Abstract
Background
Interleukin (IL)-19 and IL-20 are important members of the IL-10 cytokine family, which are known to play a role in inflammatory processes. Both anti-IL-19 and -IL-20 targeting drugs have been suggested in the treatment of inflammatory diseases such as psoriasis and rheumatoid arthritis. Recently, we presented I-kappa-B-zeta (IκBζ) as a key player in psoriasis by identifying IκBζ as a regulator of IL-17/tumor necrosis factor (TNF)α-inducible psoriasis-associated genes and proteins. Some of these genes were synergistically regulated by IL-17/TNFα.
Objective
The purpose of this study was to explore the role of IκBζ in the regulation of IL-17A/TNFα-mediated induction of IL-19 and IL-20 expression in human keratinocytes.
Methods
In vitro experiments with cultured primary humane keratinocytes were conducted and investigated by quantitative polymerase chain reaction (qPCR), Western blotting, ELISA and EMSA. For statistics, a one- or two- way repeated-measures analysis of variance (RM ANOVA) or the Friedman test (a nonparametric equivalent to the RM ANOVA) were conducted.
Results
We demonstrated that IL-19 and IL-20 mRNA and protein expressions were synergistically induced by IL-17A and TNFα, whereas IL-17A and TNFα alone had only a minor effect on the IL-19 and IL-20 expression. Moreover, we demonstrated IκBζ to be a regulator of this synergistic induction of IL-19 and IL-20. Finally, the IL-17A/TNFα-induced synergistic induction of IL-19 and IL-20 expression was found to be mediated by a p38 MAPK-, NF-κB- and JNK1/2-dependent mechanism.
Conclusion
This study demonstrates that IκBζ plays a role in the IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20 in humane keratinocytes.
Keywords: I-kappa-B-zeta, Interleukin-17A, Interleukin-19, Interleukin-20, NFKBIZ
INTRODUCTION
Interleukin (IL)-19 and IL-20 are both members of the IL-10 cytokine family, which also comprises IL-22, IL-24, and IL-26. The IL-10 family was first described in 2001 based on the similarities of the cytokines1. However, despite their similarity in regards to structure and their shared location on chromosome 1, they possess different biological functions1. IL-20 transgenic animals expose severe skin abnormalities resembling psoriatic skin2 and IL-20 plays a role in the induction and maintenance of psoriasis as demonstrated in a human xenograft transplantation model3. In contrast, IL-19 overexpressing mice show no skin phenotype4. However, IL-19 has been reported to upregulate psoriasis-associated cytokines, and effective treatment of psoriasis reduce IL-19 expression5,6. Furthermore, IL-19 and IL-20 expression are increased in psoriatic skin7, and genetic variants in the IL-10 gene cluster have been associated with psoriasis8. Moreover, anti-IL-19 and especially anti-IL-20 targeting drugs have been suggested in the treatment of inflammatory diseases including psoriasis and rheumatoid arthritis9,10, diseases where IL-17A and tumor necrosis factor alpha (TNFα) are known to play a key role11,12. Expression of IL-19 and IL-20 and their receptors have been demonstrated in human epidermis6,7,13. IL-19 and IL-20 are produced by activated monocytes, keratinocytes and to a lesser extent by B-cells1,6,9,14. IL-19 and IL-20 are each other's closest relatives within the IL-10 family when comparing sequence homology (40% sequence identity and 60% sequence similarity)1 and signal through the same receptor; the IL-20R1-IL-20R2 heterodimer. In addition, IL-20 binds to the receptor complex IL-22R1-IL-20R210. The IL-10 family of cytokines converge on the defence of several tissues from damage caused by infections or inflammation through the induction of chemokines and cytokines from keratinocytes15,16. However, the functions of both cytokines remain elusive. Especially IL-19 has been assigned both pro- and anti-inflammatory roles17,18. In this study, we aimed to characterise the molecular mechanism involved in the regulation of IL-19 and IL-20. Based on our recent studies, presenting I-kappa-B-zeta (IκBζ) as a key player in psoriasis19,20,21, we wanted to investigate the role of IκBζ in the IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20 expression in human keratinocytes. IκBζ is a nuclear localised protein encoded by the NFKBIZ gene22 and is induced by several inflammatory mediators including IL-17A, lipopolysaccharides, IL-1β, and to a lesser extent TNFα19,22. NFKBIZ functions as a primary target gene but in addition modifies the transcription of secondary genes22,23. This study contributes to the field of intracellular signalling by demonstrating that IκBζ regulates the IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20 expression through a p38 MAPK-, NF-κB-, and c-Jun N-terminal kinase (JNK) 1/2-dependent mechanism in humane keratinocytes.
MATERIALS AND METHODS
Aim
To explore the role of IκBζ in the regulation of IL-17A/TNFα-mediated induction of IL-19 and IL-20 expression in human keratinocytes.
Declarations
Ethics approval and consent to participate: The Regional Ethical Committee of Region Midtjylland, Denmark approved the experiments with cultured human keratinocytes (M-20110027). The keratinocytes were derived from healthy people undergoing reductive skin surgery and they have given consent to donate the excessive skin for research purpose.
Cell cultures
Primary human keratinocytes were obtained from healthy adults. The removed skin was trypsinised and cultured as previously described24. Second-passage keratinocytes were grown in K-SFM (included growth factors) (Gibco, Life Technologies, Austin, TX, USA) at 37℃ and 5% CO2. The medium was changed to keratinocyte basal medium without growth factors 24 hours prior to stimulation with IL-17A (100 ng/ml) and/or TNFα (10 ng/ml). The cells were harvested after 2, 6, 12, 24, or 48 hours of stimulation. In some experiments, keratinocytes were pretreated with a p38 MAPK inhibitor "SB202190" (10 µmol/L; cat. no. 559388), a NF-κB inhibitor "SC-514" (50 µmol/L; cat. no. 401479), an extracellular signal regulated kinase (ERK) 1/2 inhibitor "PD98059" (50 µmol/L; cat. no. 513000) or a JNK1/2 inhibitor "SP600125" (20 µmol/L; cat. no. 420119) (Calbiochem, La Jolla, CA, USA) for 45 minutes prior to stimulation.
siRNA transfection
Cultured human keratinocytes were grown to approximately 60%~70% confluency. Prior to transfection medium was changed to medium without growth factors. IκBζ siRNA (cat no. L-013497-00-0005) p38α siRNA (cat no. L-003512-00-0005), p38β siRNA (cat no. L-003972-00-0005), p65 siRNA (cat no. L-003533-00-0005), JNK1 siRNA (cat no. L-003514-00-0005), or JNK2 siRNA (cat no. L-003505-00-0005) (Dharmacon, Lafayette, CO, USA), were preincubated with Dharmafect-2 transfection reagent (Dharmacon) for 20 minutes. The formed transfection reagent complexes were then supplemented to the cultured cells. As a negative control (siCon), some cells were transfected with a non-targeting pool of siRNAs (cat no. D-0018101005; Dharmacon) or transfection agent alone (Mock).
Quantitative polymerase chain reaction
For quantitative polymerase chain reaction (qPCR), TaqMan reverse transcription reagents, probes and primers were bought from Life Technologies. Human NFKBIZ, IL-19, IL-20, and DEFB4 mRNA expression levels were analysed using TaqMan 20X Assay-On-Demand expression assay mix (assay ID: Hs00230071_m1, Hs00604657_m1, Hs00218888_m1, Hs00175474_m1, respectively). As a reference gene, we used RPLP0 (assay ID: Hs99999902_m1). The PCR master mix was Platinum® qPCR Supermix (Life Technologies). Genes were analysed in triplicate using a RotorGene 3000 real time PCR machine (Corbett Research, Sydney, Australia) and a standard curve of each gene was established as a 4-fold dilution of total RNA and then used to determine the relative amounts of target mRNA. The relative gene expression levels were determined by implementing a relative standard curve method as defined in User Bulletin #2 (ABI Prism 7700 sequencing detection system; Life Technologies).
Enzyme-linked immunosorbent assay
The protein levels of IL-19 and IL-20 were measured by commercial enzyme-linked immunosorbent assay (ELISA) development kits (cat no. DY1035; Bio-Techne, Abingdon, UK) and (cat no. 900-K172; PeproTech, London, UK). ELISA was conducted according to the manufacturer's protocol. An ELISA reader (Laboratory Systems iEMS Reader MF, Copenhagen, Denmark) was used to determine the results. All measurements were performed in duplicates.
Western blotting
Equivalent amounts of protein from the samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto membranes of nitrocellulose. The membranes were then incubated with anti-p65, anti-JNK1/2, anti-p38α or anti-p38β (cat no. 3034, 9252, 9218, 2339, respectively; Cell Signaling Technology, Danvers, MA, USA) or β-actin (cat no. A-1978; Sigma-Aldrich, St. Louis, MO, USA). The antibodies were afterwards detected with anti-rabbit immunoglobulin G (IgG)-HRP (cat no. 7074; Cell Signaling Technology) or with anti-mouse IgG-HRP (cat no. p0447; Dako, Glostrup, Denmark) in a standard ECL reaction (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's instructions.
Electrophoretic mobility shift assay
The oligonucleotide used for electrophoretic mobility shift assay (EMSA) was NF-κB (top strand) 5′-ATTTTCTGGGG TTTCCTGAGTC-3′. The oligonucleotide was synthesised by (DNA Technology A/S, Aarhus, Denmark) and labelled by T4 polynucleotide kinase (Promega, Madison, WI, USA), and purified on a Nick Spin column (Sephadex G-50; Pharmacia, Uppsala, Sweden). Nuclear protein (1 µg) preincubated with 32P-labelled oligonucleotides was separated on a 6% Novex® DNA retardation gel (Invitrogen, Carlsbad, CA, USA) and visualised by exposure to X-ray film. Supershift was performed by addition of 2 µg of antibodies against p65 (cat. no. sc-7151X) and against p50 (cat. no. sc-7178X), (both from Santa Cruz Biotechnology, Santa Cruz, CA, USA) to the binding reaction.
Statistical analysis
Data were tested for distribution and variance with SigmaPlot software (Systat Software, Inc., San Jose, CA, USA). If the data were normally distributed and had equal variance, a one- or two-way repeated-measures analysis of variance (RM ANOVA) was conducted according to the number of factors included. If the data were not normally distributed, the Friedman test (a nonparametric equivalent to the RM ANOVA) using ranks was conducted. For post hoc testing, multiple comparisons with the control group were made with the Holm-Sidak method when data were normally distributed or with Dunn's method when data were not normally distributed. If more conditions had to be compared, an all pairwise multiple comparisons was applied using the Student-Newman-Keuls method. A probability of p<0.05 was regarded as statistically significant.
RESULTS
IL-19 and IL-20 mRNA and protein expression are synergistically induced by IL-17A and TNFα
To examine the effects of IL-17A, TNFα or their combination on IL-19 and IL-20 mRNA and protein expression at different time points, cultured normal human keratinocytes were stimulated for 2, 6, 12, 24, and 48 hours.
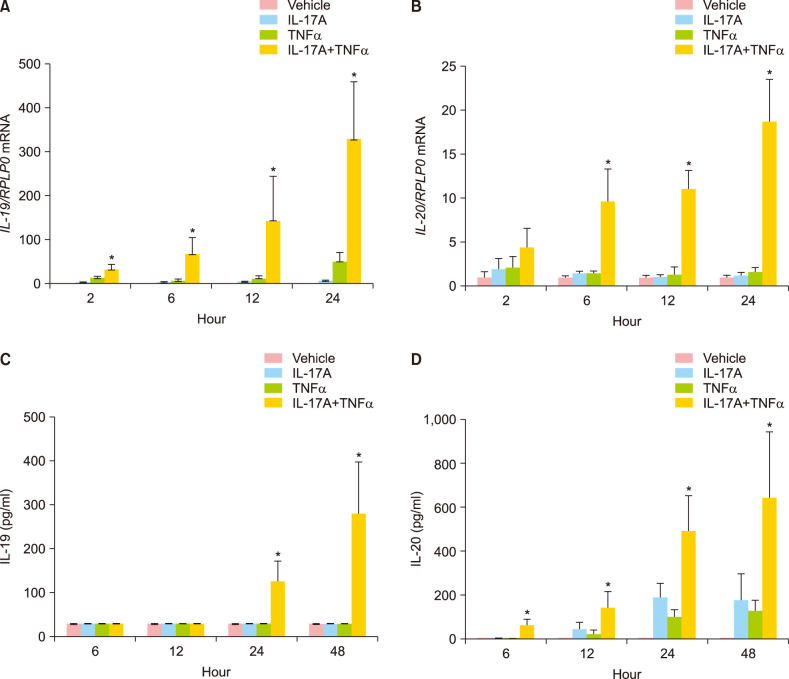
IL-17A and TNFα in combination significantly induced the mRNA expression of IL-19 already after 2 hours and induced the mRNA expression of IL-20 significantly after six hours (Fig. 1A, B). Both IL-19 and IL-20 mRNA expression were significantly induced after 12 and 24 hours of stimulation. Interestingly, IL-17A and TNFα stimulation alone had only a minor effect on the IL-19 and IL-20 mRNA expression (Fig. 1A, B). In agreement with previous results25, we found that combined IL-17A/TNFα stimulation strongly induced IL-19 and IL-20 mRNA expression at higher levels than the additive values of the individual cytokines (Fig. 1A, B).
Fig. 1. IL-19 and IL-20 mRNA and protein expression are synergistically induced by IL-17A and TNFα. Cultured human keratinocytes were stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml) or IL-17A combined with TNFα for 2, 6, 12, 24, or 48 hours (n=4). (A, B) IL-19 and IL-20 mRNA expression were analysed by qPCR, and RPLP0 was used as a reference gene for normalisation. (C, D) Protein expression was analysed by ELISA. IL: interleukin, TNFα: tumour necrosis factor alpha. *p<0.05 compared the synergistic effect against the additive effect for TNFα combined with IL-17A.
We also analysed whether the IL-17A/TNFα-induced mRNA expression of IL-19 and IL-20 was paralleled by an increased IL-19 and IL-20 protein production, respectively. The amount of IL-19 and IL-20 secreted from the keratinocytes to the medium was measured over a 48-hour period. We observed a significant synergistic induction of IL-17A/TNFα-induced protein expression of IL-19 after 24 and 48 hours, whereas we observed a significant synergistic induction of IL-17A/TNFα-induced protein expression of IL-20 after 12, 24, and 48 hours (Fig. 1C, D).
IκBζ regulates IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20
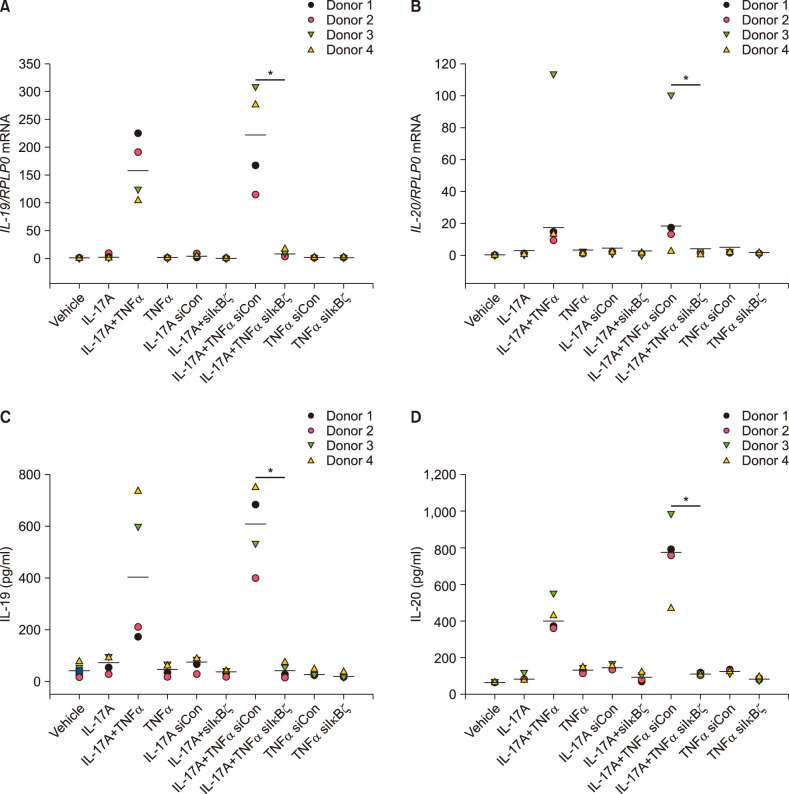
To determine whether IκBζ was involved in the synergistic induction of IL-19 and IL-20, human keratinocytes were transfected with specific siRNA directed against IκBζ. IκBζ siRNA reduced the mRNA expression of NFKBIZ by approximately 70% in IL-17A/TNFα-stimulated cells compared with cells transfected with control siRNA (Supplementary Fig. 1). Interestingly, knockdown of IκBζ by siRNA before IL-17A and TNFα stimulation in combination for 24 hours significantly reduced the mRNA and protein expression of IL-19 and IL-20 compared with control siRNA-transfected cells (Fig. 2). This demonstrates that IκBζ plays a role in the synergistic induction of IL-19 and IL-20 mediated by IL-17A and TNFα. As a control, we confirmed that IκBζ regulates IL-17A/TNFα-mediated synergistic induction of DEFB4 mRNA expression in the same cells, which has been described previously (Supplementary Fig. 2)26.
Fig. 2. IκBζ regulates IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20. Human keratinocytes were transfected with IκBζ siRNA (siIκBζ) or control siRNA (siCon) before stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml) or the combinations as indicated for 24 hours (n=4). (A, B) mRNA expression was analysed by qPCR, and RPLP0 was used as a reference gene for normalisation. (C, D) Protein level was analysed by ELISA (n=4). Horizontal lines represent medians. IL: interleukin, TNFα: tumour necrosis factor alpha, IκBζ: I-kappa-B-zeta. *p<0.05 comparing the cells transfected with IκBζ siRNA (siIκBζ) with the cells transfected with control siRNA (siCon).
Synergistic induction of IL-19 and IL-20 is regulated by a p38 MAPK-, NF-κB-, and JNK1/2-dependent mechanism in humane keratinocytes
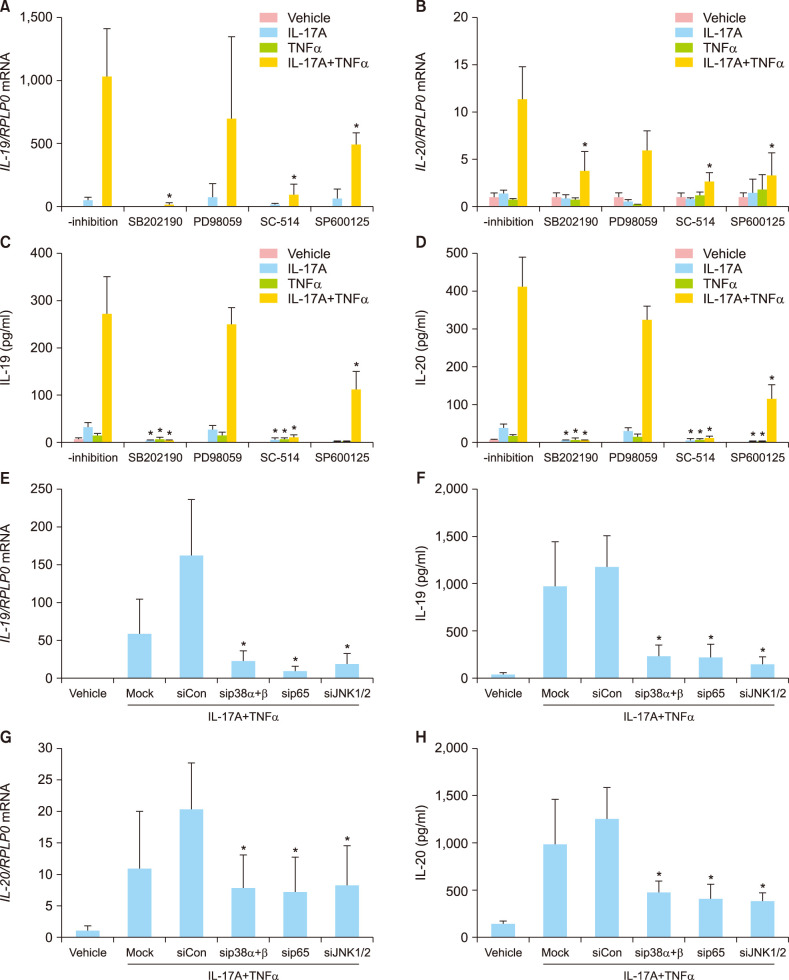
To further characterise the molecular mechanism mediating the synergistic effect of IL-17A and TNFα on the IL-19 and IL-20 mRNA and protein expression, human keratinocytes were preincubated with inhibitors targeting the p38 MAPK signalling pathway, the ERK1/2 signalling pathway, the NF-κB signalling pathway or the JNK1/2 signalling pathway before stimulation. Pretreatment with SB202190, a p38 MAPK inhibitor, SC-514, a NF-κB inhibitor or with SP600125, a JNK1/2 inhibitor, significantly reduced the IL-17A/TNFα-induced IL-19 and IL-20 mRNA expression (Fig. 3A, B). In contrast, pretreatment with PD98059, an ERK1 and 2 inhibitor, did not result in any regulation of IL-19 or IL-20 mRNA expression. We also examined the protein levels of IL-19 and IL-20 after pretreatment with the indicated inhibitors and found that the changes observed in the IL-19 and IL-20 mRNA expression correlated with the protein release from the keratinocytes (Fig. 3C, D). The involvement of p38 MAPK, JNK1/2, and NF-κB in the induction of IL-19 and IL-20 was also analysed using small interfering RNA (siRNA) to knock down p38 MAPK (p38α and p38β), JNK1/2, and NF-κB/p65. Transfection of keratinocytes with siRNA directed against p38 MAPK (p38α and p38β), JNK1/2, or NF-κB/p65 significantly reduced the IL-17A/TNFα-induced IL-19 and IL-20 mRNA and protein expression (Fig. 3E~H). Gene silencing efficiencies for p38α, p38β, JNK1, JNK2, and NF-κB/p65 were verified with Western blotting (Supplementary Fig. 3). Together, these data demonstrate that IL-17A/TNFα-induced expression of IL-19 and IL-20 is mediated by a mechanism involving the p38 MAPK, JNK1/2, and NF-κB signalling pathways.
Fig. 3. IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20 is regulated by a p38 MAPK-, NF-κB-, and JNK1/2-dependent mechanism. (A~D) Human keratinocytes were preincubated with the indicated inhibitors for 45 minutes before being stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml), or IL-17A combined with TNFα for 24 hours. (E~H) Human keratinocytes were transfected with specific siRNA as indicated. A non-targeting pool of siRNAs served as negative control (siCon). (A, B, E, G) IL-19 and IL-20 mRNA expression were measured by qPCR (n=4). RPLP0 was used for normalisation. (C, D, F, H) IL-19 and IL-20 protein levels were measured by ELISA (n=4). IL: interleukin, TNFα: tumour necrosis factor alpha. *Non-inhibited cells compared with inhibited cells; transfected cells against (sip38αβ), p65 (sip65), JNK1/2 (siJNK1/2) compared with control siRNA (siCon) (p<0.05).
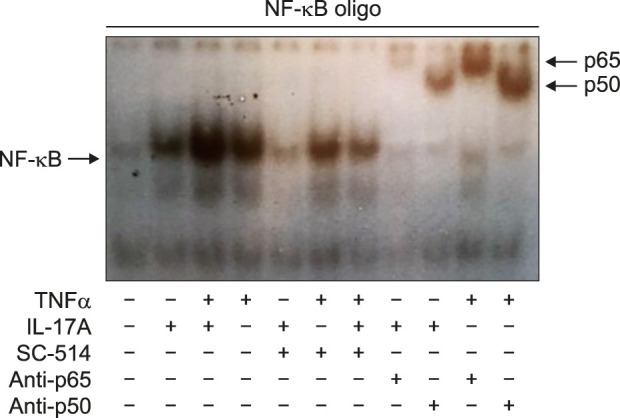
NF-κB activation upon IL-17A/TNFα stimulation was furthermore determined using EMSA. We demonstrated that stimulation with IL-17A and/or TNFα clearly increased the DNA binding activity of NF-κB with the strongest DNA binding activity resulting from TNFα alone and IL-17A/TNFα combined stimulation (Fig. 4). Incubation of the DNA-protein complexes with an anti-p50 or an anti-p65 antibody showed a shift in the migrating bands, which indicated the specificity to these proteins (Fig. 4).
Fig. 4. IL-17A and TNFα increased the DNA binding activity of NF-κB. Human keratinocytes were stimulated with IL-17A (100 ng/ml) or TNFα (10 ng/ml) or their combination for one hour before the NF-κB DNA binding activity was analysed by electrophoretic mobility shift assay. Antibodies against p50 and p65 were tested for their ability to cause a super shift of the nuclear complex associated with the probe. A representative result of three separate experiments is shown. IL: interleukin, TNFα: tumour necrosis factor alpha.
DISCUSSION
Several pro-inflammatory mediators, including IL-17A and TNFα, are known to play a major role in inflammatory diseases such as psoriasis and rheumatoid arthritis and treatments directed against these cytokines have been shown to be highly effective25. However, our understanding of the many underlying molecular mechanisms involved remains limited. The combined anti-cytokine treatments known as bispecific antibodies or antagonists of the receptors of IL-19 and IL-20 have been suggested as a novel therapeutic cocktail in inflammatory diseases such as psoriasis and rheumatoid arthritis10. However, in contrast, IL-19 has also been suggested to possess anti-inflammatory effects17, which would make it undesirable to suppress. Today it is known that the genomic alterations significant for the inflammatory transcriptome e.g. in psoriasis consist of multiple different cytokines and inflammatory elements that collaborate in creating an interactive network of numerous genes with altered expression that have to be targeted25.
In this study, we extend the current knowledge of the synergistic effects induced by IL-17A/TNFα21,26,27 with a focus on IL-19 and IL-20, yet another set of important psoriasis genes. Based on our recent studies, presenting IκBζ as a key player in psoriasis19,26 and previous studies demonstrating IL-17A/TNFα synergy26,27, we wanted to investigate the role of IκBζ in IL-17A/TNFα-mediated synergistic induction of IL-19 and IL-20. Moreover, in agreement with previous results presenting IκBζ to be involved in IL-17A/TNFα synergistic induction20,21,26, we demonstrated that IκBζ was involved in IL-17A/TNFα-mediated synergistic induction of the IL-19 and IL-20 cytokines, both at the mRNA and protein levels. Furthermore, we demonstrated that the IL-17A/TNFα-induced synergy of IL-19 and IL-20 to be regulated by a p38 MAPK-, NF-κB-, and JNK1/2-dependent mechanism in humane keratinocytes. Both IL-19 and IL-20 have previously been described involved in JAK and STAT pathways18. This study demonstrates p38 MAPK-, NF-κB-, and JNK1/2 signalling pathways to be involved, which underlines the complexity involved in the regulation. IL-17A and TNFα have previously been demonstrated to be able to activate these signalling pathways26. Furthermore, NFKBIZ/IκBζ has also been demonstrated to be regulated through some of these pathways20,21, which underlines the intertwined signalling pathways of IL-19, IL-20, IL-17A, TNFα, and IκBζ. However, this study is limited by only performing in vitro experiments. Moreover, complete knockout instead of knockdown of the players investigated could provide more clear results. However, the knockdown models present real life better as many unknown parameters interact and rarely constitute a complete knockout.
In conclusions, this study contributes to our understanding of IL-19, IL-20, and IκBζ and provide insight into IL-17A/TNFα-mediated synergistic interactions. Moreover, these results reinforce the importance of IκBζ in the pathogenesis of psoriasis by demonstrating a role of IκBζ in the expression of yet another set of psoriasis essential genes.
ACKNOWLEDGMENT
Prof. DMSc Thomas Litman kindly helped to analyse the data.
Footnotes
CONFLICTS OF INTEREST: Trine Bertelsen has served as a paid speaker for Eli Lilly. Claus Johansen has served as a paid speaker for Eli Lilly and Leo Pharma. Lars Iversen served as a consultant and/or paid speaker for and/or participated in clinical trials sponsored by: AbbVie, Almirall, Amgen, Astra Zeneca, BMS, Boehringer Ingelheim, Celgene, Centocor, Eli Lilly, Janssen Cilag, Kyowa, Leo Pharma, MSD, Novartis, Pfizer, Samsung, UCB.
FUNDING SOURCE: Funding was granted by Novartis, The Danish psoriasis foundation, and The Aage Bang Foundation. The authors have fully decided the design and focus of the study. We have applied for financial support and all funders offered their support for our research in skin-pathology.
DATA SHARING STATEMENT
The data and methods/materials used, generated and/or analysed during the current study are included in the published article and supplementary or available from the corresponding author on request.
SUPPLEMENTARY MATERIALS
Supplementary data can be found via http://anndermatol.org/src/sm/ad-33-122-s001.pdf.
Knockdown of NFKBIZ by siIκBζ. Human keratinocytes were transfected with IκBζ siRNA (siIκBζ) or control siRNA (siCon) before stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml) or the combinations as indicated for 24 hours (n=4). mRNA expression was analysed by qPCR, and RPLP0 was used as a reference gene for normalisation. Horizontal lines represent medians. IL: interleukin, TNFα: tumour necrosis factor alpha, IκBζ: I-kappa-B-zeta. *p<0.05 comparing the cells transfected with siIκBζ with the cells transfected with siCon.
DEFB4 expression was synergistically induced by IL-17A/TNFα through an IκBζ-dependent mechanism. Human keratinocytes were transfected with IκBζ siRNA (siIκBζ) or control siRNA (siCon) before stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml) or the combinations as indicated for 24 hours (n=4). mRNA expression was analysed by qPCR, and RPLP0 was used as a reference gene for normalisation. Horizontal lines represent medians. IL: interleukin, TNFα: tumour necrosis factor alpha, IκBζ: I-kappa-B-zeta. *p<0.05 comparing the cells transfected with siIκBζ with the cells transfected with siCon.
NF-κB, JNK, p38α and β were silenced by siRNA. Human keratinocytes were transfected with specific siRNAs directed against NF-κB p65, JNK1/2, p38α and β before stimulated with IL-17A (100 ng/ml) and TNFα (10 ng/ml). As controls siCon and Mock were added. Protein levels of NF-κB p65, p38α, p38β, and JNK1/2 were examined by Western blotting (n=3). β-actin was used as loading control. One representative donor is shown. IL: interleukin, TNFα: tumour necrosis factor alpha.
References
- 1.Sabat R. IL-10 family of cytokines. Cytokine Growth Factor Rev. 2010;21:315–324. doi: 10.1016/j.cytogfr.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 3.Stenderup K, Rosada C, Worsaae A, Dagnaes-Hansen F, Steiniche T, Hasselager E, et al. Interleukin-20 plays a critical role in maintenance and development of psoriasis in the human xenograft transplantation model. Br J Dermatol. 2009;160:284–296. doi: 10.1111/j.1365-2133.2008.08890.x. [DOI] [PubMed] [Google Scholar]
- 4.Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, et al. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem. 2002;277:47517–47523. doi: 10.1074/jbc.M205114200. [DOI] [PubMed] [Google Scholar]
- 5.Rømer J, Hasselager E, Nørby PL, Steiniche T, Thorn Clausen J, Kragballe K. Epidermal overexpression of interleukin-19 and -20 mRNA in psoriatic skin disappears after short-term treatment with cyclosporine a or calcipotriol. J Invest Dermatol. 2003;121:1306–1311. doi: 10.1111/j.1523-1747.2003.12626.x. [DOI] [PubMed] [Google Scholar]
- 6.Gallagher G. Interleukin-19: multiple roles in immune regulation and disease. Cytokine Growth Factor Rev. 2010;21:345–352. doi: 10.1016/j.cytogfr.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Otkjaer K, Kragballe K, Funding AT, Clausen JT, Noerby PL, Steiniche T, et al. The dynamics of gene expression of interleukin-19 and interleukin-20 and their receptors in psoriasis. Br J Dermatol. 2005;153:911–918. doi: 10.1111/j.1365-2133.2005.06800.x. [DOI] [PubMed] [Google Scholar]
- 8.Galimova E, Rätsep R, Traks T, Kingo K, Escott-Price V, Kõks S. Interleukin-10 family cytokines pathway: genetic variants and psoriasis. Br J Dermatol. 2017;176:1577–1587. doi: 10.1111/bjd.15363. [DOI] [PubMed] [Google Scholar]
- 9.Wegenka UM. IL-20: biological functions mediated through two types of receptor complexes. Cytokine Growth Factor Rev. 2010;21:353–363. doi: 10.1016/j.cytogfr.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Hsu YH, Chang MS. IL-20 in rheumatoid arthritis. Drug Discov Today. 2017;22:960–964. doi: 10.1016/j.drudis.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Nestle FO, Kaplan DH, Barker J. Mechanisms of disease: psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 12.Chapman A, El Miedany Y. Psoriasis. In: El Miedany Y, editor. Comorbidity in rheumatic diseases. Cham: Springer; 2017. pp. 81–124. [Google Scholar]
- 13.Wei CC, Chen WY, Wang YC, Chen PJ, Lee JY, Wong TW, et al. Detection of IL-20 and its receptors on psoriatic skin. Clin Immunol. 2005;117:65–72. doi: 10.1016/j.clim.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Kunz S, Wolk K, Witte E, Witte K, Doecke WD, Volk HD, et al. Interleukin (IL)-19, IL-20 and IL-24 are produced by and act on keratinocytes and are distinct from classical ILs. Exp Dermatol. 2006;15:991–1004. doi: 10.1111/j.1600-0625.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 15.Li HH, Lin YC, Chen PJ, Hsiao CH, Lee JY, Chen WC, et al. Interleukin-19 upregulates keratinocyte growth factor and is associated with psoriasis. Br J Dermatol. 2005;153:591–595. doi: 10.1111/j.1365-2133.2005.06665.x. [DOI] [PubMed] [Google Scholar]
- 16.Sa SM, Valdez PA, Wu J, Jung K, Zhong F, Hall L, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 17.Liao YC, Liang WG, Chen FW, Hsu JH, Yang JJ, Chang MS. IL-19 induces production of IL-6 and TNF-alpha and results in cell apoptosis through TNF-alpha. J Immunol. 2002;169:4288–4297. doi: 10.4049/jimmunol.169.8.4288. [DOI] [PubMed] [Google Scholar]
- 18.Autieri MV. IL-19 and other IL-20 family member cytokines in vascular inflammatory diseases. Front Immunol. 2018;9:700. doi: 10.3389/fimmu.2018.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen C, Mose M, Ommen P, Bertelsen T, Vinter H, Hailfinger S, et al. IκBζ is a key driver in the development of psoriasis. Proc Natl Acad Sci U S A. 2015;112:E5825–E5833. doi: 10.1073/pnas.1509971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertelsen T, Iversen L, Johansen C. The human IL-17A/F heterodimer regulates psoriasis-associated genes through I κBζ. Exp Dermatol. 2018;27:1048–1052. doi: 10.1111/exd.13722. [DOI] [PubMed] [Google Scholar]
- 21.Bertelsen T, Ljungberg C, Boye Kjellerup R, Iversen L, Johansen C. IL-17F regulates psoriasis-associated genes through IκBζ. Exp Dermatol. 2017;26:234–241. doi: 10.1111/exd.13182. [DOI] [PubMed] [Google Scholar]
- 22.Muta T. IkappaB-zeta: an inducible regulator of nuclear factor-kappaB. Vitam Horm. 2006;74:301–316. doi: 10.1016/S0083-6729(06)74012-2. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 24.Johansen C. Generation and culturing of primary human keratinocytes from adult skin. J Vis Exp. 2017;(130):56863. doi: 10.3791/56863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiricozzi A, Guttman-Yassky E, Suárez-Fariñas M, Nograles KE, Tian S, Cardinale I, et al. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131:677–687. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 26.Johansen C, Bertelsen T, Ljungberg C, Mose M, Iversen L. Characterization of TNF-α- and IL-17A-mediated synergistic induction of DEFB4 gene expression in human keratinocytes through IκBζ. J Invest Dermatol. 2016;136:1608–1616. doi: 10.1016/j.jid.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Russell CB, Rand H, Bigler J, Kerkof K, Timour M, Bautista E, et al. Gene expression profiles normalized in psoriatic skin by treatment with brodalumab, a human anti-IL-17 receptor monoclonal antibody. J Immunol. 2014;192:3828–3836. doi: 10.4049/jimmunol.1301737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Knockdown of NFKBIZ by siIκBζ. Human keratinocytes were transfected with IκBζ siRNA (siIκBζ) or control siRNA (siCon) before stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml) or the combinations as indicated for 24 hours (n=4). mRNA expression was analysed by qPCR, and RPLP0 was used as a reference gene for normalisation. Horizontal lines represent medians. IL: interleukin, TNFα: tumour necrosis factor alpha, IκBζ: I-kappa-B-zeta. *p<0.05 comparing the cells transfected with siIκBζ with the cells transfected with siCon.
DEFB4 expression was synergistically induced by IL-17A/TNFα through an IκBζ-dependent mechanism. Human keratinocytes were transfected with IκBζ siRNA (siIκBζ) or control siRNA (siCon) before stimulated with IL-17A (100 ng/ml), TNFα (10 ng/ml) or the combinations as indicated for 24 hours (n=4). mRNA expression was analysed by qPCR, and RPLP0 was used as a reference gene for normalisation. Horizontal lines represent medians. IL: interleukin, TNFα: tumour necrosis factor alpha, IκBζ: I-kappa-B-zeta. *p<0.05 comparing the cells transfected with siIκBζ with the cells transfected with siCon.
NF-κB, JNK, p38α and β were silenced by siRNA. Human keratinocytes were transfected with specific siRNAs directed against NF-κB p65, JNK1/2, p38α and β before stimulated with IL-17A (100 ng/ml) and TNFα (10 ng/ml). As controls siCon and Mock were added. Protein levels of NF-κB p65, p38α, p38β, and JNK1/2 were examined by Western blotting (n=3). β-actin was used as loading control. One representative donor is shown. IL: interleukin, TNFα: tumour necrosis factor alpha.
Data Availability Statement
The data and methods/materials used, generated and/or analysed during the current study are included in the published article and supplementary or available from the corresponding author on request.