA GAS5 suppresses 3T3-L1 cells adipogenesis through miR-21a-5p/PTEN signal pathway. DNA Cell Biol. 2018;37(9):767–777. doi: 10.1089/dna.2018.4264. [DOI] [PubMed] [Google Scholar]
- 60.Diakowska D., Markocka-Mączka K., Szelachowski P., Grabowski K. Serum levels of resistin, adiponectin, and apelin in gastroesophageal cancer patients. Dis. Markers. 2014;2014:619649. doi: 10.1155/2014/619649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rui C., Yunmei S., Naren Q., Guoqiang W., Yingqian W., Guiyan C., Taiyong Y., Gongshe Y., Weijun P. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta. 2018;1863(4):420–432. doi: 10.1016/j.bbalip.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 62.Jones H., Carver M., Pekala P.H. HuR binds to a single site on the C/EBPβ mRNA of 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2007;355(1):217–220. doi: 10.1016/j.bbrc.2007.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lei S., Jun H., Haiyu W., Qingyang M., Linlin C., Yuguo L., Ying F., Guohao W. Cachexia-related long noncoding RNA, CAAlnc1, suppresses adipogenesis by blocking the binding of HuR to adipogenic transcription factor mRNAs. Int. J. Canc. 2019;145(7):1809–1821. doi: 10.1002/ijc.32236. [DOI] [PubMed] [Google Scholar]
- 64.Liu H.Q., Zhou T., Wang B.Y., Li L., Ye D.W., Yu S.Y. Identification and functional analysis of a potential key lncRNA involved in fat loss of cancer cachexia. J. Cell. Biochem. 2018;119(2):1679–1688. doi: 10.1002/jcb.26328. [DOI] [PubMed] [Google Scholar]
- 65.Wan Z., Chen X., Gao X., Dong Y., Zhao Y., Wei M., Fan W., Yang G., Liu L. Chronic myeloid leukemia-derived exosomes attenuate adipogenesis of adipose derived mesenchymal stem cells via transporting miR-92a-3p. J. Cell. Physiol. 2019;234(11):21274–21283. doi: 10.1002/jcp.28732. [DOI] [PubMed] [Google Scholar]
- 66.Wu Q., Li J., Li Z., Sun S., Zhu S., Wang L., Wu J., Yuan J., Zhang Y., Sun S., Wang C. Exosomes from the tumour-adipocyte interplay stimulate beige/brown differentiation and reprogram metabolism in stromal adipocytes to promote tumour progression. J. Exp. Clin. Canc. Res. 2019;38(1):223. doi: 10.1186/s13046-019-1210-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Wu Q., Sun S., Li Z., Yang Q., Li B., Zhu S., Wang L., Wu J., Yuan J., Yang C., Li J., Sun S. Tumour-originated exosomal miR-155 triggers cancer-associated cachexia to promote tumour progression. Mol. Canc. 2018;17(1):155. doi: 10.1186/s12943-018-0899-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Kulyte A., Lorente-Cebrian S., Gao H., Mejhert N., Agustsson T., Arner P., Ryden M., Dahlman I. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 2014;306(3):E267–E274. doi: 10.1152/ajpendo.00249.2013. [DOI] [PubMed] [Google Scholar]
- 69.Qi R., Wang Q., Wang J., Huang J., Jiang S., Xiao R., Liu Z., Yang F. Expression pattern and regulatory role of microRNA-23a in conjugated linoleic acids-induced apoptosis of adipocytes. Cell. Physiol. Biochem. 2016;40(3–4):668–680. doi: 10.1159/000452579. [DOI] [PubMed] [Google Scholar]
- 70.Chen K., He H., Xie Y., Zhao L., Zhao S., Wan X., Yang W., Mo Z. miR-125a-3p and miR-483-5p promote adipogenesis via suppressing the RhoA/ROCK1/ERK1/2 pathway in multiple symmetric lipomatosis. Sci. Rep. 2015;5:11909. doi: 10.1038/srep11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang T., Zhang Z., Xia T., Liu C., Sun C. circNrxn2 promoted WAT browning via sponging miR-103 to relieve its inhibition of FGF10 in HFD mice. Mol. Ther. Nucleic Acids. 2019;17:551–562. doi: 10.1016/j.omtn.2019.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang H., Zhu L., Bai M., Liu Y., Zhan Y., Deng T., Yang H., Sun W., Wang X., Zhu K., Fan Q., Li J., Ying G., Ba Y. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Canc. 2019;144(10):2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]